Abstract
Infection with HIV or SIV often elicits a potent immune response to viral antigens. This includes T cells and antibodies specific for Gag and Env antigens. In contrast, when given as a vaccine, the same antigens have been weak immunogens, unable to elicit antibodies with comparable titer, durability, or neutralizing activity. We have used the live attenuated rubella vaccine strain RA27/3 as a viral vector to express HIV and SIV antigens. By mimicking an HIV infection, these vectors could elicit stronger and more durable immunity to HIV antigens. The vectors are based on the licensed rubella vaccine strain, which has demonstrated safety and potency in millions of children. One or two doses protect for life against rubella infection. The question was whether rubella vectors could similarly enhance the immunogenicity of a foreign vaccine insert.
We have previously reported that rubella vectors can express small protein antigens in vitro and in vivo, where they elicit a strong immune response to the vaccine insert. The vectors have now expressed larger vaccine inserts that include epitope-rich fragments of the Gag matrix and capsid proteins (aa 41–211) or the complete p27 capsid protein with p2 (aa 136–381). These vectors have elicited a robust and durable immune response to Gag in rhesus macaques. This size range also encompasses the engineered outer domain (eOD) of HIV envelope gp120 (172 amino acids). The rubella/eOD-GT6 and GT8 vectors stably expressed glycoproteins that bind germline precursors and mature forms of VRC01-class broadly neutralizing antibodies. These vectors potentially could be used as part of a sequential immunization strategy to initiate the production of broadly neutralizing antibodies.
Keywords: Live attenuated viral vectors, SIV gag, HIV env, Rubella vaccine strain RA27/3
1. Introduction
The immune response to HIV or SIV infection has revealed a number of potential vaccine targets [1–3]. These include envelope glycoprotein gp120 targeted by neutralizing antibodies [2,4,5] and Gag antigens recognized by cytolytic T cells [6,7]. However, when incorporated into vaccines, the same antigens have shown lack of potency, durability and protection [8,9]. Similar problems with other viral vaccines, including atypical reactions to subsequent infection, were solved by developing live attenuated vaccine strains with greater potency [10,11]. In the case of live HIV vaccine, however, almost no degree of attenuation would seem safe enough for human use [12,13]. Instead, we and others have created live viral vectors that present SIV and HIV antigens in a safe and immunogenic vaccine platform [14–16]. Our goal is to combine the growth and immunogenicity of the vector with the antigenicity of the vaccine insert to elicit a stronger and more durable response to the vaccine insert.
We have used the attenuated rubella vaccine stain RA27/3 as a live viral vector [14,15]. Its safety and vaccine potency have been demonstrated in millions of children [16,17]. It elicits systemic and mucosal immunity, and one or two doses protect for life against rubella infection. Rubella infects rhesus macaques as well as humans, which provides an animal model for testing immunogenicity and protection against live viral challenge. We recently reported the first successful trial of rubella/gag vectors in macaques [18]. The rubella/gag vectors grew well in vivo while expressing Gag T cell epitopes (84 amino acids insert). The vector elicited a potent and durable antibody response to Gag that was comparable to SIV infection. The antibodies were boosted strongly by reexposure to the vector. The vectors also elicited a potent T cell response to SIV Gag epitopes [19].
We have now expanded the size and variety of vaccine inserts that can be stably expressed by rubella vectors to include the complete p27 Gag protein (229 amino acids), and even larger Gag inserts (up to 324 amino acids). The large Gag inserts were immunogenic after a single dose, and the antibodies persisted for over one year. We have also expressed HIV Env domains that are large enough to fold correctly, yet small enough to fit into the vector. These were based on the engineered outer domain (eOD) constructs (eOD-GT6 and eOD-GT8) of envelope gp120 (172 amino acids) [20,21]. These are the first rubella vectors capable of expressing the CD4 binding site epitope [4] that is targeted by broadly neutralizing monoclonal antibodies like VRC01 and NIH45–46. In addition, the eOD-GT6 and GT8 constructs have been selected for binding to the VH1–2*02-inferred germline precursors of mature VRC01 antibodies [21,22]. The rubella/eOD vectors could be used in a sequential immunization strategy, to initiate the response of germline B cells, followed by boosting with other antigens, to elicit broadly neutralizing antibodies [22–24].
2. Materials and methods
2.1. Antibodies and antigens
Monoclonal antibodies (mAbs) 2F5 [25], VRC01 [4], NIH45–46 [26], 55–2F12 [27] and SIVmac251 p55 Gag recombinant protein were obtained through the NIH AIDS Reagent Program, NIAID, NIH. Germ Line-VRC01 mAb [24] was a gift of A. McGuire and L. Stamatatos (Fred Hutchinson Cancer Research Center). Polyclonal goat anti-rubella antibodies were from Fitzgerald Industries (Concord, MA). Horseradish peroxidase-conjugated goat anti-macaque and anti-human IgG antibodies were from Santa Cruz. Aldrithiol-2 inactivated SHIV virion controls for western blot were provided by Drs. Larry Arthur and Jeffrey Lifson at the AIDS Vaccine Program, NCI, NIH [28]. eOD-GT6 nanoparticles were described previously [20].
2.2. Construction of live rubella vectors
Rubella vectors were constructed by inserting the vaccine antigen into plasmid p10RA coding for a full length infectious cDNA clone of the rubella vaccine strain RA27/3 [18,29]. The Gag inserts, from SIV mac239, were expressed in frame between the transmembrane domain of rubella E2 protein and the E1 signal peptide (Fig. 1B and Table S1), for cleavage by signal peptidase. The engineered outer domain (eOD) inserts consisted of an eOD-GT construct [20] followed by a GGGGS linker, a short MPERF tag (membrane proximal external region epitope for monoclonal 2F5), the transmembrane domain of rubella E2, and the E1 signal peptide (Fig. 2B and Table S2). Synthetic DNA encoding the inserts was PCR amplified, and cloned into AvrII-NsiI or AvrII-SbfI sites in p10RA-derived plasmids. All constructs were verified by sequencing.
Fig. 1.
