Abstract
Molecular networking has become a key method to visualize and annotate the chemical space in non-targeted mass spectrometry data. We present Feature-Based Molecular Networking (FBMN) as an analysis method in the Global Natural Products Social Molecular Networking (GNPS) infrastructure that builds on chromatographic feature detection and alignment tools. The FBMN method brings quantitative analyses, isomeric resolution, including from ion-mobility spectrometry, into molecular networks.
Introduced in 20121, molecular networking has become an essential bioinformatics tool to visualize and annotate non-targeted mass spectrometry data2. The first application of molecular networking was described by Traxler and Kolter3 as holding “great promise in providing the next quantum leap in understanding the fascinating world of microbial chemical ecology”. Molecular networking goes beyond spectral matching against reference spectra, by aligning experimental spectra against one another and connecting related molecules by their spectral similarity. In a molecular network, related molecules are referred to as a “molecular family”, differing by simple transformations such as glycosylation, alkylation, and oxidation/reduction. Molecular networking became publicly accessible in 2013 through the initial release of the Global Natural Product Social Molecular Networking (GNPS), a web-enabled mass spectrometry knowledge capture and analysis platform (http://gnps.ucsd.edu)4, and has been widely applied in mass spectrometry-based metabolomics to aid in the annotation of molecular families from their fragmentation spectra (MS2).
Powered by 3,000+ CPU cores at the Center for Computational Mass Spectrometry at the University of California San Diego and the MassIVE data repository, GNPS has provided researchers from more than 150 countries with the ability to perform molecular networking. To build upon the success of the first molecular networking method (referred to as “classical” molecular networking, classical MN) which is based on the MS-Cluster algorithm5, we introduce a complementary tool named Feature-Based Molecular Networking (FBMN). FBMN leverages the capability of well-established mass spectrometry processing software and improves upon classical MN by incorporating MS1 information, such as isotope patterns and retention time, but also ion-mobility separation when performed. By relying on processed spectral information, molecular networks obtained with FBMN can 1) distinguish isomers producing similar MS2 spectra that are resolved by chromatographic or by ion mobility separation, that may have remained hidden, 2) facilitates spectral annotation, and 3) incorporates relative quantitative information which enables robust downstream metabolomics statistical analysis. Whereas users of the classical MN would have had to perform molecular networking and MS1 analysis separately before performing a cumbersome linking of the outputs, FBMN method accepts the output of feature detection and processing tools, making them directly compatible with annotation tools tand the entirety of the analysis pipeline.
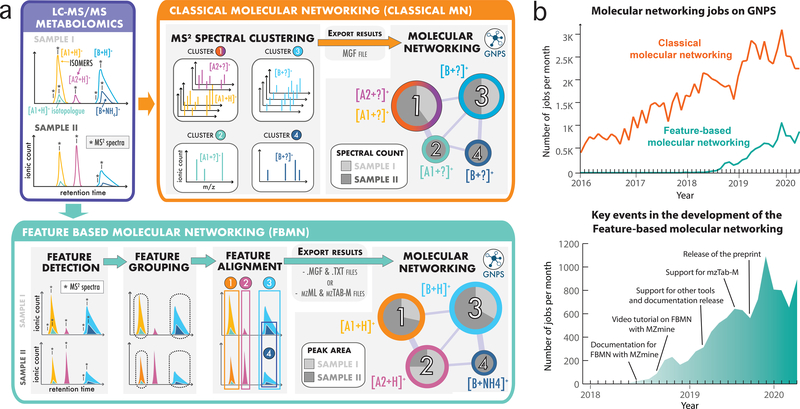
To fully utilize the MS1 and MS2 content collected during a non-targeted liquid chromatography coupled to tandem mass spectrometry data (LC-MS2) metabolomics experiment in a streamlined fashion, we have created an online workflow (Fig. 1a) infrastructure that supports the outputs of feature detection and alignment tools for FBMN analysis (https://ccms-ucsd.github.io/GNPSDocumentation/featurebasedmolecularnetworking), including the standard output format for small molecules analysis (mzTab-M)6. The diversity of supported software, each offering different functionalities/modules, serves experimentalists, bioinformaticians, and software developers. FBMN is already the second most utilized analysis tool within the GNPS environment (Fig. 1b) with more than 6,767 jobs performed in 2019 and has already been used in more than 80 publications using FBMN during its development since Nov 2017.
Fig. 1: Methods for the generation of molecular networks from non-targeted mass spectrometry data with the GNPS web platform.
a) Two methods exist for the generation of molecular networks on the GNPS web platform: classical MN and feature-based molecular networking (FBMN). For both methods, the mass spectrometry data files have first to be converted to the mzML format using tools such as Proteowizard MSConvert21. The classical MN method runs entirely on the GNPS platform. In that method, MS2 spectra are clustered with MS-Cluster and the consensus MS2 spectra obtained are used for molecular network generation. In the case of FBMN, the user first applies a feature detection and alignment tool to first process the LC-MS2 data (such as MZmine, MS-DIAL, XCMS, OpenMS, Progenesis QI, or MetaboScape) instead of using MS-Cluster (classical MN) on GNPS. Results are then exported (feature quantification table (.TXT format) along with a MS2 spectral summary (.MGF format) or an mzTab-M file) and uploaded to the GNPS web platform for molecular networking analysis with the FBMN workflow. b) Graphs showing the number of molecular networking jobs performed on GNPS. The upper graph shows the number of classical MN and FBMN jobs since 2016. The lower graph shows the number of FBMN jobs since its introduction in 2017 and key events accelerating its use.
