Abstract
Recent breakthroughs in our understanding of orthopaedic infections have come from advances in transmission electron microscopy imaging of murine models of bone infection, most notably Staphylococcus aureus invasion and colonization of osteocyte-lacuno canalicular networks of live cortical bone during the establishment of chronic osteomyelitis. To further elucidate this microbial pathogenesis and evaluate the mechanism of action of novel interventions, additional advances in transmission electron microscopy imaging are needed. Here we present detailed protocols for fixation, decalcification, and epoxy embedment of bone tissue for standard transmission electron microscopy imaging studies, as well as the application of immunoelectron microscopy to confirm S. aureus occupation within sub-micron canaliculi. We also describe the first application of the novel Automated-Tape-UltraMicrotome system with three-dimensional reconstruction and volumetric analyses to quantify S. aureus occupation within the osteocyte-lacuno canalicular networks. Reconstruction of the three-dimensional volume broadened our perspective of S. aureus colonization of the canalicular network and, surprisingly, revealed adjacent non-infected canaliculi. This observation has led us to hypothesize that viable osteocytes of the osteocyte-lacuno canalicular networks respond and resist infection, opening future research directions to explain the paradox of adjacent uninfected canaliculi and life-long deep bone infection in patients with chronic osteomyelitis.
Keywords: Transmission Electron Microscopy (TEM), Methods, Paraffin, Osteomyelitis, Automated-Tape-UltraMicrotome (ATUMtome)
INTRODUCTION
Although transmission electron microscopy (TEM) has been critical for elucidating microbial pathogenesis of human disease1, its application in bone infection research has been very limited due to the great technical challenges in generating high quality images, and the painstaking labor of mining large segments of tissue to find the bacteria at greater than x1000 magnification. Further, bone tissue is inherently difficult to prepare for TEM studies, but with proper fixation, optimization of decalcification techniques, and a slight modification to processing of bone into epoxy embedding media, excellent imaging results can be obtained. Recently, we developed methods to overcome these challenges using TEM, demonstrating Staphylococcus aureus invasion and colonization of the osteocyte-lacuno-canalicular-network (OLCN) in live cortical bone during chronic osteomyelitis2; 3. This discovery has provided insights into how S. aureus remains in a proliferative phase (not dormant) in bone, and continues to divide as elongated “rod shaped” bacteria to accommodate submicron spaces of the OLCN2; 4. Thus, S. aureus appears to be “motile” via asymmetric binary fision at the leading edge of the colony, which initiates entry into exposed canaliculi and propogation throughout the OLCN via haptotaxis and durotaxis2; 5; 6. Furthermore, we propose that bacterial secretion of lactic acid and proteases leads to demineralization of the bone and organic matrix digestion respectively, serving as an inexhaustible nutrient source supporting the proliferative phase beyond the life of the host 6; 7. To formally test these hypotheses, ultrastructural 3D interrogation of the tissue volume is required to permit a greater appreciation of the OLCN, a quantitative assessment of the OLCN bacterial load, and open up the potential inquiry into why some canaliculi appear to remain resistant to infection.
To achieve this goal, methodologies developed for serial section electron microscopy (SSEM) were employed. SSEM involves a continuous collection of ultra-thin sections onto a reel of Kapton® tape using an automated tape-collecting ultramicrotome (ATUMtome, Boeckeler Instruments, Tucson, AZ). Each section is imaged in sequential order using a scanning electron microscope (SEM) fitted with a backscatter electron (BSE) detector to produce a ‘stack’ of acquired images that can be analyzed to examine the cellular ultrastructure. Historically, the resolution capabilities of images collected with BSE detection were inferior to TEM however, this has been improved and images can now be captured at a 4 nm pixel size making it virtually indistinguishable from TEM. In published datasets 8–10, 25 nm thin sections are cut and imaged at a pixel size of 20–30 nm to produce a stack of more than 33,000 images to reconstruct 1 mm3 of cortical tissue. While SSEM has mostly been confined to the central nervous system11; 12, recent studies have modified the methodology to the regenerating peripheral nervous system13 with a view to broadening its application and determine whether the SSEM methodology could be further modified and applied the skeletal system to examine the OLCN system.
Using these techniques it is hoped that future studies will answer several long-standing questions about osteomyelitis, including: why S. aureus infection of bone is incurable?, and why the efficacy of antibiotic-loaded bone cement is limited6; 14; 15? It also begs future TEM studies on polymicrobial infections, and on the host response that must exist to allow for life-long bone infection following pediatric osteomyelitis14. This paper presents protocols for standard high contrast TEM preparation of infected mouse bone specimens and introduces methods to process paraffin embedded infected mouse bone for more rapid interrogation of bacterial localization, ultrastructural and 3-dimensional studies. Finally, the process of SSEM is introduced, and how it can be applied to interrogate the OLCN in osteomyelitis.
METHODS
The methods of this report, given its technical nature, aim to supply the reader with justification and/or explanation of the methodologies, as well as, “step-by-step” protocols described within a section. Where applicable, these detailed protocols are supplied immediately following the rationale behind the method.
Archival murine tibia tissue
EM studies were performed on archived histology blocks of S. aureus infected tibiae from previous studies in which mice were infected with a transtibial stainless-steel pin contaminated with ~105 colony forming units of UAMS-1 or USA300, and the tibiae were harvested on day 14, as previously described15. The tibiae histology blocks used to generate the data in Figures 1, 4, 6 and 7A were previously published2, and the tibiae histology blocks used to generate the data in Figures 3, 5 and 7B are from an unpublished study performed on institutional animal care and use committee protocols approved by the ethical committee of the canton of Grisons in Switzerland.
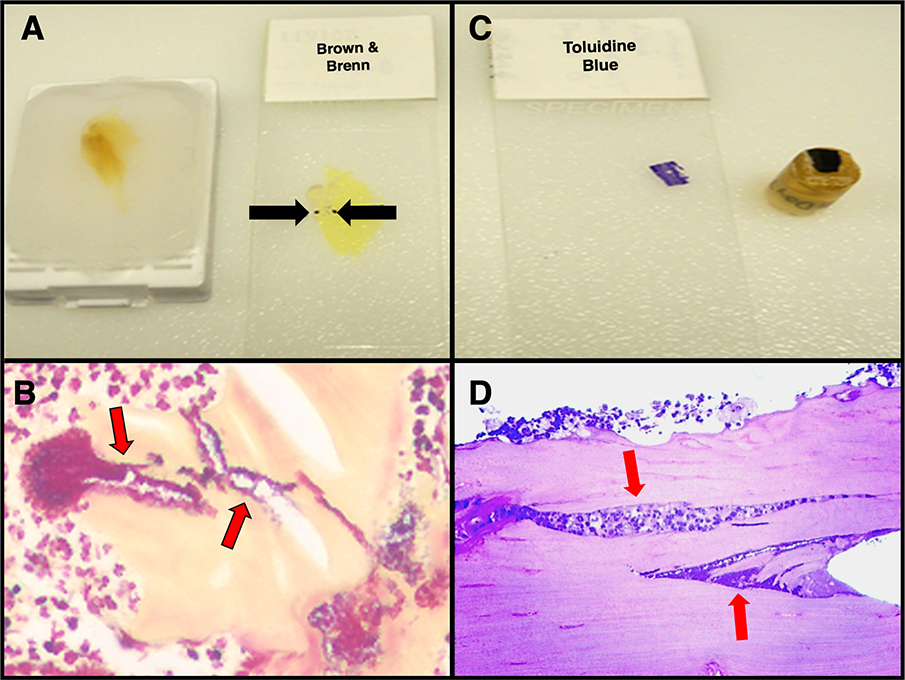
Figure 1: Rapid identification of S. aureus biofilm within infected mouse tibia sections via paraffin embedded bone special stains.
