Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has caused the global COVID-19 pandemic infecting more than 106 million people and causing 2.3 million deaths. The rapid deployment of antibody-based countermeasures has provided hope for curtailing disease and ending the pandemic1. However, the emergence of rapidly-spreading SARS-CoV-2 variants in the United Kingdom (B.1.1.7), South Africa (B.1.351), and elsewhere with mutations in the spike protein has raised concern for escape from neutralizing antibody responses and loss of vaccine efficacy based on preliminary data with pseudoviruses2–4. Here, using monoclonal antibodies (mAbs), animal immune sera, human convalescent sera, and human sera from recipients of the Pfizer-BioNTech (BNT162b2) mRNA vaccine, we report the impact on antibody neutralization of a panel of authentic SARS-CoV-2 variants including a B.1.1.7 isolate, a chimeric Washington strain with a South African spike gene (Wash SA-B.1.351), and isogenic recombinant variants with designed mutations or deletions at positions 69–70, 417, 484, 501, and/or 614 of the spike protein. Several highly neutralizing mAbs engaging the receptor binding domain (RBD) or N-terminal domain (NTD) lost inhibitory activity against Wash SA-B.1.351 or recombinant variants with an E484K spike mutation. Most convalescent sera and virtually all mRNA vaccine-induced immune sera tested showed markedly diminished neutralizing activity against the Wash SA-B.1.351 strain or recombinant viruses containing mutations at position 484 and 501. We also noted that cell line selection used for growth of virus stocks or neutralization assays can impact the potency of antibodies against different SARS-CoV-2 variants, which has implications for assay standardization and congruence of results across laboratories. As several antibodies binding specific regions of the RBD and NTD show loss-of-neutralization potency in vitro against emerging variants, updated mAb cocktails, targeting of highly conserved regions, enhancement of mAb potency, or adjustments to the spike sequences of vaccines may be needed to prevent loss of protection in vivo.
Keywords: Antibody-based Countermeasures, Vaccine Efficacy, Pseudoviruses, Chimeric Washington Strain, South African Spike Gene, Assay Standardization
To evaluate the effects of SARS-CoV-2 strain variation on antibody neutralization, we obtained or generated a panel of authentic infectious SARS-CoV-2 strains with sequence variations in the spike gene (Fig 1a–c). A B.1.1.7 isolate had signature changes in the spike gene5 including the 69–70 and 144–145 deletions, and N501Y, A570D, D614G, and P681H substitutions. We created a chimeric, fully-infectious SARS-CoV-2 strain with a South African spike gene (Wash SA-B.1.351; D80A, 242–244 deletion, R246I, K417N, E484K, N501Y, D614G, and A701V) and a panel of isogenic spike mutants (D614G, K417N/D614G, E484K/D614G, N501Y/D614G, P681H/D614G, del69–70/N501Y/D614G, E484K/N501Y/D614G, and K417N/E484K/N501Y/D614G) in the Washington strain background (2019n-CoV/USA_WA1/2020 [WA1/2020]). Recombinant viruses and B.1.1.7 were propagated in Vero-TMPRSS2 and Vero-hACE2-TMPRSS2 cells expressing transmembrane protease serine 2 (TMPRSS2) and human angiotensin converting enzyme 2 (hACE2) to prevent the development of adventitious mutations in the spike, especially at or near the furin cleavage site, which accumulate rapidly in Vero E6 cells6 and can impact entry pathways and virulence7. All viruses were used at low passage (p0 or p1) and deep sequenced to confirm mutations (Supplementary Table S1).
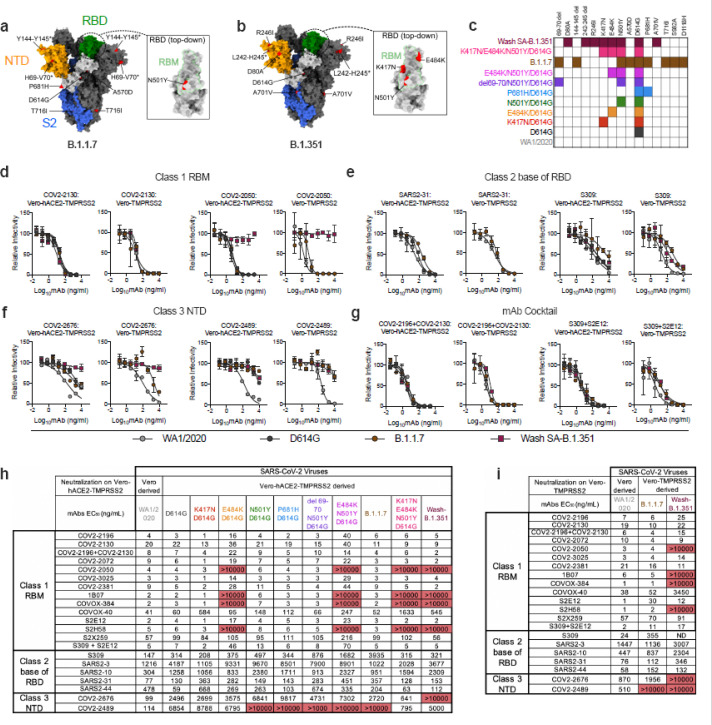
Figure 1.
Neutralization of SARS-CoV-2 viral variants by mAbs. a-b, SARS-CoV-2 spike trimer. One protomer is highlighted, showing the NTD in orange, RBD in green, and S2 portion of the molecule in blue, with N- and C-termini annotated. a, Substitutions in the B.1.1.7 variant (69–70 deletion, 144–145 deletion, A570D, D614G, P681H, and T716I) are shaded in red. Red triangle depicts approximate location of P681H, which was not included in the model. Inset shows a top-down view of the RBD showing the location of the N501Y mutation contextualized with the receptor-binding motif (RBM). b, Substitutions in the Wash-SA B.1.135 variant (242–244 deletion, D80A, R246I, D614G, and A701V) are shaded in red. The red diamond denotes approximate location of D80A, which is buried in this view. Inset shows top-down view of the RBD with Wash SA-B.1.351 substitutions K417N, E484K, and N501Y shaded red and contextualized with the receptor binding motif. For all panels, structures depicting spike protein were modeled using PDB: 7C2L. Structures depicting RBD were modeled using PDB: 6W41. All analyses and figures were generated with UCSF ChimeraX40. c, Viruses with indicated spike mutations used in this study. d-f, Neutralization curves in (left panels) Vero-hACE2-TMPRSS2 cells or (right panels) Vero-TMPRSS2 cells comparing the sensitivity of SARS-CoV-2 strains with class 1 (d, COV2–2130 and COV-2150), class 2 (e, SARS2–31 and S309), and class 3 (f, COV2–2676 and COV2–2489) mAbs and indicated viruses. Also shown are the neutralization curves for antibody cocktails (g, COV2–2196 + COV2–2130 and S309 + S2E12). One representative experiment of two is shown. h-i, Summary of EC50 values (ng/ml) of neutralization of SARS-CoV-2 viruses propagated on the indicated cells and performed in Vero-hACE2-TMPRSS2 (h) or Vero-TMPRSS2 (i) cells. Data are an average of two experiments, each performed in duplicate. Red shading of cells shows virtually complete loss of neutralizing activity: EC50 > 10,000 ng/mL.
