Abstract
The last few years have seen tremendous advances in CRISPR-mediated genome editing. Great efforts have been made to improve the efficiency, specificity, editing window, and targeting scope of CRISPR/Cas9-mediated transgene knock-in and gene correction. In this article, we comprehensively review recent progress in CRISPR-based strategies for targeted transgene knock-in and gene correction in both homology-dependent and homology-independent approaches. We cover homology-directed repair (HDR), synthesis-dependent strand annealing (SDSA), microhomology-mediated end joining (MMEJ), and homology-mediated end joining (HMEJ) pathways for a homology-dependent strategy and alternative DNA repair pathways such as non-homologous end joining (NHEJ), base excision repair (BER), and mismatch repair (MMR) for a homology-independent strategy. We also discuss base editing and prime editing that enable direct conversion of nucleotides in genomic DNA without damaging the DNA or requiring donor DNA. Notably, we illustrate the key mechanisms and design principles for each strategy, providing design guidelines for multiplex, flexible, scarless gene insertion and replacement at high efficiency and specificity. In addition, we highlight next-generation base editors that provide higher editing efficiency, fewer undesired by-products, and broader targeting scope.
Keywords: Gene correction, gene knock-in, gene replacement, genetic variant, single-nucleotide polymorphism, targeted integration
Introduction
Clustered regularly interspaced short palindromic repeats (CRISPR) has revolutionized the field of transgenic research and human gene therapy1–6. Using CRISPR, a specific genetic variant associated with human disease can be introduced into the genome, or a mutation in the genome can be replaced with a wild-type allele, at will by design in cells and organisms, respectively, for gene knock-in or gene correction7–12. The CRISPR-modified cellular and animal models then can be studied to uncover mechanisms underlying human disease and for drug discovery13–15. Therefore, CRISPR holds tremendous translational potential. CRISPR technology has been explored for in vivo gene therapy to treat a wide range of human hereditary diseases16–18 as well as ex vivo gene therapy to treat blood disorders, cancers, and immune-related diseases by genetically modifying the patient’s cells outside their body19–23. The last few years have seen enormous advances to improve the performance and maximize the potential of CRISPR-mediated gene insertion and replacement for disease modeling and gene therapeutics.
The strategies for transgene knock-in and gene correction are generally classified into homology-dependent and homology-independent. Homology-directed repair approaches require the presence of both CRISPR/Cas9 and a DNA donor repair template. The repair template can be in the form of circular double-stranded plasmid DNA24–27, single-stranded donor oligonucleotide (ssODN)28–30, linear double-stranded polymerase chain reaction (PCR) fragments31,32, or the homologous sequences of the intact sister chromatid33. Depending on the forms of repair template and CRISPR system used, homology-mediated gene insertion and replacement are carried out via specific DNA repair pathways such as homology-directed repair (HDR)24,26,33, synthesis-dependent strand annealing (SDSA)28,34, microhomology-mediated end joining (MMEJ)35,36, and homology-mediated end joining (HMEJ)27,37–39 pathways. In homology-independent approaches, transgene knock-in and gene correction are achieved with CRISPR/Cas9 in the absence of DNA donor repair templates. In this case, the error-prone non-homology end joining (NHEJ) repair pathway is commonly used to repair CRISPR-mediated double-stranded breaks and to incorporate a mutagenic sequence into the genome40,41.
As alternatives, base editing42–45 and primer editing46 are used to perform gene correction and replacement by directly converting nucleotides in genomic DNA without damaging the DNA or requiring donor DNA. Various base-editor variants have also recently been engineered to provide higher editing efficiency, fewer undesired by-products, and broader targeting scope. Simultaneous substitution of multiple nucleotides has been achieved by fusing Cas9 nickase (Cas9n) to a DNA polymerase47 or a reverse transcriptase (RT)46. To avoid permanent deleterious effects caused by unanticipated mutagenesis and complex chromosomal rearrangements to genomic DNA, CRISPR is employed to edit the endogenous RNA transcripts containing pathogenic mutations48–50.
Given the explosive popularity and rapid evolution of CRISPR technology in transgenic research and human gene therapy, the purpose of this review is to provide a comprehensive summary on various, recently developed CRISPR-based strategies for controlled and desired transgene knock-in and gene correction as a quick reference for those who are new to this fast-moving field. We illustrate and highlight the mechanisms of action and key concepts for each transgene knock-in and gene correction strategy. We elaborate design guidelines for scarless gene insertion and replacement at high efficiency and specificity. We also critically discuss the applications and pros and cons of each strategy. Finally, we briefly discuss how engineered CRISPR variants and alternate donor template designs have improved the efficiency, specificity, editing window, and targeting scope of transgene knock-in and gene correction.
Homology-dependent gene knock-in and gene correction strategies
Homology directed repair–based approaches
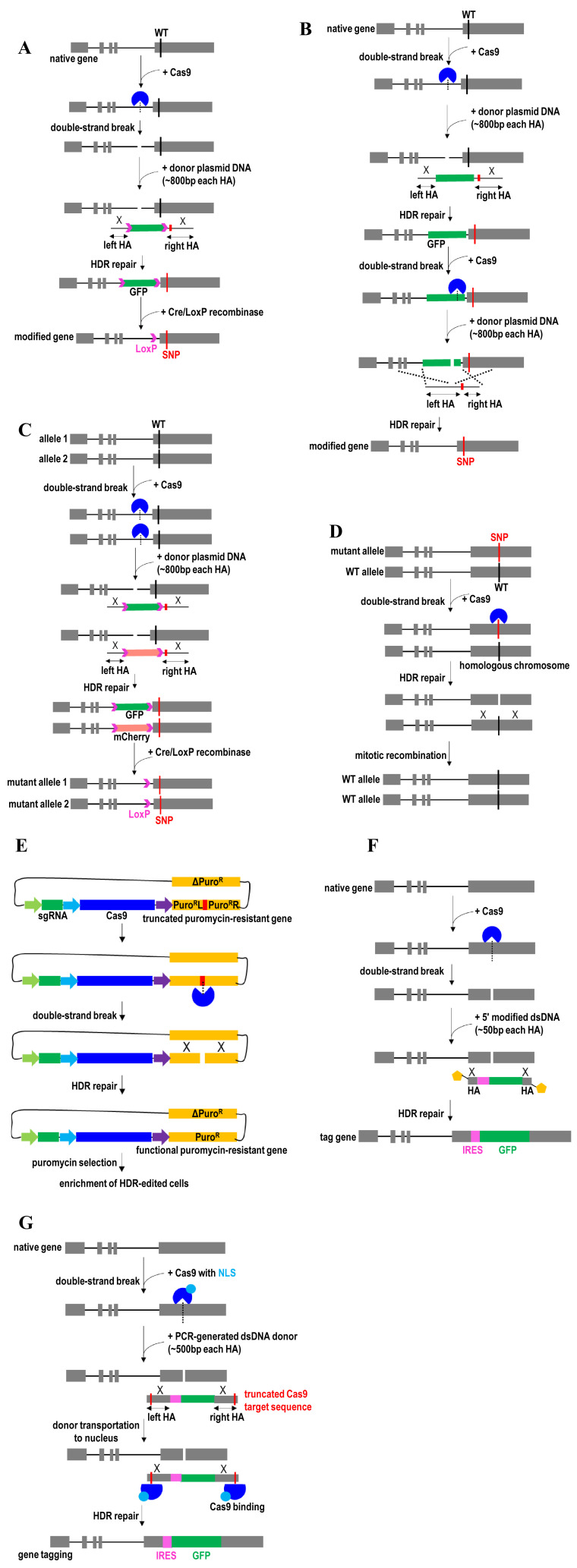
HDR mediated by homologous recombination is one of the most commonly used methods to introduce a genetic mutation into the genome (gene knock-in). It allows desired and controlled genetic modifications in the genome. This HDR approach requires the presence of a DNA repair template, usually in the form of circular plasmid DNA. However, HDR activity can be improved using the linearized plasmid with the short 5′ backbone overhang51. To introduce an exonic mutation, CRISPR/Cas9 ribonucleoprotein (RNP) complex is co-delivered with the plasmid DNA donor containing two homology arms (~800 bp each arm) flanking the mutated sequence into the cell (Figure 1A). As an option, a fluorescent tag can be added to the plasmid DNA donor to facilitate the selection of edited cells. To ensure efficient mutational knock-in, one should try to locate the mutated sequence as close as possible to the 3′ end of the left homologous arm or the 5′ end of the right homologous arm. CRISPR/Cas9 first induces a double-strand DNA break at the target intronic region, usually cleaving at 3 or 4 nucleotides upstream of the protospacer adjacent motif (PAM) sequence. The left and right homology arms should flank the CRISPR/Cas9 cleavage site. HDR-based cellular DNA repair machinery then is activated to repair the CRISPR/Cas9 cleavage site. The desired genetic variant and the fluorescent marker in the donor repair template are introduced into the genome via homologous recombination. The single-cell clones of these edited cells are flow-sorted on the basis of their fluorescent marker expressions. Finally, this fluorescent marker gene at the intronic region is removed by the overexpression of Cre/LoxP recombinase prior to functional analysis. However, a copy of a short LoxP element would remain within the intron. This knock-in approach is useful for characterizing protein-coding variants (for example, non-synonymous single-nucleotide polymorphism [SNP] or missense mutation)24,52,53. Although an intronic variant can also be introduced to the genome via this approach when the exon sequence is short, a scarless gene-editing outcome is preferred.
Figure 1. Homology directed repair (HDR)-mediated gene knock-in and gene correction strategies.
(A) Exonic SNP knock-in by CRISPR/Cas9-mediated integration of a selection marker in intron and an SNP in exon, followed by Cre/LoxP removal of the selection marker. (B) Exonic SNP knock-in by CRISPR/Cas9-mediated integration of a selection marker at intron and an SNP at exon, followed by CRISPR/Cas9-mediated removal of the selection marker. (C) Biallelic SNP knock-in by CRISPR/Cas9-mediated integration of two different fluorescent reporter genes, followed by Cre/LoxP removal of these two reporter genes. (D) Gene correction via CRISPR/Cas9-mediated mitotic recombination. (E) Enrichment of HDR-edited cells by a universal surrogate reporter plasmid. (F) Exonic SNP knock-in using Cas9 ribonucleoprotein complexes and 5′-modified linear dsDNA donors. (G) Transgene knock-in facilitated by adding truncated Cas9 target sequences to the 5′ end of left homology arm and 3′ end of right homology arm in plasmid donor. GFP, green fluorescent protein; HA, homology arm; HDR, homology-directed repair; IRES, internal ribosome entry site; NLS, nuclear localization sequence; Puro, puromycin; SNP, single-nucleotide polymorphism; WT, wild-type.
For introducing non-coding variants within the regulatory elements (for example, a mutation in the promoter, enhancer, or untranslated region), a clean mutational knock-in or scarless genome-editing approach is recommended. For example, the fluorescent marker gene at the intronic region is removed without the addition of any exogenous sequence by designing the single guide RNA (sgRNA) to target the fluorescent marker gene (Figure 1B). Upon cleavage of the CRISPR/Cas9 second target site, the edited allele uses the second plasmid donor bearing only an SNP (without fluorescent marker transgene) to mediate homologous recombination26. After the two-step editing process, the edited cells with no fluorescent protein expression are flow-sorted and single-cell-expanded for characterization. Alternatively, marker-free genome editing can be achieved by using a highly efficient HDR approach that was developed recently28,54. This approach uses electroporation to deliver adeno-associated virus (AAV) 6-mediated DNA repair template and Cas9 protein complexed with chemically modified sgRNA. As an alternative, the repair template can be delivered as an ssDNA donor. As the majority of cells are successfully edited, no selection marker (for example, fluorescent marker or antibiotic selection) is required to screen a large number of clones to identify biallelic edited clones. Therefore, this scarless genome-editing approach is particularly useful for therapeutic applications in hard-to-transfect cells such as human pluripotent stem cells54, adult stem cells28,55, and primary immune cells56.
Because some diseases are caused by the homozygous mutations57–60, efficient site-specific gene correction of both alleles is needed. Moreover, a single-allele perturbation might not be sufficient to cause significant functional alteration of a given gene, especially for disease-associated genetic variants such as non-synonymous SNPs. Thus, transgenic cells with biallelic gene manipulation are usually preferred over monoallelic, even for SNPs in a heterozygous form, to be able to detect the functional impact of the variants on the activity and molecular phenotypes of the targeted gene product. However, owing to the inefficient HDR events, it is often technically more challenging to screen transgenic cells with targeted mutational knock-in at both alleles than monoallelic gene editing. Therefore, a CRISPR/Cas9-mediated biallelic genome targeting with a dual surrogate reporter-integrated donor system was recently developed to facilitate the selection and enrichment of biallelic-modified cells61. This approach uses a CRISPR/Cas9-mediated homologous recombination system and two different sets of reporter transgene (Figure 1C). For example, CRISPR/Cas9 is first used to induce DNA double-strand breaks near the desired exonic mutation at both alleles. Then each reporter transgene (green fluorescent protein [GFP] or mCherry) is integrated into each allele. The enhanced GFP (eGFP) and mCherry double-positive cells then are sorted and expanded. The Cre/LoxP recombination system is used to remove these two fluorescent selection marker transgenes in these biallelic-modified cells prior to functional analysis. The use of this strategy can avoid time-consuming and tedious screening procedures to identify biallelic-modified cells, thereby enabling high-throughput characterization of non-synonymous mutations on gene function. A caveat of this biallelic modification strategy is that it cannot be used for essential genes that can affect cell viability. Extensive subcloning and prolonged passage of reporter-inserted clones may also lead to subtle phenotype deviations from the original parental cell line.
Gene correction has also been achieved via mitotic recombination using the endogenous wild-type allele on the homologous chromosome as a template (Figure 1D)33. In this case, the sgRNA is designed to target the mutant allele. CRISPR/Cas9-induced DNA breaks in the mutant allele then are repaired through homologous recombination using the wild-type allele on the homologous chromosome as a template. This strategy has been employed for gene drive applications in a polyploid organism62–65. In the case of gene drive, CRISPR/Cas9 possibly along with a payload transgene (desired trait) is first inserted into one of the wild-type alleles in the genome. The insertion is done using CRISPR/Cas9-mediated homologous recombination. Expressions of CRISPR/Cas9 then trigger another round of homologous recombination in the cell to modify the remaining wild-type alleles using homology sequences from the previously modified allele. Eventually, all of the wild-type alleles are converted into modified alleles. This strategy allows self-propagation of the desired trait, super-Mendelian inheritance of a transgene, and genetic modifications of specific populations or entire species.
Another way to enrich biallelic edited cells is by using a universal surrogate reporter system (HDR-USR)66. Compared with the surrogate reporter-integrated donor system61, the HDR-USR system functions by itself in an episomal manner66. Because the HDR-USR surrogate vector is not integrated into the genome, it allows scarless genome editing without introducing insertional mutagenesis and unwanted exogenous sequences to the genome. Therefore, this HDR-USR system is particularly useful for knock-in of non-coding variants to study functional regulatory elements. It also can be employed to knock-in protein-coding variants in exons. The HDR-USR system contains a truncated puromycin-resistant gene (Figure 1E). Puromycin-resistant function is restored through self-cleavage from expressed Cas9/sgRNA complex targeting truncated puromycin-resistant gene and self-repair via HDR using the full-length puromycin-resistant gene as a homologous intra-molecular template. Edited cells that have undergone HDR can be selected and enriched by co-transfecting this HDR-USR surrogate vector with the sgRNA expression cassette and integration of donor cassette into the cells. Compared with the commonly used surrogate reporter plasmids (for example, fluorescent proteins and antibiotic-resistant genes) that are used for the enrichment of transfection-positive cells, this HDR-USR surrogate vector displays better performance for the enrichment of HDR-edited cells.
It was recently shown that 5′ modification to the PCR-generated long dsDNA donors (~400 bp) strongly enhances HDR events (Figure 1F)67. Importantly, 5′ modification of the donors can tackle the problem associated with the integration of multimers due to joining together the multiple templates end to end, thereby enabling only one whole copy of the desired DNA to be inserted into the genome. This strategy also enables DNA to be edited in a more controlled way and promotes precise gene knock-in. Cas9 RNP complexes and 5′-modified linear double-stranded DNA donors with short homology arms (~50 bp) have also successfully been used to enable efficient gene knock-in68. This 5′-end modification of donor does not require long homology arms (for example, plasmid donor templates) and helps to reduce the background signal from the DNA repair templates. Modulation of targeting vectors to provide 3′ overhangs at both ends could also enhance HDR frequency by facilitating the assembly of proteins involved in strand invasion and new DNA synthesis in HDR69.
