Abstract
T cells are critical to fight pathogenic microbes and combat malignantly transformed cells in the fight against cancer. To exert their effector function, T cells produce effector molecules, such as the pro-inflammatory cytokines IFN-γ, TNF-α and IL-2. Tumors possess many inhibitory mechanisms that dampen T cell effector function, limiting the secretion of cytotoxic molecules. As a result, the control and elimination of tumors is impaired. Through recent advances in genomic editing, T cells can now be successfully modified via CRISPR/Cas9 technology. For instance, engaging (post-)transcriptional mechanisms to enhance T cell cytokine production, the retargeting of T cell antigen specificity or rendering T cells refractive to inhibitory receptor signaling can augment T cell effector function. Therefore, CRISPR/Cas9-mediated genome editing might provide novel strategies for cancer immunotherapy. Recently, the first-in-patient clinical trial was successfully performed with CRISPR/Cas9-modified human T cell therapy. In this review, a brief overview of currently available techniques is provided, and recent advances in T cell genomic engineering for the enhancement of T cell effector function for therapeutic purposes are discussed.
Keywords: CRISPR, Cas9, Genome editing, T cells, CAR T cells, T cell effector function, Cytokines
Abbreviations: AP-1, activator protein 1; ARE, AU-rich element; ARE-Del, deletion of the 3′UTR AREs from the Ifng/IFNG gene; CAR, Chimeric Antigen Receptor; Cas, CRISPR-associated; CRISPR, Clustered Regularly Interspaced Short Palindromic Repeat; CRS, cytokine release syndrome; CTLA-4, cytotoxic T-lymphocyte-associated protein 4; DGK, Diacylglycerol kinase; DHX37, DEAH-box helicase 37; FOXP3, Forkhead box P3; GATA, GATA binding protein; IFN, interferon; EBV, Epstein Barr virus; LAG-3, Lymphocyte Activating 3; IL, interleukin; NF-κB, nuclear factor of activated B cells; Pdia3, Protein Disulfide Isomerase Family A Member 3; PTPN2, Protein Tyrosine Phosphatase Non-Receptor 2; PD-1, Programmed cell Death 1; PD-L1, Programmed Death Ligand 1; RBP, RNA-binding protein; RNP, ribonuclear protein; TCR, T cell receptor; TGF, transforming growth factor; TIL, Tumor Infiltrating Lymphocyte; TLRs, Toll-like receptors; TNF, tumor necrosis factor; TRAC, TCR-α chain; TRBC, TCR-β chain; tTCR, transgenic TCR; UTR, untranslated region
1. Introduction
T cells are critical in maintaining protective immunity. As part of the adaptive immune system, T cells provide protection by eradicating infected cells and combating malignantly transformed cells. Indeed, high CD8+ T cell infiltrates in renal cell carcinoma and gallbladder tumors correlate with beneficial outcomes [1], [2]. To perform their effector function, T cells release effector molecules. These include granzymes, perforin and cytokines such as tumor necrosis factor (TNF)-α and interferon (IFN)-γ [3], [4], [5], [6], [7]. It was shown that T cell-derived TNF-α and IFN-γ are required for T-cell mediated killing of established tumors [8]. Likewise, a high IFNG gene signature is associated with beneficial clinical outcomes in patients receiving immunotherapy [9], [10], while copy number alterations of IFNG pathway genes correlate with poor immunotherapy responses [11].
Tumors possess many inhibitory mechanisms that dampen T cell effector function. Amongst others, tumor cells exploit T cell inhibitory receptors such as Programmed cell Death 1 (PD-1) by expressing their cognate ligand, i.e. Programmed Death Ligand 1 (PD-L1) [12], [13], but also by downregulating antigen presentation [14], [15], [16]. As a result, T cells lose the capacity to produce effector molecules, impairing tumor control and elimination. To circumvent this, several immunotherapy strategies have been designed to optimize T cell effector function. One form of therapy employs Tumor Infiltrating Lymphocytes (TILs) [17]. To generate T cells for adoptive TIL therapy, TILs are reprogrammed to reacquire the capacity to produce effector molecules, and are expanded in vitro for 4–5 weeks before reinfusion into patients [17], [18]. Most often, TIL therapy is used for the treatment of solid cancers [17], [19], such as melanoma [20], [21], [22], [23], [24]. Currently, the implementation of TIL therapy is being investigated for several other types of tumors [17], [25], including non-small cell lung cancer [18].
Another cellular immunotherapy approach makes use of genetically engineered T cells. By redirecting T cell antigen specificity, T cells can be specifically targeted to cells bearing a defined (subset of) antigen(s) [26], [27]. This can be achieved by the (viral) integration of a traditional T cell receptor (TCR) [28], [29], or a Chimeric Antigen Receptor (CAR) [30]. CAR-T cells are equipped with a receptor comprised of the variable region of a high affinity monoclonal antibody directed against a defined tumor antigen, e.g. CD19, fused to the signaling domain of CD3ζ and one or multiple signaling domain(s) from a costimulatory receptor for optimal T cell effector function [31], [32]. CAR T cell therapy is to date most successful in the treatment of lymphoid and myeloid tumors [33], [34], resulting in up to 90% complete remission rates in B-ALL patients treated with CD19 CAR-T cell therapy [35]. The translation to solid tumors is not yet effective due to the lack of tumor-specific antigens that can be targeted by CAR-T cells [33]. Of note, both transgenic TCRs and CARs are being investigated that target tumor neoantigens [36], [37], [38].
T cell effector function can also be enhanced through genetic editing. Several tools are available for the genetic modification of T cells, such as transcription activator-like effector nucleases, or TALENs, zinc finger nucleases, or ZFNs, and transposon-mediated genome editing [39], [40], [41], [42], [43]. Recently, also the Clustered Regularly Interspaced Short Palindromic Repeat (CRISPR) and CRISPR-associated (Cas) system was successfully applied in T cell cell-lines [44], but has also been used to genetically modify primary murine and human T cells [45], [46], [47], [48], [49]. Specifically, CRISPR/Cas9 has been employed to directly enhance T cell effector function [48], [50], [51], [52], [53], [54], [55], [56], disable inhibitory receptor expression [45], [57], [66], [67], [68], [58], [59], [60], [61], [62], [63], [64], [65], or to redirect T cell antigen specificity by targeting TCRs or CARs to the endogenous TCR-α chain (TRAC) locus [28], [29], [30], [69], [70], [71]. In fact, recently, the first in-patient trial has been performed utilizing CRISPR/Cas9-edited T cells [72]. In this review, a short overview of currently available CRISPR-based techniques for the genetic modification of T cells is provided, and recent advances in T cell genomic engineering aiming to enhance T cell effector function to improve cellular therapies are discussed.
2. CRISPR basics and different CRISPR/Cas9 tools available for T cells
CRISPR/Cas9-mediated gene-editing requires three components: the Cas9 DNA-nuclease, a targeting CRISPR RNA, or crRNA, and a trans-activating CRISPR RNA, or tracrRNA, of which the latter two can be supplied either separately or combined as a single guide RNA [73]. The targeting CRISPR RNA guides the Cas9 ribonuclear protein (RNP) complex to the cleavage site via DNA base pairing, and the trans-activating CRISPR RNA facilitates Cas9 activation [73]. However, Cas9-mediated DNA cleavage only occurs when the target site is directly adjacent to a 3′ protospacer-adjacent motif, or PAM [74], a 3–8 base pair DNA sequence [73], [75]. The CRISPR-system can therefore be targeted to virtually any DNA region of interest.
Currently, several delivery methods are available to introduce the CRISPR/Cas9 system in T cells (Table 1). Each of them has benefits, but also drawbacks. For instance, one of the first delivery methods developed were lentiviral vectors for Cas9-delivery. These have been used to target e.g. the CCR5 locus in human T cells, but resulted in low knock-out efficacies [76], [77]. Furthermore, stable expression of gene-editing factors is not desirable as this could result in increased off-target effects [78], and viral gene transfer has posed significant risks in the past [79], [80]. Furthermore, it was recently shown that the serum of a majority of healthy donors and cord bloods contains Cas9 antibodies [81], [82], which could potentially trigger immune responses upon peptide presentation or secretion by stable Cas9-expressing cells. Therefore, transient expression or presence of the CRISPR/Cas9 system is preferred to prevent unwanted side effects. This can be achieved by for instance receptor-mediated Cas9 uptake through the addition of fusion tags [83], lipid-based vector delivery [84], [85], or cell-membrane penetration via the addition of cell-penetrating peptides [86], [87], [88]. Of note, these novel methods have not been tested yet in T cells. Another non-integrative strategy makes use of membrane disruption. Through electroporation, Cas9 can be introduced as a plasmid, mRNA, Cas9 protein, or Cas9 RNPs [48], [73], [89], [90], [91]. While electroporation can be costly and laborious to optimize, several protocols describing the use thereof in T cells have been recently published [46], [92], [93]. Of note, while the electroporation of T cells was shown to be potentially cytotoxic, resulting in up to 95% loss of e.g. viable T cells in the first 24 h after electroporation [94], T cells retain their expansion potential after electroporation-based CRISPR/Cas9-mediated genome editing [48]. Thus, due to the transient presence of the CRISPR/Cas9 system and the retention of T cell expansion potential, electroporation-based strategies might be most optimal for the use of Cas9-mediated genome editing in T cells for therapeutic purposes.
Table 1.
CRISPR-tools.
| Type | Advantages | Disadvantages | Applied in primary T cells | Ref |
|---|---|---|---|---|
| Lentiviral | Inclusion of selection marker | Low knock-out efficacy Genomic integration |
Yes | [76], [77] |
| Electroporation | T cells retain expansion potential Extensive protocols are available |
Cytotoxic Costly | Yes | [48], [73], [89], [90], [91] |
| (Lipid) nanoparticles | Highly adaptable to specific need | Complex to engineer | No | [84], [85] |
| Ligand fusion tags | Cell-type specific | Cells need to express receptor | No | [83] |
| Cell-penetrating peptides | Produced in-house | Varying quality and efficacy due to batch-to-batch differences | No | [86], [87], [88] |
3. Enhancing T cell effector function via CRISPR
The CRISPR/Cas9 toolbox can be used for silencing or knocking out genes [48], [95], [96], knocking in genes [44], [90], and for the induced activation of genes in T cells [97], [98], [99], [100], [101]. Through these different approaches, various research questions have been investigated in T cells. These include diverse topics such as preventing HIV infection in T cells [90], [92], [102], regulation of T cell effector molecule production [48], TCR signaling [51], [52], redirecting T cell antigen specificity [28], [29], [30], [69], [70], [71], stabilization of regulatory T cell phenotypes [98], and investigating pathogenic cytokine receptor expression by T cells [96], [99]. Here, I focus on the different approaches that have been taken to augment T cell effector function for immunotherapy purposes by enhancing cytokine production, blocking inhibitory receptor signaling, or redirecting T cell antigen specificity through CRISPR/Cas9 induced knock-outs.
3.1. Using CRISPR to directly augment cytokine production
The CRISPR/Cas9 system has been used to directly enhance T cell function (Table 2). The production of effector molecules by T cells is regulated on multiple levels (Fig. 1). The magnitude of antigen-specific T cell activation is determined by the integration of TCR-signaling and costimulatory signals [3], [6], [103]. TCR triggering results in the engagement of downstream signaling pathways and regulatory processes [104], [105]. The threshold for T cell activation can be lowered via costimulatory signals, such as through the engagement of costimulatory receptors, e.g. CD28 and/or CD27 [103], [106], [107], triggering of cytokine receptors via cytokines such as interleukin (IL)-2 and IL-12 [108], [109], or even innate-like sensing of pathogens through pattern recognition receptors [110], such as Toll-like receptors (TLRs) [6], [110], [111]. Together, these stimulatory signals determine the cytokine output of T cells.
Table 2.
Direct enhancement of T cell effector function via CRISPR/Cas9-mediated genome editing.
| Species | Target |
CAR |
In vitro |
In vivo |
Ref | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Protein | IFN-γ | TNF-α | IL-2 | IL-17 | CD107a | GzmA | GzmB | GzmC | GzmD | Cytotoxicity | Model | IFN-γ | IL-2 | GzmB | Tumor | |||
| Human | DGKA/DGKZ | DGKα/ζ | EGFRvIII | + | + | + | Glioblastoma | + | + | R | [51] | ||||||||
| IFNG 3′UTR | IFN-γ | + | = | = | [48] | ||||||||||||||
| PTPN2 | PTPN2 | Mesothelin | + | = | = | + | [52] | ||||||||||||
| TGFBR2 | TGFBR2 | + | + | + | + | + | Lung cancer | + | R | [56] | |||||||||
| Mouse | Dhx37 | Dhx37 | + | + | + | + | + | + | E0771-OVAMammary tumor | D | [53] | ||||||||
| Gata3 | Gata3 | B16-F10 melanoma | + | + | D | [50] | |||||||||||||
| Nr2f6 | Nr2f6 | + | B16-OVA melanoma | D | [55] | ||||||||||||||
| Zcd3h12a | Regnase-1 | B16-OVA melanoma | + | + | + | D | [54] | ||||||||||||
+, increased production; =, equal production; R, tumor regression; D, tumor outgrowth delayed. Blank indicates not reported.