Identifying S. aureus in an infected bone sample is a “needle in a haystack” endeavor when interrogating traditional 1.0 μm epoxy embedded bone sections. To facilitate this, we utilize Gram and proteoglycan histochemical stains to rapidly identify infected regions of interest. A) Demineralized paraffin embedded whole mouse tibiae sectioned at 5 μm onto slides and stained with Brown & Brenn (black arrows) identifies biofilm presence. Note: This area of the paraffin block is cut out, deparaffinized and re-embedded into epoxy resin for 3D ATUMtome serial sectioning while allowing substantially more area of tissue to be examined while initially screening. B) S. aureus bacteria stained purple (red arrows) within the yellow counter stained bone. C) Traditional epoxy block of a trimmed tibial bone fragment which has been sectioned at 1.0 μm onto the glass slide and Toluidine blue stained. D) Light microscopy image of the above 1.0 μm section (in C) stained with Toluidine blue highlights the dark blue stained S. aureus bacteria (red arrows).
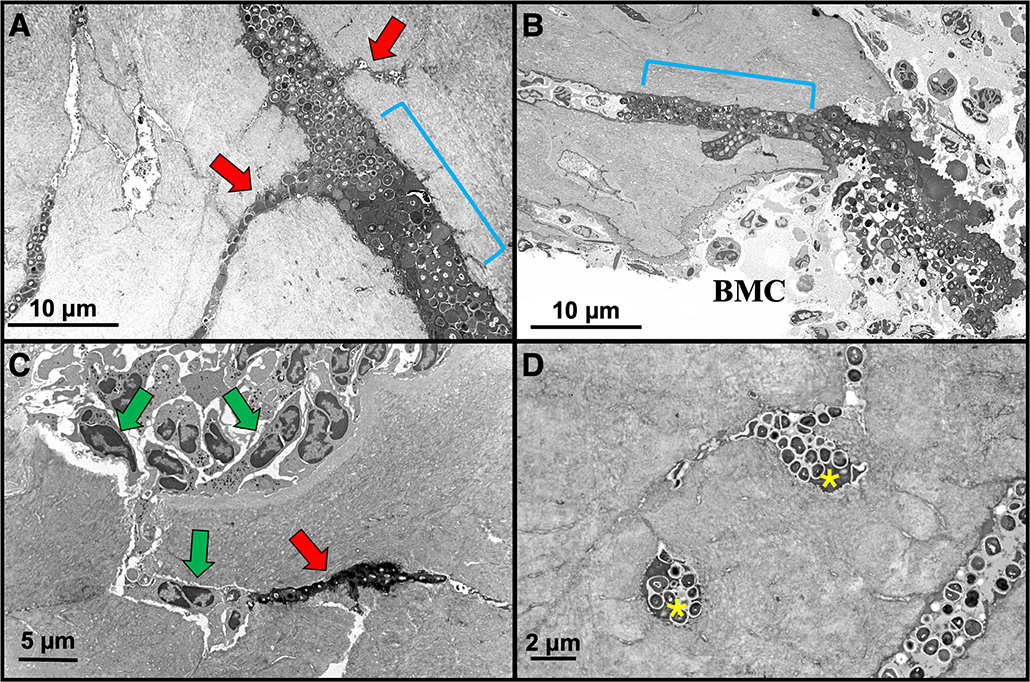
Figure 4: HC-TEM image examples of S. aureus utilizing the OLCN in mouse bone.
A) Expanded canaliculus with S. aureus biofilm (blue bracket) branches off into adjacent submicron-sized canaliculi of the OLCN (red arrows) x 4,000. B) Another digital electron micrograph displaying S. aureus biofilm invasion (blue bracket) into bone adjacent to the bone marrow compartment (BMC) x 2,000. C) During host response to the infection, neutrophils (green arrows) fill the bone marrow space and kill bone lining cells exposing canaliculi. Note neutrophils within the bone tissue attempt to track the biofilm into bone, but fail as S. aureus evades detection by invading the OLCN (red arrow) x 4,000. D) Two adjacent osteocytic lacunae (yellow astericks) are occupied by S. aureus bacteria invasion via the OLCN. Note the scalloped wall of an expanded canaliculus (lower right) x 7,000.
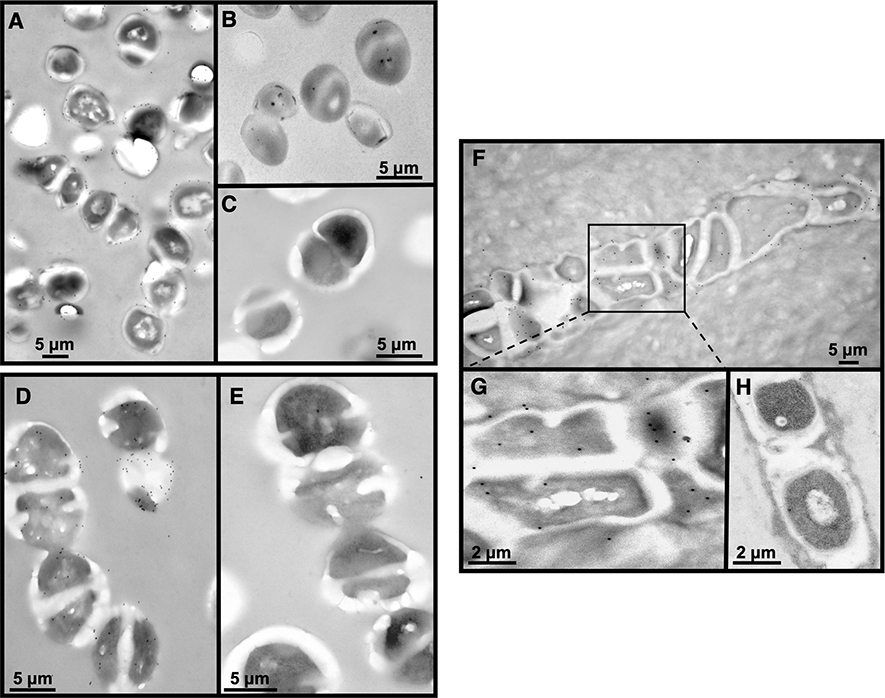
Figure 6: Immunoelectron microscopy labeling.