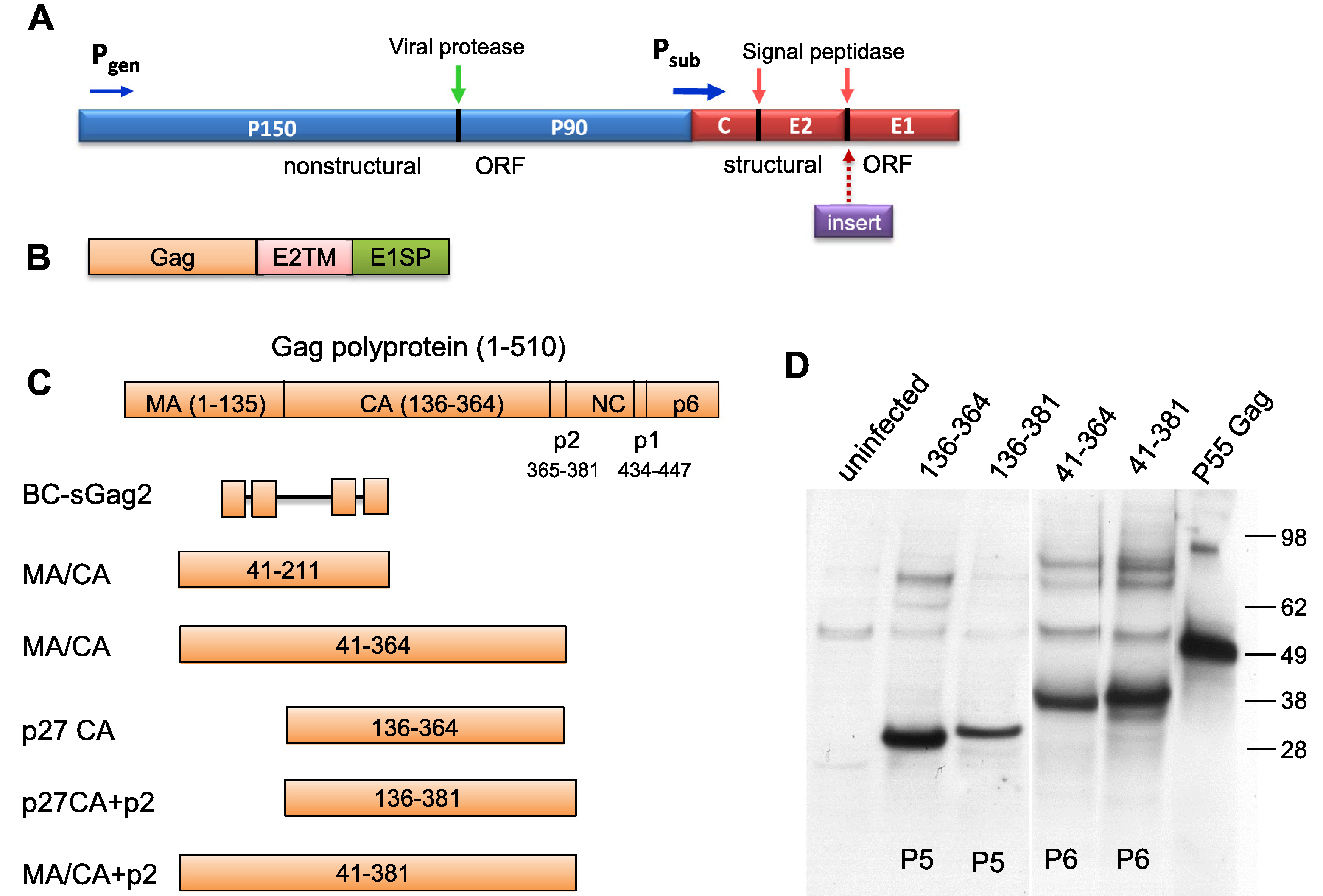
Expression of SIV Gag proteins in live rubella vectors. (A) Rubella genome organization and the structural insertion site. The non-structural (blue) and structural (red) genes are controlled by the genomic (Pgen) and strong subgenomic (Psub) promoters, respectively. The structural insertion site is located between the rubella envelope glycoproteins E2 and E1. (B). The Gag inserts were attached to the transmembrane domain of E2 glycoprotein and the E1 signal peptide, which provided membrane anchor and cleavage site. (C). Five Gag inserts of various sizes were derived from the whole Gag polyprotein (1–510 aa), spanning 41–211, 41–364, 136–364, 136–381 and 41–381 amino acids of Gag. The N and C terminal sequences of Gag inserts are shown in Table S1. BC-sGag2 contains 4 T cell epitopes in tandem and was described previously [18]. (D) Stable expression of the SIV Gag proteins was detected by western blot of Vero cell lysates with monoclonal 2F12, which is specific for the carboxyl half of CA protein. Insert expression was stable through passage P5 or P6. Controls include uninfected cell lysates and recombinant SIV p55 Gag protein.
Fig. 2.
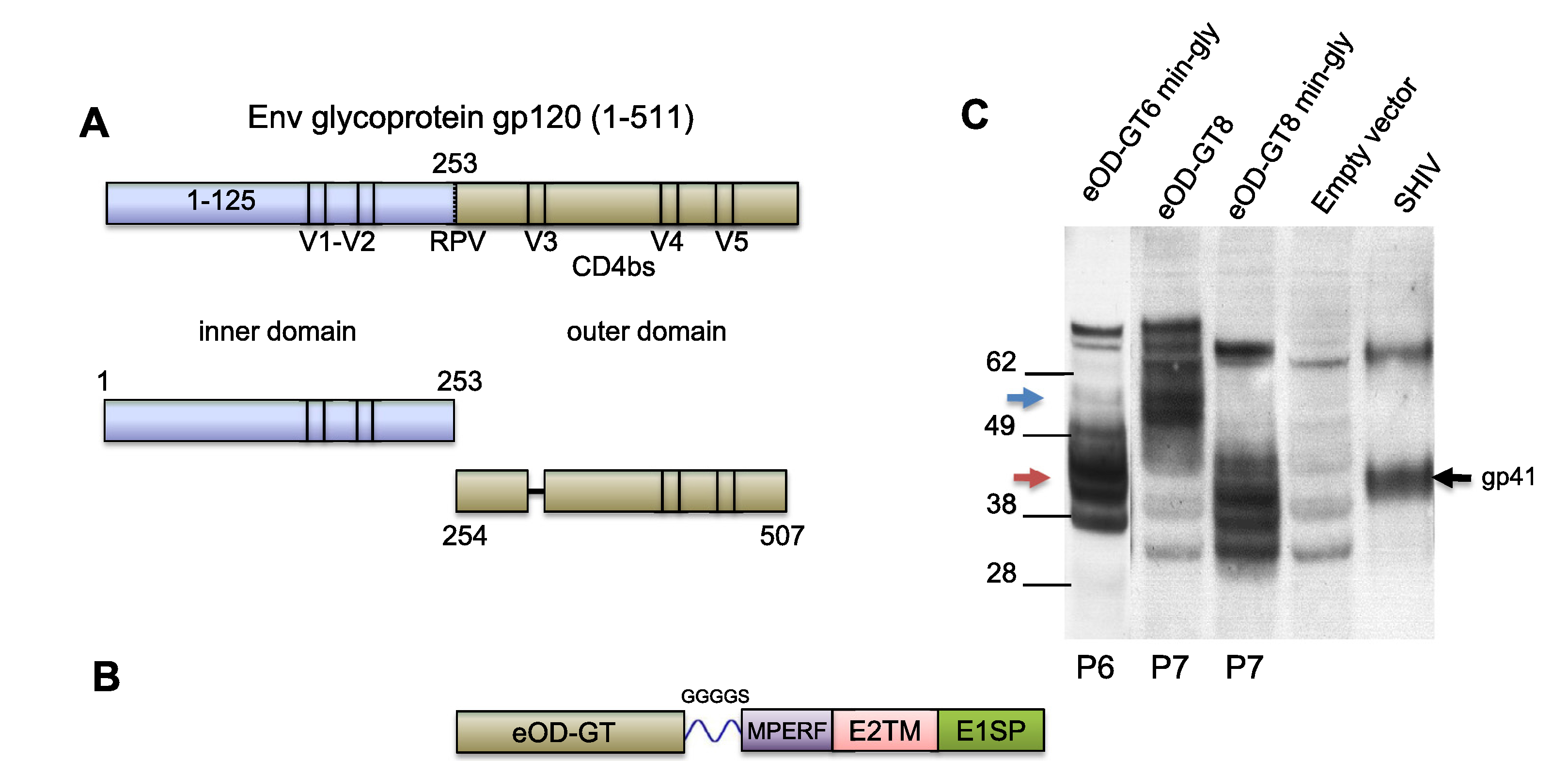
Rubella vectors expressing engineered outer domains eOD-GT6 and GT8. (A) HIV-1 envelope glycoprotein gp120 was divided into the inner domain (blue) and outer domain (olive). The engineered outer domain (eOD) sequence had a V3 deletion, and its sequence was permuted to stabilize the native conformation. (B) The eOD insert was attached via flexible linker to an MPERF tag, followed by E2 TM and rubella E1 signal peptide. (C) Expression of minimally (42 kD, red arrow) or fully (58 kD, blue arrow) glycosylated eOD by rubella vectors was detected by western blot with monoclonal 2F5. eOD-GT6 and GT8 were stably expressed through passages P6 and P7. Controls included cells infected with empty rubella virus or AT-2 inactivated SHIV virions expressing gp41 of HIV.