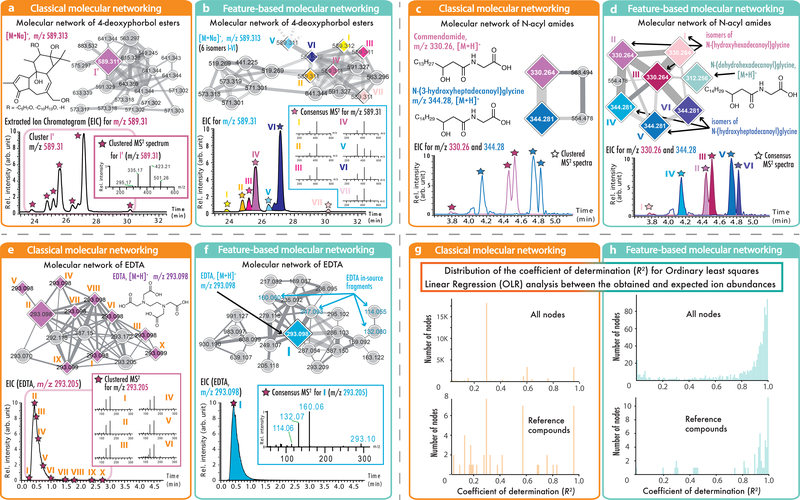
The molecular networks generated with FBMN enable the efficient visualization and annotation of isomers in LC-MS2 datasets, as demonstrated below with LC-MS2 data from a drug discovery project from Euphorbia plant extract7 (Fig. 2a–b), and the detection of human microbiome-derived lipids belonging to the commendamide family8, detected in fecal samples from the American Gut Project9 (a crowd-sourced citizen science microbiome project) (Fig. 2c–d). In both cases, FBMN resolved positional isomers/stereoisomers in the molecular networks that have similar MS2 spectra but distinct retention times, that would not have been resolved with classical MN, which facilitated the isolation of antiviral compounds7 (Fig. 2c), and the annotation of commendamide isomers9 and of a putative novel derivative, the N-(dehydrohexadecanoyl)glycine (Fig. 2d).
Fig. 2: Comparisons of classical MN and FBMN.
In these examples, the node size corresponds to the relative spectral count in classical MN (orange boxes, left) or to the sum of LC-MS peak area in FBMN (blue boxes, right); diamond shape nodes are spectra annotated by spectral library matching; the edge color gradient indicates the spectral similarity degree (the lighther the less similar). (a) displays the results from classical MN with the LC-MS2 data of Euphorbia dendroides plant samples (n = 1 LC-MS2 experiment per sample); classical MN resulted in one node for the ion at m/z 589.313, while (b) FBMN was able to detect seven isomers. (c) Classical MN with the data from the American Gut Project (n = 1 LC-MS2 experiment per sample) showed two different N-acyl amides while the use of FBMN (d) allowed the annotation of three different isomers per N-acyl amides. Classical MN (e) and FBMN (f) were used to analyse the network of EDTA in plasma (373 samples, n = 1 LC-MS2 experiment per sample). By merging MS2 spectra of EDTA eluting over 2.5 min into one best-quality MS2 spectrum, FBMN recovered the molecular similarity of in-source fragments observed for EDTA. (g and h) Evaluation of quantitative performance using multiple dilutions of a reference serum sample (3 LC-MS2 experiments per sample). The plots (g and h) are showing the distribution of the coefficient of determination (R2) from the Ordinary least squares Linear Regression (OLR) analysis between the observed and expected relative ion abundance for molecular network nodes in classical MN (g) or in FBMN (h). The upper charts present the distribution of the R2 for the network nodes with classical MN (n = 3,367) and FBMN (n = 877), and the bottom charts show the R2 distribution from the OLR analysis for the annotated reference compounds with classical MN (n = 49) and FBMN (n = 54). While classical MN uses the clustered MS2 spectral count or the sum of the precursor ions to estimate the molecular network node abundance, FBMN uses the LC-MS feature abundance (peak area or height), resulting in a more accurate estimation of the relative ion intensity.
In non-targeted LC-MS2 data acquisition, the same precursor ion is frequently fragmented multiple times during chromatographic elution. While MS-Cluster is often able to cluster these spectra into one single node in classical MN, there are cases where it will fail and produce multiple nodes representing the same compound. For example, this can happen for compounds producing mostly low intensity fragment ions, or for chimeric spectra resulting from coeluting isobaric ions are isolated and fragmented together. With FBMN, a singular representative consensus MS2 spectrum is attributed for the LC-MS feature (defined as the detected ion signal for an eluting molecule)10. The benefit of using FBMN in such a case can be illustrated with the metal chelating agent ethylenediaminetetraacetic acid (EDTA) observed in the LC-MS2 analysis of plasma samples (Fig 2e), in which it is used as an anticoagulant agent. Classical MN resulted in 13 duplicated nodes with identical precursor m/z values in one molecular family, ten of which have spectral library matches to EDTA reference MS2 data (Fig. 2e and f). On the contrary, FBMN displays a unique representative MS2 spectrum that matches EDTA spectra in the library. The reduction of redundancy within the resulting molecular network simplifies the discovery of structurally related compounds.
While classical MN uses the spectral count or the sum precursor ion count, FBMN uses the LC-MS feature abundance (peak area or peak height), resulting in a more accurate estimation of the relative ion intensity. The method of FBMN simplifies, organizes the data, and adds relative quantitative information and precursor isotope patterns. FBMN enables robust statistical analysis by providing relative ion intensities across a dataset. This capacity is demonstrated with a serial dilution series dataset of the NIST1950 serum reference standard, containing 150 spiked standards. Here, the LC-MS2 were processed with MZmine11 or OpenMS10 for FBMN (Fig 2g–h). A linear regression analysis was used to evaluate the relative quantification between classical MN and FBMN. Figure 2h shows that for FBMN, relative quantification has a coefficient of determination (R2) value distribution mostly above 0.7, while this was not the case when the precursor ion abundance was obtained from classical MN via spectral counts (Fig 2g). The improved distribution of correlation coefficients towards 1 indicates a more linear response between molecular concentration and ion abundance, which improves the accuracy and precision of quantification results. FBMN facilitates the direct application of existing statistical, visualization, and annotation tools, such as QIIME212, MetaboAnalyst13, ili14, SIRIUS15, DEREPLICATOR16, MS2LDA17, and Qemistree18.
FBMN further enables the creation of molecular networks from ion mobility spectrometry experiments coupled with LC-MS2 analysis. As an orthogonal separation method, the use of ion mobility offers additional resolving power to differentiate isomeric ions in the molecular network. The integration of ion mobility with FBMN on GNPS can currently be performed with MetaboScape, MS-DIAL19, and Progenesis QI. An example of such isomer separation using trapped ion mobility spectrometry (TIMS) coupled to LC-MS2 is shown in Supplementary Fig. 1.
