Abstract
Background
Machine learning is a broad term encompassing a number of methods that allow the investigator to learn from the data. These methods may permit large real-world databases to be more rapidly translated to applications to inform patient-provider decision making.
Methods
This systematic literature review was conducted to identify published observational research of employed machine learning to inform decision making at the patient-provider level. The search strategy was implemented and studies meeting eligibility criteria were evaluated by two independent reviewers. Relevant data related to study design, statistical methods and strengths and limitations were identified; study quality was assessed using a modified version of the Luo checklist.
Results
A total of 34 publications from January 2014 to September 2020 were identified and evaluated for this review. There were diverse methods, statistical packages and approaches used across identified studies. The most common methods included decision tree and random forest approaches. Most studies applied internal validation but only two conducted external validation. Most studies utilized one algorithm, and only eight studies applied multiple machine learning algorithms to the data. Seven items on the Luo checklist failed to be met by more than 50% of published studies.
Conclusions
A wide variety of approaches, algorithms, statistical software, and validation strategies were employed in the application of machine learning methods to inform patient-provider decision making. There is a need to ensure that multiple machine learning approaches are used, the model selection strategy is clearly defined, and both internal and external validation are necessary to be sure that decisions for patient care are being made with the highest quality evidence. Future work should routinely employ ensemble methods incorporating multiple machine learning algorithms.
Keywords: Machine learning, Decision making, Decision tree, Random forest, Automated neural network
Background
Traditional methods of analyzing large real-world databases (big data) and other observational studies are focused on the outcomes that can inform at the population-based level. The findings from real-world studies are relevant to populations as a whole, but the ability to predict or provide meaningful evidence at the patient level is much less well established due to the complexity with which clinical decision making is made and the variety of factors taken into account by the health care provider [1, 2]. Using traditional methods that produce population estimates and measures of variability, it is very challenging to accurately predict how any one patient will perform, even when applying findings from subgroup analyses. The care of patients is nuanced, and multiple non-linear, interconnected factors must be taken into account in decision making. When data are available that are only relevant at the population level, health care decision making is less informed as to the optimal course of care for a given patient.
Clinical prediction models are an approach to utilizing patient-level evidence to help inform healthcare decision makers about patient care. These models are also known as prediction rules or prognostic models and have been used for decades by health care professionals [3]. Traditionally, these models combine patient demographic, clinical and treatment characteristics in the form of a statistical or mathematical model, usually regression, classification or neural networks, but deal with a limited number of predictor variables (usually below 25). The Framingham Heart Study is a classic example of the use of longitudinal data to build a traditional decision-making model. Multiple risk calculators and estimators have been built to predict a patient’s risk of a variety of cardiovascular outcomes, such as atrial fibrillation and coronary heart disease [4–6]. In general, these studies use multivariable regression evaluating risk factors identified in the literature. Based on these findings, a scoring system is derived for each factor to predict the likelihood of an adverse outcome based on a patient’s score across all risk factors evaluated.
With the advent of more complex data collection and readily available data sets for patients in routine clinical care, both sample sizes and potential predictor variables (such as genomic data) can exceed the tens of thousands, thus establishing the need for alternative approaches to rapidly process a large amount of information. Artificial intelligence (AI), particularly machine learning methods (a subset of AI), are increasingly being utilized in clinical research for prediction models, pattern recognition and deep-learning techniques used to combine complex information for example genomic and clinical data [7–9]. In the health care sciences, these methods are applied to replace a human expert to perform tasks that would otherwise take considerable time and expertise, and likely result in potential error. The underlying concept is that a machine will learn by trial and error from the data itself, to make predictions without having a pre-defined set of rules for decision making. Simply, machine learning can simply be better understood as “learning from data.” [8].
There are two types of learning from the data, unsupervised and supervised. Unsupervised learning is a type of machine learning algorithm used to draw inferences from datasets consisting of input data without labelled responses. The most common unsupervised learning method is cluster analysis, which is used for exploratory data analysis to find hidden patterns or grouping in data. Supervised learning involves making a prediction based on a set of pre-specified input and output variables. There are a number of statistical tools used for supervised learning. Some examples include traditional statistical prediction methods like regression models (e.g. regression splines, projection pursuit regression, penalized regression) that involve fitting a model to data, evaluating the fit and estimating parameters that are later used in a predictive equation. Other tools include tree-based methods (e.g. classification and regression trees [CART] and random forests), which successively partition a data set based on the relationships between predictor variables and a target (outcome) variable. Other examples include neural networks, discriminant functions and linear classifiers, support vector classifiers and machines. Often, predictive tools are built using various forms of model aggregation (or ensemble learning) that may combine models based on resampled or re-weighted data sets. These different types of models can be fitted to the same data using model averaging.
Classical statistical regression methods used for prediction modeling are well understood in the statistical sciences and the scientific community that employs them. These methods tend to be transparent and are usually hypothesis driven but can overlook complex associations with limited flexibility when a high number of variables are investigated. In addition, when using classic regression modeling, choosing the ‘right’ model is not straightforward. Non-traditional machine learning algorithms, and machine learning approaches, may overcome some of these limitations of classical regression models in this new era of big data, but are not a complete solution as they must be considered in the context of the limitations of data used in the analysis [2].
While machine learning methods can be used for both population-based models as well as for informed patient-provider decision making, it is important to note that the data, model, and outputs used to inform the care of an individual patient must meet the highest standards of research quality, as the choice made will likely have an impact on both the long- and short-term patient outcomes. While a range of uncertainty can be expected for population-based estimates, the risk of error for patient level models must be minimized to ensure quality patient care. The risks and concerns of utilizing machine learning for individual patient decision making have been raised by ethicists [10]. The risks are not limited to the lack of transparency, limited data regarding the confidence of the findings, and the risk of reducing patient autonomy in choice by relying on data that may foster a more paternalistic model of healthcare. These are all important and valid concerns, and therefore the role of machine learning for patient care must meet the highest standards to ensure that shared, not simply informed, evidence-based decision making be supported by these methods.
A systematic literature review was published in 2018 that evaluated the statistical methods that have been used to enable large, real-world databases to be used at the patient-provider level [11]. Briefly, this study identified a total of 115 articles that evaluated the use of logistic regression (n = 52, 45.2%), Cox regression (n = 24, 20.9%), and linear regression (n = 17, 14.8%). However, an interesting observation noted several studies utilizing novel statistical approaches such as machine learning, recursive partitioning, and development of mathematical algorithms to predict patient outcomes. More recently, publications are emerging describing the use of Individualized Treatment Recommendation algorithms and Outcome Weighted Learning for personalized medicine using large observational databases [12, 13]. Therefore, this systematic literature review was designed to further pursue this observation to more comprehensively evaluate the use of machine learning methods to support patient-provider decision making, and to critically evaluate the strengths and weaknesses of these methods. For the purposes of this work, data supporting patient-provider decision making was defined as that which provided information specifically on a treatment or intervention choice; while both population-based and risk estimator data are certainly valuable for patient care and decision making, this study was designed to evaluate data that would specifically inform a choice for the patient with the provider. The overarching goal is to provide evidence of how large datasets can be used to inform decisions at the patient level using machine learning-based methods, and to evaluate the quality of such work to support informed decision making.