Methods for generating infectious rubella RNA from plasmid DNA, transfecting cells, passaging virus in Vero cells, viral stock expansion, sequencing viral RNA and determining viral RNA content were described previously [14,15,18]. Stable expression for at least 5 to 6 passages was confirmed by western blot and by sequencing the insert. Infectious virus was titered by the Reed Muench method [30].
2.3. Detection of expressed antigens and immunoprecipitation
We monitored vector replication and insert expression by western blot with polyclonal antibodies to rubella structural proteins or with monoclonal antibodies specific for the insert [14]. Gag expression was detected with murine monoclonal 2F12, and MPER-tagged eOD inserts were detected with human monoclonal 2F5. The secondary antibodies were horseradish peroxidase-conjugates. Blots were visualized with enhanced chemiluminescence (GE Healthcare) using a PXi6 image analysis system (Syngene). The association of eOD-GT6 with rubella virions was tested by sedimentation in sucrose gradients, and binding to VRC01 was demonstrated under native conditions in an ELISA assay. eOD-GT8 binding to monoclonal antibodies was detected by immune capture on antibody-coated beads (supplemental methods).
2.4. Immunizations and ELISA assays
A group of three rhesus macaques, between 5 and 12 years of age were obtained from the CBER NHP colony on the NIH campus. All experimental procedures were approved by the CBER Institutional Animal Care and Use Committee and were done in compliance with the Guide for the Care and Use of Laboratory Animals. Animals tested negative for antibodies to rubella, herpes B, STLV-1, SIV, and SRV serotypes 1, 2 and 3.
The animals were primed 2.5 years previously with three doses of DNA vaccine coding for Gag and Env antigens. The DNA vaccine was given again, followed 16 weeks later by two doses of rubella vectors given 54 weeks apart (timeline, Fig. S1). The optimized DNA vaccine [31] consisted of four plasmid DNAs: 1 mg DNA coding for SIV Gag, 1 mg each for env DNA (gp140 and gp160), and 200 μg for monkey IL-12 [32] (Table S3). The DNA vaccines were given by in vivo electroporation using the Elgen 1000 (Inovio Pharmaceuticals, Blue Bell, PA). The first rubella dose consisted of two rubella vectors: rubella/gag (aa 136–381) (0.6 × 105 PFU) and rubella/eOD-GT8 (3 × 105 PFU). The second dose was rubella/ eOD-GT8 alone at 7 × 105 PFU (Table S3). anti-rubella antibodies were detected by ELISA in rubella coated plates (BioCheck, Inc., Foster City, CA). Macaque antibodies to SIV Gag and eOD-GT8 were titered on ELISA plates coated with recombinant SIV p55 Gag or with eOD-GT6 nanoparticles at 1 μg/ml.
3. Results
3.1. Rubella/gag vectors
The design of rubella vectors expressing large SIV Gag inserts are shown in Fig. 1A–C. The insertion site is located within the rubella structural region, between envelope glycoproteins E2 and E1. At this site, Gag was expressed as part of the rubella structural polyprotein, under control of the strong subgenomic promoter. Cleavage by signal peptidase released mature rubella structural proteins and Gag. The Gag sequence was followed by two TM domains, which anchor Gag to the membrane.
Fig. 1C shows the composition of each Gag insert, as compared to the full coding potential of SIV Gag (510 amino acids). The original Gag insert BC-sGag2 consisted of four T cell epitopes linked in tandem (84 amino acids) [15]. The MA and CA protein epitopes were known targets for macaque CTLs [6]. This insert was expanded to include the sequence between epitopes (aa 41–211). We expressed the entire p27 CA protein (aa 136–364), as well as the p2 protein (aa 136–381) (Table S1). The largest Gag products consisted of aa 41–364 and aa 41–381. Insert expression was verified by western blot with monoclonal 2F12 (Fig. 1D) [27]. Each Gag protein migrated at the expected size, and expression was stable through at least passage P5 or P6, as indicated. Rubella vectors for immunization were expanded at passage P5 and tested for stable expression of the insert, correct insert sequence, and viral titer.
3.2. Rubella/eOD vectors expressing the CD4bs
The CD4 binding site (CD4bs) on gp120 is an important target of broadly neutralizing antibodies [20,21], and it depends on native folding [5]. As described previously, Jardine et al. [20,21] have shown that the native CD4bs epitope could be expressed on the smaller eOD of gp120 (172 amino acids, Fig. 2A) by permuting the outer domain sequence to stabilize the native conformation (Table S2). They also identified eOD-GT mutations that enable germline targeting of VRC01-class germline precursors. Based on our experience with larger Gag inserts, we made rubella vectors with eOD inserts and asked if they could express a well-folded CD4 binding site.
We initially expressed a minimally glycosylated (min-gly) form of eOD-GT6, lacking 6 out of 9 N-linked glycosylation sites (mutations shown in red in Table S2) to reduce the size of the expressed protein. It was the same min-gly form described by Jardine et al. [20], except for an additional N-glycosylation site preserved at N170 (Table S2). We subsequently expressed an improved version, called eOD-GT8, which binds a panel of VRC01-type germline precursors with greater breadth and affinity. Two glycoforms of eOD-GT8 were expressed: one fully glycosylated at 10 sites, and the other minimally glycosylated at 3 out of 10 sites. Western blots showed stable expression of eOD-GT6 min-gly at 42 kDa (Fig. 2C, first lane), as well as both glycoforms of eOD-GT8 at 58 kDa and 42 kDa (Fig. 2C, second and third lanes). The eOD constructs were expressed consistently through passages P6 and P7, indicating genetic stability, yet they gave broad bands on western blot. In part, this was typical of a glycoprotein with incomplete glycosylation, especially when it was over-expressed by the vector. In part, this may reflect the natural flexibility of gp120 that allows it to accommodate mutations and still fold normally. In this case, the modifications include deletion of the inner domain, permutation of the outer domain sequence, and mutation to enhance germline binding.
3.3. Antigenicity of rubella/eOD vectors
We have shown previously that smaller vaccine inserts, such as the membrane proximal external region of the HIV envelope (MPER, 22 amino acids plus two TM domains), remain associated with rubella virions after cleavage by signal peptidase [15]. We tested whether eOD-GT6-min-gly would be incorporated similarly into viral particles (Fig. 3). Rubella/eOD-GT6 infected cell lysates were sedimented in sucrose gradients, and each fraction was assayed for rubella E1 protein by western blot. The rubella peak was identified in fractions 5 to 11 (upper panel of Fig. 3). Monoclonal 2F5 detected the MPER tag on eOD-GT6 by ELISA. Similarly, VRC01 bound the CD4 binding site on eOD-GT6, indicating that it was folded in the native conformation. Both monoclonal antibodies detected eOD-GT6-min-gly in the virus peak fractions (lower vs. upper panel of Fig. 3). This result indicates that eOD-GT6-min-gly is expressed with the structural polyprotein and remains associated with rubella virions after polyprotein cleavage.