Available on the GNPS web platform at https://gnps.ucsd.edu, FBMN is ideally suited for advanced molecular networking analysis, enabling the characterization of isomers, the incorporation of relative quantification, and the integration of ion mobility data. FBMN is the recommended way to analyse a single LC-MS2 metabolomics study, but its applicability is limited when applied across multiple studies due to different experimental conditions and possible batch effects. Moreover, the use of FBMN for the analysis of very large datasets (containing several thousand samples) is limited by the scalability of most feature detection and alignment software tools. Thus, while FBMN offers an improvement upon many aspects of molecular networking analysis, classical MN remains essential for repository-scale meta-analysis large dataset processing, and is convenient for rapid analysis of LC-MS2 data with less user defined parameters: one important aspect of molecular networks obtained with FBMN is the use of adequate processing steps and parameters, which otherwise could negatively impact the resulting molecular networks. To facilitate dissemination, education of the FBMN method, and the supported processing software, we created detailed tutorials and step-by-step instructions, available at https://ccms-ucsd.github.io/GNPSDocumentation/featurebasedmolecularnetworking.
The FBMN workflow offers not only automated spectral library search and spectral entry curation, but is also integrated with other annotation tools available on GNPS environment, such as MASST20, while promoting data analysis reproducibility by saving the FBMN jobs on the user’s private online workspace. The GNPS environment conveniently enables the user to evaluate different parameters and enables the sharing of the results via a web URL for publication.
Online Methods
Development of Feature-based Molecular Networking (FBMN)
The FBMN method consists of two main steps: 1) LC-MS feature detection and alignment, then 2) a dedicated molecular networking workflow on GNPS. Our first prototype for FBMN was developed with the Optimus workflow7,14 that uses OpenMS tools10. Following step 1 (feature detection and alignment), two files are exported: a feature quantification table (.TXT format) and a MS2 spectral summary (.MGF format). The feature quantification table contains information about LC-MS features across all considered samples including a unique identifier (Feature ID) for each feature, m/z value, retention time, and intensity. The MS2 spectral summary contains a list of MS2 spectra, with one representative MS2 spectrum per feature. The mapping of information between the feature quantification table and the MS2 spectral summary is stored in these files using the feature ID and scan number, respectively. This simple mapping enables to relate LC-MS feature information or statistically derived results to the molecular network nodes. This approach was also used for the integration of other tools with FBMN, and does not require third party software like it was proposed in the past22,23. Finally, the FBMN workflow also supports the mzTab-M format6, a standardized output format designed for the report of metabolomics MS-data processing results. In this case, the mzTab-M file is used instead of feature quantification table and requires the input of the mzML files instead of the MS2 spectral summary file. Support for the mzTab-M format enables the possibility to perform FBMN with any existing and future processing tools that support this standardized format.
The FBMN workflow has been integrated into the GNPS ecosystem and thus benefits from the connection with other GNPS features, e.g. the possibility to perform automatic MS2 spectral library search, the direct addition and curation of library entries, the search of a spectrum against public datasets with MASST20, and the visualization of molecular networks directly in the web browser24 or with Cytoscape25. The FBMN workflow is available on the GNPS platform (https://gnps.ucsd.edu/) via a web interface (See Supplementary Fig. 2). Jobs are computed and stored on the computational infrastructure of the Center for Computational Mass Spectrometry at the University of California San Diego. Each finished job is saved in the private user space for future examination and has a permanent static link that enables data sharing and collaborative analyses. We strongly recommend the sharing of this static link along with publications using GNPS workflows to facilitate results accessibility and data analysis reproducibility. Instructions to perform FBMN with the supported tools and input file format requirements are provided in the GNPS documentation (https://ccms-ucsd.github.io/GNPSDocumentation/featurebasedmolecularnetworking and Supplementary Fig. 3).
Processing Mass Spectrometry Data for FBMN
FBMN supports the output from several feature detection processing softwares. Depending on the type and size of mass spectrometry data, and the intended user (e.g. bioinformatician, mass spectrometrists, biologists, etc.) different software might be more appropriate. In general, tools with a Graphical User Interface (GUI) (e.g., MZmine11, MS-DIAL19, MetaboScape, and Progenesis QI) are convenient for data visualisation and empirical parameters optimisation, but have often limited scalability that might prevent their usage for large datasets (more than 500 files). For these large datasets, tools that were designed to operate on a cluster/cloud computer should be prefered (XCMS26, OpenMS10 and MZmine to some extent). Regardless of the software or application, the processing steps and parameters should be performed as recommended by tool developers and experienced users through community feedback. Finally, automated optimisation modules can be used to finely tune parameters, which is particularly valuable when using command-line interface tools27,28. While we acknowledge that there are many tools and configurations to analyze mass spectrometry data, we provide a summary of processing steps on the supported tools in the FBMN documentation (https://ccms-ucsd.github.io/GNPSDocumentation/featurebasedmolecularnetworking). These constitute an aggregation of institutional knowledge from tool developers and experienced tool users that do not encompass all possible applications, but rather provides a starting point for new users.
FBMN after MZmine processing
MZmine11 is a popular open-source cross-platform software for mass spectrometry data processing with an advanced GUI that enables the users to visually optimize parameters and examine the results of each processing step. Moreover, MZmine allows for the export of a batch file containing all the steps and parameters used in the processing, thus enabling its reproducibility. To support FBMN in MZmine, the feature detection step (peak “Deconvolution module”) was modified to provide the ability to pair a feature with its MS2 scans using an m/z and retention time range defined by the user (Supplementary Fig. 4). Due to a new data structure and to support older projects (created with release < 2.38), an additional specific filtering module (Group MS2 scans with features) was developed to assign all MS2 scans to the features of existing peak list (see this video for instructions: https://www.youtube.com/watch?v=EL5pmFvpTFE). Moreover, a GNPS export and direct submission module was created (Supplementary Fig. 5) which offers two modes: 1) Export of the feature quantification table and the MS2 spectral summary file and 2) Direct FBMN analysis on the GNPS web platform (release 2.37+). The direct GNPS job submission generates all the files and uploads them together with an optional metadata table and default parameters (Supplementary Fig. 6) to the FBMN workflow on GNPS. By providing the user’s GNPS login credentials (optional), a new job can be created in the personal user space (https://www.youtube.com/watch?v=vFcGG7T_44E&list=PL4L2Xw5k8ITzd9hx5XIP94vFPxj1sSafB&index=4&t=0s). Otherwise, the user can be notified by email or directly redirected to the job webpage after the submission. With the option “most intense”, the GNPS Export uses the most intense MS2 spectrum as a representative spectrum for each LC-MS2 feature. When using the “merge MS/MS” spectra option (release 2.40+), a representative high quality MS2 spectrum is instead generated from all spectra and exported as a representative spectrum (Supplementary Note 1). The detailed documentation is available at https://ccms-ucsd.github.io/GNPSDocumentation/featurebasedmolecularnetworking-with-mzmine2/.
