Abstract
In recent years, marine red yeasts have been increasingly used as feed diets for larviculture of aquatic animals mainly due to their rich nutrition and immunopotentiation, however little attention is given to their other probiotic profits. In this study, a marine red yeast strain YLY01 was isolated and purified from farming water and it was identified as a member of Rhodosporidiums sphaerocarpum by the phylogeny based on 18S rDNA sequence. The strain YLY01 could effectively remove ammonia nitrogen from an initial 9.8 mg/L to 1.3 mg/L in 48 h when supplemented with slight yeast extract and glucose in water samples and the removal rate of ammonia nitrogen was up to 86%. Shrimps (Litopenaeus vannamei) in experimental group incubated with the yeast YLY01 exhibited a higher survival rate than those in blank control group and positive control group challenged by Vibrio harveyi, and it manifested that the strain has high biosecurity to at least shrimps. The strain YLY01 could inhibit the growth of Vibrio cells when a small quantity of carbon source was added into farming water. In addition, a nutrition composition assay showed the contents of protein, fatty acids, and total carotenoids of the yeast YLY01 were 30.3%, 3.2%, and 1.2 mg/g of dry cell weight, respectively. All these results indicated that the marine red yeast YLY01 has a great potential to be used as a versatile probiotic in aquaculture and to be developed as a microbial agent for high-ammonia tail water treatment.
Introduction
Intensive aquaculture has quickly expanded in recent 30 years as it is an important food resource for a growing global human population and an important way for gaining economic benefits in developing countries [1, 2]. However, the discharge of huge aquaculture wastes and the abuse of toxic chemicals and veterinary medicines have caused food security problems and environmental problems including eutrophication [3], the destruction of natural ecosystem [4], and the dispersal of aquatic pathogens and drug-resistant bacteria [5, 6]. The environmental problems caused by excessive aquaculture without tail water management greatly decreased the success rate of farming in return. Several alternative methods have been considered to improve the quality and sustainability of aquaculture production [7]. Among those methods, probiotics have been shown to have an important role in aquaculture [8].
Yeasts, as one group of probiotics, have been mainly used either as fresh baits for larva of aquatic animals or as feed supplement in aquaculture. Until now, yeast products using in aquaculture are primarily from brewer’s yeast and baker‘s yeast(Saccharomyces cerevisiae)[9] and few marine yeast products come into the market mainly because the research on the application of marine yeasts in aquaculture and the product development of marine yeasts are just beginning. Most of previous studies on marine yeasts concentrated on their abilities to enhance the immunocompetence of aquatic animals due to the existence of yeast polysaccharides (such as β-glucans) and to promote the growth of aquatic animals [10, 11], which aroused great interest on further exploring valuable marine yeasts in mariculture.
In present study, a marine red yeast strain first concentrating on the ability of ammonia removal was screened and isolated from farming water. Ammonia and nitrite removal, vibrios inhibition, and nutritional composition were further assayed to assess the potential application value in aquaculture and tail water treatment.
Materials and methods
Collection of the water samples
Farming water samples were collected from the bottoms of five different shrimp culture ponds (shrimps have been cultured for nearly three months) in Maoming, Guangdong Province, China, and the collected water samples were stored at 4°C before use.
Experimental shrimps
Healthy-looking shrimp juveniles, Litopenaeus vannamei, were collected from a farming pond of Maoming Jinyang Aquaculture Company in Maoming, Guangdong, China (150 individuals with the body length of 4 ± 1 cm and the body weight of 0.62±0.15 g). The shrimps were further randomly sampled for conventional PCR detection for WSSV [12], EHP [13], and EMS-related vibrios [14] to exclude the infection by these pandemic pathogens. The shrimps were temporarily reared in a 1000-L tank for three days in an experiment base of Maoming Jinyang Aquaculture Company. The collection of water and shrimp samples and experiments were permitted and authorized by Maoming Jinyang Aquaculture Company and South China Sea Institute of Oceanology, Chinese Academy of Sciences, Guangzhou, China.
Medium preparation and isolation for yeasts
The screening of yeasts was conducted using modified YPD (YPDM) medium containing 10 g of yeast extract, 20 g of peptone, 20 g of glucose, and 1000 mL of filtered seawater (pH was adjust to 7.0). The YPDM medium was sterilized, and supplemented with 1 ‰ (v/v) of tetracycline (12 mg/mL) and kanamycin (50 mg/mL) before use. Concentrated YPDM (10×, CYPDM) medium (Ph 7.0) was also prepared for enriching yeasts.
Isolation medium for yeasts with the ability of ammonia removal (IMAR) contains 1 g of (NH4)2SO4, 5 g of glucose, 5 g of sucrose, 0.5 g of yeast extract, and 0.5 g of NaH2PO4 in 1000mL of filtered seawater (pH was adjusted to 7.0). Concentrated IMAR (10×, CIMAR) was also prepared to enrich yeasts that have the activity of ammonia removal.
Water samples were mixed with CYPDM medium at a ratio of 9: 1 (v/v) in 250mL conical flasks and then incubated in a shaker (200 rpm) at 30°C. After 48-h incubation, each of 100-μL cell fluids was spread on fresh YPDM agar plates and incubated overnight at 30°C. Colonies were picked out based on colony morphology and further purified on YPDM agar plates for at least three times. Five yeast strains were obtained in this way, and then they were inoculated in culture tubes with 4 mL of IMAR for 12 h and 100 μL of each strain was cultured on fresh IMAR agar plates through streaking method and incubated overnight at 30°C. Single colony was picked into 4 mL of IMAR and incubated in a 200-rpm shaker for 48 h. The initial and the final concentration of ammonia nitrogen in the culture mediums were determined by a μMAC SMART hydraulics rev.3 instrument. The analysis included three replicates. The strain with the most removal of ammonia nitrogen was picked out and used for subsequent experiments.
