Abstract
Multiple nucleic acid amplification tests (NATs) are available for the detection of SARS-CoV-2 in clinical specimens, including Laboratory Developed Tests (LDT), commercial high-throughput assays and point-of-care tests. Some assays were just recently released and there is limited data on their clinical performance. We compared the Xpert® Xpress SARS-CoV-2 (Cepheid) and Vivalytic VRI Panel (Schnelltest COVID-19) (Bosch) point-of-care tests with four high-throughput assays and one LDT, the cobas® SARS-CoV-2 test (Roche), the Allplex™ 2019-nCoV Assay (Seegene), the SARS-CoV-2 AMP (Abbott) Kit, the RealStar® SARS-CoV-2 RT-PCR Kit 1.0 (altona) as well as an assay using a SARS-CoV-2 RdRP gene specific primer and probe set. Samples from patients with confirmed SARS-CoV-2 infection, samples from the first and second SARS-CoV-2-PCR External Quality Assessment (EQA) (INSTAND e.V.) and a 10-fold serial dilution of a SARS-CoV-2 cell culture (SARS-CoV-2 Frankfurt 1) supernatant were examined. We determined that the NAT assays examined had a high specificity. Assays using the N gene as target demonstrated the highest sensitivity in the serial dilution panel, while all examined NAT assays showed a comparable sensitivity when testing clinical and EQA samples.
Keywords: SARS-CoV-2, NAT, PCR, POCT
1. Introduction
Nucleic acid amplification testing (NAT) is the method of choice in diagnosing COVID-19 in the early phase of an infection with SARS-CoV-2. Laboratory Developed Tests (LDT) were applied early in the pandemic and multiple commercially developed NAT-based assays have been made available since. Testing however, is mainly performed in batches in centralized laboratories with a turn-around time of several hours, requiring well-organized sample transportation and laboratory procedures (Rabi et al., 2020; Younes et al., 2020). Point-of-care testing (POCT) can help to close this gap for time-sensitive samples. SARS-CoV-2 antigen tests are rapid, cheap and easy to handle but not generally suitable for individual testing. Examining respiratory specimens they demonstrate a low overall clinical sensitivity, primarily generating positive results for individuals with high viral concentrations (Lambert-Niclot et al., 2020; Mak et al., 2020). Serological assays also show a low clinical sensitivity at least in the early phase of infection (Deeks et al., 2020). Recently, cartridge-based NAT systems have become commercially available or are in development. They allow individual point-of-care testing of specimens and can be performed by personnel without experience with NAT testing and with minimal hands-on and short turn-around time. However, there is limited data on the performance of these assays compared to the established high-throughput assays routinely used in clinical laboratories. Aim of our study was the comparison of different commercial assays concerning sensitivity and specificity using clinical samples, samples from the first and second SARS-CoV-2-PCR External Quality Assessment (EQA) (INSTAND e.V., Düsseldorf, Germany), a dilution series of a SARS-CoV-2 positive cell culture supernatant and samples containing non-SARS-CoV-2 coronaviruses.
2. Materials and methods
2.1. Samples
We used respiratory specimens from 10 in-patients with a confirmed SARS-CoV-2 infection and a clinical course ranging from asymptomatic to severe. Six of the samples were collected from patients on an intensive care unit. Four were upper respiratory tract specimens (nasal, nasopharyngeal or pharyngeal swab), and 6 were lower respiratory tract specimens (tracheal secretion (n = 5) or sputum (n = 1)). The presence of SARS-CoV-2 was confirmed by rRT-PCR targeting the RdRp gene (Corman et al., 2020). As control, five samples from SARS-CoV-2 negative patients were used (4 outpatients and 1 inpatient), including five pharyngeal swabs and one sputum sample.
Furthermore seven samples from the first and five samples from the second SARS-CoV-2-PCR External Quality Assessment (EQA) (INSTAND e.V., Düsseldorf, Germany) containing dilutions of a SARS-CoV-2 strain, two samples with coronavirus HCoV−OC43 and HCoV-229E, respectively and a negative sample were analysed.
In addition, a dilution series (10−3, 10-4, 10-5, 10-6, 10-6.5, 10-7, 10-7.5, 10-8, 10-8.5, 10-9, 10-9.5 and 10-10)) of a SARS-CoV-2 positive cell culture supernatant (strain: SARS-CoV-2 Frankfurt 1) was used.
