Abstract
The global outbreak of coronavirus disease and rapid spread of the causative severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) represent a significant threat to human health. A key mechanism of human SARS-CoV-2 infection is initiated by the combination of human angiotensin-converting enzyme 2 (hACE2) and the receptor-binding domain (RBD) of the SARS-CoV-2-derived spike glycoprotein. Despite the importance of these protein interactions, there are still insufficient detection methods to observe their activity at the cellular level. Herein, we developed a novel fluorescence resonance energy transfer (FRET)-based hACE2 biosensor to monitor the interaction between hACE2 and SARS-CoV-2 RBD. This biosensor facilitated the visualization of hACE2-RBD activity with high spatiotemporal resolutions at the single-cell level. Further studies revealed that the FRET-based hACE2 biosensors were sensitive to both exogenous and endogenous hACE2 expression, suggesting that they might be safely applied to the early stage of SARS-CoV-2 infection without direct virus use. Therefore, our novel biosensor could potentially help develop drugs that target SARS-CoV-2 by inhibiting hACE2-RBD interaction.
Abbreviations: bg, background; CQ, chloroquine; hACE2, human angiotensin-converting enzyme 2; HCQ, hydroxychloroquine; NA, numerical aperture; RBD, receptor-binding domain; RBM, receptor-binding motif; ROI, region of interest; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; SEM, standard error of the mean
Keywords: SARS-CoV-2, FRET, Biosensor, ACE2, Live-cell imaging
1. Introduction
In December 2019, a new coronavirus (Severe acute respiratory syndrome coronavirus 2, SARS-CoV-2) was first recognized and quickly spread globally. The SARS-CoV-2 is a highly pathogenic virus that represents a global health risk and causes respiratory, cerebrovascular, heart, and lung diseases [[1], [2], [3], [4], [5]]. A key mechanism underlying SARS-CoV-2 infection is the structural recognition of human angiotensin-converting enzyme 2 (hACE2) and SARS-CoV-2-derived spike glycoprotein receptor-binding domain (RBD), which induces ACE2 downregulation [[6], [7], [8]]. hACE2 is expressed in the brain, heart, lung, kidney, liver, testis, and small intestine tissues [[9], [10], [11], [12], [13], [14]]. In a counter-regulatory manner to ACE, ACE2 mainly functions to cleave angiotensin I and angiotensin II into angiotensin-(1–9) and angiotensin-(1–7), respectively [13,15,16]. The ACE2-angiotensin fragment pathway provides a useful therapeutic target for treating cardiovascular and pulmonary diseases, heart failure, and kidney and lung fibrosis [15,[17], [18], [19], [20]]. Therefore, the regulation of ACE2 expression could exert beneficial effects on several SARS-CoV-2-related diseases [21].
In early studies, there were suggestions that drugs against SARS-CoV could probably have been used to treat the SARS-CoV-2 infection, which could shorten the time and reduce the cost. For example, chloroquine (CQ) and hydroxychloroquine (HCQ), which have been used to treat rheumatoid arthritis, malaria, and lupus erythematosus, were suggested in preventing SARS-CoV and SARS-CoV-2 infection [[22], [23], [24], [25], [26]]. In fact, CQ interferes with the terminal glycosylation of hACE2 in the treatment of SARS-CoV [25]. Due to the sequence similarity (∼76 %) between SARS-CoV and SARS-CoV-2 RBDs [27], researchers have thought that CQ and HCQ might also be used in the treatment of SARS-CoV-2 infection. Despite the structural similarity of RBD of SARS-CoV and SARS-CoV-2, they seem to exhibit different epitope features and differing immunogenicity [28]. At present, CQ and HCQ treatment trials have been stopped as they showed little or no reduction in the mortality of hospitalized COVID-19 patients versus standard of care. However, these drugs may still be considered agents to uncover the mystery of the molecular mechanism of the hACE2 pathway as an inhibitor [29,30]
Many therapeutic strategies against SARS-CoV-2 have been proposed. Most recently, Yang et al. revealed the molecular interaction of SARS-CoV-2 binding to the ACE2 receptor using atomic force microscopy [31]. This approach provides direct evidence that ACE2-derived peptides can be bound to S-glycoprotein of SARS-CoV-2 at the single-molecule level. However, there is still an insufficient detection method to visualize the dynamics of ACE2-RBD binding with high resolutions in living cells. Here, we developed a novel fluorescence resonance energy transfer (FRET)-based ACE2 biosensor by employing the RBD module derived from SARS-CoV-2, which can effectively monitor ACE2 activity with high spatiotemporal resolutions. Our study provides a new methodology to identify drugs that can inhibit ACE2-RBD interaction from preventing SARS-CoV-2 infection.