Fig. 3.
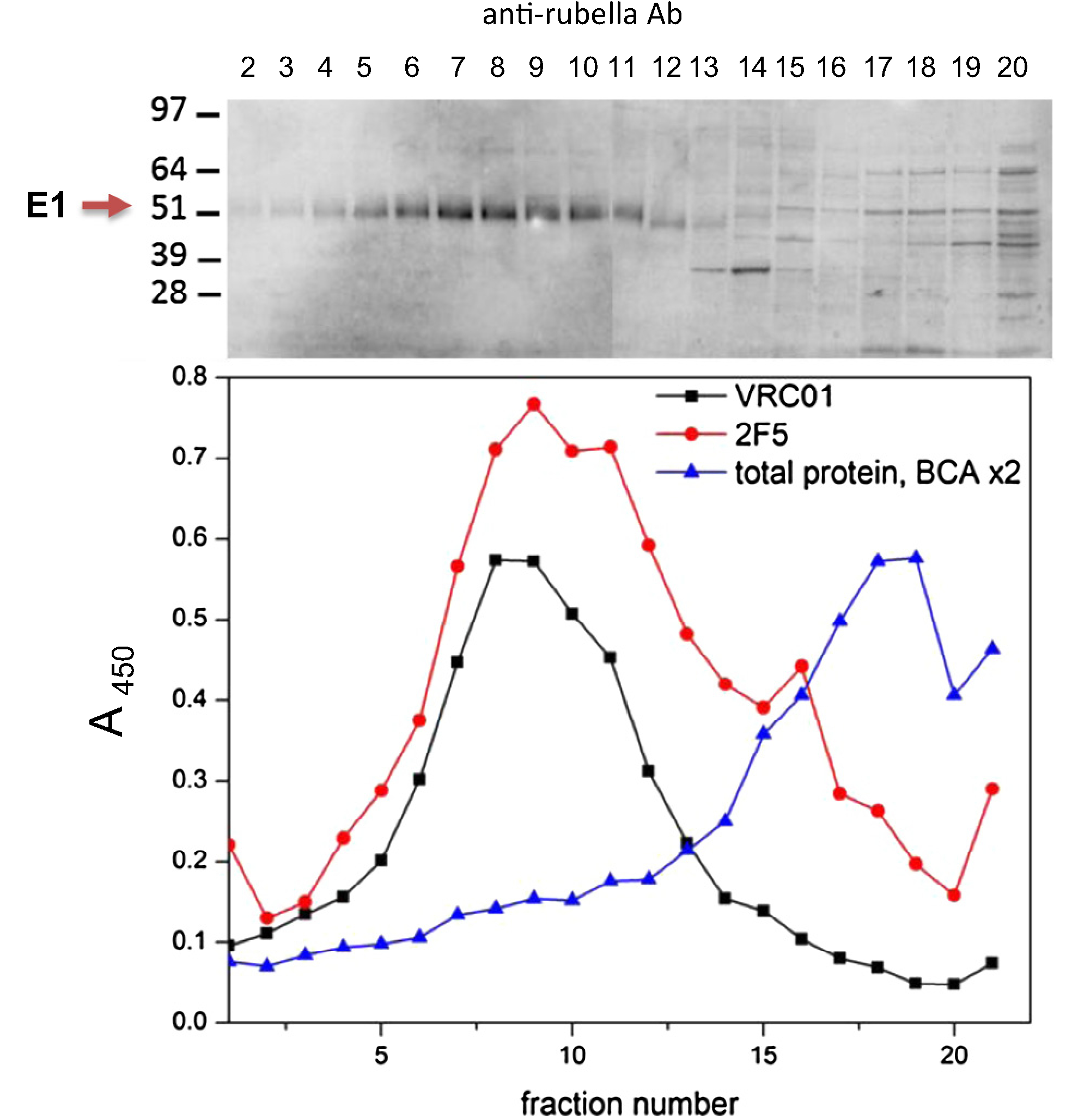
Sedimentation of rubella/eOD-GT6-min-gly virions on sucrose gradients. Cell lysates infected with eOD-GT6-min-gly vector were sedimented overnight on a 10–40% sucrose gradient. The rubella band was identified in peak fractions 5–11 by western blot with antibodies to rubella E1 protein (upper panel). The same fractions were assayed by ELISA (lower panel) with monoclonals VRC01 (black line) and 2F5 (red line). The eOD-GT6 peak coincides with the peak rubella fractions identified by western blot. Total protein content in the fractions is shown (blue line).
We measured the binding of eOD-GT8 to mature monoclonal NIH45–46 and to its VH1–2*02 inferred-germline precursor. The monoclonals were immobilized on epoxy beads. Cell lysates containing eOD-GT8, or a gp120 core control (aa 60–492 lacking variable loops V1, V2 and V3), were captured by the beads, washed, eluted, and then analyzed by western blot (Fig. 4, middle and right panels). A positive control (total IP input) consisted of the same cell lysates without adsorption (Fig. 4, left panel). The middle panel shows binding of both the gp120 core and eOD-GT8 to NIH45–46. The right panel shows binding by eOD-GT8, but not the gp120 core, to germline VRC01. These results indicate that rubella expressed eOD-GT8 binds mature and germline precursors of NIH45–46 and VRC01. The IgG heavy chains of the capture antibody (50 kDa) in the middle and right panels were also eluted from the epoxy beads, and they are distinct from the eOD- GT8 band (58 kDa).
Fig. 4.