We tested our panel of viruses for antibody-mediated neutralization in Vero-hACE2-TMPRSS2 cells and then repeated some experiments with Vero-TMPRSS2 cells to evaluate for effects of hACE2 over-expression on neutralization8. We performed high-throughput focus reduction neutralization tests (FRNTs)9 using a panel of neutralizing mAbs recognizing distinct and overlapping epitopes in the RBD including some having potential use in humans. Class 1 antibodies (e.g., COV2–2196, COV2–2072, COV2–2050, COV2–2381, COV2–2130, COVOX-384, COVOX-40, 1B07, S2E12, S2H58, and S2X259) are potently neutralizing, block soluble hACE2 binding, and bind multiple proximal sites in the receptor binding motif (RBM) of the RBD as determined by structural or escape mutation analyses (Extended Data Fig 1a)10–13; class 2 neutralizing antibodies (e.g., S309, SARS2–3, SARS2–10, SARS2–31, SARS2–44) often cross-react with SARS-CoV, bind the base of the RBD (Extended Data Fig 1b), and variably block hACE2 binding (14 and L. VanBlargan and M. Diamond, unpublished results); and class 3 neutralizing mAbs (e.g., COV2–2676 and COV2–2489) recognize the N-terminal domain (NTD) (Extended Data Fig 1c)15.
We performed neutralization tests with the different spike protein variants and the two cell types (Fig 1d–i and Extended Data Fig 2). With the parental WA1/2020 strain, which was derived in Vero CCL-81 cell cultures, neutralization by the majority of class 1 mAbs was similar in Vero-hACE2-TMPRSS2 or Vero-TMPRSS2 cells. In comparison, some class 2 mAbs (e.g., S309 and SARS2–44) showed a 4 to 6-fold loss in neutralization potency (EC50 value) on Vero-hACE2-TMPRSS2 compared to Vero-TMPRSS2 cells. Moreover, NTD-reactive mAbs (COV2–2489 and COV2–2676) neutralized Vero CCL-81 cell-derived WA1/2020 virus on Vero-hACE2-TMPRSS2 cells but lost activity on Vero-TMPRSS2 cells or when viruses were derived from Vero-hACE2-TMPRSS2 cells (Fig 1f, h–i and Extended Data Fig 2). Given that the expression of hACE2 on recipient Vero cells and the cellular source of virus both can impact the neutralizing activity of mAbs that bind principally outside of the RBM on the spike protein, virus neutralization assays being used for correlation with in vivo efficacy of mAbs and vaccines may produce variable results depending on the cell substrate used for virus propagation and infection.
We next assessed the impact of spike protein mutations on mAb neutralization using Vero-hACE2-TMPRSS2 cells (Fig 1h) and Vero-TMPRSS2 cells (Fig 1i). We observed the following patterns with the variant viruses: (a) The D614G or P681H mutations (in the C-terminal region of S1) and the 69–70 deletion (in the NTD) had marginal effects on neutralization potency for the RBM and RBD mAbs we evaluated. It was difficult to assess their impact on the NTD mAbs we tested, since the recombinant viruses were generated in Vero-hACE2-TMPRSS2 cells, and the NTD mAbs neutralized them poorly at baseline; (b) The K417N mutation resulted in ~10-fold reductions in neutralization by mAbs COVOX-40 and SARS2–44 but did not negatively affect other mAbs in our panel. If anything, several class 1 mAbs showed slightly improved inhibitory activity (P = 0.002, Wilcoxon matched-pairs signed rank test) with this mutation; (c) Mutation at N501Y reduced the neutralizing activity of COVOX-40 and SARS2–44 slightly but did not alter the potency of other mAbs substantively; this result is consistent with data showing that human convalescent sera efficiently neutralize viruses with N501Y substitutions16–18; (d) The E484K mutation negatively impacted the potency of several class 1 antibodies. Compared to the D614G virus, mAbs COV2–2196, COV2–3025, COV2–2381 and S2E12 showed 4- to 5-fold reduced activity against the E484K/D614G virus, and COV2–2050, COVOX-384, 1B07, and S2H58 lost virtually all neutralizing potential; (e) The combination of E484K/N501Y/D614G mutations, which is present in the circulating South African B.1.351 and Brazilian B.1.1.248 strains, showed even greater effects (6- to 13-fold reductions) on the activity of class 1 mAbs COV2–2196, COV2–3025, COV2–2381, and S2E12 mAbs; (f) When we tested class 1 mAbs for inhibition of the Wash SA-B.1.351 virus containing the full South African spike sequence, as expected, several mAbs (COV2–2050, COVOX-384, 1B07, and S2H58) lost activity in both Vero-hACE2-TMPRSS2 and Vero-TMPRSS2 cells. However, the reductions in neutralizing potential by other class 1 mAbs (COV2–2196, COV2–3025, COV2–2381, and S2E12) seen against the E484K/N501Y/D614G mutant virus were absent with Wash SA-B.1.351, which contains additional mutations. The K417N substitution, which is located at the edge of the RBM (Fig 1b) and enhances neutralization by some class 1 mAbs, may compensate for the negative effects on inhibition of the E484K/N501Y mutations. In comparison, we observed a distinct neutralization pattern with Wash SA-B.1.351 for class 2 and 3 mAbs. Because some of these mAbs neutralized Vero-hACE2-TMPRSS2 cell-derived virus poorly when tested in Vero-hACE2-TMPRSS2 cells, we performed parallel experiments in Vero-TMPRSS2 cells. Class 2 mAbs binding the base of the RBD showed small reductions in potency against the Wash SA-B.1.351. However, the two NTD mAbs in class 3 (COV2–2676 and COV2–2489) showed a loss of neutralizing activity against Wash SA-B.1.351 in Vero-TMPRSS2 cells, consistent with recent data with other NTD mAbs using pseudoviruses4; (g) With one exception, none of the class 1 mAbs lost activity against the B.1.1.7 isolate on Vero-TMPRSS2 or Vero-hACE2-TMPRSS2 cells. However, we observed moderately diminished neutralizing activity (3- to 11-fold reduction) of some class 2 mAbs (SARS2–31, SARS2–44, and S309) against the B.1.1.7 strain depending on the cell substrate. The reduced potency of S309 mAb against B.1.1.7 strain contrasts with data showing it binds avidly to the B.1.1.7 spike protein on the surface of cells and neutralizes a vesicular stomatitis virus (VSV) pseudotyped with B.1.1.7 spike protein in Vero E6 cells (Extended Data Fig 3a–b). Moreover, one of the NTD class 3 mAbs (COV2–2489) also showed a marked loss of inhibitory activity against the B.1.1.7 strain in both cell types, possibly due to the deletions present in the NTD (69–70 and 144–145)15. Together, these data indicate that cell line selection (for both growth of virus stocks and neutralization assays) and hACE2 receptor expression are important variables in assessing the potency of antibodies against different SARS-CoV-2 variants.
Several academic and industry groups have developed mAb cocktails to overcome possible emergence of resistance during therapy13,19. We tested two mAb combinations that have potential use in humans (COV2–2196 + COV2–2130 [Vanderbilt University Medical Center; with engineered derivatives being tested in clinical trials by AstraZeneca] and S309 + S2E12 [Vir Biotechnology] for their inhibitory activity against the SARS-CoV-2 variant viruses (Fig 1g–i). The COV2–2196 + COV2–2130 combination generally retained inhibitory activity (< 4-fold reduction) against all strains. Although the S309 + S2E12 combination showed reduced (~10-fold) potency against the E484K/N501Y/D614G strain, it performed effectively against the Wash SA-B.1.351 virus, again suggesting that additional mutations in natural variants (e.g., K417N) enable some antibodies to function better against viruses containing E484K and N501Y mutations.
