Abstract
The recent outbreak of the SARS-CoV-2 infection has affected the lives and economy of more than 200 countries. The unavailability of virus-specific drugs has created an opportunity to identify potential therapeutic agents that can control the rapid transmission of this pandemic. Here, the mechanisms of the inhibition of the RNA-dependent RNA polymerase (RdRp), responsible for the replication of the virus in host cells, are examined by different ligands, such as Remdesivir (RDV), Remdesivir monophosphate (RMP), and several artificially expanded genetic information systems (AEGISs) including their different sequences by employing molecular docking, MD simulations, and MM/GBSA techniques. It is found that the binding of RDV to RdRp may block the RNA binding site. However, RMP would acquire a partially flipped conformation and may allow the viral RNA to enter into the binding site. The internal dynamics of RNA and RdRp may help RMP to regain its original position, where it may inhibit the RNA-chain elongation reaction. Remarkably, AEGISs are found to obstruct the binding site of RNA. It is shown that dPdZ, a two-nucleotide sequence containing P and Z would bind to RdRp very strongly and may occupy the positions of two nucleotides in the RNA strand, thereby denying access of the substrate–binding site to the viral RNA. Thus, it is proposed that the AEGISs may act as novel therapeutic candidates against the SARS-CoV-2. However, in vivo evaluations of their potencies and toxicities are needed before using them against COVID-19.
Keywords: SARS-CoV-2, RdRp, AEGIS, antiviral agents, COVID-19
Graphical Abstract
AEGIS nucleotides can act as potent inhibitors of the RdRp by denying the access of the viral RNA to the replicative polymerase.
Communicated by Ramaswamy H. Sarma
1. Introduction
The recent outbreak of the COVID-19 originated from the Wuhan City of China in December 2019 (Sahin et al., 2020; Wang et al., 2020) has led to severe damage to human lives and the economy of more than 200 countries worldwide. The unavailability of potent drugs specific to COVID-19 has created a challenge and opportunity to identify potent antiviral agents for the treatment of this pandemic. Although continuous attempts are going on to develop efficient vaccines (Buchholz et al., 2004; Liu et al., 2020; Peele et al., 2020) and antiviral agents (Basit et al., 2020; Cava et al., 2020; Kadam & Wilson, 2017; Mahanta, 2020; Pant et al., 2020; Sharma, 2020; Sheahan et al., 2020; Wu et al., 2020; Zhou et al., 2020; https://www.nature.com/articles/d41573-020-00016-0; https://clinicaltrials.gov/ct2/show/NCT04252885) and to identify potent repurposed drugs for the treatment of COVID-19, not a single COVID-19 specific drug has been developed so far.
To identify better therapeutic agents for the treatment of COVID-19, it is imperative to understand the structural and dynamical roles of the key proteins of SARS-CoV-2 (Da Silva et al., 2020) that causes COVID-19. It is now believed that after the virus enters into a host cell, its proliferation is mainly controlled by the RNA-dependent RNA polymerase (RdRp) of SARS-CoV-2 that catalyzes the replication of the viral RNA (Snijder et al., 2016; Yin et al., 2020). Hence, it is desirable to block the active site of the RdRp (Gao et al., 2020; Ng et al., 2008; Venkataraman et al., 2018; Yin et al., 2020) surrounded by the Finger (residues 397–581 and 621–679), Palm (residues 582–620 and 680–815), and Thumb (residues 816–920) domains by potent inhibitors that may compete with the viral RNA.
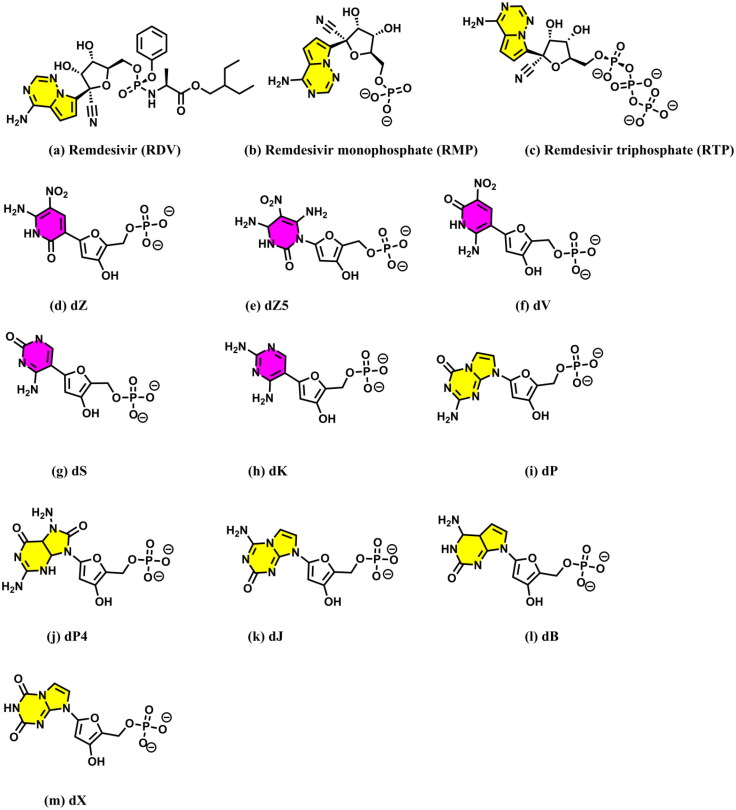
Recently, Remdesivir was shown to inhibit RdRp of SARS-CoV-2 and hence was approved for emergency use in the case of COVID-19 patients by the Food and Drug Administration (FDA) (Chaar & Makuch, 2020; Hendaus, 2020; Pruijssers et al., 2020). It was hypothesized that Remdesivir (RDV) (Figure 1a) gets converted first to Remdesivir monophosphate (RMP) (Figure 1b) and then to Remdesivir triphosphate (RTP) (Figure 1c) before binding to the protein (Jena, 2020a; Warren et al., 2016). However, as RTP does not immediately inhibit RdRp, but allows insertions of 2–4 new nucleotides before eventually halting the chain–elongation reaction (Gordon et al., 2020), it would likely make a covalent bond with the next nucleotide in RNA. For this, the conversion of RTP to RMP would be required (Yin et al., 2020). Further, recently it is proposed that RDV can induce basepair mutations in the viral RNA and for this it needs to be converted to RMP (Jena, 2020c). Hence, it is plausible that RTP gets converted back again to RMP in the presence of RdRp (Jena, 2020a, 2020c; Yin et al., 2020). However, the detailed mechanisms of these conversions are not yet fully understood. Further, whether RTP binds to RdRp first as a preinsertion complex (Zhang & Zhou, 2020) and wait for the RNA to enter into the RdRp active site to make the final postinsertion complex or RNA binds to RdRp first and then RTP is inserted into the RNA strand is not fully understood. The role of RMP in these events is also not known. Interestingly, Remdesivir (RDV) was recently proposed to bind with RdRp (Koulgi et al., 2020) and the main protease (Nayeem et al., 2021) of SARS-CoV-2. In these studies, RDV was found to act as a protein inhibitor. Hence, it is necessary to understand the roles of RDV, RTP, and RMP in inhibiting the replication of the virus by binding to either RdRp or RNA.
Figure 1.
Structures of (a) Remdesivir, (b) Remdesivir monophosphate (RMP), (c) Remdesivir triphosphate (RTP), and (d–m) the monophosphate forms of different second-generation AEGISs (dZ–dX represent the 2′-deoxynucleotide forms of AEGISs). Purines and pyrimidines are colored in yellow and purple, respectively. The IUPAC names of these AEGISs are presented in Table 1.
Because of the above, the bindings of RDV and RMP to RdRp are studied here by employing combined docking and molecular dynamics simulations. Additionally, the roles of different second-generation artificially expanded genetic information systems (AEGISs) (Table 1; Figure 1d–m) in inhibiting the active site of the RdRp are also investigated. It should be mentioned that AEGISs are synthetic nucleotides that were originally invented for the development of new aptamers (Biondi & Benner, 2018), genes (Behera et al., 2019; Benner et al., 2011; Georgiadis et al., 2015; Jena et al., 2018; 2020b; Merritt et al., 2014; Voegel & Benner, 1994; Zhang et al., 2015), and viral load assays (Glushakova et al., 2015). Interestingly, as the second generation AEGISs possesses improved hydrogen bond donors and acceptors, these are believed to form tight interactions with RdRp and RNA. Further, the binding modes of these artificial nucleotides with RdRp are compared with the corresponding results obtained for RDV and RMP to gain further insights into their mode of action. The results obtained here indicate that these artificial nucleotides can inhibit RdRp like other nucleoside analogs (Chien et al., 2020; Gordon et al., 2020; Hawman et al., 2018; Jockusch et al., 2020).
Table 1.
The list of various AEGISs, their IUPAC names, Pubchem CIDs, docking sores, and the residues of the RdRp with which they are making direct hydrogen bonding and ionic interactions as revealed by the docking studies.