Molecular identification of the selected strain
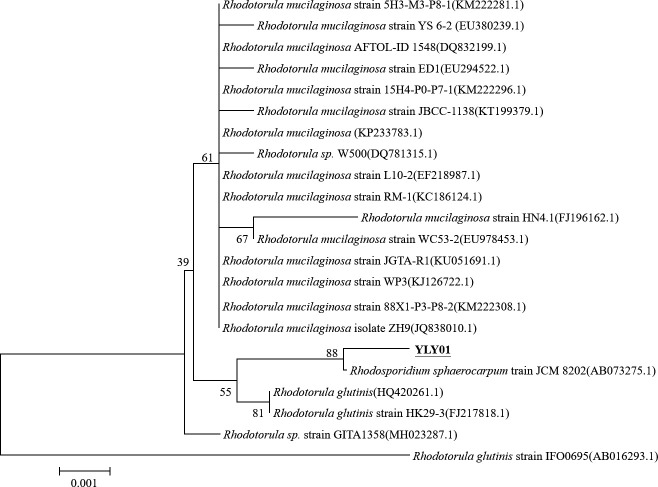
In order to identify the candidate strain, a 18S rDNA-based method was used. The genomic DNA of the strain was extracted using a Yeast Genomic DNA Extraction Kit (TianGen, China) for PCR amplification of 18S rDNA with primers EF3 and EF4 [15]. PCRs were performed as follows: 94°C for 3 min; 30 cycles at 94°C for 30 s, 56°C for 90 s, and 72°C for 1 min, and a final step of 72°C for 5 min. The purified PCR product was directly sequenced by Sangon Biotech Company (Shanghai, China). The obtained sequence was queried against the GenBank database using BLASTN (https://blast.ncbi.nlm.nih.gov/Blast.cgi). A phylogenetic tree was constructed by the neighbor-joining (NJ) method [16] based on the sequence of the strain and related sequences using MEGA 6.0 [17] with 1000 bootstrap replicates.
Observation of cell morphology by scanning electron microscopy
The yeast cells in logarithmic growth phase were collected and centrifuged at 3000 rpm for 5 min, and then the precipitation was suspended with 0.1M PBS (0.14 M NaCl, 3 mM KCl, 8 mM Na2HPO4, 1.5 mM KH2PO4, pH 7.4) and repeated for three times. Then the suspension was transferred to a new centrifugal tube and glutaraldehyde was added up to 0.5%. After inoculated at 4°C for 30 min, glutaraldehyde was added into the sample up to 2.5% at 4°C overnight. The cells were washed twice with ddH2O by centrifuging at 3000 rpm for 5 min, followed by the gradient dehydration with 30%, 50%, 70%, 80%, 90%, 95%, 100% ethanol for 20 min at each stage. The dehydrated sample was processed by critical point drying and then was deposited with gold by ion sputtering. The cell morphology was observed and photographed by scanning electron microscopy.
The effect of the selected strain on ammonia nitrogen removal in simulated farming water
Ammonia nitrogen removal of the selected strain was performed using a method [18] with a slight modification. A total of 1000 mL of the filtered and sterilized seawater (pH 8.0) containing ammonia nitrogen (10 mg/L), carbon sources (1 g/L), and yeast extract (0.5 g/L) was prepared as simulated farming water (SFW). The yeast cells were cultured overnight and adjusted to a concentration with OD600nm = 1 (approximately 2 × 107 cfu/mL). Then 3 mL of the cells was centrifuged, washed twice, and resuspended with sterilized seawater. In the experimental group, 500 μL of the suspended cells was added to each of 150-mL SFW, whilst in the control group, 500 μL of sterilized seawater was added to each of 150-mL SFW. The samples were incubated at 30°C in a 150-rpm shaker. The concentration of ammonia nitrogen was measured by the μMAC SMART hydraulics rev.3 instrument at 0, 3, 6, 9, 12, 24, 36, and 48 h. Each of experimental and control group had three replicates.
The impact of the selected strain on the growth of Vibrio Spp.
An experiment was conducted to determine whether the strain can inhibit the growth of Vibrio species in farming water [18]. A farming water sample was taken from a shrimp pond suffered by an outbreak of vibriosis, and the number of Vibrio cells was calculated by conventional TCBS plates [19]. A total of 2 L of the water sample was added with 2 g of glucose and 1 g of yeast extract. Then, 150 mL of the water sample containing glucose was added into each 250-mL conical flask. Each conical flask in the experimental group was added with 100 μL of washed and resuspended yeast cells (OD600nm = 1.0), whilst each conical flask in the control group was added with 100 μL of sterilized seawater. The experimental group and the control group each contained three replicates. After 48 h of incubation at 30°C in a 150-rpm shaker, the water samples were serially diluted, spread on fresh TCBS plates, and incubated at 30°C overnight to calculate the number of Vibrio cells [19].
To explore whether the strain can inhibit the growth of Vibrio species through secreting antibacterial substances, we also conducted a plate suppression experiment [18]. V. alginolyticus E06333, V. furnissi ATCC 33813, V. harveyi E385, and V. parahaemolyticus ATCC 17802, were used. 100 μL of Vibrio cells were spread onto LB plates and then each plate was covered with three 5-mm sterilized filter sheets. The filter sheets were dropped with 5 μL of tested cells [20]. The sizes of the inhibition zones around the filter sheets were observed and measured.
Bio-safety assessment of the selected strain to shrimp Litopenaeus vannamei
Shrimp juveniles, Litopenaeus vannamei, were used to assess the biosafety of the selected strain according to a previously reported method [18]. Healthy shrimp juveniles mentioned above were randomly grouped into tanks with 40-L filtered seawater disinfected by chlorine dioxide (South Ranch, China). Experimental group was supplemented with 50 mL of washed and resuspended yeast cells (the final concentration, approximately 1×105 cfu/mL). Blank control group was supplemented with sterilized seawater, and positive control group was supplemented with 50 mL of V. harveyi cells (the final concentration, approximately 1×105 cfu/mL). Each group contained three replicates (three tanks). Each tank contained 10 shrimps. During the period of the experiment, the shrimp were fed normally, residual feeds and feces were siphoned quickly, and sterilized seawater (approximately 0.5 L) was complemented each day. The number of survival shrimps was recorded for seven days.
Nutritional composition analysis of the selected yeast strain
Cell dry weight was calculated by measuring the mass change of 100 mL cell sediment harvested by centrifugation from initial mass to constant weight with drying at 55°C in an oven. For determining the content of carotenoids, 0.1 g of dry cells was weighed and resuspended with 2.4 mL of 3 M HCl, and then the suspension was heated in a boiling water bath for 4 min, followed by rapid cooling and centrifugation for 5 min at 4000 rpm. The cell pellets were washed twice, and then 4 mL of acetone solution was added, followed by vortex for 2 min and centrifugation for 5 min at 4000 rpm. The supernatant was harvested to measure the content of carotenoids by a BIO-RAD SmartSpecTM Plus spectrophotometer instrument at wave length of 475 nm [21, 22]. Crude protein content was determined by the Kjeldahl method [23], and fatty acids content was determined by the Bligh-Dyer method [24]. Yeast polysaccharide was extracted by pronase digestion and alcohol precipitation and was quantified by a reported method [25]. Amino acid composition of the yeast was analyzed by a Chinese national standard method (GB 5009.124–2016) using an automatic amino acid analyzer.
