Abstract
The Bronx was an early epicenter of the COVID-19 pandemic in the USA. We conducted temporal genomic surveillance of SARS-CoV-2 genomes across the Bronx from March-October 2020. Although the local structure of SARS-CoV-2 lineages mirrored those of New York City and New York State, temporal sampling revealed a dynamic and changing landscape of SARS-CoV-2 genomic diversity. Mapping the trajectories of variants, we found that while some have become ‘endemic’ to the Bronx, other, novel variants rose in prevalence in the late summer/early fall. Geographically resolved genomes enabled us to distinguish between a case of reinfection and a case of persistent infection. We propose that limited, targeted, temporal genomic surveillance has clinical and epidemiological utility in managing the ongoing COVID pandemic.
COVID-19 has had a devastating effect on the health of communities across the globe, with over 79 million reported cases and greater than 1.7 million deaths since the start of the pandemic (1). Until vaccines become widely available, understanding and interrupting SARS-CoV-2 transmission to prevent infection is the mainstay of public health efforts. The Bronx, a borough of New York City (NYC) has sustained the second highest rate of COVID-19 in New York City with 6,035 cases per 100,000 people as of January 11, 2021 (2). To track the local spread of SARS-CoV-2, we conducted a genomic epidemiologic study at Montefiore Health Systems (MHS), which offers healthcare services to two million residents throughout the Bronx, one of the most diverse and poorest urban communities in the United States.
The number of COVID-19 cases peaked in the Bronx in March–April 2020 and subsided during the late spring into summer 2020. To characterize the genetic diversity of SARS-CoV-2, we randomly selected nasopharyngeal samples that were positive for SARS-CoV-2 by RT-PCR testing at the MHS clinical laboratory between March and October 2020. Genomic viral RNA was extracted from nasopharyngeal swabs, and sequencing libraries were prepared using the ARTIC Network protocol and analyzed on an Oxford Nanopore MinION (3, 4). The ARTIC Network bioinformatics protocol was used to quality check and annotate SARS-CoV-2 genomes with default parameterization (5). We called variants with the NextClade tool and annotated lineages using the constructed PANGOLIN guide tree from 05/29/2020 (6, 7). Samples were derived from patients who required hospitalization (48%), mild disease managed as outpatients (26%) and asymptomatic carriers (8.9%) (Fig. 1A).
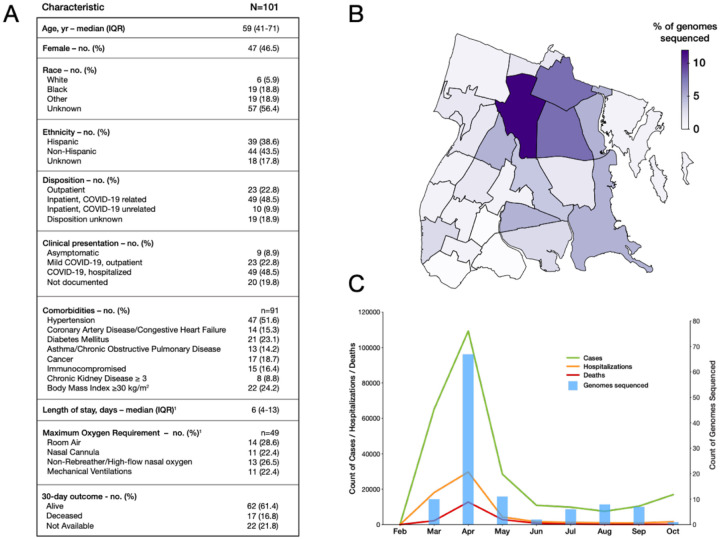
Figure 1. Surveilling SARS-CoV-2 genomes in the Bronx.
A) Table of clinical characteristics of sampled patients. B) SARS-CoV-2 genomes sequenced per Zip code in NYC. Darker colors indicate heavier sampling; C) SARS-CoV-2 genomes sequenced over time during the COVID-19 pandemic. Date is indicated on the x axis. Blue bars and the associated right hand y axis indicate the number of genomes sequenced. The left-hand y axis represents different features of COVID-19 in the Bronx; green lines indicate COVID-19 cases, the red line deaths associated with COVID-19 and the orange line hospitalizations associated with COVID-19 in the Bronx.