| S. no. | Sequence | IUPAC name | PubChem CID | Docking scores | Interacting residues |
|---|---|---|---|---|---|
| 1 | RDV | Remdesivir | 121304016 | 46.73 | Ser682, Arg553, Asp760 |
| 2 | Z | 6-amino-3-(1′-β-d-ribofuranosyl)-5-nitro-1H-pyridin-2-one | 11182729 | 46.54 | Lys545, Thr556, Arg553, Lys612 Cys222 |
| 3 | Z5 | Z5 | 27.34 | Lys545, Thr556, Arg553, Ser682 | |
| 4 | V | 6-amino-3-nitro-1H-pyridin-2-one | 71464227 | 48.40 | Lys545, Arg553, Arg624, Arg555, Ser759, Thr680, Asp623 |
| 5 | S | 2-amino-1-(1′-β-d-ribofuranosyl)-4(1H)-pyrimidinone | 11107495 | 53.43 | Lys545, Arg553, Asp623, Cys622, Lys621 |
| 6 | K | 2,4-diaminopyrimidine | 67431 | 44.30 | Lys545, Arg553, Asp623, Cys622, Lys621 |
| 7 | P | 2-amino-8-(1′-β-d-ribofuranosyl)-imidazo-[1,2a]-1,3,5-triazin-[8H]-4-one | 135600909 | 56.56 | Lys545, Arg553, Cys622, Lys621, Asp621 |
| 8 | P4 | – | 43.90 | Arg553, Cys622, Cys622, Lys621 | |
| 9 | J | 4-aminoimidazo[1,2-a][1,3,5]triazin-2(1H)-one | 10931617 | 54.43 | Lys545, Thr556, Arg553, Cys622, Lys621 |
| 10 | B | 6-amino-9-(1′-β-d-ribofuranosyl)-4-hydroxy-5-(hydroxymethyl)-oxolan-2-yl]-1H-purin-2-one | 76900 | 52.06 | Lys545, Arg553, Cys622, Asp623, Ser682, Lys621, Cys622 |
| 11 | X | imidazo[1,2-a]-1,3,5-triazine-2(8H)-4(3H)-dione | 11019049 | 35.13 | Lys545 Thr680, Asn691, Ser759 |
| 12 | PZ | – | – | 59.50 | Arg553, Thr556, Thr680, Thr687, Ser682, Ser759, |
| 13 | PZZ | – | – | 53.71 | Lys545, Arg553, Arg555, Ser682, Cys622, Asp760, Ser759 |
| 14 | PPZ | – | – | 55.40 | Lys545, Ser682, Arg553, Ser759, Lys621 |
| 15 | BBZ | – | – | 35.12 | Lys545, Thr680, Arg553, Thr687, Asn691, Ser759 |
| 16 | PPZZ | – | – | 21.14 | Lys545, Arg553, Arg555, Asp760, Lys551, Asp760, Tyr619 |
2. Computational methodology
2.1. System preparation
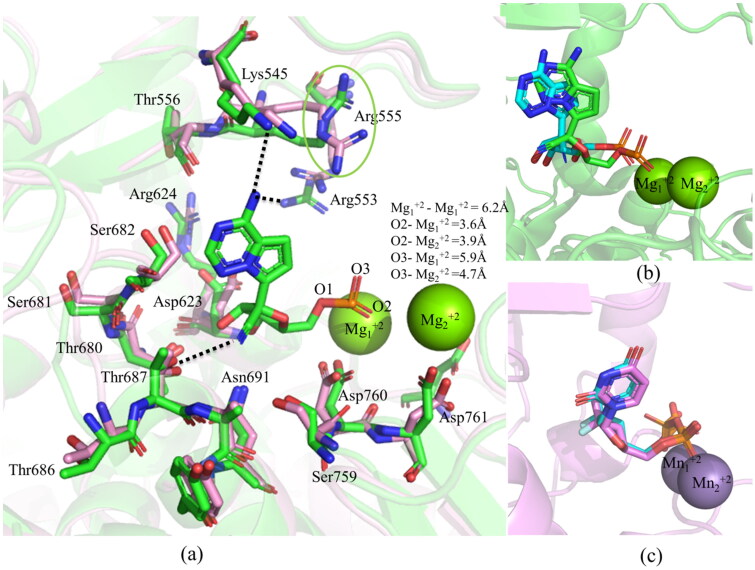
Two Cryo-EM structures of the RdRp protein complexed with NS7 and NS8 are available in the protein data bank (Gao et al., 2020; Yin et al., 2020). In one of these structures, the RdRp–NS7–NS8 complex was not bound to metal ions (PDB ID 6M71) (Gao et al., 2020). In contrast, in the other one, the RdRp–NS7–NS8 complex was bound to two Mg+2 ions (coordinated by Asp760, Asp761, Asp618, and pyrophosphate), a double-stranded RNA (14-base template strand and 11-base primer strand), and RMP (PDB ID 7BV2) (Yin et al., 2020). However, in the latter structure, Arg555 was protruding away from the active site, unlike in the former structure (Figure 2a). The superposition of RdRp without metal ions onto the RdRp with metal ions by considering the Cα atoms produced an RMSD of 0.583 Å (Figure 2a). This suggests that in these two structures, different residues of RdRp have acquired almost identical conformations. However, as the latter structure contains metal ions, which are conserved in almost all of the RdRp viral proteins (Ng et al., 2008; Venkataraman et al., 2018), this structure (RdRp-Mg+2) was considered for docking after removing coordinates of NS7, NS8, pyrophosphate, RNA, and RMP. Before docking, hydrogen atoms were added to the RdRp protein of SARS–CoV-2 by employing the UCSF Chimera (Morris et al., 2007) to maintain the neutral pH.
Figure 2.
(a) Superposition of the RdRp without metal ions (in pink, PDB ID 6M71) onto the RdRp–Mg+2–RMP complex (in green, PDB ID 7BV2). A green circle marks the Arg555 moving away from the active site in the latter structure. The hydrogen-bonding interactions (dotted lines) of RMP with different residues of the RdRp are also depicted to explain its binding mode. The bond distances between the metal ions and between the metal ions and the phosphate group of RMP are indicated. The comparison of docked (in cyan) and experimental binding modes of (b) RMP (in green, PDB ID 7BV2), and (c) SMP (in violet, PDB ID 4WTG) are also shown.
The coordinates of two highly studied artificial nucleotides, 2-amino-8-(1′-β-d-ribofuranosyl)-imidazo-[1,2a]-1,3,5-triazin-[8H]-4-one (common referred to as P) and 6-amino-3-(1′-β-d-ribofuranosyl)-5-nitro-1H-pyridin-2-one (common referred to as Z) were extracted from the protein databank (PDB ID 4XNO) (Zhang et al., 2015). Subsequently, hydrogen atoms were added to these nucleotides by using the GaussView program (Dennington et al., 2009). These nucleotides were modified to create other nucleotides such as P4 (an analogue of P) (Jena et al., 2018), Z5 (an analogue of Z) (Jena et al., 2018), 4-aminoimidazo[1,2-a][1,3,5]triazin-2(1H)-one (common referred to as J), 6-amino-3-nitro-1H-pyridin-2-one (common referred to as V), 6-amino-9-(1′-β-d-ribofuranosyl)-4-hydroxy-5-(hydroxymethyl)-oxolan-2-yl]-1H-purin-2-one (common referred to as B), 2-amino-1-(1′-β-d-ribofuranosyl)-4(1H)-pyrimidinone (common referred to as S), imidazo[1,2-a]-1,3,5-triazine-2(8H)-4(3H)-dione (common referred to as X), and 2,4-diaminopyrimidine (common referred to as K) (Hoshika et al., 2019). Subsequently, these nucleotides were optimized by using the B3LYP/6-31G** level of theory as implemented in the Gaussian 09 program (Frisch et al., 2009). It should be mentioned that as the B3LYP/6-31G** (Becke, 1993; Lee et al., 1988) level of theory produces fairly accurate geometries (Jena et al., 2018), this method was used for the geometry optimizations. Consequently, the optimized structures were used to create docking poses.
2.2. Molecular docking
Docking of RDV and various artificial nucleotides into the active site of the RdRp-Mg+2 complex was carried out by using the GOLD 5.0 program (Hartshorn et al., 2007; Jones et al., 1997; Nissink et al., 2002). The genetic algorithm (Jones et al., 1997) was used for the docking purpose to create ten different conformations of each nucleotide by keeping the protein rigid. The binding site was considered to be situated within a radius of 10 Å from the conserved Thr680. The Chemscore was used for docking, and the ChemPLP score was used to rescore the binding of different ligands with the RdRp–Mg+2 complex. It should be mentioned that the ChemScore fitness function incorporates a term dG that represents the total change in free energy, which occurs on ligand binding and was trained by regression against binding affinity data for 82 complexes. It also incorporates a protein–ligand atom clash term and an internal energy term that takes account of hydrophobic–hydrophobic contact area, hydrogen bonding, ligand flexibility, and metal interactions. Similarly, ChemPLP (piecewise linear potential) score uses ChemScore hydrogen bonding term and multiple linear potentials to model van der Waals and repulsive terms. It was found to be effective for both pose prediction and virtual screening (Hartshorn et al., 2007; Jones et al., 1997; Nissink et al., 2002). Out of the ten different poses, the one whose purine or pyrimidine interacts with the residues of motif F (Lys545, Arg553, Arg555, etc.) and the sugar–phosphate backbone interacts with the Mg+2 ions and the residues of motif C (Asp760, Asp761, etc.) like the RdRp–RMP (PDB ID 7BV2) (Yin et al., 2020) and Hepatitis C virus protein (HCV)-Sofosbuvir monophosphate (SMP) (PDB ID 4WTG) (Appleby et al., 2015) complexes is discussed herein (Table 1).
To examine if the above docking protocol can reproduce experimentally observed binding modes, the RMP was docked into the binding site of the RdRp–Mg+2 complex (PDB ID 7BV2) (Yin et al., 2020). As illustrated in Figure 2b, the above docking protocol generated the experimental binding mode accurately. It was found that the purine ring of docked RMP can make two weak hydrogen bonds with Arg553 (3.6 Å) and Lys545 (3.8 Å), while the sugar group can make a weak hydrogen bond with Thr680 (3.8 Å) as was observed in the cryo-EM structure (PDB ID 7BV2) (Figure 2a,b) (Yin et al., 2020). Similarly, the phosphate group of RMP was found to make two ionic interactions with Mg+2 ions as observed in the experimental structure (Yin et al., 2020). To re-verify the docking protocol, the SMP was docked into the active site of the HCV protein (PDB ID 4WTG) (Appleby et al., 2015) after removing all cofactors. Interestingly, the exact experimental binding mode was reproduced (Figure 2c). These results indicate that the protocol used for docking of RDV and AEGISs have generated accurate poses.
2.3. Molecular dynamics (MD) simulations and the MM/GBSA calculations
To gain deeper insights into the binding modes of RDV (Figure 1a) and RMP (Figure 1b), the docked conformation of the RdRp–RDV complex and the experimental structure of the RdRp–Mg+2–RMP complex (Yin et al., 2020) were subjected to 100 ns molecular dynamics simulations. As the cryo-EM structure does not account for the dynamical effects of the protein, it was necessary to understand the effect of protein dynamics on the binding modes of these compounds. Further, it is found that different artificial nucleotides (Figure 1d–m) and their sequences (Figure S1) in the 2′-deoxynucleotide forms can bind to RdRp strongly. Among these nucleotides, dPdZ (d stands for the 2′-deoxynucleotide form) is found to possess the highest docking score (strong binding) (Table 1). To further understand the binding mode of dPdZ and to compare the dynamics of the dPdZ–RdRp complex with those of the RDV–RdRp, and RMP–RdRp complexes, the former complex was also subjected to 100 ns molecular dynamics simulations. The Desmond 2018-4 package of the Schrodinger (Bowers et al., 2006; Schrödinger Release 2018) was used for the molecular dynamics simulations. Subsequently, the relative binding free energies of these three complexes were computed by using the MM/GBSA technique as implemented in the Schrodinger suite 2018-4 (Bowers et al., 2006; Schrödinger Release 2018). The whole 100 ns trajectories and Equation (1) were considered to compute the relative binding free energies of different complexes.
