Abstract
Interleukin 6 (IL-6) is a secreted cytokine that is an important mediator of the immune response in numerous tissues, including skeletal muscle. IL-6 is considered a myokine as it can be secreted by muscle. IL-6 is secreted following exercise, where it exerts both pro-myogenic effects as well as anti-myogenic effects such as promoting atrophy and muscle wasting. The regulation of IL-6 in skeletal muscle is not well understood. The purpose of this study was to determine if IFN-γ and TNF-ɑ stimulate IL-6 in skeletal muscle. We found that both IFN-γ and TNF-α stimulate IL-6 in skeletal muscle, but the stimulation is not cooperative as seen in monocytes. We have previously shown that the IFN-γ stimulated class II major histocompatibility complex transactivator (CIITA) mediates many of the effects of IFN-γ in skeletal muscle and we show here that CIITA directly stimulates IL-6. The regulation of IL-6 by CIITA is clearly complex, as we found that CIITA both stimulates and restrains IL-6 expression. To show that these effects could be observed in a physiological setting, mice were treated with IFN-γ and we found that both CIITA and IL-6 were upregulated in skeletal muscle.
Keywords: IFN-γ, TNF-ɑ, IL-6, CIITA, Skeletal muscle
1. Introduction
The growth and repair of skeletal muscle is regulated by a host of cytokines. IL-6 is expressed by immune cells and by many cell types, including skeletal muscle. During muscle regeneration, IL-6 is produced by infiltrating macrophages and neutrophils [1], fibroadipogenic progenitors [2] as well as the regenerating muscle [3]. Given that both inflammatory cells and skeletal muscle express IL-6, IL-6 has been proposed to have both paracrine and autocrine functions in myogenesis. Inflammatory cells infiltrating injured muscle not only produce IL-6, but also secrete other pro-inflammatory cytokines which may lead to further IL-6 expression by muscle itself, which greatly increases IL-6 concentration at the site of injury.
This local increase in IL-6 promotes muscle growth and regeneration, but is also associated with promotion of muscle atrophy and muscle wasting. Thus, an understanding of the inflammatory cytokines which regulate IL-6 in skeletal muscle is highly relevant to understanding muscle growth and repair. IL-6 is known to be regulated in specific cell types by NF-kappa B (NF-κB), which is induced by Tumor Necrosis Factor alpha (TNF-ɑ). IL-6 contains an NF-κB binding site which is required for NF-κB stimulation in Jurkat cells [4]. TNF-ɑ has also been shown to induce IL-6 in Sertoli cells through the mitogen activated protein kinase pathway (p38) [5]. NF-κB has also been shown to induce IL-6 via ERK1 activation in breast cancer cells [6]. Intriguingly, IL-1 β and TNF-ɑ have been shown to regulate IL-6 through a signaling pathway that involves MAP kinases, but not NF-κB [7].
Interferon-gamma (IFN-γ) is a pro-inflammatory cytokine with multiple physiological roles that includes stimulation of the expression of certain antiviral genes and contribution to the barrier against microbial attack. IFN-γ is known to stimulate macrophages, which play an essential role in muscle regeneration and also directly impact skeletal muscle cells. Transient IFN-γ promotes muscle repair, but we have shown that sustained IFN-γ stimulation inhibits myogenesis. In immune cells, IFN-γ has been shown to cooperate with Tumor Necrosis Factor ɑ (TNF-ɑ) to activate expression of IL-6. Monocytic cells are a primary source of IL-6 expression. Several cytokines activate monocytes to initiate the immune response. IFN-γ and TNF-ɑ have been shown to cooperate to achieve increases in immune and inflammatory responses. IFN-γ and TNF-ɑ have been shown to cooperate to regulate myogenesis as well [8]. In purified human myoblasts, IL-1β, TNF-ɑ and lipopolysaccharide (LPS) have been found to activate IL-6 [9]. In L6 cells, a rat cell line used to model myogenesis, it has also been shown that TNF-ɑ can modestly upregulate IL-6 mRNA [10]. In a monocytic cell line, it was shown that IFN-γ was required for TNF-ɑ to induce IL-6 expression [11]. TNF-ɑ and IFN-γ could not stimulate IL-6 alone, but could cooperate to induce IL-6 [11]. Priming with IFN-γ did serve to super induce IL-6 with IFN-γ and TNF-ɑ treatment, suggesting that factors induced by IFN-γ serve to aid in induction of IL-6 [11]. Treatment of cells with cycloheximide blocked the ability of IFN-γ to induce IL-6, showing that protein synthesis is required for induction of IL-6 [11]. The authors suggest that protein factors induced by IFN-γ are required for the induction of IL-6 [11]. CIITA is expressed in immune cells and indeed, in the previous study, the expression of HLA class II molecules, whose expression is governed by CIITA, was correlated with IFN-γ treatment [11].
Given the complexity of these results, we asked if IFN-γ could activate IL-6 in skeletal muscle and whether IFN-γ could cooperate with TNF-ɑ to activate IL-6 in skeletal muscle. We found that both IFN-γ and TNF-ɑ could modestly upregulate IL-6, but we saw no cooperativity between IFN-γ and TNF-ɑ. The activation of IL-6 by IFN-γ was mediated by CIITA, which we show directly regulates IL-6. Intriguingly, we show that CIITA serves to both activate IL-6 and restrain IL-6 expression. Finally, we show that the upregulation of IL-6 can be seen in vivo in mice injected with IFN-γ, confirming that IFN-γ induces IL-6 in skeletal muscle.