We collected 137 samples, and from these generated 104 high-quality genomes from 101 patients with >95% coverage (Fig. S1 and S2). Sequence data were derived from residents throughout the Bronx and were associated with 22 of 25 zip codes (Fig. 1B). Genomic sampling was greatest at the onset of the COVID-19 pandemic in March and April, but intermittent sampling continued as caseloads declined over the summer and fall (Fig. 1C).
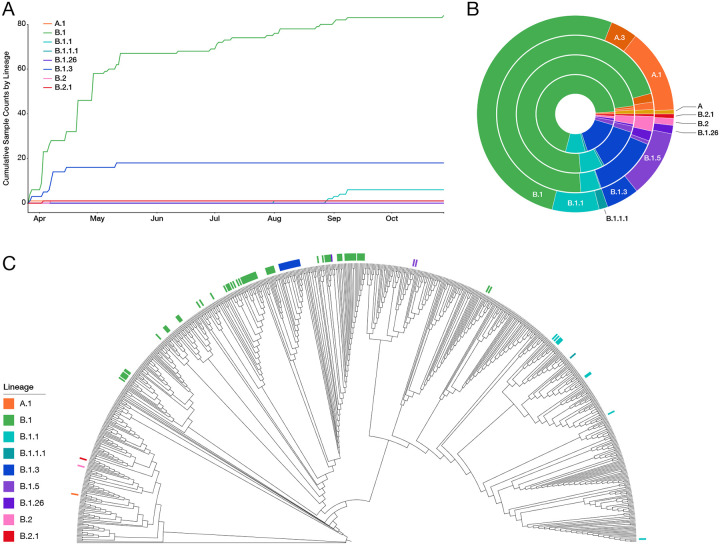
Analysis of the resulting 104 SARS-CoV-2 genome sequences revealed that B.1 and B.1.3 lineages were the most prevalent during the early months of the pandemic in the Bronx; however, several other lineages were also present at low frequencies (Fig. 2A). Although B.1.3 plateaued after the first wave, B.1 continued to be sampled, and a new lineage, B.1.1, arose in late August. We observed no major differences between Bronx SARS-CoV-2 lineages and other SARS-CoV-2 lineages in NYC and New York State (NYS) (Fig. 2B) (8, 9). We note that “A” lineage SARS-CoV-2 viruses are less prevalent in the Bronx, NYC, and NYS compared to the rest of the USA and the world. To determine how the Bronx sequences compared with those sampled across the world, we created a downsampled SARS-CoV-2 tree from 613 high-quality SARS-CoV-2 genomes deposited in GISAID with available location and collection dates. We found that Bronx SARS-CoV-2 sequences represented subsets of different clades of the global tree (Fig. 2C).
Figure 2. Bronx SARS-CoV-2 genome lineages in the context of local and global sampling.
A) Cumulative counts of PANGOLIN guide tree-based lineage assignments plotted against time; B) Prevalence of lineages seen in the Bronx compared to their prevalence in other regions. Inner to outer rings represent the Bronx, New York State, USA, the world, respectively. Lineage coloring is the same as A); C) Phylogeny of the Bronx strains in the context of SARS-CoV-2 strains from around the world. Bronx strains and their associated lineages are indicated with colored lines.