Assays specificity was estimated by testing respiratory samples from patients with a PCR-confirmed SARS-CoV (n = 1, sample from the 2003 outbreak), MERS-CoV (n = 1), HCoV−OC43 (n = 2), HCoV-NL63 (n = 1) and HCoV-229E (n = 2) infection.
2.2. Preparation of samples
Left over material from patient samples (dry swabs suspended in 1 mL phosphate buffered saline (PBS)) was diluted 1:5 in PBS in order to gain enough sample material for further testing.
When nucleic acid extraction was required (i.e. Allplex™ 2019-nCoV Assay (Seegene Inc., Seoul, South Korea) and the RealStar® SARS-CoV-2 RT-PCR Kit 1.0 (altona Diagnostics GmbH, Hamburg, Germany)), 500 μL of each sample was extracted using the QIAsymphony (Qiagen GmbH, Hilden, Germany) together with the DSP virus/pathogen midi kit (Qiagen) according to manufacturers’ instructions and eluted in a final volume of 130 μL. After extraction, the nucleic acid was stored at −80 °C until further testing.
2.3. Commercially available test systems
We examined multiple commercially available SARS-CoV-2 specific assays in this study (Table 1 ). Samples were tested with these assays according to the manufacturers’ protocol.
Table 1.
Examined commercially available SARS-Cov-2 assays.
| Assay | Target gene(s) | Company | Platform | Method | Field of application |
|---|---|---|---|---|---|
| cobas® SARS-CoV-2 | E, ORF1a gene | Roche Diagnostics International AG, Rotkreuz, Switzerland | cobas® 6800 | NAT | Laboratory (high-throughput) |
| Allplex™ 2019-nCoV Assay* | E, N, RdRP gene | Seegene Inc., Seoul, South Korea | CFX96™ (Bio-Rad) | NAT | Laboratory (high-throughput) |
| SARS-CoV-2 AMP Kit | N, RdRP gene** | Abbott GmbH, Wiesbaden, Germany | Alinity m | NAT | Laboratory (high-throughput) |
| RealStar® SARS-CoV-2 RT-PCR Kit 1.0* | E, S gene | altona Diagnostics GmbH, Hamburg, Germany | ABI Prism® 7500 (Applied Biosystems) | NAT | Laboratory (high-throughput) |
| Xpert® Xpress SARS-CoV-2 | E, N2 gene | Cepheid Inc., Sunnyvale, U.S.A. | GeneXpert® | NAT | POCT/ Laboratory*** |
| Vivalytic VRI Panel (Schnelltest COVID-19) | E, ORF1ab gene | Bosch Healthcare Solutions GmbH, Waiblingen, Germany | Vivalytic | NAT | POCT |
requires nucleic acid extraction as separate procedure before rRT-PCR testing.
not differentiating between targets.
high-throughput capable (depending on the used system).
With exception of the Allplex™ 2019-nCoV Assay (Seegene) and the RealStar® SARS-CoV-2 RT-PCR Kit 1.0 (altona), which required nucleic acid extraction as separate procedure, all commercially available NAT assays already included the nucleic acid extraction, reverse transcription of the viral RNA, amplification and detection.
For the Vivalyic VRI Panel Assay (Schnelltest COVID-19) (Bosch Healthcare Solutions GmbH, Waiblingen, Germany) and the Cepheid Xpert® Xpress SARS-CoV-2 assay (Cepheid Inc., Sunnyvale, U.S.A.), PBS is not evaluated as sample diluent.
All specimens (n = 44) were initially tested using the cobas® SARS-CoV-2 (Roche) and the Allplex™ 2019-nCoV Assay (Seegene). For the cobas® SARS-CoV-2 (Roche) three quantitative comparison samples containing 105, 106 and 107 SARS-CoV-2 (BetaCoV/Munich/ChVir984/2020) RNA copies/mL were used to generate a 3 point standard-curve and to calculate viral RNA copies/mL (Table S1/Figs. S1/S2). In total 10 aliquots of each suspension were tested on two different days (5 aliquots/day) to verify the intra- and inter-assay reproducibility. The comparison samples were provided by INSTAND e.V.