Gene knock-in and gene correction efficiency can be increased by enhancing HDR efficiency. HDR events are increased by synchronizing and enriching the cells in the G2/M cell cycle phase70. Alternatively, Cas9 expression is synchronized in S/G2/M by controlled timing of CRISPR/Cas9 RNP complex delivery71 or using a fusion of Cas9 and the N-terminal region of human Geminin25. HDR has also been successfully activated in quiescent stem cells and primary cells through controlled cycling and quiescence72. In this case, these quiescent cells are induced to briefly enter the cell cycle for HDR-based gene editing using the timed administration of a small-molecule cocktail (for example, SCF, TPO, and Flt3L). After gene editing, these HDR-edited cells are induced back into quiescence using another combination of small molecules (for example, rapamycin and CHIR99021). A modified version of Cas9 has also been employed to increase the HDR/NHEJ ratio by limiting the Cas9 nuclease activity in the G1 phase of the cell cycle73. HDR efficiency has also been improved by using small molecules that directly modulate HDR pathways, such as RS-1 (RAD51 activator)74,75, L755507 (β3-adrenergic receptor agonist)76, and nocodazole (a G2/M phase synchronizer)27. Ectopic co-expression of RAD52 and dn53BP1 could improve HDR efficacy without altering Cas9 off-target activity77. In addition, overexpression of an engineered RAD18 variant (e18) promotes HDR by suppressing the localization of the NHEJ-promoting factor 53BP1 to double-strand breaks78. To further promote HDR events, small molecules are used to inhibit the NHEJ activity by disrupting a key enzyme in the NHEJ pathway, such as DNA ligase IV79, KU7080, and 53BP181. In addition, attenuation of histone deacetylase 1 (HDAC1) and HDAC2 activities was recently shown to facilitate Cas9 access and binding to the targeted DNA, thereby enhancing HDR events and gene knock-in efficiency82. As such, a desired editing outcome can be genetically manipulated or chemically induced by altering the choice of DNA double-strand break repair to favor a specific DNA repair pathway72,83. For example, small molecules (chemical compound) and factors (genes) identified in high-throughput screens can be used to enhance or inhibit a specific DNA repair pathway for intended genome editing. To realize such applications, many other unknown small molecules and factors that potentially target the DNA damage response can be screened using a recently developed scalable CRISPR/Cas9-based fluorescent reporter assay to assess their influence on DNA double-strand break repair choice84. The underlying mechanisms by which these small molecules and factors regulate double-strand break repair then can be evaluated.
However, the use of small molecules or disruption of NHEJ and histone modifier genes can cause genome-wide changes in gene regulation and expression. Therefore, Cas9 was recently fused to CtIP, a key protein in early steps of homologous recombination, to stimulate precise transgene integration by HDR85. Directing CtIP-mediated homologous recombination to the CRISPR/Cas9 cutting site can minimize a global effect following DNA repair. On the other hand, fusions of Cas9 to DN1S (dominant-negative mutant of 53BP1) enhance HDR and precise gene knock-in by inhibiting NHEJ events specifically at Cas9 cleavage sites86. The use of this Cas9-DN1S fusion protein can circumvent the issue associated with the unwanted effects of global NHEJ inhibition. Another strategy to improve HDR efficiency is to covalently link the DNA repair templates to CRISPR/Cas9 complexes87–89. For example, to shuttle the HDR template to the nucleus, truncated Cas9 target sequences (16 bp) are added to the ends of the template for interacting with Cas9 RNPs (Figure 1G)90. Because Cas9 contains nuclear localization sequences (NLSs), Cas9 RNPs along with the associated HDR template can be transported together into the cell nucleus. To further enhance HDR efficiency, multiple copies of DNA repair templates are conjugated to multiple sites on a Cas9 via the conjugation adaptor89. Each conjugation adaptor on Cas9 bears the complementary sequence to the DNA repair template and thus multiple conjugation adaptors enable multivalent display of DNA repair template on Cas9. These conjugation strategies ensure the uptake and co-localization of both repair template and Cas9–sgRNA RNP complex in an individual cell87,88.
Machine learning has been used to further improve HDR efficiency by designing the optimal targeting strategy and repair template91. Deep learning has helped to predict editing products and genotypes such as on-target mutagenesis and off-target activities upon CRISPR-mediated gene correction of pathogenic variants in human cells92–96. In addition to aid designing an optimal DNA sequence, deep learning can also improve editing activity of CRISPR by taking into account chromatin accessibility and epigenetic features97,98.
Synthesis-dependent strand annealing (SDSA)-based approaches
ssODN30 and linear double-stranded DNA PCR fragments31 enable efficient gene knock-in via a synthesis-dependent strand annealing (SDSA)-like DNA repair mechanism. Compared with HDR-mediated knock-in using dsDNA plasmid template (>500 bp each homology arm), ssODN (50–80 bp each homology arm) and PCR fragment (~35 bp homology arms) provide higher efficiency for knock-in of small DNA modifications (for example, SNP)30,31. Given their high efficiency of a point mutation knock-in, a fluorescent tag is not required for the PCR fragment or ssODN donor. Several mechanistic models have been proposed to explain how a point mutation is introduced to the genome through DNA repair machinery mediated by a short single-stranded DNA oligonucleotide and CRISPR/Cas9 system. Two promising models are excision and corrective therapy (EXACT)99 and single-stranded template repair (SSTR)100 models. EXACT uses ssODNs to serve as both a bandage and a template for gene correction, whereas SSTR uses the Fanconi anemia DNA repair pathway for targeted mutagenesis.
The mechanism of gene repair directed by the ssODNs was elucidated in detail well before the CRISPR era101–105. In the case of single-agent gene editing, point mutations and base lesions are repaired by ssODNs in the absence of the CRISPR RNP complex106. This ssODN-mediated DNA repair machinery can be enhanced by stalling of replication forks and synchronizing the cells in the early S cell cycle phase during ssODN exposure106,107 or using anticancer drugs to induce double-strand DNA breakage108,109. Subsequently, CRISPR/Cas9 RNP complex has been used to improve point mutation repair directed by a short ssODN through induction of double-stranded breaks at the target genomic loci99. Nevertheless, the mechanism of action of DNA repair directed by an ssODN as the sole gene-editing agent is generally similar to that of ssODN and CRISPR/Cas9 working in concert. Interestingly, single-nick-induced gene editing using ssODN and Cas9n generates conversion tracts biased either mostly unidirectional or bidirectional depending on the relative strandedness of the ssODN and the nick110. In contrast, the unidirectional conversion pathway is preferentially used upon CRISPR/Cas9-induced double-strand DNA breaks. A strand bias in ssODN-mediated gene editing could be due to different accessibility of the DNA strands during DNA replication, irrespective of transcriptional status111. The sequences surrounding the targeted nucleotide also influence strand bias, while occupancy by transcriptional complexes alone does not dictate strand bias112. Notably, unmodified ssODNs are the most efficient in gene correction, while the addition of chemical modifications to the ssODN did not further improve its efficacy111.
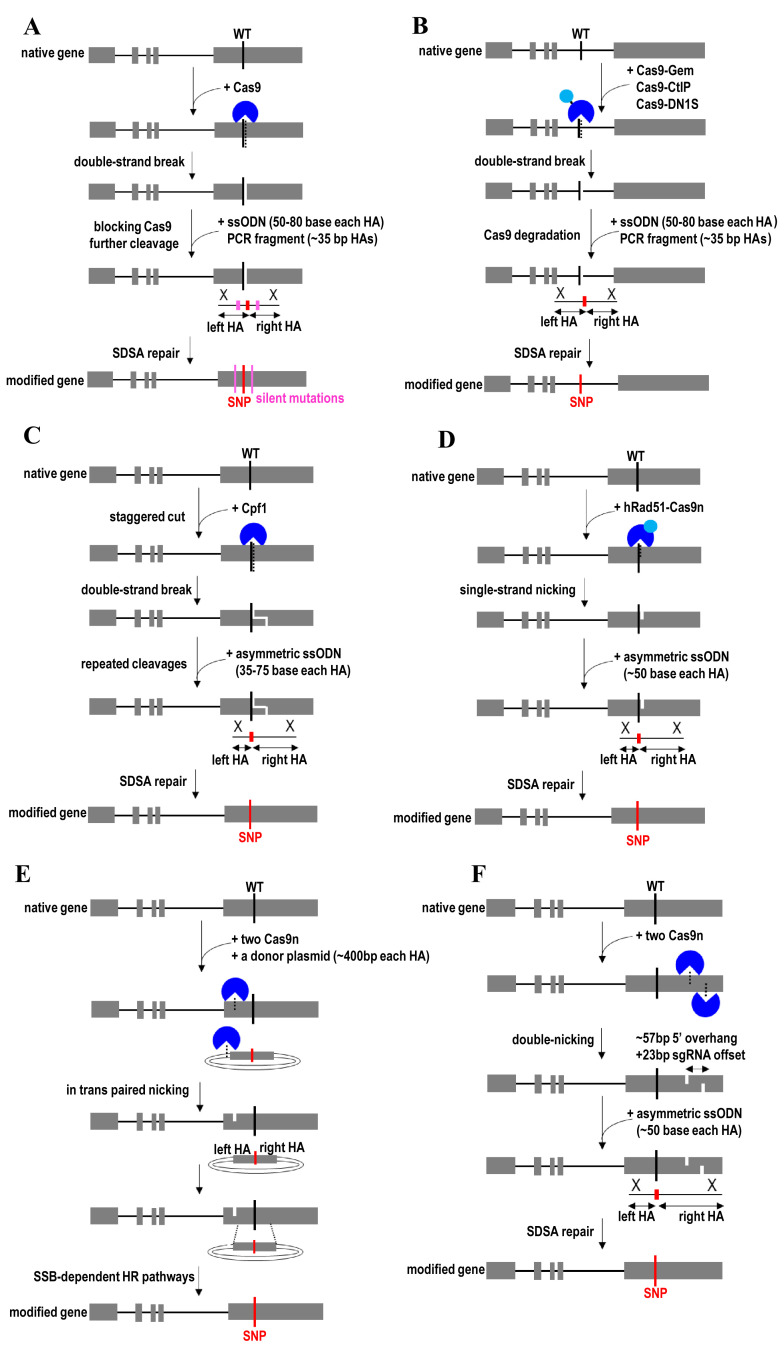
Silent mutations are usually introduced into the PAM sequence or the sgRNA seed sequence of the donor template to block further Cas9 targeting and recutting after undergoing HDR (Figure 2A). This can prevent unwanted mutations (for example, indels) introduced into the DNA upon subsequent NHEJ repair of Cas9-induced double-strand breaks. However, the insertion of silent mutations is not advisable when designing CRISPR/Cas9 to target the non-coding regions for knock-in of non-coding variants into the genome. In this case, the use of Cas9-Gem allows indel-free knock-in at the target locus by facilitating the degradation of Cas9 nuclease during the G1 phase of the cell cycle (Figure 2B)113. Gem is derived from human Geminin protein that is highly expressing during the S and G2 phases. Therefore, the fusion of Gem to Cas9 allows Cas9 to persist only during S and G2 phases. Gene correction efficiency is further increased by using CRISPR/Cpf1114 and asymmetric donor ssODN115 to enhance HDR (Figure 2C). Because the Cpf1 cleavage site is distal from the PAM sequence and sgRNA recognition site, Cpf1 enabled higher HDR rates than Cas9 by allowing repeated cleavages before indel mutations terminate targeting114. Compared with the blunt ends produced by Cas9, Cpf1 enhances gene insertion and knock-in by generating sticky ends upon DNA cleavage via a staggered DNA double-stranded break116,117. Cpf1-derived synthetic chimeric nucleases provide a broader targeting scope and a higher editing specificity than Cpf1118. Meanwhile, asymmetric donor DNA enhances gene insertion and replacement by biasing the choice of DNA repair pathway toward SDSA or increasing the local concentration of the donor115. It is also possible to use a synthetic long ssODN (1–3 kb) donor bearing two homology arms (60–300 bp each) along with CRISPR/Cas9 to integrate a long transgene into the genome119.
Figure 2. Synthesis-dependent strand annealing (SDSA)-mediated gene knock-in and gene correction strategies.
(A) Exonic SNP knock-in using CRISPR/Cas9 along with a short single-stranded donor oligonucleotide (ssODN) or a linear dsDNA PCR fragments. (B) Intronic SNP knock-in using Cas9-Gem, Cas9-CtIP, or Cas9-DN1S along with a short ssODN or a linear dsDNA PCR fragment. (C) Exonic SNP knock-in using CRISPR/Cpf1 and asymmetric donor ssODN. (D) Exonic SNP knock-in using hRad51-Cas9 nickase and asymmetric donor ssODN. (E) Exonic SNP knock-in by in trans paired nicking to target genomic DNA and donor plasmid using a pair of Cas9 nickases. (F) Exonic SNP knock-in by simultaneous nicking both DNA strands using a Cas9 nickase mutant with a pair of sgRNAs targeting opposite DNA strands. Cas9n, Cas9 nickase; CtIP, a key protein in early steps of homologous recombination; DN1S, dominant-negative mutant of 53BP1; Gem, human Geminin protein; HA, homology arm; HR, homologous recombination; hRad51, human RAD51 recombinase; SNP, single-nucleotide polymorphism; SSB, single-stranded break.
Because imprecise repair of Cas9-induced double-stranded DNA breaks can give rise to unpredictable deleterious mutations and complex chromosomal rearrangements120,121, Cas9n mutant is used to nick single-stranded DNA for seamless genome editing29. DNA nicks are repaired by the alternative HDR pathway (for example, SDSA pathway), especially when canonical RAD51/BRCA2-dependent HDR is compromised or down-regulated122. Studies have shown that Cas9 D10A (a D10A mutation on RuvC catalytic domain) is more potent in mediating HDR than Cas9 H840A (an H840A mutation on HNH catalytic domain)29. For example, Cas9n is fused to hRad51 mutants to improve HDR efficiency while minimizing undesired consequences of double-stranded DNA breaks and off-target mutagenesis (Figure 2D)123. This strategy uses ssODN as donor templates and does not require the inclusion of PAM-blocking silent mutations to prevent target recutting by Cas9n after undergoing SDSA. hRad51 participates in the repair of nicked DNA via strand invasion, and the nick-mediated HDR efficiency is improved using hRad51 mutant forms to disable its binding to BRCA2 and self-associate. Therefore, localizing hRad51 mutants to a targeted DNA nick can increase the frequency of HDR in response to a DNA nick.
In trans paired nicking was another recently developed strategy to enable efficient seamless genome editing without inducing double-stranded DNA breaks124–126. This strategy uses a pair of Cas9ns to generate coordinated single-strand breaks in donor plasmids and chromosomal target sites (Figure 2E). Recombination between nicked plasmid donor and a nicked target sequence then proceeds through SSB-dependent HR pathways. This strategy helps to avoid mutagenizing unmodified alleles and minimize large-scale chromosomal rearrangements, thereby enabling accurate gene knock-in, and maintains target protein dosages125. It also allows the editing of genes that are essential for cell function and survival. As an alternative strategy, a Cas9n mutant with a pair of sgRNAs targeting opposite DNA strands nearby (40–70 bp apart between two nicking sites) is used to induce a staggered double-strand DNA breaks with overhangs at the target locus by nicking both DNA strands simultaneously (Figure 2F)29,127. The PAM site for each Cas9n should face the outside of the target region to enable robust gene editing. While maintaining high on-target efficiencies, this double nicking strategy enhances genome-editing specificity by reducing off-target activity. Although both Cas9n must function in concert to make a double-strand break, paired Cas9n is sometimes more efficient than individual Cas9 nuclease for gene disruption128.
Microhomology-mediated end joining-based approaches
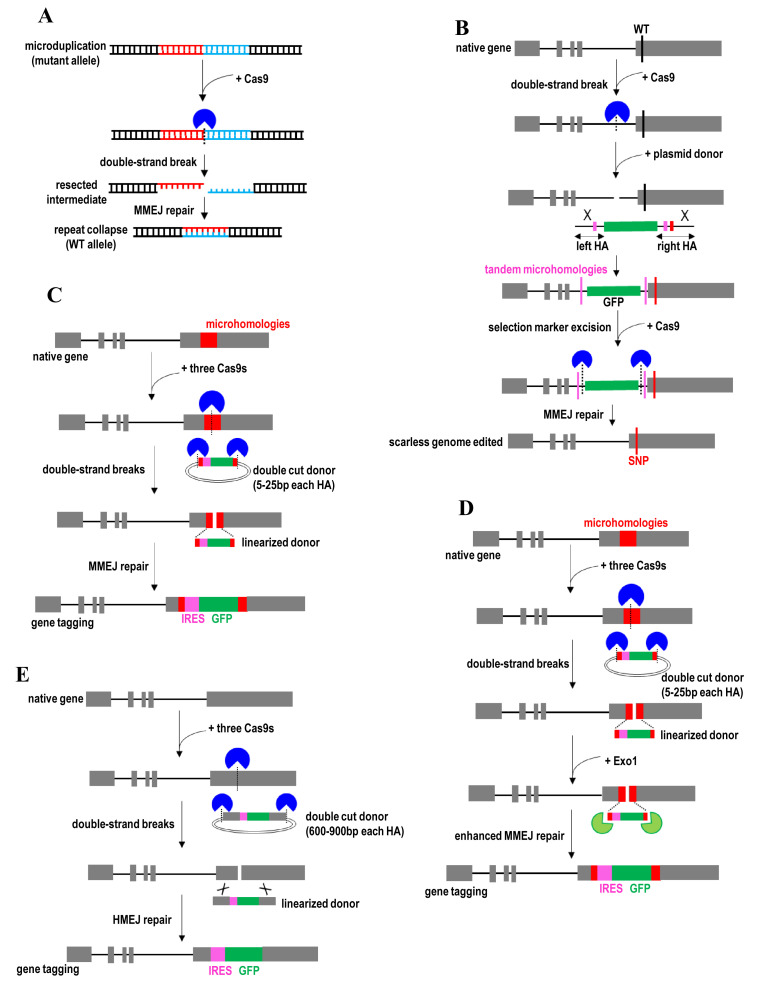
Another DNA repair pathway, namely MMEJ, has also been exploited for efficient and precise gene correction35. MMEJ pathway can be a promising alternative to classic HDR because HDR is inefficient in many cell types. MMEJ is active during most periods of the cell cycle, whereas the activity of HDR is restricted to the S/G2 phases. MMEJ uses extremely short microhomologous sequences (5–25 bp) to align the broken strands before joining129. For example, CRISPR/Cas9 is employed to induce a DNA double-stranded break near the center of a disease-causing microduplication (Figure 3A)35. This microduplication then is reverted to the wild-type genomic sequence upon MMEJ repair of double-stranded breaks. The wild-type genomic sequence is no longer targeted by the sgRNA that was used to target the microduplication, thereby avoiding another round of cleavage by Cas9. This strategy enables efficient and precise gene replacement without using exogenous DNA donors and can be used to edit a wide range of microduplication lengths. However, the application of this MMEJ-based approach is limited to this class of pathogenic mutations.