Fig. 1.
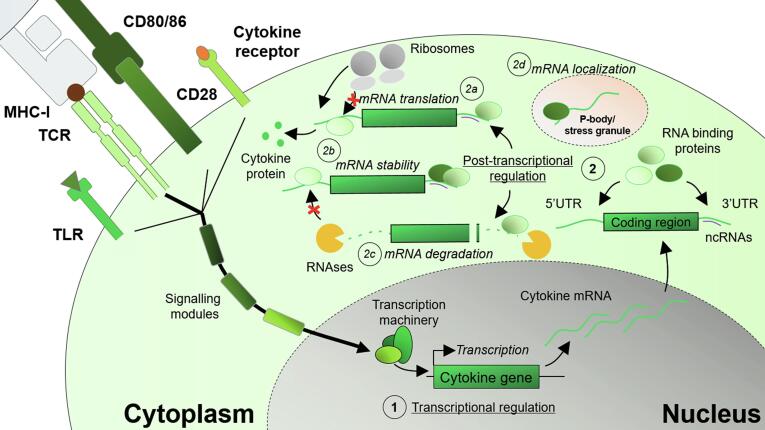
Transcriptional and post-transcriptional machinery engaged during T cell activation. Integration of stimulatory signals, i.e. engagement of the TCR, triggering of co-stimulatory receptors such as CD28, sensing of pathogens through TLRs, and stimulation via cytokine gradients through cytokine receptors results in the engagement of downstream signaling cascades and the activation of transcription factors. (1) Transcriptional regulation determines the amount of cytokine mRNA that is produced. After mRNA maturation and export to the nucleus, (2) post-transcriptional mechanisms determine the protein output. RNA binding proteins and non-coding RNAs, such as microRNAs, bind to regulatory elements present in the 5′ and 3′UTR of mRNA transcripts. Depending on the binding repertoire, (2a) mRNA translation is enhanced or hampered, (2b) mRNA stability is influenced, (2c) mRNA is actively degraded by recruited RNAses, or (2d) mRNAs are targeted to a different location, i.e. stress granules or P-bodies.
A major determinant of cytokine production is the strength of the TCR signal that T cells receive [6], [104]. Unsurprisingly, T cell activation can be amplified by direct enhancement of TCR signaling. Diacylglycerol kinases (DGKs) are enzymes that catalyze diacylglycerol metabolism, and have been shown to interact with critical signaling modules downstream of the CD3 co-receptor, such as protein kinase C and Ras activating protein [112]. Through diacylglycerol phosphorylation, DGKs inhibit CD3 signaling and T cell effector function [112]. DGKs have been suggested as potential immunotherapy targets, and it was recently shown that CRISPR-mediated DGKA and DGKZ double knock-out human T cells exhibit increased IFNG and IL2 transcription, enhanced IFN-γ and IL-2 production, and increased cytotoxicity in vitro [51]. Furthermore, in a glioblastoma xenograft model, DGKA/Z double knock-out human T cells expressing an EGFRvIII CAR provided complete protection against tumor outgrowth, in contrast to mice treated with WT EGFRvIII CAR T cells, where tumors initially shrunk but rapidly grew out again [51].
Another negative regulator of TCR signaling is protein tyrosine phosphatase non-receptor 2 (PTPN2) [113], [114]. Human PTPN2 knock-out T cells generated via CRISPR/Ca9s-mediated genome editing exhibit enhanced calcium flux and IFN-γ and IL-17 production upon TCR stimulation [52]. However, Ptpn2-/- mice exhibited CD8+ T cell mediated autoimmunity [113], therefore, PTPN2 knock-out might not be optimal for CD8+ T cell-based therapeutic purposes.
Together, these studies indicate that while there is potential for the enhancement of T cell effector function through the direct augmentation of TCR signaling, care should be taken to prevent unbridled T cell activation that might result in pathologies. Furthermore, due to the advent of CAR T cells, where multiple signaling domains from costimulatory receptors can be incorporated into one signaling module for optimal antigen-specific T cell activation [31], [32], [115], CRISPR/Cas9-mediated enhancement of T cell effector function through the modulation of TCR signaling might be suboptimal.
While CARs have been designed for optimal signal transduction, T cell cytokine production is regulated by both transcriptional and post-transcriptional events to prevent the unwanted production and release of these potentially cytotoxic mediators (Fig. 1) [105], [116], [117]. On the transcriptional level, epigenetic markers present on DNA and histones, and transcription factor availability, localization and phosphorylation determine the amount of RNA that is produced [118]. For example, demethylation of the IFNG locus only occurs in effector and in memory T cells [119]. This allows for rapid locus accessibility by transcription factors upon T cell activation to drive the transcription of IFNG RNA [119].
Multiple transcription factors are important for driving the production of T cell cytokines such as IFN-γ, including nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) and activator protein 1 (AP-1) [120], [121]. One of the interacting partners of NF-κB identified in a genome-wide CRISPR/Cas9 screen investigating immunotherapy targets is the DEAH-box helicase 37, or DHX37 [53]. DHX37 expression in TILs from breast cancer patients correlates with poor outcomes [53]. Potentially, this is due to DHX37-induced T cell dysfunction. Dhx37 knock-out murine T cells generated through CRISPR/Cas9-mediated genome editing exhibit increased expression of Gzmc and Gzmd, increased Granzyme B and IFN-γ production and increased cytotoxicity in vitro. Furthermore, Dhx37 knock-out T cells delayed tumor outgrowth in a murine mammary tumor model [53].
Similarly, DNA binding by AP-1 is hindered by the nuclear zinc-finger orphan receptor NR2F6, preventing cytokine production in T cells [122]. Indeed, CRISPR/Cas9-mediated Nr2f6 knock-out murine T cells produce more IFN-γ, and delayed tumor outgrowth in a B16-OVA model compared to WT T cells [55].
Another approach taking advantage of the transcriptional machinery is knocking out transcription factors that inhibit the production of pro-inflammatory cytokines. For instance, GATA binding protein 3 (GATA3) is regarded as the master transcription factor for inducing a type 2 phenotype in T cells [123], and has been shown to significantly inhibit IFN-γ production in T cells [124]. GATA3 was also identified as the top ranking transcription factor involved in the dysfunctional transcriptional program in T cells from melanoma patients [50]. Gata3 knock-out murine T cells generated via CRISPR/Cas9-mediated genome editing showed enhanced capacity for the production of IFN-γ and IL-2 while less Gata3 knock-out T cells produced IL-10 ex vivo in a B16-F10 melanoma model [50]. Furthermore, Gata3 knock-out T cells also delayed tumor outgrowth [50].
Similarly, also Forkhead box P3 (FOXP3), the master regulator of the regulatory T cell transcriptional profile [125], inhibits the production of cytotoxic T cell effector molecules such as IFN-γ [126]. One of the factors involved in FOXP3 upregulation is transforming growth factor (TGF)-β [127]. CRISPR/Cas9-mediated knock-out of TGFBR2, one of the TGF-β receptors, in Mesothelin CAR-T cells increased IFNG, GZMA and GZMB expression and IFN-γ and IL-2 production in vitro by preventing FOXP3 upregulation and thus reducing induced regulatory T cell formation [56]. TGFBR2 knock-out Mesothelin CAR-T cells retain their lytic capacity and exhibit enhanced in vivo tumor elimination compared to WT Mesothelin CAR-T cells in a murine lung cancer model [56].
Together, these reports identify several approaches through which transcriptional regulation can be exploited to enhance T cell effector function. Future research will determine whether modulation of transcription factor activity can be employed in treatment strategies.
The combined output of transcription factors determines the T cell transcriptome. However, another layer of regulation exists, determining the protein output of T cells (Fig. 1) [105], [117]. After RNA splicing and nuclear export of mature mRNA molecules, post-transcriptional regulatory events determine mRNA half-life, subcellular localization and translational rate, and thus the amount of protein that is produced [116]. Post-transcriptional regulation is mediated by microRNAs, (long) non-coding RNAs, RNA-binding proteins (RBPs) and mRNA modifications [116], [128], [129], [130], [131]. In the past years, it has become increasingly clear that post-transcriptional regulation plays a major role in determining the timing and magnitude of T cell responses [48], [104], [105], [116], [117], [132], [133]. These processes rely, in part, on the binding of RBPs upon recognition of cis-elements present in mRNA transcripts [116]. These cis-elements are located in either the 5′ or 3′ untranslated region (UTR) of mRNA molecules [116], [134]. Many cytokine genes contain at least one cis-regulatory element [116]. Thus, the protein production of most cytokines is, at least partially, determined by post-transcriptional events.
A critical class of cis-elements driving post-transcriptional regulatory events are AU-rich elements (AREs) encoded in the 3′UTR of many cytokine genes [105], [116], [135], [136], [137], [138], [139]. These AREs are critical to fine-tune the production of the prototypical T cell cytokines IFN-γ, TNF-α and IL-2 [135], [139], [140], [141]. For instance, while antigen-experienced T cells express Ifng mRNA in a resting state, IFN-γ protein is only produced upon T cell activation [104]. Upon deletion of the 3′UTR AREs from the Ifng gene (ARE-Del) in a murine model, ex vivo resting antigen-experienced ARE-Del T cells produce IFN-γ due to a failure to keep the Ifng mRNA translationally quiescent [139]. Furthermore, upon deletion of the AREs in the 3′UTR of IFN-γ, ARE-Del T cells produce more IFN-γ in vitro due to increased mRNA half-life [139], [142]. This enhanced mRNA stability leads to prolonged IFN-γ production [141], a feature that is critical for anti-tumor responses. Indeed, upon ARE-Del T cell therapy of B16-OVA melanoma-bearing mice, tumor outgrowth was significantly delayed compared to mice treated with wild type T cells [141].
The 3′UTR of the IFNG gene is conserved between mice and men [139]. Recently, these results were translated to human T cells [48]. By making use of CRISPR/Cas9 technology, primary human T cells with a 3′UTR ARE-deletion in the IFNG gene were generated [48]. Similar to their murine counterparts [141], also more human ARE-Del T cells produced IFN-γ, which was at least in part driven by increased IFNG mRNA stability [48]. This increased IFN-γ production capacity was also observed upon TCR engineering [48]. More MART-1 TCR-expressing ARE-Del human T cells produced IFN-γ compared to WT MART-1 TCR expressing T cells in a co-culture with patient-derived MART-1+ melanoma tumor cells [48].
TNF-α production could also be enhanced by similar exploitation of post-transcriptional regulatory mechanisms. TNF-α is also (partially) regulated via the AREs present in the 3′UTR of Tnfa, and removal of the 3′UTR AREs from the Tnfa locus in mice results in constitutive TNF-α production [135]. However, this constitutive TNF-α production elicits inflammatory diseases [135], and thus might not be optimal for use in human therapies. Furthermore, given the dual role TNF-α plays in both anti-tumor responses and promoting tumor growth [143], the exploitation of TNF-α post-transcriptional regulatory mechanisms should first be thoroughly investigated prior to use in the clinic.
A more indirect approach to interfere with post-transcriptional regulation is by modulating the expression of RBPs, which govern the effect of 5′ and 3′UTR cis-regulatory elements. For instance, while not directly binding to the 3′UTR of Ifng [144], T cells from Regnase-1 knock-out mice exhibited enhanced IFN-γ production [144]. This was due to deregulation of the Roquin family of proteins [144], known RBPs which have been previously implicated in the regulation of IFN-γ production [145]. Also murine Regnase-1 knock-out T cells generated via CRISPR/Cas9 exhibited increased IFN-γ production [54]. Additionally, also Granzyme B production was increased [54], a phenotype that was also observed in Regnase-1 knock-out mice [145].
Modulation of the post-transcriptional machinery governing cytokine production provides novel angles to enhance T cell effector function for therapeutic applications. While it has not yet been established whether the regulatory capacity of e.g. Regnase-1 is conserved in human T cells, other post-transcriptional mechanisms are conserved between mice and men and can further enhance TCR-engineered T cell effector function [48]. Potentially, this can be exploited in the future for therapeutic applications.
Together, these studies provide many opportunities for the direct augmentation of T cell effector function via CRISPR/Cas9-mediated genome editing. As regulation of T cell effector function is mediated on multiple levels [105], [118], potentially, the engagement of multiple pathways can result in T cells with the most optimal T cell effector function. This, however, remains to be determined.