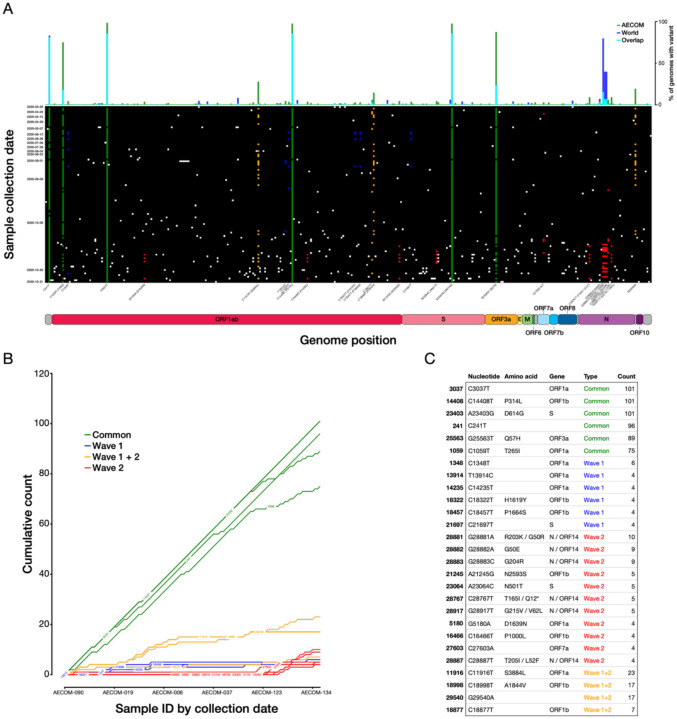
We next examined patterns in variant nucleotide positions observed in our data. We found that variation is distributed across the SARS-CoV-2 genome and that some variants are present in almost all Bronx genomes sequenced—these can be described as ‘core’ to the Bronx at present (Fig. 3A). Core variants include the spike protein variant A23403G (D614G), as well as variants C241T, C1059T (T265I), C3037T, C14408T (P314L) in Orf1ab, and G25563T (Q57H) in Orf3a. We next examined the dynamics of individual SARS-CoV-2 variants. Although the core variants continued to increase in prevalence as we sequenced new genomes, we also observed variants novel to the Bronx whose prevalence is beginning to increase, whereas others have plateaued or are in the process of plateauing (Fig. 3B).
Figure 3. SARS-CoV-2 variants and their trajectories in the Bronx.
A) Individual SARS-CoV-2 variants plotted across the viral genome (x axis), with genomes sorted by sampling date (y axis). Positions that are variable with respect to the reference SARS-CoV-2 strain are shown with a white (low-frequency), green (common), yellow (wave 1+2), blue (wave 1) or red (wave 2) squares. The histogram across the top plots the prevalence of a given variant across all Bronx SARS-CoV-2 genomes in this study relative to the world; B) Rarefaction curve of cumulative variant counts over time for variants observed at least four times in the Bronx SARS-CoV-2 genomes set; C) Table showing details for variants in 3B.
In the spike protein, we found amino acid variants D614G (core), N501T in 5 patients, and both N501Y and P681R in one patient. We note that P314L in Orf1b is also a core variant in our dataset, reflecting observations in other studies that this variant is in linkage disequilibrium with D614G (10). We did not observe the B.1.1.7 variant strain, first identified in the United Kingdom in the fall of 2020 which also contains the N501Y variant and similarly the P681H, in our samples. The N501 residue of the spike protein is part of the receptor binding domain and the receptor binding motif, and variants at this position may influence ACE2 receptor binding (11). In comparing Bronx variants to those found in the rest of the world, we find that some variants, such as the spike protein D614G variant, are prevalent both in our set and in the world; however, some ‘core’ Bronx variants such as C1059T (T265I in Orf1ab) and G25563T (Q57H in Orf3a) are not as prevalent in the rest of the world (top bar Fig. 3A, Fig. 3C, Fig S3). The geographic specificity of variants creates a fingerprint that can be useful for tracing the spread of particular variants; a lineage containing the variant C2416T, linked to the Boston Biogen COVID-19 outbreak, could be traced to infections around the world (12). The C2416T variant was also observed in three patients in our dataset. We note that rare variants are uniformly distributed throughout the sampling period (Fig. S4) and further that the functional impact of these variants is not well resolved.