A) UAMS-1 bacterial pellet trapped in agarose, embedded in LR White, ultrathin sectioned and labeled using an anti-S. aureus antibody (Abcam 37644) which specifically recognizes cell wall peptidoglycan, tagged with a 12 nm gold (small black dots around the bacteria) tagged secondary antibody x 30,000. B) negative control (no primary antibody, only secondary) x 50,000. C) S. agalactiae (GBS) bacteria do not label using the anti-S. aureus antibody x 60,000. D) S. agalactiae (GBS) bacterial pellet trapped in agarose, embedded in LR White, ultrathin sectioned displays positive labeling using a polyclonal anti-GBS antibody Abcam 53584) and a 12 nm gold tagged secondary antibody. E) negative control. F) In vivo labeling of a 14 day S. aureus infected mouse tibia embedded in LR White and ultrathin sectioned using the same antibody used for S. aureus in vitro labeling x 25,000. G) Enlargement of the boxed area in F. Note the black dots positively label peptidoglycan on the outer wall of the deformed bacteria. H) Negative control x 40,000.
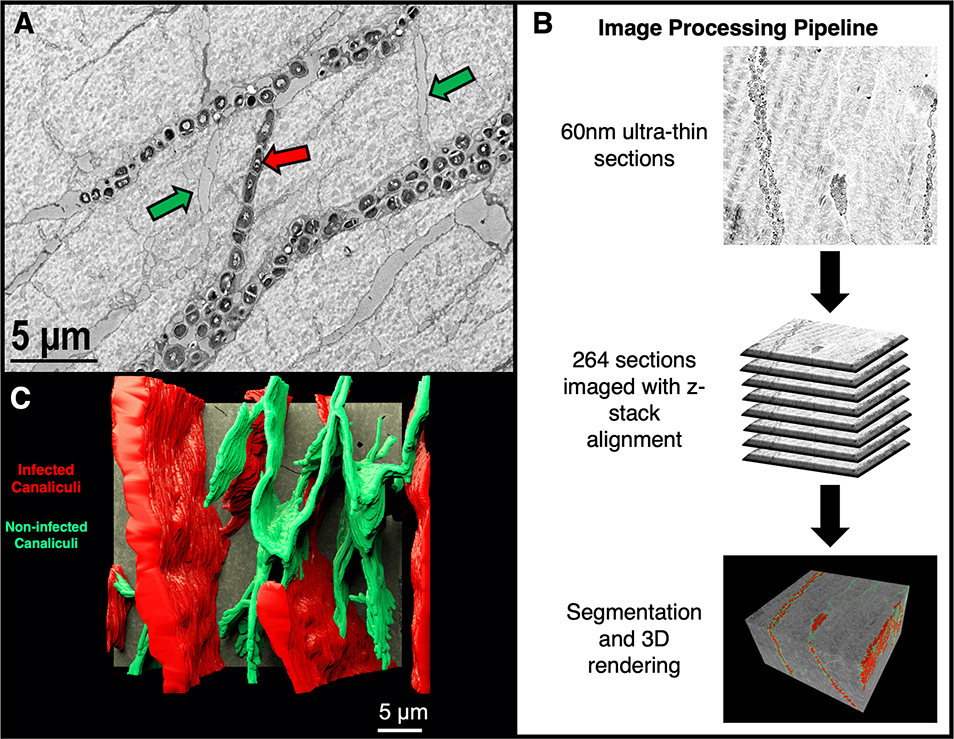
Figure 7: 3-Dimensional surveillance of S. aureus invading the Osteocyte-Lacuno-Canalicular Network (OLCN).
An archived 14 day S. aureus infected mouse tibia paraffin block was re-embedded into epoxy resin for 3D volume reconstruction of bacteria within the OLCN. A) An archived original HC-TEM image example of bacterial cross invasion from one osteocyte’s canaliculus to another (red arrow). Note: Canaliculi which are void of bacteria are denoted by green arrows x 6,000. B) The image processing pipeline for serial section imaging of an S. aureus infected mouse tibia. 60 nm ultrathin sections were collected utilizing the ATUMtome and 264 images were digitally acquired, aligned and segmented. C) A 3-dimensional rendering of the OLCN. Infected OLCN (red) represent 70% of the OLCN while the remaining non-infected canaliculi (green) appear to be resistant to infection. [Supplementary material – Figure 7-S1 & S2: Video of the overview of the OLCN (S1) and a walkthrough of the OLCN (S2)]
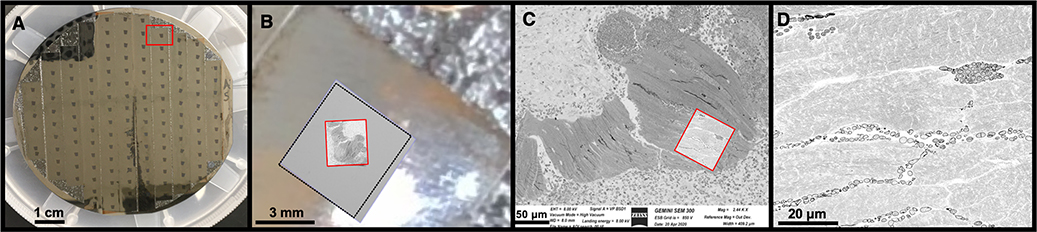
Figure 3: Locating and defining the region of interest within the serial sections.
A) Following collection on Kapton tape, the serial sections are secured onto a four inch wafer with double sided carbon tape. Up to ~150 sections can be mounted on a single wafer. The red rectangle surrounds one individual section, seen as a small black rectangle. B) A low resolution image captured from the camera mounted within the SEM chamber identifying an entire section (black dotted square) mounted on Kapton tape, this step is crucial for mapping the sections on the wafer to permit automated image acquisition. Within each section a corresponding large region of interest is defined on each section mounted on the wafer at a resolution of 75 nm/pixel (red square). C) Defining the specific region of interest to be imaged. A high resolution scan (5 nm/pixel) is completed on all sections to interrogate the specific region of interest (red square). D) The resulting SEM high resolution image demonstrating S. aureus infiltration of the OLCN. The image is 8594 × 9057 pixels (~40 μm x 45 μm area).