Methods
This study originated from a systematic literature review that was conducted in MEDLINE and PsychInfo; a refreshed search was conducted in September 2020 to obtain newer publications (Table 1). Eligible studies were those that analyzed prospective or retrospective observational data, reported quantitative results, and described statistical methods specifically applicable to patient-level decision making. Specifically, patient-level decision making referred to studies that provided data for or against a particular intervention at the patient level, so that the data could be used to inform decision making at the patient-provider level. Studies did not meet this criterion if only a population-based estimates, mortality risk predictors, or satisfaction with care were evaluated. Additionally, studies designed to improve diagnostic tools and those evaluating health care system quality indicators did not meet the patient-provider decision-making criterion. Eligible statistical methods for this study were limited to machine learning-based approaches. Eligibility was assessed by two reviewers and any discrepancies were discussed; a third reviewer was available to serve as a tie breaker in case of different opinions. The final set of eligible publications were then abstracted into a Microsoft Excel document. Study quality was evaluated using a modified Luo scale, which was developed specifically as a tool to standardize high-quality publication of machine learning models [14]. A modified version of this tool was utilized for this study; specifically, the optional item were removed, and three terms were clarified: item 6 (define the prediction problem) was redefined as “define the model,” item 7 (prepare data for model building) was renamed “model building and validation,” and item 8 (build the predictive model) was renamed “model selection” to more succinctly state what was being evaluated under each criterion. Data were abstracted and both extracted data and the Luo checklist items were reviewed and verified by a second reviewer to ensure data comprehensiveness and quality. In all cases of differences in eligibility assessment or data entry, the reviewers met and ensured agreement with the final set of data to be included in the database for data synthesis, with a third reviewer utilized as a tie breaker in case of discrepancies. Data were summarized descriptively and qualitatively, based on the following categories: publication and study characteristics; patient characteristics; statistical methodologies used, including statistical software packages; strengths and weaknesses; and interpretation of findings.
Table 1.
Search strategy
| 1 | (randomized controlled trial or controlled clinical trial).pt. or randomized.ab |
| 2 | Prospective Studies/ or observational trials.mp. or observational research.mp |
| 3 | Retrospective Studies/ or retrospective observational.mp. or retrospective database.mp |
| 4 | Cross-Sectional Studies/ or cross-sectional.mp |
| 5 | (systematic adj2 review).mp |
| 6 | 1 or 2 or 3 or 4 or 5 |
| 7 | *Neoplasms/ |
| 8 | *Cardiovascular Diseases/ |
| 9 | *Diabetes Mellitus, Type 1/ or *Diabetes Mellitus, Type 2/ or *Diabetes Mellitus/ |
| 10 | *Autoimmune Diseases/ |
| 11 | *Alzheimer Disease/ |
| 12 | 7 or 8 or 9 or 10 or 11 |
| 13 | (decision making or decision analysis).mp. or *Decision Making/ or Decision Support Techniques/ |
| 14 | Physician–Patient Relations/ or Patient-Centered Care/ or patient cent*.mp |
| 15 | nomograms.mp. or Nomograms/ |
| 16 | prediction model*.mp |
| 17 | Patient Preference/ or discrete choice.mp. or conjoint analysis.mp |
| 18 | Decision Support Techniques/ or (decision adj2 tool).mp. or decision aid.mp |
| 19 | 13 or 14 or 15 or 16 or 17 or 18 |
| 20 | 6 and 12 and 19 |
| 21 | limit 20 to (english language and humans and yr = "2000 -Current") |
| 22 | limit 20 to (english language and humans and yr = "2014 -Current") |
| 23 | "machine learning".mp. or Machine Learning/ or "Neural Networks (Computer)"/ or Computer Simulation/ or Algorithms/ |
| 24 | 22 and 23 |
| 25 | data mining.mp. or Data Mining/ or Medical Informatics/ |
| 26 | 22 and 25 |
| 27 | 24 or 26 |
| 28 | neural network*.mp. or LTSM.ab. or LTSM.ti. or memory network*.mp |
| 29 | 28 and 6 and 19 and 19 |
| 30 | limit 29 to (english language and humans and yr = "2014 -Current") |
| 31 | 27 or 30 |
Results
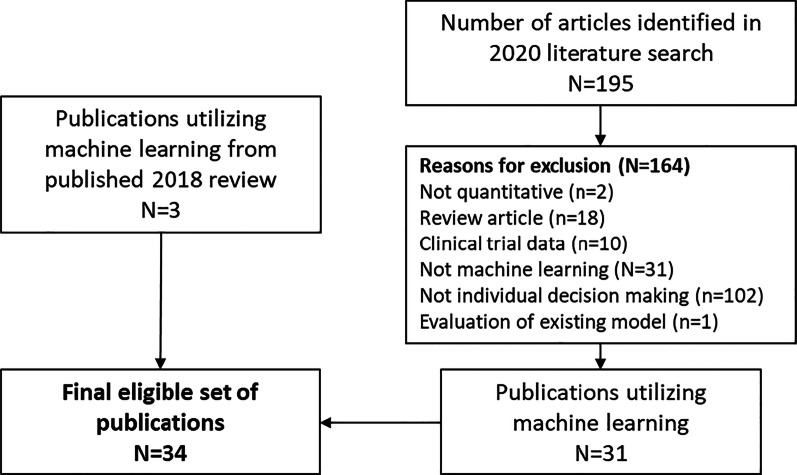
The search strategy was run on September 1, 2020 and identified a total of 34 publications that utilized machine learning methods for individual patient-level decision making (Fig. 1). The most common reason for study exclusion, as expected, was due to the study not meeting the patient-level decision making criterion. A summary of the characteristics of eligible studies and the patient data are included in Table 2. Most of the real-world data sources included retrospective databases or designs (n = 27, 79.4%), primarily utilizing electronic health records. Six analyses utilized prospective cohort studies and one utilized data from a cross sectional study.
Fig. 1.
Prisma diagram of screening and study identification
Table 2.
Summary of eligible publications
| References | Study objective | Disease state | Data source(s) | Statistical modeling method(s) | Software | Sample size | Number of different models |
|---|---|---|---|---|---|---|---|
| Alaa et al. [23] | To develop machine-learning-based risk prediction models | Cardiovascular disease | Prospective cohort study—UK Biobank | Cox proportional hazards models, linear support vector machines, random forest, neural networks, AdaBoost, and gradient boosting machines | Python | 423,604 | 6 |
| Anderson et al. [48] | To identify patient characteristics that predict progression to prediabetes and type 2 diabetes in a US adult population | Diabetes | Retrospective database—electronic health records (Humedica) | Novel analytical platform based on a Bayesian approach | Reverse Engineering and Forward Simulation (REFS™) | 24,331 | 1 |
| Azimi et al. [38] | To select patients for surgery or non-surgical options | Neurology | Retrospective database | Logistic regression, Artificial neural network | SPSS for windows (Version 17.0), STATISTICA 10.0 Neural Networks | 346 | 2 |
| Bannister et al. [22] | To determine the utility of genetic programming for the automatic development of clinical prediction models | Cardiovascular disease | Prospective observational cohort | Cox regression models, Tree-based genetic programming | R Package version 3.0.1 and 3.1.2 | 3873 | 2 |
| Baxter et al. [24] | To predict the need for surgical intervention in patients with primary open-angle glaucoma | Neurology | Retrospective database—electronic health records (EpicCare) | Logistic regression, random forest, artificial neural network | Random Forest R package; nnet package in R | 385 | 3 |
| Bertsimas et al. [21] | To predict patients at high risk of mortality before the start of treatment regimes | Oncology | Retrospective database—electronic health records and social security death index | Logistic regression, Decision tree analysis (Gradient boosted, optimal classification, and Classification and Regression Tree [CART]) | Not specified | 23,983 | 2 |
| Bowman et al. [39] | To develop and validate a comprehensive, multivariate prognostic model for carpal tunnel surgery | Neurology | Retrospective database—clinic data | Logistic regression, artificial neural network | Stata v 14, MATLAB v. 8.3.0.532 | 200 | 2 |
| Dong et al. [25] | To present and validate a novel surgical predictive model to facilitate therapeutic decision-making | Inflammatory bowel disease | Retrospective database- electronic health records | Random forest, logistic regression, decision tree, support vector machine, artificial neural network | Python 3.6 | 239 | 5 |
| Hearn et al. [40] | To assess whether the prognostication of heart failure patients using cardiopulmonary exercise test data could be improved by considering the entirety of the data generated during a cardiopulmonary exercise test, as opposed to using summary indicators alone | Cardiovascular disease | Retrospective database -electronic health records and exercise test data | Logistic regression, least absolute shrinkage and selection operator (LASSO) model, generalized additive model, feedforward neural network | R Project for Statistical Computing v3.4.2, and Python Programming Language v3.6.2 | 1156 | 4 |
| Hertroijs et al. [45] | To identify subgroups of people with newly diagnosed type 2 diabetes with distinct glycaemic trajectories; to predict trajectory membership using patient characteristics | Diabetes | Retrospective database -electronic health records | Latent growth mixture modeling, K-nearest neighbor/Parzen, Fisher, linear/quadratic discriminant classifier, supper vector machine, radial basis function, logistic regression | Mplus Version 7.1 | 14,305 | 7 |