3.2. Disabling inhibitory receptors to enhance anti-tumor T cells
Within tumors, T cells encounter many inhibitory signals in the suppressive tumor microenvironment, including the ligands for inhibitory receptors present on T cells. To date, many T cell inhibitory receptors have been identified, such as cytotoxic T-lymphocyte-associated protein 4 (CTLA-4), Lymphocyte Activating 3 (LAG-3) and PD-1 [146], [147]. Tumors exploit these checkpoint receptors to dampen T cell effector function as a means of immune escape [12], [13]. To circumvent this, inhibitory receptors are being targeted in antibody-mediated immunotherapy approaches [146]. These antibody-therapies, mostly targeting the PD-1/PD-L1 axis or CTLA-4, prevent the triggering of these receptors [12]. As a result, T cell effector function is restored, and anti-tumor responses can be mounted [148]. This type of therapy is employed in i.e. melanoma patients, resulting in an extraordinary 3-year survival of approximately 50% of patients [17]. Furthermore, clinical trials are also underway evaluating the efficacy of combined anti-PD-1/PD-L1 immunotherapy and adoptive cellular therapy with TILs [17]. CRISPR/Cas9 could also be utilized to render TILs refractive to inhibitory signals by knocking out inhibitory receptors prior to infusion. With this approach, systemic immunomodulatory antibody therapy is not required, which has been shown to be pose significant side effects [149]. A summary of the effects of CRISPR/Cas9-mediated abrogation of inhibitory receptor signaling on T cell effector function is provided in Table 3.
Table 3.
Abrogating inhibitory receptor signaling via CRISPR/Cas9-mediated genome editing to augment T cell effector function.
| Target | CAR |
In vitro |
In vivo |
Ref | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IFN-γ | TNF-α | IL-2 | CD107a | GzmA | GzmB | GzmC | GzmD | Cytotoxicity | Model | IFN-γ | IL-2 | GzmB | Tumor | |||
| PD-1 | + | + | + | + | [57], [58], [59] | |||||||||||
| + | + | Melanoma | D | [60] | ||||||||||||
| CD19 | + | + | Myeloid leukemia | R | [45] | |||||||||||
| CD133 | = | = | = | + | Glioma | D | [63] | |||||||||
| Glypican-3 | + | + | + | Hepatocellular carcinoma | + | + | D | [62] | ||||||||
| Mesothelin | + | + | + | Breast cancer | D | [64] | ||||||||||
| CTLA-4 | + | + | [66] | |||||||||||||
| + | + | Colorectal carcinoma | D | [65] | ||||||||||||
| LAG-3 | = | = | Raji Burkitt’s lymphoma | R | [67] | |||||||||||
+, increased production; =, equal production; R, tumor regression; D, tumor outgrowth delayed. Blank indicates not reported.
One of the most-studied inhibitory receptors is PD-1. The exact mode of action of inhibitory PD-1 signaling on T cell effector function remains unclear. PD-1 triggering recruits the phosphatase SHP-2 to the TCR complex [150]. SHP-2 then dephosphorylates TCR signaling molecules such as CD3ζ and ZAP70 [150]. Subsequent work showed that SHP-2 dephosphorylates the CD28 costimulatory receptor instead [151], [152]. However, yet another mode of action of PD-1 triggering was identified recently [141]. PD-1 triggering also modulates the production of IFN-γ in T cells independently from blocking the CD28 signaling pathway [141]. Neither CD28 costimulation nor blocking PD-1 signaling via antibodies enhances the Ifng mRNA transcription of murine CD8+ T cells [141]. Both rather function by acting on different post-transcriptional regulatory mechanisms [141]. Nonetheless, the therapeutic benefit of interfering with PD-1 signaling is evident [17], and therefore it is unsurprising that knocking out PD-1 via CRISPR/Cas9 technology has also been thoroughly investigated.
The effect of CRISPR/Cas9-mediated PD-1 knock-out was first assessed in primary human Epstein Barr virus (EBV)-specific T cells in healthy donors [57], [58]. Knocking out PD-1 with CRISPR/Cas9 increased the production of IFN-γ, TNF-α and IL-2 and the degranulation capacity of T cell cell-lines and EBV-specific T cells from healthy donors in vitro [57], [58]. This was replicated in T cells isolated from an EBV+ gastric cancer patient, where PD-1 knock-out in patient-derived T cells increased not only inflammatory cytokine production but also resulted in the production of less IL-10 compared to wildtype T cells upon EBV peptide stimulation [57]. Similarly, also PD-1 deficient T cells generated via CRISPR/Cas9 mediated genome editing derived from a melanoma patient produced more IFN-γ upon stimulation with several melanoma-associated peptides compared to PD-1 sufficient T cells from the same patient [59]. Furthermore, human PD-1 knock-out T cells primed by patient-derived multiple-myeloma lysate-pulsed DCs exhibited increased IFN-γ and TNF-α production in vitro and delayed tumor outgrowth in a murine xenograft model, increasing survival [60].
Not only direct genetic editing of PD-1 results in the lack of expression thereof. Post-translational modifications of proteins can affect protein function, half-life and localization [153]. For instance, glycosylation is important for the expression of surface molecules on the cell membrane [154]. This was also shown to be the case of PD-1. Fut8, a fucosyltransferase, was identified to be important for the membrane expression of PD-1 in murine T cells via a CRISPR/Cas9-based screening [61]. Furthermore, Fut8 knock-out murine T cells generated via CRISPR/Cas9-mediated genome editing also expressed less PD-1 in vivo [61]. This phenotype could be replicated via a small molecule inhibitor that inhibits the core fucosylation machinery, resulting in increased IFN-γ and IL-2 production in vitro and delayed tumor outgrowth in a murine B16-OVA melanoma model [61]. However, as noted, fucosylation is important for the location, function, and half-life of many proteins, and therefore, this approach might not be optimal for use in therapeutic applications.
Another immunotherapy approach is the use of cellular therapies, containing either TILs or genetically modified T cells. Indeed, CD19 CAR-T cells have been genetically manipulated with CRISPR/Cas9 to knock out PD-1 [45]. PD-1 knock-out CD19 CAR-T cells exhibited increased in vitro degranulation as measured by CD107a staining [45]. In a murine myeloid leukemia xenograft model, CRISPR/Cas9-generated PD-1 knock-out CD19 CAR-T cells were also able to clear the tumor, in contrast to wildtype CD19 CAR-T cells [45]. Similarly, also Glypican-3 CAR T-cells and CD133 CAR-T cells with a CRISPR/Cas9-mediated PD-1 knock-out exhibited enhanced effector function in vitro and prolonged survival in hepatocellular carcinoma and glioma xenograft models [62], [63].
Also human PD-1 knock-out Mesothelin CAR-T cells exhibited enhanced ex vivo lysis and IFN-γ production compared to PD-1+ mesothelin CAR-T cells [64]. Strikingly, PD-1 knock-out Mesothelin CAR-T cell therapy also outperformed Mesothelin CAR-T cell therapy with antibody-mediated PD-1 blockade in an in vivo mammary gland tumor xenograft model [64]. Of note, a xenograft model showed that PD-1 knock-out CD133 CAR-T cells do not proliferate uncontrollably in vivo [63].
Together, these studies show the feasibility of CRISPR/Cas9-mediated disruption of PD-1 signaling and the potent effect thereof on augmenting T cell effector function. Knocking out PD-1 can even further enhance the efficacy of CAR-T cell treatment in murine models, does not result in uncontrolled in vivo expansion and even outperforms antibody-mediated PD-1 blockade, providing in vivo evidence of the potential benefit of CRISPR-mediated augmentation of CAR-T cell function.
Less research has been performed investigating the direct knock-out of other inhibitory receptors, such as CTLA-4 and LAG-3 in T cells. CRISPR-mediated CTLA-4 knock-out has shown that CTLA-4 knock-out in human T cells increases the amount of IFN-γ and TNF-α produced in vitro [65], [66], and prolongs survival in a murine colorectal carcinoma xenograft model [65]. Likewise, also LAG-3 knock-out human CD19 CAR-T cells provide prolonged survival in a Burkitt lymphoma xenograft model [67].
Summarizing, these studies highlight the potential of CRISPR/Cas9-mediated knock-out of inhibitory receptors such as PD-1 and CTLA-4 to render T cells refractory to inhibitory receptor signaling, potently enhancing T cell effector function in vitro and in vivo.
3.3. Redirecting T cell antigen specificity
Redirecting the antigen specificity of T cells allows for the treatment of tumors expressing defined sets of antigens. The generation and effector function of TCR transgenic T cells can be further amplified by making use of CRISPR/Cas9 technology (Table 4). This has mostly been studied in the context of traditional TCR gene modification. For instance, it was shown for MLANA (MART-1, melanoma) and HER2/neu (breast cancer) TCRs that knocking the transgenic TCR into the endogenous TRAC locus enhanced TCR expression compared to TRAC WT T cells [71]. This was even further increased when also the TCR-β chain (TRBC) was also knocked out [71]. Enhanced transgenic TCR expression was also shown for the TRAC knock-in of TCRs specific for BOB (multiple myeloma), HA-1 (multiple myeloma), MPO (myeloid leukemia) and PRAME (melanoma) peptides [28], [29]. Furthermore, HA-1 TCR TRAC knock-in engineered T cells showed enhanced anti-tumor capacity in a multiple myeloma xenograft model [29].
Table 4.
Redirecting T cell antigen specificity by knocking in transgenic TCRs (tTCR) into the TRAC locus.
| TCR/CAR | Antigen | Target gene |
TRAC KI | Enhanced tTCR expression |
In vitro |
In vivo |
Ref | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TRAC | TRBC | IFN-γ | TNF-α | IL-2 | Cytotoxicity | Model | Tumor | |||||
| TCR | MLNA | Y | Y | Y | Y | [71] | ||||||
| HER2/Neu | Y | Y | Y | Y | [71] | |||||||
| BOB | Y | Y | [29] | |||||||||
| HA-1 | Y | Y | Y | + | + | Multiple myeloma | D | [29] | ||||
| MPO | Y | Y | Y | = | = | Myeloid leukeumia | = | [28] | ||||
| PRAME | Y | Y | [29] | |||||||||
| CMV | Y | Y | [29] | |||||||||
| CMV | Y | Y | Y | =/+ | = | = | [71] | |||||
| CAR | CD19 | Y | Y | Y | Leukemia | D | [30] | |||||
+, increased production; =, equal production; D, tumor outgrowth delayed; Y, yes. Blank indicates not reported.
Also CARs can be targeted to the TRAC locus after CRISPR/Cas9-mediated knock-out. CD19 CAR TRAC knock-in enhanced T cell effector function in a murine leukemia xenograft model, as CD19 CAR TRAC knock-in T cells were less vulnerable to exhaustion compared to conventional CD19 CAR-T cells [30].
Of note, several studies have also shown enhanced CMV-specific TCR expression upon knock-in into the TRAC locus [29], [71]. While used as a negative control, this could prove beneficial, as virus-specific bystander CD8+ T cells have been shown to populate tumors [69], and can be reactivated to elicit tumor control in murine models [70].
Together, these studies show the merit of using CRISPR/Cas9-mediated genome editing to enhance the anti-tumor capacity of TCR-engineered/CAR-T cells for therapeutic applications. When deemed safe for patients, this type of approach should also be considered for clinical use.
3.4. Multiplex CRISPR-editing for a multipronged approach to optimize T cell products
While CRISPR/Cas9 can be used to enhance T cell effector function directly (Table 2), to limit T cell dysfunction by abolishing the expression of inhibitory receptors (Table 3), or to redirect antigen specificity (Table 4), these approaches can also be multiplexed to maximize the effect thereof (Table 5). For instance, CD19 CAR-T cells that have been modified to lack expression of TRAC and PD-1 exhibited increased IFN-γ production and cytotoxicity in vitro compared to either TRAC-expressing or TRAC- and PD-1-expressing CD19 CAR T cells [155]. Furthermore, these PD-1-TCR-α- CAR T cells provided protective immunity in a Raji xenograft model [155] and a prostate cancer model [156].
Table 5.
Multiplex CRISPR/Cas9 approaches to maximize T cell effector function.
| TCR/CAR | Antigen | Target gene |
In vitro |
In vivo |
Ref | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TRAC | TRBC | B2M | PD-1 | Pdia3 | IFN-γ | TNF-α | IL-2 | GzmA | GzmB | GzmC | Cytotoxicity | Model | Tumor | |||
| CAR | CD19 | Y | Y | = | = | = | Leukemia | D | [158] | |||||||
| Y | Y | Y | + | + | + | Raji Burkitt’s lymphoma | R | [155] | ||||||||
| Y | Y | Y | Raji Burkitt’s lymphoma | R | [156] | |||||||||||
| EGFRvIII | Y | Y | Y | + | + | Glioblastoma | = | [161] | ||||||||
| Y | Y | + | + | + | + | Glioblastoma | D | [163] | ||||||||
| TCR | NY-ESO-1 | Y | Y | Y | [72] | |||||||||||
+, increased production; =, equal production; D, tumor outgrowth delayed; Y, yes. Blank indicates not reported.