Because of limited test kit and sample availability, only selected samples were used for the Cepheid Xpress Xpert® SARS-CoV-2 (n = 25), Bosch Vivalyic VRI Panel (Schnelltest COVID-19) (n = 31), Altona RealStar® SARS-CoV-2 RT-PCR Kit 1.0 (n = 37) and the Abbott SARS-CoV-2 AMP Kit (n = 15) (Table S2/S3).
3. Results
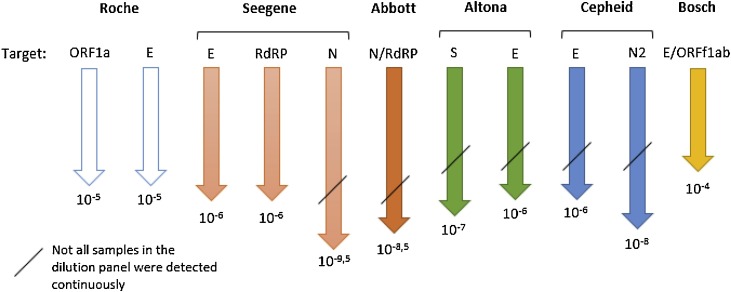
In the serial dilution panel, all examined NAT assays generated comparative results (Fig. 1 , Table 3/S2). The N gene-based assays (Allplex™ 2019-nCoV Assay (Seegene), SARS-CoV-2 AMP Kit (Abbott) and the Xpress Xpert® SARS-CoV-2 (Cepheid)), however, showed the highest sensitivity detecting nucleic acid in samples with a dilution up to 10−9.5, 10-8.5 and 10-8, respectively. The Vivalytic VRI Panel (Schnelltest COVID-19) (Bosch) showed the lowest sensitivity, but it has to be taken into account that e-NAT buffer instead of PBS is recommended by the manufacturer.
Fig. 1.
Schematic overview of each assay specific SARS-CoV-2 RT-PCR gene target examined and the maximum dilution factor where a signal was detected. The figure assumes that dilutions smaller than investigated (<10−3 and <10-4 for the Cepheid, respectively) would also have generated a reactive signal. Roche corresponds to the cobas® SARS-CoV-2, Seegene to the Allplex™ 2019-nCoV Assay, Abbott to the SARS-CoV-2 AMP Kit, Altona to the RealStar® SARS-CoV-2 RT-PCR Kit 1.0, Cepheid to the Xpert® Xpress SARS-CoV-2 and Bosch to the Vivalytic VRI Panel (Schnelltest COVID-19).
Table 3.
– Results overview of the examined assays.
| Examined samples | cobas® SARS-CoV-2 (Roche) | Allplex™ 2019-nCoV Assay (Seegene) | SARS-CoV-2 AMP Kit (Abbott) | RealStar® SARS-CoV-2 RT-PCR Kit 1.0 (altona) | Xpress Xpert® SARS-CoV-2 (Cepheid) | Vivalytic VRI Panel (Schnelltest COVID-19) (Bosch) |
|---|---|---|---|---|---|---|
| Dilution series panel (highest dilution where SARS-CoV-2-RNA was detected in) | 10−5 | 10−9,5 (N gene only) | 10−8.5 | 10−7 (S gene only) |
10−8 (N2 gene only) | 10−4 |
| Selected clinical samples (detected) | ✓(8/8) | ✓(8/8***) | – | ✓(8/8) | ✓(5/5) | (✓)(6/8) |
| EQA samples (correctly determined) | ✓(12/12) | (✓)(11/12) | ✓ (5/5) | (✓)(4/5) | ✓(7/7) | ✓(7/7**) |
| Other Coronaviruses (correctly not detected/identified)**** | ✓(4/4) | ✓(4/4) | – | ✓(4/4) | ✓(2/2) | ✓(4/4*****) |
*only the 10−3 sample was tested.
**one sample was detected as “Sarbeco-related”.
***one sample was only detected in the N gene.
****SARS-CoV-1 (2003 outbreak) excluded.
*****MERS-CoV was detected as “SARS-related”.
- not tested.
✓perfect performance (all samples were determined correctly).