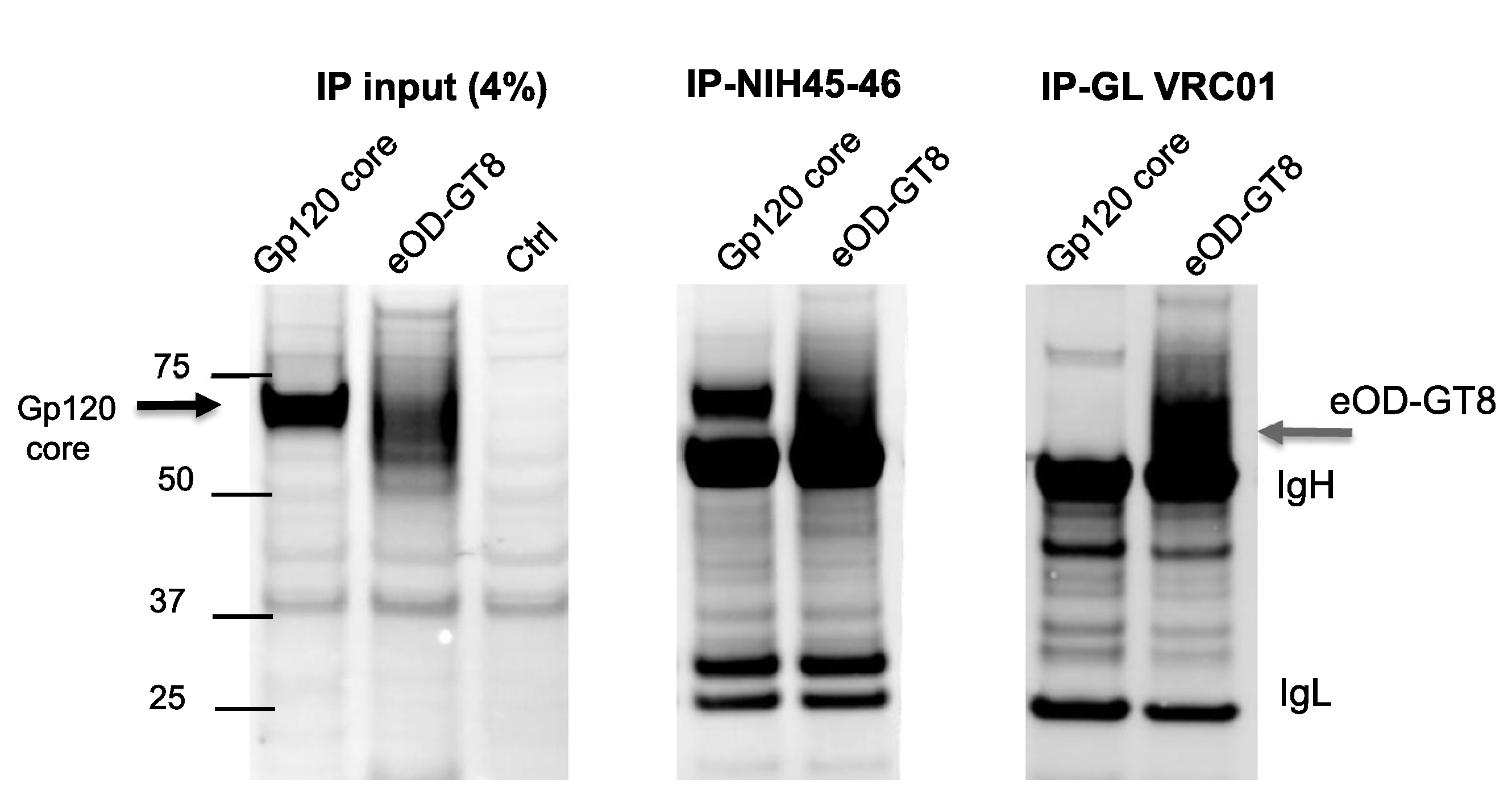
Immunoprecipitation (IP) of eOD-GT8 by broadly neutralizing monoclonal NIH45–46 and its germline VH1–2*02 precursor. Cell lysates infected with rubella/eOD-GT8 or with a control rubella/gp120 core protein were studied for total content prior to IP (left panel), after IP with mature monoclonal NIH45–46 (middle panel), or after IP with the germline precursor of VRC01 and NIH45–46 (right panel). The immobilized antibodies captured eOD-GT8 or gp120 core antigen, followed by elution and detection by western blot using monoclonal 2F5 specific for an MPER tag in both constructs. The center panel shows that both antigens bind monoclonal NIH45–46. However, the right panel shows that only eOD-GT8 can bind both the germline precursor of VRC01 and its mature form equally well. IgH and IgL bands correspond to heavy and light chains of the monoclonal antibodies used for IP.
3.4. Immunogenicity of rubella vectors
A group of three macaques were primed three times with DNA vaccines expressing SIV Gag and HIV Env with a monkey IL-12 adjuvant (Table S3). They were boosted with a pair of rubella vectors expressing SIV Gag 136–381 and eOD-GT8. A vaccine "take" was detected in all three macaques, as indicated by rising antirubella titers (not shown). Antibodies to the Gag insert (aa 136–381) were detected in all three animals by two to four weeks post immunization (Fig. 5A). The antibody response of macaque E023 peaked at 4–6 weeks, followed by a slight decline by 54 weeks (Fig. 5B). The other two macaques’ anti-Gag titers declined to 50–80% of the 4 week peak level after one year.
Fig. 5.
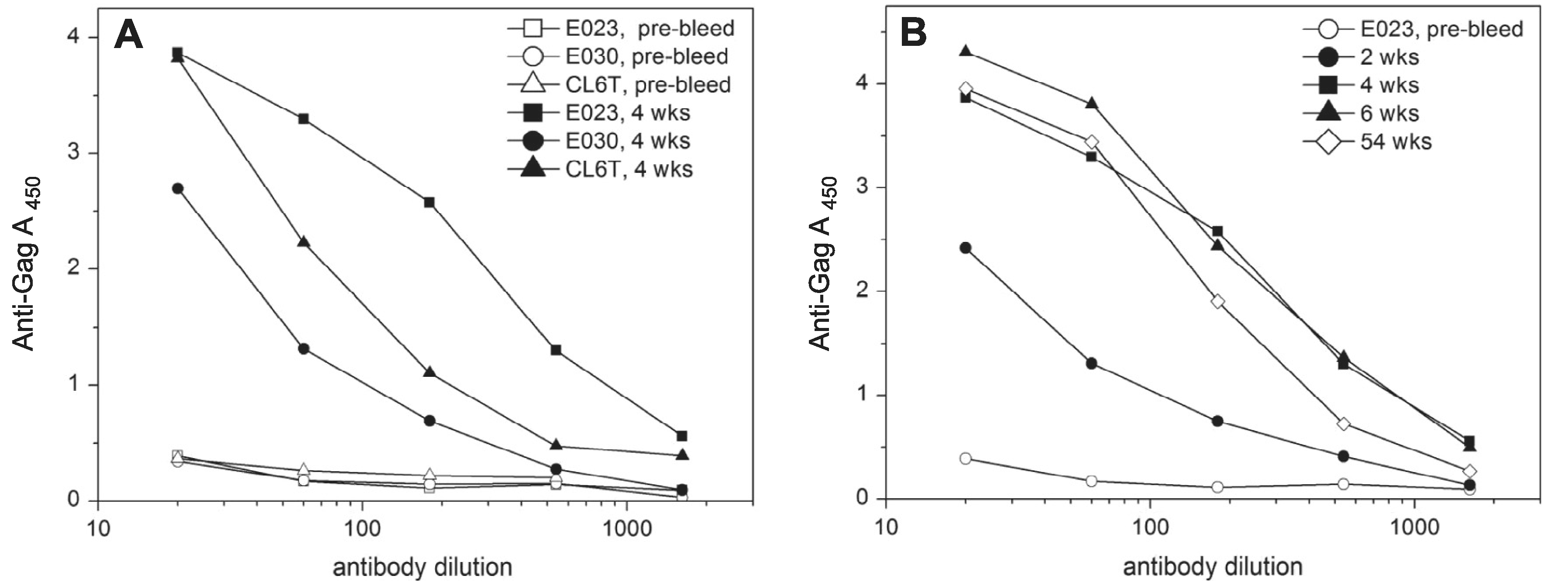
The immune response to a combined dose of rubella/gag (p27 + p2) vector plus rubella/eOD-GT8 and persistence of anti-Gag antibodies. (A) The anti-Gag response after four weeks was measured by ELISA on plates coated with recombinant SIV p55 Gag protein. Macaque E023 gave the strongest antibody response to SIV Gag. (B) The persistence of anti-Gag antibodies in macaque E023 was monitored for more than a year. The antibodies reached peak titers at 4–6 weeks post immunization and declined slightly after one year.
Antibodies to the eOD-GT8 insert were assayed on ELISA plates coated with the related GT6 60–mer nanoparticles [20]. These antibodies were not detected after the first dose of rubella/eOD-GT8 vectors. One year later, the macaques were boosted with a second dose of eOD-GT8 alone. The boost with rubella/eOD-GT8 elicited rising anti-rubella titers in all 3 macaques (Fig. 6, open symbols). One macaque (CL6T) made antibodies to eOD-GT8 that peaked at weeks 1–2 and then declined slightly by weeks 4–6 (Fig. 6, closed symbols). These antibodies lacked neutralizing activity in a pseudovirion assay based on the same IIIB envelope as used in the eOD GT8 vector.