We next assessed how spike protein mutations impacted the neutralizing activity of polyclonal sera obtained from individuals (n = 19) approximately 1 month after mild SARS-CoV-2 infection20. Based on experiments with the mAbs, we focused our testing on B.1.1.7, Wash SA-B.1.351, and WA1/2020 with mutations at D614G, K417N/D614G, E484K/N501Y/D614G, or K417N/E484K/N501Y/D614G, and used Vero-hACE2-TMPRSS2 cells for virus propagation and neutralization assays (Fig 2 and Extended Data Fig 4). These results were compared to data with similarly passaged WA1/2020 D614G and revealed the following: (a) significant differences in neutralization were not observed with the K417N/D614G or B.1.1.7 strains (Fig 2a–b), both of which lack the E484K mutation; (b) serum neutralization titers were lower against E484K/N501Y/D614G (5-fold, P < 0.0001), K417N/E484K/N501Y/D614G (3.5-fold, P < 0.0001), and Wash SA-B.1.351 (4.5-fold, < 0.0001) viruses (Fig 2c–e), all of which contain the E484K mutation. A heatmap analysis showed that most individuals lost neutralizing activity against all three viruses containing the E484K and N501Y mutations (Fig 2f).
Figure 2.
Neutralization of SARS-CoV-2 viral variants by convalescent human serum in Vero-hACE2 TMPRSS2 cells. a-e, Paired neutralization analysis of convalescent human sera (n = 19) obtained approximately 1 month after mild SARS-CoV-2 infection against WA1/2020 D614G and variant viruses in Vero-hACE2-TMPRSS2 cells: (a) B.1.1.7, (b) K417N/D614G, (c) E484K/N501Y/D614G, (d) K417N/E484K/N501Y/D614G, or (e) Wash SA-B.1.351. Results are from one experiment performed in duplicate (Wilcoxon matched-pairs signed rank test, **, P < 0.01; ****, P < 0.0001; all other comparisons, not significant). Geometric mean neutralization titers (GMT) are shown above each graph. Dotted line represents the limit of detection of the assay. f, Heat map of the relative neutralizing activity of sera from individual convalescent subjects against indicated SARS-CoV-2 viruses compared to recombinant WA1/2020 D614G. Blue, reduction; red, increase.
Given that viruses containing changes at positions 484 and 501 escape neutralization by serum from convalescent humans, we next examined the effects of vaccine-induced antibody responses. Initially, we interrogated sera from mice (n = 10), hamsters (n = 8), and non-human primates (NHP [rhesus macaques], n = 6) obtained one month after immunization with ChAd-SARS-CoV-2, a chimpanzee adenoviral vectored vaccine encoding for a prefusion stabilized form of the spike protein21–23. We assessed serum neutralization of B.1.1.7, Wash SA-B.1.351, and recombinant WA1/2020 with mutations at D614G, K417N/D614G, E484K/N501Y/D614G, or K417N/E484K/N501Y/D614G using virus derived from and tested on Vero-hACE2-TMPRSS2 cells (Extended Data Fig 5). For serum samples from mice, when comparing the GMTs of neutralization to the WA1/2020 D614G strain, we observed a slight increase (1.9-fold, P < 0.05) with K417N/D614G (Fig 3b), decreases with E484K/N501Y/D614G (9-fold, P < 0.001; Fig 3c), K417N/E484K/N501Y/D614G (5-fold, P < 0.01; Fig 3d), and Wash SA-B.1.351 (5-fold, P < 0.01; Fig 3e), yet no significant differences with B.1.1.7. (Fig 3a). In a heatmap plot (Fig 3p), 9 of the 10 mouse sera show a loss of neutralizing activity against multiple viruses containing the E484K mutation. In hamsters, the results were similar. We observed a marked decrease (10- to 12-fold, P < 0.01) in serum neutralization of E484K/N501Y/D614G, K417N/E484K/N501Y/D614G, and Wash SA-B.1.351 (Fig 3h–j). Statistically significant differences in neutralization were not observed with K417N/D614G and B.1.1.7 (Fig 3f, g). This pattern was reflected at the individual sample level (Fig 3q). In NHPs, we also observed a substantial decrease (9- to 11-fold, P < 0.05) in serum neutralization of E484K/N501Y/D614G, K417N/E484K/N501Y/D614G, and Wash SA-B.1.351 (Fig 3m–o), but no significant difference in inhibition with K417N/D614G or B.1.1.7 (Fig 3k–l). The heatmap analysis showed that all NHP sera consistently exhibited reduced neutralizing activity against viruses containing the E484K mutation (Fig 3r).
Figure 3.
Resistance of SARS-CoV-2 viral variants to neutralization by vaccine-induced serum derived from mice, hamsters, and NHPs. Paired neutralization analysis of sera from mice (a-e, n = 10), hamsters (f-j, n = 8), and NHPs (k-o, n = 6) obtained ~30 days after a single intranasal immunization with an adenoviral vectored SARS-CoV-2 vaccine (ChAd-SARS-CoV-2-S21). Neutralization data on Vero-hACE2-TMPRSS2 cells is displayed as WA1/2020 D614G versus the variant viruses: (a, f, k) B.1.1.7, (b, g, l) K417N/D614G, (c, h, m) E484K/N501Y/D614G, (d, i, n) K417N/E484K/N501Y/D614G, or (e, j, o) Wash SA-B.1.351. EC50 values were calculated from one experiment each performed in duplicate with some exceptions due to limited sample (Wilcoxon matched-pairs signed rank test, *, P < 0.05; **, P < 0.01; ***, P < 0.001; all other comparisons, not significant). GMT values are shown above each graph. Dotted line represents the limit of detection of the assay. p-r, Heat map of the relative neutralizing activity of sera from individual mice (p), hamsters (q), and NHPs (r) against indicated SARS-CoV-2 viruses compared to WA1/2020 D614G. Blue, reduction; red, increase.
Because samples from human immunization trials with ChAd-SARS-CoV-2 are not yet available, we interrogated sera from 24 individuals who received the Pfizer-BioNTech (BNT162b2) vaccine, a lipid nanoparticle encapsulated-mRNA that encodes a similar membrane-bound, prefusion stabilized form of the full-length SARS-CoV-2 spike protein24. We tested sera (Extended Data Fig 6 and 7) for neutralization of our panel of SARS-CoV-2 variants (Fig 4a–e). Compared to the WA1/2020 D614G cloned variant, we observed moderate reductions in neutralizing activity (GMTs) of B.1.1.7 (2-fold, P < 0.01; Fig 4a) and E484K/N501Y/D614G (4-fold, P < 0.0001; Fig 4c) and larger decreases in activity against Wash SA-B.1.351 (10-fold, P < 0.0001; Fig 4d), with all subjects showing substantially reduced potency (Fig 4e), results that agree with pseudovirus studies4. Significant differences in neutralizing activity were not detected with K417N/D614G (Fig 4b).
Figure 4.
Resistance of SARS-CoV-2 viral variants to neutralization by human serum from Pfizer-BioNTech BNT162b2 mRNA vaccinated individuals in Vero-hACE2-TMPRSS2 cells. Paired neutralization analysis of sera from humans (n = 24) obtained after boosting with the BNT162b2 mRNA vaccine. Neutralization data on Vero-hACE2-TMPRSS2 cells is displayed with WA1/2020 D614G versus the variant viruses: (a) B.1.1.7, (b) K417N/D614G, (c) E484K/N501Y/D614G, or (d) Wash SA-B.1.351. EC50 values were calculated from one experiment each performed in duplicate (Wilcoxon matched-pairs signed rank test, **, P < 0.01; ****, P < 0.0001; all other comparisons, not significant). GMT values are shown above each graph. Dotted line represents the limit of detection of the assay. e, Heat map of the relative neutralizing activity of sera from vaccinated individuals against indicated SARS-CoV-2 viruses compared to WA1/2020 D614G. Blue, reduction; red, increase.