2. Materials and methods
2.1. DNA plasmid construction
The hACE2 biosensor was constructed by adding the SARS-CoV-2-derived RBD to pCAGGS-6011NES. The RBD fragment was fused between N-terminal YPet and C-terminal Turquoise2-GL, a CFP variant. The RBD domain was PCR-amplified from pcDNA3-SARS-CoV-2-s-RBD-sfGFP and inserted into pCAGGS-6011NES digested by XhoⅠ/NotⅠ (Fig. S4). The RBD mutants were generated from pcDNA3-SARS-CoV-2-S-RBD-sfGFP using the EZchange™ Site-directed Mutagenesis Kit (Enzynomics Cat. No. EZ004S), following which RBD-E484 P (Fig. S5) and F486 L (Fig. S6) were PCR-amplified and inserted into pCAGGS-6011NES. pCAGGS-6011NES was a gift from Prof. Michiyuki Matsuda (Addgene plasmid # 108652). pcDNA3-SARS-CoV-2-S-RBD-sfGFP was a gift from Prof. Erik Procko (Addgene plasmid # 141184). pmScarlet-hACE2 was constructed by adding the hACE2 sequence to pmScarlet-I_C1. The hACE2 was PCR-amplified from pCEP4-myc-ACE2 and inserted into pmScarlet-I_C1 digested by BsrGⅠ/XhoI (Fig. S7). ACE2-M82 K was generated from pmScarlet-hACE2 using the EZchange™ Site-directed Mutagenesis Kit (Enzynomics Cat. No. EZ004S) (Fig. S8). The pmScarlet-I_C1 vector was provided by Dr. Jihye Seong (Korea Institute of Science and Technology, Republic of Korea). pCEP4-myc-ACE2, a truncated membrane localization sequence, was a gift from Prof. Erik Procko (Addgene plasmid # 141185). All primer pairs used in this study are listed in Table S1. All cloning design and site-directed mutagenesis procedures were simulated using the SnapGene software (Fig. S9-S10).
2.2. Cell culture and chemicals
The HEK293 T cell line was provided by Prof. Joomi Yi (Inje University, Republic of Korea). The HeLa cell line was provided by Dr. Jihye Seong (Korea Institute of Science and Technology, Republic of Korea). The HepG2 cell line was provided by Prof. Youngmi Jung (Pusan National University, Republic of Korea). The SH-SY5Y cell line was purchased from the Korean Cell Line Bank (Republic of Korea). HEK293 T, HeLa and HepG2 cells were cultured in Dulbecco’s Modified Eagle’s Medium/F12 (DMEM/F12, 11320033, Gibco) supplemented with 10 % fetal bovine serum (FBS, WB0015, Hyclone), 100 U/mL penicillin, and 100 μg/mL streptomycin (CA005, GenDepot). SH-SY5Y cells were cultured in RPMI 1640 medium (SH30027, Hyclone). The cells were cultured in a humidified incubator with 95 % air and 5 % CO2 at 37 °C in cover-glass-bottom dishes (100350, SPL). According to the manufacturer’s instructions, the DNA plasmids were transfected into the cells using Lipofectamine 3000 (L3000015, Invitrogen). Before imaging, cells were starved in DMEM/F12 or RPMI medium containing 0.5 % FBS for 6 h at 37 °C. HCQ sulfate was purchased from Sigma Aldrich (H0915). MLN-4760 was purchased from Merck(5.30616, Calbiochem).
2.3. Viability assay
The HEK293 T cells were seeded at 8 × cells/well in 96-well plates and incubated for 24 h at 37 °C before being treated with DMEM-containing control (0.5 % water) or HCQ media for 1. After washing, the cells were exposed to a 9.09 % (v/v) Cellrix Viability Assay kit in DMEM without phenol red (Cat no. 31053028, Gibco, Waltham, MA) for 2 h at 37 °C. The optical density of the solubilized formazan product was measured using a Glomax Multi + Microplate Multi Reader (9301-010, Promega, USA) at 450 nm.
2.4. Microscopy and image acquisition
During imaging, the cells were maintained in a CO2-independent medium (18045088, Gibco) containing 0.5 % FBS, 4 mM l-glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin. Images were acquired using a Leica DMi8 microscope with a 40X HC PL FLUOTAR L objective/numerical aperture (NA) 0.6 dry and 100X HC PL FLUOTAR objective/NA 1.32 oil immersion. The filter sets for CFP included a 436/20 excitation filter, 455 dichroic mirror, and 480/40 emission filter. The filter sets for FRET included a 436/20 excitation filter, 455 dichroic mirror, and 535/30 emission filter. The LAS X software was used to subtract the background of the images and acquire CFP, FRET, and ratio images.