Figure 3. Microhomology-mediated end joining (MMEJ) and homology-mediated end joining (HMEJ)-mediated gene knock-in and gene correction strategies.
(A) Gene correction of microduplications by CRISPR/Cas9-mediated DNA double-stranded break near the center of the duplication. (B) Exonic SNP knock-in by CRISPR/Cas9-mediated integration of a selection marker and two short tandem microhomologies at intron and an SNP at exon, followed by removal of the selection marker using two CRISPR/Cas9s targeting the region between the selection marker and tandem microhomologies. (C) Transgene knock-in using a double-cut donor plasmid with short microhomologies and Cas9 with three different sgRNAs. (D) Transgene knock-in facilitated by exonuclease 1 (Exo1). (E) Transgene knock-in using a double-cut donor plasmid with long homology arms. GFP, green fluorescent protein; HA, homology arm; IRES, internal ribosome entry site; SNP, single-nucleotide polymorphism; WT, wild-type.
MMEJ-based strategy has also been recruited for scarless genome editing36. In this case, a plasmid donor with two overlapped homology arms bearing two short tandem microhomologies is used (Figure 3B). These two homology arms are flanking a selection marker, while a point mutation is juxtaposed with this selection marker. CRISPR/Cas9 is first used to induce a DNA double-stranded break in the intronic region proximal to the exonic SNP site. These two engineered microhomologies, a point mutation, and the selection marker in the donor then are integrated at the CRISPR/Cas9 cleavage site. Cas9 with two different sgRNAs then is used to cleave the DNA sequences between the selection marker and engineered microhomologies. MMEJ repair pathway will be stimulated to use these two engineered microhomologies to align the broken strands before joining. This leads to the deletion of one copy of tandem homology along with intervening selection marker sequences, leaving behind only the desired point mutation at the locus. The removal of sgRNA target sites and PAM sequences enables the protection of corrected and mutant alleles from subsequent cleavage by Cas9. This strategy is useful for the scarless excision of a selectable marker and biallelic modifications.
In addition to gene correction, the MMEJ pathway has been exploited to insert large DNA fragments (for example, tags) into the genome130,131. In this case, Cas9 with three different sgRNAs is co-expressed in the cell (Figure 3C). One sgRNA targets the genomic site and two sgRNAs target the 5′ and 3′ ends of the donor sequence. One sgRNA is designed to target the center of microhomologous sequences (~20 bp) in the genome. The microhomologous sequences and exogenous DNA sequences (for example, selection marker) in the donor are flanked by two sgRNA target sites. CRISPR/Cas9 then is used to linearize donor plasmid and release the exogenous DNA sequence with microhomologous sequences. This double-cut donor vector provides a higher gene knock-in efficiency than the conventional circular donor plasmid. Because the double-cut donor plasmid contains microhomologous DNA ends corresponding to the genomic cleavage site, a copy of tandem homology along with intervening exogenous DNA sequences is integrated into the genome upon MMEJ repair. This MMEJ-based approach uses only very short microhomologies and enables flexible gene knock-in without introducing unwanted exogenous sequences (for example, vector backbone sequence)130,131. Exonuclease 1 (Exo1) was also recently used to enhance MMEJ-mediated knock-in132. Exo1 facilitates end resection of double-strand breaks at both the genomic locus and the donor vector, thereby promoting the alignment between cleavage genomic site and linearized donor using microhomology (Figure 3D).
Homology-mediated end joining (HMEJ)-based approaches
The MMEJ-based strategy was subsequently devised to an HMEJ approach by enabling more efficient targeted transgene integration using longer and more stable homology arms27,37–39. In this case, CRISPR/Cas9 is designed to cleave both the targeted genomic locus and transgene donor vector that contains long homology arms (600–900 bp each homology arm) (Figure 3E). This HMEJ-based strategy provides a higher editing efficiency and better fidelity than MMEJ, particularly in non-dividing cells and adult animals37. HMEJ-based strategy also allows efficient knock-in of multiple genes without undergoing selection and enriching processes38. HMEJ efficiency is further improved by combining the use of this double-cut donor vector with the cell cycle regulators (for example, Nocodazole and CCND1)27. Targeted transgene integration mediated by HMEJ would not alter global gene expression profiles39. Moreover, targeted integration of a large gene payload is useful for the creation of reporter cells, production of recombinant pharmaceutical proteins, and efficient biallelic gene disruption. However, similar to MMEJ-based knock-in approaches, HMEJ requires a PAM sequence close to the CRISPR/Cas9 cleavage site in the genome.
Homology-independent gene knock-in and gene correction strategies
Non-homologous end joining (NHEJ)-based approaches
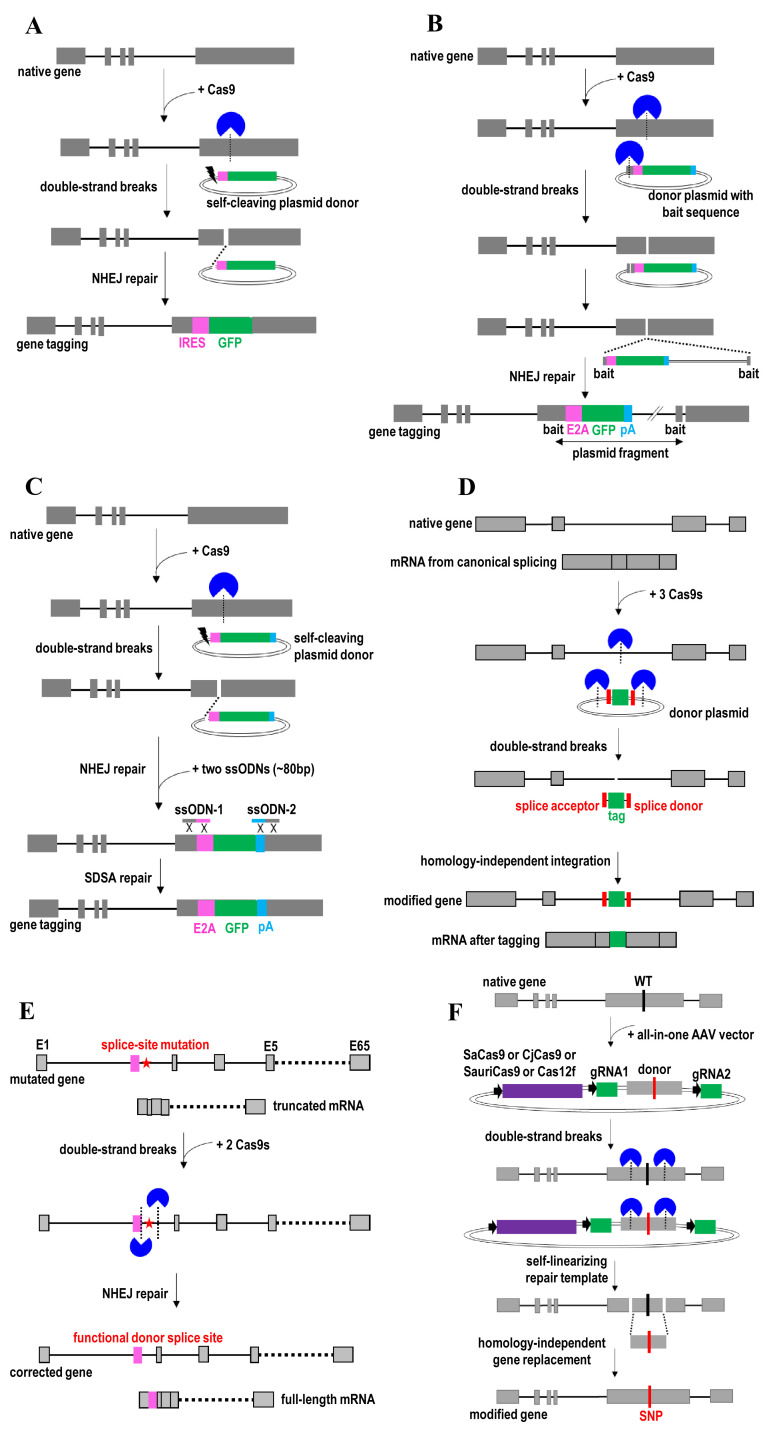
NHEJ repair occurs throughout the cell cycle, whereas the HDR event is restricted to S/G2 phase in dividing cells133. Importantly, NHEJ is a predominant DNA repair pathway in non-dividing cells and post-mitotic tissues (for example, neurons and human organoids). Therefore, the efficiency of insertional mutagenesis and gene correction is limited by NHEJ that competes with HDR. In fact, NHEJ-based knock-in was recently shown to have a higher efficiency than HDR-mediated gene targeting9,134,135. In this case, double-strand DNA breaks are introduced to both the genome and donor template for mediating transgene insertion via the NHEJ repair pathway (Figure 4A)40,135. The donor plasmid is linearized using Cas9 to cleave one sgRNA target site (for the single-cut donor) or two sgRNA target sites at both sides of the transgene (for the double-cut donor). However, double-cut donor is less efficient than the single-cut donor because the former will generate two DNA fragments that compete for genomic integration134. The linearized donor plasmid then is directly ligated to the broken genomic DNA ends upon NHEJ repair. NHEJ efficiently re-ligates DNA ends without mistakes and it does not require regions of homology for precise transgene insertion. However, the donor can be inserted in either orientation of the broken genomic DNA ends upon NHEJ repair. Off-target DNA double-strand breaks would also lead to random donor insertion to the unintended genomic sites. To circumvent this issue, a short homology DNA sequence bearing the Cas9 target sequence (bait sequence) is introduced onto a donor plasmid (Figure 4B)136. In this case, concurrent cleavage of the target genomic locus and bait plasmid sequence leads to efficient targeted integration of a large transgene via NHEJ pathway. Nevertheless, the insertion is independent from the homology sequence between the target locus and the bait in the donor plasmid. Similar NHEJ strategy can also be achieved by using Cpf1 to create sticky ends at the DNA cleavage site117.
Figure 4. Non-homologous end joining (NHEJ)-mediated gene knock-in and gene correction strategies.
(A) Transgene knock-in by introducing double-strand DNA breaks to both genome and single-cut donor. (B) Large transgene knock-in by including the bait sequence onto a donor plasmid. (C) Very large transgene knock-in using CRISPR/Cas9 and single-cut donor along with two additional short ssODNs. (D) Gene tagging by CRISPR/Cas9-mediated integration of a tag flanked by splice acceptor and donor sites. (E) Restore the function of donor splice site and full-length mRNA by CRISPR/Cas9-mediated deletion of the intervening intronic sequence that contains a pathogenic splice-site mutation. (F) Gene replacement using self-linearizing all-in-one AAV repair templates and CRISPR/Cas9s to generate two simultaneous double-strand breaks in both genomic DNA and repair template. AAV, adeno-associated virus; GFP, green fluorescent protein; IRES, internal ribosome entry site; SDSA, synthesis-dependent strand annealing; ssODN, single-stranded donor oligonucleotide.
To replace large genomic regions (for example, 58 kb) or integrate a very large transgene (for example, 200 kb) into the genome, two additional short ssODNs (~80 bp) are introduced together with self-cleaving plasmid donor and CRISPR/Cas9 targeting the genomic locus into the cell (Figure 4C)119. Upon double-strand breaks, these two ssODNs ligate each cut end to join the genomic DNA and the plasmid donor via the SDSA repair pathway. This integrated approach based on NHEJ and SDSA is useful for protein tagging, labeling of cellular structures, generating reporter lines, creating loss-of-function alleles, and lineage-tracing experiments119,134,135. It eliminates the need of attaching homology arms to the donor vector. It is also more efficient than traditional HDR-based knock-in strategies to assess and edit the silenced genomic loci or compact chromatin areas134.
The NHEJ-based knock-in strategy has also been employed to program exon skipping and gene tagging by modulation of gene splicing137. Gene splicing is manipulated by mutating specific DNA bases within splice acceptor sites or the addition of splice acceptor and donor sites to the donor DNA. For example, to add a tag to the endogenous mRNA, a donor plasmid with a tag flanked by splice acceptor and donor sites is used (Figure 4D)137. Two sgRNA target sites are located at both ends of the tag with splicing sites. CRISPR/Cas9 is first used to generate double-strand breaks in both the intron and donor. A tag together with splicing sites in this donor then is introduced to the intronic region via a homology-independent integration pathway. Through alternative splicing, the tag will be added to the coding sequence upon mRNA expression. Similarly, exon inclusion can be achieved by replacing the tag with a synthetic exon. Gene tagging is useful for interrogation of protein function, localization, and interactions in their native context within living cells. Therefore, it can avoid artifacts associated with overexpression of exogenous transgenes. Meanwhile, exon inclusion is useful to derive a protein with a new function and property. Actually, a similar strategy has been previously employed to tag at the C and N terminus of an exon using a generic donor plasmid138,139. This NHEJ-based gene tagging approach is scalable for tagging endogenous loci because of the use of the universal donor DNA. However, compared with the N- and C-terminal tagging approach, this intron-tagging strategy can prevent the disruptive or deleterious impact on the coding sequence. Another efficient way to tag the endogenous genes is by mutating specific DNA bases within splice acceptor sites using CRISPR technology41. For example, a pathogenic splice-site mutation causes the exclusion of exon 2 from an mRNA and generates the truncated protein (Figure 4E). CRISPR/Cas9 is used to excise an intronic region containing this mutation41. Deletion of this mutation will restore the function of donor splice site through NHEJ repair. The edited gene can now produce the full-length mRNA and restore the protein function. This strategy is useful for correcting non-coding mutations and creating a functional splice donor site by deleting the intervening intronic sequence that contains a splice-site mutation.
An all-in-one AAV vector with two sgRNAs, SaCas9140,141 (or other small Cas variants such as CjCas9142, SauriCas9143, Cas12f144, St1Cas9145, and CRISPR-CasPhi146), and a self-linearizing repair template was recently used for gene replacement (Figure 4F). In this case, CRISPR/Cas9 is first used to generate two simultaneous double-strand breaks in both genomic DNA and repair template. This leads to the replacement of the intervening sequence in genomic DNA with a repair template containing the desired mutation or wild-type sequence via a homology-independent DNA repair pathway. An integrated use of single AAV vector platform and homology-independent gene replacement approach enables robust gene correction of pathogenic variants in vivo. The use of this all-in-one AAV vector enables delivery of CRISPR/Cas9 and repair template using only a single AAV vector, thereby simplifying AAV production and improving delivery efficiency and CRISPR editing potency140.
Other gene knock-in and gene correction strategies
Base editing
Because the imprecise repairing of Cas9-induced DNA double-stranded breaks can lead to on-target mutagenesis and chromosomal rearrangements147, base editing provides a safer way to perform gene correction. Base editor, a CRISPR-based synthetic biology tool, was recently repurposed to enable programmable editing of a target base in genomic DNA by harnessing the base excision repair (BER) ability42,43. BER accurately repairs base lesions and single-strand breaks throughout the cell cycle148. Base editor is favorable to Cas9 because it enables clean mutational knock-in by direct conversion of nucleotides in genomic DNA without inducing double-strand DNA breaks that can generate unwanted indel mutations. Nevertheless, designing multiple guide sequences of base editor to target several genomic sites in trans may still be able to lead to double-strand DNA breaks and result in the deletion of the intervening segment between two sgRNA target sites. Importantly, base editing is particularly useful for manipulating point mutations and correcting pathogenic variants in postmitotic cells and most of the somatic tissue in vivo149–152. Nucleotide substitutions with base editor generated much excitation partly because the HDR event is highly inefficient in non-dividing cells and the NHEJ-based editing can lead to on-target indel formation. The use of base editors also can simplify the delivery of editing machinery because no homologous DNA repair template is required to edit the genomic DNA sequence.