We also evaluated the impact of cell substrate and hACE2 receptor expression on neutralizing activity of serum samples from convalescent adults enrolled at approximately one month after infection (Fig 5a–c) and from BNT162b2 mRNA-vaccinated individuals (Fig 5d–f). Given the limited remaining serum quantities, we performed neutralization experiments only with WA1/2020, B.1.1.7, and Wash SA-B.1.351 viruses using Vero-TMPRSS2 cells. Using next-generation sequence analysis of viral genomes, we confirmed additional mutations did not occur during passage in Vero-TMPRSS2 cells (Supplementary Table S1). These experiments (Extended Data Fig 8) revealed the following: (a) Convalescent and vaccine sera showed reductions in neutralizing activity of B.1.1.7 compared to the WA1/2020 virus. Whereas sera from the vaccinated individuals inhibited B.1.1.7 virus infection less efficiently (3.6-fold, P < 0.01; Fig 5d), convalescent sera showed a trend towards reduced neutralization that did not attain statistical significance (2.4-fold, P = 0.08; Fig 5a). (b) In comparison, sera from both convalescent and vaccinated individuals showed a marked 10- to 13-fold reduction (P < 0.01) in neutralizing potency against the Wash SA-B.1.351 virus (Fig 5b, e). The results for the vaccine sera were similar in magnitude between Vero-hACE2-TMPRSS2 and Vero-TMPRSS2 cells (see also Fig 4a, d) and suggest that cellular expression of hACE2 does not markedly impact functional outcome. However, we generally observed greater decreases in neutralizing activity against the B.1.1.7 strain with convalescent sera in Vero-TMPRSS2 cells than in Vero-hACE2-TMPRSS2 cells (see Fig 2a), possibly because NTD antibodies are produced at higher levels during natural infection than vaccination and lose binding to B.1.1.7 viruses because of deletions in the NTD25.
Figure 5.
Resistance of SARS-CoV-2 viral variants to neutralization by human serum from convalescent and vaccinated individuals in Vero-TMPRSS2 cells. Sera from individuals who had been infected with SARS-CoV-2 (a-c; n = 20, ~1-month post-infection) or vaccinated with the Pfizer-BioNTech mRNA vaccine (d-f; n = 10) were tested for neutralization of the indicated SARS-CoV-2 strains (WA1/2020 (a, b, d, e), B.1.1.7 (a, d), or Wash SA-B.1.351 (b, e) using a FRNT in Vero-TMPRSS2 cells. EC50 values were calculated from one experiment performed in duplicate (Wilcoxon matched-pairs signed rank test, **, P < 0.01). GMT values are shown above each graph. Dotted line represents the limit of detection of the assay. c, f, Heat maps of the relative neutralizing activity of sera from convalescent (c) or vaccinated (f) individuals against indicated SARS-CoV-2 viruses compared to WA1/2020. Blue, reduction; red, increase.
Our in vitro experiments using a B.1.1.7 isolate and engineered variants in the backbone of the WA1/2020 strain establish that mutations in the spike can impact the potency of antibody neutralization. Some neutralizing mAbs targeting the base of the RBD or NTD showed reduced activity against the B.1.1.7 isolate, whereas others targeting the RBM or NTD failed to inhibit infection of Wash SA-B.1.351 or variants containing the E484K mutation. These finding are potentially important because the RBM has functional plasticity26,27, and additional mutations in this region that occur as the pandemic evolves could further impact the efficacy of mAb therapies or vaccines. Our results establishing the E484K substitution as a vulnerability for multiple neutralizing mAbs are consistent with deep mutational scanning or VSV-SARS-CoV-2-based neutralization escape screening campaigns26,28,29. However, several other highly neutralizing mAbs (e.g., COV2–2196, COV2–2381, COV2–3025, and S2E12) showed intact or only slightly diminished inhibitory activity against the suite of variant viruses we tested. Moreover, cocktails of mAbs binding different epitopes in spike protein overcame virus resistance to individual mAbs. Alternative approaches to addressing the diminished mAb neutralization activity by variant SARS-CoV-2 lineages include targeting of conserved regions of the spike and identifying clonal mAb variants with greater potency, such that a given dose of mAb can protect against a range of variants despite some decrease in neutralization activity.
Our studies with human sera from convalescent subjects and recipients of the BNT162b2 mRNA vaccine, and animal sera after immunization with a vaccine encoding a similar spike gene, demonstrate a lower potency of neutralization against E484K and N501Y-containing viruses (note: we did not perform studies with the single-mutation viruses, due to limited serum availability). This observation is unexpected given that antibody responses in animals and humans are polyclonal and in theory, should overcome resistance associated with individual mutations and loss of activity of particular B cell clones.
Our analyses agree with some studies showing substantial or complete escape against spike proteins corresponding to the South African lineage (B.1.351 or 501Y.V2) by antibodies in convalescent or vaccine-immune plasma using lentiviral-based pseudotype neutralization assays2,3,17. Moreover, they are consistent with studies showing loss of neutralization potency of human convalescent serum against VSV-SARS-CoV-2 chimeric virus variants containing the E484K mutation30 and selection of escape E484K mutants under serial passage of convalescent COVID-19 plasma31. These findings may have therapeutic implications, as immune plasma derived from individuals infected early during the pandemic might fail to protect patients infected with more recent isolates containing the E484K mutation.
Our studies focused exclusively on the impact of sequence changes in the spike protein on antibody neutralization in cell culture. Despite observing marked differences in serum neutralizing activity against authentic SARS-CoV-2 variant viruses, it remains unclear how this finding translates into effects on protection in the context of secondary infection or infection after vaccination with platforms using historical spike gene sequences. Although serum neutralizing titers are an anticipated correlate of protection32, this measurement does not account for Fc effector functions; Fcg receptor or complement protein engagement by non-, weakly-, or strongly- neutralizing antibodies that bind the SARS-CoV-2 spike protein on the surface of infected cells could confer substantial protection33–35. Also, the role of memory T or B cells in protection against variant viruses is unknown and could prevent severe infection even in the setting of compromised serum antibody responses36–38.
Moreover, the field still does not know whether Vero or other cell-based neutralization assays predict antibody-mediated protection. Indeed, primary cells targeted by SARS-CoV-2 in vivo can express unique sets of attachment and entry factors39, which could impact receptor and entry blockade by specific antibodies. We observed that cell line used for growth of viral stocks or neutralization assays affects the potency of monoclonal or serum-derived antibodies against different SARS-CoV-2 variants. Such results may impact the congruity of data across laboratories and interpretation of effects of viral variants on vaccine efficacy. As an example, a recent study with Vero E6 cell-derived SARS-CoV-2 with a spike protein containing some of the South African mutations (E484K, N501Y, and D614G) showed only a small 1.2-fold decrease in neutralization potency by BNT162b2 mRNA vaccine-elicited human sera16. When we compared neutralization of deep-sequenced confirmed p0 (Vero E6 cell-produced) and p1 (Vero-hACE2-TMPRSS2 cell-produced) K417N/E484K/N501Y/D614G viruses by immune serum from vaccinated mice, hamsters, or NHPs, or naturally infected humans in recipient Vero-hACE2-TMPRSS2 cells, the p0 viruses produced in Vero E6 cells were neutralized more efficiently (~3-fold, P < 0.05) than the p1 viruses produced in Vero-hACE2-TMPRSS2 cells (Extended Data Fig 9). We speculate that TMPRSS2 might modify the spike protein of authentic SARS-CoV-2 in the producer cell such that some classes of antibodies no longer efficiently block infection of the recipient cell.