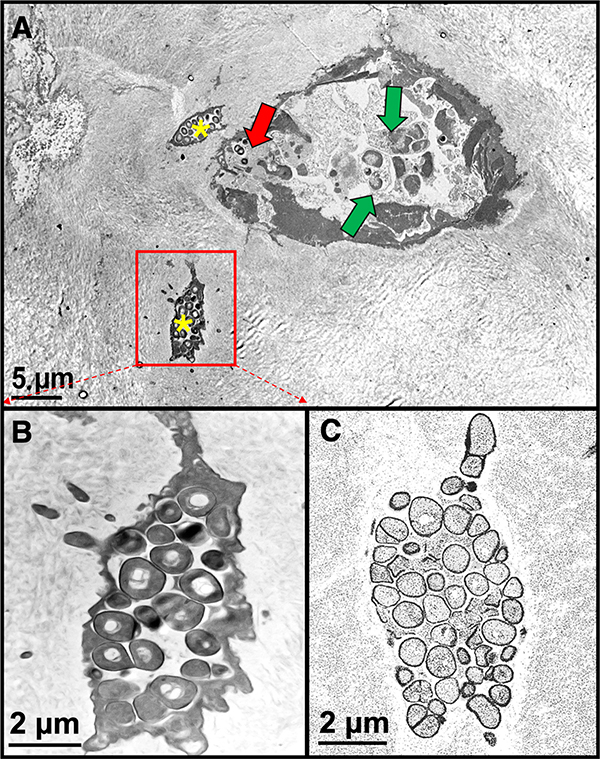
Figure 5: Epoxy re-embedded paraffin bone TEM versus SEM comparison.
A) The large ovoid structure represents a pseudo-inclusion lined by dead osteoblasts and bone lining cells. In the center are several dead neutrophils with apoptotic nuclei (green arrows). Note cocci (red arrow). Two osteocytes (yellow asterisks) have been invaded by S. aureus bacteria. B) An enlargement of the lower left osteocyte in (A). Note the bacteria are electron dense using standard TEM imaging. C) Same osteocyte imaged using a SEM from the ATUMtome sections on a wafer. Note the bacteria are comparable to (B) but are electron lucent due to the SEM set up in backscatter mode.
Fixation and Decalcification
In our mouse tibia implant model of S. aureus osteomyelitis15; 16 we have routinely used immersion fixation with excellent preservation of the ultrastructural integrity of both soft and hard tissue compartments. This fixation method works well, as the integrity of the tibia cortical bone wall is compromised during implant insertion, thus after sacrifice a combination fixative made of buffered 2.5% glutaraldehyde (GA) in 4.0% paraformaldehyde (PFA) diffuses readily into the bone tissue over a period of 24 hours. Specifically, PFA penetrates quickly to stabilize bone and soft tissue, followed by permanent cross linking with slower penetrating GA. After fixation we have ascertained the most suitable decalcification method is with Webb Jee buffer, a buffered 14% ethylenediaminetetraacetic acid (EDTA) solution, followed by implant removal. To produce routine High Contrast-TEM (HC-TEM) images, the bone must be trimmed down to the implant site for post-fixation of lipids and lipoproteins by osmium tetroxide where it reduces, blackening the tissue, as it is deposited as metallic osmium. However, no osmium is used for tibias destined for bacterial species identification of either mono- or polymicrobial infected mouse bone as preservation of antigenicity for immunoelectron microscopy (IEM) is best using 4.0% PFA and not more than 8 hours of fixation, prior to decalcification.
Fixative Preparation:
Heat 400 ml of distilled H2O to approximately 60oC.
Add 40 g E.M. grade PFA (Cat# 19210, Electron Microscopy Sciences, Hatfield, PA) to distilled H2O at 60oC and bring the volume up to 500 ml.
Add 10N sodium hydroxide solution dropwise until the PFA solution clears. Cool to room temperature.
Add 500 ml of 0.2M sodium cacodylate buffer (EMS Cat. #11653), mix and pH to 7.4. A 1:1 dilution with the 8% PFA.
Add glutaraldehyde to 2.5%. A 50% concentrate of glutaraldehyde (EMS Cat. #16310) is preferred to reduce the volume added.
Decalcification with Webb Jee Buffer (14% EDTA pH 7.4):
24 hours post fixation (8–12 hours for bone destined for immunolabeling) bone sample(s) are decalcified in labeled tissue cassette(s).
Wash samples, 3 × 5 minutes with 1X phosphate buffered saline (PBS).
Webb Jee buffer is changed daily for 7 days. Agitate buffer using a stir bar or by placing the cassettes into a large beaker onto a rocker platform.
[Note: Samples should be stripped of most soft tissue and the volume should be 50 times the volume of the specimen, minimally.]
Post-Fixation
Prepare a 1.0% osmium tetroxide solution in 0.1M sodium cacodylate buffer
Incubate at room temperature on a shaker with gentle agitation for 60 minutes
Wash 2X 10 minutes in dH2O
Resin Embedment of Bone
Subsequent choice of an embedding resin optimizes the intended ultrastructural result. For example, EPON/Araldite epoxy resin is routinely used to produce HC-TEM ultrastructural images and for serial sectioning 3D volume reconstruction. LR White acrylic resin is ideal for IEM labeling experiments, as it is a hydrophilic resin. However, either resin type requires preliminary 1 μm sectioning to locate bacteria invasion into the OLCN by light microscopy. Both types of resin can be ultrathin sectioned onto and contrast enhanced with heavy metal staining. The processing into either resin requires preliminary dehydration and infiltration steps, although the two processes vary slightly, details are outlined below. In the case of EPON/araldite resin, the ratio of individual components can be altered to optimize the properties of the resin.
EPON/Araldite Processing
Dehydrate stepwise in a graded series of ethanol (50%, 65%, 80%, 95%, 100%) in 30 minute steps on a rocking platform. Repeat the 100% ethanol incubation for a total of 3 changes.
Transition to propylene oxide (PO) (EMS Cat# 20401) for infiltration. Incubate in 1:1 100% ethanol: PO 30 minutes, then 100% PO twice for 30 minutes.
Mix EPON/Araldite resin components at the following ratio; Embed 812 (EMS Cat# 14900) 14.7g, Araldite 502 (EMS Cat# 10900) 9.0g, DDSA (EMS Cat# 13710) 30.2g and stir vigorously. Lastly add 1 mL accelerator DMP-30 (EMS Cat# 13600) and stir to ensure through mixing.
Infiltrate in 1:1, PO: EPON/araldite resin a minimum of 2.5 hours and as long as overnight.
Transfer to 100% EPON/araldite resin for 12–24 hours with the lid off the processing vials. Prior to embedment place in fresh EPON/araldite resin for a minimum of 2 hours.
Polymerize tissue in EPON/araldite at 60–65° C for 48 hours.
LR White Processing
[Note: Processing for LR White embedment proceeds without osmication.]
Wash tissue three times in base buffer following removal from fixative for 15 minutes each.
Dehydrate stepwise in a graded series of ethanol (50%, 65%, 80%, 95%,100%) in 20 minute steps on a rocking platform.
Infiltrate in 1:1, 100% ethanol.:LR White resin (EMS Cat# 14381) for 90 minutes and then into 1:2 100% ethanol:LR White resin for a minimum of 60 minutes.