2.5. FRET image analysis
The fluorescence intensity of the background region was selected and quantified to subtract the signals from the region of interest (ROI) of the FRET and CFP channels. The pixel-by-pixel ratio images of FRET/CFP were calculated based on the background (bg)-subtracted fluorescence intensity images of FRET and CFP as follows:
| I (FRETROI) - I (FRETbg) / I (CFPROI) – I (CFPbg), |
where I represents the intensity of each region from each channel. The ratio images were displayed in the intensity modified display mode, where the color and brightness of the pixel are determined by the FRET/CFP ratio.
2.6. Statistical analysis
All statistical analyses were performed using GraphPad 7.0. The statistical data are expressed as the mean ± standard error of the mean (SEM), and the statistical evaluation was performed by an unpaired t-test using GraphPad 7.0. P values at **<0.01, ***<0.001 and ****<0.0001 were considered statistically significant.
3. Results and discussion
3.1. ACE2 biosensor design and characterization
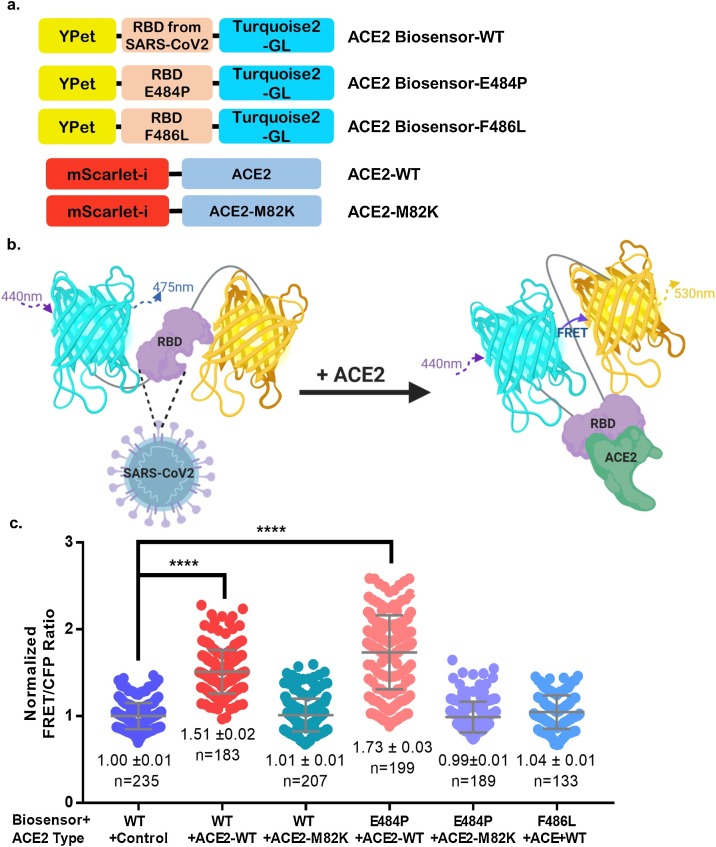
It has been previously demonstrated that the SARS-CoV-2-derived RBD binds to the N-terminal peptidase domain of ACE2, which initially mediates viral infection. To visualize RBD-ACE2 binding activity, we designed a FRET-based ACE2 biosensor, with SARS-CoV-2-derived RBD concatenated between YPet and Turquoise2-GL (Fig. 1 ). According to previous SARS-CoV-2 spike and ACE2 structural interaction studies, a critical receptor-binding motif (RBM) is present between residues 481–487, and ACE2-WT binding affinities are measured through mutations in the critical RBM domain [7]. A similar study has revealed critical amino acid residues to maintain its binding activity with hACE2 unveiled by site mutagenesis scanning. The results demonstrated that the E484 P mutant enhanced receptor binding affinity more than twice, whereas F486 L markedly diminished its binding affinity [32]. For cross-validation, the E484 P and F486 L biosensor mutants have been constructed based on SARS-CoV-2 and SARS-like virus RBD sequence alignments [32,33]. SARS-CoV-2 RBD 481-487 residues include the ACE2-binding ridge, which affects two disulfide-bond-forming RBD cysteines 480 and 488. The E484 P mutant, substituted from SAR-CoV-2 (Glu, E) to SARS-CoV (Pro, P), is expected to increase the ACE2 binding affinity of the biosensor [32]. The F486 L mutant, also substituted from SAR-CoV-2 (Phe, F) to SARS-CoV (Leu, L), is expected to decrease the ACE2-WT binding affinity of the biosensor [32]. The ACE2 hydrophobic pocket, interacting with the SARS-CoV-2-derived RBD F486, constitutes L79, M82, and Y83 [7]. As the ACE2 affinity change of the hydrophobic region was also imperative, we mutated the ACE2 Met82 (M) residue into lysine (K) of ACEs in the testis-specific form and Drosophila melanogaster that have homology with ACE2 (similarities 61 % and 55 %, respectively) [34].
Fig. 1.