FBMN after OpenMS processing
OpenMS is an open-source cross-platform software specifically designed for the flexible and reproducible analysis of high-throughput MS data analysis, including more than 200 tools for common mass spectrometric data processing tasks10. Building on our experience with the Optimus development, the integration of OpenMS and FBMN was achieved by creating a GNPSExport tool (TOPP tool) as a part of the OpenMS tool collection (https://github.com/Bioinformatic-squad-DorresteinLab/OpenMS). A detailed description of the GNPSExport module and how to use it for FBMN is available at the following webpage https://ccms-ucsd.github.io/GNPSDocumentation/featurebasedmolecularnetworking-with-openms/. In brief, after running an OpenMS non-targeted metabolomics pipeline, the GNPSExport TOPP tool can be applied to the consensusXML file resulting from FeatureLinkerUnlabeledKD or FeatureLinkerUnlabeledQT tools (alignment step), and the corresponding mzML files. For each consensusElement (LC-MS2 feature) in the consensusXML file, the GNPSExport generates one representative consensus MS2 spectrum that will be exported in the MS2 spectral summary file (using either the option “most intense” or “merged spectra”, see Supplementary Note 1). The TextExport tool is applied to the same consensusXML file to generate the feature quantification table. Note that the GNPSExport requires the use of the IDMapper tool on the featureXML files (from the feature detection step) prior to feature linking, in order to associate MS2 scans [peptide annotation in OpenMS terminology] with each feature. These MS2 scans are used by the GNPSExport for the generation of the representative MS2 spectrum. Additionally, the FileFilter has to be run on the consensusXML file, prior to the GNPSExport, in order to remove consensus Elements without associated MS2 scans. The two files exported (feature quantification table and MS2 spectral summary) can be directly used for FBMN analysis on GNPS. The OpenMS-GNPS workflow for metabolomics data processing was implemented as a python wrapper around OpenMS TOPP tools (https://github.com/Bioinformatic-squad-DorresteinLab/openms-gnps-tools), and released as a workflow (https://github.com/Bioinformatic-squad-DorresteinLab/openms-gnps-workflow) on the GNPS/MassIVE web platform. OpenMS version 2.4.0 was used10. The OpenMS + GNPS workflow can be accessed and run here: https://proteomics2.ucsd.edu/ProteoSAFe/.
FBMN after XCMS processing
The XCMS package (https://github.com/sneumann/xcms for the most recent version) is one of the most widely used software for processing of mass spectrometry-based metabolomics data26. The integration of XCMS and FBMN is currently possible using a custom utility function “formatSpectraForGNPS” creating the MS2 spectral summary. This function is available on the following GitHub repository https://github.com/jorainer/xcms-gnps-tools and is compatible with the CAMERA algorithm for isotopes and adduct annotation29. Representative XCMS R scripts in markdown and Jupyter notebook formats are available in the following GitHub repository https://github.com/DorresteinLaboratory/XCMS3_FeatureBasedMN. The two exported files (feature quantification table and MS2 spectral summary) can be directly used for FBMN analysis on GNPS. The detailed documentation is available at https://ccms-ucsd.github.io/GNPSDocumentation/featurebasedmolecularnetworking-with-xcms3/.
FBMN after MS-DIAL processing
MS-DIAL is an open-source mass spectrometry data processing software19 (available for Windows only, http://prime.psc.riken.jp/Metabolomics_Software/MS-DIAL/). The integration of MS-DIAL and FBMN was made possible since ver. 2.68 by exporting the “Alignment results” using the “GNPS export” option. In addition to LC-MS2 data processing, MS-DIAL can process data from SWATH-MS2 (data-independent LC-MS2 acquisition), and ion mobility spectrometry coupled to LC-MS 30. The two files exported (feature quantification table and MS2 spectral summary) can be directly used for FBMN analysis on GNPS. A video tutorial on the use of MS-DIAL for FBMN is available at https://www.youtube.com/watch?v=hxk40jwAkcc&t=7s. The detailed documentation is available at https://ccms-ucsd.github.io/GNPSDocumentation/featurebasedmolecularnetworking-with-ms-dial/.
FBMN after MetaboScape processing
MetaboScape is a proprietary mass spectrometry metabolomics data processing software commercialized by Bruker and available on Windows. MetaboScape can perform feature detection, alignment and annotation of non-targeted LC-MS2 data acquired on Bruker mass spectrometers. Support for the processing of trapped ion mobility spectrometry (TIMS) coupled to non-targeted LC-MS2 (LC-TIMS-MS2) was added in MetaboScape 4.0, which results in LC-TIMS-MS features. Feature-based molecular networking can be performed on LC-MS2 or LC-TIMS-MS2 data by exporting the feature quantification table and MS2 spectral summary from the “bucket table” using the “Export to GNPS format” function. These files can be uploaded to GNPS for FBMN analysis. Information from MetaboScape, such as the Collision Cross Section values, or other spectral annotations can be mapped into the molecular networks using Cytoscape25. The detailed documentation is available at https://ccms-ucsd.github.io/GNPSDocumentation/featurebasedmolecularnetworking-with-metaboscape/
FBMN after Progenesis QI processing
Progenesis QI is a proprietary feature detection and alignment software developed by Nonlinear Dynamics (Waters) that is compatible with various proprietary and open mass spectrometry data formats. Progenesis QI can perform feature detection, alignment and annotation of non-targeted LC-MS2 data acquired either in data-dependent acquisition (DDA) or data independent analysis (DIA, such as MSE), and can also utilize the ion mobility spectrometry (IMS) dimension. FBMN can be performed on any of these data types processed with Progenesis QI (ver 4.0), by exporting the feature quantification table (.CSV format) and the MS2 spectral summary (.MSP format). These two files can be exported from the “Identify Compounds” submenu by using the function “Export compound measurement” and “Export fragment database”, respectively. These files can be uploaded to GNPS for FBMN analysis. Information from Progenesis QI, such as the Collision Cross Section values, or other spectral annotations can be mapped into the molecular networks using Cytoscape25. The detailed documentation is available at https://ccms-ucsd.github.io/GNPSDocumentation/featurebasedmolecularnetworking-with-progenesisQI/.