2. Materials and methods
2.1. Cell culture
C2C12 cells (ATCC) were grown in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (Hyclone) according to standard protocols. Proliferating C2C12 myoblasts were grown in DMEM supplemented with 10% fetal bovine serum (FBS, Hyclone) and 1XPenicillin-Streptomycin (Corning). Primary myoblasts were isolated according to standard protocols [18]. Briefly, hindlimb muscle of the neonate mice were isolated, digested with Collagenase Type II (Worthington). The cells were then filtered through a sterile 70-µm filter and plated on gelatin-coated plates in 20% FBS in F-10 basal media with 1X Penicillin-Streptomycin (Corning) and 2.5 ng/ml bFGF (gift of D. Cornelison, University of Missouri). Primary myoblasts were enriched on every passage afterward by pre-platting cells on to uncoated plates for 30 min before transferring the myoblast suspensions onto collagen-coated plates. It was repeated until the majority of the cells were primary myoblasts. Myoblast identity was confirmed by expression analysis of MRFs, differentiation assay, and staining. Both C2C12 and primary myoblast cells were seeded in a new plate for 24 h before treatment with the indicated amount of cytokines for the indicated duration. All mouse procedures were approved by the SIU Institutional Animal Care and Use Committee.
2.2. Overexpression and shRNA knock down of CIITA
The Myc-CIITA plasmid (provided by Jeremy Boss, Emory University) was used for expressing CIITA with a Myc epitope on the N-terminus. The Myc-CIITA plasmid was linearized with the ScaI restriction enzyme (New England Biolab) and stably transfected in C2C12 cells. Clones were selected with neomycin (G418) (400 µg/ml), propagated and confirmed by mRNA and protein analysis. CIITA was depleted with shRNA constructs designed by the RNAi Consortium in the pLOK.1 plasmid (Open Biosystems) as described [12]. A construct targeting murine CIITA and a scrambled control were transiently transfected into C2C12 cells. For all transient transfections, plasmids were not linearized and drug selection was not used.
2.3. Western blot analysis
Cell extracts were made by lysing PBS washed cell pellets in radio-immunoprecipitation assay buffer (RIPA) supplemented with protease inhibitors (Complete protease inhibitor, Roche Diagnostics). Following incubation on ice, clear lysates were obtained by centrifugation. Protein concentrations were determined by Bradford’s assay (Bio-Rad). For each sample, 30 µg of protein was loaded into each lane of the gel. Proteins were transferred onto a PVDF membrane using a tank blotter (Bio-Rad). The membranes were then blocked with 5% milk in 1X Tris-buffered saline plus tween 20 (TBST) and incubated with primary antibody overnight at 4 °C. Membranes were then washed with 1X TBST and incubated with the corresponding secondary antibody. Membranes were again washed with 1X TBST, incubated with chemiluminescent substrate according to manufacturer's protocol (SuperSignal, Pierce) and visualized by and iBright Imaging System. The antibodies used include anti-IL-6 (10E5, SCBT) and anti-GAPDH (Millipore). Protein expression levels were quantified using iBright analysis software on at least three independent experiments. Representative images are shown.
2.4. Mouse procedures
All procedures were approved by the SIU Animal Care and Use Committee. ND4 Swiss mice (n = 12) were anesthetized by isoflurane gas. IFN-γ in sterile buffer was injected into the tail vein (IV) of each mouse and sterile liposomes containing IFN-γ were injected intraperitoneally (IP) to total 1.7x105 units IFN-γ per mouse. Control mice received buffer IV and control liposomes IP. Three control and 3 IFN-γ treated mice were killed at each 10 and 24 h after treatment. Plasma and muscle samples were collected at each time point. Plasma was assayed for IFN-γ by ELISA and muscle was assayed for gene expression by qRT-PCR.
2.5. Chromatin immunoprecipitation assays
ChIP assays were performed as described previously [12]. The following antibodies were used: anti-CIITA (7-1H, SCBT) and Rabbit IgG (SCBT) was used as a non-specific control. Primers are described in Supplemental Table 1. The real-time PCR was performed in triplicate. The results were represented as the percentage of IP over input signal (% Input). All ChIP assays shown are representative of at least three individual experiments. Standard error from the mean was calculated and plotted as the error bar.
2.6. Plasma harvest
Mice were humanely killed (approved protocol) and blood was collected. The blood was refrigerated for 30–45 min and centrifuged to yield plasma. A commercial ELSIA kit (Thermo Scientific) was used to assay mouse IFN-γ levels in plasma and liposomes
2.7. Encapsulation
Lipids (1,2-dipalmitoyl-sn-glycero-3-phosphocholine, DPPC; 1,2-dipalmitoly-sn-glycero-3-phospho-rac-(1-glycerol) sodium salt, DPPG; and cholesterol, Chol) were mixed in a 10 DPPC:1 DPPG:10 Chol ratio in ethanol and dried onto a round bottom flask with a rotary evaporator. The flask was flushed for 30 min with argon. The film was then hydrated with IFN-γ in buffer or with buffer alone for control. This was agitated lightly for 30 min at 45 °C. The resultant suspensions were tested to ensure encapsulation and particle sizes were determined [13], [14], [15]. The hydrodynamics radius was 118 ± 47.