| Hill et al. [26] | To develop a clinically applicable risk prediction model to identify associations between baseline and time-varying factors and the identification of atrial fibrillation | Cardiovascular disease | Retrospective database—electronic health records (Clinical Practice Research Datalink, CPRD) | Logistic least absolute shrinkage and selector operator (LASSO), random forests, support vector machines, neural networks | R v3.3.1 | 2,994,837—baseline model; 162,672—time varying model | 4 |
| Hische et al. [20] | To create a simple and reliable tool to identify individuals with impaired glucose metabolism | Diabetes | Cross-sectional study | Decision tree analysis | Quinlan C4.5 | 1737 | 1 |
| Isma'eel et al. [41] | To investigate the use of artificial neural networks to improve risk stratification and prediction of myocardial perfusion imaging and angiographic results | Cardiovascular disease | Retrospective medical records | Artificial neural network | Not specified | 5354 | 1 |
| Isma'eel et al. [42] | To compare artificial neural network-based prediction models to other risk models that are being used in clinical practice | Cardiovascular disease | Prospective cohort | Artificial neural network | Not specified | 486 | 1 |
| Jovanovic et al. [43] | to determine whether an artificial neural network model could be constructed to accurately predict the need for therapeutic ERCP in patients with a firm clinical suspicion of having common bile duct stones and to compare it with our previously reported predictive model | Choledocholithiasis | Prospective cohort | Artificial neural network | SPSS v20.0 | 291 | 1 |
| Kang et al. [27] | To investigate the feasibility of developing a machine-learning model to predict postinduction hypotension | Cardiovascular disease | Retrospective database—electronic health records | Developed Naïve Bayes, logistic regression, random forest, and artificial neural network | caret R package | 222 | 4 |
| Karhade et al. [28] | To develop algorithms for prediction of prolonged opioid prescription after surgery for lumbar disc herniation | Lumbar disk herniation | Retrospective chart review | Random forest, stochastic gradient boosting, neural network, support vector machine, elastic-net penalized logistic regression | Anaconda Distribution, R version 3.5.0, RStudio version 1.1.453, and Python version 3.6 | 5413 | 5 |
| Kebede et al. [29] | to predict CD4 count changes and to identify the predictors of CD4 count changes among patients on ART | HIV/AIDS | Retrospective database and chart review | J48 decision tree/random forest, neural network | WEKA 3.8 | 3104 | 2 |
| Khanji et al. [47] | To identify an effective method to build prediction models and assess predictive validity of pre-defined indicators | Cardiovascular disease | Observational trial, cluster randomization | Logistic regression, LASSO regression, Hybrid approach (combination of both approaches) | SAS version 9.1, SPSS version 24, and R version 3.3.2 | 759 | 2 |
| Kim et al. [30] | to develop a prediction tool using machine learning for high- or low-risk oncotype dx criteria | Oncology | Retrospective chart review | Two-class Decision Forest, Two-class Decision Jungle, Two-class Bayes Point Machine, Two-class Support Vector Machine, Two-class Neural Network | SAS 9.4; Azure Machine Learning Platform | 284 | 4 |
| Kwon et al. [32] | to predict cardiac arrest using deep learning | Cardiovascular disease | Retrospective database—electronic health records | Random forest, logistic regression, recurrent neural network | Not specified | 52,131 | 3 |
| Kwon et al. [31] | to predict prognosis of out-of-hospital cardiac arrest using deep-learning | Cardiovascular disease | Retrospective database – registry | Logistic regression, support vector machine, random forest | Not specified | 36,190 | 3 |
| Lopez-de-Andres et al. [34] | To estimate predictive factors of in-hospital mortality in patients with type 2 diabetes after major lower extremity amputation | Diabetes | Retrospective database—hospital discharge database | Artificial neural network | Neural Designer; Stata MP version 10.1 | 40,857 lower extremity amputation events | 1 |
| Mubeen et al. [19] | To assess risk of developing Alzheimer’s Disease in mildly cognitively impaired subjects; to classify subjects in two groups: those who would remain stable and those who would progress to develop Alzheimer’s disease | Alzheimer’s Disease | Retrospective database – (Alzheimer’s Disease Neuroimaging Project) | Random Forest algorithm | Random Forest R package | 247 | 1 |
| Neefjes et al. [18] | To develop a prediction model to identify patients with cancer at high risk for delirium | Oncology | Retrospective database –hospital inpatient data | Decision tree analysis | R program Rpart version 3.1; Statistical Package for the Social Sciences (SPSS) v20.0 | 574 | 1 |
| Ng et al. [36] | To create a clinical decision support tool to predict survival in cancer patients beyond 120 days after palliative chemotherapy | Oncology | Retrospective database—electronic health records and case notes | Naïve Bayes, neural network, and support vector machine | SIMCA-P + version 12.0.1; SPSS version 19.0; RapidMiner version 5.0.010 | 325 | 3 |
| Oviedo et al. [46] | To focus on patient-specific prediction of hypoglycemic events | Diabetes | Retrospective database – hospital clinic data | Support vector classifier | Python | 10 | 1 |
| Pei et al. [17] | To identify individuals with potential diabetes | Diabetes | Retrospective medical records review | Decision tree analysis | WEKA 3.8.1 and SPSS version 20.0 | 10,436 | 1 |
| Perez-Gandia et al. [37] | To predict future glucose concentration levels from continuous glucose monitoring data | Diabetes | Retrospective database—device dataset | Artificial neural network | Not specified | 15 | 1 |
| Ramezankhani et al. [16] | To gain more information on interactions between factors contributing to the incidence of type 2 diabetes | Diabetes | Prospective cohort | Decision tree analysis (CART, Quick Unbiased Efficient Statistical Tree [QUEST], and commercial version [C5.0]) | IBM SPSS modeler 14.2 | 6647 | 1 |
| Rau et al. [35] | To predict the development of liver cancer within 6 years of diagnosis with type 2 diabetes | Diabetes and Oncology | Retrospective database—claims linked to registry data | Logistic regression, artificial neural network, support vector machine, and decision tree analysis | STATISTICA, version 10 | 2060 | 4 |
| Scheer et al. [33] | To develop a model based on baseline demographic, radiographic, and surgical factors that can predict if patients will sustain an intraoperative or perioperative major complication | Spinal deformity | Retrospective database | Decision tree analysis | SPSS version 22; SPSS modeler version 16 | 557 | 1 |
| Toussi et al. [15] | To identify knowledge gaps in guidelines and to explore physicians' therapeutic decisions using data mining techniques to fill these knowledge gaps | Diabetes | Retrospective database—electronic health records | Decision tree analysis | Quinlan’s C5.0 decision-tree learning algorithm; SPSS Clementine software version 10.1 | 463 | 1 |
| Zhou et al. [44] | To assess pre-procedural independent risk factors and to establish a “Risk Prediction for Early Biliary Infection” nomogram for patients with malignant biliary obstruction who underwent percutaneous transhepatic biliary stent | Oncology | Retrospective medical record and trial data | Logistic regression, artificial neural network | SPSS version 22; R package (version 3.4.3) | 243 | 2 |
General approaches to machine learning
The types of classification or prediction machine learning algorithms are reported in Table 2. These included decision tree/random forest analyses (19 studies) [15–33] and neural networks (19 studies) [24–30, 32, 34–44]. Other approaches included latent growth mixture modeling [45], support vector machine classifiers [46], LASSO regression [47], boosting methods [23], and a novel Bayesian approach [26, 40, 48]. Within the analytical approaches to support machine learning, a variety of methods were used to evaluate model fit, such as Akaike Information Criterion, Bayesian Information Criterion, and the Lo-Mendel-Rubin likelihood ratio test [22, 45, 47], and while most studies included the area under the curve (AUC) of receiver-operator characteristic (ROC) curves (Table 3), analyses also included sensitivity/specificity [16, 19, 24, 30, 41–43], positive predictive value [21, 26, 32, 38, 40–43], and a variety of less common approaches such as the geometric mean [16], use of the Matthews correlation coefficient (ranges from -1.0, completely erroneous information, to + 1.0, perfect prediction) [46], defining true/false negatives/positives by means of a confusion matrix [17], calculating the root mean square error of the predicted versus original outcome profiles [37], or identifying the model with the best average performance training and performance cross validation [36].