Results
Isolation and identification of the candidate yeast strains
Totally, five strains that could grow in IMAR containing ammonia nitrogen were selected and purified on YPDM agar plates. After 48 h incubation, all of them showed the ability to remove ammonia nitrogen but there were obvious discrepancies in the removal rates of ammonia nitrogen by them (Fig 1). The strain YLY01 showed the strongest ability to remove ammonia nitrogen, and totally 81.9% of ammonia nitrogen in IMAR medium was removed by it in 48 h (from 213.5 mg/L to 38.7 mg/L) (Fig 1), which is 4.8 times removal rate than the strain YLY05 (17.0%). As a result, the strain YLY01 was screened out for the subsequent experiments.
Fig 1. Ammonia nitrogen removal in IMAR medium by five marine yeast strains after 48-h culture.
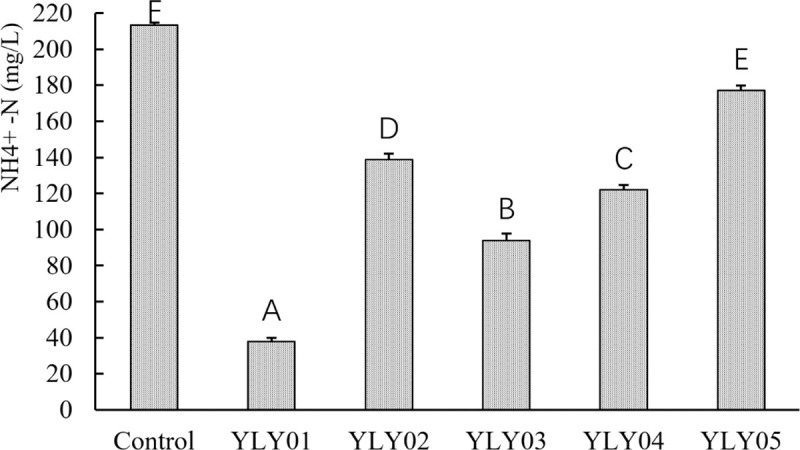
Control: blank IMAR medium. YLY01-YLY05: five candidate yeast strains tested. The values at the top of each column represent the concentration of ammonia nitrogen in IMAR medium after 48-h culture. Data are given as mean ± SD (n = 3). Different letters above bars indicate significant difference among treatments (p < 0.05).
A BLASTN search indicated that 18S rDNA sequence of YLY01 strain had a 99% identity with a Rhodosporidium sphaerocarpum strain JCM 8202 (GenBank: AB073275). The results of the phylogenetic tree based on 18S rDNA sequences of the strain YLY01 and other related yeast strains clearly indicated that the strain YLY01 and other strains from R. sphaerocarpum were clustered into one branch (Fig 2). Therefore, phylogeny based on 18S rDNA sequences confirmed that the strain YLY01 is one member of R. sphaerocarpum. The 18S rDNA sequence of the strain R. sphaerocarpum YLY01was deposited in GenBank under the accession number, MK583688.
Fig 2. A phylogenetic tree based on 18S rDNA sequences of the strain YLY01 and related strains.
Bootstrap values were obtained after 1000 repetitions. Scale bar indicates 0.1% sequence dissimilarity.
Cell morphology of the yeast strain YLY01 under SEM
Similar to most yeast cells [26], YLY01 cells are round or elliptical with an average diameter of 2 μm, and the surface of the cells is smooth and flagellum-free (Fig 3). The germinating and proliferating cells and the bud marks on the surface of the cells could be clearly observed under SEM.
Fig 3. The morphology of the yeast YLY01cells under SEM.
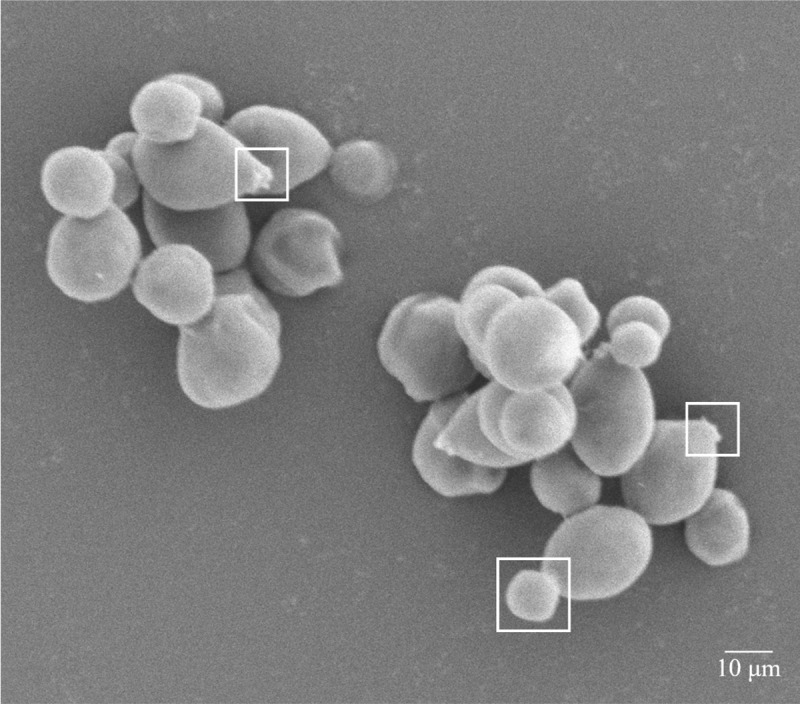
The bud marks (shown in write boxes) on the surface of some cells could be clearly observed.
Ammonia nitrogen removal in SFW by the yeast strain YLY01
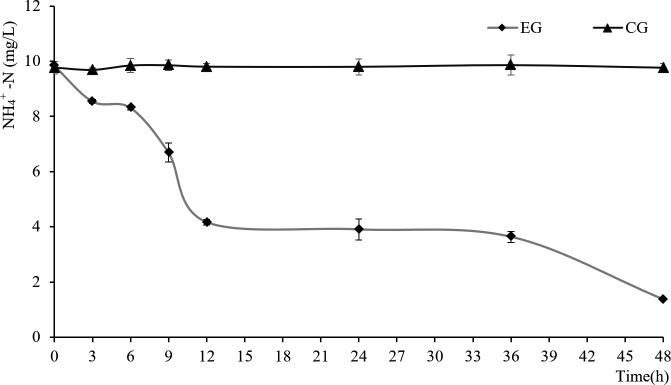
After 48-h treatment with the yeast YLY01 in the experimental group, the ammonia nitrogen concentration in SFW declined from initial 9.8mg/L to 1.3 mg/L, with a removal rate of 86.7% (Fig 4). However, in the control group, the ammonia nitrogen concentration in SFW nearly remained unchanged (Fig 4). It demonstrated that the yeast YLY01 has a strong ability to remove ammonia nitrogen in water when little carbon source was added. During this process, no nitrite was detected in the experimental group or the control group.