(✓) good performance (nearly all samples were determined correctly).
Ten of the 15 clinical samples were initially tested positive (confirmed by RdRP-gene specific rRT-PCR) (Table 2 ). After 1:5 dilution in PBS, two initially low positive samples showed negative results in all NAT assays (samples 8 and 9). Of the remaining 8 positive samples, nearly all could be detected with all NAT assays (Tables 2/3). However, the cobas® SARS-CoV-2 (Roche) and the RealStar® SARS-CoV-2 (altona) assay detected all eight samples with a positive reaction for both gene regions (E, ORF1a or S, respectively) whereas the Allplex™ 2019-nCoV Assay (Seegene) assay showed a positive result for one region only (N-protein) in one sample. Interestingly, the Vivalytic VRI Panel (Schnelltest COVID-19 (Bosch) was able to detect 6 of these 8 samples despite the use of diluent that is not recommended for use in the assay by the manufacturer. For samples No. 2 and 7 the Vivalytic VRI Panel (Schnelltest COVID-19 (Bosch) generated a negative result whereas the other examined assays generated relatively weak positive results. In contrast, the assay generated a positive result for sample No. 10, where the other assays, as far as examined, even generated more weak positive results. The five negative samples showed negative results in all assays (if tested).
Table 2.
Examined clinical samples and assay results.
| No. | Clinical sample | cobas® SARS-CoV-2 (Roche) |
Allplex™ 2019-nCoV Assay (Seegene) |
RealStar® SARS-CoV-2 RT-PCR Kit 1.0 (altona) |
Xpress Xpert® SARS-CoV-2 (Cepheid) |
Vivalytic VRI Panel (Schnelltest COVID-19) (Bosch) | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Initial CT-value (undiluted)* | ORF1a gene CT-value (log10 RNA copies/mL) | E gene CT-value (log10 RNA copies/mL) | E gene (CT-value) | RdRP gene (CT-value) | N gene (CT-value) | S gene (CT-value) | E gene (CT-value) | E gene (CT-value) | N2 gene (CT-value) | E/ORFf1ab gene | |
| 1 | 24.5 (T) ▮ | 25.65 (6.03) | 26.31 (5.58) | 22.88 | 25.11 | 25.88 | 21.74 | 2180 | – | – | positive |
| 2 | 30.2 (T) ▮ | 31.27 (4.31) | 33.64 (3.35) | 32.07 | 35.65 | 34.27 | 31.07 | 3199 | 33.7 | 36.1 | ø |
| 3 | 33.8 (T) ▮ | 31.94 (4.12) | 34.19 (3.18) | 31.89 | 34.81 | 34.61 | 30.99 | 3167 | 32.4 | 35.4 | positive |
| 4 | 29.2 (S) ▮ | 30.21 (4.63) | 31.31 (4.06) | 29.63 | 31.09 | 32.64 | 28.42 | 2848 | 30.6 | 33.1 | positive |
| 5 | 25.2 (T) ▮ | 30.23 (4.63) | 31.06 (4.13) | 2808 | 30.20 | 30.86 | 26.80 | 2685 | 29.1 | 31.5 | positive |
| 6 | 22.1 (T) ▮ | 27.1 (5.58) | 27.81 (5.12) | 24.28 | 26.56 | 272 | 22.70 | 22.91 | – | – | positive |
| 7 | 29.37 (N) ▮ | 33.81 (5.58) | 35.67 (2.73) | 31.53 | 33.03 | 33.66 | 30.60 | 31.34 | 32.5 | 34.4 | ø |
| 8 | 35.6 (P) △ | ø | ø | ø | ø | ø | ø | ø | – | – | ø |
| 9 | 35.65 (P) □ | ø | ø | ø | ø | ø | ø | ø | – | – | ø |
| 10 | 32.12 (N) ▲ | 36.91 (2.59) | 37.42 (2.19) | ø | ø | 37.24 | 34.4 | 37.05 | – | – | positive |
| 11 | ø (S) | ø | ø | ø | ø | ø | ø | ø | ø | ø | ø |
| 12 | ø (P) | ø | ø | ø | ø | ø | ø | ø | ø | ø | ø |
| 13 | ø (P) | ø | ø | ø | ø | ø | ø | ø | ø | ø | ø |
| 14 | ø (P) | ø | ø | ø | ø | ø | ø | ø | – | – | ø |
| 15 | ø (P) | ø | ø | ø | ø | ø | ø | ø | – | – | ø |
All samples where a signal was detected were considered SARS-CoV-2 positive according to each manufacturer’s interpretation algorithm.