Table 3.
Details of methods applied to the analysis in eligible studies
| References | Internal validation | Evaluation of model fit/performance | Handling of missing values |
|---|---|---|---|
| Alaa et al. [23] | Training set corresponding to 90% of the sample; ten-fold cross validation | Area under the receiver operating characteristic curve and 95% confidence intervals (Wilson score intervals), Brier score | Retained as an informative variable, and only variables missing for 85% or more of participants were excluded |
| Anderson et al. [48] | Split the data based on site of care | Bayesian Information Criterion, prediction model ensembles, ß estimates, predicted probabilities, and area under the receiver operating characteristic curve estimates | Missing covariate values were included in models as a discrete category |
| Azimi et al. [38] | 2:1:1 ratio to generate training, testing, and validation cohorts | Receiver-operating characteristic curves, positive predictive value, negative predictive value, area under the curve from the receiver operating curve analysis, Homer-Lemeshow statistic | Cases with missing outcome data were excluded |
| Bannister et al. [22] | Derivation set of approximately 66.67% and a validation set of approximately 33.33% as noted in text (abstract states 70:30 split) | Akaike Information Criterion | Single imputation, followed by multiple imputation in the final model to evaluate differences in model parameters |
| Baxter et al. [24] | Leave-one-out cross-validation (LOOCV) approach, also known as the jackknife method | Area under the receiver operating characteristic curve, sensitivity, specificity, accuracy, the Youden index | Not mentioned |
| Bowman [39] | Re-analysis of models using clinic data during a different time period | Area under the receiver operating characteristic curve | Not mentioned |
| Bertsimas et al. [21] | 60/40 split | Positive predictive value, area under the curve, accuracy | Imputed using an optimal-impute algorithm |
| Dong et al. [25] | 9/1 random split, ten-fold cross validation | Accuracy, precision, F1 score, true negative rate, area under the receiver operating characteristic curve | Missing values were filled based on the mean value of data from the same attribute of the same patient. If a patient had fewer than 3 records, imputed the mean value of that attribute from all patients |
| Hearn et al. [40] | 100-iteration Monte Carlo cross validation, 75/25 split, five-fold cross validation | Mean area under the receiver operating characteristic curve, true- and false-positive rates, true- and false-negative rates, positive and negative predictive values | Variables with > 10% of values missing were discarded from the analysis, whereas the remaining missing values were filled in using multiple imputation by chained random forests (maximum number of iterations = 5, number of trees = 10). The sole exception to the 10% cutoff was heart rate recovery, which featured 32% missing values but was kept in the data set and imputed with the above procedure because of its wide usage in prognostication from cardiopulmonary exercise test |
| Hertroijs et al. [45] | Five-fold cross validation | Akaike Information Criterion, Bayesian Information Criterion, Lo-Mendel-Rubin likelihood ratio test | The full information maximum likelihood method was used for estimating model parameters in the presence of missing data for the development of the model, but patients with missing covariate values at baseline were excluded from the validation of the model |
| Hill et al. [26] | 2/1 split | Area under the receiver operating characteristic curve, positive predictive value, potential number reeded-to-screen | Imputed with last-observation-carried-forward |
| Hische et al. [20] | Ten-fold cross validation | Models with a mean sensitivity above 90% after cross-validation were selected, specificity, positive predictive value, negative predictive value | Not mentioned |
| Isma'eel et al. [41] | For myocardial perfusion imaging: 120 out the 479 patients who tested positive were added randomly to the derivation cohort; 120 of the remaining 4875 patients who tested negative were also added randomly to the derivation cohort. The remaining 5114 patients were all added to the validation cohort. For coronary artery disease: the derivation cohort was randomly chosen as follows: 93 out of the 278 patients who tested positive were added randomly to the derivation cohort and 93 out of the remaining 5076 patients who tested negative were also added randomly to the derivation cohort. The remaining 5169 patients are all added to the validation cohort | Sensitivity and specificity, discriminatory power and 95% confidence interval, number of tests avoided, negative predictive value, positive predictive value | Not mentioned |
| Isma'eel et al. [42] | The derivation cohort was randomly chosen 30 out of the 59 patients who tested positive were added randomly to the derivation cohort, and 30 out of the remaining 427 patients who tested negative were also added randomly to the derivation cohort. The remaining 426 patients (29 positive, 397 negative) were all added to the testing cohort; during the training phase, the 60 patients that are used for training were split 80% for pure training and 20% for validation | Negative and positive predictive values, descriminatory power, percentage of avoided tests, sensitivity and specificity | Not mentioned |
| Jovanovic et al. [43] | The sample was randomly divided into 3 parts: training, testing, and validation sample | Area under the receiver operating curve, sensitivity, specificity, and positive and negative predictive values | Not mentioned |
| Kang et al. [27] | Four-fold cross validation, 75/25 split (training/validation) | Area under the receiver operating curve, accuracy, precision, recall | Not mentioned |
| Karhade et al. [28] | tenfold cross validation | Discrimination (c-statistic or area under the receiver operating curve), calibration (calibration slope, calibration intercept), and overall performance (Brier score) | Multiple imputation with the missForest methodology was undertaken for variables with less than 30% missing data |
| Kebede et al. [29] | 10% cross validation, 90/10 split (training/testing) | Area under the receiver operating curve; classification accuracy-true positive, false positive, precision, recall | If information is incomplete, un-readable or their manual record is lost, patients were excluded from the study |
| Khanji et al. [47] | Ten-fold cross validation | Akaike Information Criterion, area under the receiver operating curve | Excluded patients with missing data at the end of the study (± 6 months) |
| Kim et al. [30] | 70/30 split (training/testing) | Area under the receiver operating curve, sensitivity, specificity, precision, accuracy | Not mentioned |
| Kwon et al. [32] | Derivation set (June 2010-July 2016) and validation set (Aug 2016–2017) split by date | Receiver operating characteristic curve, the area under the precision–recall curve, net reclassification index, sensitivity, positive predictive value, negative predictive value, net reclassification index, F-measure | Not mentioned |
| Kwon et al. [31] | Split into derivation and validation datasets according to the year. The derivation data was the patient data for 2012–2015, and the validation data was the patient data for 2016 | Area under the receiver operating characteristic curve | Excluded patients with missing values |
| Lopez-de-Andres et al. [34] | Random 60/20/20 split, where the third group was selected for model selection purposes prior to validation | Area under the receiver operating characteristic curve, accuracy rate, error rate, sensitivity, specificity, precision, positive likelihood, negative likelihood, F1 score, false positive rate, false negative rate, false discovery rate, positive predictive value, negative predictive value, Matthews correlation, informedness, markedness | Not mentioned |
| Mubeen et al. [19] | Ten-fold cross validation | Accuracy, sensitivity, specificity based on prediction of out-of-bag samples, area under the receiver operating characteristic curve | Not mentioned |
| Neefjes et al. [18] | Five-fold cross validation | Area under the receiver operating characteristic curve | Not mentioned |
| Ng et al. [36] | 60/40 split, five-fold cross validation | Area under the curve, sensitivity, specificity, accuracy | Excluded patients with missing data |
| Oviedo et al. [46] | 80/20 split (training/testing) | Matthews correlation coefficient | Removed records from the model with missing data |
| Pei et al. [17] | 70/30 split (training/testing) | True positives, true negatives, false negatives, false positives, area under the receiver operating characteristic curve | Removed patients from the model with missing data |
| Perez-Gandia et al. [37] | Three subjects (each with two daily profiles) were used for training, the remaining patients were used for validation | Accuracy | Spline techniques were used to impute missing data in the training set |
| Ramezankhani et al. [16] | 70/30 split (training/validation) | Sensitivity, specificity, positive predictive value, negative predictive value, geometric mean, F-measure, area under the curve | Single imputation was used, for continuous variables- CART, for categorical variables- weighted K-Nearest Neighbor approach |
| Rau et al. [35] | 70/30 split, ten-fold cross validation | Sensitivity, specificity, area under the curve | Not mentioned |
| Scheer et al. [33] | 70/30 split (training/testing) | Accuracy, area under the receiver operating characteristic curve, predictor importance | Missing values within the database were imputed using standard techniques such as mean and median imputation |
| Toussi et al. [15] | Ten-fold cross validation | Precision (proportion of true positive records to the proportion of true positive and false positive records) | imputed using a model-based approach |
| Zhou et al. [44] | Training cohort: 142 patients were part of a prior phase III randomized controlled trial (Oct 2013–Mar 2016) + 182 eligible consecutive patients (Jan 2012–Dec 2016). Validation cohort: 61 eligible consecutive patients (Jan 2017–Aug 2017) | The concordance index (c-index) was calculated as the area under the receiver operating characteristic curve, calibration plot | Patients were excluded if clinical data were missing before or 30 days after percutaneous transhepatic biliary drainage |
Statistical software packages
The statistical programs used to perform machine learning varied widely across these studies, no consistencies were observed (Table 2). As noted above, one study using decision tree analysis used Quinlan’s C5.0 decision tree algorithm [15] while a second used an earlier version of this program (C4.5) [20]. Other decision tree analyses utilized various versions of R [18, 19, 22, 24, 27, 47], International Business Machines (IBM) Statistical Package for the Social Sciences (SPSS) [16, 17, 33, 47], the Azure Machine Learning Platform [30], or programmed the model using Python [23, 25, 46]. Artificial neural network analyses used Neural Designer [34] or Statistica V10 [35]. Six studies did not report the software used for analysis [21, 31, 32, 37, 41, 42].