Here, ΔΔGbind is the calculated change in relative binding free energy, ΔGcomplex (minimized) is the change in free energy of the minimized complex, ΔGprotein (unbound, minimized) is the change in free energy of the minimized protein after separating it from its bound ligand and ΔGlig (unbound, minimized) is the change in free energy of the ligand after separating it from the complex and allowing it to relax. The ΔH is the change in enthalpy, T is the room temperature, and ΔS is the change in entropy. As entropy calculations are complex and time consuming, these were not computed. Hence, the ΔG term contains only the enthalpy term, which is a combination of various interaction energies, such as electrostatic, hydrogen bonding, van der Waal, hydrophobic, and solvation energies (Table 2).
Table 2.
The relative total binding free energies of different inhibitors and the contributions of different energies to the total binding free energies.
| Inhibitors | ΔGBind | ΔEele | ΔEvdW | ΔEcovalent | ΔEhb | ΔElipo | ΔEpi–pi | ΔEGB |
|---|---|---|---|---|---|---|---|---|
| dPdZ | –6.69 | 127.20 | –48.79 | 7.07 | –4.73 | –2.59 | –0.11 | –84.74 |
| RDV | –32.31 | –40.37 | –38.23 | 3.25 | –2.56 | –7.59 | –0.51 | 53.71 |
| RMP | 22.11 | –70.96 | –15.25 | 3.25 | –1.93 | –2.10 | –0.05 | 109.16 |
ΔGBind=total relative binding free energy, ΔEele=electrostatic energy, ΔEvdW=van der Waals energy, ΔEcov=covalent energy, ΔEhb=hydrogen–bonding energy, ΔElipo=lipophilic energy, ΔEpi–pi=pi–pi packing energy, and ΔEGB=generalized born electrostatic solvation energy.
Before molecular dynamics simulations, these three complexes were solvated by placing them in an explicit water box of size 10 Å. The OPLS3e force field (Roos et al., 2019) was used to model the protein and ligand, while the single-point charge (SPC) model (Berendsen et al., 1987; Toukan Rahman, 1985) was used to account for the implicit water molecules. It should be mentioned that the use of the OPLS3e force field for proteins and organic ligands and the SPC model for water molecules were found to produce reliable results (Gahtori et al., 2019). Sufficient numbers of ions were added to make the solvated complexes neutral. Subsequently, these complexes were energy minimized. The minimized complexes were slowly heated to maintain a temperature of 300 kelvin by using the Nose–Hoover thermostatic algorithm (Posch et al., 1986). The Martina–Tobias–Klein method (Martyna et al., 1994) was used to maintain constant pressure throughout the simulations. Consequently, the production run of each system was carried out for 100 ns by considering the NPT ensemble. The long-range electrostatic interactions were calculated by using the particle-mesh Ewald (PME) (Peterson, 1995) method with a grid spacing of 0.8 Å. The cutoff radius for Coulomb interactions was set at 9.0 Å. The equations of motion were integrated using the multistep RESPA integrator with an inner time step of 2.0 fs for both the bonded and nonbonded interactions within the short-range cutoff. To incorporate nonbonded interactions beyond the cutoff, an outer time step of 6.0 fs was also used. The periodic boundary condition (PBC) was considered for all of the simulations. The simulation interaction diagram tool implemented in the Desmond 2018-4 package (Bowers et al., 2006; Schrödinger Release 2018) was used to analyze the detailed interactions between the ligands and protein.
3. Results and discussions
3.1. Docking binding mode of remdesivir (RDV)
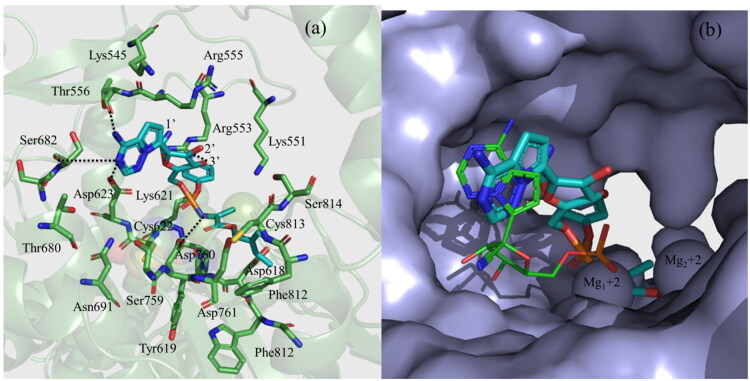
The docked conformation of RDV is shown in Figure 3. From this figure, it is clear that the purine ring of RDV is making two hydrogen bonds with each of Ser682 and Thr556. Similarly, the 1′-CN group of the sugar moiety is pointing toward Arg553 (4.5 Å) and Arg555 (4.5 Å), while the 3′-OH group is making a hydrogen bond with Arg553 (3.1 Å). The phenoxy group of the extended backbone is pointing toward the Finger domain residues, such as Lys551, Arg553, Lys621, Cys622, and Asp623 and the 2-ethyl butyl acetate group is pointing toward the Palm domain residues, such asTrp617, Asp618, Tyr619, Phe812, Cys813, and Ser814 (Figure 3a). The NH group of the extended backbone is making a hydrogen bond with Asp760 (3.2 Å). In addition to these, the phosphate group is found to make an ionic interaction with one of the Mg+2 ions. If we compare the binding mode of RDV with the experimental binding mode of RMP (PDB ID 7BV2) (Yin et al., 2020), it is clear that the purine ring of both ligands binds to RdRp in a similar way. However, the sugar group of RDV interacts with the Finger residues (Figure 3b), while in RMP it interacts with the Palm residues. This is presumably due to the bulky and extended group attached to the sugar moiety of RDV makes it rigid and restrains to freely rotate. However, the phosphate group in both cases binds to the Mg+2 ions (Figure 3b).
Figure 3.
The docking binding mode of Remdesivir (RDV). (a) The interactions of RDV with different residues of RdRp. (b) The surface representation of the docking binding mode of RDV and its comparison with that of RMP (PDB ID 7BV2) (shown as a line) obtained by the cryo-EM study (Yin et al., 2020).
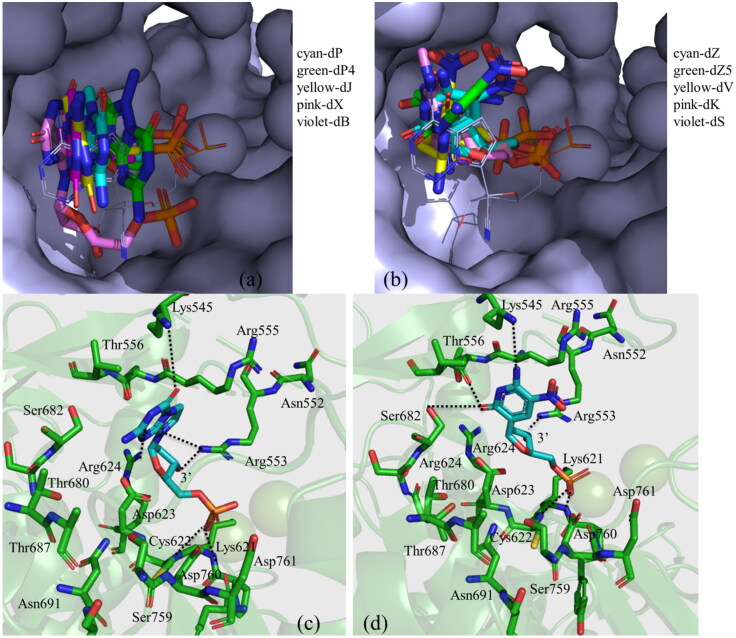
3.2. Docking binding modes of AEGISs
The docking scores and protein residues with which different AEGISs are making hydrogen bonding and ionic interactions (as revealed by docking studies) are presented in Table 1. A higher docking score indicates a better docking pose and binding energy. The binding modes of these artificial nucleotides are illustrated in Figure 4. As can be seen from this figure, the 2′-deoxynucleotide forms of different purines, such as dP, dP4, dJ, dB, and dX possess a similar binding mode (Figure 4a). Similarly, the 2′-deoxynucleotide forms of different pyrimidines, such as dZ, dZ5, dV, dS, and dK follow a similar binding mode (Figure 4b). To better understand the binding modes of these AEGISs, the detailed interactions of a purine (dP) and a pyrimidine (dZ) with RdRp are depicted in Figure 4c and 4d, respectively. As can be seen from Figure 4c, the purine ring of dP makes two hydrogen bonds with Lys545 and Arg624, while the 3′-OH group of the sugar moiety makes a hydrogen bond with Arg553. Similarly, the purine ring of dZ makes three hydrogen bonds, one each with Lys545, Thr556, and Ser682, and the 3′-OH group makes a hydrogen bond with Arg553 (Figure 4c). The phosphate groups of these AEGISs are also found to make tight ionic interactions with the Mg+2 ions. Similar trends are also noticed for other purines and pyrimidines (Table 1). If we compare the number of hydrogen bonds, salt bridge, and other interactions and docking scores of various AEGISs with those of RDV, it is evident that the artificial nucleotides can make stronger interactions with RdRp and hence may act as better inhibitors of the RdRp.
Figure 4.
The binding modes of different (a) purine-based (in the stick) and (b) pyrimidine-based (in the stick) artificial nucleotides with RdRp (in surface representation). The two Mn+2 ions are shown as two spherical balls. The detailed interactions of (c) dP and (d) dZ with different amino acid residues of the RdRp are also shown. Dotted lines represent the hydrogen bonding interactions.