*RdRP-gene specific rRT-PCR.
(T) = tracheal secretion.
(S) = sputum.
(N) = nasal swab.
(P) = pharyngeal swab.
▮ = severe clinical course.
▲ = moderate clinical course.
△ = mild clinical course.
□ = asymptomatic clinical course.
Ø = negative.
- not tested.
All NAT assays detected the SARS-CoV-2 positive EQA samples (Table 3/S3). Only the Vivalytic VRI Panel (Schnelltest COVID-19) (Bosch) generated the result “Sarbeco-related” for one sample. With the exception of one HCoV-NL63 sample, which was detected by the Vivalytic VRI Panel (Schnelltest COVID-19) (Bosch) as HCoV-229E/HCoV-NL63, the negative samples and the samples containing not SARS-related coronaviruses (i.e. HCoV-OC43 and HCoV-229E), showed negative results in all assays. The Vivalytic VRI Panel (Schnelltest COVID-19) (Bosch) failed to detect two HCoV-229E and HCoV-OC43 samples (Table S4). Assays using the Sarbeco virus E gene as target detected the SARS-CoV sample (from the 2003 outbreak) as positive.
4. Discussion
All examined NAT assays are eligible for the detection of SARS-CoV-2-RNA. With exception of the samples of the serial dilution panel, where the N gene based assays demonstrated the highest sensitivity, the equal performance of the Xpert® Xpress SARS-CoV-2 (Cepheid) and the cobas® SARS-CoV-2 (Roche) for the EQA and clinical samples in our study, was also demonstrated in a study by Moran et al., where they showed an agreement of 99 % for generated positive results between the Xpert® Xpress SARS-CoV-2 (Cepheid) and the cobas® SARS-CoV-2 (Roche) (Moran et al., 2020). Two multi-center studies, one by Wolter et al. and one by Loeffelholz et al., demonstrated equal performance or a positive agreement of ≥ 92.3 %, respectively, for the Xpert® Xpress SARS-CoV-2 (Cepheid) compared to multiple RT-PCR tests (Loeffelholz et al., 2020; Wolters et al., 2020).
The good overall performance of the cobas® SARS-CoV-2 (Roche) was also demonstrated in two studies, in which it correlated well with two LDTs using the Centers for Disease Control and Prevention 2019‐nCoV primers and probes (Lieberman et al., 2020; Pujadas et al., 2020). Although the SARS-CoV-2 AMP Kit (Abbott), cobas® SARS-CoV-2 (Roche) and the Allplex™ 2019-nCoV Assay (Seegene) showed a comparable sensitivity for the clinical and EQA samples (if tested), the N gene based assays (together with the N2 gene of the Xpert® Xpress SARS-CoV-2 (Cepheid)) showed the highest sensitivity within the serial dilution panel. A study by Merindol et al. demonstrated a similar efficiency of the Allplex™ 2019-nCoV Assay (Seegene) compared to the RealStar® SARS-CoV-2 RT-PCR kit (Altona Diagnostics, Germany). The Allplex™ 2019-nCoV Assay (Seegene) even generated equivalent CT-means for swabs stored in UTM™ whether or not RNA was extracted before the rRT-PCR. This is an interesting finding, as the overall turn-around time could be further reduced (Merindol et al., 2020). All positive clinical samples derived from patients with confirmed SARS-CoV-2-infection. The weak positive clinical samples (No. 8 and 9) were follow up samples. The swabs were stored for several days at 4 °C and washed a second time in 5 mL PBS buffer and diluted accordingly. In addition, there was still a freeze and thawing step in between. This might be the reason that some samples were negative in all further tests. The correlation between viral load and transmissibility is not entirely clear, however, several studies showed that samples with viral loads ≥ 6 log10 SARS-CoV-2 RNA copies/mL likely correlate with infectivity in cell culture models (Kohmer et al., 2021; La Scola et al., 2020; Perera et al., 2020; WHO, 2020; Wölfel et al., 2020). As far as examined in our study, the assays were able to detect clinical sample 1 [> 6 log10 RNA/copies/mL for the ORF1a gene of the cobas® SARS-CoV-2 (Roche)] and from the dilution series panel the sample with the 10−4 dilution [<< 6 log10 RNA copies/mL for the ORF1a and E gene of the cobas® SARS-CoV-2 (Roche)]. These observations demonstrate on the one hand that the examined assays may be able to detect potential infectious individuals (when cell culture infectivity is used as surrogate for human-to-human transmission), but on the other hand, that they may be too sensitive for this approach, underlining the need of a defined threshold for potential transmissibility.