Fig. 6.
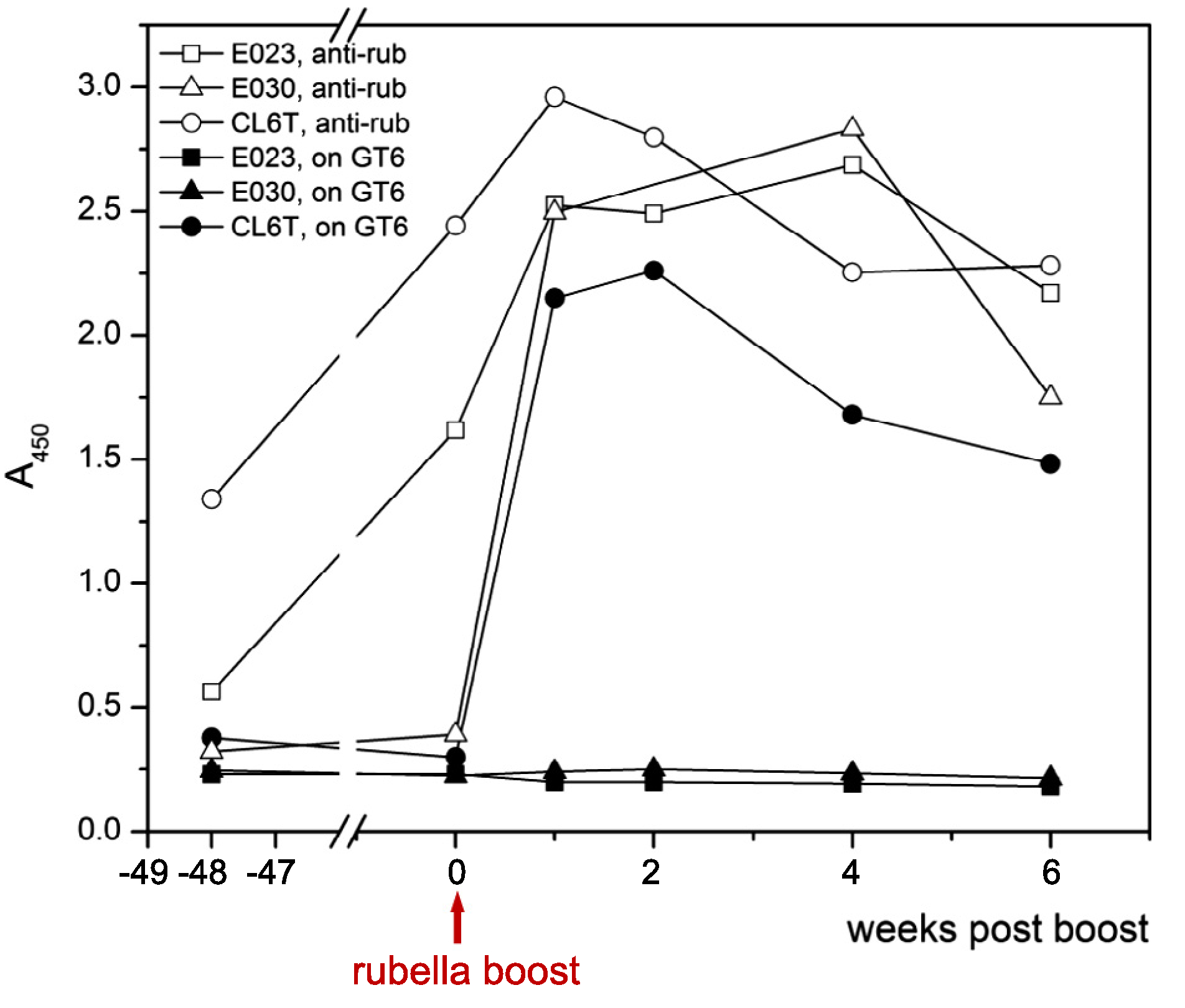
The immune response to a rubella/eOD-GT8 vector prime and boost. antirubella antibodies (open symbols) and anti-GT8 antibodies (solid symbols) were measured by ELISA after the rubella/eOD-GT8 prime ( –54 weeks) and boost (0 weeks). Anti-rubella antibodies (1:40 dilution) showed a rapid response to the boost in all animals. Anti-eOD-GT8 antibodies showed a response in only one of three animals. These antibodies (at 1:20 dilution) were assayed on plates coated with purified eOD-GT6 nanoparticles.
The comparison of responder and non-responder animals does not suggest interference by anti-rubella antibodies. The macaque CL6T with the highest anti-rubella titers before the boost made the best anti-eOD response, while the macaque with the lowest anti-rubella titers (E030) made no anti-eOD antibodies after the boost. This pattern suggests that a strong primary immunization may favor the response to the rubella/eOD-GT8 boost.
4. Discussion
4.1. Live, attenuated viral vectors for SIV and HIV immunogens
Live attenuated rubella vectors provide a safe and immunogenic vaccine platform for expressing viral antigens such as SIV Gag and HIV Env. Rubella vectors can accommodate the complete p27 capsid protein (245 amino acids and larger) or the engineered outer domain of gp120 (eOD, 172 amino acids). The rubella/eOD vectors are the first rubella vectors to express vaccine antigens capable of binding broadly neutralizing monoclonal antibodies like VRC01 that target the CD4 binding site. The eOD-GT8 construct was also capable of binding germline precursors of VRC01. These vectors may enhance vaccine potency by initiating the response of germline VH1–2*02 B cells, followed by additional stimulation to produce mature VRC01-like antibodies with broad neutralizing activity.
The goal of live attenuated vaccines is to separate growth and immunogenicity of the virus from its pathogenicity. For HIV, the risks of reversion to wild type and aberrant integration seem too great for an attenuated HIV vaccine to be used in humans. Instead, a number of attenuated vectors expressing individual HIV antigens are under study including: DNA viruses, like adenovirus [8,9,33], MVA [34], ALVAC [35] and CMV [36,37]. RNA vectors include VEE replicons [38], as well as live attenuated vaccine strains such as: yellow fever 17D [39], VSV [40], and measles vaccine [41]. DNA viral vectors can generally hold larger inserts, however, some of them may lead to chronic infection [36,37] or actively interfere with the host immune response. RNA vectors tend to have limitations on insert size and stability [39,42]. Some attenuated RNA viral vaccines, like rubella, are even recommended for HIV infected children [16].
Rubella/gag vectors elicited a potent anti-Gag response that persisted for over 1 year. Rubella/eOD-GT8 vectors elicited a weaker response that may need additional help to achieve high antibody titers with neutralizing activity. This could be accomplished in several ways: (a) through a carrier effect, by linking Env to a more potent immunogen, such as Gag, in a rubella/envgag vector; or (b) through a prime and boost strategy with a rubella/eOD-GT8 prime and a native Env protein boost or a succession of increasingly native-like Env boosts [43–45]. If rubella vectors could elicit neutralizing antibodies in macaques, the overlapping host range of both viruses would allow us to test protection against a live viral challenge with SIV or SHIV.
