The introduction in 2005 of the genome-wide association study (GWAS) ushered in an era of disease gene discovery at an unprecedented pace and scale, touching every field in medicine. This is certainly true for asthma, in which in fewer than 15 years, more than 100 asthma risk loci have been identified, implicating a wide range of previously unrecognized biologic processes to asthma pathogenesis. Among the most notable is a common haplotype on chromosome 17q12-2, which has emerged as the most consistently reproducible locus for childhood asthma, conferring risk in populations of varying geographic and ethnic origin (1, 2). Although conferring substantive main effects, the locus also interacts with key environmental factors, including tobacco smoke exposure, viral respiratory illness, vitamin D, and others, establishing 17q21 among the most impactful asthma loci identified to date (3).
But how does this locus confer genetic risk? An inherent limitation of GWASs is that the association often cannot be narrowed to a specific disease-causing variant but rather to a broad region of strong linkage disequilibrium (LD), where alleles at multiple variants in close proximity to one another are highly correlated. This is the case for the 17q21 locus, a region spanning more than 100 kb, containing numerous highly correlated variants that exhibit statistically similar association with asthma, and containing multiple candidate genes. Which SNPs and genes are most relevant?
A broad armamentarium of genomic and experimental approaches has been called on to answer these questions. Expression quantitative trait locus mapping found strong association of the risk haplotype with increased expression of the following two 17q21 genes: ORMDL3 (ORMDL sphingolipid biosynthesis regulator 3) and GSDMB (gasdermin B) (1, 4). Hierarchical functional fine mapping employing multiple techniques identified a causal functional variant (rs12936231) that increases ORMDL3 and GSDMB expression by disrupting the binding of the insulator protein CTCF (5). Chromatin immunoprecipitation sequencing (6) and CpG methylation studies (7, 8) found that the risk haplotype also confers broad epigenetic modification that contributes to the regulation of both genes (and others), providing linkage to the previously noted environmental factors.
Definitive evidence implicating both genes in asthma pathogenesis came from transgenic mouse models developed by David Broide. ORMDL3 transgenic mice spontaneously exhibit physiologic and histologic features of asthma, including airway hyperresponsiveness, airway smooth muscle hypertrophy, and airway wall remodeling, all in the absence of airway inflammation (9). ORMDL3 plays a role in several asthma-relevant biologic processes, including intracellular calcium flux, the unfolded protein response, and sphingolipid metabolism (10, 11).
What about GSDMB? Like the other five members of the gasdermin family of pore-forming proteins, GSDMB is activated after caspase-mediated cleavage and is a potent inducer of both inflammatory cell death (pyroptosis) and extracellular inflammatory cytokine release (11). Although GSDMB does not naturally exist in mice, Broide’s group demonstrated that, like the ORMDL3 mouse, mice expressing a human GSDMB transgene display airway hyperresponsiveness and airway remodeling (12). Also like with ORMDL3, these changes were observed in the absence of accompanying airway inflammation. Though additional studies suggested a role for TGF-β1 in promoting the noninflammatory manifestations, the lack of airway inflammation this model, given the prominent role of the gasdermins in inducing epithelial inflammation, is curious. Why would this be so?