FBMN makes it possible to resolve isomers in a drug lead discovery effort
The examination of the LC-MS2 data (MSV000080502) from the Euphorbia dendroides plant extract showed the presence of numerous chromatographic peaks for ions in the range m/z 500–900, corresponding to diterpene ester derivatives. These specialized metabolites consist of a polyhydroxylated diterpene core acylated with various acidic moieties, that are typically found as positional isomers based on their acylation pattern. The extracted ion chromatogram (EIC) for the ion m/z 589.31 in the Euphorbia dendroides extract data (Supplementary Fig. 7) shows the presence of at least seven distinct LC-MS peaks between 24.5 and 27.3 min, including five peaks with an associated MS2 spectra. The analysis of the extract and the fractions where these molecules were originally isolated (fractions 13 and 14) with classical MN resulted in a molecular network with two nodes for the m/z 589.31 ions (Fig. 2a and Supplementary Fig. 8). These MS2 spectra (cluster index 5352 and 5354) resulted from merging 96 fragmentation spectra spanning from 23.6 to 26.5 min by MS-Cluster (Fig. 2b and Supplementary Fig. 9). Close examination of the clustered spectra revealed that while all MS2 spectra for the precursor m/z 589.31 present fragment ions m/z 501.26, 423.21, 335.16, and 295.17, three distinct spectral types could be established based on the ions relative intensities (Supplementary Fig. 10). FBMN of the dataset with MZmine processing (see the GNPS job) enabled the differentiation of the MS2 spectra of seven isomers (Figure 2b and Supplementary Fig. 11 for the molecular network view). A detailed discussion on the differences observed between the two methods can be found in the Supporting Information (Supplementary Note 2 and Supplementary Table 1). Interestingly, in the original study7 OpenMS was used for FBMN and resulted in the observation of three different positional isomers instead of seven, which shows that different processing methods and/or parameters can lead to different results with FBMN. These three isomers were subsequently isolated and differed by the position of one double bond on the C-12 acyl chain, or from carbon C-4 configuration7. Because FBMN connects the accurate relative abundance of the ions across the fractions and the molecular networks, it allowed to create bioactivity-based molecular networks7, which were used to predict and target potentially antiviral compounds. For detailed description of the extraction, mass spectrometry analysis, and structural elucidation, see the original manuscript7. The MZmine project and parameters used can be accessed on the MassIVE submission (MSV000080502).
FBMN resolves isomers in large scale metabolomics studies
FBMN was applied on a cohort of the American Gut Project (AGP), a citizen-scientist research project that enabled the observation of the commendamide in humans, along with other new N-acyl amide derivatives using molecular networking9. Commendamide is a recently discovered bacterial N-acyl amide that was shown to modulate host metabolism via G-protein-coupled receptors (GPCRs) in the murine intestinal tract31.
The use of FBMN for the AGP data (Figure 2d) made possible to observe the presence of two additional commendamide isomers (m/z 330.26) and of an analogue N-(hydroxyheptadecanoyl)glycine (m/z 344.28), while classical MN resulted in the observation of one single consensus spectrum for each compound (Figure 2c). In addition, FBMN allowed to observe a putative commendamide derivative N-(dehydrohexadecanoyl)glycine (CCMSLIB00005436498 and Supplementary Fig. 12) in the commendamide molecular network. The sample collection and mass spectrometry acquisition methods are described in the original manuscript9. The data were downloaded from MassIVE (MSV000080186) and processed with MZmine (2.37). The MZmine project along with parameters and export files were deposited to the MassIVE repository (MSV000084095). The chromatograms for m/z 330.26 and m/z 344.28 displayed in Figure 2c–d are from samples 43076_P3_RB9_01_314.mzML and 38131_P5_RA4_01_538.mzML, respectively. Chromatograms were exported with MZmine. The results were exported with the “Export for/Submit to GNPS” module for FBMN analysis on GNPS. The corresponding job can be accessed here: https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=0a8432b5891a48d7ad8459ba4a89969f (only logged users can see all the input files). The mzML files were used for the classical MN job can be accessed here: https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=3c27e43d908c4044bace405cc394cd25.
FBMN reduces spectral redundancy and deobfuscates spectral similarity relationships: the case of EDTA
The benefit of using FBMN can be illustrated with the metal chelating agent ethylenediaminetetraacetic acid (EDTA), widely used in beauty products, food, and scientific protocols. A search for its occurrences in public spectral datasets with the mass spectrometry search tool (MASST)32 showed that it is frequently observed in plasma samples where it is used during the sample preparation. We took one of the public human plasma datasets (MSV00008263) where EDTA was observed. For a detailed description of the protocol and mass spectrometry parameters, see Supplementary Note 3. The analysis of the data with classical MN showed that the EDTA ions are found in two molecular networks. One network consists of [M+H]+ spectra and the other of [M+Na]+ spectra. Interestingly, each of these networks have one node with a large number of clustered spectra (node 91,205 for 4,655 spectra, and node 116,470 for 571 spectra, respectively), but yet EDTA ions are represented by multiple nodes although these nodes have the same precursor ion mass and retention time. Detailed analysis showed that while the median pairwise cosine values between EDTA spectra are high (median value of 0.93 and 0.94), the spectra are not clustering into a single node. Examination of the multiple fragmentation spectra for EDTA ions showed that some 1) are chimeric spectra “contaminated” by fragment ions produced by co-eluting isobaric ions, and 2) that other spectra were dominated by low intensity fragment ions resulting from MS2 spectra acquired at low intensity. The method of FBMN was applied on that same dataset using the OpenMS-GNPS workflow (see the job), and the results showed that it efficiently reduces the appearance of these redundant node patterns from the same molecule (see the FBMN job, Figure 2f), both for the molecular networks containing the [M+H]+ and [M+Na]+ spectra. FBMN recovers the molecular similarity of in-source fragments observed for EDTA, which were not displayed with classical MN, as they now fall within the top-K rank (typically set to 10) of MS2 spectral similarity considered in the network topology. The parameters used for OpenMS tools can be accessed in the OpenMS-GNPS job (see the job). OpenMS ver. 2.4.0 was used10.