Fig 4. The changes of ammonia nitrogen content of SFW in 48-h incubation.
EG: The experimental group treated with the yeast YLY01; CG: blank control group treated sterilized seawater. Data are given as mean ± SD (n = 3).
Inhibition of the yeast YLY01 to the growth of Vibrio spp.
Two methods were used to assay the inhibitory activity of the strain YLY01 against Vibrio spp.. The number of Vibrio cells in farming water of the control group was approximately 4.6×104 cfu/mL at the start (T0)and slightly decreased to 3.8×104 cfu/mL at 48 h, while the number of Vibrio cells in farming water of the experiment group (EG) decreased from 4.6×104 cfu/mL at the start to 6.8×103 cfu/mL at 48 hr, which was only 18% of the Vibrio cells in the control group (CG) at the same time point (Fig 5). This result demonstrated that the existence of the yeast YLY01 cells inhibited the survival of Vibrio cells in farming water.
Fig 5. The number of Vibrio cells in farming water after 48-h incubation.
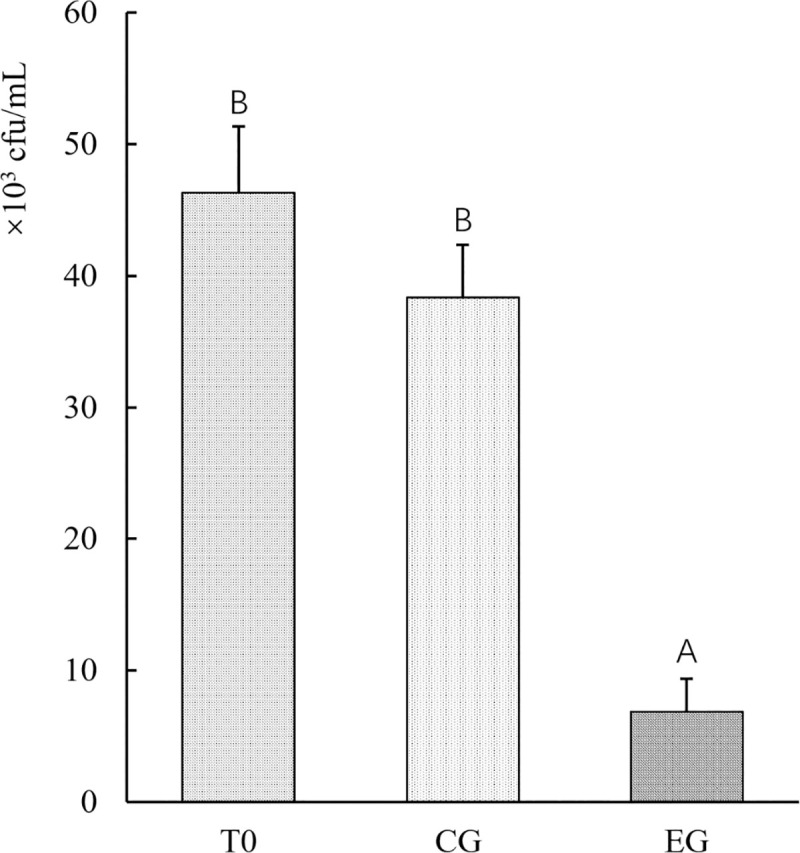
T0: the start of experiment at 0 h; CG: the control group without the supplement of the yeast YLY01; EG: the experimental group supplemented by the yeast YLY01. Data are given as mean ± SD (n = 3). Different letters above bars indicate significant difference among treatments (p < 0.05).
To further explore what caused the inhibition to the growth of Vibrio spp., the plate suppression experiment of the strain YLY01 was also conducted. No inhibition zones were observed when the YLY01 cells were applied whilst inhibition zones in the positive control group (added with chloramphenicol) were clear and obvious (Fig 6). This result clearly indicated that the YLY01 cells could not generate antimicrobial substances against Vibrio cells. Therefore, the obvious decrease in numbers of Vibrio cells during incubating with the YLY01 cells in farming water was probably caused by other mechanisms.
Fig 6. Inhibition assay of the yeast YLY01against four Vibrio species.
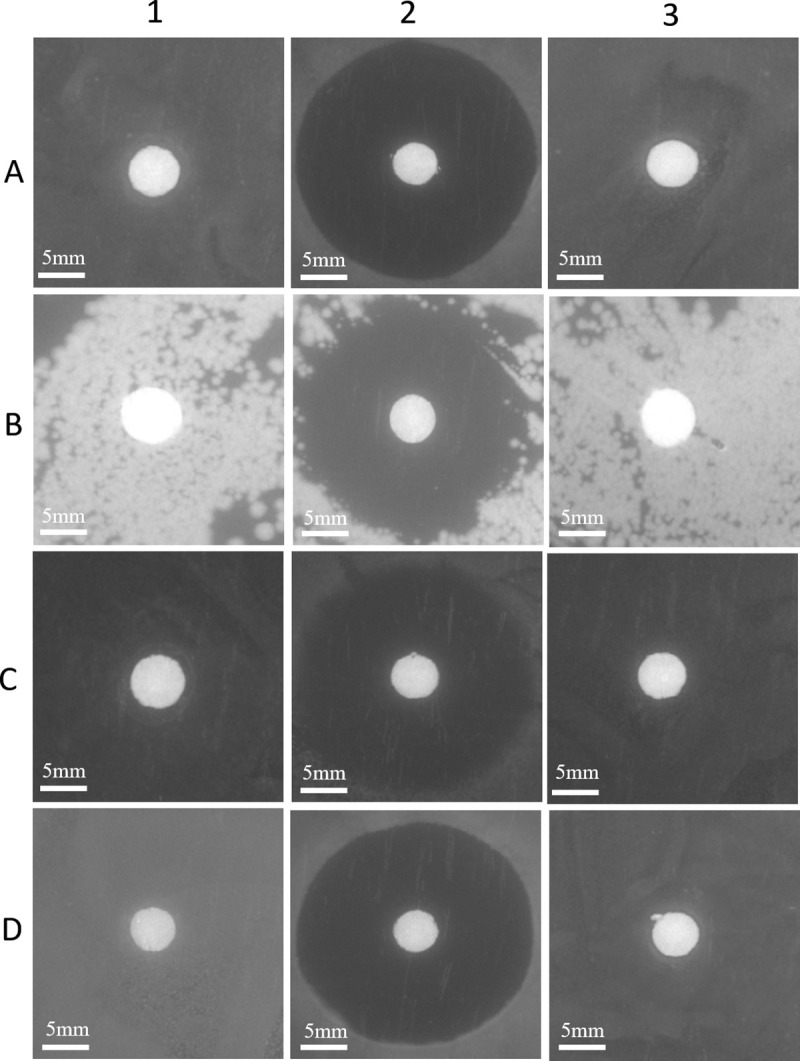
A: V. alginolyticus E06333; B: V. furnissi ATCC 33813; C: V. harveyi E385; D: V. parahaemolyticus ATCC 17802; 1: experimental group (added with the YLY01 cell culture); 2: positive control (added with chloramphenicol); 3: negative control (added with the medium).
