Abstract
Wheat flour is one of the most important food ingredients containing several essential nutrients including proteins. Gluten is one of the major protein components of wheat consisted of glutenin (encoded on chromosome 1) and gliadin (encoded on chromosome 1 and 6) and there are around hundred genes encoding it in wheat. Gluten proteins have the ability of eliciting the pathogenic immune responses and hypersensitivity reactions in susceptible individuals called “gluten-related disorders (GRDs)”, which include celiac disease (CD), wheat allergy (WA), and non-celiac gluten sensitivity (NCGS). Currently removing gluten from the diet is the only effective treatment for mentioned GRDs and studies for the appropriate and alternative therapeutic approaches are ongoing. Accordingly, several genetic studies have focused on breeding wheat with low immunological properties through gene editing methods. The present review considers genetic characteristics of gluten protein components, focusing on their role in the incidence of gluten-related diseases, and genetic modifications conducted to produce wheat with less immunological properties.
Keywords: gliadin, glutenin, genetic loci, wheat allergy, celiac disease, non-celiac gluten sensitivity
Introduction
Gluten-containing grains are essential food ingredients, consumed in most parts of the world.1,2 Owing to their importance and thanks to their ability to grow in different climatic areas, wheat cereals were among the first crops to be cultivated (established in the “Fertile Crescent”, such as modern Turkey, Iraq and Iran) and their importance has increased significantly over time.3–6 The complex genome of Triticum aestivum L. is arranged into three subgenomes, A, B, and D, each contains seven pairs of chromosomes.7,8 However, the ability of wheat to adapt to different eco-climatic conditions and deliberate breeding for specific traits have led to the emergence of varieties with different characteristics.8 Accordingly, evidence showed that changes in the environmental conditions (such as temperature, water, and fertilizer situations) could influence the expression of gluten genes in wheat.9
Wheat (Triticum aestivum L. 2n = 6 × = 42) flour is composed of starch (~70–75%: main component), proteins (~10–15%), lipids (~2%), minerals (~2%), that convey substantial nutritional benefits to humans.10 Gluten is one of the major protein components of wheat (~80% of the total proteins), which is specifically expressed in the developing grains and provides a source of nitrogen for germination and seedling growth.4,11–13 Gluten is composed of storage proteins including glutenin and gliadin and is the term applied to the viscoelastic matrix formed when these proteins are mixed with water. There are around hundred of genes encoding gluten proteins in wheat.14–16 In general, wheat proteins are divided into water/salt-soluble and insoluble categories, of which gliadins and glutenins are insoluble components.17 As gliadins influence the extensibility and viscous nature and glutenins are responsible for the elasticity and strength of dough, gluten is known as the main factor in determining the quality of the baked products and processed foods' texture and flavor.18–20 Due to the high amount of proline (P) and glutamine (Q) residues in gluten, T.B. Osborne, the father of plant protein chemistry, called it “prolamine”.4,11,21 Accordingly, gliadin and glutenin are known as prolamin I and II, respectively.22 Prolamines in other cereals include secalin in rye, hordein in barley, avenins in oats, zeins in corn, but the medical use of the term “gluten” has evolved to include only those prolamines implicated in human disease.23
Despite the numerous benefits of wheat, gluten proteins have the ability of eliciting the pathogenic immune responses and hypersensitivity reactions in susceptible individuals known as “gluten-related disorders (GRDs)”.4,23,24 GRDs encompasses three major types of diseases: celiac disease (CD), wheat allergy (WA) and non-celiac gluten sensitivity (NCGS) that affect around 1–7% of people worldwide.4,25–27 These are, biologically, different diseases with distinct immune, allergic, and possibly non-immune etiologies for which gluten or wheat flour is a common triggering factor that will be discussed in detail below.28
Nevertheless, numerous studies, based on findings showing that avenins alone do not induce immune responses in most CD patients and symptomatically tolerated by them, point to the safety of adding oats to their diet.29–32 In this regard, however, Hardy et al33 in their in vivo study on 73 biopsy-confirmed HLA-DQ2.5+ CD patients showed that the ingestion of oats (100 g/day) for 3 days mobilizes polyclonal avenin-specific T-cells in blood in fewer than 10% of studied patients. Half of the patients had at least one digestive symptom during this challenge, which was due to a high daily intake of oats (100 g) and a high amount of fiber in them. Moreover, they reported that these T-cells were cross-reactive against avenin and hordein, and oral challenge with barley (and not wheat or rye) could stimulate these T-cells more efficiently than oats. They concluded that daily consumption of up to 100 g uncontaminated oats is insufficient to cause clinical relapse in CD patients.33 A plausible explanation for oats having low immune-toxicity is their low proline content and lack of proteolytically resistant peptides with more than 10 amino acid residues.33,34
This review aims to provide a thorough overview of genetic characteristics of gluten protein components, their role in the incidence of various gluten-associated diseases, and genetic modifications that could reduce the immunogenic properties of gluten and lead to wheat improvement.
Method
In general, searches are developed in PubMed, Google Scholar, MEDLINE, and SCOPUS databases from September 1987 to September 2020. The following terms, alone or in combination, were searched: “gluten content”, “gliadin chromosomal locations”, “glutenin chromosomal locations”, “celiac immunogenic peptides”, “wheat allergy immunogenic peptides”, “immunogenic peptides and non-celiac gluten sensitivity”, “toxic gluten”, and “gluten genetic manipulations”.