Transfer to 100% LR White resin for 60 minutes and then fresh 100% LR White resin for overnight. Prior to embedment incubate twice in fresh changes of resin for 30 minutes each.
Polymerize tissue in the resin for 24 hours in an oven at 55°C.
Immunogold Labeling for TEM
[Note: Use LR White embedded tissue.]
-
Ultrathin sections and grids: Collect 80–90 nm ultrathin sections onto nickel 150 mesh formvar/carbon coated grids for the post-embedding immunogold labeling procedure.
[Note: let grids dry overnight before starting the labeling procedure.]
-
Blocking: Use 1.0% serum diluted in 1.0% bovine serum albumin/PBS. Use serum to match the secondary antibody. Incubate for 30 minutes.
[Note: The grids must not dry during the entire labeling procedure.]
-
Primary Antibody: Incubate sections on droplets of primary antibody (1:20 for both anti-S. aureus (ab37644) and anti- S. agalactiae (GBS) (ab53584) (Abcam, Cambridge, MA) diluted in 1.0% BSA/PBS overnight at 40C. NOTE: The primary antibody dilution is concentrated 10 times for immunogold labeling procedures (determine the dilution after testing the antibody on paraffin sections at the light microscopy level).
[Note: You must use an antibody compatible with immunohistochemistry/paraffin (IHC-P).]
Rinsing of grids: Using squeeze bottle of PBS, stream droplets from the nozzle for one minute directly over the surface of the each grid (held by a reverse action tweezers), then place grids on droplets of PBS 5 × 2 minutes each.
Secondary Antibody: Incubate sections at room temperature for 1 hour on droplets of 12nm gold tagged secondary antibody (1:25 Jackson Immunoresearch Laboratories, West Grove, PA ) diluted in 10% BSA/PBS. Rinse with squeeze bottle of PBS, then distilled water, then droplets of distilled water (4 × 1 minute each) to thoroughly rinse.
Rinse grids using distilled water and stain with aqueous uranyl acetate 0.3% lead citrate 5 minutes each.
Examine under the TEM for digital photography.
Most EM research laboratories would agree that sectioning epoxy embedded tissue at 1 μm to find a “needle in a haystack” region of interest (ROI) (with Toluidine blue staining) is extremely labor intensive. Thus, we present a new methodology to rapidly interrogate mouse tibiae using formalin fixed paraffin embedded tibiae. In general, 5 μm paraffin sections are cut and Brown & Brenn stained to identify Gram positive bacteria and presumed invasion into the OLCN (Figure 1). Once the ROI is identified, the targeted area is cut from the paraffin block, deparaffinized, post-fixed and reprocessed into epoxy resin for thin sectioning and TEM imaging. Specifically, to deparaffinize the embedded bone the excised fragment was taken through 3 incubations of fresh xylene followed by a rehydration in an ethanol series from 100% to 50% in 30 minute steps and eventually transferred to deionized water for post-fixation in GA and osmium. This significantly expedites the interrogation of infected mouse bone while still providing excellent ultrastructural morphology.
Serial section electron microscopy
To our knowledge, all electron microscopy studies of infected bone to date have been restricted to interpretation of 2D ultrastructure. This approach was implemented for the initial investigations and subsequent discovery of S. aureus invasion and colonization of the OLCN2; 3. However, upon discovering that S. aureus propagates deeply into bone via the OLCN, a 3D volumetric analysis of infected murine tibiae was initiated. To accomplish this, the ATUMtome system was selected (Figure 2). The resulting digital images are similar in resolution and contrast to traditional HC-TEM digital micrographs.
Figure 2: Comparing the workflow of traditional sectioning and HC-TEM imaging with serial sectioning and SEM imaging.
In traditional HC-TEM only a single grid can be loaded at a time for image collection as opposed to >100 sections placed onto a 4 inch silicon wafer for imaging using a SEM. Left: Standard ultrathin sectioning for routine TEM requires single sections be placed onto individual slot grids. Note: A traditional ultramicrotome and diamond knife setup produces individual floating ultrathin sections on the surface of the water in the diamond knife reservoir. Right: The ATUMtome setup for collection of large numbers of serial sections. Note: The ATUMtome system feeds Kapton tape into the water reservoir of the diamond knife, directly adjacent to the diamond knife edge, loading ultrathin 50–60 nm sections sequentially onto the Kapton tape. [Supplementary material - Figure 2-S: A video demonstrating the process of sections being collected onto the ATUMtome.]
ATUMtome Microtomy
After the Brown and Brenn stained ROI is matched to the paraffin blockface, it is excised and re-embedded blockface down into epoxy resin and surveyed initially by light microscopy. A 45° Histo Diamond Knife with a 6 mm blade (DiATOME, Hatfield, PA) was used at 0.5 μm levels to pinpoint the previously identified bacteria from the block face of the original paraffin interrogated bone tissue. Once the ROI is re-identified, the block is trimmed and subsequently ultrathin sectioned using a 35° Ultra Diamond Knife (DiATOME) at a thickness of 50–60 nm. Prior to the set-up of the ATUMtome for serial sectioning event, we collect 2–3 sections for mounting onto 2 × 1 mm formvar carbon coated nickel slot grids (EMS Cat# G102-Ni) for standard 2D HC-TEM imaging using a Hitachi 7650 TEM (Tokyo, Japan) to assess the ultrastructure of the re-embedded bone. This step is also necessary to target the ROI once serial sections on wafers are placed into the SEM. The ATUMtome is set-up to generate serial sections, and has been previously described in detail17. Sections are generally cut at a speed of 1 mm/sec through the cutting window (see supplementary Figure 2-S) which is matched by the tape collection reel of the ATUMtome and using these parameters ~14 sections are cut per minute. At this rate, up to 840 sections can be cut and collected in an hour, through ~ 50 μm of tissue. Following serial sectioning, the ATUM collection apparatus is removed from the diamond knife boat and an additional 2–3 ultrathin sections are collected as before for standard HC-TEM image analysis.
Wafer mounting, post-staining and carbon coating
264 sections from the sample were collected onto Kapton® ¼” tape (Sheldahl, Northfield, MN). 12 mm width double sided conductive carbon tape (EMS Cat# 77817–12) was applied to 100 mm diameter silicon wafers (EMS Cat# 71893–01). The serial sections were mounted onto the wafer by cutting the Kapton® into lengths corresponding to the diameter of the wafer. To enhance the signal for electron microscopy, the sections were post-stained with 2% uranyl acetate dissolved in deionized water for two minutes and rinsed with 18.2 MΩ deionized water before secondary rinsing with Photo-Flo 200 solution (Eastman Kodak Company, Rochester, NY). Sections were dried with an air gun to prevent the formation of water marks. To improve contrast, a secondary post-stain was applied using 0.3% lead citrate for two minutes before rinsing with deionized water and Photo-Flo as described above. To prevent the sections from becoming electrically charged during electron microscopy imaging, grounding is required. To achieve this, the sections were coated in a thin film of carbon using a Ladd vacuum evaporator (Ladd Research, Williston, VT). 3 mm carbon rods (EMS Cat# 70210) were sharpened and placed between the two electrodes, and the wafer-mounted specimens were inserted beneath the rods into the vacuum dome and pumped down until a vacuum of −8 E-5 torr was achieved. Current was applied across the carbon rods until the wafer was finely coated.