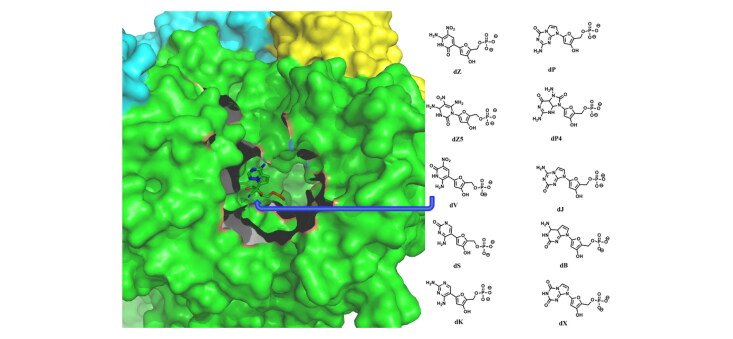
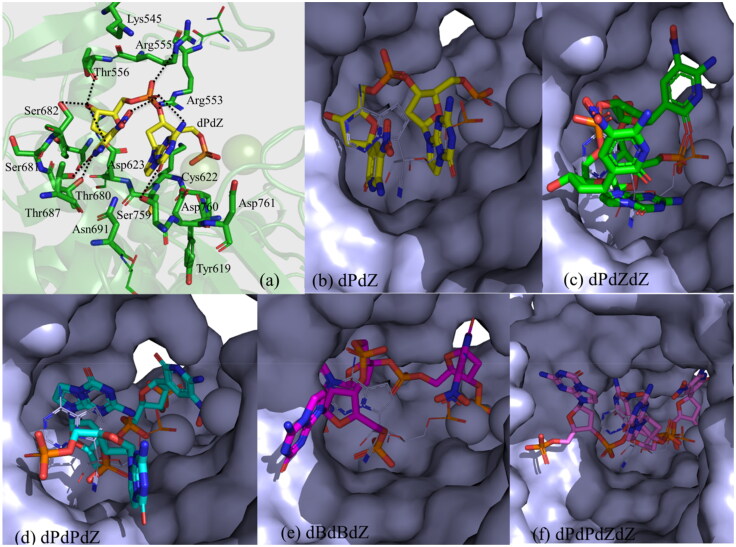
To further evaluate the binding modes of more than one artificial nucleotides with RdRp (Table 1), different sequences of these nucleotides (Figure S1) in the 2′-deoxynucleotide forms were docked into the active site of RdRp. The docking scores and interacting residues as revealed by the docking studies are presented in Table 1. The binding modes of these nucleotides are shown in Figure 5. It is found that among these nucleotides, dPdZ would make the strongest interactions with RdRp (Table 1). For example, the purine ring of dZ in the dPdZ can make two hydrogen bonds, one each with Thr680 and Thr687. The 3′-OH group can make three hydrogen bonds, one each with Thr556, Asp623, and Ser682. Its phosphate group can make two hydrogen bonds, one each with Arg553 and the NO2 group of dZ (Figure 5a). Similarly, the purine ring of dP can make two hydrogen bonds, one each with Ser759 and the phosphate group of dZ. Its phosphate group makes two ionic interactions with the Mg+2 ions. Interestingly, in this conformation, dP can also make a stacking interaction with dZ (Figure 5a). However, the interbase stacking interaction gets weak when more than two nucleotides are present in a sequence (Figure 5c–f and Figure S2) and it gets completely lost when four nucleotides are present in a sequence (Figure 5f and Figure S2). Further, in large sequences, the phosphate–Mg+2 interactions became weak (Figure S3). This is quite evident from Table 1, wherein nucleotides in higher sequences are associated with the least docking score (Table 1). These results suggest that more than three nucleotide sequences may reduce the inhibitory activity of the artificial nucleotides.
Figure 5.
(a) The interaction of dPdZ (yellow) with different residues of RdRp (green). (b–f) The binding modes of different sequences containing various AEGISs (in surface representation). The two Mn+2 ions are shown as two spherical balls.
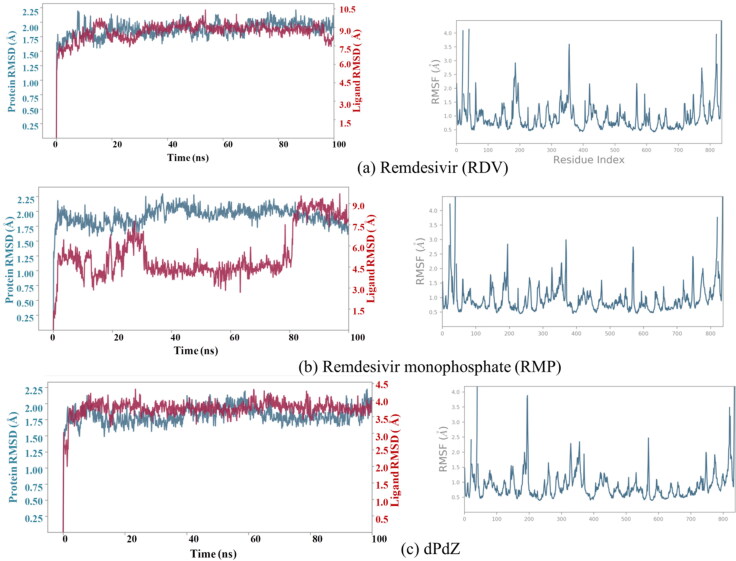
3.3. MD-simulations of remdesivir (RDV)
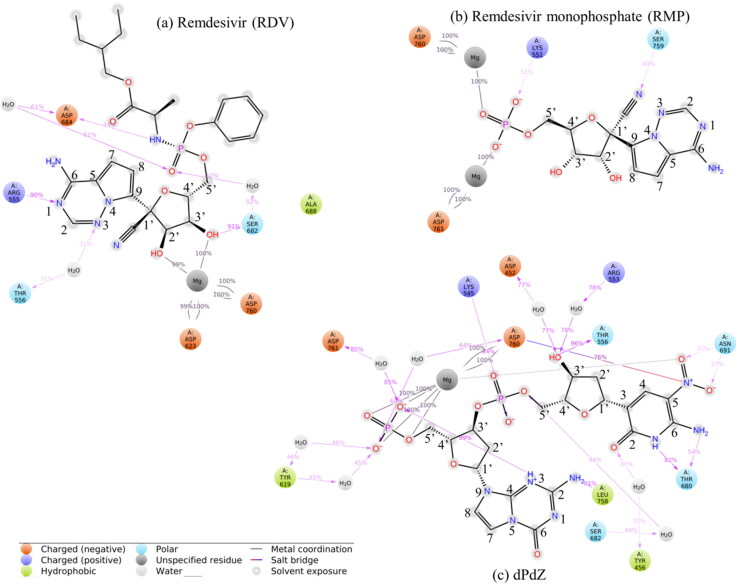
As illustrated in Figure 6a, the root mean square deviation (RMSD) of the Cα atoms of RdRp computed with respect to the minimized structure is less than 2.25 Å. This suggests that RdRp does not undergo any major structural changes in the RDV–RdRp complex. This is quite evident from the root mean square fluctuations (RMSF) of different amino acids of the RdRp. The stability of the protein can be accounted for two main reasons. (1) The coordinates of RdRp were extracted from the ligand-bound conformation and (2) in the docked conformation; RDV was tightly bound to RdRp. Similarly, RDV is also found to be stable during the simulations (Figure 6a). The interaction diagram showing the detailed interactions of RDV with RdRp is depicted in Figure 7a. The hydrogen-bonding interactions calculated by considering the donor–acceptor bond distance of 2.5 Å (D–H···A), a donor angle of ≥120° between the donor–hydrogen–acceptor atoms (D–H···A) and an acceptor angle of ≥90° between the hydrogen–acceptor-bonded atoms (H···A–X) are shown in this figure. The ionic and hydrophobic interactions are also shown in this figure. It should be noted that protein–ligand ionic interactions were calculated between two oppositely charged atoms that are present within a distance of 3.7 Å from each other and do not involve a hydrogen bond. Similarly, the protein–metal and ligand–metal ionic interactions were calculated by considering a metal ion coordinated within 3.4 Å of protein’s and ligand’s heavy atoms (except carbon). Hydrophobic interactions were categorized as (1) the π–cation interaction, which was formed between aromatic and charged groups situated within a distance of 4.5 Å, (2) the π–π interaction, where two aromatic groups were stacked face-to-face or face-to-edge, and (3) other interaction, where a nonspecific hydrophobic side chain was present within 3.6 Å of a ligand’s aromatic or aliphatic carbon atoms. The percentage occupation of each interaction is also shown in this figure.
Figure 6.
The root mean square deviation (RMSD) of the atoms of the protein and heavy atoms of the ligand (left side) and the root mean square fluctuations (RMSF) of different amino acid residues of the protein (right side) computed with respect to the corresponding minimized structure.
Figure 7.
Schematic representation of detailed protein–ligand direct and indirect interactions (through Mg+2 ions, and water molecules). These interactions lasted for more than 30% of the simulation time. The atomic numbering scheme followed in each molecule is also shown.
From Figure 7a, it is evident that the purine ring of RDV can make a direct hydrogen bond with Arg555 (90% occupancy) and an indirect water-mediated hydrogen bond with Thr556 (31% occupancy). Similarly, the 3′-OH group of sugar is found to make a strong hydrogen bond with Ser682 (91% occupancy). Remarkably, the 2′-OH group is found not to be engaged with protein residues. However, both the 3′-OH and 2′-OH groups are coordinated by one of the Mg+2 ions (Mg1+2). In this conformation, Mg1+2 is found to make two salt–bridge interactions with Asp623 and Asp760 (100% occupancy) and the NH-backbone makes one hydrogen bond with Asp684 (61% occupancy).
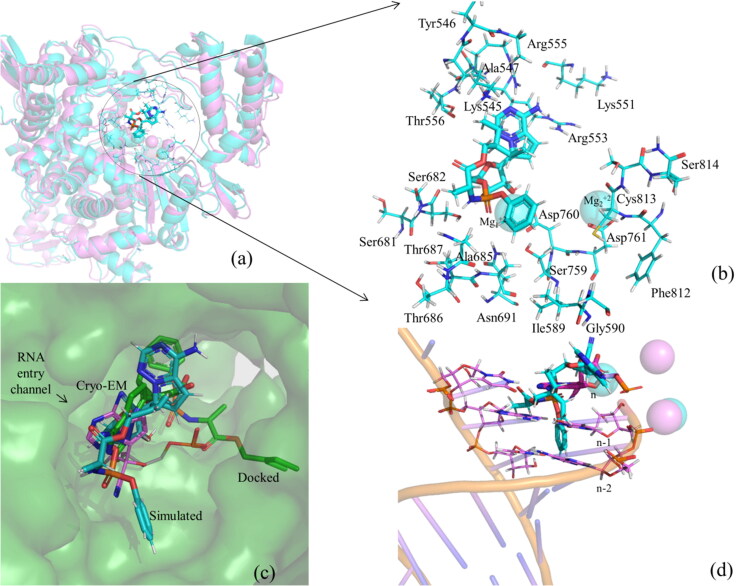
Interestingly, the superposition of the average simulated structure of the RDV–RdRp complex onto the cryo-EM structure of RMP–RdRp (PDB ID 7BV2) (Yin et al., 2020) (RMSD of Cα atoms = 1.50 Å) and its comparison with the initial docked RDV–RdRp complex suggest that the protein did not change too much during the simulation (Figure 8a). However, the purine ring of RDV rotated slightly toward the F-block residues (Figure 8b) and the extended backbone of RDV rotated significantly toward the Palm domain. As a result, the 1′-CN-group, which was pointing toward Arg553 and Arg555 in the docked conformation (Figures 3a and 8c), points toward Thr556 in the simulated structure and makes a weak hydrogen bond with it (3.9 Å) (Figure 8b). However, it was pointing toward Thr680, Thr687, and Ala688 in the experimental structure (Figure 2a). Similarly, the phenoxy group of RDV, which was pointing toward the F-block residues in the docking study (Figures 3a and 8c), is found to be rotated toward the Palm domain residues, such as Ile589, Asp684, Ala685, Thr687, Ala688, Ser759, and Cys813 (Figure 8b). The 2-ethyl butyl acetate group, which was originally pointing toward the Thumb domain, has also rotated toward the Finger domain residues, such as Lys545 and Ala547, and is making a stacking interaction with the purine ring of RDV (Figure 8b,c). Interestingly, Mg1+2 is found to adjust its position to maintain its interaction with RDV, while Mg2+2 was almost unperturbed during the simulations and maintained its interaction with the protein (Asp761). It should be mentioned that Mg1+2 in the cryo-EM structure (Yin et al., 2020) was coordinated with the phosphate group of RMP tightly, while Mg2+2 was strongly associated with the phosphate group of the n-1 nucleotide of the RNA (n is the insertion site of the antiviral drug). However, as RNA is not considered in the present study, Mg2+2 remains bound with the protein.