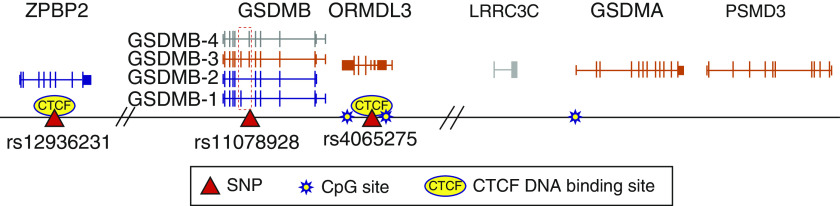
In this issue of the Journal, Gui and colleagues (pp. 424–436) seem to provide an explanation (13). They took a fresh look at the 17q21 locus by performing a comprehensive next-generation DNA-sequencing association study in 5,630 African American children. Genetic association studies in populations of African ancestry, in which genetic diversity is much greater compared with that observed in European populations (both in terms of the total number of variants and the extent of LD), offer two important advantages. First, the greater diversity in total variation ensures that a larger number of variants will be discovered by sequencing, with the potential of identifying novel disease-causing variants not observed in European populations. More importantly, because LD is considerably narrower in African American individuals, fine mapping in this population can help isolate functional variants to shorter DNA segments. Indeed, by using this approach, Gui and colleagues found that the asthma association localized most convincingly to a single variant (rs11078928) situated in a consensus splice site of exon 6 of GSDMB (Figure 1). Although this variant is also present in Europeans, the shorter LD in this African American cohort around rs11078928 (4 kb) focused the association with asthma more narrowly to this variant over others. Subsequent peripheral blood RNA sequencing and expression quantitative trait locus studies found rs11078928 to be associated with alternative splicing of GSDMB, with the allele conferring lower asthma risk (the protective “C” allele) associated with increased expression of a GSDMB isoform lacking exons 6 and 7 (isoform 2). Panganiban and colleagues (14) previously found that this asthma-protective C allele induces the skipping of exon 6 in human bronchial epithelial cells and results in the production of a GSDMB protein (isoform 1) that is resistant to caspase-mediated activation and lacks pyroptotic activity. It is isoform 1 that was used to construct the GSDMB transgenic mouse, providing the following explanation for the absence of inflammation in that model: they evaluated an isoform that does not promote inflammation. From the totality of the evidence, two things are now clear. First, despite being asthma-protective relative to other isoforms, increased expression of GSDMB isoforms that lack pyroptotic potential nonetheless promote the development of noninflammatory airway manifestations of asthma, suggesting alternative GSDMB functions. More importantly, however, they argue for the evaluation of additional murine models that employ the GSDMB isoforms associated with increased asthma risk (i.e., those containing exon 6, whose expression is increased in the presence of asthma risk alleles) to adequately assess the role of GSDMB in asthma pathobiology.
Figure 1.
The 17q21 asthma susceptibility locus. Depicted are the positions of known functional asthma susceptibility SNPs and key CTCF (CCCTC-binding factor) binding sites and CpG methylation sites relative to the canonical exon/intron sequences of candidate genes at the 17q21 asthma susceptibility locus, including the four translated isoforms of GSDMB (gasdermin-B). Transcript color corresponds with whether expression is increased (orange), decreased (blue), or unchanged/unknown (gray) by asthma risk alleles. The dashed box denotes GSDMB exons 6 and 7, which are skipped in the presence of the asthma-protective rs11078928-C allele. Hash marks denote regions of noncoding DNA omitted from the figure for formatting purposes.
In addition to furthering of our understanding of the 17q21 locus, Gui and colleagues offer two timely lessons. First, they provide a glimpse of what is to come as we leverage the full power of next-generation sequencing. The ability to assess all genetic variation at genome scale promises to propel the use of genetic approaches in pulmonary medicine to even greater heights than those achieved by GWASs. Second, Gui and colleagues remind us of the tremendous value of ethnic diversity in population genetic research. Although race is a purely social (not biologic) construct, divergent genealogical histories and mating patterns have resulted in important differences in variant distribution, allele frequency, and LD patterns. As shown here, these differences can be leveraged to facilitate gene discovery. Moreover, in a time when our society is attempting to confront the ills of racial discrimination, including the ongoing racial disparities in health care, it is imperative that future studies are more inclusive to ensure that all peoples benefit equally in the postgenome era.
Supplementary Material
Footnotes
The authors’ research on the genetics and genomics of the 17q21 asthma locus is supported through grants R01 HL123546 and P01 HL132825 from the NHLBI of the NIH.