2.8. Quantitative real-time PCR
RNA was isolated from cells by Trizol extractions (Invitrogen). Following treatment with DNase (Promega), two micrograms of total RNA was reversed transcribed with MultiScribe™ MuLV reverse transcriptase (Applied Biosystems). cDNA equivalent to 40 ng was used for quantitative polymerase chain reaction (qPCR) amplification (Applied Biosystems) with SYBR green PCR master mix (Applied Biosystems). Samples in which no reverse transcriptase was added (no RT) were included for each RNA sample. qPCR data were calculated using the comparative Ct method (Applied Biosystems). Standard deviations from the mean of the ΔCt values were calculated from three independent RNA samples. Primers are described in Supplemental Table 1. Where possible, intron spanning primers were used. All quantitative PCR was performed in triplicate, and three independent RNA samples were assayed for each time point. For measurements of relative gene expression, a fold change was calculated for each sample pair and then normalized to the fold change observed at HPRT and/or 18S rRNA.
2.9. Statistics
Data are presented as means ± standard errors (S.E.). The dots on the bar graphs represents individual data points. Statistical comparisons were performed using unpaired two-tailed Student's t tests or ANOVA followed by Tukey's multiple comparisons test. For ANOVA analysis, all means not sharing the same letter were significantly different from one another statistically. Probability value of < 0.05 was taken to indicate significance. All statistical analyses and graphs were made in GraphPad Prism 8.0 software.
3. Results
3.1. IFN-γ and TNF-ɑ stimulate IL-6 in C2C12 cells
To determine if IFN-γ could contribute to the regulation of IL-6 in skeletal muscle, we first asked if IFN- could activate IL-6 mRNA in C2C12 myoblasts, a commonly used model for myogenesis. Treatment of C2C12 cells in vitro with 1000 units (U)/ml IFN-γ induced an approximately two-fold expression increase of the IL-6 gene (Fig. 1A). We have previously shown that IFN-γ activates CIITA in skeletal muscle [16] and to confirm that IFN-γ upregulated CIITA in these experiments, the mRNA expression of CIITA was assayed as well. We found that CIITA was upregulated as anticipated (Fig. 1A). The upregulation of IL-6 by TNF-ɑ has previously been shown in L6 rat myoblasts [10], another cell based model for myogenesis. To determine if TNF-ɑ could also upregulate IL-6 in C2C12 cells, we treated C2C12 cells with either IFN-γ or TNF-ɑ and in combination and assayed for the expression of IL-6 and CIITA. The concentration of IFNγ was reduced to 500U/ml to better reflect the physiological level of IFNγ. TNF-ɑ was also used at 500U/ml. We found that TNF-ɑ also upregulated IL-6 approximately two-fold (Fig. 1B). The upregulation induced by IFN-γ and TNF-α were very similar and the upregulation induced by either cytokine at this time point were not statistically distinct (Fig. 1B). To determine if IFN-γ and TNF-ɑ could synergize to upregulate IL-6, C2C12 cells were treated with both IFN-γ and TNF-ɑ. We found that IL-6 was modestly upregulated, but the upregulation was not statistically distinct from the induction seen by either cytokine alone, indicating that IFN-γ and TNFα do not synergize to activate IL-6 (Fig. 1B). We also examined the expression of CIITA and found that TNF-ɑ did not upregulate CIITA and only exposure to IFN-γ upregulated CIITA (Fig. 1B). Interestingly, we also noted that the upregulation of CIITA was blunted in the presence of TNF-α.
Fig. 1.
IFN-γ and TNF-α activate IL-6. (A) mRNA expression of CIITA and IL-6 were assayed by qRT-PCR in C2C12 cells treated with either vehicle control or 1000U/ml of IFN-γ for 24 h. Data plotted are mean (±SEM) (Student t.test, *p < 0.05 and **p < 0.01, n = 3 biological replicates). (B) C2C12 cells were treated with either vehicle control, 500U/ml IFN-γ or 500U/ml TNFα or combination for 12 h. mRNA expression of IL6 and CIITA were assayed by qRT-PCR. Data plotted are mean (±SEM). Bars not sharing same letter represents statistical significance (ANOVA with Tukey’s multiple comparisons test; p < 0.05, n = 6–9 biological replicates). (C) C2C12 cells were treated with either vehicle control or 500U/ml IFN-γ or 500U/ml TNFα or combination for 6 h. mRNA expression of IL6 and CIITA were assayed by qRT-PCR. Data plotted are mean (±SEM). Bars not sharing same letter represents statistical significance (ANOVA with Tukey’s multiple comparisons test; p < 0.05, n = 3–5 biological replicates). (D) mRNA expression of CIITA and IL-6 were assayed in C2C12 cells either primed with 500U/ml of IFN-γ or vehicle treated for 12 h before treating with a combination of 500U/ml of IFN-γ and TNF-α each for 6 h. (E) mRNA expression of CIITA and IL-6 were assayed in C2C12 cells either primed with 500U/ml of TNF-ɑ or vehicle treated for 6 h before treating with a combination of 500U/ml of IFN-γ and TNF-α each for 12 h. Data plotted are mean (±SEM). (Student t.test ; *p < 0.05, **p < 0.01 and ***p < 0.001, ns represents ‘not significant’, n = 3–6 biological replicates).