Figure 8.
(a) The superposition of the average structure of the RDV–RdRp complex (in cyan) onto the experimental structure of the RMP–RdRp complex (Yin et al., 2020) (in violet). The interactions of RDV with different residues of RdRp as obtained in the average simulated structure. (c) The surface representation of the binding mode of RDV (cyan) in the average structure and its comparison with that of RMP (PDB ID 7BV2, shown in violet color and line representation) and the docked conformation of RDV (green) obtained after superposing the average simulated and docked structures of the RDV–RdRp complexes onto the experimental structure of RMP–RdRp complex (Yin et al., 2020). This illustrates the fitting of RDV in the shallow binding pocket of RdRp. (d) The interference of RDV with the RNA strand in the superposed structure and the locations of the Mg+2 ions in the simulated and experimental structures.
Interestingly, it is found that in the average simulated structure, both the phenoxy and 2-ethyl butyl acetate groups are placed in the RNA entry channel (Figure 8c,d) and hence would clash with the RNA. For example, the phenoxy group is found to collide with the n-1 and n-2 nucleotides in the RNA primer strand, while the 2-ethyl butyl acetate group is found to collide with the complementary nucleotides in the template strand (Figure 8d). This would ultimately perturb the RNA structure. Further, as the purine ring of RDV has moved slightly away from the nucleotide insertion site, it may not base pair with the complementary nucleotide on the template strand (Figure 8c). These results imply that RDV bound to RdRp would not only occupy the nucleotide triphosphate (NTP) channel (ligand insertion site) of the RNA binding site but also block the entry channel of RNA. This would ultimately hinder the replication of the viral RNA. However, as it was presumed that RDV would be converted to the RMP before it can get inserted into the RNA strand (Jena, 2020a, 2020c; Yin et al., 2020), it would be interesting to understand, the detailed conversion mechanism of RDV to RMP in the presence and absence of RdRp.
In an earlier study (Koulgi et al., 2020), three different replicates of the RDV–RdRp complex were simulated (two for 50 ns each and one for 100 ns) by using the AMBER 16 program. In these simulations, RDV was found to undergo large conformational changes. In two of the replicates, the purine ring of RDV was pointing toward the Palm domain (Leu758, Cys813, and Gln815). Similarly, the extended backbone was pointing toward the F-block residues (Ala550, Lys551, Ile548, Ser549, and Arg555). Similar conformation was also found in the third replicate (largest cluster) (Koulgi et al., 2020). If we compare the binding mode of RDV obtained here with the earlier study (Koulgi et al., 2020), it appears that in the present study, RDV makes tight interactions with RdRp and does not move away too much from the experimental conformation of RMP (Yin et al., 2020) and hence the present study represents the true binding mode of RDV.
3.4. MD-simulations of remdesivir monophosphate (RMP)
The RMP–RdRp structure considered for the molecular dynamics simulations corresponds to the Cryo-EM structure of the RdRp–RMP complex without the presence of RNA and other cofactors (Yin et al., 2020). As the docked conformation of RMP in the active site of RdRp is structurally similar to the Cryo-EM structure of the RMP–RdRp conformation (Yin et al., 2020), it is expected that RMP will bind to RdRp independent of RNA. Hence, it is necessary to understand the behavior of RMP without RNA under the influence of protein dynamics. This will help to understand the binding mode of RMP with RdRp, which in turn may help to understand the role of Remdesivir in inhibiting the replication of viral RNA. The advantage of considering the experimental RMP–RdRp conformation but not the docked conformation for molecular dynamics simulations lies in the fact that the experimental structure normally corresponds to a local minimum on the potential energy surface.
The RMSD of RMP as illustrated in Figure 6b shows that the ligand undergoes a significant conformational change during the simulation. However, the protein is found to be stable (RMSD < 2.25 Å) The RMSF of different residues of RdRp confirms this (Figure 6b). The detailed interactions between different residues of RdRp and inhibitors are illustrated in Figure 7b. The interaction histogram is depicted in Figure S3. The percentage occupations of different interactions throughout the simulations (100 ns) are also shown in these figures (Figure 7b and Figure S3). From these figures, it is evident that, after the inclusion of protein dynamics, the observed hydrogen bonds between the purine ring of RMP and Lys545 and Arg555 (Figure 2a) (Sharma, 2020) disappeared. Similarly, the observed hydrogen bond between the 1′-CN group of RMP and Thr680 disappeared. Instead, the 1′-CN group is found to make a new hydrogen bond with Ser759 (40% occupancy) (Figure 7b). It should be mentioned that despite this rearrangement, the 1′-CN group is still facing toward the Palm domain of the RdRp (Thr680 and Ser759 are on the same side of the Palm domain) (Figure 7b). However, the 2′-OH and 3′-OH groups lost their interactions with RdRp. Interestingly, the ionic interactions between the phosphate group of RMP and Mg+2 ions remained intact and a new, but, transient ionic interaction between the phosphate group and Lys551 (32% occupancy) is formed (Figure 7b and Figure S3b). Similarly, the salt–bridge interactions between the Mg+2 ions and Asp760 and Asp761 are maintained throughout the simulations (100% occupancy) (Figure 7b).
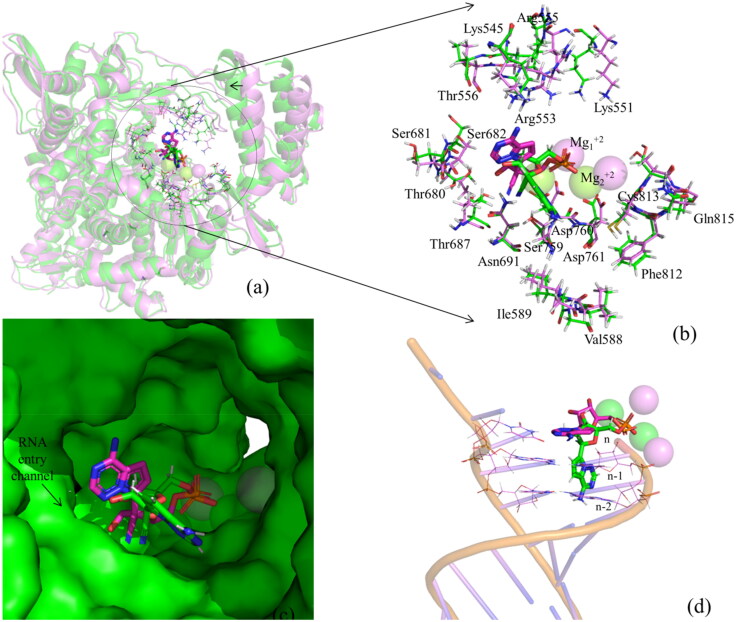
The average structure of the simulated RdRp–RMP complex superposed onto the cryo-EM structure (PDB ID 7bV2) (Yin et al., 2020) (RMSD of Cα atoms= 1.37 Å) suggests that during the simulation, the purine ring along with the sugar group rotated away from the binding site to acquire a partially flipped state as illustrated in Figure 9. However, the protein did not undergo any significant structural changes at the binding site except the movements of side chains of Arg551, Arg553, and Arg555 and the Thumb domain (moved slightly toward the ligand to make the binding site more compact) (Figure 9a,b). Remarkably, in the partially flipped conformation, the phosphate group of RMP is found to be located near its original position (experimental conformation). As the PO4 group did not undergo any major structural change, it constrained RMP to leave the active site of RdRp (Figure 9c). However, in this conformation, the purine ring of RMP is pointing toward the RNA entry channel and may sterically clash with the n-1 and n-2 nucleotides in the RNA primer strand (Figure 9d). As the rotation of RMP away from the NTP channel occurred during the simulations, its association with RdRp became weak. This weak binding may be recognized by the incoming RNA, which would push RMP back to its original position. Further, as the location of the phosphate group of simulated RMP coincides with that of experimental RMP, the rotation of RMP from partially flipped conformation to the original position may occur in the presence of RNA. It should be mentioned that DNA polymerases often repair modified nucleotides by flipping them away from the double helix. After repair, these nucleotides are sent back to their original positions, mainly because of the internal dynamics of DNA and DNA–polymerase complex (Banerjee et al., 2005; Jena, 2012; Peterson, 1995; Dizdaroglu et al., 2017).
Figure 9.
(a) The superposition of the RMP–RdRp simulated complex structure (in green) onto the RMP–RdRp experimental complex structure (PDB ID 7BV2) (in violet). The arrow sign indicates that the Thumb domain slightly moved toward the ligand to make the binding site more compact. (b) The comparison of the binding modes of RMP with different residues of RdRp in the simulated and experimental structures. This shows that the active site residues did not undergo any significant conformational changes (except the side chains of Arg551, Arg553, and Arg555) during the simulation. (c) Surface representations of experimental (violet) and simulated (cyan) structures of RMP. This clearly illustrates the rotation of RMP from its initial conformation. (d) The interference of simulated RMP with the experimental RNA strand and the positions of Mg+2 ions in these complex structures.