Design and characterization of the FRET-based ACE2 biosensor. (a) Schematic diagram of the ACE2 biosensor. The ACE2 biosensor is composed of a YPet acceptor, SARS-CoV-2-derived RBD domain, and truncated Turquoise2-GL donor. The ACE2 biosensor mutants are E484 P (positive control) and F486 L (negative control). The ACE2-WT truncated membrane localization signal was inserted into the pmScarlet-I vector, and the ACE2-M82 K construct was developed as a negative control. (b) The ACE2 biosensor shows the conformational change by binding to ACE2, leading to an increase in the FRET ratio. This diagram was produced by Biorender (http://biorender.com/) (c) The normalized FRET/CFP emission ratios of the ACE2 biosensors (WT, E484 P, and F486 L) in HEK293 T cells co-transfected with ACE2-WT, ACE2-M82 K, or control pmScarlet-I vector (****p < 0.0001).
It is expected that RBD-ACE2 binding could be modified by RBD or ACE2 inhibitors, causing a conformational change and decrease in FRET (Fig. 1b). HCQ, an ACE2 N-terminal glycosylation inhibitor, was an option to use in the current study due to the lack of known RBD inhibitors. The FRET/CFP emission ratios of the ACE2 biosensor were significantly higher in HEK293 T cells co-transfected with pmScarlet-hACE2 than in control (Fig. 1c) in which ACE2 expression was absent. This is consistent with a previous study which reported that HEK293 T cells scarcely express ACE2 [35]. As shown in Fig. 1c, the changes in FRET compared to the control value were rarely observed in HEK293 T cells co-transfected with ACE2-M82 K and ACE2 biosensor-WT or -E484 P, while the FRET/CFP ratios were high in HEK293 T cells co-transfected with ACE2-WT and ACE2 biosensor-E484 P. These results indicate that the RBD specifically binds to exogenous ACE2 in human cells.
3.2. ACE2 biosensor specificity with exogenous and endogenous ACE2
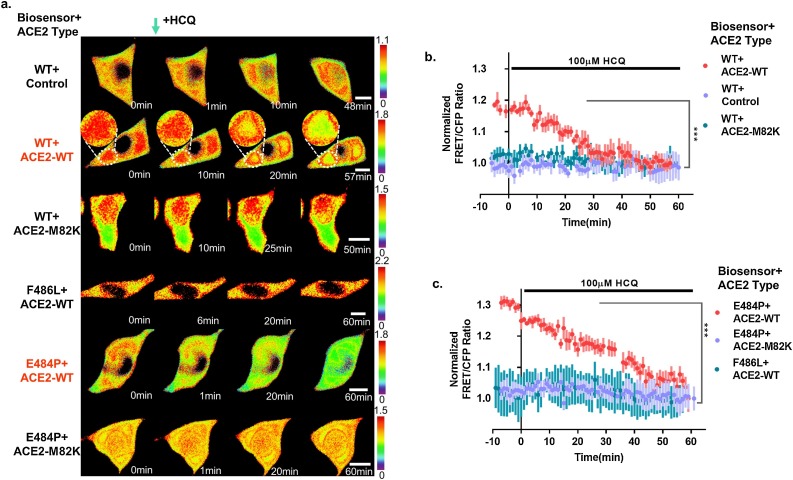
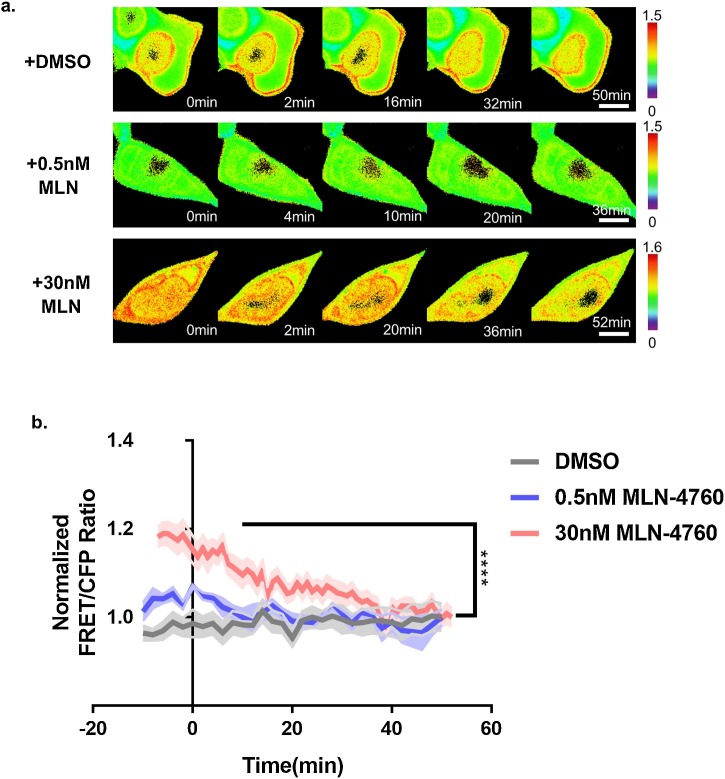
To observe the affinity of ACE2 and ACE2 biosensors through real-time imaging, we determined the HCQ concentration using a viability assay and FRET analysis (Fig. S1 and S2). Since a previous study reported that the cytotoxic concentration (CC50) of HCQ was 249.5 μM in Vero cells [26], and the HCQ treatment within 2 h had minimal effect on HEK293 cell viability [36], we decided to use a concentration of 100 μM. Indeed, our experimental results did not show any significant cytotoxicity under this condition (Fig. S1). As shown in Fig. 2 , there were few changes in the FRET/CFP ratios in HEK293 T cells transfected with the ACE2 biosensor-WT and control vector/ACE2-M82 K or the ACE2 biosensor-F486 L and ACE2-WT using a 100 μM HCQ treatment (Fig. 2a). However, there were