Our initial experiments were performed 12 h after exposure to the cytokines so we next asked if the upregulation of IL-6 could be observed 6 h after cytokine stimulation. C2C12 cells were treated with IFN-γ and TNF-α alone and in combination and assayed for expression of IL-6 and CIITA. We found that IFN-γ did not produce a statically significant increase in IL-6 expression at 6 h (Fig. 1C). However, TNF-α produced an approximately four-fold increase in IL-6 expression, which was higher than the activation seen at 12 h. Combined treatment with IFN-γ and TNF-α lead to modestly enhanced expression of IL-6 (Fig. 1C). CIITA was activated by IFN-γ, but we note that the activation of CIITA was much less robust at 6 h compared to the activation seen at 12 h. TNF-α did not activate CIITA, but TNF-ɑ also did not suppress CIITA expression at 6 h (Fig. 1C). Finally, we asked if IFN-γ could prime IL-6 for activation by TNF-α as had been seen in monocytes [11]. C2C12 cells were treated with IFN-γ or vehicle control for 12 h and then stimulated with IFN-γ and TNF-α for 6 h. We saw no upregulation in IL-6 expression with pre-treatment and instead saw a decrease in expression of IL-6 with pre-treatment (Fig. 1D). We next asked if TNF-ɑ could prime IL-6 for activation by IFN-γ. C2C12 cells were treated with TNF-α or vehicle control for 6 h and then stimulated with IFN-γ and TNF-α for 12 h. We saw no statistically significant priming effect for TNF-ɑ on IL-6 expression (Fig. 1E). Together, our data show that both IFN-γ and TNFα activate IL-6, but they appear to do so through independent mechanisms with different kinetics and do not appear to significantly synergize with each other.
3.2. CIITA directly activates and restrain Il-6 expression
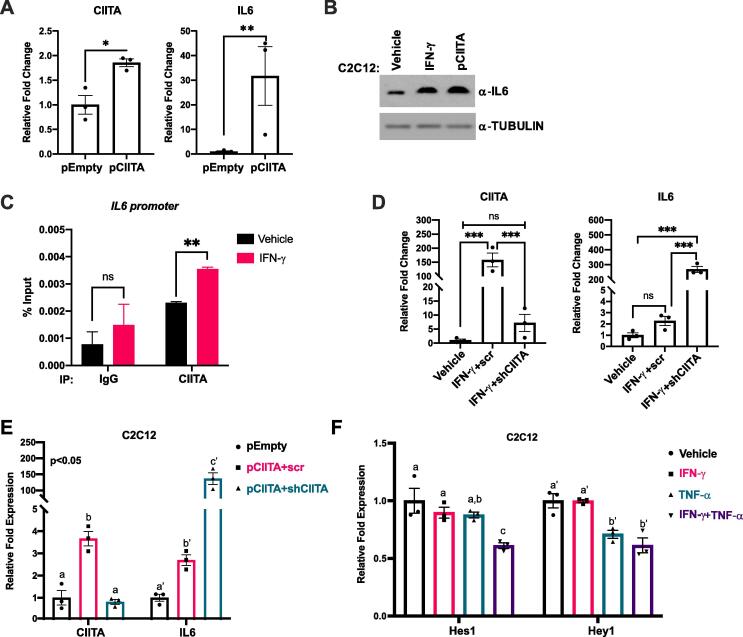
Given the results in monocytes that showed that IFN-γ stimulation of IL-6 involved translation and that MHC class II genes were activated [11], we sought to determine if CIITA played a direct role in IL-6 induction. C2C12 cells were transiently transfected with an expression construct for CIITA and we found that CIITA and IL-6 were upregulated, confirming that CIITA activates IL-6 (Fig. 2A). Importantly, this experiment was performed in the absence of IFN-γ, confirming that CIITA alone can upregulate IL-6. To confirm that these results could be confirmed at the protein level as well, we examined IL-6 expression by western blot following either stimulation by IFN-γ or exogenous expression of CIITA. We found that IL-6 expression was stimulated by both IFN-γ and exogenous CIITA to comparable levels (Fig. 2B). To confirm that this was a direct regulation, we performed chromatin immunoprecipitation assays (ChIP) for CIITA on the Il6 promoter. We found that CIITA bound to the Il6 promoter and this binding was enhanced upon IFN-γ stimulation (Fig. 2C).
Fig. 2.