While our analysis of neutralizing antibody responses with authentic infectious SARS-CoV-2 variants on Vero-hACE2-TMPRSS2 and Vero-TMPRSS2 cells suggests that adjustments to some therapeutic antibody cocktails or existing spike sequences in vaccines might be necessary, corroborating in vivo studies are needed. Sequential infection and/or vaccination/infection studies in animals and analysis of vaccine efficacy in the setting of new variant infections ultimately will determine the impact of emerging SARS-CoV-2 lineages, especially those containing E484K mutations.
Methods
Cells.
Vero E6 (CRL-1586, American Type Culture Collection (ATCC), Vero-TMPRSS2, and Vero-hACE2-TMPRSS2 cells were cultured at 37°C in Dulbecco’s Modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 10 mM HEPES pH 7.3, 1 mM sodium pyruvate, 1× non-essential amino acids, and 100 U/ml of penicillin–streptomycin. Vero-TMPRSS2 were generated after lentivirus transduction. Briefly, human TMPRSS2 was cloned into a pLX304 lentiviral vector (gift of S. Ding, Washington University) with a C-terminal V5 tag and a blasticidin selection marker. TMPRSS2-V5-encoding vectors were packaged as lentiviruses and Vero E6 cells were transduced. Vero E6 cells stably expressing TMPRSS2 were selected under blasticidin (5 mg/mL), and surface TMPRSS2 expression was confirmed using an anti-V5 antibody (Thermo Fisher 2F11F7) or anti-TMPRSS2 mAb (Abnova, Clone 2F4) and APC-conjugated goat anti-mouse IgG (BioLegend, 405308). Vero-hACE2-TMPRSS2 were obtained as a generous gift (A. Creanga and B. Graham, NIH).
Viruses.
The 2019n-CoV/USA_WA1/2020 isolate of SARS-CoV-2 was obtained from the US Centers for Disease Control (CDC). The B.1.1.7 isolate was obtained from an infected individual. Individual point mutations in the spike gene (D614G, K417N/D614G, E484K/D614G, N501Y/D614G, P681H/D614G, del69–70/N501Y/D614G, and E484K/N501Y/D614G) were introduced into an infectious cDNA clone of the 2019n-CoV/USA_WA1/2020 (WA1/2020) strain as described previously41. Nucleotide substitutions were introduced into a subclone puc57-CoV-2-F5–7 containing the spike gene of the SARS-CoV-2 wild-type infectious clone42. The South African variant spike gene (B.1.351) was produced synthetically by Gibson assembly. The full-length infectious cDNA clones of the variant SARS-CoV-2 viruses were assembled by in vitro ligation of seven contiguous cDNA fragments following the previously described protocol42. In vitro transcription was then performed to synthesize full-length genomic RNA. To recover the mutant viruses, the RNA transcripts were electroporated into Vero E6 cells. The viruses from the supernatant of cells were collected 40-h later and served as p0 stocks43. All viruses were passaged once in Vero-hACE2-TMPRSS2 or Vero-TMPRSS2 cells and subjected to deep sequencing after RNA extraction to confirm the introduction and stability of substitutions (Supplementary Table 1). Viral RNA from cell culture supernatants was used to generate next generation sequencing libraries using either the Illumina TruSeq Stranded Total RNA Library Prep with Ribo-Zero kit or the Illumina Stranded Total RNA Prep, Ligation with Ribo-Zero Plus kit per the manufacturer’s protocol. The final indexed libraries were quantified using Agilent’s Bioanalyzer and pooled at an equal molar concentration. Illumina’s NextSeq sequencer was used to generate paired end 150 base pair reads. Raw sequencing data was processed using fastp44 v.0.20.1 (https://github.com/OpenGene/fastp) to trim adapters and filter out sequence with < Q30. Alignment to the SARS-CoV-2 reference genome (MN908947.3) was performed using BWA45 v0.7.17-r1188 (http://bio-bwa.sourceforge.net). DeepVariant46 v1.1.0 (https://github.com/google/deepvariant) was used to call variants with an allele frequency >= 50%. Variants were annotated using SNPEff47 5.0c (https://sourceforge.net/projects/snpeff/). All virus preparation and experiments were performed in an approved Biosafety level 3 (BSL-3) facility.
Monoclonal antibodies.
The human mAbs studied in this paper (COV2–2196, COV2–2072, COV2–2050, COV2–2381, COV2–2130, COVOX-384, COVOX-40, S309, S2E12, S2H58, S2X333, VIR-7381, and S2X259) were isolated from blood samples from individuals in North America or Europe with previous laboratory-confirmed symptomatic SARS-CoV or SARS-CoV-2 infection. The original clinical studies to obtain specimens after written informed consent were previously described10,13,14,25 and approved by the Institutional Review Board of Vanderbilt University Medical Center, the Institutional Review Board of the University of Washington, the Research Ethics Board of the University of Toronto, and the Canton Ticino Ethics Committee (Switzerland). Chimeric mAb 1B07 with a murine Fv and human Fc (human IgG1) were isolated from C57BL/6 mice immunized with recombinant spike and RBD proteins and described previously12. Murine mAbs were generated in BALB/c or C57BL/6 mice immunized with recombinant spike and RBD proteins and described previously28,30.
Human immune sera.
Multiple sources of human serum samples were used in this study: Convalescent serum samples were obtained from a cohort recruited from the St. Louis metropolitan area who experienced mild SARS-CoV-2 infection. None of those patients required intubation, and the study was approved by Washington University School of Medicine Institutional Review Board (202003186 (WU353)). The serum samples from individuals immunized with the Pfizer-BioNTech (BNT162b2) mRNA vaccine were obtained prior to primary immunization or one week after boosting from young adults, and the studies were approved by Washington University School of Medicine Institutional Review Board (202012081 (WU368) and 202012084 (COVaRiPAD))
Mouse, hamster, and NHP immune sera.
The mouse, hamster, and NHP immune sera were obtained one month after intranasal immunization with ChAd-SARS-CoV-2, a chimpanzee adenoviral vectored vaccine encoding for a prefusion stabilized form of the spike protein. Details of the immunization protocol and functional analyses have been described elsewhere21–23.
Focus reduction neutralization test.
Serial dilutions of mAbs or serum were incubated with 102 focus-forming units (FFU) of different strains or variants of SARS-CoV-2 for 1 h at 37°C. Antibody-virus complexes were added to Vero-hACE2-TMPRSS2 or Vero-TMPRSS2 cell monolayers in 96-well plates and incubated at 37°C for 1 h. Subsequently, cells were overlaid with 1% (w/v) methylcellulose in MEM supplemented with 2% FBS. Plates were harvested 24 h later by removing overlays and fixed with 4% PFA in PBS for 20 min at room temperature. Plates were washed and sequentially incubated with an oligoclonal pool of SARS2–2, SARS2–11, SARS2–16, SARS2–31, SARS2–38, SARS2–57, and SARS2–71 anti-S antibodies and HRP-conjugated goat anti-mouse IgG in PBS supplemented with 0.1% saponin and 0.1% bovine serum albumin. SARS-CoV-2-infected cell foci were visualized using TrueBlue peroxidase substrate (KPL) and quantitated on an ImmunoSpot microanalyzer (Cellular Technologies).
ELISA.
Assays were performed in 96-well plates (MaxiSorp; Thermo) coated with 100 μL of recombinant spike or RBD protein12 in PBS, and plates were incubated at 4 °C overnight. Plates were then blocked with 10% FBS and 0.05% Tween20 in PBS. Serum were serially diluted in blocking buffer and added to the plates. Plates were incubated for 90 min at room temperature and then washed 3 times with 0.05% Tween-20 in PBS. Goat anti-human IgG-HRP (Jackson ImmunoResearch, 1:2,500) was diluted in blocking buffer before adding to wells and incubating for 60 min at room temperature. Plates were washed 3 times with 0.05% Tween-20 in PBS, and then washed 3 times with PBS before the addition of peroxidase substrate (SigmaFAST o-Phenylenediamine dihydrochloride, Sigma-Aldrich). Reactions were stopped by the addition of 1 M HCl. Optical density measurements were taken at 490 nm. The half-maximal binding dilution for each serum or plasma sample was calculated using nonlinear regression. The limit of detection was defined as 1:30.