FBMN enables the use of relative quantification in the molecular networks
While classical MN uses the spectral count or the sum of precursor ion intensity to estimate the ion abundance, FBMN uses the accurate ion intensities obtained from LC-MS feature detection. The FBMN method brings in ion abundance across all samples by using the value of the chromatographic peak area or peak height as determined by the LC-MS feature detection and alignment software. Using multiple dilutions (n = 5) of the NIST 1950 serum reference metabolome sample33 analyzed by LC-MS2 (3 independent experiments per sample) on an Orbitrap mass spectrometer (Q Exactive, ThermoFisher) and processed with MZmine or OpenMS, we show the higher linearity of the relative quantification with FBMN and the improvement compared to classical MN (Figure 2h, Supplementary Note 4, and Supplementary Figures 13–19) using Ordinary least squares Linear Regression (OLR) analysis between the feature intensity and expected relative abundance in samples of known dilution factor (serial dilution). The sample preparation and mass spectrometry methods are described in Supplementary Note 4. The files along with the parameters for MZmine are available on the following MassIVE repository (MSV000084092). The OLR analysis was performed with python 2 (ver. 2.7.15) with the LinearRegression function of the sklearn package (ver. 0.20.1)33. The analysis is available as Jupyter notebook at https://github.com/lfnothias/FeatureBasedMolecularNetworking_RelativeQuantEval. The molecular networking jobs and parameters can be accessed here: classical MN (https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=daf3f0d7cec94104b2c9001739964c31), FBMN with MZmine (https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=f443cad083be4979aedd2af0f97b9fe9), GNPS-OpenMS job and parameters (https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=d6a430cc6da2458f8135ae76126eb763) and FBMN job with OpenMS https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=53bcfa39fa674c749b4da0b613df1b8d).
FBMN enables molecular networking with ion mobility spectrometry
The sample NIST 1950 serum33 was analyzed using a timsTOF Pro (Bruker Daltonics, Bremen) in data-dependent acquisition mode using PASEF (Parallel Accumulation-Serial Fragmentation)34. The data were then processed with MetaboScape (ver. 5.0) and the results were exported for FBMN analysis on GNPS. The mass spectrometry acquisition method, data, and parameters used for the processing were deposited on MassIVE (MSV000084402). Classical MN were annotated with the GNPS4, NIST17 and LipidBlast35 spectral libraries https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=f2adc2cf33c646548798d0e285197a96). Lipid annotation in MetaboScape was performed using SimLipid (ver. 6.04, Premier Biosoft, Palo Alto) and mapped to the FBMN (https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=0d89db67b0974939a91cb7d5bfe87072). The molecular networks were visualized with Cytoscape ver. 3.7.125, and the results are presented in the Supplementary Information (Fig. S1).
Integration with other computational mass spectrometry annotation tools
The MGF file format is accepted by numerous computational mass spectrometry annotation tools. The use of these tools with the MS2 spectral summary file enables 1) to reduce the spectral load inputted in the annotation tool compare to when using the unprocessed mass spectrometry files, 2) to allow the subsequent mapping of these annotations to the molecular networks produced by the FBMN method. Some of these are directly available in the GNPS environment, including SIRIUS15, DEREPLICATOR16, NAP36, MS2LDA17, MolNetEnhancer37, and Qemistree18 (see Supplementary Note 5), as well as other software such as MetWork38, CFM-ID39, MetFrag40.
Running time and scalability of the FBMN method
While the molecular networking computation of the FBMN method is performed online on the GNPS web-server (runtime = 5 min to several hours depending on the number of features and job parameters), the data processing part of the method has to be performed with the computational resources available to the researcher (laptop/desktop computer, workstation, cluster/cloud infrastructure). The computational cost of the data processing part depends on 1) the software employed, 2) the number of samples in the dataset, and 3) the parameters set. For this reason, the computational cost of the method in all scenarios cannot be established comprehensively. Nevertheless, our experience and feedback from the FBMN community with open source tools such as MZmine or MS-DIAL showed that small size datasets (< 50 samples) can be processed in 10–60 minutes with a desktop/laptop computer equipped with 8–16 GB of RAM. Medium size datasets (few hundred(s) samples) require the use of a workstation equipped with 16–32 GB of RAM memory, and large size datasets (> 500 samples) need 32–64 GB of RAM. For very large datasets (more than a thousand samples), it is currently recommended to use OpenMS or XCMS on a cluster/cloud infrastructure.
Large dataset processing with OpenMS and XCMS
The processing of large metabolomics datasets (more than a thousand samples) is limited by the scalability of existing LC-MS feature detection tools, especially those based on a GUI (such as MZmine and MS-DIAL). We showed that with specific peak picking parameters the use of XCMS or OpenMS enables using FBMN for large metabolomics study (MSV000080030, approximately 2,000 samples). See the Supplementary Note 6, Supplementary Table 2, and Supplementary Fig. 19.
Code availability
The FBMN workflow is available as a web-interface on the GNPS web platform (https://gnps-quickstart.ucsd.edu/featurebasednetworking). The workflow code is open source and available on GitHub (https://github.com/CCMS-UCSD/GNPS_Workflows/tree/master/feature-based-molecular-networking). It is released under the licence of The Regents of the University of California and free for non-profit research (https://github.com/CCMS-UCSD/GNPS_Workflows/blob/master/LICENSE). The workflow was written in Python (ver. 3.7) and deployed with the ProteoSAFE workflow manager employed by GNPS (http://proteomics.ucsd.edu/Software/ProteoSAFe/). We also provide documentation, support, example files, and additional information on the GNPS documentation website (https://ccms-ucsd.github.io/GNPSDocumentation/featurebasedmolecularnetworking/). The source code of the GNPSExport module in MZmine is available at (https://github.com/mzmine/mzmine2) under the GNU General Public License. The source code of the GNPSExport tool in OpenMS is available at (https://github.com/Bioinformatic-squad-DorresteinLab/OpenMS) under the BSD licence. The source code for the GNPSExport custom function for XCMS is available at https://github.com/jorainer/xcms-gnps-tools under the GNU General Public License.
Data availability
The LC-MS2 data for the Euphorbia dendroides dataset, along with the MZmine project and parameters used can be accessed on the MassIVE submission (MSV000080502, Creative Commons CC0 1.0 Universal license). The classical MN and FBMN jobs can be accessed via the GNPS website at https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=189e8bf16af145758b0a900f1c44ff4a and https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=672d0a5372384cff8c47297c2048d789, respectively.
The LC-MS2 data for the American Gut Project (AGP) were downloaded from MassIVE (MSV000080186 Creative Commons CC0 1.0 Universal license) and processed with MZmine (2.37). The MZmine project along with parameters and export files were deposited (MSV000084095, Creative Commons CC0 1.0 Universal license). The classical MN and FBMN jobs can be accessed at https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=3c27e43d908c4044bace405cc394cd25 and https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=0a8432b5891a48d7ad8459ba4a89969f, respectively.