CIITA activates IL-6 and restrains its expression. (A) C2C12 cells were transiently transfected with either empty plasmid (pEmpty) or the CIITA expression plasmid (pCIITA). mRNA expression of CIITA and IL-6 were assayed by qRT-PCR. Data plotted are mean (±SEM) (Student t.test, *p < 0.05 and **p < 0.01, n = 3 biological replicates). (B) C2C12 cells treated with 500U/ml IFN-γ for 12 h or transiently transfected with pCIITA were assayed by western blot. Tubulin was used as the loading control (C) CIITA directly regulates expression of the Il6 gene. ChIP assays were performed on C2C12 cells treated with either vehicle control or 500U/ml of IFN-γ using antibodies against CIITA and primers spanning Il6 proximal promoter. IgG is used as a non-specific background control. Data plotted are mean (±SEM). (Student t.test, **p < 0.01, ns represents ‘not significant’, n = 3 biological replicates). (D) mRNA expression of CIITA and IL-6 were assayed by qRT-PCR in C2C12 cells stimulated with either vehicle control or IFN-γ. The IFN-γ treated cells were also transiently transfected with either non-specific scramble control (IFN-γ + scr) or CIITA targeting shRNA (IFN-γ + shCIITA). Cells were transfected, allowed to recover for 24 h post transfection and were then stimulated with IFN-γ for 24 h. Data plotted are mean (±SEM). (ANOVA with Tukey’s multiple comparisons test; ***p < 0.001, ns represents ‘not significant’, n = 3 biological replicates). (E) C2C12 cells were stably transfected with either empty plasmid (pEmpty) or the CIITA expression plasmid (pCIITA). CIITA overexpression clones were further transiently transfected with either non-specific scramble control (pCIITA + scr) or CIITA targeting shRNA (pCIITA + shCIITA). mRNA expression of CIITA and IL-6 were assayed by qRT-PCR, 48 h post-transient transfection of shRNAs. Data plotted are mean (±SEM). Bars not sharing same letter represents statistical significance (ANOVA with Tukey’s multiple comparisons test; p < 0.05, n = 3 biological replicates). (F) C2C12 cells were treated with vehicle control or 500U/ml of IFN-γ or TNF-α or combination for 6 h. mRNA expression of two Notch signaling effector genes, Hes1 and Hey1 were assayed by qRT PCR. Data plotted are mean (±SEM). Bars not sharing same alphabet represents statistical significance (ANOVA with Tukey’s multiple comparisons test; p < 0.05, n = 3 biological replicates).
We next asked if CIITA was required for the IFN-γ activation of IL-6. We used shRNA constructs that we have previously characterized [16] to deplete CIITA C2C12 cells were transfected with shRNA against CIITA or scr control and stimulated with IFN-γ. To our surprise, we found that IFN-γ stimulated very high levels of IL-6 mRNA when CIITA was depleted (Fig. 2D). To confirm this unexpected result, we utilized a C2C12 cell line which contains a stable integration of an expression construct for CIITA [16]. We transiently transfected these cells with a shRNA construct for CIITA and a scr control. In the scr control, we could see that CIITA, as anticipated, was over expressed in the cells and we found that IL-6 mRNA was modestly upregulated as well (Fig. 2E). Upon CIITA depletion, we found that CIITA was down regulated and IL-6 was highly upregulated (Fig. 2E). Thus, our results show that CIITA serves to both stimulate IL-6, but also serves to restrain the expression of IL-6. Our results also show that IFN-γ must have a CIITA independent mechanism to stimulate IL-6. However, it is clear that CIITA plays an essential role in the normal physiological expression of IL-6.
In macrophages, it has also been shown that IFN-γ blocks the toll-like receptor (TLR) activation of Notch2 signaling [17], The block to Notch signaling leads to the repression of the Notch target genes Hes1 and Hey1, which repress Il6 expression [17]. Hes1 and Hey1 are known to function is skeletal muscle as well [18], so we asked if expression of Hes1 or Hey1 were altered by IFN-γ or TNF-ɑ in our experiments. We found that IFN-γ treatment did not result in a statistically significant downregulation of Hes1 or Hey1 mRNA (Fig. 2F). However, TNF-ɑ did show a significant downregulation of Hey1 and the combined treatment with both IFN-γ and TNF-ɑ resulted in a significant downregulation of both Hes1 and Hey1 (Fig. 2F). Thus, the repression of Hes1 and Hey1 may contribute to IL-6 upregulation in skeletal muscle as well.
3.3. IFN-γ stimulates IL-6 in primary myoblasts
To confirm the regulation of IL-6 by IFN-γ in skeletal muscle, we treated freshly isolated primary myoblasts with IFN-γ and TNF-α and examined the expression of IL-6. We found that IL-6 mRNA was upregulated two-fold by IFN-γ at 500 U/ml and increasing the IFN-γ concentration led to a modest additional upregulation (Fig. 3A). Treatment with TNF-α also lead to a two-fold activation of IL-6, similar to what was observed with C2C12 cells (Fig. 3A). Stimulation by IFN-γ activated CIITA in primary myoblasts and the activation of CIITA was stimulated by increasing the concentration of IFN-γ (Fig. 3A). We also confirmed the IFN-γ stimulation of IL-6 at the protein level as well (Fig. 3B). We also examined the mRNA expression of Hes1 and Hey1 in primary myoblasts stimulated with IFN-γ. Here, we saw that IFN-γ treatment did significantly down regulate both Hes1 and Hey1 (Fig. 3C), indicating that repression of these factors may indeed play a role in IL-6 activation in skeletal muscle.
Fig. 3.
IFN-γ and TNF-α activate IL-6 in primary myoblasts. (A) Freshly isolated primary myoblast cells were treated with either vehicle control or 500U/ml of either IFN-γ or TNF-α for 12 h. mRNA expression of IL6 and CIITA were assayed by qRT-PCR. Data plotted are mean (±SEM). Bars not sharing same letter represents statistical significance (ANOVA with Tukey’s multiple comparisons test; p < 0.05, n = 3–4 biological replicates). (B) Cells as in A were assayed for the expression of IL-6 protein by western blotting. GAPDH was used as a loading control. Blots were quantified, normalized to GAPDH and plotted (lower panel). (C) Primary myoblasts were treated with vehicle control or either 500U/ml or 1000U/ml of IFN-γ for 12 h. mRNA expression of Hes1 and Hey1 were assayed by qRT-PCR. Data plotted are mean (±SEM) Bars not sharing same letter represents statistical significance (ANOVA with Tukey’s multiple comparisons test; p < 0.05, n = 4 biological replicates).