This multiplex approach can also be utilized to produce “universal T cells”. Currently, approved CAR-T cell therapies are produced on a custom-made basis from patient-derived PBMCs. This is a costly and time-consuming process for patients who are at a high risk. Additionally, cellular products might not meet release criteria, even further endangering patients. One way to circumvent this would be to make use of universal T cells, which have been genetically engineered to lack expression of MHC-I molecules, such as B2M, and/or TCR α and/or β chains, limiting graft versus host disease and graft rejection [157]. Universal T cells can be generated via CRISPR/Cas9-mediated multiplex genome editing [158], [159]. By knocking out the TRAC [160], or TRAC and B2M [158], high levels of CD19 CAR-expressing T cells could be generated that exhibited robust anti-tumor responses in murine xenograft models [158], [160]. Furthermore, multiplex editing could also be employed to generate “off-the-shelve” inhibitory signaling-refractory CAR-T cells. For instance, CD19 CAR-T cells lacking the expression of both B2M and PD-1 have been generated [161], [162] . Similarly, also EGFRvIII CAR+TRAC-PD-1-B2M- CAR T cells were generated via CRISPR/Cas9 mediated genome editing. These cells exhibited enhanced IFN-γ and TNF-α production in vitro, but did not enhance survival of mice in a murine xenograft glioblastoma model compared to mice treated with WT EGFRvIII CAR-T cells [161].These studies highlight the potential of CRISPR/Cas9-mediated genome editing to supplement and further enhance T cell therapies, paving the way to an “on-the-shelf” solution for patients.
Also the direct enhancement of T cell effector function can be combined in a multiplexed CRISPR-approach. For instance, Protein Disulfide Isomerase Family A Member 3 (Pdia3) was identified in an in vivo CRISPR/Cas9 screen to regulate T cell effector function in a glioblastoma model [163]. Pdia3 knock-out combined with EGFRvIII CAR TRAC knock-in via CRISPR/Cas9-mediated genome editing lowered the TCR threshold required for T cell activation, resulting in higher IFN-γ production upon TCR activation compared to Pdia3+ T cells [163]. EGFRvIII CAR TRAC knock-in Pdia3- T cells also protected against tumor outgrowth in a glioblastoma model [163].
Recently, CRISPR/Cas9-modified T cells have been applied as a therapeutic agent [72]. While TIL therapy is highly successful for the treatment of melanoma [17], attempts are still being made to further enhance T cell therapy. As previously discussed, one possibility currently under investigation is the combination of TIL therapy and anti-PD-1 therapy. Also the recent first-in-patient-trial where CRISPR/Cas9-mediated gene-edited T cells were administered investigated the targeting of PD-1. Stadtmauer et al. (2020) utilized CRISPR/Cas9 gene editing to abolish the expression of PD-1 and to target a NY-ESO-1 TCR to the TRAC locus in human T cells [72]. These multiplex-edited T cells were administered to 3 patients with refractory advanced cancer, and none of the patients experienced cytokine release syndrome (CRS) [72]. CRS is one of the main adverse events reported during TIL and CAR-T cell therapy [164], [165], with up to 100 percent of patients treated with CD19 CAR-T cells experiencing CRS [165]. While symptoms of CRS can be mild, including mild flu-like symptoms, CRS can also result in severe life-threatening inflammatory conditions [165]. None of the three patients receiving CRISPR/Cas9-mediated gene edited T cells experienced CRS [72], highlighting the safety of this treatment. Furthermore, at the end of the trial, 2/3 patients achieved stable disease [72].
Summarizing, multiplexed CRISPR-approaches to augment T cell effector function is feasible and has even been employed as a therapeutic application. While this trial was only the first of its kind, the results are hopeful, and indicate that CRISPR/Cas9-mediated genome editing has a place in augmenting the efficacy of T cell therapies.
4. Conclusion and outlook
Since the introduction of the CRISPR/Cas9 toolbox, genome editing in T cells has taken flight. Through Cas9-mediated genome editing, the manipulation of large numbers of cells is feasible, a feature that is critical for therapeutic applications. With the novel opportunities provided by the ease of CRISPR/Cas9-mediated genome editing, new ideas have been explored to enhance T cell effector function directly, prevent T cell dysfunction through interfering with inhibitory receptor signaling and redirect T cell antigen specificity. Together, these approaches have led to T cells with a redirected antigen-specificity and enhanced effector function, as measured by cytokine production, granular content, cytotoxicity or protection in murine models. These advances have led to the recent first-in-patient clinical trial with CRISPR/Cas9-modified human T cells. While only three patients were treated, the adoptive therapy with genetically engineered T cells was deemed safe, providing the first results for the clinical use of CIRPS/Cas9-engineered T cells in patients. Nonetheless, rigorous testing is necessary to determine the efficacy and safety of CRISPR/Cas9-enhanced T cells for adoptive therapy, with a focus on safety. In particular, off-target genetic editing, in-patient T cell expansion and survival, and the potential induction of CRS need to be thoroughly investigated. Lastly, while the current CRISPR/Cas9 methods are suitable for the manipulation of small numbers of cells, the editing of the large numbers of cells required to for patient treatment might still be sub-optimal. Thus, for the clinical use of CRISPR/Cas9-mediated genome editing in for instance TIL-products, novel CRISPR/Cas9 approaches should be developed. Once these hurdles have been overcome, multiplex approaches incorporating both the maximization of T cell effector function and the use of antigen-specificity redirection via transgenic TCRs or CARs for optimized T cell activation and targeting with the CRISPR/Cas9 toolbox will allow for the production of universal T cells with augmented effector function for therapeutic applications.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Availability of data and materials
Not applicable.
Funding
Author has no funding to declare.
Authors contributions
JJFH conceived the manuscript topic, performed literature research, and wrote the manuscript.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Author would like to thank AE Oja (Sanquin Research, Amsterdam, The Netherlands) for critical input.
References
- 1.Fluxá P., Rojas-Sepúlveda D., Gleisner M.A., Tittarelli A., Villegas P., Tapia L., Rivera M.T., López M.N., Catán F., Uribe M., Salazar-Onfray F. High CD8+ and absence of Foxp3+ T lymphocytes infiltration in gallbladder tumors correlate with prolonged patients survival. BMC Cancer. 2018;18 doi: 10.1186/s12885-018-4147-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ghatalia P., Gordetsky J., Kuo F., Dulaimi E., Cai K.Q., Devarajan K., Bae S., Naik G., Chan T.A., Uzzo R., Hakimi A.A., Sonpavde G., Plimack E. Prognostic impact of immune gene expression signature and tumor infiltrating immune cells in localized clear cell renal cell carcinoma. J. Immunother. Cancer. 2019;7 doi: 10.1186/s40425-019-0735-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pennock N.D., White J.T., Cross E.W., Cheney E.E., Tamburini B.A., Kedl R.M. T cell responses: naïve to memory and everything in between. Adv. Psyiology Educ. 2013;37:273–283. doi: 10.1152/advan.00066.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Voskoboinik I., Whisstock J.C., Trapani J.A. Perforin and granzymes: function, dysfunction and human pathology. Nat. Rev. Immunol. 2015;15:388–400. doi: 10.1038/nri3839. [DOI] [PubMed] [Google Scholar]
- 5.Golubovskaya V., Wu L. Different subsets of T cells, memory, effector functions, and CAR-T immunotherapy. Cancers (Basel). 2016;8 doi: 10.3390/cancers8030036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salerno F., Freen-van Heeren J.J., Guislain A., Nicolet B.P., Wolkers M.C. Costimulation through TLR2 drives polyfunctional CD8+ T cell responses. J. Immunol. 2019;202:714–723. doi: 10.4049/jimmunol.1801026. [DOI] [PubMed] [Google Scholar]
- 7.Nicolet B.P., Guislain A., van Alphen F.P.J., Gomez-Eerland R., Schumacher T.N., van den Biggelaar M., Wolkers M. CD29 marks superior cytotoxic human CD8+ T cells. Proc. Natl. Acad. Sci. 2020;117:6686–6696. doi: 10.1101/562512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang B., Karrison T., Rowley D.A., Schreiber H. IFN-γ- and TNF-dependent bystander eradication of antigen-loss variants in established mouse cancers. J. Clin. Invest. 2008;118:1398–1404. doi: 10.1172/JCI33522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patel S.J., Sanjana N.E., Kishton R.J., Eidizadeh A., Vodnala S.K., Cam M., Gartner J.J., Jia L., Steinberg S.M., Yamamoto T.N., Merchant A.S., Mehta G.U., Chichura A., Shalem O., Tran E., Eil R., Sukumar M., Guijarro E.P., Day C.P., Robbins P., Feldman S., Merlino G., Zhang F., Restifo N.P. Identification of essential genes for cancer immunotherapy. Nature. 2017;548:537–542. doi: 10.1038/nature23477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ayers M., Lunceford J., Nebozhyn M., Murphy E., Loboda A., Kaufman D.R., Albright A., Cheng J.D., Kang S.P., Shankaran V., Piha-paul S.A., Yearley J., Seiwert T.Y., Ribas A., Mcclanahan T.K. IFN-γ–related mRNA profile predicts clinical response to PD-1 blockade. J. Clin. Invest. 2017;127:2930–2940. doi: 10.1172/JCI91190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao J., Shi L.Z., Zhao H., Chen J., Xiong L., He Q., Chen T., Roszik J., Bernatchez C., Woodman S.E., Chen P., Hwu P., Allison J.P., Futreal A., Wargo J.A., Sharma P. Loss of IFN-γ pathway genes in tumor cells as a mechanism of resistance to anti-CTLA-4 therapy. Cell. 2016;167:397–404. doi: 10.1016/j.physbeh.2017.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seidel J.A., Otsuka A., Kabashima K. Anti-PD-1 and Anti-CTLA-4 Therapies in Cancer: Mechanisms of Action, Efficacy, and Limitations. Front. Oncol. 2018;8 doi: 10.3389/fonc.2018.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang F., Zheng P. Tumor cells versus host immune cells: whose PD-L1 contributes to PD-1/PD-L1 blockade mediated cancer immunotherapy? Cell Biosci. 2018;8 doi: 10.1186/s13578-018-0232-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garrido F., Aptsiauri N., Doorduijn E.M., Lora A.M.G., Van Hall T. The urgent need to recover MHC class I in cancers for effective immunotherapy. Curr. Opin. Immunol. 2016;39:44–51. doi: 10.1016/j.coi.2015.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang L., Malu S., McKenzie J.A., Andrews M.C., Talukder A.H., Tieu T., Karpinets T., Haymaker C., Forget M.A., Williams L.J., Wang Z., Mbofung R.M., Wang Z.Q., Davis R.E., Lo R.S., Wargo J.A., Davies M.A., Bernatchez C., Heffernan T., Amaria R.N., Korkut A., Peng W., Roszik J., Lizee G., Woodman S.E., Hwu P. The RNA-binding Protein MEX3B Mediates Resistance to Cancer Immunotherapy by Downregulating HLA-A Expression. Clin. Cancer Res. 2018;24:3366–3376. doi: 10.1158/1078-0432.CCR-17-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morrison B.J., Steel J.C., Morris J.C. Reduction of MHC-I expression limits T- lymphocyte-mediated killing of Cancer- initiating cells. BMC Cancer. 