Families of machine learning algorithms
Also as summarized in Table 2, more than one third of all publications (n = 13, 38.2%) applied only one family of machine learning algorithm to model development [16–20, 34, 37, 41–43, 46, 48]; and only four studies utilized five or more methods [23, 25, 28, 45]. One applied an ensemble of six different algorithms and the software was set to run 200 iterations [23], and another ran seven algorithms [45].
Internal and external validation
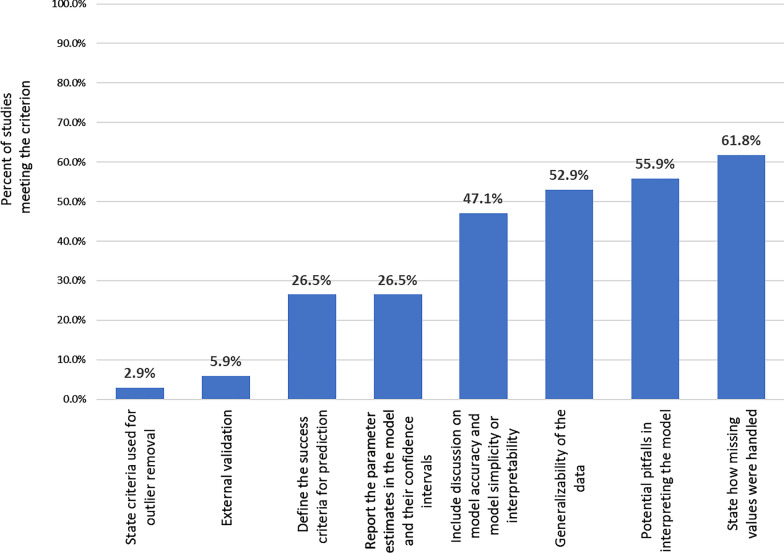
Evaluation of study publication quality identified the most common gap in publications as the lack of external validation, which was conducted by only two studies [15, 20]. Seven studies predefined the success criteria for model performance [20, 21, 23, 35, 36, 46, 47], and five studies discussed the generalizability of the model [20, 23, 34, 45, 48]. Six studies [17, 18, 21, 22, 35, 36] discussed the balance between model accuracy and model simplicity or interpretability, which was also a criterion of quality publication in the Luo scale [14]. The items on the checklist that were least frequently met are presented in Fig. 2. The complete quality assessment evaluation for each item in the checklist is included in Additional file 1: Table S1.
Fig. 2.
Least frequently met study quality items, modified Luo Scale [14]
There were a variety of approaches taken to validate the models developed (Table 3). Internal validation with splitting into a testing and validation dataset was performed in all studies. The cohort splitting approach was conducted in multiple ways, using a 2:1 split [26], 60/40 split [21, 36], a 70/30 split [16, 17, 22, 30, 33, 35], 75/25 split [27, 40], 80/20 split [46], 90/10 split [25, 29], splitting the data based on site of care [48], a 2/1/1 split for training, testing and validation [38], and splitting 60/20/20, where the third group was selected for model selection purposes prior to validation [34]. Nine studies did not specifically mention the form of splitting approach used [15, 18–20, 24, 29, 39, 45, 47], but most of those noted the use of k fold cross validation. One training set corresponded to 90% of the sample [23], whereas a second study was less clear, as input data were at the observation level with multiple observations per patient, and 3 of the 15 patients were included in the training set [37]. The remaining studies did not specifically state splitting the data into testing and validation samples, but most specified they performed five-fold cross validation (including one that generally mentioned cohort splitting) [18, 45] or ten-fold cross validation strategies [15, 19, 20, 28].
External validation was conducted by only two studies (5.9%). Hische and colleagues conducted a decision tree analysis, which was designed to identify patients with impaired fasting glucose [20]. Their model was developed in a cohort study of patients from the Berlin Potsdam Cohort Study (n = 1527) and was found to have a positive predictive value of 56.2% and a negative predictive value of 89.1%. The model was then tested on an independent from the Dresden Cohort (n = 1998) with a family history of type II diabetes. In external validation, positive predictive value was 43.9% and negative predictive value was 90.4% [20]. Toussi and colleagues conducted both internal and external validation in their decision tree analysis to evaluate individual physician prescribing behaviors using a database of 463 patient electronic medical records [15]. For the internal validation step, the cross-validation option was used from Quinlan’s C5.0 decision tree learning algorithm as their study sample was too small to split into a testing and validation sample, and external validation was conducted by comparing outcomes to published treatment guidelines. Unfortunately, they found little concordance between physician behavior and guidelines potentially due to the timing of the data not matching the time period in which guidelines were implemented, emphasizing the need for a contemporaneous external control [15].
Handling of missing values
Missing values were addressed in most studies (n = 21, 61.8%) in this review, but there were thirteen remaining studies that did not mention if there were missing data or how they were handled (Table 3). For those that reported methods related to missing data, there were a wide variety of approaches used in real-world datasets. The full information maximum likelihood method was used for estimating model parameters in the presence of missing data for the development of the model by Hertroijs and colleagues, but patients with missing covariate values at baseline were excluded from the validation of the model [45]. Missing covariate values were included in models as a discrete category [48]. Four studies removed patients from the model with missing data [46], resulting in the loss of 16%-41% of samples in three studies [17, 36, 47]. Missing data from primary outcome variables were reported among with 59% (men) and 70% (women) within a study of diabetes [16]. In this study, single imputation was used; for continuous variables CART (IBM SPSS modeler V14.2.03) and for categorical variables the authors used the weighted K-Nearest Neighbor approach using RapidMiner (V.5) [16]. Other studies reported exclusion but not specifically the impact on sample size [29, 31, 38, 44]. Imputation was conducted in a variety of ways for studies with missing data [22, 25, 28, 33]. Single imputation was used in the study by Bannister and colleagues, but followed by multiple imputation in the final model to evaluate differences in model parameters [22]. One study imputed with a standard last-imputation-forward approach [26]. Spline techniques were used to impute missing data in the training set of one study [37]. Missingness was largely retained as an informative variable, and only variables missing for 85% or more of participants were excluded by Alaa et al. [23] while Hearn et al. used a combination of imputation and exclusion strategies [40]. Lastly, missing or incomplete data were imputed using a model-based approach by Toussi et al. [15] and using an optimal-impute algorithm by Bertsimas et al. [21].