3.4. IFN-γ stimulates CIITA and IL-6 in skeletal muscle in vivo
To determine if IFN-γ could upregulate IL-6 in vivo, we treated six mice with 1.69x105 units of IFN-γ. IFN-γ is highly labile, so we chose to inject IFN-γ directly into the blood stream to rapidly increase the IFN-γ concentration. To potentially sustain the effect, we also injected encapsulated IFN-γ to slow the release. 1.9x104 units of IFN-γ were injected by tail vein injection and 1.5x105 units encapsulated within the inner aqueous cavity of liposomes were injected intraperitoneally. Six mice were treated with vehicle as control. The experimental strategy is summarized in Fig. 4A. We harvested plasma and tissue including tibialis anterior (TA) muscle, heart and brain after 10 (n = 6; 3 each per control and IFN-γ group) and 24 (n = 6; 3 per group) hours. ELISA assays demonstrated that plasma IFN-γ levels averaged 3560 ± 1167 SEM pg/ml after 10 h while no enhanced IFN-γ was detected after 24 h (Fig. 4B). IL-6 and CIITA mRNA expression was assayed in tissue. In TA muscles, we found that 10 h of IFN-γ treatment induced a 15-fold increase in CIITA expression and a two-fold increase in IL-6 expression (Fig. 4C). At 24 h after treatment, we detected a four-fold increase in CIITA expression while IL-6 expression was back to pre-treatment levels (Fig. 4C). To determine if this effect was specific to skeletal muscle, we also examined heart and brain tissue. We found that both CIITA and IL-6 were stimulated in the heart (Fig. 4D). These effects were robust at 10 h, but largely returned to baseline 24 h after treatment. No stimulation of either CIITA or IL-6 was seen in brain tissue at either time point (Fig. 4E). Thus, our data confirm that CIITA and IL-6 are activated by IFN-γ in vivo. A proposed model regulation of IL-6 by IFN-γ is shown in Fig. 4E.
Fig. 4.
IFN-γ stimulates CIITA and IL-6 in vivo. (A) Experimental strategy for the in vivo encapsulated IFN-γ injection study. (B) IFN-γ was estimated in plasma isolated from the mice that were injected with either vehicle control or IFN-γ after 10 h or 24 h post-injections. ELISA was performed for IFN-γ estimation. Data plotted are mean (±SEM) (Student t.test, *p < 0.05, ns represents’ not significant’, n = 3 biological replicates). (C-E) mRNA expression of IL-6 and CIITA were assayed in tibialis anterior (TA) muscle (C), heart (D) or brain (E) of the animals from B using qRT-PCR. Data plotted are mean (±SEM) (Student t.test, ***p < 0.001, **p < 0.01, ns represents ’not significant’, n = 3 biological replicates). (F) A schematic model of IL-6 gene regulation by interferon-γ (IFN-γ) in skeletal muscle. IFN-γ activates CIITA, which is recruited to the Il6 promoter to both activate and restrain expression of the Il6 gene. IFN-γ also promotes the activation of Il6 expression through inhibition of the Notch signaling pathway that inhibits the expression of IL-6.
4. Discussion
IL-6 is a myokine with both known beneficial and deleterious effects on skeletal muscle. We show that either IFN-γ or TNF-ɑ can modestly stimulate IL-6 expression in skeletal muscle, but these cytokines do not appear to cooperate to induce IL-6 in skeletal muscle. The regulation of IL-6 by IFN-γ involves CIITA, which acts both to activate IL-6 and to restrain IL-6 activation. Intriguingly, our results also indicate that inhibition of the Notch signaling pathway could also partially contribute to the IFN-γ dependent upregulation of IL-6, although these results need to be confirmed at the protein level and further characterized in future experiments. Given the known effects of IL-6 on skeletal muscle, it is not unexpected that skeletal muscle cells would tightly restrict expression of IL-6. CIITA is known to be both a co-activator and a co-repressor, but it is interesting that it appears to show both activities on the Il6 promoter. Our results also indicate that IFN-γ activates Il6 in a CIITA independent manner as well. IFN-γ stimulates through the JAK/STAT pathway. Phosphorylation of STAT1 induces translocation to the nucleus where STAT1 activates IFN-γ dependent genes such as CIITA. We have previously shown that CIITA acts to inhibit the expression and activity of myogenin, a myogenic regulatory factor required for myoblast fusion and terminal differentiation [16]. It is interesting that both TFN-α and IFN-γ activate IL-6 through alternative mechanisms and different kinetics. TNF-α activates IL-6 by 6 h in a CIITA independent manner. IFN-γ requires additional time to fully activate IL-6, indicating that the activation mechanism may require the transcription of effector genes. This is certainly the case for CIITA, but it will be interesting to understand the effector proteins for the CIITA independent mechanism as well.