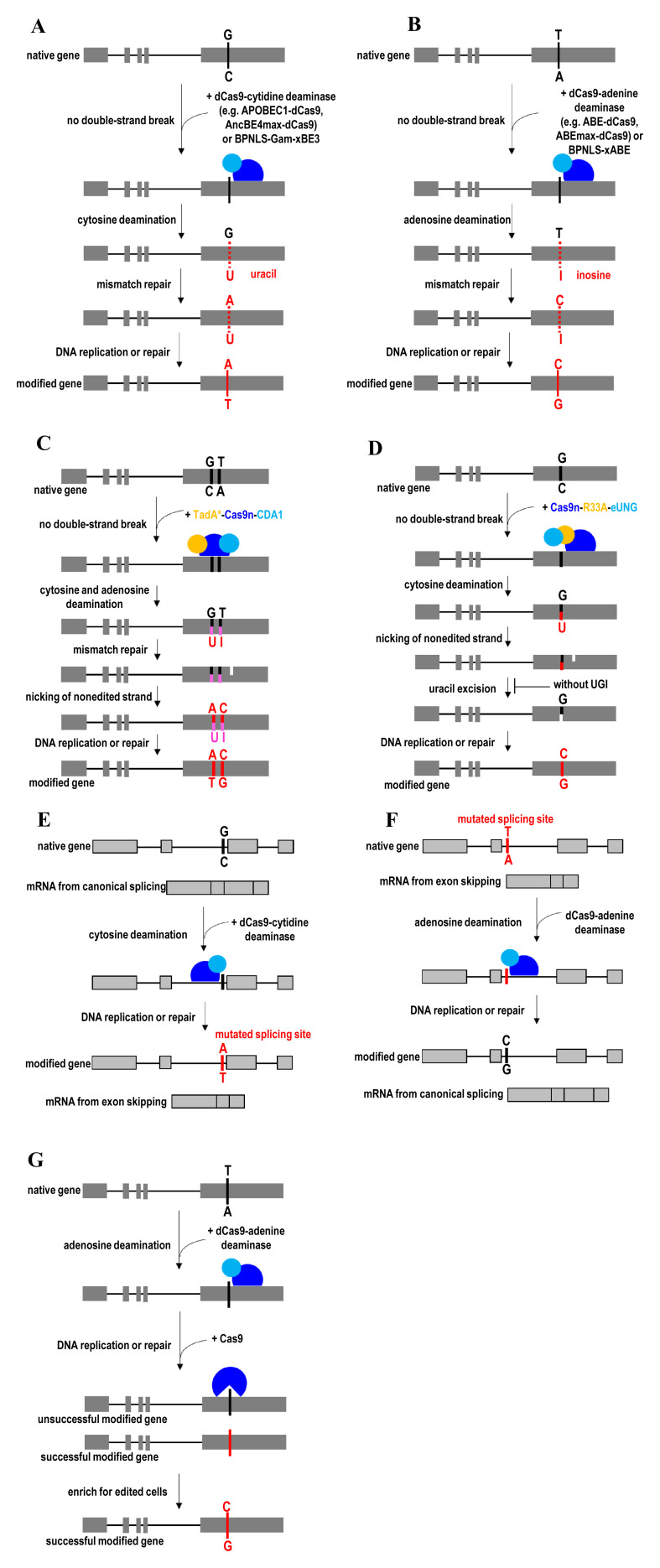
In general, base editors are classified into two broad classes: cytidine base editors and adenine base editors. Cytidine base editor (for example, APOBEC1-dCas9) mediates the direct conversion of cytidine to uridine, which eventually leads to a C-to-T (or G-to-A) substitution (Figure 5A)42. Meanwhile, adenine base editor (for example, ABE-dCas9) mediates the direct conversion of adenine to inosine, which eventually leads to an A-to-G (or T-to-C) substitution (Figure 5B)43. Concurrent adenine and cytosine editing has also been achieved by using a dual-deaminase CRISPR base editor153 or an engineered TadA* enzyme of adenine base editors154. The dual-deaminase CRISPR base editor can concurrently introduce A-to-G and C-to-T substitutions to the target DNA in the genome, thereby enabling creation or correction of multi-nucleotide variants (Figure 5C). In addition to DNA base transitions (pyrimidine-to-pyrimidine or purine-to-purine conversions), targeted C-to-G base transversions have been achieved by using a Cas9n-R33A-eUNG155. This C-to-G base editor consists of a Cas9n, an Escherichia coli–derived uracil DNA N-glycosylase (eUNG), and a rat APOBEC1 cytidine deaminase variant (R33A) (Figure 5D). The specificity of the C-to-G base editor was improved by removing the eUNG domain from it to yield Cas9n-R33A. C-to-G editing permits nucleotide substitutions and mutational correction at the desired genomic sites not possible with cytidine and adenine base editors. Compared with transition mutations, introduction of transversion mutations into the transcription factor binding sites may exert stronger effects on transcription factor binding and gene expression.
Figure 5. Base excision repair (BER)-mediated gene knock-in and gene correction strategies.
(A) C-to-T (or G-to-A) substitution by direct conversion of cytidine to uridine using cytidine base editors. (B) A-to-G (or T-to-C) substitution by direct conversion of adenine to inosine using adenine base editors. (C) Concurrent adenine and cytosine editing by a dual-deaminase CRISPR base editor. (D) Targeted C-to-G base transversions by a Cas9n-R33A-eUNG base editor. (E) Program exon skipping and (F) restore full-length mRNA by mutating target DNA bases within splice acceptor sites. (G) Enrich base-edited cells by eradicating non-edited cells using an inducible active Cas9 with the same sgRNA as the base editor. ABE, adenine base editor; APOBEC1, apolipoprotein B mRNA-editing enzyme, catalytic polypeptide 1; BPNLS, biparticle nucleus localization signal; eUNG, an Escherichia coli–derived uracil DNA N-glycosylase; R33A, a rat APOBEC1 cytidine deaminase variant; TadA, adenosine deaminase; UGI, uracil glycosylase inhibitor.
Base editors have also successfully been used to program exon skipping156 (Figure 5E) and restore full-length mRNA157 (Figure 5F) by modulation of gene splicing through mutating target DNA bases within splice acceptor sites. Base conversion efficiency mediated by both cytidine42 and adenine43 base editors is the highest when the target cytidine or adenine is at protospacer position 5. Nevertheless, the base editing can occur within a window of about 5 nucleotides (at protospacer positions 4–8). To broaden the targeting scope, the editing window of base editors was recently expanded from 4 or 5 nucleotides to 8 or 9 nucleotides by circularly permuting the Cas9n domain of base editors158. To narrow the editing window for single-nucleotide replacement at a specific position, high-precision base editors were engineered by optimizing the length of the linker between the deaminase domain and the Cas domain of the editor159,160.
Although the base editors enable direct conversion of C-to-G base transversions and all four transition mutations (C to T, A to G, T to C, and G to A) at target loci in living cells, base editing encounters several limitations. The application of base editing is restricted to point mutation substitutions. Base editing also has a limited targeting scope, low editing efficiency, and high off-target activities161–163. It is a concern that adenine base editors can display unexpected on-target cytosine deamination activity by converting cytosine to guanine or thymine154. Base editors also can induce unwanted bystander nucleotide conversions when repeated sequences (for example, multiple Cs) are present in the editing window. Therefore, several next-generation base editors were recently engineered to address these issues. These next-generation base editors introduce desired point mutations at a target locus at significantly higher efficiency and fewer undesired by-products than a CRISPR/Cas9-mediated HDR approach42,43. The genome-targeting scope of these next-generation base editors is also broader than the dCas9- or Cas9n-derived base editor164.
Next-generation cytidine base editors enhance desired base editing and minimized on-target indel formation by fusing uracil glycosylase inhibitor to the base editor (for example, APOBEC1-dCas9-UGI) and employing a Cas9n (for example, APOBEC1-Cas9n-UGI) targeting the non-edited strand42. Uracil glycosylase inhibitor (UGI) blocks uracil DNA glycosylase from reversing the U:G pair to a C:G pair, thereby improving base-editing efficiency. Replacing dCas9 with Cas9n improves base-editing efficiency by inducing mismatch repair pathway to resolve the U:G mismatch into desired U:A and T:A products. To stimulate mismatch repair, Cas9n is used to nick the non-edited strand containing a G opposite the edited U. Later, APOBEC1-dCas9-UGI and APOBEC1-Cas9n-UGI were further evolved into BE4max or AncBE4max by improving nuclear localization and optimizing codon usage for maximum expression of base editors44. The BE4max and AncBE4max provide higher base-editing efficiency than the older cytidine base editors. Nevertheless, the editing window remains similar to the previously established cytidine base editors. With the phage-assisted continuous evolution of the base-editor method, these cytosine base editors were subsequently evolved into evoAPOBEC1-BE4max, evoFERNY-BE4max, and evoCDA1-BE4max165. Compared with BE4max and AncBE4max, these three evolved base editors are more efficient at editing cytosine in the GC context. EvoFERNY is also smaller than APOBEC1, thereby enabling the packaging of EvoFERNY-BE4max into the AAV vector for in vivo applications. EvoCDA1-BE4max is useful for a difficult-to-edit site or cell type. It should be noted that their editing windows and targeting scope are different from the original BE4max. Human APOBEC3G-Cas9n fusion was another recently engineered cytidine base editor to reduce unwanted bystander activities and improve precision of base editing in the CC context (repeated Cs)166. To minimize unwanted RNA-editing activity without sacrificing on-target DNA-editing activity, Cas9n-UGI was fused to non-APOBEC1 cytidine deaminases, such as an enhanced human APOBEC3A (eA3A) and human activation-induced cytidine deaminase (hAID)167. These base-editor variants also did not self-edit their own transcripts, thereby avoiding heterogeneity in base-editor coding sequences.
Similar to cytidine base editor, next-generation adenine base editor (for example, ABE7.10) was engineered to mediate the conversion of A/T to G/C in genomic DNA43. ABE7.10 was derived from TadA*6-dCas9 variants that harbor W23L/R, P48A, and R152H/P mutations after seven rounds of evolution and engineering. These mutations abrogate base-specific enzyme–DNA interactions to broaden target sequence compatibility, thereby enhancing ABE7.10-mediated base-editing efficiency at targets that contain multiple A residues. Subsequently, ABE7.10 editor was evolved into ABEmax for more efficient base editing by improving nuclear localization, maximizing protein expression, and ancestral reconstruction of the deaminase component44. To reduce off-target RNA-editing activity, miniABEmax was engineered by removing the wild-type TadA domain and altering amino acid residues (K20A/R21A or V82G) within the mutant TadA domain of ABEmax to reduce the RNA-recognition capability167. Compared with ABEmax, miniABEmax has lower Cas9-independent RNA-editing activity but maintains Cas9-assisted on-target DNA-editing function. Notably, the self-editing activity of miniABEmax was significantly lower than ABEmax. Self-editing by base editors can lead to the generation of truncated Cas9n with intact deaminase activity, thereby inducing off-target RNA editing but not on-target DNA editing.
The recent development of xCas9-derived base editors that recognize a broad range of PAM sequences has expanded the targeting scope in the genome168,169. Despite its broadened PAM compatibility, xCas9 retains high DNA specificity with minimal off-target activity. Subsequently, the base-editing efficiency of the xCas9-derived base editors (for example, xBE3 and xABE) was improved by fusing the BPNLS or Gam (or both) to the N terminus of xCas9-derived base editors (for example, BPNLS-Gam-xBE3 and BPNLS-xABE)45. Biparticle nucleus localization signal (BPNLS) enables efficient localization of base editors to the nucleus, while the Gam protein of bacteriophage Mu minimizes the production of undesired by-products upon base editing. More recently, NG-BE4max and NG-ABEmax base editors were generated by fusing a newly engineered SpCas9-NG variant to BE4max or ABEmax base editor170. SpCas9-NG variant recognizes minimal NG PAMs more efficiently than the xCas9 variant, thereby enabling efficient base editing with expanded targeting scope. The targeting scope and editing window of base editors have also been expanded using circularly permuted and PAM-modified Cas9n variants such as CP-CBEmax and CP-ABEmax158. In 2020, SpRY, a near-PAMless SpCas9 variant, was successfully engineered to target almost all PAMs (NRN>NYN PAMs), thereby eliminating PAM recognition constrains171. To overcome potential undesirable off-target effects due to the relaxation of the PAM requirement, a high-fidelity SpRY variant (SpRY-HF1) with improved DNA targeting specificity was subsequently engineered. Compared with dCas9, xCas9, and SpCas9-NG, SpRY-derived base editors enabled robust editing of the vast majority of disease-relevant genetic variants in the genome.
To enrich base-edited cells and increase the base-editing efficiency, a double-check base-editing approach is employed to exert a selective pressure against non-edited cells (Figure 5G)172. In this case, an inducible active Cas9 with the same sgRNA as the base editor is employed. The cells with non-edited target bases remain vulnerable to Cas9 cleavage. Cas9-mediated double-strand breaks lead to the death of non-edited cells in prokaryotic cells. However, the NHEJ is the major outcome instead of cell death in mammalian cells. To enrich base-edited cells in eukaryote cells, several alternative reporter systems have been developed173,174. For example, a transient reporter that converts blue fluorescent protein (BFP) to GFP upon a base substitution was employed173. The GFP-positive cells can be subsequently sorted for enrichment of base-edited cells. Compared with reporters of transfection that reflect only transfection efficiency, this BFP-to-GFP conversion reporter provides more accurate information about base-editing efficiency. In another reporter system, an inactivated eGFP reporter was placed downstream of wild-type mCherry and a T2A site174. The eGFP fluorescence is restored by correcting a point mutation in eGFP transgene via base editing. In this way, mCherry allows the identification of successfully transfected cells, while eGFP enables the quantification of single base-editing efficiency. The eGFP- and mCherry-positive cells then can be sorted. These two fluorescent-based reporter systems also allow high-throughput evaluation and identification of small molecules and factors that influence base-editing efficiency. To generalize the applications of the reporter to different cell types and systems, a functional reporter system (GO system) was subsequently developed for enrichment of base-edited cells175. GO works by initiating protein expression and affecting protein translation of different reporter proteins through correction of a mutated start codon immediately downstream of a kozak sequence using a base editor. This flexible GO system has helped to expand the base-editing reporter toolbox because it can be used to induce the translation of an array of different reporters, including fluorescent proteins, antibiotic resistance, and luciferase.
Prime editing
In 2019, a prime-editing strategy was developed to overcome the limitations of homology-directed gene correction and base editing46. Prime editing has lower off-target editing than Cas9 nuclease and displays higher editing efficiency with fewer by-products than the CRISPR/Cas9-mediated homologous recombination approach. Compared with homology-dependent approaches that introduce edits within 10 bp from the Cas9 cleavage site, prime editor can install point mutations at distances of more than 30 bp from the Cas9n nicked site176. Therefore, prime editor offers greater targeting flexibility than the homology-dependent approaches. Most importantly, prime editing enables correction of all types of substitutions such as transitions and transversions as well as small insertions and deletions without requiring double-strand breaks or exogenous donor DNA repair templates177,178. Prime editing could also complement base editing in the case of unwanted bystander edits from the presence of multiple cytidine or adenine bases within the editing window of base editors176. Furthermore, prime editing outperformed the base editor for bases positioned outside the center of the base-editing window and could be an alternative tool when the desired genomic DNA site is not targetable by the base editor.
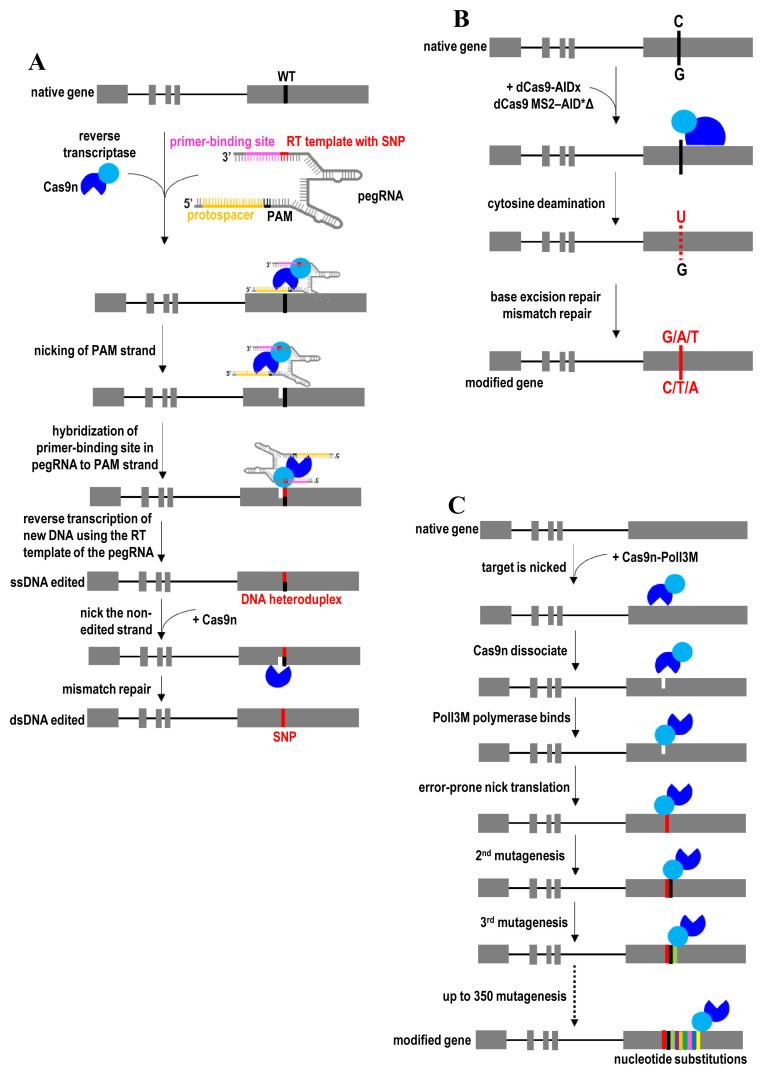
Prime editing uses a fusion protein consisting of Cas9n and an engineered RT. Importantly, a prime-editing guide RNA (pegRNA) that both specifies the target site of sgRNA and encodes the desired edit of the RT template is used to form complexes with Cas9n–RT. As a general principle, Cas9n–RT complexed with the pegRNA first binds and nicks the target DNA (Figure 6A). The resulting 3′ end of the nicked DNA strand hybridizes to the primer binding site in pegRNA. Then the RT template of the pegRNA is used to prime reverse transcription of new DNA bearing the desired edit. The desired genetic information is directly copied from an extension on the pegRNA to replace the target DNA in the genome. Equilibration between the edited 3′ flap and the unedited 5′ flap, cellular 5′ flap cleavage and ligation, and base excision repair will eventually lead to nucleotide substitutions at the target DNA site. Prime-editing efficiency is further increased by nicking the non-edited strand to induce DNA repair of that strand. This repair leads to the generation of duplex DNA containing the desired edit. The prime-editing strategy has been successfully applied in post-mitotic, terminally differentiated primary cells46. The efficiency of prime editing is determined largely by the design of the pegRNA such as the length of primer binding site and RT template as well as the GC content, primary sequence motifs, and secondary structures of the pegRNA176. Manipulation of DNA repair machinery to favor the replacement of the non-edited strand in the DNA heteroduplex could also improve the desired editing outcome.