All assays examined in this study demonstrated a high specificity, however more samples need to be tested to get a clearer picture. As SARS-CoV from the 2003 outbreak is known to be eradicated, its detection in the E gene targets should be negligible.
The Vivalytic VRI Panel (Schnelltest COVID-19) (Bosch) demonstrated to be a POCT with potential. However, more data on its performance when testing specimens according to the manufacturers’ specifications are needed as we deviated in our study from the manufacturers specifications: PBS was used as sample diluent for the Vivalytic VRI Panel (Schnelltest COVID-19) (Bosch) and for the Xpert® Xpress SARS-CoV-2 (Cepheid). We cannot exclude that a non-reproducible influence of the used dilution buffer might be the reason for the unexpected result constellation (samples No. 2 and 7 versus sample No. 10).
In summary, all commercially available NATs, especially the Xpert® Xpress SARS-CoV-2 (Cepheid), are eligible in the detection of SARS-CoV-2 RNA and demonstrated a high specificity. NAT assays using an N gene target demonstrated the highest sensitivity within the serial dilution panel, while all examined NAT assays showed a comparable sensitivity when testing clinical and EQA samples.
Contributor ship
All the authors contributed to this work by performing experiments, analyzing the data and writing the manuscript. NK drafted the first version.
Funding
Bosch Healthcare Solutions GmbH provided the assays free of charge. Otherwise this research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Declaration of Competing Interest
None of the authors have competing interests related to this work.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.jviromet.2021.114102.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.-L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3) doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeks J.J., Dinnes J., Takwoingi Y., Davenport C., Spijker R., Taylor-Phillips S., Adriano A., Beese S., Dretzke J., Di Ferrante Ruffano L., Harris I.M., Price M.J., Dittrich S., Emperador D., Hooft L., Leeflang M.M., van den Bruel A. Antibody tests for identification of current and past infection with SARS-CoV-2. Cochrane Database Syst. Rev. 2020;6 doi: 10.1002/14651858.CD013652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohmer N., Toptan T., Pallas C., Karaca O., Pfeiffer A., Westhaus S., Widera M., Berger A., Hoehl S., Kammel M., Ciesek S., Rabenau H.F. The Comparative Clinical Performance of Four SARS-CoV-2 Rapid Antigen Tests and Their Correlation to Infectivity In Vitro. J. Clin. Med. 2021;10(2):328. doi: 10.3390/jcm10020328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Scola B., Le Bideau M., Andreani J., van Hoang T., Grimaldier C., Colson P., Gautret P., Raoult D. Viral RNA load as determined by cell culture as a management tool for discharge of SARS-CoV-2 patients from infectious disease wards. Eur. J. Clin. Microbiol. Infect. Dis. 2020;39(6):1059–1061. doi: 10.1007/s10096-020-03913-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert-Niclot S., Cuffel A., Le Pape S., Vauloup-Fellous C., Morand-Joubert L., Roque-Afonso A.-M., Le Goff J., Delaugerre C. Evaluation of a rapid diagnostic assay for detection of SARS CoV-2 antigen in nasopharyngeal swab. J. Clin. Microbiol. 2020;58(8) doi: 10.1128/JCM.00977-20. e00977-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman J.A., Pepper G., Naccache S.N., Huang M.-L., Jerome K.R., Greninger A.L. Comparison of Commercially Available and Laboratory Developed Assays for in vitro Detection of SARS-CoV-2 in Clinical Laboratories. J. Clin. Microbiol. 