Assessment of biological safety on shrimp Litopenaeus vannamei
Under normal culture conditions, the survival rates of shrimps in different treatments were shown in the Fig 7. The result indicated that the survival rate of shrimps in the blank control group (C0, treated with sterilized seawater) slightly decreased to 86.7% after 7-day culture. The survival of rate of shrimps in the experimental group (EG, challenged by the strain YLY01) reach up to 96.7%, which was even higher than the survival rate of shrimp in the blank control group. The survival rate of shrimps in the positive control group (C1, challenged by Vibrio harveyi) gradually declined, and after seven days the survival rate of shrimps in the group was only 40.0%. Considering a big dosage of the strain YLY01 in the experimental group (approximately 1 × 105 cfu/mL), the result indicated that the strain YLY01 not only has high biosafety for L. vannamei but also has a potential to reduce the death rate of the cultured shrimps. Notably, much clearer water was observed in the experimental group than in blank control group and in the positive control group after the seven-day experiment, and obvious flocculent precipitate generated on the bottom of tanks in the experimental group.
Fig 7. Survival rates of shrimp Litopenaeus vannamei after the immersion challenge in seven days.
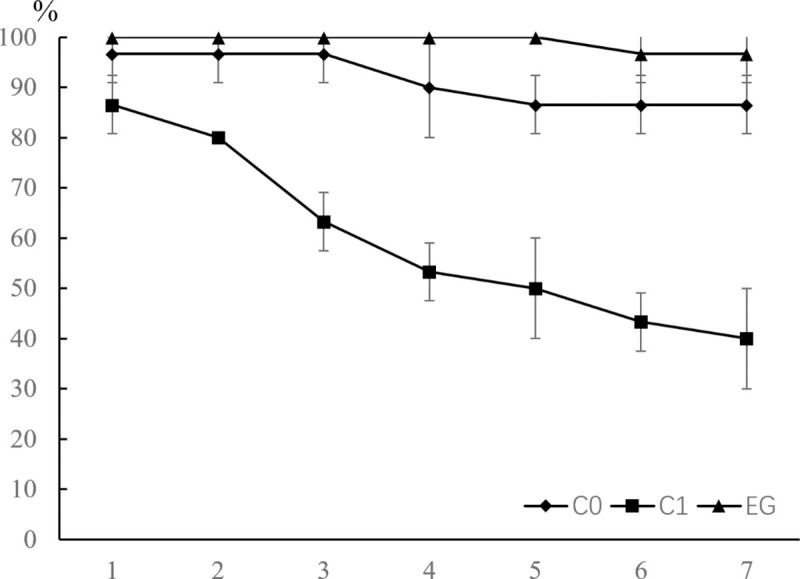
EG: the experimental group challenged by the yeast YLY01; C0: the blank control group; C1: the positive control group challenged by Vibrio harveyi. Data are given as mean ± SD (n = 3).
Nutritional components of the yeast YLY01
The nutritional composition of the yeast YLY01 is shown in Table 1. The biomass of the yeast YLY01 could reach up to in 26.5 g/L after 72-h culture, and the free water content of the cells was about 78%. The protein content of the yeast accounted for 30.3% of dry cell weight, and 16 hydrolyzed amino acids were detected and compared with counterparts in a marine red yeast, R. paludigenum [27], and brewer’s yeast, Saccharomyces cerevisiae [28] (Table 2). There was no obvious difference in terms of the contents of nine essential amino acids for crustaceans [29] among the yeast YLY01 (50.2%), a marine red yeast R. paludigenum (52.2%), and S. cerevisiae (49.2%). The yeast YLY01, R. paludigenum, and S. cerevisiae also had similar total contents of six flavor amino acids (accounting for 43.1%, 45.6%, and 43.5%, respectively). The profile of amino acid composition of the yeast YLY01 was also compared with these of R. paludigenum and S. cerevisiae and they were exhibited in a radar plot (S1 Fig). Similar shapes in the plot manifested that three kinds of yeasts had roughly similar variation trends in terms of relative amino acid contents except the contents of histidine (His) and lysine (Lys), wherein the yeast YLY01 had almost twice the lysine (Lys) content of R. paludigenum and S. cerevisiae.
Table 1. The nutritional components of the yeast R. sphaercarpum YLY01.
| Nutrition ingredients | Protein | Crude Polysaccharides | Saturated Fatty Acids | Unsaturated fatty acids | Carotenoid | B vitamins |
|---|---|---|---|---|---|---|
| Proportion in dry weight (%) | 30.3 | 16.8 | 0.48 | 2.76 | 0.125 | 0.015 |
Table 2. The composition of hydrolyzed amino acids of the yeast R. sphaercarpum YLY01 and other two yeasts.
| Groups (%) | Amino acids | Proportion (%) | ||
|---|---|---|---|---|
| YLY01 | 1 | 2 | ||
| EAA | Phe* | 4.32 | 6.10 | 3.85 |
| Met | 1.99 | 1.87 | 1.67 | |
| Arg | 7.89 | 8.45 | 6.92 | |
| Lys | 5.66 | 7.20 | 8.85 | |
| Leu | 8.24 | 8.95 | 7.69 | |
| Val | 5.96 | 6.64 | 6.28 | |
| Ile | 4.27 | 4.48 | 5.51 | |
| His | 6.60 | 3.53 | 3.21 | |
| Thr | 5.26 | 5.41 | 5.90 | |
| NEAA | Ala* | 7.79 | 8.16 | 7.05 |
| Pro | 5.16 | 2.83 | 5.13 | |
| Gly* | 5.56 | 6.14 | 5.13 | |
| Glu* | 12.41 | 11.62 | 13.97 | |
| Tyr* | 2.38 | 3.88 | 4.23 | |
| Ser | 5.86 | 5.03 | 5.38 | |
| Asp* | 10.67 | 9.70 | 9.23 | |
In addition, the fatty acids content was 3.24% of dry cell weight, and the unsaturated fatty acids could reach up to 85.2% of total fatty acids. The crude polysaccharides accounted for 16.8% of cell dry weight. Moreover, the carotenoid content was 1.25 mg/g. The yeast YLY01 was also detected to contain a small amount of B vitamins (0.15 mg/g).