Strengths and weaknesses noted by authors
Publications summarized the strengths and weaknesses of the machine learning methods employed. Low complexity and simplicity of machine-based learning models were noted as strengths of this approach [15, 20]. Machine learning approaches were both powerful and efficient methods to apply to large datasets [19]. It was noted that parameters in this study that were significant at the patient level were included, even if at the broader population-based level using traditional regression analysis model development they would have not been significant and therefore would have been otherwise excluded using traditional approaches [34]. One publication noted the value of machine learning being highly dependent on the model selection strategy and parameter optimization, and that machine learning in and of itself will not provide better estimates unless these steps are conducted properly [23].
Even when properly planned, machine learning approaches are not without issues that deserve attention in future studies that employ these techniques. Within the eligible publications, weaknesses included overfitting the model with the inclusion of too much detail [15]. Additional limitations are based on the data sources used for machine learning, such as the lack of availability of all desired variables and missing data that can affect the development and performance of these models [16, 34, 36, 48]. The lack of all relevant variables was noted as a particular concern for retrospective database studies, where the investigator is limited to what has been recorded [26, 28, 29, 38, 40]. Importantly and as observed in the studies included in this review, the lack of external validation was stated as a limitation of studies included in this review [28, 30, 38, 42].
Limitations can also be on the part of the research team, as the need for both clinical and statistical expertise in the development and execution of studies using machine learning-based methodology, and users are warned against applying these methods blindly [22]. The importance of the role of clinical and statistical experts in the research team was noted in one study and highlighted as a strength of their work [21].
Discussion
This study systematically reviewed and summarized the methods and approaches used for machine learning as applied to observational datasets that can inform patient-provider decision making. Machine learning methods have been applied much more broadly across observational studies than in the context of individual decision making, so the summary of this work does not necessarily apply to all machine learning-based studies. The focus of this work is on an area that remains largely unexplored, which is how to use large datasets in a manner that can inform and improve patient care in a way that supports shared decision making with reliable evidence that is applicable to the individual patient. Multiple publications cite the limitations of using population-based estimates for individual decisions [49–51]. Specifically, a summary statistic at the population level does not apply to each person in that cohort. Population estimates represent a point on a potentially wide distribution, and any one patient could fall anywhere within that distribution and be far from the point estimate value. On the other extreme, case reports or case series provide very specific individual-level data, but are not generalizable to other patients [52]. This review and summary provides guidance and suggestions of best practices to improve and hopefully increase the use of these methods to provide data and models to inform patient-provider decision making.
It was common for single modeling strategies to be employed within the identified publications. It has long been known that single algorithms to estimation can produce a fair amount of uncertainty and variability [53]. To overcome this limitation, there is a need for multiple algorithms and multiple iterations of the models to be performed. This, combined with more powerful analytics in recent years, provides a new standard for machine learning algorithm choice and development. While in some cases, a single model may fit the data well and provide an accurate answer, the certainty of the model can be supported through novel approaches, such as model averaging [54]. Few studies in this review combined multiple families of modeling strategies along with multiple iterations of the models. This should become a best practice in the future and is recommended as an additional criterion to assess study quality among machine learning-based modeling [54].
External validation is critical to ensure model accuracy, but was rarely conducted in the publications included in this review. The reasons for this could be many, such as lack of appropriate datasets or due to the lack of awareness of the importance of external validation [55]. As model development using machine learning increases, there is a need for external validation prior to application of models in any patient-provider setting. The generalizability of models is largely unknown without these data. Publications that did not conduct external validation also did not note the need for this to be completed, as generalizability was discussed in only five studies, one of which had also conducted the external validation. Of the remaining four studies, the role of generalizability was noted in terms of the need for future external validation in only one study [48]. Other reviews that were more broadly conducted to evaluate machine learning methods similarly found a low rate of external validation (6.6% versus 5.9% in this study) [56]. It was shown that there was lower prediction accuracy by external validation than simply by cross validation alone. The current review, with a focus on machine learning to support decision making at a practical level, suggests external validation is an important gap that should be filled prior to using these models for patient-provider decision making.
Luo and others suggest that k-fold validation may be used with proper stratification of the response variable as part of the model selection strategy [14, 55]. The studies identified in this review generally conducted 5- or tenfold validation. There is no formal rule for the selection for the value of k, which is typically based on the size of the dataset; as k increases, bias will be reduced, but in turn variance will increase. While the tradeoff has to be accounted for, k = 5–10 has been found to be reasonable for most study purposes [57].
The evidence from identified publications suggests that the ethical concerns of lack of transparency and failure to report confidence in the findings are largely warranted. These limitations can be addressed through the use of multiple modeling approaches (to clarify the ‘black box’ nature of these approaches) and by including both external and high k-fold validation (to demonstrate the confidence in findings). To ensure these methods are used in a manner that improves patient care, the expectations of population-based risk prediction models of the past are no longer sufficient. It is essential that the right data, the right set of models, and appropriate validation are employed to ensure that the resulting data meet standards for high quality patient care.
This study did not evaluate the quality of the underlying real-world data used to develop, test or validate the algorithms. While not directly part of the evaluation in this review, researchers should be aware that all limitations of real-world data sources apply regardless of the methodology employed. However, when observational datasets are used for machine learning-based research, the investigator should be aware of the extent to which the methods they are using depend on the data structure and availability, and should evaluate a proposed data source to ensure it is appropriate for the machine learning project [45]. Importantly, databases should be evaluated to fully understand the variables included, as well as those variables that may have prognostic or predictive value, but may not be included in the dataset. The lack of important variables remains a concern with the use of retrospective databases for machine learning. The concerns with confounding (particularly unmeasured confounding), bias (including immortal time bias), and patient selection criteria to be in the database must also be evaluated [58, 59]. These are factors that should be considered prior to implementing these methods, and not always at the forefront of consideration when applying machine learning approaches. The Luo checklist is a valuable tool to ensure that any machine-learning study meets high research standards for patient care, and importantly includes the evaluation of missing or potentially incorrect data (i.e. outliers) and generalizability [14]. This should be supplemented by a thorough evaluation of the potential data to inform the modeling work prior to its implementation, and ensuring that multiple modeling methods are applied.
Conclusions
This review found a wide variety of approaches, methods, statistical software and validation strategies that were employed in the application of machine learning methods to inform patient-provider decision making. Based on these findings, there is a need to ensure that multiple modeling approaches are employed in the development of machine learning-based models for patient care, which requires the highest research standards to reliably support shared evidence-based decision making. Models should be evaluated with clear criteria for model selection, and both internal and external validation are needed prior to applying these models to inform patient care. Few studies have yet to reach that bar of evidence to inform patient-provider decision making.
Supplementary Information
Additional file 1. Table S1. Study quality of eligible publications, modified Luo scale [14].
Acknowledgements
Not applicable.
Abbreviations
- AI
Artificial intelligence
- AUC
Area under the curve
- CART
Classification and regression trees
- LASSO
Logistic least absolute shrinkage and selector operator
Authors’ contributions
AB and LMH contributed to the design, implementation, analysis and interpretation of the data included in this study. AB and LMH wrote, revised and finalized the manuscript for submission. AB and LMH have both read and approved the final manuscript.
Funding
No funding was received for the conduct of this study.
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
Authors are employees of Eli Lilly and Company and receive salary support in that role.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12911-021-01403-2.
References
- 1.Steyerberg EW, Claggett B. Towards personalized therapy for multiple sclerosis: limitations of observational data. Brain. 2018;141(5):e38-e. [DOI] [PubMed]
- 2.Fröhlich H, Balling R, Beerenwinkel N, Kohlbacher O, Kumar S, Lengauer T, et al. From hype to reality: data science enabling personalized medicine. BMC Med. 2018;16(1):150. doi: 10.1186/s12916-018-1122-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Steyerberg EW. Clinical prediction models. Berlin: Springer; 2019. [Google Scholar]
- 4.Schnabel RB, Sullivan LM, Levy D, Pencina MJ, Massaro JM, D'Agostino RB, Sr, et al. Development of a risk score for atrial fibrillation (Framingham Heart Study): a community-based cohort study. Lancet. 2009;373(9665):739–745. doi: 10.1016/S0140-6736(09)60443-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.D'Agostino RB, Wolf PA, Belanger AJ, Kannel WB. Stroke risk profile: adjustment for antihypertensive medication. Framingham Study Stroke. 1994;25(1):40–43. doi: 10.1161/01.STR.25.1.40. [DOI] [PubMed] [Google Scholar]
- 6.Framingham Heart Study: Risk Functions 2020. https://www.framinghamheartstudy.org/.