Transient expression of recombinant SARS-CoV-2 spike proteins and flow cytometry.
The full-length S gene of SARS-CoV-2 strain (SARS-CoV-2-S) isolate BetaCoV/Wuhan-Hu-1/2019 (accession number MN908947) carrying D614G was codon-optimized for expression in hamster cells and cloned into the pcDNA3 expression vector. Amino acid substitutions for B.1.1.7, P.1 (Brazilian lineage: L18F, T20N, P26S, D138Y, R190S, K417T, E484K, N501Y, H655Y, T1027I, and V1167F), and B.1.351 variants were introduced by overlap extension PCR. Briefly, DNA fragments with overlap sequences were amplified by PCR (step 1). Mutations were introduced by amplification with primers with similar melting points (Tm). Deletion of the C-terminal 21 amino acids was introduced to increase surface expression of the recombinant spike. Next, three contiguous overlapping fragments were fused by a first overlap PCR (step 2) using the utmost external primers of each set, resulting in three larger fragments with overlapping sequences. A final overlap PCR (step 3) was performed on the three large fragments using the utmost external primers to amplify the S gene and the flanking sequences including the restriction sites KpnI and NotI. This fragment was digested and cloned into the expression plasmid phCMV1. For all PCR reactions, the Q5 Hot Start High fidelity DNA polymerase was used (New England Biolabs), according to the manufacturer’s instructions and adapting the elongation time to the size of the amplicon. After each PCR step, the amplified regions were separated on agarose gel and purified using Illustra GFX™ PCR DNA and Gel Band Purification Kit (Merck KGaA).
Expi-CHO cells were transiently transfected with SARS-CoV-2-S expression vectors using Expifectamine CHO Enhancer. Two days later, cells were collected for immunostaining with mAbs. An Alexa647-labelled secondary antibody anti-human IgG Fc was used for detection. Binding of mAbs to transfected cells was analyzed by flow-cytometry using a ZE5 Cell Analyzer (Biorard) and FlowJo software (TreeStar). Positive binding was defined by differential staining of CoV-S-transfectants versus mock-transfectants.
SARS-CoV-2 pseudotyped virus production.
293T/17 cells were seeded in 10-cm dishes for 80% next day confluency. The next day, cells were transfected with the plasmid pcDNA3.1(+)-spike-D19 (encoding the SARS-CoV-2 spike protein) or pcDNA3.1(+)-spike-D19 variants using the transfection reagent TransIT-Lenti according to the manufacturer’s instructions. One day post-transfection, cells were infected with VSV-luc(VSV-G) at an MOI of 3. The cell supernatant containing SARS-CoV-2 pseudotyped virus was collected at day 2 post-transfection, centrifuged at 1,000 x g for 5 min to remove cellular debris, aliquoted and frozen at −80°C. The SARS-CoV-2 pseudotyped virus preparation was quantified using Vero E6 cells seeded at 20,000 cells/well in clear bottom black 96 well plates the previous day. Cells were inoculated with 1:10 dilution series of pseudotyped virus in 50 μL DMEM for 1 h at 37°C. An additional 50 μL of DMEM was added, cells were incubated overnight at 37°C. Luciferase activity was quantified with Bio-Glo reagent by adding 100 μL of Bio-Glo (diluted 1:1 in PBS), incubated at room temperature for 5 min, and relative light units (RLU) were read on an EnSight or EnVision plate reader.
Neutralization of SARS-CoV-2 pseudotyped virus.
Vero E6 cells were seeded into clear bottom black-walled 96-well plates at 20,000 cells/well in 100 μL medium and cultured overnight at 37°C. Twenty-four hours later, 1:3 8-point serial dilutions of mAb were prepared in medium, with each dilution tested in duplicate on each plate (range: 10 μg/mL to 4 ng/mL final concentration). Pseudovirus was diluted 1:25 in medium and added 1:1 to 110 μL of each antibody dilution. Pseudovirus:antibody mixtures were incubated for 1 h at 37°C. Media was removed from the Vero E6 cells and 50 μL of pseudovirus:antibody mixtures were added to the cells. One hour post-infection, 100 μL of medium was added to wells containing pseudovirus:antibody mixtures and incubated for 17 h at 37°C. Media then was removed and 100 μL of Bio-Glo reagent (diluted 1:1 in DPBS) was added to each well. The plate was shaken on a plate shaker at 300 RPM at room temperature for 20 min, and RLUs were read on an EnSight or EnVision plate reader.
Data availability.
All data supporting the findings of this study are available within the paper and are available from the corresponding author upon request. Deep sequencing datasets of viral stocks are available at NCBI BioProject PRJNA698378 (https://dataview.ncbi.nlm.nih.gov/object/PRJNA698378?reviewer=g0mic4v5t4e1tpssk63p990suu).
Statistical analysis.
All statistical tests were performed as described in the indicated figure legends. Non-linear regression curve fitting was performed to calculate EC50 values. Statistical significance was calculated using a non-parametric Wilcoxon matched-pairs signed rank test and Prism 8.0. The number of independent experiments used are indicated in the relevant Figure legends.
Supplementary Material
Table 1.
Monoclonal antibodies used in this study
| MAb | Region | Species | EC50 value (ng/ml) a | block ACE2 binding | Binds RBM | Structural contacts | Functionally important residues b | Other mapping data | Reference | |
|---|---|---|---|---|---|---|---|---|---|---|
| CLASS 1 | COV2-2196 | RBD - RBM | human | 15 | yes | yes | F486A, N487A (LOB) | (Dong et al., 2021; Zost et al., 2020a; Zost et al., 2020b) | ||
| COV2-2130 | RBD - RBM | human | 107 | yes | yes | K444A, G447R (LOB); and R346I, K444R, K444E (NE) | (Dong et al., 2021; Zost et al., 2020a; Zost et al., 2020b) | |||
| COV2-2072 | RBD - RBM | human | 26 | yes | yes | COV2-2196 competitive | (Zost et al., 2020a; Zost et al., 2020b) | |||
| COV2-2050 | RBD - RBM | human | 80 | yes | yes | E484K (LOB and NE) | COV2-2196 and COV2-2130 competitive | (Greaney et al., 2021; Zost et al., 2020a; Zost et al., 2020b) | ||
| COV2-3025 | RBD - RBM | human | 37 | yes | yes | COV2-2196 competitive | (Zost et al., 2020a; Zost et al., 2020b) | |||
| COV2-2381 | RBD - RBM | human | 42 | yes | yes | COV2-2196 competitive | (Zost et al., 2020a; Zost et al., 2020b) | |||
| 1B07 | RBD - RBM | mouse-human chimera | 279 | yes | yes | E484A/D/G/K, F486Y (NE) | (Alsoussi et al., 2020; Liu et al., 2020) | |||
| S2E12 | RBD - RBM | human | 4.2 | yes | yes | 455 to 458, 473 to 493 | G476S, F486A (LOB) | (Tortorici et al., 2020), Starr, Corti et al. unpublished | ||
| COVOX-384 | RBD - RBM | human | 2 | yes | yes | L455, F456, G482-F486 | Dejnirattisai and Screaton, unpublished | |||
| COVOX-40 | RBD - RBM | human | 24 | yes | yes | K417, Q409, Y505 | Dejnirattisai and Screaton, unpublished | |||
| S2H58 | RBD-RBM | human | 5 | yes | yes | E484K, F490L, S494P (LOB) | Starr, Corti et al. unpublished | |||
| S2X259 | RBD | human | 55 | yes | no | G504D (LOB) | McCallum, Corti et al, unpublished | |||
| CLASS 2 | S309 | RBD-BASE | human | 79 | no | no | T333-L335,P337,G339-V341,N343R346,N354,K356-C361,N440L441,K444,R509 | (Pinto et al., 2020) | ||
| SARS2-31 | RBD-BASE | mouse | 246 | yes | no | K378E/Q, R408K, G504D (NE) | CR3022 competitive | VanBlargan and Diamond, unpublished | ||
| SARS2-10 | RBD-BASE | mouse | 694 | yes | no | CR3022 competitive | VanBlargan and Diamond, unpublished | |||
| SARS2-54 | RBD-BASE | mouse | 32 | yes | no | CR3022 competitive | VanBlargan and Diamond, unpublished | |||
| SARS2-44 | RBD-BASE | mouse | 258 | yes | no | CR3022 competitive | VanBlargan and Diamond, unpublished | |||
| SARS2-3 | RBD-BASE | mouse | 670 | no | no | CR3022 competitive | VanBlargan and Diamond, unpublished | |||
| SARS2-65 | RBD-BASE | mouse | 145 | no | no | CR3022 competitive | VanBlargan and Diamond, unpublished | |||
| CLASS 3 | COV2-2676 | NTD | human | 501 | no | no | Y144A, N164A (LOB) and F140S (NE) | (Suryadevara et al., 2021; Zost et al., 2020a; Zost et al., 2020b) | ||
| COV2-2489 | NTD | human | 199 | no | no | G142A, Y144A, F157A, N164A (LOB); and G142D, R158S (NE) | (Suryadevara et al., 2021; Zost et al., 2020a; Zost et al., 2020b) |
Neutralization potency determined by FRNT assay with WA1/2020 isolate.