Originally Published in Press as DOI: 10.1164/rccm.202010-3802ED on October 27, 2020
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1. Moffatt MF, Kabesch M, Liang L, Dixon AL, Strachan D, Heath S, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448:470–473. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 2. Torgerson DG, Ampleford EJ, Chiu GY, Gauderman WJ, Gignoux CR, Graves PE, et al. Mexico City Childhood Asthma Study (MCAAS); Children’s Health Study (CHS) and HARBORS study; Genetics of Asthma in Latino Americans (GALA) Study, Study of Genes-Environment and Admixture in Latino Americans (GALA2) and Study of African Americans, Asthma, Genes & Environments (SAGE); Childhood Asthma Research and Education (CARE) Network; Childhood Asthma Management Program (CAMP); Study of Asthma Phenotypes and Pharmacogenomic Interactions by Race-Ethnicity (SAPPHIRE); Genetic Research on Asthma in African Diaspora (GRAAD) Study. Meta-analysis of genome-wide association studies of asthma in ethnically diverse North American populations. Nat Genet. 2011;43:887–892. doi: 10.1038/ng.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Stein MM, Thompson EE, Schoettler N, Helling BA, Magnaye KM, Stanhope C, et al. A decade of research on the 17q12-21 asthma locus: piecing together the puzzle. J Allergy Clin Immunol. 2018;142:749–764, e3. doi: 10.1016/j.jaci.2017.12.974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sharma S, Zhou X, Thibault DM, Himes BE, Liu A, Szefler SJ, et al. A genome-wide survey of CD4(+) lymphocyte regulatory genetic variants identifies novel asthma genes. J Allergy Clin Immunol. 2014;134:1153–1162. doi: 10.1016/j.jaci.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Verlaan DJ, Berlivet S, Hunninghake GM, Madore A-M, Larivière M, Moussette S, et al. Allele-specific chromatin remodeling in the ZPBP2/GSDMB/ORMDL3 locus associated with the risk of asthma and autoimmune disease. Am J Hum Genet. 2009;85:377–393. doi: 10.1016/j.ajhg.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schmiedel BJ, Seumois G, Samaniego-Castruita D, Cayford J, Schulten V, Chavez L, et al. 17q21 asthma-risk variants switch CTCF binding and regulate IL-2 production by T cells. Nat Commun. 2016;7:13426. doi: 10.1038/ncomms13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nicodemus-Johnson J, Myers RA, Sakabe NJ, Sobreira DR, Hogarth DK, Naureckas ET, et al. DNA methylation in lung cells is associated with asthma endotypes and genetic risk. JCI Insight. 2016;1:e90151. doi: 10.1172/jci.insight.90151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kothari PH, Qiu W, Croteau-Chonka DC, Martinez FD, Liu AH, Lemanske RF, Jr, et al. Asthma BioRepository for Integrative Genomic Exploration (Asthma BRIDGE) Consortium. Role of local CpG DNA methylation in mediating the 17q21 asthma susceptibility gasdermin B (GSDMB)/ORMDL sphingolipid biosynthesis regulator 3 (ORMDL3) expression quantitative trait locus. J Allergy Clin Immunol. 2018;141:2282–2286, e6. doi: 10.1016/j.jaci.2017.11.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Miller M, Rosenthal P, Beppu A, Gordillo R, Broide DH. Oroscomucoid like protein 3 (ORMDL3) transgenic mice have reduced levels of sphingolipids including sphingosine-1-phosphate and ceramide. J Allergy Clin Immunol. 2017;139:1373–1376, e4. doi: 10.1016/j.jaci.2016.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Miller M, Tam AB, Cho JY, Doherty TA, Pham A, Khorram N, et al. ORMDL3 is an inducible lung epithelial gene regulating metalloproteases, chemokines, OAS, and ATF6. Proc Natl Acad Sci USA. 2012;109:16648–16653. doi: 10.1073/pnas.1204151109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen Q, Shi P, Wang Y, Zou D, Wu X, Wang D, et al. GSDMB promotes non-canonical pyroptosis by enhancing caspase-4 activity. J Mol Cell Biol. 2019;11:496–508. doi: 10.1093/jmcb/mjy056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Das S, Miller M, Beppu AK, Mueller J, McGeough MD, Vuong C, et al. GSDMB induces an asthma phenotype characterized by increased airway responsiveness and remodeling without lung inflammation. Proc Natl Acad Sci USA. 2016;113:13132–13137. doi: 10.1073/pnas.1610433113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gui H, Levin AM, Hu D, Sleiman P, Xiao S, Mak ACY, et al. Mapping the 17q12–21.1 locus for variants associated with early-onset asthma in African Americans Am J Respir Crit Care Med 2021203424–436.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Panganiban RA, Sun M, Dahlin A, Park H-R, Kan M, Himes BE, et al. A functional splice variant associated with decreased asthma risk abolishes the ability of gasdermin B to induce epithelial cell pyroptosis. J Allergy Clin Immunol. 2018;142:1469–1478, e2. doi: 10.1016/j.jaci.2017.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.