The LC-MS2 data for the EDTA case are available on the MassIVE submission (MSV00008263, Creative Commons CC0 1.0 Universal license). The classical MN job can be accessed at https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=fbac1a5061ba4ad683a284ef55d45df6). The OpenMS and the FBMN job at https://proteomics2.ucsd.edu/ProteoSAFe/status.jsp?task=83a0a417a49b4b76b61e9a8191a6ea2d at https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=8f40420c11694cf9ab06fdf7a5a4c53b, respectively.
The mass spectrometry acquisition method, data, and parameters used for the processing of the serum analysis with the timsTOF mass spectrometer were deposited (MSV000084402). Classical MN and FBMN jobs can be accessed here: https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=f2adc2cf33c646548798d0e285197a96, and https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=0d89db67b0974939a91cb7d5bfe87072, respectively.
Supplementary Material
Acknowledgements
We gratefully acknowledge financial support by: the U.S. National Institutes of Health (NIH) for the Center for Computational Mass Spectrometry grant (P41 GM103484), the reuse of metabolomics data (R03 CA211211), and the tools for rapid and accurate structure elucidation of natural products (R01 GM107550) to P.D.; the NIH R24GM127667 and the National Science Foundation (NSF) award ABI 175998; the European Union’s Horizon 2020 grants 704786 (MSCA-GF to L.F.N) 634402, 777222 (T.A. and I.P.) and the ERC Consolidator grant METACELL (T.A.); L.F.N was supported by the Centre for Microbiome Innovation from the UC San Diego (Support Program award); the German Research Foundation (DFG) with grant number PE 2600/1 to D.P.; S.N. acknowledges funding from Bundesministerium für Bildung und Forschung (FKZ 031L0107) and the European Commission (EC654241); R.S. acknowledges funding by the German Chemical Industry Fund (FCI) fellowship; H.T. was supported by KAKENHI (18H02432, 18K19155); AMCR was supported by the NSF grant IOS-1656481 to PCD; O.A., acknowledges the funding from: the Bundesministerium für Ernährung und Landwirtschaft (FKZ 2816501214), the Bundesministerium für Wirtschaft und Energie (FKZ AiF18475N), the Bundesministerium für Bildung und Forschung (FKZ 031A430C), and the European Commission (823839) that also benefited to F.A. and O.K.; S.B. acknowledges funding from Deutsche Forschungsgemeinschaft (BO 1910/20); M.L. was supported by the Deutsche Forschungsgemeinschaft [BO 1910/20-1]. J.J.J.v.d.H. was supported by an Accelerating Scientific Discoveries Grant funded by the Netherlands eScience Center [NLeSC] (No. ASDI.2017.030). S.N. acknowledges BMBF funding under grant number 031L0107 and SN acknowledges funding from the European Commission PhenoMeNal Grant EC654241. F.V. was funded by the Department of Navy, Office of Naval Research Multidisciplinary University Research Initiative (MURI) Award (N00014-15-1-2809).
V.V.P acknowledges the ALSAM Foundation (Therapeutic Innovation Award and L.S. Skaggs Professorship) and the NIH (R35 GM128690). T.P. is a Simons Foundation Fellow of the Helen Hay Whitney Foundation. Z.K. was supported by the project International Mobility of Researchers (CZ.02.2.69/0.0/0.0/16_027/0007990). A.K.J was supported by the American Society for Mass Spectrometry (Postdoctoral Career Development Award). K.B.K. was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (No. NRF-2019R1F1A1058068). H.Y. was supported by the Basic Science Research Program through the NRF grant (NRF-2018R1C1B6002574). A.L.G was supported by Vaincre la mucoviscidose and Association Grégory Lemarchal. The authors would like to thank Nils Hoffman for maintaining the mzTab-M format. Finally, we would like to acknowledge the continuous feedback from the GNPS community, and the contribution of all researchers/institutions who are commited to deposit their mass spectrometry data on public repositories.
Footnotes
Ethics/COI declaration
Pieter C. Dorrestein is a scientific advisor for Sirenas LLC and Cybele.
Mingxun Wang is a consultant for Sirenas LLC and the founder of Ometa labs LLC.
Tomáš Pluskal is a consultant for Ginkgo Bioworks.
Alexander Aksenov is a consultant for Ometa labs LLC.
Theodore Alexandrov is on the Scientific Advisory Board of SCiLS, a Bruker company.
Kai Dührkop, Marcus Ludwig, Markus Fleischauer and Sebastian Böcker are founders of Bright Giant GmbH.
Aiko Barsch, Sven W. Meyer, Heiko Neuweger and Florian Zubeil are employes of Bruker Daltonics GmbH.
Giorgis Isaac, Jonathan McSayles, and Bindesh Shrestha are employees of Waters Corporation.