Figure 6. Prime editing and genetic diversification.
(A) Gene correction by prime editing using Cas9n-RT complexed with the pegRNA. (B) Targeted mutagenesis by converting cytidines or guanines to the other three bases at desired loci using dCas9-AIDx. (C) Continuous diversifying all nucleotides within a tunable window length at a target locus using Cas9n-PolI3M. AID, activation-induced cytidine deaminase; Cas9n, Cas9 nickase; MS2, bacteriophage coat proteins; pegRNA, prime-editing guide RNA; PolI3M, fidelity-reduced variant of Escherichia coli DNA polymerase I (PolI) harboring the mutations D424A, I709N, and A759R; RT, reverse transcription; SNP, single-nucleotide polymorphism.
Since prime editing and base editing do not employ any selection marker, their precise, scarless genome-editing events can be identified through the recently developed dinucleotide signature capture (DTECT) detection method179. The application of primer editor, similar to that of base editors, is limited to correcting point mutations or small genetic alterations. HDR- and NHEJ-based approaches are preferred to confer large DNA insertions or deletions. The addition of the desired edit of the RT template to the sgRNA may destabilize the extended sgRNA through the activity of intracellular RNA-degrading enzymes180. Therefore, it remains impractical to add a very long RT template to the sgRNA for large genetic alterations. Moreover, the RT template in pegRNA can be incorporated in the genome during reverse transcription. It is also a challenge to co-package Cas9n-RT and its pegRNA into a single AAV vector for prime editing in vivo because of large sizes of Cas9n-RT and the lengthened sgRNA. This AAV packaging issue can be addressed by using smaller Cas9n and RT enzymes.
Genetic diversification
CRISPR technology has recently been harnessed for modifying multiple DNA bases simultaneously and enabling genetic diversification in living cells through targeted mutagenesis in the genome. Genetic diversification is useful for establishing complex libraries of genetic variants, enabling high-throughput screening of functional variants, evolving protein structure and function in situ, engineering antibodies, mapping protein–drug interactions, and cell-lineage tracking.
In 2016, two independent research groups developed the dCas9-AIDx fusion protein for converting cytidines or guanines to the other three bases at the desired loci (Figure 6B)181,182. When dCas9-AIDx is used together with multiple sgRNAs, it expands the editing window and generates more mutations in the protospacers. The dCas9-AIDx is created by fusing dCas9 to the activation-induced cytidine deaminase (AID). AID directs somatic hypermutation to a given genomic locus by deamination of cytosine to uracil. After deamination, point mutations are generated through misread of the uracil–guanine mismatch, base-excision repair of the uracil, or mismatch-repair pathway to generate transitions and transversions near the lesion182. Targeted mutagenesis efficiency is further improved by incorporating two MS2 hairpin-binding sites to the sgRNA for recruiting multiple copies of the hyperactive AID variant (AID*Δ) to each dCas9182. Besides, targeted mutagenesis specificity of dCas9-AIDx is improved by blocking uracil DNA glycosylase181. Coupled with uracil-DNA glycosylase inhibitor, dCas9-AIDx specifically converts cytidines to thymines in the protospacer. AIDx has also been fused to Cas9 nickase for optimal nucleotide conversion181. Therefore, these targeted mutagenesis strategies create a diverse repertoire of point mutations in situ, dismissing the need to use error-prone PCR to create mutations in vitro or design a large number of DNA oligos for incorporation into a genomic locus. It also introduces a large spectrum of point mutations without generating insertions and deletions observed with active Cas9. Nevertheless, Cas9n-AIDx can induce double-strand DNA breaks when designing multiple sgRNAs to target different target DNA sites in the genome181. To improve the efficiency of base editing in GC contexts and reduce bystander activity, AID-Cas9 was later evolved into the enhanced AID-Cas9 fusion (eAID-BE4max)183.
In 2018, another strategy that uses CRISPR-guided DNA polymerases (Cas9n-PolI3M) was developed for genetic diversification in living cells47. Compared with dCas9-AIDx-based targeted mutagenesis approaches (editing window within five nucleotides)181,182, Cas9n-PolI3M is capable of continuously diversifying all nucleotides within a tunable window length (up to 350 nucleotides) at user-defined loci47. Therefore, Cas9n-PolI3M generates a larger spectrum of substitution mutations than dCas9-AIDx for large-scale genetic screenings. Cas9n-PolI3M consists of a CRISPR-guided nickase (Cas9n) and a fused nick-translating DNA polymerase (PolI3M)47. The targeted mutation rate of Cas9n-PolI3M was enhanced by creating three mutations (K848A, K1003A, and R1060A) to Cas9n to promote the dissociation of Cas9n from DNA after nicking the target locus. In addition, a more processive DNA polymerase variant, PolI3M, was engineered to increase the length of the editing window and provide a higher targeted mutation rate than the wild-type DNA polymerase. As a general principle, Cas9n first nicks the target locus, resulting in a single-strand break in DNA (Figure 6C). Then Cas9n from the Cas9n-PolI3M complex dissociates, followed by the binding of PolI3M to the nicking site. Using this nicked DNA as an initiation site, PolI3M performs error-prone nick translation via strand displacement synthesis and cleavage of the displaced strand by the flap endonuclease domain. The resulting ligatable nick at the start site can undergo targeted mutagenesis. Finally, PolI3M continues to mutagenesis other nucleotides in the vicinity of the start site or nick site with an editing window of up to 350 nucleotides. Therefore, Cas9n specifies the start site of the editing window for PolI3M, while the mutagenesis window length and mutation rate are determined by PolI3M. This strategy enables multiplexing and continuously diversifying all nucleotides within user-defined genomic loci. Also, this strategy is more efficient than the homology-directed oligonucleotide integration approach. Simultaneously diversifying multiple loci through co-expression of multiple gRNAs is useful for studying epistatic interactions. In this case, multiple gRNAs targeting the same strand are used to nick the same strand to avoid double-strand breaks.
RNA editing
Pathogenic point mutations can be directly corrected in RNA transcripts instead of mutagenesis of the genomic DNA49,184,185. Cas9 activity poses the risk of unanticipated chromosomal rearrangement at the targeted sites and permanent, off-target mutagenesis to DNA120. In this case, RNA base-editing technology offers a safer strategy by directly editing the pathogenic transcripts without the loss of genomic information. Because RNA editing is transient, it ensures that no by-products or unwanted mutations are generated in genomic DNA that can be passed on to daughter cells during mitosis. It also useful for curing diseases without a genetic origin and mimicking genetic variants that provide a health advantage186. Because dCas13 has no targeting sequence constraints and no motif preference surrounding the target base, RNA base-editing technology helps to circumvent the issue associated with the limited targeting sites of DNA base editors. RNA base-editing technology does not rely on endogenous DNA repair pathways; therefore, it mediates efficient RNA editing even in post-mitotic cells.
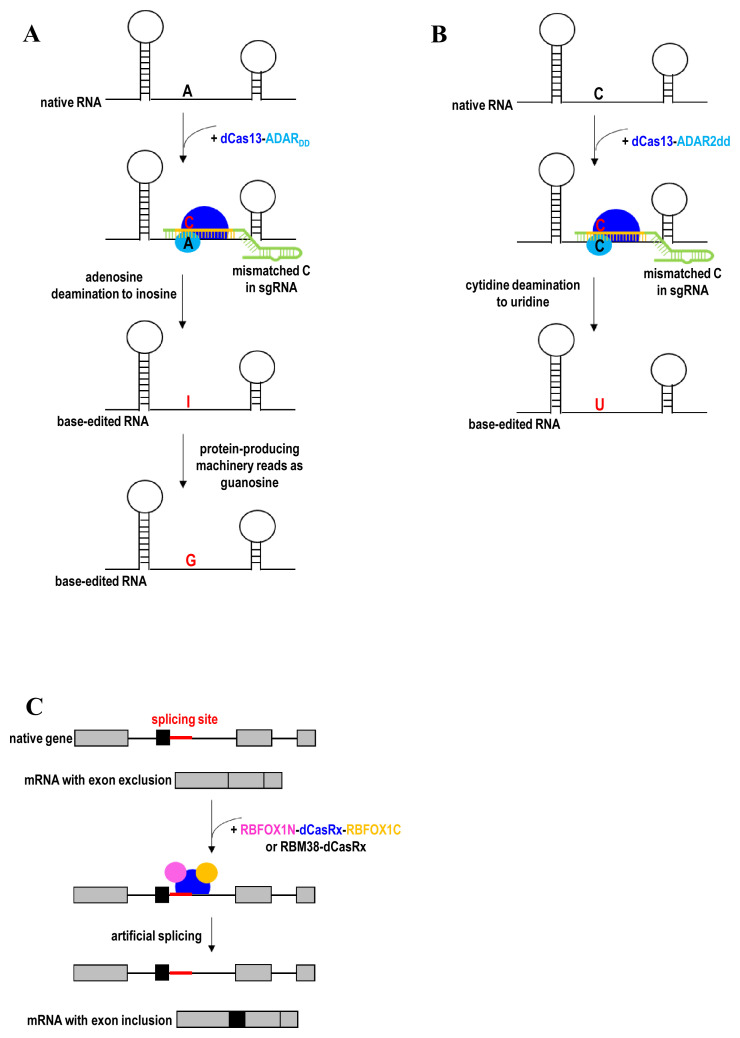
dCas13-ADAR2 has been employed to direct adenosine-to-inosine deaminase activity to endogenous RNA transcripts (Figure 7A)49. dCas13 is capable of binding mRNA in a nucleic acid–programmed manner, while ADAR2 (adenosine deaminase acting on RNA type 2) deaminates adenosines mispaired with cytidine bases in RNA duplexes without any protein co-factors. The deaminase domain of ADAR2-containing hyperactivating mutations (ADARDD) was later fused to dCas13b to enhance RNA-editing efficiency. The editing specificity of dCas13b-ADAR2DD was improved by structure-guided protein engineering of ADAR2DD (E488Q/T375G) to destabilize its binding to off-target RNA. Owing to the exogenous nature of dCas13b-ADAR2DD fusion proteins, host immune responses may compromise the efficacy of the RNA editing and cause side effects in vivo. The dCas13b-ADAR2DD fusion protein is also too large for packaging into the limited payload capacity of AAV vector. About two years later, dCas13b-ADAR2DD was refined to improve RNA-editing specificity and overcome immunogenicity issues associated with ectopic overexpression of ADAR by recruiting endogenous ADAR using short engineered ADAR-recruiting RNAs (arRNAs)48. The arRNA is a long-guide RNA (≥71 nucleotides) that could anneal with the target transcript to form dsRNA substrate. The dsRNA substrate then recruits endogenous ADAR proteins for targeted editing. Placing the A-C mismatch in the middle region of arRNAs results in the highest RNA-editing efficiency, while the addition of multiple A-G mismatches to each arRNA minimizes off-target editing. To further enhance RNA-editing efficiency, arRNA is fused with an ADAR-recruiting scaffold to recruit multiple copies of ADAR protein to target RNAs. A similar arRNA scaffold can also be used to recruit endogenous ADAR proteins to edit the genetic materials of RNA virus such as coronavirus in order to restrict viral propagation in the host. In fact, there was evidence for host-dependent RNA editing in the transcriptome of coronaviruses from coronavirus-infected patients187.
Figure 7. RNA editing.
(A) A-to-G substitution by directing adenosine-to-inosine deaminase activity to endogenous RNA transcripts using dCas13-ADARDD. (B) C-to-U substitution by directing cytidine-to-uridine deaminase activity to endogenous RNA transcripts using dCas13-ADAR2dd. (C) Exon inclusion to the mRNA by using a fusion of dCasRx and splicing regulatory domains. ADAR, adenosine deaminase; RBFOX1, RNA-binding protein fox-1 homolog; RBM38, RNA-binding motif protein 38.
To complement adenosine-to-inosine RNA-editing technology, a cytidine-to-uridine RNA editor was subsequently developed by directly evolving the adenine deaminase domain of ADAR2 (ADAR2dd) into a cytidine deaminase50. ADAR2dd then was fused to dCas13 for targeted RNA editing (Figure 7B). The specificity of dCas13-ADAR2dd was further improved via rational mutagenesis of ADAR2dd at residues (S375A) interacting with the RNA target. The RNA-editing specificity was also improved by introducing disfavorable guanine mismatches to the guide RNA. Apart from expanding the RNA-editing toolbox, the cytidine-to-uridine RNA editor is capable of multiplex RNA editing by having the capability of both adenosine-to-inosine and cytidine-to-uridine conversions.
More recently, CRISPR artificial splicing factors (CASFx) were developed for manipulation of gene splicing188. Although dCasRx alone was sufficient to promote exon exclusion by blocking access of splicing machinery189, dCasRx requires splicing regulatory domains to induce exon inclusion. Alternative slicing can be programmed using RBFOX1N-dCasRx-C or RBM38-dCasRx, a fusion of dCasRx and splicing factors (RBFOX1 or RBM38)188. The RNA-binding domain of splicing factors was substituted by dCasRx to derive the CASFx. For example, RBFOX1N-dCasRx-C or RBM38-dCasRx induces exon inclusion when bound downstream of the target exon (Figure 7C). In contrast, exon exclusion is induced when CASFx bind within a target exon. Therefore, this strategy can induce exon inclusion and exclusion to the mRNA without the need to modify the DNA sequence or introduce permanent changes in the genome. CASFx can also be applied transiently such that the alternative splicing is reversible.
Despite the reactiveness of RNA editing for research and therapeutic potential, RNA editing is much less efficient than gene editing186. Moreover, endogenous ADAR proteins are usually present in small amounts in most tissues for arRNA-based recruitment, thereby undermining RNA-editing efficiency in complex tissue186. The efficiency of RNA editing can be improved by using chemically modified sgRNA and a next-generation viral delivery vector or nanoparticle. For example, AAV delivery of exogenous RNA editor and guide sequence has enabled in vivo repair of mutant RNAs in mouse models of neurological disease190. Compared with gene editing, RNA editing also makes limited kinds of change to RNA. However, RNA-editing capabilities could be expanded through protein engineering to derive new RNA editors.
Summary and conclusions
Various CRISPR-mediated homology-dependent and -independent gene knock-in and gene correction strategies have been developed. Multiplex, flexible, scarless gene insertion and replacement—together with the emerging tools for genetic diversification, prime editing, base editing, and RNA editing—can be achieved at high efficiency and specificity. We highlight structure-guided engineering of Cas9 variants that have greatly improved the gene-editing efficacy, specificity, editing window, and targeting scope. In addition, we comprehensively discuss factors involved in various DNA repair pathways enabling transgene integration and gene correction, such as Cas9 variants (for example, Cas9 nuclease, Cas9 nickase, Cpf1 nuclease, CasX, dCas9, or dCas13), fusion domains (for example, HDR enhancer, base editor, splicing effector, polymerase, or RT), donor type (for example, plasmid DNA, ssODN, PCR fragments, or homologous chromosome), length of homology arms (for example, none, 5–80 bp, or 500–1000 bp), number of sgRNAs (for example, single-cut or double-cut on donor and genomic DNA), DNA cleavage (for example, double-strand break, single-strand nick, or no cleavage), and cleavage pattern (for example, blunt or staggered cut).