2018;18 doi: 10.1186/s12885-018-4389-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rohaan M.W., Van Den Berg J.H., Kvistborg P., Haanen J.B.A.G. Adoptive transfer of tumor-infiltrating lymphocytes in melanoma: a viable treatment option. J. Immunother. Cancer. 2018;6 doi: 10.1186/s40425-018-0391-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Groot R., van Loenen M.M., Guislain A., Nicolet B.P., Freen-van Heeren J.J., van den Heuvel M.M., Burger P., van der Schoot E.C., Spaapen R.M., Amsen D., Haanen J.B., Monkhorst K., Hartemink K.J., Wolkers M.C. Polyfunctional tumor-reactive T cells are effectively expanded from non-small cell lung cancers, and correlate with an immune-engaged T cell profile. Oncoimmunology. 2019;8 doi: 10.1080/2162402X.2019.1648170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stevanovic S., Draper L.M., Langhan M.M., Campbell T.E., Kwong M.L., Wunderlich J.R., Dudley M.E., Yang J.C., Sherry R.M., Kammula U.S., Restifo N.P., Rosenberg S.A., Hinrichs C.S. Complete Regression of Metastatic Cervical Cancer After Treatment With Human Papillomavirus-Targeted Tumor-Infiltrating T Cells. J. Clin. Oncol. 2015;33:1543–1550. doi: 10.1200/JCO.2014.58.9093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dudly M.E., Wunderlich J.R., Yang J.C., Sherry R.M., Topalian S.L., Restifo N.O., Royal R.E., Kammula U., White D.E., Mavroukakis S.A., Rogers L.J., Gracia G.J., Jones S.A., Mangiameli D.P., Pelletier M.M., Gea-Banachloche J., Robinson M.R., Berman D.M., Filie A.C., Abati A., Rosenberg S.A. Adoptive Cell Transfer Therapy Following Non-Myeloablative but Lymphodepleting Chemotherapy for the Treatment of Patients With Refractory Metastatic Melanoma. J. Clin. Oncol. 2005;23:2346–2357. doi: 10.1200/JCO.2005.00.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosenberg S.A., Yang J.C., Sherry R.M., Kammula U.S., Marybeth S., Phan G.Q., Citrin D.E., Restifo N.P., Robbins P.F., John R., Morton K.E., Laurencot C.M., Steinberg S.M., Donald E., Dudley M.E. Durable Complete Responses in Heavily Pretreated Patients with Metastatic Melanoma Using T Cell Transfer Immunotherapy. Clin. Cancer Res. 2011;17:4550–4557. doi: 10.1158/1078-0432.CCR-11-0116.Durable. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Radvanyi L.G., Bernatchez C., Zhang M., Fox P.S., Chacon J., Wu R., Lizee G., Mahoney S., Glass M., Johnson V.E., Mcmannis J.D., Shpall E., Prieto V., Papadopoulos N., Kim K., Homsi J., Bedikian A., Hwu J., Patel S., Ross M.I., Lee J.E., Gershenwald J.E., Lucci A., Royal R., Cormier J.N., Davies M.A., Fulbright O.J., Toth C., Ramachandran R., Gonzalez A., Hwu P. Specific lymphocyte subsets predict response to adoptive cell therapy using expanded autologous tumor-infiltrating lymphocytes in metastatic melanoma patients. Clin. Cancer Res. 2012;18:6758–6770. doi: 10.1158/1078-0432.CCR-12-1177.Specific. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Besser M.J., Shapira-frommer R., Itzhaki O., Treves A.J., Zippel D.B., Levy D., Kubi A., Shoshani N., Zikich D., Ohayon Y., Ohayon D. Adoptive Transfer of Tumor-Infiltrating Lymphocytes in Patients with Metastatic Melanoma: Intent-to-Treat Analysis and Efficacy after Failure to Prior Immunotherapies. Clin. Cancer Res. 2013;19:4792–4801. doi: 10.1158/1078-0432.CCR-13-0380. [DOI] [PubMed] [Google Scholar]
- 24.Andersen R., Donia M., Ellebaek E., Borch T.H., Kongsted P., Hendel H.W. Long-Lasting Complete Responses in Patients with Metastatic Melanoma after Adoptive Cell Therapy with Tumor-In fi ltrating Lymphocytes and an Attenuated IL2 Regimen. Clin. Cancer Res. 2016;22:3734–3745. doi: 10.1158/1078-0432.CCR-15-1879. [DOI] [PubMed] [Google Scholar]
- 25.Leko V., Mcduffie L.A., Zheng Z., Gartner J.J., Prickett T.D., Apolo A.B., Agarwal P.K., Rosenberg S.A., Lu Y. Identification of Neoantigen-Reactive Tumor-Infiltrating Lymphocytes in Primary Bladder Cancer. J. Immunol. 2019;202:3458–3467. doi: 10.4049/jimmunol.1801022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gomez-Eerland R., Nuijen B., Heemskerk B., van Rooij N., van den Berg J.H., Beijnen J.H., Uckert W., Kvistborg P., Schumacher T.N., Haanen J.B.A.G., Jorritsma A. Manufacture of Gene-Modified Human T-Cells with a Memory Stem/Central Memory Phenotype. Hum. Gene Ther. Methods. 2014;25:277–287. doi: 10.1089/hgtb.2014.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fousek K., Watanabe J., Joseph S.K., George A., An X., Byrd T.T., Morris J.S., Luong A., Martínez-Paniagua M.A., Sanber K., Navai S.A., Gad A.Z., Salsman V.S., Mathew P.R., Kim H.N., Wagner D.L., Brunetti L., Jang A., Baker M.L., Varadarajan N., Hegde M., Kim Y.M., Heisterkamp N., Abdel-Azim H., Ahmed N. CAR T-cells that target acute B-lineage leukemia irrespective of CD19 expression. Leukemia. 2020 doi: 10.1038/s41375-020-0792-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Albers J.J., Ammon T., Gosmann D., Audehm S., Thoene S., Winter C., Secci R., Wolf A., Stelzl A., Steiger K., Ruland J., Bassermann F., Kupatt C., Anton M., Krackhardt A.M. Gene editing enables T-cell engineering to redirect antigen specificity for potent tumor rejection. Life Sci. Alliance. 2019;2 doi: 10.26508/lsa.201900367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morton L.T., Reijmers R.M., Wouters A.K., Kweekel C., Remst D.F.G., Pothast C.R., Falkenburg J.H.F., Heemskerk M.H.M. Simultaneous Deletion of Endogenous TCRαβ for TCR Gene Therapy Creates an Improved and Safe Cellular Therapeutic. Mol. Ther. 2020;28:64–74. doi: 10.1016/j.ymthe.2019.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eyquem J., Mansilla-soto J., Giavridis T., Hamieh M., Cunanan K.M., Odak A., Sadelain M. Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nature. 2017;543:113–117. doi: 10.1038/nature21405.Targeting. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pettitt D., Arshad Z., Smith J., Stanic T., Holländer G., Brindley D. CAR-T Cells : A Systematic Review and Mixed Methods Analysis of the Clinical Trial Landscape. Mol. Ther. 2018;26:342–353. doi: 10.1016/j.ymthe.2017.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weinkove R., George P., Dasyam N., McLellan A.D. Selecting costimulatory domains for chimeric antigen receptors: functional and clinical considerations. Clin. Transl. Immunol. 2019;8 doi: 10.1002/cti2.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Newick K., O’Brien S., Moon E., Albelda S.M. CAR T Cell Therapy for Solid Tumors. Annu. Rev. Med. 2017;68:139–152. doi: 10.1146/annurev-med-062315-120245. [DOI] [PubMed] [Google Scholar]
- 34.Majzner R.G., Mackall C.L. Clinical lessons learned from the first leg of the CAR T cell journey. Nat. Med. 2019;25:1341–1355. doi: 10.1038/s41591-019-0564-6. [DOI] [PubMed] [Google Scholar]
- 35.Mohanty R., Chowdhury C.R., Arega S., Sen P., Ganguly P., Ganguly N. CAR T cell therapy: A new era for cancer treatment. Oncol. Rep. 2019;42:2183–2195. doi: 10.3892/or.2019.7335. [DOI] [PubMed] [Google Scholar]
- 36.Schumacher T.N., Scheper W., Kvistborg P. Cancer Neoantigens. Annu. Rev. Immunol. 2019;37:173–200. doi: 10.1146/annurev-immunol-042617-053402. [DOI] [PubMed] [Google Scholar]
- 37.Yamamoto T.N., Kishton R.J., Restifo N.P. Developing neoantigen-targeted T cell–based treatments for solid tumors. Nat. Med. 2019;25:1488–1499. doi: 10.1038/s41591-019-0596-y. [DOI] [PubMed] [Google Scholar]
- 38.Wang Z., Cao Y.J. Adoptive Cell Therapy Targeting Neoantigens: A Frontier for Cancer Research. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.00176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu J., Gaj T., Wallen M.C., Barbas C.F. Improved cell-penetrating zinc-finger nuclease proteins for precision genome engineering. Mol. Ther. - Nucleic Acids. 2015;4 doi: 10.1038/mtna.2015.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Knipping F., Osborn M.J., Petri K., Tolar J., Glimm H., von Kalle C., Schmidt M., Gabriel R. Genome-wide Specificity of Highly Efficient TALENs and CRISPR/Cas9 for T Cell Receptor Modification. Mol. Ther. - Methods Clin. Dev. 2017;4:213–224. doi: 10.1016/j.omtm.2017.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qasim W., Zhan H., Samarasinghe S., Adams S., Amrolia P., Stafford S., Butler K., Rivat C., Wright G., Somana K., Ghorashian S., Pinner D., Ahsan G., Gilmour K., Lucchini G., Inglott S., Mifsud W., Chiesa R., Peggs K.S., Chan L., Farzeneh F., Thrasher A.J., Vora A., Pule M., Veys P. Molecular remission of infant B-ALL after infusion of universal TALEN gene-edited CAR T cells. Sci. Transl. Med. 2017;9:1–9. doi: 10.1126/scitranslmed.aaj2013. [DOI] [PubMed] [Google Scholar]
- 42.O’Neil R.T., Saha S., Veach R.A., Welch R.C., Woodard L.E., Rooney C.M., Wilson M.H. Transposon-modified antigen-specific T lymphocytes for sustained therapeutic protein delivery in vivo. Nat. Commun. 2018;9:1–10. doi: 10.1038/s41467-018-03787-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Werner Sunderland M., Peggs K.S. Successful translation and future prospects of TALEN editing for leukemia patients. Expert Opin. Biol. Ther. 2018;18:725–726. doi: 10.1080/14712598.2018.1484105. [DOI] [PubMed] [Google Scholar]
- 44.Borowicz P., Chan H., Medina D., Gumpelmair S., Kjelstrup H., Spurkland A. A simple and efficient workflow for generation of knock-in mutations in Jurkat T cells using CRISPR/Cas9, Scand. J. Immunol. 2019:1–14. doi: 10.1111/sji.12862. [DOI] [PubMed] [Google Scholar]
- 45.Rupp L.J., Schumann K., Roybal K.T., Gate R.E., Ye C.J., Lim W.A., Marson A. CRISPR/Cas9-mediated PD-1 disruption enhances anti-Tumor efficacy of human chimeric antigen receptor T cells. Sci. Rep. 2017;7:1–10. doi: 10.1038/s41598-017-00462-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seki A., Rutz S. Optimized RNP transfection for highly efficient CRISPR/Cas9-mediated gene knockout in primary T cells. J. Exp. Med. 2018;215:985–997. doi: 10.1084/jem.20171626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shifrut E., Carnevale J., Tobin V., Roth T.L., Woo J.M., Bui C.T., Li P.J., Diolaiti M.E., Ashworth A., Marson A. Genome-wide CRISPR Screens in Primary Human T Cells Reveal Key Regulators of Immune Function. Cell. 2018;175:1958–1971.e15. doi: 10.1016/j.cell.2018.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Freen-van Heeren J.J., Popović B., Guislain A., Monika C. Human T cells employ conserved AU-rich elements to fine-tune IFN-γ production. Eur. J. Immunol. 2020;50:949–958. doi: 10.1002/eji.201948458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nüssing S., House I.G., Kearney C.J., Chen A.X.Y., Vervoort S.J., Beavis P.A., Oliaro J., Johnstone R.W., Trapani J.A., Parish I.A. Efficient CRISPR/Cas9 gene editing in uncultured naive mouse T cells for in vivo studies. J. Immunol. 2020:ji1901396. doi: 10.4049/jimmunol.1901396. [DOI] [PubMed] [Google Scholar]