significant changes in cells co-transfected with the ACE2-WT and ACE2 biosensor-WT or -E484 P after 50–60 min of HCQ treatment (Fig. 2a–c). Similarly, the infection with SARS-CoV was regulated by ACE2 expression in HEK293 cells [35]. To further validate the specificity of the ACE2 biosensor, we used MLN-4760, a specific ACE2 inhibitor. Our data revealed a decrease of the FRET/CFP ratio of the ACE2 biosensor in HEK293 T cells treated with MLN-4760 (Fig. 3 ). It was also confirmed that the FRET/CFP ratio of the ACE2 biosensor decreased (∼30 %) in response to HCQ treatment in HeLa cells that do not express intrinsic ACE2 (Fig. S3).
Fig. 2.
Dynamic visualization of the ACE2 biosensor in response to the ACE2 inhibitor. (a) Time-lapse FRET/CFP ratio images of the ACE2 biosensors (WT, E484 P, and F486 L) in HEK293 T cells co-transfected with control, ACE2-WT, or ACE2-M82 K treated with 100 μM HCQ. Hot and cold colors indicate high and low affinities between ACE2s and RBDs, respectively (scale bar = 10 μm). (b) Time courses of the normalized FRET/CFP ratio of the ACE2 biosensor-WT in HEK293 T cells co-transfected with ACE2-WT (red, n = 4), control (blue-violet, n = 5), or ACE2-M82 K (turquoise, n = 9) treated with 100 μM HCQ. All error bars represent SEM. (***p < 0.001). (c) Time courses of the normalized FRET/CFP ratios of the ACE2 mutant biosensors (E484P-positive control and F486L-negative control) in HEK293 T cells co-transfected with ACE2-WT or ACE2-M82 K treated with 100 μM HCQ (red, blue-violet, and turquoise; n = 5,5, and 4, respectively). All error bars represent SEM (***p < 0.001).
Fig. 3.
Validation of the ACE2 biosensor in response to MLN-4760, a specific ACE2 inhibitor. (a) Time-lapse FRET/CFP ratio images of the ACE2 biosensor in HEK293 T co-transfected with ACE2-WT treated with DMSO, 0.5 nM, and 30 nM MLN-4760, respectively. Hot and cold colors indicate high and low affinities between exogenous ACE2 and RBD, respectively (scale bar = 10 μm). (b) Time courses of the normalized FRET/CFP ratios of the ACE2 biosensor in HEK293 T co-transfected with ACE2-WT treated with DMSO, 0.5 nM, and 30 nM MLN-4760 (gray, blue, and red; n = 4,4, and 7, respectively). All error bars represent SEM (****p < 0.0001).
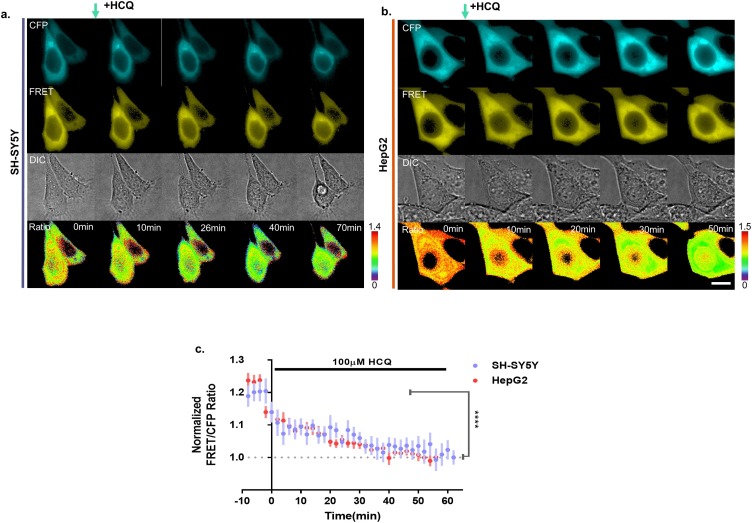
Next, we designed additional experiments to detect endogenous ACE2 with the ACE2 biosensor, and thus, we examined SH-SY5Y and HepG2 cells that express hACE2 [37]. The FRET/CFP ratios of the ACE2 biosensor were 18.9 % and 21.0 % in SH-SY5Y and HepG2 cells, respectively, which decreased gradually (Fig. 4 ). Thus, our results indicate that the ACE2 biosensor visualizes the interaction of the endogenous or exogenous ACE2 and SARS-CoV-2-derived RBD.