4.2. Sequential immunization strategy
Successful immunization against HIV will most likely require the production of broadly neutralizing antibodies against key Env determinants. When these antibodies are elicited during chronic HIV infection, they are highly evolved from germline precursors [22]. This process requires relatively intact germinal center (GC) function over years of persistent exposure to viral antigens. A successful vaccine would have to accomplish the same process with a few doses over several months to a year. Live rubella vectors can stimulate GC function without damaging them, as shown by the durable immune response to the vector itself [18].
A significant obstacle for HIV specific B cells is that VH1–2*02 germline precursors do not bind the same antigenic form of gp120 as the mature VRC01-like neutralizing antibodies derived from these precursors [24]. This suggests that sequential immunization with distinct forms of gp120 will be required for successful immunization: to initiate the response of germline B cells, and then to guide the response toward mature antibodies with broad neutralizing activity [21,45]. Consistent with Jardine et al. [20], we have found that rubella/eOD-GT8 binds both the germline VH1–2*02 precursor and mature VRC01. This vector could initiate the germline B cell response leading to VRC01-like antibodies.
In this study, however, boosting with rubella/eOD-GT8 vectors was not sufficient to complete the maturation of neutralizing antibodies. The lack of response to eOD-GT8 in two macaques in our study could be due to the fact that the closest analog of VH1–2*02 in the rhesus macaque lacks one out of three amino acids critical for a VRC01 response [20]. Alternatively, nanoparticles expressing eOD/GT8 were tested in transgenic mice expressing rearranged human VH1–2*02 [43] and in transgenic mice carrying the human IgG locus [46]. In both cases, the mice showed priming of VRC01 germline precursors, although they did not mature into VRC01-like neutralizing antibodies. To elicit these antibodies in rubella/eOD primed macaques, it may be necessary to boost with rubella/env vectors expressing a more natural form of gp120 or with native SOSIP protein trimers, possibly stabilized in the prefusion state [47,48].
4.3. Therapeutic potential
Live attenuated rubella/gag vectors, as potent inducers of Gag specific CD8 + T cells [19], could be used for immunotherapy of recent HIV or SIV infection. Therapy could combine antiretroviral therapy (ART) to control active viral replication and T cell immunity to target the latent viral reservoir. Immunotherapy could be effective in treating infections of newborns, when the latent viral reservoir is small and the immune system is still intact. In the rare case of the Mississippi baby [49,50], early ART therapy alone led to viral control for 27 months after ART was withdrawn. More commonly, it may be possible to control the virus by immunizing with rubella/gag vectors while on ART. A potent T cell response could reset the balance between the virus and the host, reduce viral reservoirs, and maintain immune control of the infection long after stopping ART.
Supplementary Material
Acknowledgements
The following reagents were obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: SIVmac p27 monoclonal antibody (55–2F12) from Dr Niels Pedersen; anti-HIV-1 gp41 monoclonal (2F5) from Dr. Hermann Katinger; anti-HIV-1 gp120 monoclonal (VRC01), from Dr. John Mascola; SIVmac251 BK28 pr55 Gag recombinant protein from the NIAID Vaccine Research and Development Branch, Division of AIDS, NIAID. We are grateful to Dr. Suzanne Epstein of CBER for insightful comments and review of the manuscript. We thank Dr. Jill Ascher, Lewis Shankle, and Faith Sentz for excellent animal care that made these studies possible.
Footnotes
Conflicts of interest
None.
Appendix A. Supplementary material
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.vaccine.2017.04.047.
References
- [1].Burton DR, Mascola JR. Antibody responses to envelope glycoproteins in HIV-1 infection. Nat Immunol 2015;16:571–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Scheid JF, Mouquet H, Feldhahn N, Seaman MS, Velinzon K, Pietzsch J, et al. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature 2009;458:636–40. [DOI] [PubMed] [Google Scholar]
- [3].McElrath MJ, Haynes BF. Induction of immunity to human immunodeficiency virus type-1 by vaccination. Immunity 2010;33:542–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Wu XL, Yang ZY, Li YX, Hogerkorp CM, Schief WR, Seaman MS, et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 2010;329:856–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Berkower I, Murphy D, Smith CC, Smith GE. A predominant group-specific neutralizing epitope of human-immunodeficiency-virus type-1 maps to residue-342 to residue-511 of the envelope glycoprotein-Gp120. J Virol 1991;65:5983–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Allen TM, Mothe BR, Sidney J, Jing PC, Dzuris JL, Liebl ME, et al. CD8(+) lymphocytes from simian immunodeficiency virus-infected rhesus macaques recognize 14 different epitopes bound by the major histocompatibility complex class I molecule Mamu-A*01: implications for vaccine design and testing. J Virol 2001;75:738–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Migueles SA, Connors M. Success and failure of the cellular immune response against HIV-1. Nat Immunol 2015;16:563–70. [DOI] [PubMed] [Google Scholar]
- [8].Barouch DH, Korber B. HIV-1 vaccine development after STEP. Annu Rev Med 2010;61:153–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hammer SM, Sobieszczyk ME, Janes H, Karuna ST, Mulligan MJ, Grove D, et al. Efficacy trial of a DNA/rAd5 HIV-1 preventive vaccine. New Engl J Med 2013;369:2083–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Krugman S, Giles JP, Jacobs AM. Studies on an attenuated measles-virus vaccine. 6. Clinical, antigenic and prophylactic effects of vaccine in institutionalized children. New Engl J Med 1960;263:174–7. [DOI] [PubMed] [Google Scholar]
- [11].Karzon DT, Rush D, Winkelstein W. Immunization with inactivated measles virus vaccine - effect of booster dose and response to natural challenge. Pediatrics 1965;36:40–50. [PubMed] [Google Scholar]
- [12].Daniel MD, Kirchhoff F, Czajak SC, Sehgal PK, Desrosiers RC. Protective effects of a live attenuated SIV vaccine with a deletion in the nef gene. Science 1992;258:1938–41. [DOI] [PubMed] [Google Scholar]
- [13].Baba TW, Liska V, Khimani AH, Ray NB, Dailey PJ, Penninck D, et al. Live attenuated, multiply deleted simian immunodeficiency virus causes AIDS in infant and adult macaques. Nat Med 1999;5:194–203. [DOI] [PubMed] [Google Scholar]
- [14].Virnik K, Ni Y, Berkower I. Live attenuated rubella viral vectors stably express HIV and SIV vaccine antigens while reaching high titers. Vaccine 2012;30:5453–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Virnik K, Ni Y, Berkower I. Enhanced expression of HIV and SIV vaccine antigens in the structural gene region of live attenuated rubella viral vectors and their incorporation into virions. Vaccine 2013;31:2119–25. [DOI] [PubMed] [Google Scholar]
- [16].Plotkin SA, Reef S. Rubella vaccine In: Plotkin SA, Orenstein WA, editors. Vaccines. Philadelphia: Saunders; 2013. p. 688–717. [Google Scholar]
- [17].Parkman PD, Phillips PE, Kirschstein RL, Meyer HM Jr. Experimental rubella virus infection in the rhesus monkey. J Immunol 1965;95:743–52. [PubMed] [Google Scholar]