We discuss the choice of gene correction approaches depending on the nature of the mutations (coding, non-coding, or splicing variants), types of mutation (substitution, deletion, or duplication), copy of alleles for modification (monoallelic or biallelic), and editing outcome (scarless modification or insertion of the selection marker to the genome for the enrichment of edited clones). We also discuss the choice of transgene knock-in approaches determined by the transgene size, donor design (the length of homology arms), plasmid donor conformation (circular, single-cut linearized, or double-cut linearized), chromatin accessibility of insertion site (open or closed chromatin), and the purpose of transgene knock-in (for overexpression, gene tagging, exon skipping, exon inclusion, gene knockout, gene replacement, or study of functional regulatory elements).
Finally, we discuss various strategies to address the potential immunogenicity associated with exogenous CRISPR fusion proteins. These include strategies to recruit endogenous catalytic domains (for example, base editor, HDR enhancer, RNA editor, or chromatin modifier) for gene knock-in and gene correction and controlled timing of CRISPR/Cas9 delivery and expression to minimize the off-target effects, toxicity, and immunogenicity associated with CRISPR components. Immunogenicity issue can also be circumvented by transient modulation of the genes involved in immunity by using a CRISPR-based synthetic repressor191. Other strategies to minimize undesired off-target mutagenesis are using light-activated, chemical-inducible, or multiple-input logic gate genetic circuits to spatiotemporal and conditional control of CRISPR/Cas9 activities192; cell type- or tissue-specific promoters and delivery vehicles (for example, specific viral serotypes16,17,193 or ligand-targeted cargos192), engineered high-fidelity Cas9 variants194–200, and rational chemical modifications to the sgRNA192; a chimeric DNA-RNA guide201; and engineered truncated sgRNA202,203 and sgRNA secondary structures204. Onsite mutagenesis and heterogeneity can also occur in the uncorrected population of cells following CRISPR/Cas9-induced double-stranded DNA breaks99. We also emphasize the importance of examining the potential unwanted head-to-tail insertions of donor DNA templates into the genome by HDR or NHEJ mechanisms (or both) before precisely edited alleles can be correctly identified205 and how chemical modification of both ends of the donor can help overcome this issue by enabling only one whole copy of the desired DNA to be inserted into the genome. In this fast-moving field of CRISPR-mediated gene editing, we hope to provide comprehensive information on recent developments to a broad scientific community for facilitating the application of this versatile technology.
The peer reviewers who approve this article are:
Ophir Shalem, Department of Genetics, Perelman School of Medicine, University of Pennsylvania, Philadelphia, USA
Eric B Kmiec, Gene Editing Institute, Helen F. Graham Cancer Center & Research Institute, Christianacare, Newark, DE, USA; Department of Medical and Molecular Sciences, University of Delaware, Newark, DE, USA; The Wistar Institute, Philadelphia, PA, USA
Funding Statement
This work was supported by National Institutes of Health grants to YS (AG069750, DK127778, AG055501, HL150521, HG008153, AG061521, AG017242, AG056278, AG057341, AG057433, AG057706), and Global Consortium for Reproductive Longevity and Equality (GCRLE-1320).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Contributor Information
Cia-Hin Lau, Email: lauciahin_4275@yahoo.com.
Chung Tin, Email: chungtin@um.cityu.edu.hk.
Yousin Suh, Email: ys3214@cumc.columbia.edu.
References
- 1. Gasiunas G, Barrangou R, Horvath P, et al. : Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci U S A. 2012; 109(39): E2579–86. 10.1073/pnas.1208507109 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 2. Jinek M, Chylinski K, Fonfara I, et al. : A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012; 337(6096): 816–21. 10.1126/science.1225829 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 3. Cong L, Ran FA, Cox D, et al. : Multiplex genome engineering using CRISPR/Cas systems. Science. 2013; 339(6121): 819–23. 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 4. Mali P, Yang L, Esvelt KM, et al. : RNA-guided human genome engineering via Cas9. Science. 2013; 339(6121): 823–6. 10.1126/science.1232033 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 5. Cho SW, Kim S, Kim JM, et al. : Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013; 31(3): 230–2. 10.1038/nbt.2507 [DOI] [PubMed] [Google Scholar]
- 6. Lau CH: Applications of CRISPR-Cas in Bioengineering, Biotechnology, and Translational Research. CRISPR J. 2018; 1(6): 379–404. 10.1089/crispr.2018.0026 [DOI] [PubMed] [Google Scholar]
- 7. Ma H, Marti-Gutierrez N, Park SW, et al. : Correction of a pathogenic gene mutation in human embryos. Nature. 2017; 548(7668): 413–9. 10.1038/nature23305 [DOI] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 8. Yin H, Xue W, Chen S, et al. : Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat Biotechnol. 2014; 32(6): 551–3. 10.1038/nbt.2884 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 9. Román-Rodríguez FJ, Ugalde L, Álvarez L, et al. : NHEJ-Mediated Repair of CRISPR-Cas9-Induced DNA Breaks Efficiently Corrects Mutations in HSPCs from Patients with Fanconi Anemia. Cell Stem Cell. 2019; 25(5): 607–621.e7. 10.1016/j.stem.2019.08.016 [DOI] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 10. Beyret E, Liao HK, Yamamoto M, et al. : Single-dose CRISPR-Cas9 therapy extends lifespan of mice with Hutchinson-Gilford progeria syndrome. Nat Med. 2019; 25(3): 419–22. 10.1038/s41591-019-0343-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Santiago-Fernández O, Osorio FG, Quesada V, et al. : Development of a CRISPR/Cas9-based therapy for Hutchinson-Gilford progeria syndrome. Nat Med. 2019; 25(3): 423–6. 10.1038/s41591-018-0338-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ledford H: CRISPR treatment inserted directly into the body for first time. Nature. 2020; 579(7798): 185. 10.1038/d41586-020-00655-8 [DOI] [PubMed] [Google Scholar]
- 13. Sharon E, Chen SAA, Khosla NM, et al. : Functional Genetic Variants Revealed by Massively Parallel Precise Genome Editing. Cell. 2018; 175(2): 544–557.e16. 10.1016/j.cell.2018.08.057 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 14. Sadhu MJ, Bloom JS, Day L, et al. : Highly parallel genome variant engineering with CRISPR-Cas9. Nat Genet. 2018; 50(4): 510–4. 10.1038/s41588-018-0087-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Soldner F, Stelzer Y, Shivalila CS, et al. : Parkinson-associated risk variant in distal enhancer of α-synuclein modulates target gene expression. Nature. 2016; 533(7601): 95–9. 10.1038/nature17939 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 16. Wang D, Zhang F, Gao G: CRISPR-Based Therapeutic Genome Editing: Strategies and In Vivo Delivery by AAV Vectors. Cell. 2020; 181(1): 136–50. 10.1016/j.cell.2020.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lau CH, Suh Y: In vivo genome editing in animals using AAV-CRISPR system: applications to translational research of human disease [version 1; peer review: 2 approved]. F1000Res. 2017; 6: 2153. 10.12688/f1000research.11243.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tay LS, Palmer N, Panwala R, et al. : Translating CRISPR-Cas Therapeutics: Approaches and Challenges. CRISPR J. 2020; 3(4): 253–75. 10.1089/crispr.2020.0025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dever DP, Bak RO, Reinisch A, et al. : CRISPR/Cas9 β-globin gene targeting in human haematopoietic stem cells. Nature. 2016; 539(7629): 384–9. 10.1038/nature20134 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 20. Kim MY, Yu KR, Kenderian SS, et al. : Genetic Inactivation of CD33 in Hematopoietic Stem Cells to Enable CAR T Cell Immunotherapy for Acute Myeloid Leukemia. Cell. 2018; 173(6): 1439–1453.e19. 10.1016/j.cell.2018.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 21. Eyquem J, Mansilla-Soto J, Giavridis T, et al. : Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nature. 2017; 543(7643): 113–7. 10.1038/nature21405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cannon PM: Gene Editing Expands the Donor Pool for CCR5-Negative Stem Cell Transplants. Cell Stem Cell. 2019; 25(6): 735–6. 10.1016/j.stem.2019.11.006 [DOI] [PubMed] [Google Scholar]
- 23. Xu L, Wang J, Liu Y, et al. : CRISPR-Edited Stem Cells in a Patient with HIV and Acute Lymphocytic Leukemia. N Engl J Med. 2019; 381(13): 1240–7. 10.1056/NEJMoa1817426 [DOI] [PubMed] [Google Scholar]
- 24. Selvaraj S, Dhoke NR, Kiley J, et al. : Gene Correction of LGMD2A Patient-Specific iPSCs for the Development of Targeted Autologous Cell Therapy. Mol Ther. 2019; 27(12): 2147–57. 10.1016/j.ymthe.2019.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gutschner T, Haemmerle M, Genovese G, et al. : Post-translational Regulation of Cas9 during G1 Enhances Homology-Directed Repair. Cell Rep. 2016; 14(6): 1555–66. 10.1016/j.celrep.2016.01.019 [DOI] [PubMed] [Google Scholar]
- 26. Ikeda K, Uchida N, Nishimura T, et al. : Efficient scarless genome editing in human pluripotent stem cells. Nat Methods. 2018; 15(12): 1045–7. 10.1038/s41592-018-0212-y [DOI] [PubMed] [Google Scholar]
- 27. Zhang JP, Li XL, Li GH, et al. : Efficient precise knockin with a double cut HDR donor after CRISPR/Cas9-mediated double-stranded DNA cleavage. Genome Biol. 2017; 18(1): 35. 10.1186/s13059-017-1164-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vaidyanathan S, Salahudeen AA, Sellers ZM, et al. : High-Efficiency, Selection-free Gene Repair in Airway Stem Cells from Cystic Fibrosis Patients Rescues CFTR Function in Differentiated Epithelia. Cell Stem Cell. 2020; 26(2): 161–171.e4. 10.1016/j.stem.2019.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 29. Ran FA, Hsu PD, Lin CY, et al. : Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013; 154(6): 1380–9. 10.1016/j.cell.2013.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 30. Paquet D, Kwart D, Chen A, et al. : Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature. 2016; 533(7601): 125–9. 10.1038/nature17664 [DOI] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 31. Paix A, Folkmann A, Goldman DH, et al. : Precision genome editing using synthesis-dependent repair of Cas9-induced DNA breaks. Proc Natl Acad Sci U S A. 2017; 114(50): E10745–E10754. 10.1073/pnas.1711979114 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 32. Böttcher R, Hollmann M, Merk K, et al. : Efficient chromosomal gene modification with CRISPR/cas9 and PCR-based homologous recombination donors in cultured Drosophila cells. Nucleic Acids Res. 2014; 42(11): e89. 10.1093/nar/gku289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wu Y, Liang D, Wang Y, et al. : Correction of a genetic disease in mouse via use of CRISPR-Cas9. Cell Stem Cell. 2013; 13(6): 659–62. 10.1016/j.stem.2013.10.016 [DOI] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 34. Kwart D, Paquet D, Teo S, et al. : Precise and efficient scarless genome editing in stem cells using CORRECT. Nat Protoc. 2017; 12(2): 329–54. 10.1038/nprot.2016.171 [DOI] [PubMed] [Google Scholar]
- 35. Iyer S, Suresh S, Guo D, et al. : Precise therapeutic gene correction by a simple nuclease-induced double-stranded break. Nature. 2019; 568(7753): 561–5. 10.1038/s41586-019-1076-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kim SI, Matsumoto T, Kagawa H, et al. : Microhomology-assisted scarless genome editing in human iPSCs. Nat Commun. 2018; 9(1): 939. 10.1038/s41467-018-03044-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yao X, Wang X, Hu X, et al. : Homology-mediated end joining-based targeted integration using CRISPR/Cas9. Cell Res. 2017; 27(6): 801–14. 10.1038/cr.2017.76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shin SW, Lee JS: Optimized CRISPR/Cas9 strategy for homology-directed multiple targeted integration of transgenes in CHO cells. Biotechnol Bioeng. 2020; 117(6): 1895–903. 10.1002/bit.27315 [DOI] [PubMed] [Google Scholar]
- 39. Xie L, Sun J, Mo L, et al. : HMEJ-mediated efficient site-specific gene integration in chicken cells. J Biol Eng. 2019; 13: 90. 10.1186/s13036-019-0217-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Suzuki K, Tsunekawa Y, Hernandez-Benitez R, et al. : In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature. 2016; 540(7631): 144–9. 10.1038/nature20565 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 41. Kemaladewi DU, Maino E, Hyatt E, et al. : Correction of a splicing defect in a mouse model of congenital muscular dystrophy type 1A using a homology-directed-repair-independent mechanism. Nat Med. 2017; 23(8): 984–9. 10.1038/nm.4367 [DOI] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 42. Komor AC, Kim YB, Packer MS, et al. : Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016; 533(7603): 420–4. 10.1038/nature17946 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 43. Gaudelli NM, Komor AC, Rees HA, et al. : Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature. 2017; 551(7681): 464–71. 10.1038/nature24644 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 44. Koblan LW, Doman JL, Wilson C, et al. : Improving cytidine and adenine base editors by expression optimization and ancestral reconstruction. Nat Biotechnol. 2018; 36(9): 843–6. 10.1038/nbt.4172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Liu X, Li G, Zhou X, et al. : Improving Editing Efficiency for the Sequences with NGH PAM Using xCas9-Derived Base Editors. Mol Ther Nucleic Acids. 2019; 17: 626–35. 10.1016/j.omtn.2019.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 46. Anzalone AV, Randolph PB, Davis JR, et al. : Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. 2019; 576(7785): 149–57. 10.1038/s41586-019-1711-4 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 47. Halperin SO, Tou CJ, Wong EB, et al. : CRISPR-guided DNA polymerases enable diversification of all nucleotides in a tunable window. Nature. 2018; 560(7717): 248–52. 10.1038/s41586-018-0384-8 [DOI] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 48. Qu L, Yi Z, Zhu S, et al. : Programmable RNA editing by recruiting endogenous ADAR using engineered RNAs. Nat Biotechnol. 2019; 37(9): 1059–69. 10.1038/s41587-019-0178-z [DOI] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 49. Cox DBT, Gootenberg JS, Abudayyeh OO, et al. : RNA editing with CRISPR-Cas13. Science. 2017; 358(6366): 1019–27. 10.1126/science.aaq0180 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 50. Abudayyeh OO, Gootenberg JS, Franklin B, et al. : A cytosine deaminase for programmable single-base RNA editing. Science. 2019; 365(6451): 382–6. 10.1126/science.aax7063 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 51. Song F, Stieger K: Optimizing the DNA Donor Template for Homology-Directed Repair of Double-Strand Breaks. Mol Ther Nucleic Acids. 2017; 7: 53–60. 10.1016/j.omtn.2017.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Crowley EH, Arena S, Lamba S, et al. : Targeted knock-in of the polymorphism rs61764370 does not affect KRAS expression but reduces let-7 levels. Hum Mutat. 2014; 35(2): 208–14. 10.1002/humu.22487 [DOI] [PubMed] [Google Scholar]
- 53. Burnight ER, Gupta M, Wiley LA, et al. : Using CRISPR-Cas9 to Generate Gene-Corrected Autologous iPSCs for the Treatment of Inherited Retinal Degeneration. Mol Ther. 2017; 25(9): 1999–2013. 10.1016/j.ymthe.2017.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Martin RM, Ikeda K, Cromer MK, et al. : Highly Efficient and Marker-free Genome Editing of Human Pluripotent Stem Cells by CRISPR-Cas9 RNP and AAV6 Donor-Mediated Homologous Recombination. Cell Stem Cell. 2019; 24(5): 821–828.e5. 10.1016/j.stem.2019.04.001 [DOI] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 55. Tran NT, Graf R, Wulf-Goldenberg A, et al. : CRISPR-Cas9-Mediated ELANE Mutation Correction in Hematopoietic Stem and Progenitor Cells to Treat Severe Congenital Neutropenia. Mol Ther. 2020; S1525-0016(20)30414-7. 10.1016/j.ymthe.2020.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pomeroy EJ, Hunzeker JT, Kluesner MG, et al. : A Genetically Engineered Primary Human Natural Killer Cell Platform for Cancer Immunotherapy. Mol Ther. 2020; 28(1): 52–63. 10.1016/j.ymthe.2019.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lin Y, Zhang Q, Zhong ZA, et al. : Whole Genome Sequence Identified a Rare Homozygous Pathogenic Mutation of the DSG2 Gene in a Familial Arrhythmogenic Cardiomyopathy Involving Both Ventricles. Cardiology. 2017; 138(1): 41–54. 10.1159/000462962 [DOI] [PubMed] [Google Scholar]
- 58. Borck G, de Vries L, Wu HJ, et al. : Homozygous truncating PTPRF mutation causes athelia. Hum Genet. 2014; 133(8): 1041–7. 10.1007/s00439-014-1445-1 [DOI] [PubMed] [Google Scholar]
- 59. Figueiredo T, Melo US, Pessoa ALS, et al. : A homozygous loss-of-function mutation in inositol monophosphatase 1 (IMPA1) causes severe intellectual disability. Mol Psychiatry. 2016; 21(8): 1125–9. 10.1038/mp.2015.150 [DOI] [PubMed] [Google Scholar]