Fig. 4.
ACE2 biosensor specificity in SY5Y and HepG2 cells. (a-b) Time-lapse CFP, FRET, DIC, and FRET/CFP ratio images of the ACE2 biosensor in SH-SY5Y and HepG2 cells treated with 100 μM HCQ, respectively. Hot and cold colors indicate high and low endogenous ACE2 and RBD affinities, respectively (scale bar = 10 μm). (c) Time courses of the normalized FRET/CFP ratios of the ACE2 biosensor in SH-SY5Y (blue-violet, n = 7) and HepG2 (red, n = 10) cells treated with 100 μM HCQ. All error bars represent SEM (****p < 0.0001).
In this study, we developed an ACE2 biosensor to visualize the ACE2-RBD interaction and monitored its activity at the single-cell level. As we targeted early-stage viral infection and were inspired to design the ACE2 biosensor with ACE2-RBD interaction, several processes specified that HCQ might be useful for treating early-stage SARS-CoV-2 infection. Genetically encoded biosensors can potentially visualize another viral infection mechanism, such as the TMPRSS2- or cathepsin L-derived RBD cleavage step, when targeting the priming stage.
RBD was substantially suitable for composing the biosensor because 1) the weight and size of the domain were low and compact for performing gene cloning. 2) The three-dimensional distance between the N- and C-terminal sites, designed to connect to YPet and Turquoise2-GL, respectively, was short. Thus, we assumed that in the biosensor, the space between YPet- Turquoise2-GL would be shorter than 100 Å (10 nm), and the two fluorescence proteins could form the FRET pair. Although we made an effort to apply the ACE2 protein for developing biosensors, we encountered certain limitations during its generation: 1) If we introduced a full ACE2 sequence into the FRET pair, the FRET phenomenon would not occur due to the considerable distance between the N-terminal extracellular and C-terminal intracellular domains of ACE2 linked to YPet and Turquoise2-GL, respectively. Thus, there would be less possibility of shaping the FRET pair. 2) When researchers design biosensors, they do not use full-sequence proteins but specific domains or short polypeptides. In the case of ACE2, which has complex secondary and tertiary structures interacting with one another, it remains a challenge to elucidate whether the biosensors would be translated and function appropriately when composed of short peptide fragments or ACE2 domains.
Thus, if researchers establish and apply a process to develop the drug-screening biosensor, it would help visualize sophisticated receptor-ligand interactions and shorten the time to identify candidate drugs when the next novel viral disease outbreak occurs.
4. Conclusions
In summary, we have developed a novel hACE2 biosensor capable of monitoring the early interaction between ACE2 and SARS-CoV-2-derived RBD. In the cell-based assay, our biosensor displayed significant FRET change in response to exogenous or endogenous ACE2 expression in the presence of pharmacological inhibition. Therefore, we believe that our biosensor could be applied safely to study SARS-CoV-2 for drug candidate screening purposes without direct virus use.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
CRediT authorship contribution statement
Jung-Soo Suh: Conceptualization, Methodology, Investigation, Validation, Data curation, Writing - original draft. Heon-Su Kim: Conceptualization, Methodology, Investigation, Formal analysis, Writing - original draft. Tae-Jin Kim: Conceptualization, Resources, Data curation, Writing - review & editing, Supervision.
Acknowledgments
This research was funded by the National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) (2020R1C1C1010107). This work was supported by the Busan Innovation Institute of Industry, Science & Technology Planning (BISTEP) grant funded by the Busan Metropolitan City (Project: R&BD for regional outstanding researchers, startups, and innovative firms).