- 50.Singer M., Wang C., Cong L., Marjanovic N.D., Kowalczyk M.S., Zhang H., Nyman J., Sakuishi K., Kurtulus S., David G., Xia J., Kwon J.Y.H., Nevin J., Herbst R.H., Yanai I., Rozenblatt-Rosen O., Kucroo V.K., Regev A., Anderson A.C. A distinct gene module for dysfunction uncoupled from activation in tumor-infiltrating T cells. Cell. 2016;166:1500–1511. doi: 10.1016/j.physbeh.2017.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jung I.Y., Kim Y.Y., Yu H.S., Lee M., Kim S., Lee J. CRISPR/Cas9-Mediated Knockout of DGK Improves Antitumor Activities of Human T Cells. Cancer Res. 2018;78:4692–4703. doi: 10.1158/0008-5472.CAN-18-0030. [DOI] [PubMed] [Google Scholar]
- 52.Anderson W., Thorpe J., Long S.A., Rawlings D.J. Efficient CRISPR/Cas9 Disruption of Autoimmune-Associated Genes Reveals Key Signaling Programs in Primary Human T Cells. J. Immunol. 2019;203:3166–3178. doi: 10.4049/jimmunol.1900848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dong M.B., Wang G., Chow R.D., Ye L., Zhu L., Dai X., Park J.J., Kim H.R., Errami Y., Guzman C.D., Zhou X., Chen K.Y., Renauer P.A., Du Y., Shen J., Lam S.Z., Zhou J.J., Lannin D.R., Herbst R.S., Chen S. Systematic Immunotherapy Target Discovery Using Genome-Scale In Vivo CRISPR Screens in CD8 T Cells. Cell. 2019;178:1189–1204. doi: 10.1016/j.cell.2019.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wei J., Long L., Zheng W., Dhungana Y., Lim S.A., Guy C., Wang Y., Wang Y.D., Qian C., Xu B., Kc A., Saravia J., Huang H., Yu J., Doench J.G., Geiger T.L., Chi H. Targeting REGNASE-1 programs long-lived effector T cells for cancer therapy. Nature. 2019;576:471–476. doi: 10.1038/s41586-019-1821-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Klepsch V., Pommermayr M., Humer D., Brigo N., Hermann-Kleiter N., Baier G. Targeting the orphan nuclear receptor NR2F6 in T cells primes tumors for immune checkpoint therapy. Cell Commun. Signal. 2020;18 doi: 10.1186/s12964-019-0454-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tang N., Cheng C., Zhang X., Qiao M., Li N., Mu W., Wei X.F., Han W., Wang H. TGF-β inhibition via CRISPR promotes the long-term efficacy of CAR T cells against solid tumors. JCI Insight. 2020;5 doi: 10.1172/jci.insight.133977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Su S., Zou Z., Chen F., Ding N., Du J., Shao J., Li L., Fu Y., Hu B., Yang Y., Sha H., Meng F., Wei J., Huang X., Liu B. CRISPR-cas9-mediated disruption of PD-1 on human T cells for adoptive cellular therapies of EBV positive gastric cancer. Oncoimmunology. 2017;6 doi: 10.1080/2162402X.2016.1249558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang C., Peng Y., Hublitz P., Zhang H., Dong T. Genetic abrogation of immune checkpoints in antigen-specific cytotoxic T-lymphocyte as a potential alternative to blockade immunotherapy. Sci. Rep. 2018;8:5549. doi: 10.1038/s41598-018-23803-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Su S., Hu B., Shao J., Shen B., Du J., Du Y., Zhou J., Yu L., Zhang L., Chen F., Sha H., Cheng L., Meng F., Zou Z., Huang X., Liu B. CRISPR-Cas9 mediated efficient PD-1 disruption on human primary T cells from cancer patients. Sci. Rep. 2016;6:20070. doi: 10.1038/srep20070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhao Z., Shi L., Zhang W., Han J., Zhang S., Fu Z., Cai J. CRISPR knock out of programmed cell death protein 1 enhances anti-tumor activity of cytotoxic T lymphocytes. Oncotarget. 2018;9:5208–5215. doi: 10.18632/oncotarget.23730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Okada M., Chikuma S., Kondo T., Hibino S., Machiyama H., Yokosuka T., Nakano M., Yoshimura A. Blockage of Core Fucosylation Reduces Cell-Surface Expression of PD-1 and Promotes Anti-tumor Immune Responses of T Cells. Cell Rep. 2017;20:1017–1028. doi: 10.1016/j.celrep.2017.07.027. [DOI] [PubMed] [Google Scholar]
- 62.Guo X., Jiang H., Shi B., Zhou M., Zhang H., Shi Z., Du G., Luo H., Wu X., Wang Y., Sun R., Li Z. Disruption of PD-1 enhanced the anti-tumor activity of chimeric antigen receptor T cells against hepatocellular carcinoma. Front. Pharmacol. 2018;9 doi: 10.3389/fphar.2018.01118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hu B., Zou Y., Zhang L., Tang J., Niedermann G., Firat E., Huang X., Zhu X. Nucleofection with Plasmid DNA for CRISPR/Cas9-Mediated Inactivation of Programmed Cell Death Protein 1 in CD133-Specific CAR T Cells. Hum. Gene Ther. 2019;30:446–458. doi: 10.1089/hum.2017.234. [DOI] [PubMed] [Google Scholar]
- 64.Hu W., Zi Z., Jin Y., Li G., Shao K., Cai Q., Ma X., Wei F. CRISPR/Cas9-mediated PD-1 disruption enhances human mesothelin-targeted CAR T cell effector functions. Cancer Immunol. Immunother. 2019;68:365–377. doi: 10.1007/s00262-018-2281-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shi L., Meng T., Zhao Z., Han J., Zhang W., Gao F., Cai J. CRISPR knock out CTLA-4 enhances the anti-tumor activity of cytotoxic T lymphocytes. Gene. 2017;636:36–41. doi: 10.1016/j.gene.2017.09.010. [DOI] [PubMed] [Google Scholar]
- 66.Zhang W., Shi L., Zhao Z., Du P., Ye X., Li D., Cai Z., Han J., Cai J. Disruption of CTLA-4 expression on peripheral blood CD8+ T cell enhances anti-tumor efficacy in bladder cancer. Cancer Chemother. Pharmacol. 2019;83:911–920. doi: 10.1007/s00280-019-03800-x. [DOI] [PubMed] [Google Scholar]
- 67.Zhang Y., Zhang X., Cheng C., Mu W., Liu X., Li N., Wei X., Liu X., Xia C., Wang H. CRISPR-Cas9 mediated LAG-3 disruption in CAR-T cells. Front. Med. 2017;11:554–562. doi: 10.1007/s11684-017-0543-6. [DOI] [PubMed] [Google Scholar]
- 68.Batra S.A., Rathi P., Guo L., Courtney A.N., Fleurence J., Balzeau J., Shaik R.S., Nguyen T.P., Wu M.F., Bulsara S., Mamonkin M., Metelitsa L.S., Heczey A. Glypican-3-Specific CAR T Cells Coexpressing IL15 and IL21 Have Superior Expansion and Antitumor Activity against Hepatocellular Carcinoma. Cancer Immunol. Res. 2020;8:309–320. doi: 10.1158/2326-6066.CIR-19-0293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Simoni Y., Becht E., Fehlings M., Loh C.Y., Teng K.W.W., Yeong J.P.S., Nahar R., Zhang T., Kared H., Duan K., Ang N., Poidinger M., Lee Y.Y., Larbi A., Khng A.J., Tan E., Fu C., Mathew R., Teo M., Lim W.T., Toh C.K., Ong B., Koh T., Hillmer A.M., Takano A., Lim T.K.H., Tan E.H., Zhai W., Tan D.S.W., Tan I.B., Newell E.W. Bystander CD8+ T cells are abundant and phenotypically distinct in human tumour infiltrates. Nature. 2018;557:575–580. doi: 10.1038/s41586-018-0130-2. [DOI] [PubMed] [Google Scholar]
- 70.Rosato P.C., Wijeyesinghe S., Stolley J.M., Nelson C.E., Davis R.L., Manlove L.S., Pennell C.A., Blazar B.R., Chen C.C., Geller M.A., Vezys V., Masopust D. Virus-specific memory T cells populate tumors and can be repurposed for tumor immunotherapy. Nat. Commun. 2019;10 doi: 10.1038/s41467-019-08534-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schober K., Müller T.R., Gökmen F., Grassmann S., Effenberger M., Poltorak M., Stemberger C., Schumann K., Roth T.L., Marson A., Busch D.H. Orthotopic replacement of T-cell receptor α- and β-chains with preservation of near-physiological T-cell function. Nat. Biomed. Eng. 2019;3:974–984. doi: 10.1038/s41551-019-0409-0. [DOI] [PubMed] [Google Scholar]
- 72.Stadtmauer E.A., Fraietta J.A., Davis M.M., Cohen A.D., Weber K.L., Lancaster E., Mangan P.A., Kulikovskaya I., Gupta M., Chen F., Tian L., Gonzalez V.E., Xu J., Jung I., Melenhorst J., Plesa G., Shea J., Matlawski T., Cervini A., Gaymon A.L., Desjardins S., Lamontagne A., Salas-Mckee J., Fesnak A., Siegel D.L., Levine B.L., Jadlowsky J.K., Young R.M., Chew A., Hwang W.T., Hexner E.O., Carreno B.M., Nobles C.L., Bushman F.D., Parker K.R., Qi Y., Satpathy A.T., Chang H.Y., Zhao Y., Lacey S.F., June C.H. CRISPR-engineered T cells in patients with refractory cancer. Science. 2020;367 doi: 10.1126/science.aba7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Adli M. The CRISPR tool kit for genome editing and beyond. Nat. Commun. 2018 doi: 10.1038/s41467-018-04252-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang W., Ye C., Liu J., Zhang D., Kimata J.T. CCR5 gene disruption via lentiviral vectors expressing Cas9 and single guided RNA renders cells resistant to HIV-1 infection. PLoS One. 2014:1–26. doi: 10.1371/journal.pone.0115987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li C., Guan X., Du T., Jin W., Wu B., Liu Y., Wang P., Hu B., Griffin G.E., Shattock R.J., Hu Q. Inhibition of HIV-1 infection of primary CD4+ T-cells by gene editing of CCR5 using CRISPR/Cas9. J. Gen. Virol. 2015;96:2381–2393. doi: 10.1099/vir.0.000139. [DOI] [PubMed] [Google Scholar]
- 78.Kim S., Kim D., Cho S., Kim J., Kim J.-S. Highly Efficient RNA-guide genome editing. Genome Res. 2014;24:1012–1019. doi: 10.1101/gr.171322.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schagen F.H.E., Rademaker H.J., Fallaux F.J., Hoeben R.C. Insertion vectors for gene therapy. Gene Ther. 2000;7:271–272. doi: 10.1038/sj.gt.3301121. [DOI] [PubMed] [Google Scholar]
- 80.Poletti V., Mavilio F. Interactions between Retroviruses and the Host Cell Genome. Mol. Ther. - Methods Clin. Dev. 2018;8:31–41. doi: 10.1016/j.omtm.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Simhadri V.L., McGill J., McMahon S., Wang J., Jiang H., Sauna Z.E. Prevalence of Pre-existing Antibodies to CRISPR-Associated Nuclease Cas9 in the USA Population. Mol. Ther. - Methods Clin. Dev. 2018;10:105–112. doi: 10.1016/j.omtm.2018.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Charlesworth C.T., Deshpande P.S., Dever D.P., Camarena J., Lemgart V.T., Cromer M.K., Vakulskas C.A., Collingwood M.A., Zhang L., Bode N.M., Behlke M.A., Dejene B., Cieniewicz B., Romano R., Lesch B.J., Gomez-Ospina N., Mantri S., Pavel-Dinu M., Weinberg K.I., Porteus M.H. Identification of preexisting adaptive immunity to Cas9 proteins in humans. Nat. Med. 2019;25:249–254. doi: 10.1038/s41591-018-0326-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rouet R., Thuma B.A., Roy M.D., Lintner N.G., Rubitski D.M., Finley J.E., Wisniewska H.M., Mendonsa R., Hirsh A., De Oñate L., Compte Barrón J., McLellan T.J., Bellenger J., Feng X., Varghese A., Chrunyk B.A., Borzilleri K., Hesp K.D., Zhou K., Ma N., Tu M., Dullea R., McClure K.F., Wilson R.C., Liras S., Mascitti V., Doudna J.A. Receptor-mediated delivery of CRISPR-Cas9 endonuclease for cell-type-specific gene editing. J. Am. Chem. Soc. 2018;140:6596–6603. doi: 10.1021/jacs.8b01551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Qiao J., Sun W., Lin S., Jin R., Ma L., Liu Y. Cytosolic delivery of CRISPR/Cas9 ribonucleoproteins for genome editing using chitosan-coated red fluorescent protein. Chem. Commun. 2019;55:4707–4710. doi: 10.1039/c9cc00010k. [DOI] [PubMed] [Google Scholar]
- 85.Zhen S., Li X. Liposomal delivery of CRISPR/Cas9. Cancer Gene Ther. 2019 doi: 10.1038/s41417-019-0141-7. [DOI] [PubMed] [Google Scholar]
- 86.Ramakrishna S., Kwaku Dad A.B., Beloor J., Gopalappa R., Lee S.K., Kim H. Gene disruption by cell-penetrating peptide-mediated delivery of Cas9 protein and guide RNA. Genome Res. 2014;24:1020–1027. doi: 10.1101/gr.171264.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Del’Guidice T., Lepetit-Stoffaes J.P., Bordeleau L.J., Roberge J., Théberge V., Lauvaux C., Barbeau X., Trottier J., Dave V., Roy D.C., Gaillet B., Garnier A., Guay D. Membrane permeabilizing amphiphilic peptide delivers recombinant transcription factor and CRISPR-Cas9/Cpf1 ribonucleoproteins in hard-to-modify cells. PLoS One. 2018;13 doi: 10.1371/journal.pone.0195558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang H.X., Song Z., Lao Y.H., Xu X., Gong J., Cheng D., Chakraborty S., Park J.S., Li M., Huang D., Yin L., Cheng J., Leong K.W. Nonviral gene editing via CRISPR/Cas9 delivery by membrane-disruptive and endosomolytic helical polypeptide. Proc. Natl. Acad. Sci. U. S. A. 2018;115:4903–4908. doi: 10.1073/pnas.1712963115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Liang X., Potter J., Kumar S., Zou Y., Quintanilla R., Sridharan M., Carte J., Chen W., Roark N., Ranganathan S., Ravinder N., Chesnut J.D. Rapid and highly efficient mammalian cell engineering via Cas9 protein transfection. J. Biotechnol. 2015;208:44–53. doi: 10.1016/j.jbiotec.2015.04.024. [DOI] [PubMed] [Google Scholar]