A phylogenetic tree of SARS-CoV-2 shows that strains collected earlier in the pandemic are distinguishable from strains collected later, suggesting that new strains are being continuously introduced into the Bronx (Fig. 4, inner ring, red indicates earlier samples, green newer samples). In considering the lineages of SARS-CoV-2, there was evidence of ongoing presence of some B.1 lineage-associated strains, throughout the study period, starting from the onset of pandemic until the end of the study period (Fig. 4, outer ring indicates lineage). We found that the B.1.1 lineage had increasing presence in the latter part of the study period and that newer B.1 strains, which cluster away from older B.1 sequences, appear at later sampling dates. Newer B.1 and B.1.1 lineages form a distinct clade from older B.1 lineages in the Bronx SARS-CoV-2 tree. We posit that these two clades reflect two different types of SARS-CoV-2 isolates: those that are circulating locally and those that were newly introduced. We consider SARS-CoV-2 isolates that group on the downsampled global tree and group on the local Bronx tree with our first wave pandemic sampling to be ‘circulating.’ We continue to observe isolates that fall into this ‘first wave’ clade during the summer, post-first wave, and therefore consider these to have persisted in the Bronx. We consider ‘introduced’ isolates those that are newer sequences in the local Bronx tree that are also spread out in different clades across the global tree; it remains to be seen if these introduced isolates will form the basis of a second set of circulating SARS-CoV-2 strains during a new wave of COVID-19 in the Bronx (Fig. 2C and 4).
Figure 4. Clinical relevance of the changing genomic landscape of SARS-CoV-2 in the Bronx.
Phylogenetic tree based on whole genome alignments of Bronx isolates. Colored rings around the tree indicate SARS-Cov-2 lineage (outer ring) and the date of sampling (inner ring, red=earlier, green=later). Samples from the same patient are indicated with symbols; a reinfection case is indicated with purple arrows and a putative persistent infection case is indicated with orange arrows. Grey circles on the branches indicate bootstrap values of 85 or greater. Tree was generated with TimeTree and visualized with iTOL (13, 21).
This local phylogenetic framework of SARS-CoV-2 strains in the Bronx enabled us to distinguish between a case of reinfection and a case of persistent infection in two pediatric patients. The first case is a 10–15-y.o. female who was initially seen in April 2020 in the emergency department with 3 days of fever, sore throat, anosmia, and ageusia. SARS-CoV-2 infection in this patient was confirmed by RT-PCR. She had a total of 6 days of symptoms and was in general good health until the second presentation. In August 2020, she presented again to the emergency department with two days of fever, severe postprandial abdominal cramps, watery diarrhea and generalized body aches. All other reviews of symptoms were negative. The patient had no known COVID-19 exposures and limited outside exposure. A respiratory pathogen panel was negative but her SARS-CoV-2 RT-PCR was positive, as was her SARS-COV-2 IgM Immune Status Ratio (ISR) (2.1, with less than 1 considered negative). Her IgG ISR was negative, 8.7 (normal range ISR < 9). The patient had a total of three days of fever with complete resolution of all other symptoms by day four of illness.
The two SARS-CoV-2 genomes sequenced from this patient were 142 days apart and differed in nucleotide sequence at 17 different positions. The first and second samples from this patient fall in different local phylogenetic clades in the Bronx phylogenetic tree, supporting the hypothesis that we are observing a new infection and not prolonged shedding from the original SARS-CoV-2 infection (Fig. 4, purple arrows). To our knowledge, this is the first case of symptomatic reinfection in a child who had prior symptomatic SARS-CoV-2 infection. Given the history of limited exposures to high-risk activities for this patient between the two episodes and the overall low incidence of SARS-CoV-2 infection in New York at the time of the second presentation in August, genomic and phylogenetic analysis provided key confirmatory evidence in support of the clinical inference of a reinfection.
The second case involved a 15–20-y.o. female with an incompletely characterized immunodeficiency who presented in July 2020 with an oral lesion. She had no fever, or respiratory or gastrointestinal symptoms, and had neutropenia (absolute neutrophil count 700 cells/ul). After admission for further evaluation, she was found to be SARS-CoV-2 positive. During the admission, she was intermittently febrile and neutropenic and was treated with broad spectrum antibiotics. She developed a buttock lesion that was biopsied, revealing a thrombotic vasculopathy with infarction. Due to concern that the lesion could represent COVID-19–associated vasculopathy, and in the setting of persistent fever and intermittent neutropenia, she was treated with a 10-day course of remdesivir. The patient continued to have positive nasopharyngeal swabs for SARS-CoV-2 from early July to the end of September (Table S1 and Fig. S5). Her SARS-CoV-2 IgG (Abbott) was negative in mid-August.