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.snb.2021.129663.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- 1.Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., Ren R., Leung K.S.M., Lau E.H.Y., Wong J.Y., Xing X., Xiang N., Wu Y., Li C., Chen Q., Li D., Liu T., Zhao J., Liu M., Tu W., Chen C., Jin L., Yang R., Wang Q., Zhou S., Wang R., Liu H., Luo Y., Liu Y., Shao G., Li H., Tao Z., Yang Y., Deng Z., Liu B., Ma Z., Zhang Y., Shi G., Lam T.T.Y., Wu J.T., Gao G.F., Cowling B.J., Yang B., Leung G.M., Feng Z. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C., Zhang Y.Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020 doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li Y., Wang M., Zhou Y., Chang J., Xian Y., Mao L., Hong C., Chen S., Wang Y., Wang H., Li M., Jin H., Hu B. Acute cerebrovascular disease following COVID-19: a single center, retrospective, observational study. SSRN Electron. J. 2020 doi: 10.2139/ssrn.3550025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen L., Li X., Chen M., Feng Y., Xiong C. The ACE2 expression in human heart indicates new potential mechanism of heart injury among patients infected with SARS-CoV-2. Cardiovasc. Res. 2020 doi: 10.1093/cvr/cvaa078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu X., Yu C., Qu J., Zhang L., Jiang S., Huang D., Chen B., Zhang Z., Guan W., Ling Z., Jiang R., Hu T., Ding Y., Lin L., Gan Q., Luo L., Tang X., Liu J. Imaging and clinical features of patients with 2019 novel coronavirus SARS-CoV-2. Eur. J. Nucl. Med. Mol. Imaging. 2020 doi: 10.1007/s00259-020-04735-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fu Y., Cheng Y., Wu Y. Understanding SARS-CoV-2-Mediated inflammatory responses: from mechanisms to potential therapeutic tools. Virol. Sin. 2020 doi: 10.1007/s12250-020-00207-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020 doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science (80-.) 2020 doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xia H., Lazartigues E. Angiotensin-converting enzyme 2 in the brain: properties and future directions. J. Neurochem. 2008 doi: 10.1111/j.1471-4159.2008.05723.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crackower M.A., Sarao R., Oliveira-dos-Santos A.J., Da Costa J., Zhang L. Angiotensin-converting enzyme 2 is an essential regulator of heart function. Nature. 2002 doi: 10.1038/nature00786. [DOI] [PubMed] [Google Scholar]
- 11.Komatsu T., Suzuki Y., Imai J., Sugano S., Hida M., Tanigami A., Muroi S., Yamada Y., Hanaoka K. Molecular cloning, mRNA expression and chromosomal localization of mouse angiotensin-converting enzyme-related carboxypeptidase (mACE2) Mitochondrial DNA. 2002 doi: 10.1080/1042517021000021608. [DOI] [PubMed] [Google Scholar]
- 12.Herath C.B., Warner F.J., Lubel J.S., Dean R.G., Jia Z., Lew R.A., Smith A.I., Burrell L.M., Angus P.W. Upregulation of hepatic angiotensin-converting enzyme 2 (ACE2) and angiotensin-(1-7) levels in experimental biliary fibrosis. J. Hepatol. 2007 doi: 10.1016/j.jhep.2007.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donoghue M., Hsieh F., Baronas E., Godbout K., Gosselin M., Stagliano N., Donovan M., Woolf B., Robison K., Jeyaseelan R., Breitbart R.E., Acton S. A novel angiotensin-converting enzyme-related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1-9. Circ. Res. 2000 doi: 10.1161/01.res.87.5.e1. [DOI] [PubMed] [Google Scholar]
- 14.Hamming I., Timens W., Bulthuis M.L.C., Lely A.T., Navis G.J., van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 2004 doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferrario C.M., Trask A.J., Jessup J.A. Advances in biochemical and functional roles of angiotensin-converting enzyme 2 and angiotensin-(1-7) in regulation of cardiovascular function. Am. J. Physiol. - Hear. Circ. Physiol. 2005 doi: 10.1152/ajpheart.00618.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferrario C.M. Angiotensin-converting enzyme 2 and angiotensin-(1-7): an evolving story in cardiovascular regulation. Hypertension. 2006 doi: 10.1161/01.HYP.0000196268.08909.fb. [DOI] [PubMed] [Google Scholar]
- 17.Jiang F., Yang J., Zhang Y., Dong M., Wang S., Zhang Q., Liu F.F., Zhang K., Zhang C. Angiotensin-converting enzyme 2 and angiotensin 1-7: novel therapeutic targets. Nat. Rev. Cardiol. 2014 doi: 10.1038/nrcardio.2014.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shenoy V., Qi Y., Katovich M.J., Raizada M.K. ACE2, a promising therapeutic target for pulmonary hypertension. Curr. Opin. Pharmacol. 2011 doi: 10.1016/j.coph.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chamsi-Pasha M.A.R., Shao Z., Tang W.H.W. Angiotensin-converting enzyme 2 as a therapeutic target for heart failure. Curr. Heart Fail. Rep. 2014 doi: 10.1007/s11897-013-0178-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simões e Silva A.C., Teixeira M.M. ACE inhibition, ACE2 and angiotensin-(1-7) axis in kidney and cardiac inflammation and fibrosis. Pharmacol. Res. 2016 doi: 10.1016/j.phrs.2016.03.018. [DOI] [PubMed] [Google Scholar]