IL-6 is known to be upregulated during differentiation of C2C12 cells where it is thought to promote skeletal muscle differentiation [19]. Thus, the CIITA dependent upregulation and restraint of IL-6 in skeletal muscle may serve as a switch during skeletal muscle differentiation. Skeletal muscle proliferation and differentiation are tightly coordinated but mutually exclusive processes with a temporal switch from Notch signaling to the Wnt signaling pathway. The switch from Notch signaling to Wnt signaling enables myoblasts to exit from the proliferative stage and enter the differentiation phase [20]. The upregulation of IL-6 through the inhibition of Notch signaling is in agreement with the proliferation to differentiation switch, indicating a possible pro-differentiation function of IFN-γ during this process. However, further investigation would be needed to further characterize the role of the CIITA/IL-6 axis in skeletal muscle differentiation.
A significant recent study has shown that an evolutionary recent IFN/IL-6/CEBP axis is linked to monocyte expansion and tuberculosis (TB) severity in humans [21]. TB is caused by Mycobacterium tuberculosis (Mtb), which can direct CD34- cells to differentiate into monocytes/macrophages [21]. Neutralizing IL-6 inhibited the Mtb enhanced myeloid differentiation by CD34- cultures [21]. Large genome wide association studies (GWAS) showed that type I IFN pathway genes were associated with IL6/STAT3 pathway genes, indicating that type I IFN regulates the expression of IL-6 signaling [21]. IFN-γ is a type II IFN, but stimulates expression of many of the same genes.
It will be interesting to understand if CIITA contributes to IL-6 regulation in immune cells as well. While we show that IFN-γ does not have priming effect for the activation of IL-6 by TNF-ɑ in skeletal muscle as was observed in monocytes [11], we do show that CIITA both activates and restrains IL-6 expression in skeletal muscle. IFN-γ stimulates CIITA in both immune cells and skeletal muscle and it is very likely that CIITA plays a role in the activation of IL-6 in both systems. The observed upregulation of both CIITA and IL-6 in the heart is intriguing and warrants future study. CIITA was thought to be restricted to immune cells, but we have shown that it plays an important role in skeletal muscle as well [16], [22]. Our results suggest that CIITA may also function in the heart, but it will be important to establish that cardiac cells express CIITA.
The ability of CIITA to both activate and restrain IL-6 expression is intriguing given the known deleterious effects of chronic elevation of IL-6 in skeletal muscle. Our work suggests that CIITA serves to protect skeletal muscle from excessive IL-6 expression in response to inflammation, further reinforcing the important role of CIITA in skeletal muscle.
CRediT authorship contribution statement
Abhinav Adhikari: Methodology, Investigation, Validation, Formal analysis, Writing - original draft, Writing - review & editing. Brittan Cobb: Investigation, Validation. Seth Eddington: . Nathalie Becerra: Methodology, Investigation. Punit Kohli: Methodology, Writing - review & editing. Amber Pond: Methodology, Investigation, Validation, Writing - review & editing, Funding acquisition, Project administration. Judith Davie: Methodology, Investigation, Validation, Formal analysis, Writing - original draft, Writing - review & editing, Funding acquisition, Project administration.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Funding and Acknowledgements
This work was funded in part by Southern Illinois University start up finds to ALP and in part by the Department of Defense Office of the Congressionally Directed Medical Research Programs in the form of a Discovery Award (PR170326) to ALP. No DOD funds were used to purchase or house the animals used in this work. JKD is supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health under Award Number RAR068622. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cytox.2020.100023.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Zhang C., Li Y., Wu Y., Wang L., Wang X., Du J. Interleukin-6/signal transducer and activator of transcription 3 (STAT3) pathway is essential for macrophage infiltration and myoblast proliferation during muscle regeneration. J. Biol. Chem. 2013;288:1489–1499. doi: 10.1074/jbc.M112.419788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Joe A.W., Yi L., Natarajan A., Le Grand F., So L., Wang J., Rudnicki M.A., Rossi F.M. Muscle injury activates resident fibro/adipogenic progenitors that facilitate myogenesis. Nat. Cell Biol. 2010;12:153–163. doi: 10.1038/ncb2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kami K., Senba E. Localization of leukemia inhibitory factor and interleukin-6 messenger ribonucleic acids in regenerating rat skeletal muscle. Muscle Nerve. 1998;21:819–822. doi: 10.1002/(sici)1097-4598(199806)21:6<819::aid-mus20>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 4.Libermann T.A., Baltimore D. Activation of interleukin-6 gene expression through the NF-kappa B transcription factor. Mol. Cell Biol. 1990;10:2327–2334. doi: 10.1128/mcb.10.5.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Cesaris P., Starace D., Riccioli A., Padula F., Filippini A., Ziparo E. Tumor necrosis factor-alpha induces interleukin-6 production and integrin ligand expression by distinct transduction pathways. J. Biol. Chem. 1998;273:7566–7571. doi: 10.1074/jbc.273.13.7566. [DOI] [PubMed] [Google Scholar]