- 90.Schumann K., Lin S., Boyer E., Simeonov D.R., Subramaniam M., Gate R.E., Haliburton G.E., Ye C.J., Bluestone J.A., Doudna J.A., Marson A. Generation of knock-in primary human T cells using Cas9 ribonucleoproteins. Proc. Natl. Acad. Sci. 2015;112:10437–10442. doi: 10.1073/pnas.1512503112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kornete M., Marone R., Jeker L.T. Highly Efficient and Versatile Plasmid-Based Gene Editing in Primary T Cells. J. Immunol. 2018;200:2489–2501. doi: 10.4049/jimmunol.1701121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hultquist J., Schumann K., Woo J., Manganaro L., McGregor M., Doudna J., Simon V., Krogan N., Marson A. A Cas9 Ribonucleoprotein Platform for Functional Genetic Studies of HIV-Host Interactions in Primary Human T Cells Judd. Cell Rep. 2016;17:1438–1452. doi: 10.1016/j.celrep.2016.09.080.A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Oh S.A., Seki A., Rutz S. Ribonucleoprotein Transfection for CRISPR/Cas9-Mediated Gene Knockout in Primary T Cells. Curr. Protoc. Immunol. 2019;124 doi: 10.1002/cpim.69. [DOI] [PubMed] [Google Scholar]
- 94.Chicaybam L., Sodre A.L., Curzio B.A., Bonamino M.H. An Efficient Low Cost Method for Gene Transfer to T Lymphocytes. PLoS One. 2013;8:1–11. doi: 10.1371/journal.pone.0060298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mezzadra R., Hollenstein A., Gomez-Eerland R., Schumacher T.N. A Traceless Selection: Counter-selection System That Allows Efficient Generation of Transposon and CRISPR-modified T-cell Products. Mol. Ther. - Nucleic Acids. 2016;5 doi: 10.1038/mtna.2016.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Roth T.L., Puig-Saus C., Yu R., Shifrut E., Carnevale J., Li P.J., Hiatt J., Saco J., Krystofinski P., Li H., Tobin V., Nguyen D.N., Lee M.R., Putnam A.L., Ferris A.L., Chen J.W., Schickel J.N., Pellerin L., Carmody D., Alkorta-Aranburu G., Del Gaudio D., Matsumoto H., Morell M., Mao Y., Cho M., Quadros R.M., Gurumurthy C.B., Smith B., Haugwitz M., Hughes S.H., Weissman J.S., Schumann K., Esensten J.H., May A.P., Ashworth A., Kupfer G.M., Greeley S.A.W., Bacchetta R., Meffre E., Roncarolo M.G., Romberg N., Herold K.C., Ribas A., Leonetti M.D., Marson A. Reprogramming human T cell function and specificity with non-viral genome targeting. Nature. 2018;559:405–409. doi: 10.1038/s41586-018-0326-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chi S., Weiss A., Wang H. A CRISPR-Based Toolbox for Studying T Cell Signal Transduction. Biomed Res. Int. 2016;2016 doi: 10.1155/2016/5052369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Okada M., Kanamori M., Someya K., Nakatsukasa H., Yoshimura A. Stabilization of Foxp3 expression by CRISPR-dCas9-based epigenome editing in mouse primary T cells. Epigenetics and Chromatin. 2017;10:24. doi: 10.1186/s13072-017-0129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Simeonov D.R., Gowen B.G., Boontanrart M., Roth T.L., Gagnon J.D., Mumbach M.R., Satpathy A.T., Lee Y., Bray N.L., Chan A.Y., Lituiev D.S., Nguyen M.L., Gate R.E., Subramaniam M., Li Z., Woo J.M., Mitros T., Ray G.J., Curie G.L., Naddaf N., Chu J.S., Ma H., Boyer E., Van Gool F., Huang H., Liu R., Tobin V.R., Schumann K., Daly M.J., Farh K.K., Ansel K.M., Ye C.J., Greenleaf W.J., Anderson M.S., Bluestone J.A., Chang H.Y., Corn J.E., Marson A. Discovery of stimulation-responsive immune enhancers with CRISPR activation. Nature. 2017;549:111–115. doi: 10.1038/nature23875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chen X., Kozhaya L., Tastan C., Placek L., Dogan M., Horne M., Abblett R., Karhan E., Vaeth M., Feske S., Unutmaz D. Functional interrogation of primary human T cells via CRISPR genetic editing. J. Immunol. 2018;201:1586–1598. doi: 10.4049/jimmunol.1701616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.C.R. Rankin, J. Treger, E. Faure-Kumar, J. Benhammou, D. Anisman-Posner, A.E. Bollinger, C. Pothoulakis, M. Padua, Overexpressing Long Noncoding RNAs Using Gene-activating CRISPR, J. Vis. Exp. (2019) e59233. [DOI] [PMC free article] [PubMed]
- 102.Hultquist J.F., Hiatt J., Schumann K., McGregor M.J., Roth T.L., Haas P., Doudna J.A., Marson A., Krogan N.J. CRISPR–Cas9 genome engineering of primary CD4 + T cells for the interrogation of HIV–host factor interactions. Nat. Protoc. 2019;14 doi: 10.1038/s41596-018-0069-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Schmitz M.L., Krappmann D. Controlling NF-κB activation in T cells by costimulatory receptors. Cell Death Differ. 2006;13:834–842. doi: 10.1038/sj.cdd.4401845. [DOI] [PubMed] [Google Scholar]
- 104.Salerno F., Paolini N.A., Stark R., von Lindern M., Wolkers M.C. Distinct PKC-mediated posttranscriptional events set cytokine production kinetics in CD8+ T cells. Proc. Natl. Acad. Sci. 2017;114:9677–9682. doi: 10.1073/pnas.1704227114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Salerno F., Turner M., Wolkers M.C. Dynamic Post-Transcriptional Events Governing CD8+ T Cell Homeostasis and Effector Function. Trends Immunol. 2020;41:240–254. doi: 10.1016/j.it.2020.01.001. [DOI] [PubMed] [Google Scholar]
- 106.Mittrücker H., Kursar M., Köhler A., Kaufmann S.H.E., Hurwitz R. Role of CD28 for the Generation and Expansion of Antigen-Specific CD8+ T Lymphocytes During Infection with Listeria monocytogenes. J. Immunol. 2001;167:5620–5627. doi: 10.4049/jimmunol.167.10.5620. [DOI] [PubMed] [Google Scholar]
- 107.Welten S.P.M., Redeker A., Franken K.L., Benedict C.A., Yagita H., Wensveen F.M., Borst J., Melief C.J.M., van Lier R.A.W., van Gisbergen K.P.J.M., Arens R. CD27-CD70 costimulation controls T cell immunity during acute and persistent cytomegalovirus infection. J. Virol. 2013;87:6851–6865. doi: 10.1128/JVI.03305-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Au-Yeung B., Smith G.A., Mueller J.L., Heyn C.S., Jaszczak R.G., Weiss A., Zikherman J. IL-2 modulates the TCR signaling threshold for CD8 but not CD4 T cell proliferation on a single cell level. J. Immunol. 2017;198:2445–2456. doi: 10.1016/j.physbeh.2017.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Stark R., Hartung A., Zehn D., Frentsch M., Thiel A. IL-12-mediated STAT4 signaling and TCR signal strength cooperate in the induction of CD40L in human and mouse CD8+ T cells. Eur. J. Immunol. 2013;43:1511–1517. doi: 10.1002/eji.201243218. [DOI] [PubMed] [Google Scholar]
- 110.Imanishi T., Saito T. T Cell Co-stimulation and Functional Modulation by Innate Signals. Trends Immunol. 2020;41:200–212. doi: 10.1016/j.it.2020.01.003. [DOI] [PubMed] [Google Scholar]
- 111.Cottalorda A., Verschelde C., Marçais A., Tomkowiak M., Musette P., Uematsu S., Akira S., Marvel J., Bonnefoy-Berard N. TLR2 engagement on CD8 T cells lowers the thresholdfor optimal antigen-induced T cell activation. Eur. J. Immunol. 2006;36:1684–1693. doi: 10.1002/eji.200636181. [DOI] [PubMed] [Google Scholar]
- 112.Riese M.J., Moon E.K., Johnson B.D., Albelda S.M. Diacylglycerol kinases (DGKs): Novel targets for improving T cell activity in cancer. Front. Cell Dev. Biol. 2016;4 doi: 10.3389/fcell.2016.00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wiede F., Shields B.J., Chew S.H., Kyparissoudis K., Van Vliet C., Galic S., Tremblay M.L., Russell S.M., Godfrey D.I., Tiganis T. T cell protein tyrosine phosphatase attenuates T cell signaling to maintain tolerance in mice. J. Clin. Invest. 2011;121:4758–4774. doi: 10.1172/JCI59492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.LaFleur M.W., Nguyen T.H., Coxe M.A., Yates K.B., Trombley J.D., Weiss S.A., Brown F.D., Gillis J.E., Coxe D.J., Doench J.G., Haining W.N., Sharpe A.H. A CRISPR-Cas9 delivery system for in vivo screening of genes in the immune system. Nat. Commun. 2019;10 doi: 10.1038/s41467-019-09656-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lai Y., Weng J., Wei X., Qin L., Lai P., Zhao R., Jiang Z., Li B., Lin S., Wang S., Wu Q., Tang Z., Liu P., Pei D., Yao Y., Du X., Li P. Toll-like receptor 2 costimulation potentiates the antitumor efficacy of CAR T Cells. Leukemia. 2018;32:801–808. doi: 10.1038/leu.2017.249. [DOI] [PubMed] [Google Scholar]
- 116.Salerno F., Wolkers M.C. T-cells require post-transcriptional regulation for accurate immune responses. Biochem. Soc. Trans. 2015;43:1201–1207. doi: 10.1042/BST20150154. [DOI] [PubMed] [Google Scholar]
- 117.Turner M., Díaz-Muñoz M.D. RNA-binding proteins control gene expression and cell fate in the immune system. Nat. Immunol. 2018;19:120–129. doi: 10.1038/s41590-017-0028-4. [DOI] [PubMed] [Google Scholar]
- 118.Gray S.M., Kaech S.M., Staron M.M. The interface between transcriptional and epigenetic control of effector and memory. Immunol. Rev. 2014;261:157–168. doi: 10.1111/imr.12205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Jones B., Chen J. Inhibition of IFN-gamma transcription by site-specific methylation during T helper cell development. EMBO J. 2006;25:2443–2452. doi: 10.1038/sj.emboj.7601148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Samten B., Townsend J.C., Weis S.E., Bhoumik A., Shams H., Barnes P.F. Creb, ATF and AP-1 Transcription Factors Regulate IFN-γ Secretion by Human T Cells in Response to Mycobacterial Antigen. J. Immunol. 2008;181:2056–2064. doi: 10.4049/jimmunol.181.3.2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sica A., Dorman L., Viggiano V., Cippitelli M., Ghosh P., Rice N., Young H.A. Interaction of NF-kB and NFAT with the Interferon-g Promoter. J. Biol. Chem. 1997;272:30412–30420. doi: 10.1074/jbc.272.48.30412. [DOI] [PubMed] [Google Scholar]
- 122.Hermann-Kleiter N., Gruber T., Lutz-Nicoladoni C., Thuille N., Fresser F., Labi V., Schiefermeier N., Warnecke M., Huber L., Villunger A., Eichele G., Kiminski S., Baier G. Group The Nuclear Orphan Receptor NR2F6 Suppresses Lymphocyte Activation and T Helper 17-Dependent Autoimmunity. Immunity. 2008;29:205–216. doi: 10.1016/j.immuni.2008.06.008.The. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ho I., Tai T., Pai S. GATA3 and the T-cell lineage: essential functions before and after T-helper-2-cell differentiation. Nat. Rev. Immunol. 2009;9:125–135. doi: 10.1038/nri2476.GATA3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Usui T., Nishikomori R., Kitani A., Strober W. GATA-3 suppresses Th1 development by downregulation of Stat4 and not through effects on IL-12Rβ2 chain or T-bet. Immunity. 2003;18:415–428. doi: 10.1016/S1074-7613(03)00057-8. [DOI] [PubMed] [Google Scholar]