Discussion
Marine red yeasts are saprophytic organisms with a strong ability to resist adversity and they act as decomposer through converting plant and animal organics to yeast biomass in the natural environment [30], which enables them to have great potential to be explored as probiotics. So far as now, marine red yeasts have never been reported to clarify farming water and control Vibrio spp. in farming water though the ability of some marine red yeasts to utilize inorganic nitrogen has long been noticed or utilized by several researchers. Saenge et al. [31] revealed that the accumulation of lipids and carotenoids in Rhodotorula glutinis increased when ammonium sulfate was used as nitrogen source to change the C/N ratio in medium. Inorganic nitrogen is also used as a nitrogen source in some isolation media of Rhodotorula spp. [32, 33]. These findings implied that some marine red yeasts have the potential to remove ammonia nitrogen in farming water through ammonium assimilation, which is one important way to utilize inorganic nitrogen source by common yeast species such as Saccharomyces cerevisiae [34, 35], Candida utilis [34], and Kluyveromyces marxianus [36]. In this study, R. sphaerocarpum YLY01 was found to remove ammonia nitrogen with a high efficiency when carbon source was added and other nitrogen sources were limited, and meanwhile no nitrite was detected during this process. It indicated R. sphaerocarpum YLY01 can utilize ammonia nitrogen for growth through ammonium assimilation as other common yeast species do. Therefore, it is the first report that a marine red yeast can be used to remove ammonia nitrogen in farming water. In addition, we observed much clearer water and obvious flocculent precipitate in the experiment group of the biosafety assay, and it also indicated that R. sphaerocarpum YLY01 has an excellent flocculation ability. Yeast flocculation has been explored for long time [37] and the advantage of yeast flocculation has been taken to develop microbial flocculant for wastewater treatment [38]. It is worth to further explore the detailed flocculant profiles of R. sphaerocarpum YLY01 in future focusing on sewage (including aquaculture tail water) treatment.
In this study, R. sphaerospora YLY01 exhibited the strong inhibition ability to the survival of Vibrio cells in farming water when little carbon source was supplied, and the inhibition to Vibrio cells was not likely caused by generating antibiotic substances as no inhibition zones occurred. Therefore, R. sphaerospora YLY01 should have other antibacterial mechanisms. Anyway, this is the first report to confirm that a marine red yeast has inhibition ability to Vibrio cells in farming water, which will largely expand the usage of marine red yeasts.
The yeast R. sphaerospora YLY01 contains 16 hydrolyzed amino acids, which is consist with the kinds of amino acids found in R. paludigenum [27] and brewer’s yeast S. cerevisiae [28]. The yeast YLY01 also had high total content of six flavor amino acids (up to 43.1%) compared with that of R. paludigenum [27] and brewer’s yeast [28]. Researchers suggest that high content of flavor amino acids can greatly improve the palatability and food attractiveness of feed or live bacteria preparations [39]. The yeast YLY01 in this assay had a moderate lipid content (3.24%), and the ratio of unsaturated fatty acids (85.2%) was higher than that (approximately 74.2%) of the representative species, R. toruloides, in genus Rhodosporidium [40]. An obvious feature of members in genus Rhodosporidium is that they have prominent ability to accumulate lipids, which makes them to have a great potential in microbial lipid industry [41, 42], and thus we speculated that R. sphaerospora YLY01 can accumulate rich lipids under certain growing conditions. Unsaturated fatty acids play an important role in providing energy, forming high bioactive substances, regulating lipid metabolism, and immune function in aquatic animals [43]. Coyle et al. [44] found that unsaturated fatty acids supplied in the diet reduce the metabolic energy expenditure and improve growth rate or diet efficiency of largemouth bass. It is worthy to exploit the fatty acid composition and potential application value of lipids of the yeast YLY01 in future. The production capacity of carotenoids in the yeast YLY01 is 1.25 mg/g dry weight under the conventional cultivation condition. A high yield of carotenoids (35 mg/g dry weight) in R. mucilaginosa was recorded [45], it suggests that the yield of carotenoids in R. sphaerocarpum YLY01 can be largely improved under suitable conditions if needed. Besides, R. sphaerocarpum YLY01 contained 16.79% crude polysaccharides, which are considered to stimulate the immune system and antioxidant systems of cultured animals and thus to have positive effects on reducing the risk of disease outbreaks and improving the resistance to adverse circumstances [11, 46]. Rhodosporidium is one of common marine red yeasts frequently isolated from various marine environments [30], however the yeasts in Rhodosporidium were exploited as probiotics for animals just in recent years. The yeast, R. paludigenum, enhanced the growth performance and antioxidant competence of L. vannamei when it was added into the diet in forms of dried yeast or live bait [47]. The dietary addition of Rhodotorula (Rhodosporidium) benthica D30 could increase growth performance and some digestive enzyme activities, improve immunity and disease resistance of sea cucumber A. japonicus [33]. These rigorous and informative evidences from congeneric yeast species and the assays on nutrition and biosecurity of R. sphaerocarpum YLY01 provide our impetus to further explore probiotic functions of R. sphaerocarpum YLY01 and exploit it as a promising aquatic probiotics.
In aquaculture, Bacillus spp., lactic acid bacteria, and nitrifying bacteria are most common probiotics. Bacillus spp. generally improve animal growth/ feed utilization due to high activities of extracellular enzymes screened by them [48]. Lactic acid bacteria are mainly used to adjust regulate intestinal flora balance and animal immunity [49]. Nitrifying bacteria (mainly refer to ammonia-oxidizing bacteria [AOB] and nitrite-oxidizing bacteria [NOB]) were adopted to transform toxic ammonia nitrogen or nitrite into nitrate, and the metabolites and the growth speed of AOB and NOB are much lower than heterotrophic bacteria [50, 51], which leads to low efficiency of ammonia or nitrite removal by them in farming ponds. Compared to these conventional probiotics that only have one prominent function, the red yeast YLY01 exhibited versatile functions including high-efficiency ammonia removal without its transformation to nitrite, clarifying water, inhibition on Vibrio spp., and rich nutrition, which confers the red yeast YLY01 a good prospect to be developed as aquatic probiotics. Besides, it also shed some light on exploring new applications of marine red yeasts.
Conclusion
In summary, the yeast R. sphaerocarpum YLY01 exhibits multiple prominent advantages including efficiently removing ammonia nitrogen without transforming it to nitrite, clarifying farming water, the inhibition to Vibrio spp., and rich nutrition, which makes it has a great potential to be developed as a versatile aquatic probiotic and as an effective microbial agent for high-ammonia sewage treatment.