- 60. Barnett CP, Nataren NJ, Klingler-Hoffmann M, et al. : Ectrodactyly and Lethal Pulmonary Acinar Dysplasia Associated with Homozygous FGFR2 Mutations Identified by Exome Sequencing. Hum Mutat. 2016; 37(9): 955–63. 10.1002/humu.23032 [DOI] [PubMed] [Google Scholar]
- 61. Wu Y, Xu K, Ren C, et al. : Enhanced CRISPR/Cas9-mediated biallelic genome targeting with dual surrogate reporter-integrated donors. FEBS Lett. 2017; 591(6): 903–13. 10.1002/1873-3468.12599 [DOI] [PubMed] [Google Scholar]
- 62. Champer J, Liu J, Oh SY, et al. : Reducing resistance allele formation in CRISPR gene drive. Proc Natl Acad Sci U S A. 2018; 115(21): 5522–7. 10.1073/pnas.1720354115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Gantz VM, Jasinskiene N, Tatarenkova O, et al. : Highly efficient Cas9-mediated gene drive for population modification of the malaria vector mosquito Anopheles stephensi. Proc Natl Acad Sci U S A. 2015; 112(49): E6736-43. 10.1073/pnas.1521077112 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 64. Hammond A, Galizi R, Kyrou K, et al. : A CRISPR-Cas9 gene drive system targeting female reproduction in the malaria mosquito vector Anopheles gambiae. Nat Biotechnol. 2016; 34(1): 78–83. 10.1038/nbt.3439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kyrou K, Hammond AM, Galizi R, et al. : A CRISPR-Cas9 gene drive targeting doublesex causes complete population suppression in caged Anopheles gambiae mosquitoes. Nat Biotechnol. 2018; 36(11): 1062–6. 10.1038/nbt.4245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Yan N, Sun Y, Fang Y, et al. : A Universal Surrogate Reporter for Efficient Enrichment of CRISPR/Cas9-Mediated Homology-Directed Repair in Mammalian Cells. Mol Ther Nucleic Acids. 2020; 19: 775–89. 10.1016/j.omtn.2019.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 67. Gutierrez-Triana JA, Tavhelidse T, Thumberger T, et al. : Efficient single-copy HDR by 5' modified long dsDNA donors. eLife. 2018; 7: e39468. 10.7554/eLife.39468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Yu Y, Guo Y, Tian Q, et al. : An efficient gene knock-in strategy using 5'-modified double-stranded DNA donors with short homology arms. Nat Chem Biol. 2020; 16(4): 387–90. 10.1038/s41589-019-0432-1 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 69. Hirotsune S, Kiyonari H, Jin M, et al. : Enhanced homologous recombination by the modulation of targeting vector ends. Sci Rep. 2020; 10(1): 2518. 10.1038/s41598-020-58893-9 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 70. Yang D, Scavuzzo MA, Chmielowiec J, et al. : Enrichment of G2/M cell cycle phase in human pluripotent stem cells enhances HDR-mediated gene repair with customizable endonucleases. Sci Rep. 2016; 6: 21264. 10.1038/srep21264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lin S, Staahl BT, Alla RK, et al. : Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. eLife. 2014; 3: e04766. 10.7554/eLife.04766 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 72. Shin JJ, Schröder MS, Caiado F, et al. : Controlled Cycling and Quiescence Enables Efficient HDR in Engraftment-Enriched Adult Hematopoietic Stem and Progenitor Cells. Cell Rep. 2020; 32(9): 108093. 10.1016/j.celrep.2020.108093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Lomova A, Clark DN, Campo-Fernandez B, et al. : Improving Gene Editing Outcomes in Human Hematopoietic Stem and Progenitor Cells by Temporal Control of DNA Repair. Stem Cells. 2019; 37(2): 284–94. 10.1002/stem.2935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Pinder J, Salsman J, Dellaire G: Nuclear domain 'knock-in' screen for the evaluation and identification of small molecule enhancers of CRISPR-based genome editing. Nucleic Acids Res. 2015; 43(19): 9379–92. 10.1093/nar/gkv993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Song J, Yang D, Xu J, et al. : RS-1 enhances CRISPR/Cas9-and TALEN-mediated knock-in efficiency. Nat Commun. 2016; 7: 10548. 10.1038/ncomms10548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Yu C, Liu Y, Ma T, et al. : Small molecules enhance CRISPR genome editing in pluripotent stem cells. Cell Stem Cell. 2015; 16(2): 142–7. 10.1016/j.stem.2015.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 77. Paulsen BS, Mandal PK, Frock RL, et al. : Ectopic expression of RAD52 and dn53BP1 improves homology-directed repair during CRISPR-Cas9 genome editing. Nat Biomed Eng. 2017; 1(11): 878–88. 10.1038/s41551-017-0145-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Nambiar TS, Billon P, Diedenhofen G, et al. : Stimulation of CRISPR-mediated homology-directed repair by an engineered RAD18 variant. Nat Commun. 2019; 10(1): 3395. 10.1038/s41467-019-11105-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Maruyama T, Dougan SK, Truttmann MC, et al. : Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat Biotechnol. 2015; 33(5): 538–42. 10.1038/nbt.3190 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 80. Chu VT, Weber T, Wefers B, et al. : Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat Biotechnol. 2015; 33(5): 543–8. 10.1038/nbt.3198 [DOI] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 81. Canny MD, Moatti N, Wan LCK, et al. : Inhibition of 53BP1 favors homology-dependent DNA repair and increases CRISPR-Cas9 genome-editing efficiency. Nat Biotechnol. 2018; 36(1): 95–102. 10.1038/nbt.4021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Liu B, Chen S, La Rose A, et al. : Inhibition of histone deacetylase 1 (HDAC1) and HDAC2 enhances CRISPR/Cas9 genome editing. Nucleic Acids Res. 2020; 48(2): 517–32. 10.1093/nar/gkz1136 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 83. Ray U, Raghavan SC: Modulation of DNA double-strand break repair as a strategy to improve precise genome editing. Oncogene. 2020; 39(41): 6393–6405. 10.1038/s41388-020-01445-2 [DOI] [PubMed] [Google Scholar]
- 84. Roidos P, Sungalee S, Benfatto S, et al. : A scalable CRISPR/Cas9-based fluorescent reporter assay to study DNA double-strand break repair choice. Nat Commun. 2020; 11(1): 4077. 10.1038/s41467-020-17962-3 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 85. Charpentier M, Khedher AHY, Menoret S, et al. : CtIP fusion to Cas9 enhances transgene integration by homology-dependent repair. Nat Commun. 2018; 9(1): 1133. 10.1038/s41467-018-03475-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Jayavaradhan R, Pillis DM, Goodman M, et al. : CRISPR-Cas9 fusion to dominant-negative 53BP1 enhances HDR and inhibits NHEJ specifically at Cas9 target sites. Nat Commun. 2019; 10(1): 2866. 10.1038/s41467-019-10735-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Savić N, Ringnalda FC, Berk C, et al. : In vitro Generation of CRISPR-Cas9 Complexes with Covalently Bound Repair Templates for Genome Editing in Mammalian Cells. Bio Protoc. 2019; 9(1): e3136. 10.21769/BioProtoc.3136 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 88. Savic N, Ringnalda FC, Lindsay H, et al. : Covalent linkage of the DNA repair template to the CRISPR-Cas9 nuclease enhances homology-directed repair. eLife. 2018; 7: e33761. 10.7554/eLife.33761 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 89. Lim D, Sreekanth V, Cox KJ, et al. : Engineering designer beta cells with a CRISPR-Cas9 conjugation platform. Nat Commun. 2020; 11(1): 4043. 10.1038/s41467-020-17725-0 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 90. Nguyen DN, Roth TL, Li PJ, et al. : Polymer-stabilized Cas9 nanoparticles and modified repair templates increase genome editing efficiency. Nat Biotechnol. 2020; 38(1): 44–9. 10.1038/s41587-019-0325-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. O'Brien AR, Wilson LOW, Burgio G, et al. : Unlocking HDR-mediated nucleotide editing by identifying high-efficiency target sites using machine learning. Sci Rep. 2019; 9(1): 2788. 10.1038/s41598-019-39142-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Shen MW, Arbab M, Hsu JY, et al. : Predictable and precise template-free CRISPR editing of pathogenic variants. Nature. 2018; 563(7733): 646–51. 10.1038/s41586-018-0686-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Listgarten J, Weinstein M, Kleinstiver BP, et al. : Prediction of off-target activities for the end-to-end design of CRISPR guide RNAs. Nat Biomed Eng. 2018; 2(1): 38–47. 10.1038/s41551-017-0178-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Kim HK, Kim Y, Lee S, et al. : SpCas9 activity prediction by DeepSpCas9, a deep learning-based model with high generalization performance. Sci Adv. 2019; 5(11): eaax9249. 10.1126/sciadv.aax9249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Wang D, Zhang C, Wang B, et al. : Optimized CRISPR guide RNA design for two high-fidelity Cas9 variants by deep learning. Nat Commun. 2019; 10(1): 4284. 10.1038/s41467-019-12281-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Lin J, Wong KC: Off-target predictions in CRISPR-Cas9 gene editing using deep learning. Bioinformatics. 2018; 34(17): i656–i663. 10.1093/bioinformatics/bty554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Kim HK, Min S, Song M, et al. : Deep learning improves prediction of CRISPR-Cpf1 guide RNA activity. Nat Biotechnol. 2018; 36(3): 239–41. 10.1038/nbt.4061 [DOI] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 98. Chuai G, Ma H, Yan J, et al. : DeepCRISPR: Optimized CRISPR guide RNA design by deep learning. Genome Biol. 2018; 19(1): 80. 10.1186/s13059-018-1459-4 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 99. Rivera-Torres N, Banas K, Bialk P, et al. : Insertional Mutagenesis by CRISPR/Cas9 Ribonucleoprotein Gene Editing in Cells Targeted for Point Mutation Repair Directed by Short Single-Stranded DNA Oligonucleotides. PLoS One. 2017; 12(1): e0169350. 10.1371/journal.pone.0169350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Richardson CD, Kazane KR, Feng SJ, et al. : CRISPR-Cas9 genome editing in human cells occurs via the Fanconi anemia pathway. Nat Genet. 2018; 50(8): 1132–9. 10.1038/s41588-018-0174-0 [DOI] [PubMed] [Google Scholar]
- 101. Radecke F, Radecke S, Schwarz K: Unmodified oligodeoxynucleotides require single-strandedness to induce targeted repair of a chromosomal EGFP gene. J Gene Med. 2004; 6(11): 1257–71. [DOI] [PubMed] [Google Scholar]
- 102. Radecke S, Radecke F, Peter I, et al. : Physical incorporation of a single-stranded oligodeoxynucleotide during targeted repair of a human chromosomal locus. J Gene Med. 2006; 8(2): 217–28. [DOI] [PubMed] [Google Scholar]
- 103. Olsen PA, Randøl M, Luna L, et al. : Genomic sequence correction by single-stranded DNA oligonucleotides: Role of DNA synthesis and chemical modifications of the oligonucleotide ends. J Gene Med. 2005; 7(12): 1534–44. 10.1002/jgm.804 [DOI] [PubMed] [Google Scholar]
- 104. Aarts M, te Riele H: Progress and prospects: Oligonucleotide-directed gene modification in mouse embryonic stem cells: a route to therapeutic application. Gene Ther. 2011; 18(3): 213–9. 10.1038/gt.2010.161 [DOI] [PubMed] [Google Scholar]
- 105. Andersen MS, Sørensen CB, Bolund L, et al. : Mechanisms underlying targeted gene correction using chimeric RNA/DNA and single-stranded DNA oligonucleotides. J Mol Med (Berl). 2002; 80(12): 770–81. 10.1007/s00109-002-0393-8 [DOI] [PubMed] [Google Scholar]
- 106. Olsen PA, Randol M, Krauss S: Implications of cell cycle progression on functional sequence correction by short single-stranded DNA oligonucleotides. Gene Ther. 2005; 12(6): 546–51. 10.1038/sj.gt.3302454 [DOI] [PubMed] [Google Scholar]
- 107. Brachman EE, Kmiec EB: Gene repair in mammalian cells is stimulated by the elongation of S phase and transient stalling of replication forks. DNA Repair (Amst ). 2005; 4(4): 445–57. 10.1016/j.dnarep.2004.11.007 [DOI] [PubMed] [Google Scholar]
- 108. Ferrara L, Kmiec EB: Camptothecin enhances the frequency of oligonucleotide-directed gene repair in mammalian cells by inducing DNA damage and activating homologous recombination. Nucleic Acids Res. 2004; 32(17): 5239–48. 10.1093/nar/gkh822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Ferrara L, Parekh-Olmedo H, Kmiec EB: Enhanced oligonucleotide-directed gene targeting in mammalian cells following treatment with DNA damaging agents. Exp Cell Res. 2004; 300(1): 170–9. 10.1016/j.yexcr.2004.06.021 [DOI] [PubMed] [Google Scholar]
- 110. Kan Y, Ruis B, Takasugi T, et al. : Mechanisms of precise genome editing using oligonucleotide donors. Genome Res. 2017; 27(7): 1099–111. 10.1101/gr.214775.116 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 111. Aarts M, te Riele H: Parameters of oligonucleotide-mediated gene modification in mouse ES cells. J Cell Mol Med. 2010; 14(6B): 1657–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Sørensen CB, Krogsdam AM, Andersen MS, et al. : Site-specific strand bias in gene correction using single-stranded oligonucleotides. J Mol Med. 2005; 83(1): 39–49. 10.1007/s00109-004-0592-6 [DOI] [PubMed] [Google Scholar]
- 113. Howden SE, McColl B, Glaser A, et al. : A Cas9 Variant for Efficient Generation of Indel-Free Knockin or Gene-Corrected Human Pluripotent Stem Cells. Stem Cell Reports. 2016; 7(3): 508–17. 10.1016/j.stemcr.2016.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Moreno-Mateos MA, Fernandez JP, Rouet R, et al. : CRISPR-Cpf1 mediates efficient homology-directed repair and temperature-controlled genome editing. Nat Commun. 2017; 8(1): 2024. 10.1038/s41467-017-01836-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Richardson CD, Ray GJ, Dewitt MA, et al. : Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat Biotechnol. 2016; 34(3): 339–44. 10.1038/nbt.3481 [DOI] [PubMed] [Google Scholar]
- 116. Yamano T, Nishimasu H, Zetsche B, et al. : Crystal Structure of Cpf1 in Complex with Guide RNA and Target DNA. Cell. 2016; 165(4): 949–62. 10.1016/j.cell.2016.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Zetsche B, Gootenberg JS, Abudayyeh OO, et al. : Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015; 163(3): 759–71. 10.1016/j.cell.2015.09.038 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 118. Liu RM, Liang LL, Freed E, et al. : Synthetic chimeric nucleases function for efficient genome editing. Nat Commun. 2019; 10(1): 5524. 10.1038/s41467-019-13500-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Yoshimi K, Kunihiro Y, Kaneko T, et al. : ssODN-mediated knock-in with CRISPR-Cas for large genomic regions in zygotes. Nat Commun. 2016; 7: 10431. 10.1038/ncomms10431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Kosicki M, Tomberg K, Bradley A: Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat Biotechnol. 2018; 36(8): 765–71. 10.1038/nbt.4192 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 121. Adikusuma F, Piltz S, Corbett MA, et al. : Large deletions induced by Cas9 cleavage. Nature. 2018; 560(7717): E8–E9. 10.1038/s41586-018-0380-z [DOI] [PubMed] [Google Scholar]
- 122. Davis L, Maizels N: Homology-directed repair of DNA nicks via pathways distinct from canonical double-strand break repair. Proc Natl Acad Sci U S A. 2014; 111(10): E924–32. 10.1073/pnas.1400236111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Rees HA, Yeh WH, Liu DR: Development of hRad51-Cas9 nickase fusions that mediate HDR without double-stranded breaks. Nat Commun. 2019; 10(1): 2212. 10.1038/s41467-019-09983-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Chen X, Janssen JM, Liu J, et al. : In trans paired nicking triggers seamless genome editing without double-stranded DNA cutting. Nat Commun. 2017; 8(1): 657. 10.1038/s41467-017-00687-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Chen X, Tasca F, Wang Q, et al. : Expanding the editable genome and CRISPR-Cas9 versatility using DNA cutting-free gene targeting based on in trans paired nicking. Nucleic Acids Res. 2020; 48(2): 974–95. 10.1093/nar/gkz1121 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 126. Nakajima K, Zhou Y, Tomita A, et al. : Precise and efficient nucleotide substitution near genomic nick via noncanonical homology-directed repair. Genome Res. 2018; 28(2): 223–30. 10.1101/gr.226027.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Kocher T, Peking P, Klausegger A, et al. : Cut and Paste: Efficient Homology-Directed Repair of a Dominant Negative KRT14 Mutation via CRISPR/Cas9 Nickases. Mol Ther. 2017; 25(11): 2585–98. 10.1016/j.ymthe.2017.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Gopalappa R, Suresh B, Ramakrishna S, et al. : Paired D10A Cas9 nickases are sometimes more efficient than individual nucleases for gene disruption. Nucleic Acids Res. 2018; 46(12): e71. 10.1093/nar/gky222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Sfeir A, Symington LS: Microhomology-Mediated End Joining: A Back-up Survival Mechanism or Dedicated Pathway? Trends Biochem Sci. 2015; 40(11): 701–14. 10.1016/j.tibs.2015.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Nakade S, Tsubota T, Sakane Y, et al. : Microhomology-mediated end-joining-dependent integration of donor DNA in cells and animals using TALENs and CRISPR/Cas9. Nat Commun. 2014; 5: 5560. 10.1038/ncomms6560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Sakuma T, Nakade S, Sakane Y, et al. : MMEJ-assisted gene knock-in using TALENs and CRISPR-Cas9 with the PITCh systems. Nat Protoc. 2016; 11(1): 118–33. 10.1038/nprot.2015.140 [DOI] [PubMed] [Google Scholar]