- 125.Josefowicz S.Z., Lu L., Rudensky A.Y. Regulatory T Cells: Mechanisms of Differentiation and Function Steven. Annu. Rev. Immunol. 2012;30:531–564. doi: 10.1016/j.physbeh.2017.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Cavatorta D.J., Erb H.N., Felippe M.J. Activation-induced FoxP3 expression regulates cytokine production in conventional T cells stimulated with autologous dendritic cells. Clin. Vaccine Immunol. 2012;19:1583–1592. doi: 10.1128/CVI.00308-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Fu S., Zhang N., Yopp A.C., Chen D., Mao M., Chen D., Zhang H., Ding Y., Bromberg J.S. TGF-β induces Foxp3 + T-regulatory cells from CD4 + CD25 - precursors. Am. J. Transplant. 2004;4:1614–1627. doi: 10.1111/j.1600-6143.2004.00566.x. [DOI] [PubMed] [Google Scholar]
- 128.Cohen-Chalamish S., Hasson A., Weinberg D., Namer L.S., Banai Y., Osman F., Kaempfer R. Dynamic refolding of IFN-gamma mRNA enables it to function as PKR activator and translation template. Nat. Chem. Biol. 2009;5:896–903. doi: 10.1038/nchembio.234. [DOI] [PubMed] [Google Scholar]
- 129.Kafasla P., Skliris A., Kontoyiannis D.L. Post-transcriptional coordination of immunological responses by RNA-binding proteins. Nat. Immunol. 2014;15:492–502. doi: 10.1038/ni.2884. [DOI] [PubMed] [Google Scholar]
- 130.Turner M., Galloway A., Vigorito E. Noncoding RNA and its associated proteins as regulatory elements of the immune system. Nat. Immunol. 2014;15:484–491. doi: 10.1038/ni.2887. [DOI] [PubMed] [Google Scholar]
- 131.Yue Y., Liu J., He C. RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev. 2015;29:1343–1355. doi: 10.1101/gad.262766.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.González-Feliciano J., Hernández-Pérez M., Estrella L., Colón-López D.D., López A., Martínez M., Maurás-Rivera K.R., Lasalde C., Martínez D., Araujo-Pérez F., González C.I. The role of HuR in the post-transcriptional regulation of interleukin-3 in T cells. PLoS One. 2014;9 doi: 10.1371/journal.pone.0092457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ganguly K., Giddaluru J., August A., Khan N. Post-transcriptional regulation of immunological responses through riboclustering. Front. Immunol. 2016;7 doi: 10.3389/fimmu.2016.00161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Moore K.S., Von Lindern M. RNA Binding Proteins and Regulation of mRNA Translation in Erythropoiesis. Front. Physiol. 2018;9 doi: 10.3389/fphys.2018.00910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kontoyiannis D., Pasparakis M., Pizarro T.T., Cominelli F., Kollias G. Impaired On/Off Regulation of TNF Biosynthesis in Mice Lacking TNF AU-Rich Elements: Implications for Joint and Gut-Associated Immunopathologies. Immunity. 1999;10:387–398. doi: 10.1016/S1074-7613(00)80038-2. [DOI] [PubMed] [Google Scholar]
- 136.Beisang D., Bohjanen P. Perspectives on the ARE as it turns 25 years old. Wiley Interdiscip Rev RNA. 2012;3:719–731. doi: 10.1002/wrna.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hodge D.L., Berthet C., Coppola V., Kastenmüller W., Buschman M.D., Schaughency P.M., Shirota H., Scarzello A.J., Subleski J.J., Anver M.R., Ortaldo J.R., Lin F., Reynolds D.A., Sanford M.E., Kaldis P., Tessarollo L., Klinman D.M., Young H.A. IFN-gamma AU-rich element removal promotes chronic IFN-gamma expression and autoimmunity in mice. J. Autoimmun. 2014;53:33–45. doi: 10.1016/j.jaut.2014.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Louis I. Vlasova-St., Bohjanen P.R. Post-transcriptional regulation of cytokine signaling by AU-rich and GU-rich elements. J. Interf. Cytokine Res. 2014;34:233–241. doi: 10.1089/jir.2013.0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Salerno F., Engels S., van den Biggelaar M., Van Alphen F.P.J., Guislain A., Zhao W., Hodge D.L., Bell S.E., Medema J.P., Von Lindern M., Turner M., Young H.A., Wolkers M.C. Translational repression of pre-formed cytokine-encoding mRNA prevents chronic activation of memory T cells. Nat. Immunol. 2018;19:828–837. doi: 10.1038/s41590-018-0155-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ogilvie R.L., Abelson M., Hau H.H., Vlasova I., Blackshear P.J., Bohjanen P.R. Tristetraprolin down-regulates IL-2 gene expression through AU-rich element-mediated mRNA decay. J. Immunol. 2005;174:953–961. doi: 10.4049/jimmunol.174.2.953. [DOI] [PubMed] [Google Scholar]
- 141.Salerno F., Guislain A., Freen-van Heeren J.J., Benoit P., Young H.A., Wolkers M.C. Critical role of post-transcriptional regulation for IFN-γ in tumor-infiltrating T cells. Oncoimmunology. 2018;8 doi: 10.1080/2162402X.2018.1532762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.J.J. Freen-van Heeren, B.P. Nicolet, M.C. Wolkers, Combined Single-Cell Measurement of Cytokine mRNA and Protein in Immune Cells, Methods Mol. Biol. 2108 (2020) 259–271. [DOI] [PubMed]
- 143.Aggarwal B.B. Signalling pathways of the TNF superfamily: A double-edged sword. Nat. Rev. Immunol. 2003;3:745–756. doi: 10.1038/nri1184. [DOI] [PubMed] [Google Scholar]
- 144.Matsushita K., Takeuchi O., Standley D.M., Kumagai Y., Kawagoe T., Miyake T., Satoh T., Kato H., Tsujimura T., Nakamura H., Akira S. Zc3h12a is an RNase essential for controlling immune responses by regulating mRNA decay. Nature. 2009;458:1185–1190. doi: 10.1038/nature07924. [DOI] [PubMed] [Google Scholar]
- 145.Chang P.-P., Lee S.K., Hu X., Davey G., Duan G., Cho J.-H., Karupiah G., Sprent J., Heath W.R., Bertram E.M., Vinuesa C.G. Breakdown in repression of IFN-γ mRNA leads to accumulation of self-reactive effector CD8+ T cells. J. Immunol. 2012;189:701–710. doi: 10.4049/jimmunol.1102432. [DOI] [PubMed] [Google Scholar]
- 146.Turnis M.E., Andrews L.P., Vignali D.A.A. Inhibitory receptors as targets for cancer immunotherapy. Eur. J. Immunol. 2015;45:1892–1905. doi: 10.1002/eji.201344413.Inhibitory. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wolf Y., Anderson A.C., Kuchroo V.K. TIM3 comes of age as an inhibitory receptor. Nat. Rev. Immunol. 2020;20:173–185. doi: 10.1038/s41577-019-0224-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Xia A., Zhang Y., Xu J., Yin T., Lu X.J. T Cell Dysfunction in Cancer Immunity and Immunotherapy. Front. Immunol. 2019;10 doi: 10.3389/fimmu.2019.01719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Bajwa R., Cheema A., Khan T., Amirpour A., Paul A., Chaughtai S., Patel S., Patel T., Bramson J., Gupta V., Levitt M., Asif A., Hossain M.A. Adverse Effects of Immune Checkpoint Inhibitors (Programmed Death-1 Inhibitors and Cytotoxic T-Lymphocyte-Associated Protein-4 Inhibitors): Results of a Retrospective Study. J. Clin. Med. Res. 2019;11:225–236. doi: 10.14740/jocmr3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Sheppard K.A., Fitz L.J., Lee J.M., Benander C., George J.A., Wooters J., Qiu Y., Jussif J.M., Carter L.L., Wood C.R., Chaudhary D. PD-1 inhibits T-cell receptor induced phosphorylation of the ZAP70/CD3ζ signalosome and downstream signaling to PKCθ. FEBS Lett. 2004;574:37–41. doi: 10.1016/j.febslet.2004.07.083. [DOI] [PubMed] [Google Scholar]
- 151.Hui E., Cheung J., Zhu J., Su X., Taylor M.J., Wallweber H.A., Sasmal D.K., Huang J., Kim J.M., Mellman I., Vale R.D. T cell costimulatory receptor CD28 is a primary target for PD-1–mediated inhibition. Science. 2017;355:1428–1433. doi: 10.1126/science.aaf1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kamphorst A.O., Wieland A., Nasti T., Yang S., Zhang R., Barber D.L., Konieczny B.T., Daugherty C.Z., Koenig L., Yu K., Sica G.L., Sharpe A.H., Freeman G.J., Blazar B.R., Turka L.A., Owonikoko T.K., Pillai R.N., Ramalingam S.S., Araki K., Ahmed R. Rescue of exhausted CD8 T cells by PD-1–targeted therapies is CD28-dependent. Science. 2017;355:1423–1427. doi: 10.1126/science.aaf0683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Liu J., Qian C., Cao X. Post-Translational Modification Control of Innate Immunity. Immunity. 2016;45:15–30. doi: 10.1016/j.immuni.2016.06.020. [DOI] [PubMed] [Google Scholar]
- 154.Reily C., Stewart T.J., Renfrow M.B., Novak J. Glycosylation in health and disease. Nat. Rev. Nephrol. 2019;15:346–366. doi: 10.1038/s41581-019-0129-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Liu X., Zhang Y., Cheng C., Cheng A.W., Zhang X., Li N., Xia C., Wei X., Liu X., Wang H. CRISPR-Cas9-mediated multiplex gene editing in CAR-T cells. Cell Res. 2017;27:154–157. doi: 10.1038/cr.2016.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ren J., Liu X., Fang C., Jiang S., June C.H., Zhao Y. Multiplex genome editing to generate universal CAR T cells resistant to PD1 inhibition. Clin. Cancer Res. 2017;23:2255–2266. doi: 10.1158/1078-0432.CCR-16-1300.Multiplex. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Zhao J., Lin Q., Song Y., Liu D. Universal CARs, universal T cells, and universal CAR T cells. J. Hematol. Oncol. 2018;11:132. doi: 10.1186/s13045-018-0677-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Ren J., Zhang X., Liu X., Fang C., Jiang S., June C.H., Zhao Y. A versatile system for rapid multiplex genome-edited CAR T cell generation. Oncotarget. 2017;8:17002–17011. doi: 10.18632/oncotarget.15218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Preece R., Georgiadis C., Gkazi S.A., Etuk A., Christi A., Qasim W. Mini’ U6 Pol III promoter exhibits nucleosome redundancy and supports multiplexed coupling of CRISPR/Cas9 effects. Gene Ther. 2020:1–8. doi: 10.1038/s41434-020-0142-z. [DOI] [PubMed] [Google Scholar]
- 160.Georgiadis C., Preece R., Nickolay L., Etuk A., Petrova A., Ladon D., Danyi A., Humphryes-Kirilov N., Ajetunmobi A., Kim D., Kim J.S., Qasim W. Long Terminal Repeat CRISPR-CAR-Coupled “Universal” T Cells Mediate Potent Anti-leukemic Effects. Mol. Ther. 2018;26:1215–1227. doi: 10.1016/j.ymthe.2018.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Choi B.D., Yu X., Castano A.P., Darr H., Henderson D.B., Bouffard A.A., Larson R.C., Scarfò I., Bailey S.R., Gerhard G.M., Frigault M.J., Leick M.B., Schmidts A., Sagert J.G., Curry W.T., Carter B.S., Maus M.V. CRISPR-Cas9 disruption of PD-1 enhances activity of universal EGFRvIII CAR T cells in a preclinical model of human glioblastoma. J. Immunother. Cancer. 2019;7 doi: 10.1186/s40425-019-0806-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Webber B.R., Lonetree C. lin, Kluesner M.G., Johnson M.J., Pomeroy E.J., Diers M.D., Lahr W.S., Draper G.M., Slipek N.J., Smeester B.S., Lovendahl K.N., McElroy A.N., Gordon W.R., Osborn M.J., Moriarity B.S. Highly efficient multiplex human T cell engineering without double-strand breaks using Cas9 base editors. Nat. Commun. 2019;10 doi: 10.1038/s41467-019-13007-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Ye L., Park J., Dong M., Yang Q., Chow R., Peng L., Du Y., Guo J., Dai X., Wang G., Errami Y., Chen S. In vivo CRISPR screening in CD8 T cells with AAV-Sleeping Beauty hybrid vectors identifies membrane targets for improving immunotherapy for glioblastoma. Nat. Biotech. 2019;37:1302–1313. doi: 10.1016/j.physbeh.2017.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Yang J.C. Toxicities Associated with Adoptive T-Cell Transfer for Cancer James. Cancer J. 2015;21:506–509. doi: 10.1016/j.physbeh.2017.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Shimabukuro-Vornhagen A., Gödel P., Subklewe M., Stemmler H.J., Schlösser H.A., Schlaak M., Kochanek M., Böll B., von Bergwelt-Baildon M.S. Cytokine release syndrome. J. Immunother. Cancer. 2018;6:56. doi: 10.1016/j.jons.2018.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.