Supporting information
(DOCX)
Acknowledgments
We thank the manager Huo Li from Maoming Jinyang Aquaculture Company for provide an experimental base and samples.
Data Availability
The 18S rDNA sequence of the strain was deposited in GenBank under accession number MK583688.
Funding Statement
This work was supported by Natural Science Foundation of Guangdong Province, China (2019A1515011492), National key Research and development program: Blue Granary Scientific and Technological Innovation (2020YFD0901104), and Key Special Project for Introduced Talents Team of Southern Marine Science and Engineering Guangdong Laboratory (GML2019ZD0402).
References
- 1.Martinez-Porchas M, Martinez-Cordova LR. World aquaculture: environmental impacts and troubleshooting alternatives. Sci. World J. 2012; 2012: 389623 10.1100/2012/389623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hai NV. The use of probiotics in aquaculture. J. Appl. Microbiol. 2015; 119(4): 917–935. 10.1111/jam.12886 [DOI] [PubMed] [Google Scholar]
- 3.Talbot C, Hole R. Fish diets and the control of eutrophication resulting from aquaculture. J. Appl. Ichthyol. 1994; 10(4):258–270. [Google Scholar]
- 4.Amirkolaie AK. Reduction in the environmental impact of waste discharged by fish farms through feed and feeding. Rev. Aqua. 2011; 3(1):19–26. [Google Scholar]
- 5.Baquero F, José-Luis Martínez, Rafael Cantón. Antibiotics and antibiotic resistance in water environments. Curr. Opin. Biotechnol. 2008; 19(3):260–265. 10.1016/j.copbio.2008.05.006 [DOI] [PubMed] [Google Scholar]
- 6.De Schryver P, Vadstein O. Ecological theory as a foundation to control pathogenic invasion in aquaculture. ISME J. 2014; 8(12): 2360–2368. 10.1038/ismej.2014.84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li J, Tan B, Mai K, Ai Q, Zhang W, Xu W, et al. Comparative study between probiotic bacterium Arthrobacter XE-7 and chloramphenicol on protection of Penaeus chinensis post-larvae from pathogenic vibrios. Aquaculture 2006; 253(1–4):140–147. [Google Scholar]
- 8.Skjermo J, Vadstein O. Techniques for microbial control in the intensive rearing of marine larvae. Aquaculture 1999; 177(1–4):333–343. [Google Scholar]
- 9.Sarlin PJ, Philip R. Marine yeasts as feed supplement for Indian white prawn Fenneropenaeus indicus: screening and testing the efficacy. Int. J. Curr. Microbiol. Appl. Sci. 2016; 5(1): 55–70. [Google Scholar]
- 10.Chi ZM, Liu ZQ, Gao LM, Gong F, Ma CL, Wang XH, et al. Marine yeasts and their applications in mariculture. J. Ocean U. China 2006; 5(3):251–256. [Google Scholar]
- 11.Navarrete P, Tovar-Ramírez D. Use of yeasts as probiotics in fish aquaculture In: Hernandez-Vergara MP, Perez-Rostro CI, editors. Sustainable aquaculture techniques. London: IntechOpen; 2014. pp.136–172. [Google Scholar]
- 12.Zhan W, Xing J, Wang Y, Suziki S, Fukuda H. Detection of white spot syndrome virus (WSSV) in cultured shrimp by PCR. J. Fish. Sci. China (In Chinese) 2000; 7(1):51–54. [Google Scholar]
- 13.Tang KFJ, Han JE, Aranguren LF, White-Noble B, Schmidt MM, Piamsomboon P, et al. Dense populations of the microsporidian Enterocytozoon hepatopenaei (EHP) in feces of Penaeus vannamei exhibiting white feces syndrome and pathways of their transmission to healthy shrimp. J. Invertebr. Pathol. 2016; 140:1–7. 10.1016/j.jip.2016.08.004 [DOI] [PubMed] [Google Scholar]
- 14.Lee CT, Chen IT, Yang YT, Ko TP, Huang YT, Huang JY, et al. The opportunistic marine pathogen Vibrio parahaemolyticus becomes virulent by acquiring a plasmid that expresses a deadly toxin. PNAS 2015; 112(34):10798–10803. 10.1073/pnas.1503129112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smit E, Leeflang P, Gl B, Elsas JDV, Wernars K, Smit E, et al. Analysis of fungal diversity in the wheat rhizosphere by sequencing of cloned PCR-amplified genes encoding 18S rRNA and temperature gradient gel electrophoresis. Appl. Environ. Microbiol. 1999; 65(6): 2614–2621. 10.1128/AEM.65.6.2614-2621.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987; 4(4):406–425. 10.1093/oxfordjournals.molbev.a040454 [DOI] [PubMed] [Google Scholar]
- 17.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013; 30(12): 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yun L, Yu ZH, Li YY, Luo P, Jiang X, Tian YS, et al. Ammonia nitrogen and nitrite removal by a heterotrophic Sphingomonas sp. strain LPN080 and its potential application in aquaculture. Aquaculture 2019; 500:477–484. [Google Scholar]
- 19.Oliver JD, Warner RA, Cleland DR. Distribution of Vibrio vulnificus and other lactose-fermenting vibrios in the marine environment. Appl. Environ. Microbiol. 1983; 45(3):985–998. 10.1128/AEM.45.3.985-998.1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barry AL, Coyle MB, Thornsberry C, Gerlach EH, Hawkinson RW. Methods of measuring zones of inhibition with the Bauer-Kirby disk susceptibility test. J. Clin. Microbiol. 1979; 10(6): 885–889. 10.1128/JCM.10.6.885-889.1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hasegawa J, Ogura M, Tsuda S, Maemoto SI, Kutsuki H, Ohashi T. High-yield production of optically active 1, 2-diols from the corresponding racemates by microbial stereoinversion. Agric. Biol. Chem. 1990; 54(7):1819–1827. [Google Scholar]
- 22.Nasrabadi MRN, Razavi SH. Optimization of β-carotene production by a mutant of the lactose-positive yeast Rhodotorula acheniorum from whey ultrafiltrate. Food Sci. Biotechnol. 2011; 20(2):445–454. [Google Scholar]
- 23.Brown MR, Barrett SM, Volkman JK, Nearhos SP, Nell JA, Allan GL. Biochemical composition of new yeasts and bacteria evaluated as food for bivalve aquaculture. Aquaculture 1996; 143(3–4):341–360. [Google Scholar]