- 7.Gawehn E, Hiss JA, Schneider G. Deep learning in drug discovery. Mol Inf. 2016;35:3–14. doi: 10.1002/minf.201501008. [DOI] [PubMed] [Google Scholar]
- 8.Vamathevan J, Clark D, Czodrowski P, Dunham I, Ferran E, Lee G, et al. Applications of machine learning in drug discovery and development. Nat Rev Drug Discov. 2019;18(6):463–477. doi: 10.1038/s41573-019-0024-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marcus G. Deep learning: A critical appraisal. arXiv preprint arXiv:180100631. 2018.
- 10.Grote T, Berens P. On the ethics of algorithmic decision-making in healthcare. J Med Ethics. 2020;46(3):205–211. doi: 10.1136/medethics-2019-105586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brnabic A, Hess L, Carter GC, Robinson R, Araujo A, Swindle R. Methods used for the applicability of real-world data sources to individual patient decision making. Value Health. 2018;21:S102. doi: 10.1016/j.jval.2018.07.773. [DOI] [Google Scholar]
- 12.Fu H, Zhou J, Faries DE. Estimating optimal treatment regimes via subgroup identification in randomized control trials and observational studies. Stat Med. 2016;35(19):3285–3302. doi: 10.1002/sim.6920. [DOI] [PubMed] [Google Scholar]
- 13.Liang M, Ye T, Fu H. Estimating individualized optimal combination therapies through outcome weighted deep learning algorithms. Stat Med. 2018;37(27):3869–3886. doi: 10.1002/sim.7902. [DOI] [PubMed] [Google Scholar]
- 14.Luo W, Phung D, Tran T, Gupta S, Rana S, Karmakar C, et al. Guidelines for developing and reporting machine learning predictive models in biomedical research: a multidisciplinary view. J Med Internet Res. 2016;18(12):e323. doi: 10.2196/jmir.5870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Toussi M, Lamy J-B, Le Toumelin P, Venot A. Using data mining techniques to explore physicians' therapeutic decisions when clinical guidelines do not provide recommendations: methods and example for type 2 diabetes. BMC Med Inform Decis Mak. 2009;9(1):28. doi: 10.1186/1472-6947-9-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramezankhani A, Hadavandi E, Pournik O, Shahrabi J, Azizi F, Hadaegh F. Decision tree-based modelling for identification of potential interactions between type 2 diabetes risk factors: a decade follow-up in a Middle East prospective cohort study. BMJ Open. 2016;6(12):e013336. doi: 10.1136/bmjopen-2016-013336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pei D, Zhang C, Quan Y, Guo Q. Identification of potential type II diabetes in a Chinese population with a sensitive decision tree approach. J Diabetes Res. 2019;2019:4248218. doi: 10.1155/2019/4248218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neefjes EC, van der Vorst MJ, Verdegaal BA, Beekman AT, Berkhof J, Verheul HM. Identification of patients with cancer with a high risk to develop delirium. Cancer Med. 2017;6(8):1861–1870. doi: 10.1002/cam4.1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mubeen AM, Asaei A, Bachman AH, Sidtis JJ, Ardekani BA, Initiative AsDN. A six-month longitudinal evaluation significantly improves accuracy of predicting incipient Alzheimer's disease in mild cognitive impairment. J Neuroradiol. 2017;44(6):381–387. doi: 10.1016/j.neurad.2017.05.008. [DOI] [PubMed] [Google Scholar]
- 20.Hische M, Luis-Dominguez O, Pfeiffer AF, Schwarz PE, Selbig J, Spranger J. Decision trees as a simple-to-use and reliable tool to identify individuals with impaired glucose metabolism or type 2 diabetes mellitus. Eur J Endocrinol. 2010;163(4):565. doi: 10.1530/EJE-10-0649. [DOI] [PubMed] [Google Scholar]
- 21.Bertsimas D, Dunn J, Pawlowski C, Silberholz J, Weinstein A, Zhuo YD, et al. Applied informatics decision support tool for mortality predictions in patients with cancer. JCO Clin Cancer Inform. 2018;2:1–11. doi: 10.1200/CCI.18.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bannister CA, Halcox JP, Currie CJ, Preece A, Spasic I. A genetic programming approach to development of clinical prediction models: a case study in symptomatic cardiovascular disease. PLoS ONE. 2018;13(9):e0202685. doi: 10.1371/journal.pone.0202685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alaa AM, Bolton T, Di Angelantonio E, Rudd JHF, van der Schaar M. Cardiovascular disease risk prediction using automated machine learning: a prospective study of 423,604 UK Biobank participants. PLoS ONE. 2019;14(5):e0213653. doi: 10.1371/journal.pone.0213653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baxter SL, Marks C, Kuo TT, Ohno-Machado L, Weinreb RN. Machine learning-based predictive modeling of surgical intervention in glaucoma using systemic data from electronic health records. Am J Ophthalmol. 2019;208:30–40. doi: 10.1016/j.ajo.2019.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dong Y, Xu L, Fan Y, Xiang P, Gao X, Chen Y, et al. A novel surgical predictive model for Chinese Crohn's disease patients. Medicine (Baltimore) 2019;98(46):e17510. doi: 10.1097/MD.0000000000017510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hill NR, Ayoubkhani D, McEwan P, Sugrue DM, Farooqui U, Lister S, et al. Predicting atrial fibrillation in primary care using machine learning. PLoS ONE. 2019;14(11):e0224582. doi: 10.1371/journal.pone.0224582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kang AR, Lee J, Jung W, Lee M, Park SY, Woo J, et al. Development of a prediction model for hypotension after induction of anesthesia using machine learning. PLoS ONE. 2020;15(4):e0231172. doi: 10.1371/journal.pone.0231172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karhade AV, Ogink PT, Thio Q, Cha TD, Gormley WB, Hershman SH, et al. Development of machine learning algorithms for prediction of prolonged opioid prescription after surgery for lumbar disc herniation. Spine J. 2019;19(11):1764–1771. doi: 10.1016/j.spinee.2019.06.002. [DOI] [PubMed] [Google Scholar]
- 29.Kebede M, Zegeye DT, Zeleke BM. Predicting CD4 count changes among patients on antiretroviral treatment: Application of data mining techniques. Comput Methods Programs Biomed. 2017;152:149–157. doi: 10.1016/j.cmpb.2017.09.017. [DOI] [PubMed] [Google Scholar]
- 30.Kim I, Choi HJ, Ryu JM, Lee SK, Yu JH, Kim SW, et al. A predictive model for high/low risk group according to oncotype DX recurrence score using machine learning. Eur J Surg Oncol. 2019;45(2):134–140. doi: 10.1016/j.ejso.2018.09.011. [DOI] [PubMed] [Google Scholar]
- 31.Kwon JM, Jeon KH, Kim HM, Kim MJ, Lim S, Kim KH, et al. Deep-learning-based out-of-hospital cardiac arrest prognostic system to predict clinical outcomes. Resuscitation. 2019;139:84–91. doi: 10.1016/j.resuscitation.2019.04.007. [DOI] [PubMed] [Google Scholar]
- 32.Kwon JM, Lee Y, Lee Y, Lee S, Park J. An algorithm based on deep learning for predicting in-hospital cardiac arrest. J Am Heart Assoc. 2018;7(13):26. doi: 10.1161/JAHA.118.008678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scheer JK, Smith JS, Schwab F, Lafage V, Shaffrey CI, Bess S, et al. Development of a preoperative predictive model for major complications following adult spinal deformity surgery. J Neurosurg Spine. 2017;26(6):736–743. doi: 10.3171/2016.10.SPINE16197. [DOI] [PubMed] [Google Scholar]