Interestingly, in an earlier MD study of RdRp–RTP complex (Zhang & Zhou, 2020), the RTP was found to be placed within the active site of RdRp, although at a different position than that of ATP. Contrary to the results obtained here and in the earlier cryo-EM study (Yin et al., 2020), the CN-group of RTP was pointing toward the Finger domain bound by Lys545, Tyr546, and Ala547 (Zhang & Zhou, 2020). The purine ring was also pointing toward the Finger domain bound by the above residues (situated within a distance of 5 Å), and the triphosphate group was pointing toward the Finger (Lys551, Arg553, Arg555, Lys621), Palm (Lys798), and Thumb (Arg836) domains. However, the only one Mg+2ion present in the RdRp–RTP complex was bound to Ser549 but not to Asp760 and Asp761 (Zhang & Zhou, 2020). In this conformation, RTP was found to partially block the active site of the RNA (Zhang & Zhou, 2020). However, as RMP acquires a partially flipped conformation, it may not hinder the binding of RNA to RdRp. Further, as RTP would be converted to RMP, understanding of the binding mode of the latter is more important than that of the former.
Based on these results, the following mechanism may be proposed for the blocking of the chain-elongation reaction of the viral RNA by Remdesivir (RDV). (1) Initially, RDV may get converted to RTP, most likely in the absence of RdRp. (2) RTP would bind to RdRp and subsequently get catalyzed to RMP by RdRp. Alternatively, RTP may get converted to RMP before biding to RdRp. (3) RMP would acquire a partially flipped conformation in the active site of RdRp till the RNA enters into the active site. (4) The incoming of RNA may induce further conformational changes in the RMP helping it to get flipped back to the antiviral insertion site. At this position, RMP may stop the elongation of the RNA strand by delaying RNA synthesis. However, experimental structural studies would be required to confirm this mechanism.
3.5. MD simulations of dPdZ
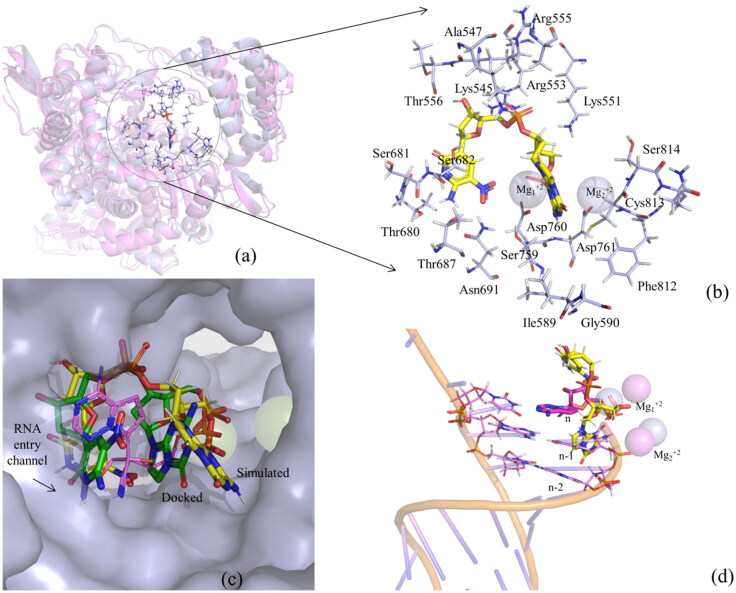
The RMSD of the protein and ligand in the dPdZ–RdRp complex is found to be quite stable. The protein stability is also evident from the RMSF of different protein residues as illustrated in Figure 6c). The interaction diagrams shown in Figures 7c and S3c suggest that dPdZ makes the highest interactions with RdRp. For example, the purine ring of dZ makes two hydrogen bonds with Thr680 (82% and 52% occupancies) and two water-mediated hydrogen bonds, one each with Ser682 and Tyr456. Its NO2 group also makes two hydrogen bonds with Asn691 (31% and 37% occupancies) and a salt–bridge interaction with Asp760. Its 3′-OH group can make a direct hydrogen bond with Thr556 (96% occupancy) and two water-mediated hydrogen bonds, one each with Arg553 and Asp452. Its phosphate group is also making a hydrogen bond with Lys545 (84% occupancy). Similarly, the purine ring of dP is found to make a hydrogen bond with Leu758 (81% occupancy). Its phosphate group makes an ionic interaction with one of the Mg+2 ions and three water-mediated hydrogen bonds, one each with Asp760, Asp761, and Tyr619. Further, if we compare the total number of protein–ligand contacts throughout the simulations of all of the simulated complexes, it is clear that dPdZ makes the maximum contacts with the protein (Figure S4).
The superposition of the average structure of the dPdZ–RdRp complex onto the RMP–RdRp complex (PDB ID 7BV2) (RMSD of Cα atoms = 1.38 Å) and its comparison with the docked dPdZ–RdRp complex suggest that the protein does not undergo any significant changes during the simulations except the minor movement of the Thumb domain toward the ligand (Figure 10a). However, the ligand changed its conformation slightly. As a result, the initial stacking interaction between dP and dZ became weak in the average simulated structure (Figure 10b,c). This helped dZ to get buried deep into the Finger domain (Figure 10b,c). Similarly, the purine ring of dP is found to be extended toward the Palm domain and interacts with Asp761, Cys813, and Ser814 (lie between 3 and 4 Å) (Figure 10b). Remarkably, in this conformation, dP is found to point toward the RNA-entry channel and would occupy the position of n-1 nucleotide (Figure 10d). This indicates that dPdZ is not only buried deep into the active site and is strongly engaged with several conserved residues of RdRp, but also adopts a conformation to block the entry channel of RNA. Hence, it would obstruct the RNA binding site, thereby inhibiting the replication of the viral RNA.
Figure 10.
(a) The superposition of the dPdZ–RdRp complex (light blue) onto the experimental structure of RMP–RdRp complex (violet) (Yin et al., 2020). (b) The binding of dPdZ (in yellow) with different residues of RdRp (in light blue). (c) The comparison of the simulated (in yellow) and docked (in green) binding modes of dPdZ with the experimental binding mode of RMP (PDB ID 7BV2) (in violet). This illustratesthe movement of dPdZ during the simulations. As can be seen from this figure, the space left behind RMP is occupied by dZ. (d) The interference of dPdZ with the RNA strand and the positions of Mg+2 ions in the simulated and experimental structures.
A comparison of simulated average structures of all complexes suggests that the extended backbone of RDV and the purine rings of RMP and dP (in dPdZ) are pointing toward the RNA-entry channel (Figure S5). It should be mentioned that two channels, such as the nucleotide triphosphate (NTP) entry channel formed by a set of hydrophilic residues, such as Lys545, Arg553, and Arg555 of motif F and the RNA entry channel composed of motifs A (residues 611–626) and C (residues 753–767) through a groove clamped by motifs F and G of the Finger domain (Gao et al., 2020) play vital roles in the binding of RNA and insertion of antiviral agents (e.g. Remdesivir) into the RNA strand. As RDV and dPdZ bind strongly to these channels, their inhibitory activity would be higher than that of RMP. However, as RDV gets converted to RMP (Jena, 2020a; 2020c; Yin et al., 2020) as the final metabolite and its binding with RdRp is weaker compared to dPdZ, the latter would act as a strong inhibitor of RdRp.
3.6. Relative binding free energies of ligands
The relative binding free energies of all of the simulated complexes are presented in Table 2. From this table, it is evident that the binding of RDV would be about 27 kcal/mol more stable than that of dPdZ and about 54 kcal/mol more stable than that of RMP. As RMP acquires a partially flipped conformation, its binding energy is positive, which is an indication of its poor binding to RdRp. However, the less stability of dPdZ compared to RDV is surprising as dPdZ makes more hydrogen bonding, ionic, and van der Walls interactions with the protein than that of RDV. The analysis of all components of energies that contribute to the enthalpy and hence to the relative free energy indicates that in contrary to RDV, dPdZ has nonfavorable electrostatic energy, which is also surprising as it makes more salt–bridge interactions with RdRp than that of RDV (Figure 7a,c). Similarly, the GBSA energy is found to be negative for dPdZ, while it is positive for RDV and RMP. To understand the free energy changes during the simulations, free energy values are plotted against the simulation time in Figure S6. From this figure, it is clear that the free energy changes vary significantly for RMP due to its large conformational changes and remain positive for most of the simulation time. Similarly, it remained less negative for dPdZ and more negative for RDV. It should be mentioned that in the earlier study (Koulgi et al., 2020), despite the flipped conformation of RDV, a relative binding free energy of −25 kcal/mol was computed for the RDV–RdRp complex by employing the MM/PBSA technique (without entropy calculations). In another study (Zhang & Zhou, 2020), the free energy of −7.68 ± 0.57 kcal/mol was obtained for the RTP–RdRp complex by using the free energy perturbation calculation. These wide differences in computed free energies suggest that MM/GBSA results are not reliable. To verify this, induced fit docking (IFD) method was used to calculate the IFD scores of each complex by employing the Glide program of Schrodinger 2018-4 (Bowers et al., 2006; Schrödinger Release 2018) package. The main aim was to use a different energy function to compute the binding energy (IFD score). The protein and ligands used for the induced fit docking were extracted from the corresponding average simulated structures. The docked conformations that possess the lowest RMSD values computed with respect to the average simulated structures were considered for the comparison of IFD scores. As presented in Table S1, dPdZ possesses the lowest IFD score, which is indicative of its strong binding with RdRp. Interestingly, the IFD score of RMP is found to be negative unlike the MM/GBSA result. This confirms that MM/GBSA results are not reliable. Hence, based on maximum contacts and favorable hydrogen and ionic interactions, it can be presumed that dPdZ would act as a better inhibitor of RdRp.
4. Conclusions
It is revealed that the RDV can make strong interactions with RdRp in such a way that it would block the entry of RNA into the active site of RdRp. However, as it is believed that RDV is converted to RTP and RMP before binding to RdRp, it is unlikely that RDV would inhibit the replication of the viral RNA. It is further found that RMP would acquire a partially flipped conformation in the active site of RdRp. In this conformation, it would collide with n-1 and n-2 nucleotides of RNA. Although in this conformation, RMP is protruded into the RNA entry channel, it may not block the entry of RNA into the binding pocket of RdRp due to its loose association with RdRp. Hence, it is plausible that the entry of RNA would push RMP back to its original position where it can stop the synthesis of the RNA strand by employing a delayed chain-termination mechanism. The dPdZ, a two nucleotide sequence of the AEGIS nucleotide, is found to make the maximum number of interactions with RdRp. It is not only buried deep into the binding pocket but also is extended toward the RNA entry channel to block the entry of RNA into the active site of RdRp. Based on these results, it can be proposed that dPdZ may act as a novel inhibitor of RdRp and may potentially help in the treatment of COVID-19. Due to the better docking scores of other AEGIS nucleotides, they may also act as efficient inhibitors of RdRp. However, in vivo evaluation of these nucleotides as SARS-CoV-2 specific inhibitors and measurement of their toxicities are needed to establish them as potent therapeutic candidates.