- 21.Hamming I., Cooper M.E., Haagmans B.L., Hooper N.M., Korstanje R., Osterhaus A.D.M.E., Timens W., Turner A.J., Navis G., van Goor H. The emerging role of ACE2 in physiology and disease. J. Pathol. 2007 doi: 10.1002/path.2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clark P., Casas E., Tugwell P., Medina C., Gheno C., Tenorio G., Orozco J.A. Hydroxychloroquine compared with placebo in rheumatoid arthritis: a randomized controlled trial. Ann. Intern. Med. 1993 doi: 10.7326/0003-4819-119-11-199312010-00002. [DOI] [PubMed] [Google Scholar]
- 23.Ben-Zvi I., Kivity S., Langevitz P., Shoenfeld Y. Hydroxychloroquine: From malaria to autoimmunity. Clin. Rev. Allergy Immunol. 2012;42:145–153. doi: 10.1007/s12016-010-8243-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Page F. Treatment of Lupus Erythematosus with MEPACRINE. Lancet. 1951 doi: 10.1016/S0140-6736(51)91643-1. [DOI] [PubMed] [Google Scholar]
- 25.Vincent M.J., Bergeron E., Benjannet S., Erickson B.R., Rollin P.E., Ksiazek T.G., Seidah N.G., Nichol S.T. Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virol. J. 2005 doi: 10.1186/1743-422X-2-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu J., Cao R., Xu M., Wang X., Zhang H., Hu H., Li Y., Hu Z., Zhong W., Wang M. Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Discov. 2020 doi: 10.1038/s41421-020-0156-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 2020 doi: 10.1128/jvi.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Witkowska D. 2020. Mass Spectrometry and Structural Biology Techniques in the Studies on the Coronavirus-Receptor Interaction; pp. 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang N., Han S., Liu R., Meng L., He H. 2020. Chloroquine and Hydroxychloroquine As ACE2 Blockers to Inhibit Viropexis of 2019-nCoV Spike Pseudotyped Virus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pahan P., Pahan K. Smooth or Risky Revisit of an Old Malaria Drug for COVID-19? J. Neuroimmune Pharmacol. 2020;19:174–180. doi: 10.1007/s11481-020-09923-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.J. Yang, S.J.L. Petitjean, M. Koehler, W. Chen, S. Derclaye, S.P. Vincent, Q. Zhang, A.C. Dumitru, P. Soumillion, D. Alsteens, Molecular interaction and inhibition of SARS-CoV-2 binding to the ACE2 receptor, Nat. Commun. (n.d.). 10.1038/s41467-020-18319-6. [DOI] [PMC free article] [PubMed]
- 32.Yi C., Sun X., Ye J., Ding L., Liu M., Yang Z., Lu X., Zhang Y. Key residues of the receptor binding motif in the spike protein of SARS-CoV-2 that interact with ACE2 and neutralizing antibodies. Cell. Mol. Immunol. 2020 doi: 10.1038/s41423-020-0458-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Othman H., Bouslama Z., Brandenburg J.T., da Rocha J., Hamdi Y., Ghedira K., Srairi-Abid N., Hazelhurst S. Interaction of the spike protein RBD from SARS-CoV-2 with ACE2: similarity with SARS-CoV, hot-spot analysis and effect of the receptor polymorphism. Biochem. Biophys. Res. Commun. 2020 doi: 10.1016/j.bbrc.2020.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prabakaran P., Xiao X., Dimitrov D.S. A model of the ACE2 structure and function as a SARS-CoV receptor. Biochem. Biophys. Res. Commun. 2004 doi: 10.1016/j.bbrc.2003.12.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hong P.J., Look D.C., Tan P., Shi L., Hickey M., Gakhar L., Chappell M.C., Wohlford-Lenane C., McCray P.B. Ectodomain shedding of angiotensin converting enzyme 2 in human airway epithelia. Am. J. Physiol. - Lung Cell. Mol. Physiol. 2009 doi: 10.1152/ajplung.00071.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang J., Wu M., Liu X., Liu Q., Guo Z., Yao X., Liu Y., Cui C., Li H., Song C., Liu D., Xue L. Cytotoxicity evaluation of chloroquine and hydroxychloroquine in multiple cell lines and tissues by dynamic imaging system and PBPK model. BioRxiv. 2020 doi: 10.1101/2020.04.22.056762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma D., Chen C.B., Jhanji V., Xu C., Yuan X.L., Liang J.J., Huang Y., Cen L.P., Ng T.K. Expression of SARS-CoV-2 receptor ACE2 and TMPRSS2 in human primary conjunctival and pterygium cell lines and in mouse cornea. Eye. 2020 doi: 10.1038/s41433-020-0939-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.