Image acquisition
Once the specimens had been prepared for imaging, they were transferred to a ZEISS GeminiSEM 300 scanning electron microscope fitted with an electron backscatter diffusion detector (Carl Zeiss Microscopy Gmbh, Germany). Utilizing the ZEISS Atlas 5 software (Carl Zeiss), the x,y locations of the wafers were mapped to stage coordinates. The region of interest was determined after low magnification, low resolution overview images were acquired (Figure 3). Sections were imaged individually using the inbuilt auto-focus and auto-contrast functions. The image parameters used included 5 nm pixel size using a 8594 × 9057 pixel field of view (approximately 40 μm x 45 μm area). The beam was set to use 8kV with a 60 μm aperture to give approximately 290 pA of current, 6.4 us dwell time, and 7.8 mm working distance.
Alignment and segmentation of data
During automated tape-collection, the ATUMtome cannot control the orientation of specimens as they are collected from the water reservoir onto the Kapton® tape, image stacks require alignment adjustment. Alignment was achieved utilizing ImageJ open source imaging software18 and the alignment plugin Linear Stack Alignment with SIFT19. The aligned data set was manually segmented using the open source software VAST Lite20. Two types of segmentation were undertaken, firstly, a saturated segmentation in which all S. aureus bacteria were segmented within the OCLN system and secondly, infected and not infected canaliculi were segmented separately. Once segmentation had been completed, data was exported as .OBJ files utilizing VAST Tools (included with VAST Lite) that are typically recognized by 3D rendering software. Autodesk 3ds Max (Autodesk Inc., San Rafael, CA) was used to render three-dimensional reconstructions of the segmented data.
RESULTS
Transmission electron microscopy of re-embedded paraffin tissue
Given the high resolution micrographs generated by EM, there is a legitimate concern for the loss of ultrastructural integrity with re-embedment of the tissue. For comparison, we present archived images from standard HC-TEM (Figure 4) for reference. Specifically in all four electron micrographs, from previous HC-TEM studies, the decalcified bone presents as a gray stippled matrix and host inflammatory cells located either in the bone or marrow cavity are identifiable as neutrophils, due to their well preserved multi-lobed nuclear profiles and cytoplasmic granules (Figure 4B & C). The S. aureus bacteria are distinctive as electron dense (dark gray) 1 μm cocci (Figure 4A–D). The identification of osteocyte lacunae with (Figure 4D) and without bacteria (Figure 4A) are architecturally distinctive. Most remarkable is the preservation of canaliculi in the matrix of bone, some of which are occupied by propagating S. aureus.
In comparison, a thin section taken prior to the SSEM event was imaged by TEM (Figure 5A & B) and we were able to observe all ultrastructural components (as in Figure 4) of bone tissue, host neutrophils and S. aureus cocci near and within osteocytic lacunar spaces (Figure 5A). The textural matrix of bone of is consistent with the quality of HC-TEM tissue, as well as the nuclear features of neutrophils and bone marrow cells identifiable at the ultrastructural level. Note how the ultrastructural preservation of bacteria within osteocytic lacunar spaces maintains the same distinctive electron density making them comparable to the HC-TEM reference images. The large ovoid structure in Figure 5A, represents a pseudo-inclusion lined by dead osteoblasts and bone lining cells which have condensed nuclear and cytoplasmic features. Of note, the same osteocyte is present in both the TEM image and the SSEM image (a micron or so from one another), and are morphologically similar (Figure 5B–C). Note that in standard TEM images, bacteria appear electron dense as the electrons have been both absorbed and transmitted at varying degrees to produce the contrast seen in a 60 nm ultrathin section. In contrast, using an SEM in backscatter mode, the bacteria appear electron lucent as the electrons are reflected back after elastic interactions occur between the electron beam and the sections on the wafers.
Overall the bacteria and tissue architecture are very well preserved and identifiable, despite the treatment with xylene to remove paraffin and rehydration down to water. To ensure high quality preservation of ultrastructure of re-embedded bone, it is necessary to perform a two-step post-fixation method; 24 hours in 2.5% GA followed by 1 hour incubation in 1.0% osmium tetroxide prior to processing into epoxy resin.
Immunoelectron microscopy (IEM) labeling
Patients presenting with diabetic foot ulcers typically have either mono- or polymicrobial infections. Most monomicrobial infections involve S. aureus21, but S. agalactiae22 (GBS) has also been recognized to be an important pathogen. To establish IEM labeling protocols to distinguish these different pathogenes in infected bone, pure cultures of both bacterial strains were embedded into LR White resin for in vitro IEM labeling using anti-S. aureus and anti-GBS antibodies (Figure 6A–E). Furthermore, IEM labeling of bacteria occupying the OLCN was performed on archived LR White resin-embedded bone blocks of S. aureus 14 day infected mouse bone (Figure 6F–H). Positive IEM labeling was observed for both S. aureus (Figure 6A) and S. agalactiae (Figure 6D), as evidenced by the 12 nm gold particles coating of the cell wall, while no reactivity was observed in no primary antibody controls (Figure 6B & E). Importantly a lack of cross-reactivity was demonstrated by the inability of the anti-S. aureus antibody to label cultured GBS (Figure 6C). While the controlled labeling of pure culture was successful, the efficacy of the antibody requires testing with samples from in vivo infection. Therefore, we sought to evaluate the efficacy of the S. aureus antibody on archived LR White resin embedded bone blocks of S. aureus 14 day infected mouse bone. In vivo we observed S. aureus positive labeling in widened canaliculi (Figure 6F&G), while the secondary antibody only control had no labeling (Figure 6H) of the elongated S. aureus aligned in a canaliculus.
Serial section electron microscopy (SSEM) and quantitation of infected OLCN volume
The total volume of infected and non-infected canaliculi was calculated utilizing in-built VAST Lite computational capabilities using cubic pixels (voxels). The total volume of segmented canaliculi was 1,524,285,936 voxels of which 1,070,046,752 (70.2%) demonstrated infiltration by S. aureus in contrast to 454,239,184 (29.8%) that appeared not to be infected (Figure 7).
DISCUSSION
Traditionally, high magnification TEM imaging requires specific fixation, processing and embedding protocols for high quality preservation of tissue ultrastructure. Most diagnostic EM laboratories believe excising tissue from formalin fixed paraffin embedded soft tissue blocks often results in loss of fine structural details. However, since bone is a rigid connective tissue type, sparsely populated by cells, it is more resistant to loss of the fine structural qualities of the OLCN even after re-embedding. A protocol that includes post-fixation GA and post-staining with osmium tetroxide, can optimize the ultrastructural aspects of not only the bone tissue but also bacterial morphology within the OLCN; this greatly enhances the structural detail captured in this 3D volume imaging study.