Supplementary Material
Acknowledgements
SP is thankful to the Department of Pharmaceuticals, Ministry of Chemical and Fertilizers, Govt. of India for providing a research fellowship. We also thank IIITDMJ, NIPER-K, and NIPER-G for various facilities.
Funding Statement
NRJ and HKS thank the Science and Engineering Research Board (SERB), Department of Science and Technology (DST, New Delhi) for the financial supports.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Appleby, T. C., Perry, J. K., Murakami, E., Barauskas, O., Feng, J., Cho, A., Fox, D., Wetmore, D. R., McGrath, M. E., Ray, A. S., Sofia, M. J., Swaminathan, S., & Edwards, T. E. (2015). Viral replication. Structural basis for RNA replication by the hepatitis C virus polymerase. Science (New York, NY), 347(6223), 771–775. 10.1126/science.1259210 [DOI] [PubMed] [Google Scholar]
- Banerjee, A., Yang, W., Karplus, M., & Gregory, L. V. (2005). Structure of a repair enzyme interrogating undamaged DNA elucidates recognition of damaged DNA. Nature, 434(7033), 612–618. 10.1038/nature03458 [DOI] [PubMed] [Google Scholar]
- Basit, A., Ali, T., & Rehman, S. U. (2020). Truncated human angiotensin converting enzyme 2; a potential inhibitor of SARS-CoV-2 spike glycoprotein and potent COVID-19 therapeutic agent. Journal of Biomolecular Structural Dynamics. 10.1080/07391102.2020.1768150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becke, A. D. (1993). A new mixing of Hartree–Fock and local density‐functional theories. Journal of Chemical Physics, 98, 5648–5652. [Google Scholar]
- Behera, B., Das, P., & Jena, N. R. (2019). Accurate base pair energies of artificially expanded genetic information systems (AEGIS): Clues for their mutagenic characteristics. The Journal of Physical Chemistry B, 123(31), 6728–6739. 10.1021/acs.jpcb.9b04653 [DOI] [PubMed] [Google Scholar]
- Benner, S. A., Yang, Z., & Chen, F. (2011). Synthetic biology, tinkering biology, and artificial biology. What are we learning? Comptes Rendus: Chimie (Print), 14(4), 372–387. 10.1016/j.crci.2010.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendsen, H. J. C., Grigera, J. R., & Straatsma, T. P. (1987). The missing term in effective pair potentials. The Journal of Physical Chemistry, 91(24), 6269–6271. 10.1021/j100308a038 [DOI] [Google Scholar]
- Biondi, E., & Benner, S. A. (2018). Artificially expanded genetic information systems for new aptamer technologies. Biomedicines, 6(2), 53. 10.3390/biomedicines6020053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers, K. J., Xu, E. C. H., Dror, R. O., Eastwood, M. P., Gregersen, B. A., Klepeis, J. L., Kolossvary, I., Moraes, M. A., Sacerdoti, F. D., Salmon, J. K., Shan, Y., Shaw, D. E. Scalable algorithms for molecular dynamics simulations on commodity clusters. In Proceedings of the ACM/IEEE conference on supercomputing (SC06), Tampa, Florida, 2006, November, pp. 11–17.
- Buchholz, U. J., Bukreyev, A., Yang, L., Lamirande, E. W., Murphy, B. R., Subbarao, K., & Collins, P. L. (2004). Contributions of the structural proteins of severe acute respiratory syndrome coronavirus to protective immunity. Proceedings of the National Academy of Sciences of the United States of America, 101(26), 9804–9809. 10.1073/pnas.0403492101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cava, C., Bertoli, G., & Castiglioni, I. (2020). In silico discovery of candidate drugs against COVID-19. Viruses, 12(4), 404. 10.3390/v12040404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaar, C. I. O., & Makuch, R. (2020). Emergency use authorization for remdesivir and its potential implications. Therapeutic Innovation and Regulatory Science. 10.1007/s43441-020-00212-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien, M., Anderson, T. K., Jockusch, S., Tao, C., Li, X., Kumar, S., Russo, J. J., Kirchdoerfer, R. N., & Ju, J. (2020). Nucleotide analogues as inhibitors of SARS-CoV-2 polymerase, a key drug target for COVID-19. Journal of Proteome Research, 19(11), 4690–4697. 10.1021/acs.jproteome.0c00392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Silva, S. J. R., Da Silva, C. T. A., Mendes, R. P. G., & Pena, L. (2020). Role of non-structural proteins in the pathogenesis of SARS-CoV-2. Journal of Medical Virology. 10.1002/jmv.25858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennington, R., Keith, T., & Millam, J. (2009). GaussView,Version 5. Semichem Inc. Shawnee Mission.
- Dizdaroglu, M., Coskun, E., & Jaruga, P. (2017). Repair of oxidatively induced DNA damage by DNA glycosylases: Mechanisms of action, substrate specificities and excision kinetics. Mutation Research/Reviews in Mutation Research, 771, 99–127. 10.1016/j.mrrev.2017.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisch, M. J., Trucks, G. W., Schlegel, H. B., Scuseria, G. E., Robb, M. A., Cheeseman, J. R., Scalmani, G., Barone, V., Mennucci, B., Petersson, G. A., Nakatsuji, H., Caricato, M., Li, X., Hratchian, H. P., Izmaylov, A. F., Bloino, J., Zheng, G., Sonnenberg, J. L., Hada, M., Cioslowski, J., & Fox, D. J. (2009). Gaussian09, Revision A.1. Gaussian, Inc. [Google Scholar]
- Gahtori, J., Pant, S., & Srivastava, H. K. (2019). Modeling antimalarial and antihuman African trypanosomiasis compounds: A ligand- and structure-based approaches. Molecular Diversity. 10.1007/s11030-019-10015-y. [DOI] [PubMed] [Google Scholar]
- Gao, Y., Yan, L., Huang, Y., Liu, F., Zhao, Y., Cao, L., Wang, T., Sun, Q., Ming, Z., Zhang, L., Ge, J., Zheng, L., Zhang, Y., Wang, H., Zhu, Y., Zhu, C., Hu, T., Hua, T., Zhang, B., & Rao, Z. (2020). Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science (New York, NY), 368(6492), 779–782. 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiadis, M. M., Singh, I., Kellett, W. F., Hoshika, S., Benner, S. A., & Richards, N. G. (2015). Structural basis for a six nucleotide genetic alphabet. Journal of the American Chemical Society, 137(21), 6947–6955. 10.1021/jacs.5b03482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glushakova, L., Sharma, N., Hoshika, S., Bradley, A. C., Bradley, K. M., Yang, Z., & Benner, S. A. (2015). Detecting respiratory viral RNA using expanded genetic alphabets and self-avoiding DNA. Analytical Biochemistry, 489, 62–72. 10.1016/j.ab.2015.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon, C. J., Tchesnokov, E. P., Woolner, E., Perry, J. K., Feng, J. Y., Porter, D. P., & Götte, M. (2020). Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. Journal of Biological Chemistry, 295(20), 6785–6797. 10.1074/jbc.RA120.013679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartshorn, M. J., Verdonk, M. L., Chessari, G., Brewerton, S. C., Mooij, W. T. M., Mortenson, P. N., & Murray, C. W. (2007). Diverse, high-quality test set for the validation of protein–ligand docking performance. Journal of Medicinal Chemistry, 50(4), 726–741. 10.1021/jm061277y [DOI] [PubMed] [Google Scholar]
- Hawman, D. W., Haddock, E., Meade-White, K., Williamson, B., Hanley, P. W., Rosenke, K., Komeno, K., T., Furuta, Y., Gowen, B. B., & Feldmann, H. (2018). Favipiravir (T-705) but not ribavirin is effective against two distinct strains of Crimean–Congo hemorrhagic fever virus in mice. Antiviral Research, 157, 18–26. 10.1016/j.antiviral.2018.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendaus, M. A. (2020). Remdesivir in the treatment of coronavirus disease 2019 (COVID-19): A simplified summary. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1767691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshika, S., Leal, N. A., Kim, M.-J., Kim, M.-S., Karalkar, N. B., Kim, H.-J., Bates, A. M., Watkins, N. E., SantaLucia, H. A., Meyer, A. J., DasGupta, S., Piccirilli, J. A., Ellington, A. D., SantaLucia, J., Georgiadis, M. M., & Benner, S. A. (2019). Hachimoji DNA and RNA: A genetic system with eight building blocks. Science (New York, NY), 363(6429), 884–887. 10.1126/science.aat0971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jena, N. R. (2012). DNA damage by reactive species: Mechanisms, mutation and repair. Journal of Biosciences, 37(3), 503–517. 10.1007/s12038-012-9218-2 [DOI] [PubMed] [Google Scholar]
- Jena, N. R. (2020a). Drug targets, mechanisms of drug action, and therapeutics against SARS-CoV-2. Chemical Physics Impact, 2, 100011, 10.1016/j.chphi.2021.100011 [DOI] [Google Scholar]
- Jena, N. R. (2020b). Electron and hole interactions with P, Z, and P:Z and the formation of mutagenic products by proton transfer reactions. Physical Chemistry Chemical Physics, 22(2), 919–931. 10.1039/C9CP05367K [DOI] [PubMed] [Google Scholar]
- Jena, N. R. (2020c). Role of different tautomers on the base-pairing abilities of some of the antiviral drugs used against COVID-19. Physical Chemistry Chemical Physics: PCCP, 22(48), 28115–28122. 10.1039/D0CP05297C [DOI] [PubMed] [Google Scholar]
- Jena, N. R., Das, P., Behera, B., & Mishra, P. C. (2018). Analogues of P and Z as efficient artificially expanded genetic information system. The Journal of Physical Chemistry. B, 122(34), 8134–8146. 10.1021/acs.jpcb.8b04207 [DOI] [PubMed] [Google Scholar]
- Jockusch, S., Tao, C., Li, X., Anderson, T. K., Chien, M., Kumar, S., Russo, J. J., Kirchdoerfer, R. K., & Ju, J. (2020). A library of nucleotide analogues terminate RNA synthesis catalyzed by polymerases of coronaviruses that cause SARS and COVID-19. Antiviral Research, 180, 104857. 10.1016/j.antiviral.2020.104857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, G., Willett, P., Glen, R. C., Leach, A. R., & Taylor, R. (1997). Development and validation of a genetic algorithm for flexible docking. Journal of Molecular Biology, 267(3), 727–748. 10.1006/jmbi.1996.0897 [DOI] [PubMed] [Google Scholar]