For this patient, the three sequenced SARS-CoV-2 genomes sampled in July, August and October fall in the same clade (Fig. 4, orange arrows). This clade is polytomic by TimeTree, meaning that it is not possible to resolve the relationships between sequences within this clade, but the clade itself is supported by a bootstrap value of 870/1000 for (SH-aLRT replicates) (13, 14). We therefore posit that the three strains sequenced from this patient, despite having some variation, are more likely to represent a single SARS-CoV-2 infection rather than multiple infections. Together, these genomic, phylogenetic, and clinical observations strongly suggest that this patient has been unable to clear a single infection of SARS-CoV-2, as opposed to being reinfected with a distinct strain. Other examples of persistent infection with SARS-CoV-2 have been reported, but not, to our knowledge, in children (15–17). A woman diagnosed with chronic lymphocytic leukemia who was sampled 5 times, had SARS-CoV-2 sequences displaying intrahost variation despite the SARS-CoV-2 being polytomic, similar to what we observe here (18). The polytomy that encompasses this persistent case also contains independent local strains of SARS-CoV-2 that do not separate on the global tree, suggesting that some variants seen in this patient are also shared locally in the Bronx (Fig. 2C and 4).
Our work supports guiding principles for practical and clinical applications of SARS-CoV-2 sequencing in the COVID-19 pandemic. How many genomes do you need to sequence for a local community to resolve clinical questions? In our case, ~100 genomes were sufficient to place new patients into the context of the variability of SARS-CoV-2 during the pandemic and to be able to answer coarse-grained questions to determine reinfection vs. persistent infection and community-level observations of older vs. newly introduced strains. The targeted utilization of small numbers of stored swabs for temporally resolved viral genomic surveillance could thus resolve clinical questions related to persistent vs. reinfection. With the introduction and spread of recently identified United Kingdom and South African SARS-Cov-2 variants with potentially different epidemiological features from existing strains, there has been speculation that we may observe a selective sweep of existing viral genotypes in the months to follow (19, 20). Temporally and geographically resolved sequencing of SARS-CoV-2 genotypes provides a background against which introduction of these or other new genotypes into our local community can be observed in real time. Given the lack of a national sequencing effort, we suggest that decentralized, small-scale sequencing coupled with rapid data sharing to public databases provides an alternative and more practical tool to monitor and curtail the introduction and spread of SARS-CoV-2.
Supplementary Material
Significance:
The ongoing emergence of novel SARS-CoV-2 variants has highlighted the need for continual genomic surveillance in order to track their spread and limit introductions into new areas. An understanding of circulating viral strains also provides a powerful tool that can be used to make clinical inferences. Here, we employ temporally and geographically resolved sequencing of SARS-CoV-2 samples in order to describe the local landscape of viral variants in the Bronx and to differentiate between cases of re-infection and persistent infection. We propose that local and targeted sequencing of viral isolates is an underutilized approach for managing the COVID pandemic.
Acknowledgements:
We thank Isabel Gutierrez, Estefania Valencia, and Laura Polanco for laboratory management and technical assistance and the Chandran and Kelly labs for helpful comments on the manuscript. We thank NextStrain, GISAID, and all labs who contributed SARS-CoV-2 sequences for public access. We thank the healthcare workers and patients of the Montefiore Healthcare System.
Funding: L.K. is supported in part by a Peer Reviewed Cancer Research Program Career Development Award from the United States Department of Defense (CA171019). S.K. is supported by the Einstein Medical Scientist Training Program (2T32GM007288-45) and by a National Institutes of Health T32 Fellowship in Geographic Medicine and Emerging Infectious Diseases (2T32AI070117-13). K.S. is supported by a National Institutes of Health F30 Fellowship (F30CA200411) and T32 Fellowship (T32GM007288).