To date, the production of 3D images in the musculoskeletal system has been achieved using CT and MRI imaging modalities. However, these are limited by resolution parameters and cannot capture ultrastructural detail. Other 3D technologies exist and are generally grouped into SBF or SSEM imaging. To use SBF imaging, an ultramicrotome is inserted into the SEM chamber and the block face of the sample is imaged. Subsequently, a 25–50 nm thin section is shaved from the block face, discarded and then the new block face of the specimen is imaged. This process ensures that the image acquisition is well aligned and reduces the post-processing effort required in contrast to the ATUMtome. As described above, sections are collected onto the tape system that can allow a changed orientation whilst floating in the water reservoir as they are cut on the diamond knife. The process of alignment of the data stack is both time and computationally involved. There are several advantages of the ATUMtome system, but the most notable is that sections can be stored indefinitely and may be re-imaged at any time, permitting continued analyses without the need to repeat experiments thus has clear advantages over any other methodology. A full comparison of the approaches for serial section imaging is presented in Table 1.
Table 1:
Comparison of serial section imaging modalities.
| System | Classification | Specifications | Advantages | Disadvantages |
|---|---|---|---|---|
| Thermofisher Volumescope | SBF | The limited block height (2–3 mm) | ||
| Widest knife for serial block face | Enbloc staining is required | |||
| Knife width 2 mm | The knife travel | Cutting blind - block may be non-uniformly fixed or stained. Impossible to screen before cutting | ||
| Knife travel 2–3 mm | Setup and running is all in one step | Monopolizing the microscope-- Once the cutting and imaging has started, it must continue until the project is complete | ||
| Block height 2–3 mm | Serial section alignment is reasonably easy | Possible catastrophic failure at any point in the cutting/imaging phase | ||
| Too large of an electron dose per pixel can change the mechanical properties of the block and make cutting difficult | ||||
| Gatan 3View | SBF | Knife width is limited to less than 2 mm | ||
| Knife travel is about 1 mm (this means that the block face cannot be larger than 1 mm) | ||||
| Knife width <2 mm | Add-on to any existing SEM | Maximum block height is too small | ||
| Easy to set up and run | Enbloc staining is required | |||
| Knife travel ~1 mm | Section thickness of 15 nm or greater | Cutting blind - block may be non-uniformly fixed or stained. Impossible to screen before cutting | ||
| Block height 600 μm | Serial section alignment is reasonably easy | Monopolizing the microscope-- Once the cutting and imaging has started, it must continue until the project is complete | ||
| Possible catastrophic failure at any point in the cutting/imaging phase | ||||
| Too large of an electron dose per pixel can change the mechanical properties of the block and make cutting difficult | ||||
| ATUMtome System | SSEM | Can be used with either Leica or RMC Boeckeler ultramicrotomes | ||
| Independent of the SEM type | ||||
| Virtually unlimited depth of cut (z-direction) | ||||
| Knife width 4 mm | Can cut longitudinally (parallel to the fiber axis) or transverse (diameter cross section) | Multi-step process: (1) cutting and collecting, (2) wafer making, (3) post-staining, and (4) wafer mapping |
||
| Knife travel 6 mm | Sections can be post-stained for better signal to noise | 2D stitching may be required | ||
| Sections are collected and mounted on silicon wafers | 3D alignment (alignment from section to section) can be difficult | |||
| Block height 6 mm | Each wafer can be imaged independently and at the microscope’s convenience | Section cutting and collection may generate wrinkles | ||
| Sections can be reimaged, in the case of focus or other problems or a change in the region of interest | ||||
| Can perform Enbloc gold or other immuno-labeling | ||||
| Catastrophic failure occurs early in the workflow (long before the imaging phase) | ||||
| Serial TEM | SSEM | N/A | Fast data acquisition | Requirement of grid tape, expense and difficulty |
| Post-staining is limited | ||||
| 2D stitching and 3D alignment is extremely challenging | ||||
| Transmission SEM | SSEM | N/A | Attaches to any SEM | Slow data acquisition |
| Limited field of view (200 μm x 200 μm) | ||||
| Slotted grids required for section collection | ||||
| Requires additional detector | ||||
There are limitations to the SSEM data presented. SSEM is a labor-intensive process that is dependent on user-expertise, availability of instrumentation and staff, and computer processing personel and resources and furthermore the data being collected and analyzed. The most predictable components are the tissue preparation and sectioning as outlined above. The time taken to section a volume of data is related to section thickness; the data we present here was sectioned at 60 nm, whereas other reports sectioned at 20 nm10; 17 and collected the same volume of tissue which would take orders of magnitude longer with a concurrent increase in the volume of data. Image acquisition is similarly difficult to predict without imaging a limited sample to determine optimal parameters. Our dataset was acquired utilizing a pixel size of 5 nm and a field of view (FOV) of 8594 × 9057 pixels (approximately 40 μm x 45 μm area). Some datasets do not require such high resolution13 and individual sections can be imaged more quickly however, other parameters also impact acquisition time such as the dwell time, the time taken for auto-focus and auto-contrast. To provide some guidance, and overview of the time taken for each step for the OLCN data collected is listed in Table 2.
Table 2.
An approximation for the time and labor intensity required to complete the 3D reconstruction of the S. aureus infected bone sample
| Processing Step | Hands on Involvement | Time taken | |
|---|---|---|---|
| Tissue preparation | Fixation | Full | 1 day |
| Decalcification | Minimal | 7 days | |
| Resin embedding | Full | 4 days | |
| Serial Sectioning | Block face preparation | Full | 1 hour |
| ATUMtome setup | Full | 1–2 hours | |
| Serial sectioning | Minimal | 1 hour | |
| Wafer Preparation (time taken per wafer) | Mounting sections | Full | 30 minutes |
| Post-staining | Full | 15 minutes | |
| Carbon coating | Full | 15 minutes | |
| Data Acquisition (time taken per wafer) | Defining ROI | Full | 1–2 hours |
| Defining imaging parameters | Full | 2–4 hours | |
| Wafer mapping | Minimal | 1 hour | |
| Image acquisition | Minimal | 2–3 days | |
| Post Processing (264 image stack) | Data-stack alignment | Minimal | 1–2 days |
| Manual segmentation | Full | 5 days | |
| 3D Rendering/Modelling | Moderate | 5 days | |
| TOTAL | 28 days |
Another important limitation of SSEM is that quantitation of the infected OLCN volume cannot be validated with traditional CFU assays, as the tissue destructive experimental methods for each are incompatible with the other. Thus, we propose the primary use of the volume outcome is to gain a general assessment of the extent of OLCN infection. Consistent with this, our results have successfully demonstrated that SSEM can be applied to the skeletal system, and provide a new solution to not only investigate the OLCN system but also bacterial propagation and antibiotic resistance. The preliminary results demonstrate that 70% of canaliculi examined within this tissue volume were occupied by S. aureus. With the application of this methodology, future studies could reveal how osteocytes (and their canaliculi) may resist colonization through cell signaling, eventually succumbing to bacterial persistence, as evidenced by patient reports of recurrence decades later. Potentially, this could open up new directions for discovery of osteocyte protective mechanisms in future osteomyelitis research23; 24. Finally, in order to make this a realistic target, automated segmentation methodologies utilizing machine learning must be developed and applied to the bone tissue datasets to increase the efficiency of 3D image analyses.