- Kadam, R. U., & Wilson, I. A. (2017). Structural basis of influenza virus fusion inhibition by the antiviral drug Arbidol. Proceedings of the National Academy of Sciences of the United States of America, 114(2), 206–214. 10.1073/pnas.1617020114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koulgi, S., Jani, V., Uppuladinne, M. V. N., Sonavane, U., & Joshi, R. (2020). Remdesivir-bound and ligand-free simulations reveal the probable mechanism of inhibiting the RNA dependent RNA polymerase of severe acute respiratory syndrome coronavirus 2. RSC Advances, 10(45), 26792–26803. 10.1039/D0RA04743K [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, C., Yang, W., & Parr, R. G. (1988). Development of the Colle–Salvetti correlation-energy formula into a functional of the electron density. Physical Review B: Condensed Matter, 37(2), 785–789. 10.1103/physrevb.37.785 [DOI] [PubMed] [Google Scholar]
- Liu, C., Zhou, Q., Li, Y., Garner, L. V., Watkins, S. P., Carter, L. J., Smoot, J., Gregg, A. C., Daniels, A. D., Jervey, S., & Albaiu, D. (2020). Research and development on therapeutic agents and vaccines for COVID-19 and related human coronavirus diseases. ACS Central Science, 6(3), 315–331. 10.1021/acscentsci.0c00272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahanta, S. (2020). Potential anti-viral activity of approved repurposed drug against main protease of SARS-CoV-2: An in silico based approach. Journal of Biomolecular Structural Dynamics. 10.1080/07391102.2020.1768902 [DOI] [PubMed] [Google Scholar]
- Martyna, G. J., Tobias, D. J., & Klein, M. L. (1994). Constant pressure molecular dynamics algorithms. Journal of Chemical Physics, 101(5), 4177–4189. 10.1063/1.467468 [DOI] [Google Scholar]
- Merritt, K. K., Bradley, K. M., Hutter, D., Matsuura, M. F., Rowold, D. J., & Benner, S. A. (2014). Autonomous assembly of synthetic oligonucleotides built from an expanded DNA alphabet. total synthesis of a gene encoding kanamycin resistance. Beilstein Journal of Organic Chemistry, 10, 2348–2360. 10.3762/bjoc.10.245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, J. H., Huang, C. C., Babbitt, P. C., & Ferrin, T. E. (2007). StructureViz: Linking cytoscape and UCSF chimera. Bioinformatics (Oxford, England), 23(17), 2345–2347. 10.1093/bioinformatics/btm329 [DOI] [PubMed] [Google Scholar]
- Nayeem, S. M., Sohail, E. M., Sudhir, G. P., & Reddy, M. S. (2021). Computational and theoretical exploration for clinical suitability of Remdesivir drug to SARS-CoV-2. European Journal of Pharmacology, 890, 173642. 10.1016/j.ejphar.2020.173642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, K. K. S., Arnold, J. E., & Cameron, C. E. (2008). Structure–function relationships among RNA-dependent RNA polymerases. Current Topics in Microbiology & Immunology, 320, 137–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissink, J. W. M., Murray, C., Hartshorn, M., Verdonk, M. L., Cole, J. C., & Taylor, R. (2002). A new test set for validating predictions of protein–ligand interaction. Proteins, 49(4), 457–471. 10.1002/prot.10232 [DOI] [PubMed] [Google Scholar]
- Pant, S., Singh, M., Ravichandran, V., Murty, U. S. N., & Srivastava, H. (2020). Peptide-like and small-molecule inhibitors against COVID-19. Journal of Biomolecular Structural Dynamics. 10.1080/07391102.2020.1757510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peele, K. A., Krupanidhi, T. S. S., Sai, A. V., & Venketeswarulu, T. C. (2020). Design of multi-epitope vaccine candidate against SARS-CoV-2: A in-silico study. Journal of Biomolecular Structural Dynamics. 10.1080/07391102.2020.1770127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson, H. G. (1995). Accuracy and efficiency of the particle mesh Ewald method. Journal of Chemical Physics, 103, 3668. [Google Scholar]
- Posch, H. A., Hoover, W. G., & Vesely, F. J. (1986). Canonical dynamics of the Nosé oscillator: Stability, order, and chaos. Physical Review A: General Physics, 33(6), 4253–4265. 10.1103/physreva.33.4253 [DOI] [PubMed] [Google Scholar]
- Pruijssers, A. J., George, A. S., Schäfer, A., Leist, S. R., Gralinksi, L. E., Dinnon, K. H., Yount, B. L., Agostini, M. L., Stevens, L. J., Chappell, J. D., Lu, X., Hughes, T. M., Gully, K., Martinez, D. R., Brown, A. J., Graham, R. L., Perry, J. K., Du Pont, V., Pitts, J., & Sheahan, T. P. (2020). Remdesivir inhibits SARS-CoV-2 in human lung cells and chimeric SARS-CoV expressing the SARS-CoV-2 RNA polymerase in mice. Cell Reports, 32(3), 107940. 10.1016/j.celrep.2020.107940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos, K., Wu, C., Damm, W., Reboul, M., Stevenson, J. M., Lu, C., Dahlgren, M. K., Mondal, S., Chen, W., Wang, L., Abel, R., Friesner, R. A., & Harder, E. D. (2019). OPLS3e: Extending force field coverage for drug-like small molecules. Journal of Chemical Theory and Computation, 15(3), 1863–1874. 10.1021/acs.jctc.8b01026 [DOI] [PubMed] [Google Scholar]
- Sahin, A. R., Erdogan, A., Agaolglu, P. M., Dineri, Y., Cakirci, A. Y., Senel; R, M., Okyay, E. A., & Tasdogan, A. M. (2020). 2019 Novel coronavirus (COVID-19) outbreak: A review of the current literature. European Journal of Medical Oncology, 4, 1–7. [Google Scholar]
- Schrödinger Release 2018-4: Desmond Molecular Dynamics System, D. E. Shaw Research, New York, NY, 2018. (2018). Maestro-Desmond interoperability tools. Schrödinger. [Google Scholar]
- Sharma, P. (2020). In-silico homology assisted identification of inhibitor of RNA binding against 2019-nCoV N-protein (N terminal domain). Biomolecular Structural Dynamics. 10.1080/07391102.2020.1753580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan, T. P., Sims, A. C., Leist, S. R., Schafer, A., Won, J., Brown, A. J., Montgomery, S. A., Hogg, A., Babusis, D., Clarke, M. O., Spahn, J. E., Bauer, L., Sellers, S., Porter, D., Feng, J. Y., Cihlar, T., Jordan, R., Denison, M. R., & Baric, R. S. (2020). Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nature Communications, 11, 222 . 10.1038/s41467-019-13940-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder, E. J., Decroly, E., & Ziebuhr, J. (2016). The non-structural proteins directing coronavirus RNA synthesis and processing. Advanced Virus Research, 96, 59–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The efficacy of LopinavirPlus Ritonavir and Arbidol against novel coronavirus Infection (ELACOI) . https://clinicaltrials.gov/ct2/show/NCT04252885.
- Therapeutic options for the 2019 novel coronavirus (2019-nCoV). https://www.nature.com/articles/d41573-020-00016-0. [DOI] [PubMed]
- Toukan Rahman, K. A. (1985). A molecular-dynamics study of atomic motions in water. Physical Review: B, Condensed Matter, 31(5), 2643–2648. 10.1103/physrevb.31.2643 [DOI] [PubMed] [Google Scholar]
- Venkataraman, S., Prasad, B. V. L. S., & Selvarajan, R. (2018). RNA dependent RNA polymerases: Insights from structure, function and evolution. Viruses, 10(2), 76. 10.3390/v10020076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voegel, J. J., & Benner, S. A. (1994). Nonstandard hydrogen bonding in duplex oligonucleotides. The base pair between an acceptor–donor–donor pyrimidine analog and a donor–acceptor–acceptor purine analog. Journal of the American Chemical Society, 116(15), 6929–6930. 10.1021/ja00094a055 [DOI] [Google Scholar]
- Wang, D., Hu, B., Hu, C., Zhu, F., Liu, X., Zhang, J., Wang, B., Xiang, H., Cheng, Z., Xiong, Y., Zhao, Y., Li, Y., Wang, X., & Peng, Z. (2020). Clinical characteristics of 138 hospitalized patients with 2019 coronavirus-infected pneumonia in Wuhan, China. JAMA, 323(11), 1061–1069. 10.1001/jama.2020.1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren, T. K., Jordan, R., Lo, M. K., Ray, A. S., Mackman, R. L., Soloveva, V., Siegel, D., Perron, M., Bannister, R., Hui, H. C., Larson, N., Strickley, R., Wells, J., Stuthman, K. S., Van Tongeren, S. A., Garza, N. L., Donnelly, G., Shurtleff, A. C., Retterer, C. J., & Bavari, S. (2016). Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature, 531(7594), 381–385. 10.1038/nature17180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, R., Wang, L., Kuo, H. C. D., Shannar, A., Peter, R., Chou, P. J., Li, S., Hudlikar, R., Liu, X., Liu, Z., Poiani, G. J., Amorosa, L., Brunetti, L., & Kong, A. N. (2020). An update on current drugs treating COVID-19. Current Pharmacology Reports, 6(3), 56–70. 10.1007/s40495-020-00216-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, W., Mao, C., Luan, X., Shen, D.-D., Shen, Q., Su, H., Wang, X., Zhou, F., Zhao, W., Gao, M., Chang, S., Xie, Y.-C., Tian, G., Jiang, H.-W., Tao, S.-C., Shen, J., Jiang, Y., Jiang, H., Xu, Y., & Xu, H. E. (2020). Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by Remdesivir. Science (New York, NY), 368(6498), 1499–1504. 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L., & Zhou, R. (2020). Structural basis of the potential binding mechanism of remdesivir to SARS-CoV-2 RNA-dependent RNA polymerase. The Journal of Physical Chemistry: B, 124(32), 6955–6962. 10.1021/acs.jpcb.0c04198 [DOI] [PubMed] [Google Scholar]
- Zhang, L., Yang, Z., Sefah, K., Bradley, K. M., Hoshika, S., Kim, M.-J., Kim, H.-J., Zhu, G., Jiménez, E., Cansiz, S., Teng, I.-T., Champanhac, C., McLendon, C., Liu, C., Zhang, W., Gerloff, D. L., Huang, Z., Tan, W., & Benner, S. A. (2015). Evolution of functional six-nucleotide DNA. Journal of the American Chemical Society, 137(21), 6734–6737. 10.1021/jacs.5b02251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Y., Hou, Y., Shen, J., Huang, Y., Martin, W., & Cheng, F. (2020). Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discovery, 6, 14. 10.1038/s41421-020-0153-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.