- 24.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959; 37(8):911–917. 10.1139/o59-099 [DOI] [PubMed] [Google Scholar]
- 25.Lee TH, Arai M, Murao S. Structure of fucogalactomannan of red yeast cell wall. Agric. Biol. Chem. 1981; 45(6):1301–1309. [Google Scholar]
- 26.Feldmann H. Yeast: molecular and cell Biology. 2nd ed. Weinheim: Wiley-VCH Verlag GmbH & Co. KGaA; 2012. [Google Scholar]
- 27.Yang SP, Wu ZH, Jian JC. Analysis of nutrition compositions of marine red yeast, R. paludigenum. Feed Industry (in Chinese). 2011;10. [Google Scholar]
- 28.Wasserman AE. Amino acid and vitamin composition of Saccharomyces fragilis grown in whey. J. Dairy. Sci. 1961; 44(3):379–386. [Google Scholar]
- 29.Teshima SI, Kanazawa A, Yamashita M. Dietary value of several proteins and supplemental amino acids for larvae of the prawn Penaeus japonicus. Aquaculture 1986; 51(3–4):225–235. [Google Scholar]
- 30.Kutty SN, Philip R. Marine yeasts—a review. Yeast 2008; 25(7):465–483. 10.1002/yea.1599 [DOI] [PubMed] [Google Scholar]
- 31.Saenge C, Cheirsilp B, Suksaroge TT, Bourtoom T. Potential use of oleaginous red yeast Rhodotorula glutinis for the bioconversion of crude glycerol from biodiesel plant to lipids and carotenoids. Proc. Biochem. 2011; 46(1):210–218. [Google Scholar]
- 32.Zhang X, Tan TW, Gong GP, Guo GY. Effect of ammonium-N on malic enzyme and lipid production in Rhodotorula glutinis grown on monosodium glutamate wastewater. Biocatal Biotransfor. 2016; 34(1):18–23. [Google Scholar]
- 33.Wang JH, Zhao LQ, Liu JF, Wang H, Xiao S. Effect of potential probiotic Rhodotorula benthica D30 on the growth performance, digestive enzyme activity and immunity in juvenile sea cucumber Apostichopus japonicus. Fish shellfish Immunol. 2015; 43(2):330–336. 10.1016/j.fsi.2014.12.028 [DOI] [PubMed] [Google Scholar]
- 34.Choudary PV, Rao GR. Molecular analysis of inorganic nitrogen assimilation in yeasts. Arch. Microbiol. 1984; 138(3):183–186. [Google Scholar]
- 35.Magasanik B. Ammonia assimilation by Saccharomyces cerevisiae. Eukaryot. cell. 2003; 2(5):827–829. 10.1128/ec.2.5.827-829.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gethins L, Guneser O, Demirkol A, Rea MC, Stanton C, Ross RP, et al. Influence of carbon and nitrogen source on production of volatile fragrance and flavour metabolites by the yeast Kluyveromyces marxianus. Yeast 2015; 32(1):67–76. 10.1002/yea.3047 [DOI] [PubMed] [Google Scholar]
- 37.Speers RA, Tung MA, Durance TD, Stewart GG. Biochemical aspects of yeast flocculation and its measurement: a review. J. I. Brewing 1992; 98(4):293–300. [Google Scholar]
- 38.Jia BJ, Yu JM. The research status and development trend of microbial flocculant. Physics Procedia 2012; 24:425–428. [Google Scholar]
- 39.Kader MA, Bulbul M, Koshio S, Ishikawa M, Yokoyama S, Nguyen BT, et al. Effect of complete replacement of fishmeal by dehulled soybean meal with crude attractants supplementation in diets for red sea bream, Pagrus major. Aquaculture 2012; 350:109–116. [Google Scholar]
- 40.Pham KD, Shida Y, Miyata A, Takamizawa T, Suzuki Y, Ara S, et al. Effect of light on carotenoid and lipid production in the oleaginous yeast Rhodosporidium toruloides. Biosci. Biotech. Biochem. 2020;1–12. 10.1080/09168451.2020.1740581 [DOI] [PubMed] [Google Scholar]
- 41.Xu J, Liu D. Exploitation of genus Rhodosporidium for microbial lipid production. World J. Microbiol. Biotechnol. 2017; 33(3):54 10.1007/s11274-017-2225-6 [DOI] [PubMed] [Google Scholar]
- 42.Lyman M, Urbin S, Strout C, Rubinfeld B. The oleaginous red yeast Rhodotorula/Rhodosporidium: a factory for industrial bioproducts In: Basso TP, editor. Yeasts in Biotechnology. London: IntechOpen; 2019. [Google Scholar]
- 43.De Silva SS, Anderson TA. Fish nutrition in aquaculture. Springer Science & Business Media. 1994. [Google Scholar]
- 44.Coyle SD, Tidwell JH, Webster CD. Response of largemouth bass Micropterus salmoides to dietary supplementation of lysine, methionine, and highly unsaturated fatty acids. J. World Aquac. Soc. 2000; 31(1):89–95. [Google Scholar]
- 45.Aksu Z, Eren AT. Carotenoids production by the yeast Rhodotorula mucilaginosa: use of agricultural wastes as a carbon source. Proc. Biochem. 2005; 40(9):2985–2991. [Google Scholar]
- 46.Gatesoupe FJ. Live yeasts in the gut: natural occurrence, dietary introduction, and their effects on fish health and development. Aquaculture 2007; 267(1–4):20–30. [Google Scholar]
- 47.Yang SP, Wu ZH, Jian JC, Zhang XZ. Effect of marine red yeast Rhodosporidium paludigenum on growth and antioxidant competence of Litopenaeus vannamei. Aquaculture 2010; 309(1–4):62–65. [Google Scholar]
- 48.Kuebutornye FKA, Abarike ED, Lu Y. A review on the application of Bacillus as probiotics in aquaculture. Fish Shellfish Immunol. 2019; 87:820–828. 10.1016/j.fsi.2019.02.010 [DOI] [PubMed] [Google Scholar]
- 49.Vieco-Saiz N, Belguesmia Y, Raspoet R, Auclair E, Frédérique Gancel, Kempf I, et al. Benefits and inputs from lactic acid bacteria and their bacteriocins as alternatives to antibiotic growth promoters during food-animal production. Front. Microbiol 2019; 10:57 10.3389/fmicb.2019.00057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mota C, Ridenoure J, Cheng J, de los Reyes FL. High levels of nitrifying bacteria in intermittently aerated reactors treating high ammonia wastewater. FEMS Microbiol. Ecol. 2005; 54(3):391–400. 10.1016/j.femsec.2005.05.001 [DOI] [PubMed] [Google Scholar]
- 51.Sepehri A, Sarrafzadeh MH. Activity enhancement of ammonia-oxidizing bacteria and nitrite-oxidizing bacteria in activated sludge process: metabolite reduction and CO2 mitigation intensification process. Appl. Water Sci. 2019; 9(5):131. [Google Scholar]