CONCLUSION:
Electron microscopy remains an invaluable tool for the interrogation of osteomyelitis. Through manipulation of established methodologies and incorporation of emerging technology, a comprehensive strategy for the investigation of S. aureus osteomyelitis is presented. Based on the results of this study, SSEM utilizing the ATUMtome system can successfully interrogate the OLCN and provide 3D volumetric analysis.
Future studies focused on polymicrobial bone infections, specifically combined S. aureus and S. agalactiae (GBS) infections, are required to determine why some osteocytic lacunae within the OLCN are resistant to invasion and colonization. These studies will also include IEM labeling to answer questions pertaining to mono vs. polymicrobial infections, and if other microorganisms are capable of invasion of and propagation within the OLCN.
Supplementary Material
ACKNOWLEDGMENTS:
This work was supported by research grants from the National Institutes of Health (P30 AR069655 and P50 AR72000), and AOTrauma Clinical Priority Program. The processing and sectioning, mounting and post processing prior to imaging was conducted in the Electron Microscopy Shared Resource Laboratory at the University of Rochester. The authors would like to thank Boeckeler, Instruments of Tucson, AZ for loaning the ATUMtome system to complete this project and also the Zeiss imaging facility at Pleasanton, CA, for completing the data acquisition remotely during the current pandemic.
REFERENCES:
- 1.Curry A, Appleton H, Dowsett B. 2006. Application of transmission electron microscopy to the clinical study of viral and bacterial infections: present and future. Micron 37:91–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Mesy Bentley KL, MacDonald A, Schwarz EM, et al. 2018. Chronic Osteomyelitis with Staphylococcus aureus Deformation in Submicron Canaliculi of Osteocytes: A Case Report. JBJS Case Connect 8:e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Mesy Bentley KL, Trombetta R, Nishitani K, et al. 2017. Evidence of Staphylococcus Aureus Deformation, Proliferation, and Migration in Canaliculi of Live Cortical Bone in Murine Models of Osteomyelitis. J Bone Miner Res 32:985–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Philip Kollmannsberger MK, Felix Repp, Wolfgang Wagermaier, Richard Weinkamer, Peter Fratzl. 2017. The small world of osteocytes: connectomics of the lacunocanalicular network in bone. New J Phys 19. [Google Scholar]
- 5.Masters EA, Salminen AT, Begolo S, et al. 2019. An in vitro platform for elucidating the molecular genetics of S. aureus invasion of the osteocyte lacuno-canalicular network during chronic osteomyelitis. Nanomedicine 21:102039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Masters EA, Trombetta RP, de Mesy Bentley KL, et al. 2019. Evolving concepts in bone infection: redefining “biofilm”, “acute vs. chronic osteomyelitis”, “the immune proteome” and “local antibiotic therapy”. Bone Res 7:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muthukrishnan G, Masters EA, Daiss JL, et al. 2019. Mechanisms of Immune Evasion and Bone Tissue Colonization That Make Staphylococcus aureus the Primary Pathogen in Osteomyelitis. Curr Osteoporos Rep 17:395–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaynig V, Vazquez-Reina A, Knowles-Barley S, et al. 2015. Large-scale automatic reconstruction of neuronal processes from electron microscopy images. Med Image Anal 22:77–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Joesch M, Mankus D, Yamagata M, et al. 2016. Reconstruction of genetically identified neurons imaged by serial-section electron microscopy. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kasthuri N, Hayworth KJ, Berger DR, et al. 2015. Saturated Reconstruction of a Volume of Neocortex. Cell 162:648–661. [DOI] [PubMed] [Google Scholar]
- 11.McCarthy M 2013. US to launch major brain research initiative. BMJ 346:f2156. [DOI] [PubMed] [Google Scholar]
- 12.Morgan JL, Lichtman JW. 2013. Why not connectomics? Nature methods 10:494–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leckenby JI, Chacon MA, Grobbelaar AO, et al. 2019. Imaging Peripheral Nerve Regeneration: A New Technique for 3D Visualization of Axonal Behavior. J Surg Res 242:207–213. [DOI] [PubMed] [Google Scholar]
- 14.Libraty DH, Patkar C, Torres B. 2012. Staphylococcus aureus reactivation osteomyelitis after 75 years. N Engl J Med 366:481–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nishitani K, Sutipornpalangkul W, de Mesy Bentley KL, et al. 2015. Quantifying the natural history of biofilm formation in vivo during the establishment of chronic implant-associated Staphylococcus aureus osteomyelitis in mice to identify critical pathogen and host factors. J Orthop Res 33:1311–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li D, Gromov K, Soballe K, et al. 2008. Quantitative mouse model of implant-associated osteomyelitis and the kinetics of microbial growth, osteolysis, and humoral immunity. Journal of orthopaedic research : official publication of the Orthopaedic Research Society 26:96–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baena V, Schalek RL, Lichtman JW, et al. 2019. Serial-section electron microscopy using automated tape-collecting ultramicrotome (ATUM). Methods Cell Biol 152:41–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schindelin J, Arganda-Carreras I, Frise E, et al. 2012. Fiji: an open-source platform for biological-image analysis. Nature methods 9:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lowe DG. 2004. Distinctive image features from scale-invariant keypoints. International journal of computer vision 60:91–110. [Google Scholar]
- 20.Berger DR, Seung HS, Lichtman JW. 2018. VAST (Volume Annotation and Segmentation Tool): Efficient Manual and Semi-Automatic Labeling of Large 3D Image Stacks. Front Neural Circuits 12:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pulido L, Ghanem E, Joshi A, et al. 2008. Periprosthetic joint infection: the incidence, timing, and predisposing factors. Clin Orthop Relat Res 466:1710–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaplan SL. 2014. Recent lessons for the management of bone and joint infections. J Infect 68 Suppl 1:S51–56. [DOI] [PubMed] [Google Scholar]
- 23.Dallas SL, Prideaux M, Bonewald LF. 2013. The osteocyte: an endocrine cell … and more. Endocr Rev 34:658–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang D, Wijenayaka AR, Solomon LB, et al. 2018. Novel Insights into Staphylococcus aureus Deep Bone Infections: the Involvement of Osteocytes. mBio 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.