2020;58(8) doi: 10.1128/JCM.00821-20. e00821-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeffelholz M.J., Alland D., Butler-Wu S.M., Pandey U., Perno C.F., Nava A., Carroll K.C., Mostafa H., Davies E., McEwan A., Rakeman J.L., Fowler R.C., Pawlotsky J.-M., Fourati S., Banik S., Banada P.P., Swaminathan S., Chakravorty S., Kwiatkowski R.W., Chu V.C., Kop J., Gaur R., Sin M.L.Y., Nguyen D., Singh S., Zhang N., Persing D.H. Multicenter Evaluation of the Cepheid Xpert Xpress SARS-CoV-2 Test. J. Clin. Microbiol. 2020;58(8) doi: 10.1128/JCM.00926-20. e00926-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak G.C., Cheng P.K., Lau S.S., Wong K.K., Lau C.S., Lam E.T., Chan R.C., Tsang D.N. Evaluation of rapid antigen test for detection of SARS-CoV-2 virus. J. Clin. Virol. 2020;129 doi: 10.1016/j.jcv.2020.104500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merindol N., Pépin G., Marchand C., Rheault M., Peterson C., Poirier A., Houle C., Germain H., Danylo A. SARS-CoV-2 detection by direct rRT-PCR without RNA extraction. J. Clin. Virol. 2020;128 doi: 10.1016/j.jcv.2020.104423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran A., Beavis K.G., Matushek S.M., Ciaglia C., Francois N., Tesic V., Love N. The Detection of SARS-CoV-2 using the Cepheid Xpert Xpress SARS-CoV-2 and Roche cobas SARS-CoV-2 Assays. J. Clin. Microbiol. 2020;58(8) doi: 10.1128/JCM.00772-20. e00772-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera R.A.P.M., Tso E., Tsang O.T.Y., Tsang D.N.C., Fung K., Leung Y.W.Y., Chin A.W.H., Chu D.K.W., Cheng S.M.S., Poon L.L.M., Chuang V.W.M., Peiris M. SARS-CoV-2 Virus Culture and Subgenomic RNA for Respiratory Specimens from Patients with Mild Coronavirus Disease. Emerging Infect. Dis. 2020;26(11):2701–2704. doi: 10.3201/eid2611.203219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujadas E., Ibeh N., Hernandez M.M., Waluszko A., Sidorenko T., Flores V., Shiffrin B., Chiu N., Young-Francois A., Nowak M.D., Paniz-Mondolfi A.E., Sordillo E.M., Cordon-Cardo C., Houldsworth J., Gitman M.R. Comparison of SARS-CoV-2 detection from nasopharyngeal swab samples by the Roche cobas 6800 SARS-CoV-2 test and a laboratory-developed real-time RT-PCR test. J. Med. Virol. 2020;92(9):1695–1698. doi: 10.1002/jmv.25988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabi F.A., Al Zoubi M.S., Kasasbeh G.A., Salameh D.M., Al-Nasser A.D. SARS-CoV-2 and coronavirus disease 2019: what we know so far. Pathogens. 2020;9(3) doi: 10.3390/pathogens9030231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . 2020. Target Product Profiles for Priority Diagnostics to Support Response to the COVID-19 Pandemic v.1.0.https://www.who.int/publications/m/item/covid-19-target-product-profiles-for-priority-diagnostics-to-support-response-to-the-covid-19-pandemic-v.0.1 [Google Scholar]
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Brünink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581(7809):465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Wolters F., van de Bovenkamp J., van den Bosch B., van den Brink S., Broeders M., Chung N.H., Favié B., Goderski G., Kuijpers J., Overdevest I., Rahamat-Langedoen J., Wijsman L., Melchers W.J., Meijer A. Multi-center evaluation of cepheid xpert® xpress SARS-CoV-2 point-of-care test during the SARS-CoV-2 pandemic. J. Clin. Virol. 2020;128 doi: 10.1016/j.jcv.2020.104426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younes N., Al-Sadeq D.W., Al-Jighefee H., Younes S., Al-Jamal O., Daas H.I., Yassine H.M., Nasrallah G.K. Challenges in laboratory diagnosis of the novel coronavirus SARS-CoV-2. Viruses. 2020;12(6) doi: 10.3390/v12060582. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.