- 34.Lopez-de-Andres A, Hernandez-Barrera V, Lopez R, Martin-Junco P, Jimenez-Trujillo I, Alvaro-Meca A, et al. Predictors of in-hospital mortality following major lower extremity amputations in type 2 diabetic patients using artificial neural networks. BMC Med Res Methodol. 2016;16(1):160. doi: 10.1186/s12874-016-0265-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rau H-H, Hsu C-Y, Lin Y-A, Atique S, Fuad A, Wei L-M, et al. Development of a web-based liver cancer prediction model for type II diabetes patients by using an artificial neural network. Comput Methods Programs Biomed. 2016;125:58–65. doi: 10.1016/j.cmpb.2015.11.009. [DOI] [PubMed] [Google Scholar]
- 36.Ng T, Chew L, Yap CW. A clinical decision support tool to predict survival in cancer patients beyond 120 days after palliative chemotherapy. J Palliat Med. 2012;15(8):863–869. doi: 10.1089/jpm.2011.0417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pérez-Gandía C, Facchinetti A, Sparacino G, Cobelli C, Gómez E, Rigla M, et al. Artificial neural network algorithm for online glucose prediction from continuous glucose monitoring. Diabetes Technol Therapeut. 2010;12(1):81–88. doi: 10.1089/dia.2009.0076. [DOI] [PubMed] [Google Scholar]
- 38.Azimi P, Mohammadi HR, Benzel EC, Shahzadi S, Azhari S. Use of artificial neural networks to decision making in patients with lumbar spinal canal stenosis. J Neurosurg Sci. 2017;61(6):603–611. doi: 10.23736/S0390-5616.16.03078-2. [DOI] [PubMed] [Google Scholar]
- 39.Bowman A, Rudolfer S, Weller P, Bland JDP. A prognostic model for the patient-reported outcome of surgical treatment of carpal tunnel syndrome. Muscle Nerve. 2018;58(6):784–789. doi: 10.1002/mus.26297. [DOI] [PubMed] [Google Scholar]
- 40.Hearn J, Ross HJ, Mueller B, Fan CP, Crowdy E, Duhamel J, et al. Neural networks for prognostication of patients with heart failure. Circ. 2018;11(8):e005193. doi: 10.1161/CIRCHEARTFAILURE.118.005193. [DOI] [PubMed] [Google Scholar]
- 41.Isma'eel HA, Cremer PC, Khalaf S, Almedawar MM, Elhajj IH, Sakr GE, et al. Artificial neural network modeling enhances risk stratification and can reduce downstream testing for patients with suspected acute coronary syndromes, negative cardiac biomarkers, and normal ECGs. Int J Cardiovasc Imaging. 2016;32(4):687–696. doi: 10.1007/s10554-015-0821-9. [DOI] [PubMed] [Google Scholar]
- 42.Isma'eel HA, Sakr GE, Serhan M, Lamaa N, Hakim A, Cremer PC, et al. Artificial neural network-based model enhances risk stratification and reduces non-invasive cardiac stress imaging compared to Diamond-Forrester and Morise risk assessment models: a prospective study. J Nucl Cardiol. 2018;25(5):1601–1609. doi: 10.1007/s12350-017-0823-1. [DOI] [PubMed] [Google Scholar]
- 43.Jovanovic P, Salkic NN, Zerem E. Artificial neural network predicts the need for therapeutic ERCP in patients with suspected choledocholithiasis. Gastrointest Endosc. 2014;80(2):260–268. doi: 10.1016/j.gie.2014.01.023. [DOI] [PubMed] [Google Scholar]
- 44.Zhou HF, Huang M, Ji JS, Zhu HD, Lu J, Guo JH, et al. Risk prediction for early biliary infection after percutaneous transhepatic biliary stent placement in malignant biliary obstruction. J Vasc Interv Radiol. 2019;30(8):1233–41.e1. doi: 10.1016/j.jvir.2019.03.001. [DOI] [PubMed] [Google Scholar]
- 45.Hertroijs DF, Elissen AM, Brouwers MC, Schaper NC, Köhler S, Popa MC, et al. A risk score including body mass index, glycated haemoglobin and triglycerides predicts future glycaemic control in people with type 2 diabetes. Diabetes Obes Metab. 2018;20(3):681–688. doi: 10.1111/dom.13148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oviedo S, Contreras I, Quiros C, Gimenez M, Conget I, Vehi J. Risk-based postprandial hypoglycemia forecasting using supervised learning. Int J Med Inf. 2019;126:1–8. doi: 10.1016/j.ijmedinf.2019.03.008. [DOI] [PubMed] [Google Scholar]
- 47.Khanji C, Lalonde L, Bareil C, Lussier MT, Perreault S, Schnitzer ME. Lasso regression for the prediction of intermediate outcomes related to cardiovascular disease prevention using the TRANSIT quality indicators. Med Care. 2019;57(1):63–72. doi: 10.1097/MLR.0000000000001014. [DOI] [PubMed] [Google Scholar]
- 48.Anderson JP, Parikh JR, Shenfeld DK, Ivanov V, Marks C, Church BW, et al. Reverse engineering and evaluation of prediction models for progression to type 2 diabetes: an application of machine learning using electronic health records. J Diabetes Sci Technol. 2016;10(1):6–18. doi: 10.1177/1932296815620200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Patsopoulos NA. A pragmatic view on pragmatic trials. Dialogues Clin Neurosci. 2011;13(2):217–224. doi: 10.31887/DCNS.2011.13.2/npatsopoulos. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lu CY. Observational studies: a review of study designs, challenges and strategies to reduce confounding. Int J Clin Pract. 2009;63(5):691–697. doi: 10.1111/j.1742-1241.2009.02056.x. [DOI] [PubMed] [Google Scholar]
- 51.Morgenstern H. Ecologic studies in epidemiology: concepts, principles, and methods. Annu Rev Public Health. 1995;16(1):61–81. doi: 10.1146/annurev.pu.16.050195.000425. [DOI] [PubMed] [Google Scholar]
- 52.Vandenbroucke JP. In defense of case reports and case series. Ann Intern Med. 2001;134(4):330–334. doi: 10.7326/0003-4819-134-4-200102200-00017. [DOI] [PubMed] [Google Scholar]
- 53.Buckland ST, Burnham KP, Augustin NH. Model selection: an integral part of inference. Biometrics. 1997;53:603–618. doi: 10.2307/2533961. [DOI] [Google Scholar]
- 54.Zagar A, Kadziola Z, Lipkovich I, Madigan D, Faries D. Evaluating bias control strategies in observational studies using frequentist model averaging 2020 (submitted). [DOI] [PubMed]
- 55.Kang J, Schwartz R, Flickinger J, Beriwal S. Machine learning approaches for predicting radiation therapy outcomes: a clinician's perspective. Int J Radiat Oncol Biol Phys. 2015;93(5):1127–1135. doi: 10.1016/j.ijrobp.2015.07.2286. [DOI] [PubMed] [Google Scholar]
- 56.Scott IM, Lin W, Liakata M, Wood J, Vermeer CP, Allaway D, et al. Merits of random forests emerge in evaluation of chemometric classifiers by external validation. Anal Chim Acta. 2013;801:22–33. doi: 10.1016/j.aca.2013.09.027. [DOI] [PubMed] [Google Scholar]
- 57.Kuhn M, Johnson K. Applied predictive modeling. Berlin: Springer; 2013. [Google Scholar]
- 58.Hess L, Winfree K, Muehlenbein C, Zhu Y, Oton A, Princic N. Debunking Myths While Understanding Limitations. Am J Public Health. 2020;110(5):E2-E. [DOI] [PMC free article] [PubMed]
- 59.Thesmar D, Sraer D, Pinheiro L, Dadson N, Veliche R, Greenberg P. Combining the power of artificial intelligence with the richness of healthcare claims data: Opportunities and challenges. PharmacoEconomics. 2019;37(6):745–752. doi: 10.1007/s40273-019-00777-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Table S1. Study quality of eligible publications, modified Luo scale [14].
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files.