Footnotes
Competing interests: KC is a member of the scientific advisory boards of Integrum Scientific, LLC and the Pandemic Security Initiative of Celdara Medical, LLC..
Data availability: All sequences generated in this study have been made publicly available through the GISAID hCoV-19 sequence database. The source code used for sequencing, analysis, and figure generation, is hosted on Github at https://github.com/kellylab/genomic-surveillance-of-the-bronx.
Supplementary materials:
Materials and Methods
Figs. S1 to S5
Table S1
References and notes
- 1.World Health Organization, Weekly epidemiological update - 29 December 2020 (2020), (available at https://www.who.int/publications/m/item/weekly-epidemiological-update-−−29-december-2020).
- 2.Statista, Rates of COVID-19 cases in New York City as of January 11, 2021, by borough (2021), (available at https://www.statista.com/statistics/1109817/coronavirus-cases-rates-by-borough-new-york-city/).
- 3.Quick J. et al. , Multiplex PCR method for MinION and Illumina sequencing of Zika and other virus genomes directly from clinical samples. Nat. Protoc. 12, 1261–1276 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.ARTIC Network, ARTIC Network SARS-CoV-2 sequencing, (available at https://artic.network/ncov-2019).
- 5.ARTIC Network, nCoV-2019 novel coronavirus bioinformatics protocol (2020), (available at https://artic.network/ncov-2019/ncov2019-bioinformatics-sop.html).
- 6.Hadfield J. et al. , Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. 34, 4121–4123 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rambaut A. et al. , A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 5, 1403–1407 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gonzalez-Reiche A. S. et al. , Introductions and early spread of SARS-CoV-2 in the New York City area. Science. 369, 297–301 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maurano M. T. et al. , Sequencing identifies multiple early introductions of SARS-CoV-2 to the New York City region. Genome Res. 30, 1781–1788 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ogawa J. et al. , The D614G mutation in the SARS-CoV2 Spike protein increases infectivity in an ACE2 receptor dependent manner. BioRxiv (2020), doi: 10.1101/2020.07.21.214932. [DOI] [Google Scholar]
- 11.Wan Y., Shang J., Graham R., Baric R. S., Li F., Receptor Recognition by the Novel Coronavirus from Wuhan: an Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. J. Virol. 94 (2020), doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lemieux J. E. et al. , Phylogenetic analysis of SARS-CoV-2 in Boston highlights the impact of superspreading events. Science (2020), doi: 10.1126/science.abe3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sagulenko P., Puller V., Neher R. A., TreeTime: Maximum-likelihood phylodynamic analysis. Virus Evol. 4, vex042 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nguyen L.-T., Schmidt H. A., von Haeseler A., Minh B. Q., IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.To K. K.-W. et al. , COVID-19 re-infection by a phylogenetically distinct SARS-coronavirus-2 strain confirmed by whole genome sequencing. Clin. Infect. Dis. (2020), doi: 10.1093/cid/ciaa1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abu-Raddad L. J. et al. , Assessment of the risk of SARS-CoV-2 reinfection in an intense re-exposure setting. Clin. Infect. Dis. (2020), doi: 10.1093/cid/ciaa1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tillett R. L. et al. , Genomic evidence for reinfection with SARS-CoV-2: a case study. Lancet Infect. Dis. 21, 52–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Avanzato V. A. et al. , Case Study: Prolonged Infectious SARS-CoV-2 Shedding from an Asymptomatic Immunocompromised Individual with Cancer. Cell. 183, 1901–1912.e9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tegally H. et al. , Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. medRxiv (2020), doi: 10.1101/2020.12.21.20248640. [DOI] [Google Scholar]
- 20.Davies N. G. et al. , Estimated transmissibility and severity of novel SARS-CoV-2 Variant of Concern 202012/01 in England. medRxiv (2020), doi: 10.1101/2020.12.24.20248822. [DOI] [Google Scholar]
- 21.Letunic I., Bork P., Interactive tree of life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47, W256–W259 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katoh K., Standley D. M., MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.