- 6.Suarez-Cuervo C., Harris K.W., Kallman L., Vaananen H.K., Selander K.S. Tumor necrosis factor-alpha induces interleukin-6 production via extracellular-regulated kinase 1 activation in breast cancer cells. Breast Cancer Res. Treat. 2003;80:71–78. doi: 10.1023/a:1024443303436. [DOI] [PubMed] [Google Scholar]
- 7.Palmqvist P., Lundberg P., Lundgren I., Hanstrom L., Lerner U.H. IL-1beta and TNF-alpha regulate IL-6-type cytokines in gingival fibroblasts. J. Dent Res. 2008;87:558–563. doi: 10.1177/154405910808700614. [DOI] [PubMed] [Google Scholar]
- 8.Guttridge D.C., Mayo M.W., Madrid L.V., Wang C.Y., Baldwin A.S., Jr. NF-kappaB-induced loss of MyoD messenger RNA: possible role in muscle decay and cachexia. Science. 2000;289:2363–2366. doi: 10.1126/science.289.5488.2363. [DOI] [PubMed] [Google Scholar]
- 9.Gallucci S., Provenzano C., Mazzarelli P., Scuderi F., Bartoccioni E. Myoblasts produce IL-6 in response to inflammatory stimuli. Int. Immunol. 1998;10:267–273. doi: 10.1093/intimm/10.3.267. [DOI] [PubMed] [Google Scholar]
- 10.Roher N., Samokhvalov V., Diaz M., MacKenzie S., Klip A., Planas J.V. The proinflammatory cytokine tumor necrosis factor-alpha increases the amount of glucose transporter-4 at the surface of muscle cells independently of changes in interleukin-6. Endocrinology. 2008;149:1880–1889. doi: 10.1210/en.2007-1045. [DOI] [PubMed] [Google Scholar]
- 11.Sanceau J., Wijdenes J., Revel M., Wietzerbin J. IL-6 and IL-6 receptor modulation by IFN-gamma and tumor necrosis factor-alpha in human monocytic cell line (THP-1). Priming effect of IFN-gamma. J. Immunol. 1991;147:2630–2637. [PubMed] [Google Scholar]
- 12.Londhe P., Davie J.K. Sequential association of myogenic regulatory factors and E proteins at muscle-specific genes. Skelet. Muscle. 2011:1–14. doi: 10.1186/2044-5040-1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang L., Yang W., Bi D., Zeng Q. A novel method to prepare highly encapsulated interferon-alpha-2b containing liposomes for intramuscular sustained release. Eur. J. Pharm. Biopharm. 2006;64:9–15. doi: 10.1016/j.ejpb.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 14.Van Slooten M.L., Boerman O., Romoren K., Kedar E., Crommelin D.J., Storm G. Liposomes as sustained release system for human interferon-gamma: biopharmaceutical aspects. Biochim. Biophys. Acta. 2001;1530:134–145. doi: 10.1016/s1388-1981(00)00174-8. [DOI] [PubMed] [Google Scholar]
- 15.Hopkins S., DiMassimo B., Rusnak P., Heuman D., Lalezari J., Sluder A., Scorneaux B., Mosier S., Kowalczyk P., Ribeill Y., Baugh J., Gallay P. The cyclophilin inhibitor SCY-635 suppresses viral replication and induces endogenous interferons in patients with chronic HCV genotype 1 infection. J. Hepatol. 2012;57:47–54. doi: 10.1016/j.jhep.2012.02.024. [DOI] [PubMed] [Google Scholar]
- 16.Londhe P., Davie J.K. Gamma interferon modulates myogenesis through the major histocompatibility complex class II transactivator, CIITA. Mol. Cell. Biol. 2011;31:2854–2866. doi: 10.1128/MCB.05397-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu X., Chung A.Y., Wu I., Foldi J., Chen J., Ji J.D., Tateya T., Kang Y.J., Han J., Gessler M., Kageyama R., Ivashkiv L.B. Integrated regulation of Toll-like receptor responses by Notch and interferon-gamma pathways. Immunity. 2008;29:691–703. doi: 10.1016/j.immuni.2008.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buas M.F., Kabak S., Kadesch T. The Notch effector Hey1 associates with myogenic target genes to repress myogenesis. J. Biol. Chem. 2010;285:1249–1258. doi: 10.1074/jbc.M109.046441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoene M., Runge H., Haring H.U., Schleicher E.D., Weigert C. Interleukin-6 promotes myogenic differentiation of mouse skeletal muscle cells: role of the STAT3 pathway. Am. J. Physiol. Cell Physiol. 2013;304:C128–C136. doi: 10.1152/ajpcell.00025.2012. [DOI] [PubMed] [Google Scholar]
- 20.Brack A.S., Conboy I.M., Conboy M.J., Shen J., Rando T.A. A temporal switch from notch to Wnt signaling in muscle stem cells is necessary for normal adult myogenesis. Cell Stem Cell. 2008;2:50–59. doi: 10.1016/j.stem.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 21.Delgobo M., Mendes D.A., Kozlova E., Rocha E.L., Rodrigues-Luiz G.F., Mascarin L., Dias G., Patricio D.O., Dierckx T., Bicca M.A., Bretton G., Tenorio de Menezes Y.K., Starick M.R., Rovaris D., Del Moral J., Mansur D.S., Van Weyenbergh J., Bafica A. An evolutionary recent IFN/IL-6/CEBP axis is linked to monocyte expansion and tuberculosis severity in humans. Elife. 2019;8 doi: 10.7554/eLife.47013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Londhe P., Davie J.K. Interferon-gamma resets muscle cell fate by stimulating the sequential recruitment of JARID2 and PRC2 to promoters to repress myogenesis. Sci. Signal. 2013;6:ra107. doi: 10.1126/scisignal.2004633. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.