- [18].Virnik K, Hockenbury M, Ni Y, Beren J, Pavlakis GN, Felber BK, et al. Live attenuated rubella vectors expressing SIV and HIV vaccine antigens replicate and elicit durable immune responses in rhesus macaques. Retrovirology 2013;10:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Rosati M, Alicea C, Kulkarni V, Virnik K, Hockenbury M, Sardesai NY, et al. Recombinant rubella vectors elicit SIV Gag-specific T cell responses with cytotoxic potential in rhesus macaques. Vaccine 2015;33:2167–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Jardine J, Julien JP, Menis S, Ota T, Kalyuzhniy O, McGuire A, et al. Rational HIV immunogen design to target specific germline B cell receptors. Science 2013;340:711–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Jardine JG, Kulp DW, Havenar-Daughton C, Sarkar A, Briney B, Sok D, et al. HIV-1 broadly neutralizing antibody precursor B cells revealed by germline-targeting immunogen. Science 2016;351:1458–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wu X, Zhou T, Zhu J, Zhang B, Georgiev I, Wang C, et al. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science 2011;333:1593–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Haynes BF, Kelsoe G, Harrison SC, Kepler TB. B-cell-lineage immunogen design in vaccine development with HIV-1 as a case study. Nat Biotechnol 2012;30:423–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].McGuire AT, Hoot S, Dreyer AM, Lippy A, Stuart A, Cohen KW, et al. Engineering HIV envelope protein to activate germline B cell receptors of broadly neutralizing anti-CD4 binding site antibodies. J Exp Med 2013;210:655–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Purtscher M, Trkola A, Gruber G, Buchacher A, Predl R, Steindl F, et al. A broadly neutralizing human monoclonal antibody against gp41 of human immunodeficiency virus type 1. AIDS Res Hum Retroviruses 1994;10:1651–8. [DOI] [PubMed] [Google Scholar]
- [26].Diskin R, Scheid JF, Marcovecchio PM, West AP Jr, Klein F, Gao H, et al. Increasing the potency and breadth of an HIV antibody by using structure-based rational design. Science 2011;334:1289–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Higgins JR, Sutjipto S, Marx PA, Pedersen NC. Shared antigenic epitopes of the major core proteins of human and simian immunodeficiency virus isolates. J Med Primatol 1992;21:265–9. [PubMed] [Google Scholar]
- [28].Arthur LO, Bess JW Jr, Chertova EN, Rossio JL, Esser MT, Benveniste RE, et al. Chemical inactivation of retroviral infectivity by targeting nucleocapsid protein zinc fingers: a candidate SIV vaccine. AIDS Res Hum Retroviruses 1998;14(Suppl. 3):S311–9. [PubMed] [Google Scholar]
- [29].Pugachev KV, Galinski MS, Frey TK. Infectious cDNA clone of the RA27/3 vaccine strain of Rubella virus. Virology 2000;273:189–97. [DOI] [PubMed] [Google Scholar]
- [30].Reed LJ, Muench H. A simple method of estimating fifty percent endpoints. Am J Hygiene 1938;27:493–7. [Google Scholar]
- [31].Felber BK, Valentin A, Rosati M, Bergamaschi C, Pavlakis GN. HIV DNA vaccine: stepwise improvements make a difference. Vaccines (Basel) 2014;2:354–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Jalah R, Rosati M, Ganneru B, Pilkington GR, Valentin A, Kulkarni V, et al. The p40 Subunit of Interleukin (IL)-12 promotes stabilization and export of the p35 subunit: Implications for improved IL-12 cytokine production. J Biol Chem 2013;288:6763–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Casimiro DR, Wang F, Schleif WA, Liang X, Zhang ZQ, Tobery TW, et al. Attenuation of simian immunodeficiency virus SIVmac239 infection by prophylactic immunization with dna and recombinant adenoviral vaccine vectors expressing Gag. J Virol 2005;79:15547–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Cox JH, Ferrari MG, Earl P, Lane JR, Jagodzinski LL, Polonis VR, et al. Inclusion of a CRF01_AE HIV envelope protein boost with a DNA/MVA prime-boost vaccine: Impact on humoral and cellular immunogenicity and viral load reduction after SHIV-E challenge. Vaccine 2012;30:1830–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, et al. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. New Eng J Med 2009;361:2209–20. [DOI] [PubMed] [Google Scholar]
- [36].Hansen SG, Ford JC, Lewis MS, Ventura AB, Hughes CM, Coyne-Johnson L, et al. Profound early control of highly pathogenic SIV by an effector memory T-cell vaccine. Nature 2011;473:523–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Hansen SG, Sacha JB, Hughes CM, Ford JC, Burwitz BJ, Scholz I, et al. Cytomegalovirus vectors violate CD8+ T cell epitope recognition paradigms. Science 2013;340:1237874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Johnston RE, Johnson PR, Connell MJ, Montefiori DC, West A, Collier ML, et al. Vaccination of macaques with SIV immunogens delivered by Venezuelan equine encephalitis virus replicon particle vectors followed by a mucosal challenge with SIVsmE660. Vaccine 2005;23:4969–79. [DOI] [PubMed] [Google Scholar]
- [39].Bonaldo MC, Martins MA, Rudersdorf R, Mudd PA, Sacha JB, Piaskowski SM, et al. Recombinant yellow fever vaccine virus 17D expressing simian immunodeficiency virus SIVmac239 gag induces SIV-specific CD8+ T-cell responses in rhesus macaques. J Virol 2010;84:3699–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Rose NF, Marx PA, Luckay A, Nixon DF, Moretto WJ, Donahoe SM, et al. An effective AIDS vaccine based on live attenuated vesicular stomatitis virus recombinants. Cell 2001;106:539–49. [DOI] [PubMed] [Google Scholar]
- [41].Liniger M, Zuniga A, Morin TN, Combardiere B, Marty R, Wiegand M, et al. Recombinant measles viruses expressing single or multiple antigens of human immunodeficiency virus (HIV-1) induce cellular and humoral immune responses. Vaccine 2009;27:3299–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Pugachev KV, Tzeng WP, Frey TK. Development of a rubella virus vaccine expression vector: use of a picornavirus internal ribosome entry site increases stability of expression. J Virol 2000;74:10811–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Jardine JG, Ota T, Sok D, Pauthner M, Kulp DW, Kalyuzhniy O, et al. HIV-1 VACCINES. Priming a broadly neutralizing antibody response to HIV-1 using a germline-targeting immunogen. Science 2015;349:156–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Jardine JG, Sok D, Julien JP, Briney B, Sarkar A, Liang CH, et al. Minimally mutated HIV-1 broadly neutralizing antibodies to guide reductionist vaccine design. PLoS Pathog 2016;12:e1005815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Briney B, Sok D, Jardine JG, Kulp DW, Skog P, Menis S, et al. Tailored immunogens direct affinity maturation toward HIV neutralizing antibodies. Cell 2016;166(1459–70):e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Sok D, Briney B, Jardine JG, Kulp DW, Menis S, Pauthner M, et al. Priming HIV-1 broadly neutralizing antibody precursors in human Ig loci transgenic mice. Science 2016;353(6307):1557–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Sanders RW, Derking R, Cupo A, Julien JP, Yasmeen A, de Val N, et al. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog 2013;9:e1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Kwon YD, Pancera M, Acharya P, Georgiev IS, Crooks ET, Gorman J, et al. Crystal structure, conformational fixation and entry-related interactions of mature ligand-free HIV-1 Env. Nat Struct Mol Biol 2015;22:522–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Persaud D, Gay H, Ziemniak C, Chen YH, Piatak M Jr, Chun TW, et al. Absence of detectable HIV-1 viremia after treatment cessation in an infant. New Eng J Med 2013;369:1828–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Luzuriaga K, Gay H, Ziemniak C, Sanborn KB, Somasundaran M, Rainwater-Lovett K, et al. Viremic relapse after HIV-1 remission in a perinatally infected child. New Eng J Med 2015;372:786–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