References
- 1.Watrous J et al. Mass spectral molecular networking of living microbial colonies. Proc. Natl. Acad. Sci. 109, E1743–52 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Quinn RA et al. Molecular Networking As a Drug Discovery, Drug Metabolism, and Precision Medicine Strategy. Trends Pharmacol. Sci. 38, 143–154 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Traxler MF & Kolter R A massively spectacular view of the chemical lives of microbes. Proceedings of the National Academy of Sciences of the United States of America vol. 109 10128–10129 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang M et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 34, 828–837 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frank AM et al. Clustering Millions of Tandem Mass Spectra. J. Proteome Res. 7, 113–122 (01/2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoffmann N et al. mzTab-M: A Data Standard for Sharing Quantitative Results in Mass Spectrometry Metabolomics. Anal. Chem. 91, 3302–3310 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nothias L-F et al. Bioactivity-Based Molecular Networking for the Discovery of Drug Leads in Natural Product Bioassay-Guided Fractionation. J. Nat. Prod. 81, 758–767 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Cohen LJ et al. Functional metagenomic discovery of bacterial effectors in the human microbiome and isolation of commendamide, a GPCR G2A/132 agonist. Proc. Natl. Acad. Sci. U. S. A. 112, E4825–E4834 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McDonald D et al. American Gut: an Open Platform for Citizen Science Microbiome Research. mSystems 3, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Röst HL et al. OpenMS: a flexible open-source software platform for mass spectrometry data analysis. Nat. Methods 13, 741–748 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Pluskal T, Castillo S, Villar-Briones A & Oresic M MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics 11, 395 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bolyen E et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. (2019) doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xia J, Sinelnikov IV, Han B & Wishart DS MetaboAnalyst 3.0—making metabolomics more meaningful. Nucleic Acids Res. 43, W251–W257 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Protsyuk I, Melnik AV, Nothias LF & Rappez L 3D molecular cartography using LC–MS facilitated by Optimus and’ili software. Nat. Protoc. (2018). [DOI] [PubMed] [Google Scholar]
- 15.Dührkop K et al. SIRIUS 4: a rapid tool for turning tandem mass spectra into metabolite structure information. Nat. Methods 16, 299–302 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Mohimani H et al. Dereplication of peptidic natural products through database search of mass spectra. Nat. Chem. Biol. 13, 30–37 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van der Hooft JJJ, Wandy J, Barrett MP, Burgess KEV & Rogers S Topic modeling for untargeted substructure exploration in metabolomics. Proc. Natl. Acad. Sci. U. S. A. 113, 13738–13743 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tripathi A et al. Chemically-informed Analyses of Metabolomics Mass Spectrometry Data with Qemistree. bioRxiv 2020.05.04.077636 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsugawa H et al. MS-DIAL: data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat. Methods 12, 523–526 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang M et al. Mass spectrometry searches using MASST. Nat. Biotechnol. 38, 23–26 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chambers MC et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 30, 918–920 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Winnikoff JR, Glukhov E, Watrous J, Dorrestein PC & Gerwick WH Quantitative molecular networking to profile marine cyanobacterial metabolomes. J. Antibiot. 67, 105–112 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olivon F, Grelier G, Roussi F, Litaudon M & Touboul D MZmine 2 Data-Preprocessing To Enhance Molecular Networking Reliability. Anal. Chem. (2017) doi: 10.1021/acs.analchem.7b01563. [DOI] [PubMed] [Google Scholar]
- 24.Ono K, Demchak B & Ideker T Cytoscape tools for the web age: D3.js and Cytoscape.js exporters. F1000Res. 3, 143 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shannon P et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tautenhahn R, Böttcher C & Neumann S Highly sensitive feature detection for high resolution LC/MS. BMC Bioinformatics 9, 504 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Libiseller G et al. IPO: a tool for automated optimization of XCMS parameters. BMC Bioinformatics 16, 118 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McLean C & Kujawinski EB AutoTuner: High Fidelity and Robust Parameter Selection for Metabolomics Data Processing. Anal. Chem. 92, 5724–5732 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuhl C, Tautenhahn R, Böttcher C, Larson TR & Neumann S CAMERA: an integrated strategy for compound spectra extraction and annotation of liquid chromatography/mass spectrometry data sets. Anal. Chem. 84, 283–289 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsugawa H et al. MS-DIAL 4: accelerating lipidomics using an MS/MS, CCS, and retention time atlas. bioRxiv 2020.02.11.944900 (2020) doi: 10.1101/2020.02.11.944900. [DOI] [Google Scholar]
- 31.Cohen LJ et al. Commensal bacteria make GPCR ligands that mimic human signalling molecules. Nature 549, 48–53 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang M et al. MASST: A Web-based Basic Mass Spectrometry Search Tool for Molecules to Search Public Data. bioRxiv 591016 (2019) doi: 10.1101/591016. [DOI] [Google Scholar]
- 33.Simón-Manso Y et al. Metabolite profiling of a NIST Standard Reference Material for human plasma (SRM 1950): GC-MS, LC-MS, NMR, and clinical laboratory analyses, libraries, and web-based resources. Anal. Chem. 85, 11725–11731 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Meier F et al. Online Parallel Accumulation-Serial Fragmentation (PASEF) with a Novel Trapped Ion Mobility Mass Spectrometer. Mol. Cell. Proteomics 17, 2534–2545 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kind T et al. LipidBlast in silico tandem mass spectrometry database for lipid identification. Nat. Methods 10, 755–758 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.da Silva RR et al. Propagating annotations of molecular networks using in silico fragmentation. PLoS Comput. Biol. 14, e1006089 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ernst M et al. MolNetEnhancer: Enhanced Molecular Networks by Integrating Metabolome Mining and Annotation Tools. Metabolites 9, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beauxis Y & Genta-Jouve G Metwork: a web server for natural products anticipation. Bioinformatics (2018) doi: 10.1093/bioinformatics/bty864. [DOI] [PubMed] [Google Scholar]
- 39.Allen F, Greiner R & Wishart D Competitive fragmentation modeling of ESI-MS/MS spectra for putative metabolite identification. Metabolomics 11, 98–110 (2015). [Google Scholar]
- 40.Ruttkies C, Schymanski EL, Wolf S, Hollender J & Neumann S MetFrag relaunched: incorporating strategies beyond in silico fragmentation. J. Cheminform. 8, 1–16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The LC-MS2 data for the Euphorbia dendroides dataset, along with the MZmine project and parameters used can be accessed on the MassIVE submission (MSV000080502, Creative Commons CC0 1.0 Universal license). The classical MN and FBMN jobs can be accessed via the GNPS website at https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=189e8bf16af145758b0a900f1c44ff4a and https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=672d0a5372384cff8c47297c2048d789, respectively.
The LC-MS2 data for the American Gut Project (AGP) were downloaded from MassIVE (MSV000080186 Creative Commons CC0 1.0 Universal license) and processed with MZmine (2.37). The MZmine project along with parameters and export files were deposited (MSV000084095, Creative Commons CC0 1.0 Universal license). The classical MN and FBMN jobs can be accessed at https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=3c27e43d908c4044bace405cc394cd25 and https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=0a8432b5891a48d7ad8459ba4a89969f, respectively.
The LC-MS2 data for the EDTA case are available on the MassIVE submission (MSV00008263, Creative Commons CC0 1.0 Universal license). The classical MN job can be accessed at https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=fbac1a5061ba4ad683a284ef55d45df6). The OpenMS and the FBMN job at https://proteomics2.ucsd.edu/ProteoSAFe/status.jsp?task=83a0a417a49b4b76b61e9a8191a6ea2d at https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=8f40420c11694cf9ab06fdf7a5a4c53b, respectively.
The mass spectrometry acquisition method, data, and parameters used for the processing of the serum analysis with the timsTOF mass spectrometer were deposited (MSV000084402). Classical MN and FBMN jobs can be accessed here: https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=f2adc2cf33c646548798d0e285197a96, and https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=0d89db67b0974939a91cb7d5bfe87072, respectively.