LOB, loss-of-binding to spike protein, and NE, neutralization escape mutants.
ACKNOWLEDGEMENTS
This study was supported by contracts and grants from NIH (75N93019C00062, 75N93019C00051, 75N93019C00074, HHSN272201400006C, HHSN272201400008C, R01 AI157155, U01 AI151810, R01 AI142759, R01 AI134907, R01 AI145617, UL1 TR001439, P30 AR073752, U01AI141990), and the Defense Advanced Research Project Agency (HR001117S0019), the Dolly Parton COVID-19 Research Fund at Vanderbilt University, Fast Grants (Mercatus Center, George Mason University), and the Future Insight Prize (Merck KGaA; to J.E.C). J.B.C. is supported by a Helen Hay Whitney Foundation postdoctoral fellowship, E.S.W. is supported by F30 AI152327, and J.S.T. is supported by 5T32CA009547. P-Y.S. is supported by awards from the Sealy & Smith Foundation, the Kleberg Foundation, the John S. Dunn Foundation, the Amon G. Carter Foundation, the Gilson Longenbaugh Foundation, and the Summerfield Robert Foundation. P.D. is supported by a Junior Faculty Development Award from the American College of Gastroenterology. We thank Rachel Nargi and Robert Carnahan for assistance with mAb purification, Lisa Purcell for critical comments on experiments and the manuscript, Adrian Creanga and Barney Graham for the Vero-hACE2-TMPRSS2 cells, and the Laboratory of Virology, Division of Intramural Research, NIAID, NIH for the NHP immune sera.
M.S.D. is a consultant for Inbios, Vir Biotechnology, NGM Biopharmaceuticals, and Carnival Corporation, and on the Scientific Advisory Boards of Moderna and Immunome. The Diamond laboratory has received funding support in sponsored research agreements from Moderna, Vir Biotechnology, and Emergent BioSolutions. J.E.C. has served as a consultant for Eli Lilly and Luna Biologics, is a member of the Scientific Advisory Boards of CompuVax and Meissa Vaccines and is Founder of IDBiologics. The Crowe laboratory at Vanderbilt University Medical Center has received sponsored research agreements from AstraZeneca and IDBiologics. Vanderbilt University (J.E.C.) and Washington University (A.H.E., A.C.M.B., M.S.D., and D.H.F.) have applied for patents related to antibodies described in this paper. The Ellebedy laboratory has received funding support in sponsored research agreements from AbbVie and Emergent BioSolutions. The Boon laboratory has received funding support in sponsored research agreements from AI Therapeutics, GreenLight Biosciences Inc., AbbVie Inc., and Nano targeting & Therapy Biopharma Inc. The Shi laboratory has received sponsored research agreements from Pfizer, Gilead, Merck, and IGM Sciences Inc. D.P., D.C., and H.W.V. are employees of Vir Biotechnology and may hold equity in Vir Biotechnology. H.W.V. is a founder of Casma Therapeutics and PierianDx. P.D. has served as an advisory board member for Janssen, Pfizer, Prometheus Biosciences, and Arena Pharmaceuticals and has received funding support in sponsored research agreements from Takeda Pharmaceuticals.
Footnotes
COMPETING FINANCIAL INTERESTS
Declarations
Contributor Information
Michael Diamond, Washington University School of Medicine.
Rita Chen, Washington University School of Medicine.
Xuping Xie, University of Texas Medical Branch.
James Case, Washington University School of Medicine.
Xianwen Zhang, UTMB.
Laura VanBlargan, Washington University.
Yang Liu, University of Texas Medical Branch.
Jianying Liu, University of Texas Medical Branch.
John Errico, Washington University.
Emma Winkler, Washington University School of Medicine.
Naveenchandra Suryadevara, Vanderbilt University Medical Center.
Stephen Tahan, Washington University.
Jackson Turner, Washington University School of Medicine.
Wooseob Kim, Washington University School of Medicine.
Aaron Schmitz, Washington University School of Medicine.
Mahima Thapa, Washington University.
David Wang, Washington University in St. Louis.
Andrianus Boon, Washington University School of Medicine.
Dora Pinto, Humabs BioMed SA, a subsidiary of Vir Biotechnology, Inc.
Rachel Presti, Washington University School of Medicine.
Jane O’Halloran, Washington University School of Medicine.
Alfred Kim, Washington University.
Parakkal Deepak, Washington University.
Daved Fremont, Washington University School of Medicine.
Davide Corti, VIR.
Herbert Virgin, Vir Biotechnology, Washington University School of Medicine.
James Crowe, Vanderbilt University Medical Center.
Lindsay Droit, Washington University School of Medicine.
Ali Ellebedy, Washington University School of Medicine.
Pei-Yong Shi, The University of Texas Medical Branch at Galveston.
Pavlo Gilchuk, Vanderbilt University Medical Center.