- 132. Aida T, Nakade S, Sakuma T, et al. : Gene cassette knock-in in mammalian cells and zygotes by enhanced MMEJ. BMC Genomics. 2016; 17(1): 979. 10.1186/s12864-016-3331-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Heyer WD, Ehmsen KT, Liu J: Regulation of homologous recombination in eukaryotes. Annu Rev Genet. 2010; 44: 113–39. 10.1146/annurev-genet-051710-150955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. He X, Tan C, Wang F, et al. : Knock-in of large reporter genes in human cells via CRISPR/Cas9-induced homology-dependent and independent DNA repair. Nucleic Acids Res. 2016; 44(9): e85. 10.1093/nar/gkw064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Artegiani B, Hendriks D, Beumer J, et al. : Fast and efficient generation of knock-in human organoids using homology-independent CRISPR-Cas9 precision genome editing. Nat Cell Biol. 2020; 22(3): 321–31. 10.1038/s41556-020-0472-5 [DOI] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 136. Auer TO, Duroure K, de Cian A, et al. : Highly efficient CRISPR/Cas9-mediated knock-in in zebrafish by homology-independent DNA repair. Genome Res. 2014; 24(1): 142–53. 10.1101/gr.161638.113 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 137. Serebrenik YV, Sansbury SE, Kumar SS, et al. : Efficient and flexible tagging of endogenous genes by homology-independent intron targeting. Genome Res. 2019; 29(8): 1322–8. 10.1101/gr.246413.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Lackner DH, Carré A, Guzzardo PM, et al. : A generic strategy for CRISPR-Cas9-mediated gene tagging. Nat Commun. 2015; 6: 10237. 10.1038/ncomms10237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Schmid-Burgk JL, Höning K, Ebert TS, et al. : CRISPaint allows modular base-specific gene tagging using a ligase-4-dependent mechanism. Nat Commun. 2016; 7: 12338. 10.1038/ncomms12338 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 140. Krooss SA, Dai Z, Schmidt F, et al. : Ex Vivo/In vivo Gene Editing in Hepatocytes Using "All-in-One" CRISPR-Adeno-Associated Virus Vectors with a Self-Linearizing Repair Template. iScience. 2020; 23(1): 100764. 10.1016/j.isci.2019.100764 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 141. Ran FA, Cong L, Yan WX, et al. : In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015; 520(7546): 186–91. 10.1038/nature14299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Kim E, Koo T, Park SW, et al. : In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat Commun. 2017; 8: 14500. 10.1038/ncomms14500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Hu Z, Wang S, Zhang C, et al. : A compact Cas9 ortholog from Staphylococcus Auricularis (SauriCas9) expands the DNA targeting scope. PLoS Biol. 2020; 18(3): e3000686. 10.1371/journal.pbio.3000686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Karvelis T, Bigelyte G, Young JK, et al. : PAM recognition by miniature CRISPR-Cas12f nucleases triggers programmable double-stranded DNA target cleavage. Nucleic Acids Res. 2020; 48(9): 5016–23. 10.1093/nar/gkaa208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Agudelo D, Carter S, Velimirovic M, et al. : Versatile and robust genome editing with Streptococcus thermophilus CRISPR1-Cas9. Genome Res. 2020; 30(1): 107–17. 10.1101/gr.255414.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Pausch P, Al-Shayeb B, Bisom-Rapp E, et al. : CRISPR-CasΦ from huge phages is a hypercompact genome editor. Science. 2020; 369(6501): 333–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Rezza A, Jacquet C, Le Pillouer A, et al. : Unexpected genomic rearrangements at targeted loci associated with CRISPR/Cas9-mediated knock-in. Sci Rep. 2019; 9(1): 3486. 10.1038/s41598-019-40181-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Liu Y, Prasad R, Beard WA, et al. : Coordination of steps in single-nucleotide base excision repair mediated by apurinic/apyrimidinic endonuclease 1 and DNA polymerase beta. J Biol Chem. 2007; 282(18): 13532–41. 10.1074/jbc.M611295200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Villiger L, Grisch-Chan HM, Lindsay H, et al. : Treatment of a metabolic liver disease by in vivo genome base editing in adult mice. Nat Med. 2018; 24(10): 1519–25. 10.1038/s41591-018-0209-1 [DOI] [PubMed] [Google Scholar]
- 150. Yeh WH, Chiang H, Rees HA, et al. : In vivo base editing of post-mitotic sensory cells. Nat Commun. 2018; 9(1): 2184. 10.1038/s41467-018-04580-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Yeh WH, Shubina-Oleinik O, Levy JM, et al. : In vivo base editing restores sensory transduction and transiently improves auditory function in a mouse model of recessive deafness. Sci Transl Med. 2020; 12. 10.1126/scitranslmed.aay9101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Lim CKW, Gapinske M, Brooks AK, et al. : Treatment of a Mouse Model of ALS by In Vivo Base Editing. Mol Ther. 2020; 28(4): 1177–89. 10.1016/j.ymthe.2020.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Grünewald J, Zhou R, Lareau CA, et al. : A dual-deaminase CRISPR base editor enables concurrent adenine and cytosine editing. Nat Biotechnol. 2020; 38(7): 861–4. 10.1038/s41587-020-0535-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Kim HS, Jeong YK, Hur JK, et al. : Adenine base editors catalyze cytosine conversions in human cells. Nat Biotechnol. 2019; 37(10): 1145–8. 10.1038/s41587-019-0254-4 [DOI] [PubMed] [Google Scholar]
- 155. Kurt IC, Zhou R, Iyer S, et al. : CRISPR C-to-G base editors for inducing targeted DNA transversions in human cells. Nat Biotechnol. 2020. 10.1038/s41587-020-0609-x [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 156. Gapinske M, Luu A, Winter J, et al. : CRISPR-SKIP: programmable gene splicing with single base editors. Genome Biol. 2018; 19(1): 107. 10.1186/s13059-018-1482-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Song CQ, Jiang T, Richter M, et al. : Adenine base editing in an adult mouse model of tyrosinaemia. Nat Biomed Eng. 2020; 4(1): 125–30. 10.1038/s41551-019-0357-8 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 158. Huang TP, Zhao KT, Miller SM, et al. : Circularly permuted and PAM-modified Cas9 variants broaden the targeting scope of base editors. Nat Biotechnol. 2019; 37(6): 626–31. 10.1038/s41587-019-0134-y [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 159. Tan J, Zhang F, Karcher D, et al. : Engineering of high-precision base editors for site-specific single nucleotide replacement. Nat Commun. 2019; 10(1): 439. 10.1038/s41467-018-08034-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Tan J, Zhang F, Karcher D, et al. : Expanding the genome-targeting scope and the site selectivity of high-precision base editors. Nat Commun. 2020; 11(1): 629. 10.1038/s41467-020-14465-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Zuo E, Sun Y, Wei W, et al. : Cytosine base editor generates substantial off-target single-nucleotide variants in mouse embryos. Science. 2019; 364(6437): 289–92. 10.1126/science.aav9973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Jin S, Zong Y, Gao Q, et al. : Cytosine, but not adenine, base editors induce genome-wide off-target mutations in rice. Science. 2019; 364(6437): 292–5. 10.1126/science.aaw7166 [DOI] [PubMed] [Google Scholar]
- 163. Grünewald J, Zhou R, Garcia SP, et al. : Transcriptome-wide off-target RNA editing induced by CRISPR-guided DNA base editors. Nature. 2019; 569(7756): 433–7. 10.1038/s41586-019-1161-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Cheng TL, Li S, Yuan B, et al. : Expanding C-T base editing toolkit with diversified cytidine deaminases. Nat Commun. 2019; 10(1): 3612. 10.1038/s41467-019-11562-6 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 165. Thuronyi BW, Koblan LW, Levy JM, et al. : Continuous evolution of base editors with expanded target compatibility and improved activity. Nat Biotechnol. 2019; 37(9): 1070–9. 10.1038/s41587-019-0193-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Liu Z, Chen S, Shan H, et al. : Precise base editing with CC context-specificity using engineered human APOBEC3G-nCas9 fusions. BMC Biol. 2020; 18(1): 111. 10.1186/s12915-020-00849-6 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 167. Grünewald J, Zhou R, Iyer S, et al. : CRISPR DNA base editors with reduced RNA off-target and self-editing activities. Nat Biotechnol. 2019; 37(9): 1041–8. 10.1038/s41587-019-0236-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Hu JH, Miller SM, Geurts MH, et al. : Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature. 2018; 556(7699): 57–63. 10.1038/nature26155 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 169. Geurts MH, de Poel E, Amatngalim GD, et al. : CRISPR-Based Adenine Editors Correct Nonsense Mutations in a Cystic Fibrosis Organoid Biobank. Cell Stem Cell. 2020; 26(4): 503-510.e7. 10.1016/j.stem.2020.01.019 [DOI] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 170. Liu Z, Shan H, Chen S, et al. : Highly efficient base editing with expanded targeting scope using SpCas9-NG in rabbits. FASEB J. 2020; 34(9): 588–96. 10.1096/fj.201901587R [DOI] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 171. Walton RT, Christie KA, Whittaker MN, et al. : Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants. Science. 2020; 368(6488): 290–6. 10.1126/science.aba8853 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 172. Xin X, Li J, Zhao D, et al. : Double-Check Base Editing for Efficient A to G Conversions. ACS Synth Biol. 2019; 8(12): 2629–34. 10.1021/acssynbio.9b00284 [DOI] [PubMed] [Google Scholar]
- 173. Standage-Beier K, Tekel SJ, Brookhouser N, et al. : A transient reporter for editing enrichment (TREE) in human cells. Nucleic Acids Res. 2020; 47(19): e120. 10.1093/nar/gkz713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. St Martin A, Salamango D, Serebrenik A, et al. : A fluorescent reporter for quantification and enrichment of DNA editing by APOBEC-Cas9 or cleavage by Cas9 in living cells. Nucleic Acids Res. 2018; 46(14): e84. 10.1093/nar/gky332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Katti A, Foronda M, Zimmerman J, et al. : GO: A functional reporter system to identify and enrich base editing activity. Nucleic Acids Res. 2020; 48(6): 2841–52. 10.1093/nar/gkaa124 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 176. Anzalone AV, Koblan LW, Liu DR: Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors. Nat Biotechnol. 2020; 38(7): 824–44. 10.1038/s41587-020-0561-9 [DOI] [PubMed] [Google Scholar]
- 177. Yang L, Yang B, Chen J: One Prime for All Editing. Cell. 2019; 179(7): 1448–50. 10.1016/j.cell.2019.11.030 [DOI] [PubMed] [Google Scholar]
- 178. Kantor A, McClements ME, MacLaren RE: CRISPR-Cas9 DNA Base-Editing and Prime-Editing. Int J Mol Sci. 2020; 21(17): 6240. 10.3390/ijms21176240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Billon P, Nambiar TS, Hayward SB, et al. : Detection of Marker-Free Precision Genome Editing and Genetic Variation through the Capture of Genomic Signatures. Cell Rep. 2020; 30(10): 3280-3295.e6. 10.1016/j.celrep.2020.02.068 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 180. Matsoukas IG: Prime Editing: Genome Editing for Rare Genetic Diseases Without Double-Strand Breaks or Donor DNA. Front Genet. 2020; 11: 528. 10.3389/fgene.2020.00528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Ma Y, Zhang J, Yin W, et al. : Targeted AID-mediated mutagenesis (TAM) enables efficient genomic diversification in mammalian cells. Nat Methods. 2016; 13(12): 1029–35. 10.1038/nmeth.4027 [DOI] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 182. Hess GT, Frésard L, Han K, et al. : Directed evolution using dCas9-targeted somatic hypermutation in mammalian cells. Nat Methods. 2016; 13(12): 1036–42. 10.1038/nmeth.4038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Liu Z, Shan H, Chen S, et al. : Improved base editor for efficient editing in GC contexts in rabbits with an optimized AID-Cas9 fusion. FASEB J. 2019; 33(8): 9210–9. 10.1096/fj.201900476RR [DOI] [PubMed] [Google Scholar]
- 184. Aquino-Jarquin G: Novel Engineered Programmable Systems for ADAR-Mediated RNA Editing. Mol Ther Nucleic Acids. 2020; 19: 1065–72. 10.1016/j.omtn.2019.12.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Schaffer AA, Kopel E, Hendel A, et al. : The cell line A-to-I RNA editing catalogue. Nucleic Acids Res. 2020; 48(11): 5849–58. 10.1093/nar/gkaa305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Reardon S: Step aside CRISPR, RNA editing is taking off. Nature. 2020; 578(7793): 24–7. 10.1038/d41586-020-00272-5 [DOI] [PubMed] [Google Scholar]
- 187. Di Giorgio S, Martignano F, Torcia MG, et al. : Evidence for host-dependent RNA editing in the transcriptome of SARS-CoV-2. Sci Adv. 2020; 6(25): eabb5813. 10.1126/sciadv.abb5813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Du M, Jillette N, Zhu JJ, et al. : CRISPR artificial splicing factors. Nat Commun. 2020; 11(1): 2973. 10.1038/s41467-020-16806-4 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 189. Konermann S, Lotfy P, Brideau NJ, et al. : Transcriptome Engineering with RNA-Targeting Type VI-D CRISPR Effectors. Cell. 2018; 173(3): 665–676.e14. 10.1016/j.cell.2018.02.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Sinnamon JR, Kim SY, Fisk JR, et al. : In Vivo Repair of a Protein Underlying a Neurological Disorder by Programmable RNA Editing. Cell Rep. 2020; 32(2): 107878. 10.1016/j.celrep.2020.107878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Moghadam F, LeGraw R, Velazquez JJ, et al. : Synthetic immunomodulation with a CRISPR super-repressor In Vivo. Nat Cell Biol. 2020; 22(9): 1143–54. 10.1038/s41556-020-0563-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Lau CH, Tin C: The Synergy between CRISPR and Chemical Engineering. Curr Gene Ther. 2019; 19(3): 147–71. 10.2174/1566523219666190701100556 [DOI] [PubMed] [Google Scholar]
- 193. Lau CH, Ho JWT, Lo PK, et al. : Targeted Transgene Activation in the Brain Tissue by Systemic Delivery of Engineered AAV1 Expressing CRISPRa. Mol Ther Nucleic Acids. 2019; 16: 637–49. 10.1016/j.omtn.2019.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Tan Y, Chu AHY, Bao S, et al. : Rationally engineered Staphylococcus aureus Cas9 nucleases with high genome-wide specificity. Proc Natl Acad Sci U S A. 2019; 116(42): 20969–76. 10.1073/pnas.1906843116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Chen JS, Dagdas YS, Kleinstiver BP, et al. : Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature. 2017; 550(7676): 407–10. 10.1038/nature24268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Kleinstiver BP, Pattanayak V, Prew MS, et al. : High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016; 529(7587): 490–5. 10.1038/nature16526 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 197. Slaymaker IM, Gao L, Zetsche B, et al. : Rationally engineered Cas9 nucleases with improved specificity. Science. 2016; 351(6268): 84–8. 10.1126/science.aad5227 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 198. Xie H, Ge X, Yang F, et al. : High-fidelity SaCas9 identified by directional screening in human cells. PLoS Biol. 2020; 18(7): e3000747. 10.1371/journal.pbio.3000747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Strecker J, Jones S, Koopal B, et al. : Engineering of CRISPR-Cas12b for human genome editing. Nat Commun. 2019; 10(1): 212. 10.1038/s41467-018-08224-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Schmid-Burgk JL, Gao L, Li D, et al. : Highly Parallel Profiling of Cas9 Variant Specificity. Mol Cell. 2020; 78(4): 794–800.e8. 10.1016/j.molcel.2020.02.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Kim H, Lee WY, Oh Y, et al. : Enhancement of target specificity of CRISPR-Cas12a by using a chimeric DNA-RNA guide. Nucleic Acids Res. 2020; 48(15): 8601–16. 10.1093/nar/gkaa605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Fu Y, Sander JD, Reyon D, et al. : Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol. 2014; 32(3): 279–84. 10.1038/nbt.2808 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 203. Coelho MA, de Braekeleer E, Firth M, et al. : CRISPR GUARD protects off-target sites from Cas9 nuclease activity using short guide RNAs. Nat Commun. 2020; 11(1): 4132. 10.1038/s41467-020-17952-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Kocak DD, Josephs EA, Bhandarkar V, et al. : Increasing the specificity of CRISPR systems with engineered RNA secondary structures. Nat Biotechnol. 2019; 37(6): 657–66. 10.1038/s41587-019-0095-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Skryabin BV, Kummerfeld D-M, Gubar L, et al. : Pervasive head-to-tail insertions of DNA templates mask desired CRISPR-Cas9-mediated genome editing events. Sci Adv. 2020; 6(7): eaax2941. 10.1126/sciadv.aax2941 [DOI] [PMC free article] [PubMed] [Google Scholar]