References
- 1.Sempowski G. D., Saunders K. O., Acharya P., Wiehe K. J. & Haynes B. F. Pandemic Preparedness: Developing Vaccines and Therapeutic Antibodies For COVID-19. Cell 181, 1458–1463, doi: 10.1016/j.cell.2020.05.041 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wibmer C. K. et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. bioRxiv, doi: 10.1101/2021.01.18.427166 (2021). [DOI] [PubMed] [Google Scholar]
- 3.Wang Z. et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. bioRxiv, doi: 10.1101/2021.01.15.426911 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang P. et al. Increased Resistance of SARS-CoV-2 Variants B.1.351 and B.1.1.7 to Antibody Neutralization. bioRxiv, doi: 10.1101/2021.01.25.428137 (2021). [DOI] [Google Scholar]
- 5.Leung K., Shum M. H., Leung G. M., Lam T. T. & Wu J. T. Early transmissibility assessment of the N501Y mutant strains of SARS-CoV-2 in the United Kingdom, October to November 2020. Euro Surveill 26, doi: 10.2807/1560-7917.es.2020.26.1.2002106 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klimstra W. B. et al. SARS-CoV-2 growth, furin-cleavage-site adaptation and neutralization using serum from acutely infected hospitalized COVID-19 patients. J Gen Virol 101, 1156–1169, doi: 10.1099/jgv.0.001481 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson B. A. et al. Loss of furin cleavage site attenuates SARS-CoV-2 pathogenesis. Nature, doi: 10.1038/s41586-021-03237-4 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rappazzo C. G. et al. Broad and potent activity against SARS-like viruses by an engineered human monoclonal antibody. Science, doi: 10.1126/science.abf4830 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Case J. B. et al. Neutralizing antibody and soluble ACE2 inhibition of a replication-competent VSV-SARS-CoV-2 and a clinical isolate of SARS-CoV-2. Cell Host and Microbe 28, 475–485 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zost S. J. et al. Rapid isolation and profiling of a diverse panel of human monoclonal antibodies targeting the SARS-CoV-2 spike protein. Nat Med 26, 1422–1427, doi: 10.1038/s41591-020-0998-x (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zost S. J. et al. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature 584, 443–449, doi: 10.1038/s41586-020-2548-6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alsoussi W. B. et al. A Potently Neutralizing Antibody Protects Mice against SARS-CoV-2 Infection. J Immunol, doi: 10.4049/jimmunol.2000583 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tortorici M. A. et al. Ultrapotent human antibodies protect against SARS-CoV-2 challenge via multiple mechanisms. Science 370, 950–957, doi: 10.1126/science.abe3354 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pinto D. et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature 583, 290–295, doi: 10.1038/s41586-020-2349-y (2020). [DOI] [PubMed] [Google Scholar]
- 15.Suryadevara N. et al. Neutralizing and protective human monoclonal antibodies recognizing the N-terminal domain of the SARS-CoV-2 spike protein. bioRxiv, doi: 10.1101/2021.01.19.427324 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xie X. et al. Neutralization of SARS-CoV-2 spike 69/70 deletion, E484K and N501Y variants by BNT162b2 vaccine-elicited sera. Nature Medicine, In press (2021). [DOI] [PubMed] [Google Scholar]
- 17.Wu K. et al. mRNA-1273 vaccine induces neutralizing antibodies against spike mutants from global SARS-CoV-2 variants. bioRxiv, doi: 10.1101/2021.01.25.427948 (2021). [DOI] [Google Scholar]
- 18.Rathnasinghe R. et al. The N501Y mutation in SARS-CoV-2 spike leads to morbidity in obese and aged mice and is neutralized by convalescent and post-vaccination human sera. medRxiv : the preprint server for health sciences, doi: 10.1101/2021.01.19.21249592 (2021). [DOI] [Google Scholar]
- 19.Baum A. et al. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science, doi: 10.1126/science.abd0831 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ellebedy A. et al. SARS-CoV-2 infection induces long-lived bone marrow plasma cells in humans. Research square, doi: 10.21203/rs.3.rs-132821/v1 (2020). [DOI] [PubMed] [Google Scholar]
- 21.Hassan A. O. et al. A Single-Dose Intranasal ChAd Vaccine Protects Upper and Lower Respiratory Tracts against SARS-CoV-2. Cell 183, 169–184.e113, doi: 10.1016/j.cell.2020.08.026 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bricker T. L. et al. A single intranasal or intramuscular immunization with chimpanzee adenovirus vectored SARS-CoV-2 vaccine protects against pneumonia in hamsters. bioRxiv, doi: 10.1101/2020.12.02.408823 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hassan A. O. et al. A single intranasal dose of chimpanzee adenovirus-vectored vaccine protects against SARS-CoV-2 infection in rhesus macaques. bioRxiv, doi: 10.1101/2021.01.26.428251 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Polack F. P. et al. Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N Engl J Med 383, 2603–2615, doi: 10.1056/NEJMoa2034577 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCallum M. et al. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. bioRxiv, doi: 10.1101/2021.01.14.426475 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greaney A. J. et al. Complete Mapping of Mutations to the SARS-CoV-2 Spike Receptor-Binding Domain that Escape Antibody Recognition. Cell Host Microbe 29, 44–57.e49, doi: 10.1016/j.chom.2020.11.007 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Piccoli L. et al. Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology. Cell 183, 1024–1042.e1021, doi: 10.1016/j.cell.2020.09.037 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Z. et al. Landscape analysis of escape variants identifies SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. bioRxiv, doi: 10.1101/2020.11.06.372037 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weisblum Y. et al. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. Elife 9, doi: 10.7554/eLife.61312 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Z. et al. Identification of SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. Cell Host Microbe, doi: 10.1016/j.chom.2021.01.014 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Andreano E. et al. SARS-CoV-2 escape in vitro from a highly neutralizing COVID-19 convalescent plasma. bioRxiv, doi: 10.1101/2020.12.28.424451 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim J. H., Marks F. & Clemens J. D. Looking beyond COVID-19 vaccine phase 3 trials. Nat Med, doi: 10.1038/s41591-021-01230-y (2021). [DOI] [PubMed] [Google Scholar]
- 33.Schäfer A. et al. Antibody potency, effector function, and combinations in protection and therapy for SARS-CoV-2 infection in vivo. J Exp Med 218, doi: 10.1084/jem.20201993 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Winkler E. S. et al. Human neutralizing antibodies against SARS-CoV-2 require intact Fc effector functions and monocytes for optimal therapeutic protection. bioRxiv, doi: 10.1101/2020.12.28.424554 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zohar T. et al. Compromised Humoral Functional Evolution Tracks with SARS-CoV-2 Mortality. Cell 183, 1508–1519.e1512, doi: 10.1016/j.cell.2020.10.052 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dan J. M. et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science, doi: 10.1126/science.abf4063 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lipsitch M., Grad Y. H., Sette A. & Crotty S. Cross-reactive memory T cells and herd immunity to SARS-CoV-2. Nat Rev Immunol 20, 709–713, doi: 10.1038/s41577-020-00460-4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sette A. & Crotty S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell, doi: 10.1016/j.cell.2021.01.007 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bailey A. L. & Diamond M. S. A Crisp(r) New Perspective on SARS-CoV-2 Biology. Cell 184, 15–17, doi: 10.1016/j.cell.2020.12.003 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goddard T. D. et al. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci 27, 14–25, doi: 10.1002/pro.3235 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Plante J. A. et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature, doi: 10.1038/s41586-020-2895-3 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xie X. et al. An Infectious cDNA Clone of SARS-CoV-2. Cell Host Microbe 27, 841–848.e843, doi: 10.1016/j.chom.2020.04.004 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xie X. et al. Engineering SARS-CoV-2 using a reverse genetic system. Nat Protoc, doi: 10.1038/s41596-021-00491-8 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen S., Zhou Y., Chen Y. & Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890, doi: 10.1093/bioinformatics/bty560 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li H. & Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760, doi: 10.1093/bioinformatics/btp324 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Poplin R. et al. A universal SNP and small-indel variant caller using deep neural networks. Nat Biotechnol 36, 983–987, doi: 10.1038/nbt.4235 (2018). [DOI] [PubMed] [Google Scholar]
- 47.Cingolani P. et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 6, 80–92, doi: 10.4161/fly.19695 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this study are available within the paper and are available from the corresponding author upon request. Deep sequencing datasets of viral stocks are available at NCBI BioProject PRJNA698378 (https://dataview.ncbi.nlm.nih.gov/object/PRJNA698378?reviewer=g0mic4v5t4e1tpssk63p990suu).