Gliadin Components and Genetic Characteristics
Gliadin is a combination of monomeric proteins that makes up about 30% of total flour proteins.4,35,36 Polyacrylamide gel electrophoresis at acidic pH (pH = 3.1) shows four major groups called α- (25–35 kDa), β- (30–35 kDa), γ- (35–40 kDa), and ω- (55–75 kDa) gliadins.2,4,8,37 As α- and β-gliadins have several similarities in their structure and number of amino acid residues, they are usually grouped and collectively named as α-gliadins.8,38 α/β- and γ-subunits are considered to be the major components of gliadins, and ω-gliadin is lower.39 In addition, the ω- gliadin differs in amino acid composition from those of the α- and γ-gliadins.4,11 Moreover, according to the Shewry classification based on the presence of sulfur-containing amino acids, gliadin subunits are divided into S-rich (α/β- and γ-) and S-poor (ω-) gliadins.11
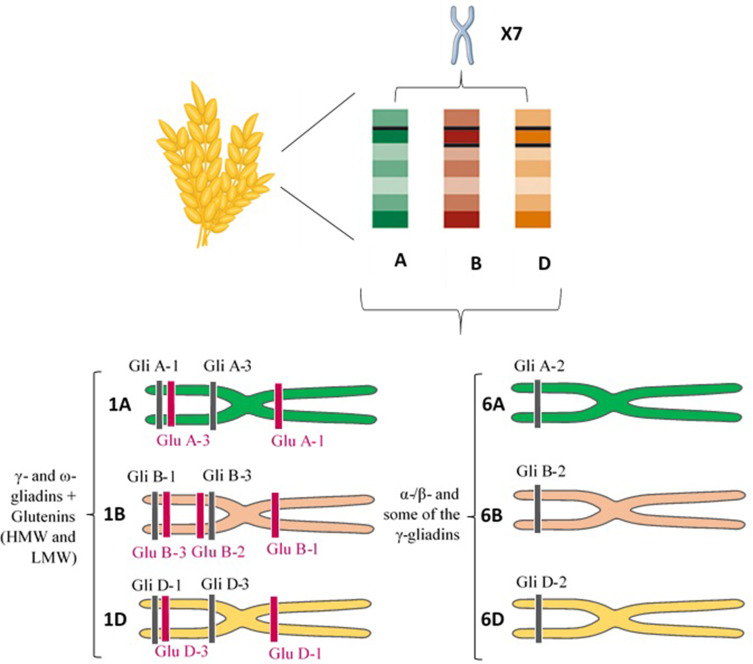
The gliadin is encoded by multigene families.40 Different reports on the chromosomal location showed that gliadin encoding genes are found on the short arm of the homoeologous group 1 (Gli-A1, -B1 and -D1 loci) and 6 (Gli-A2, -B2 and -D2 loci) chromosomes.2 Each of these loci contains multiple alleles and so far, more than 30 allelic variants have been identified for some Gli loci.2,37 Gli-1 genes encode γ - and ω-gliadins and Gli-2 genes encode the α-/β- and some of the γ-gliadins (Figure 1).34 There are also some minor gliadin loci located on 1AS (Gli-A3, -A5 and -A6), 1BS (Gli-B3 and -B5) and 1DS (Gli-D4 and -D6).41
Figure 1.
Chromosome site of different gluten constituents. Short arm of the homoeologous group 1 (Gli-A1, -B1 and -D1 loci) and 6 (Gli-A2, -B2 and -D2 loci) chromosomes encode γ - and ω-gliadins, and α-/β- and some of the γ-gliadins, respectively. Glu-A1, B1, and D1 loci (long arm) and Glu-A3, Glu-B3, and Glu-D3 loci (short arm) of the homoeologous group 1 chromosomes also encode the HMW-GS and LMW-GS subunits of glutenin, respectively.
Abbreviations: HMW, high molecular weight; LMW, low molecular weight.
α-gliadins are the most abundant storage proteins in cereal and several gene copy numbers (from 25 to 150) have been reported for them in haploid genome.4,42–45 The α-gliadin genes originated from the D sub-genome of wheat and contribute the most immunogenic T-cell stimulatory peptides in wheat gluten for the 90% of CD patients who are positive for the HLA-DQ2.5 genotype.42,46,47 However, the base substitution of glutamine codon (CAA) to a stop codon (TAA) can potentially cause inactivation of almost 50% of the α-gliadin genes.20
Glutenin Components and Genetic Characteristics
The glutenin, which represents 50% of total flour proteins, consists of huge polymeric proteins linked through inter- and intramolecular disulfide bonds and are among the largest protein molecules in nature.2,14,36,48,49 Its separation by Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) shows high and low molecular weight (HMW-GS vs LMW-GS) subunits, which are 75 to 120 kDa and 30 to 74 kDa, respectively.4,36,50 LMW-GS accounts for ~60% of the glutenins and has a greater and more favorable impact on the properties of the dough than HMW-GS.51,52 HMW-GS is grouped into x- and y-type subunits based on its electrophoretic mobility and molecular mass.4,53 LMW-GS, which is similar to γ- gliadins in size and structure, is subdivided into B-, C-, and D-type subunits (differ from A, B and D genomes of wheat) basis on their isoelectric point (PI) and electrophoretic mobility (B-type is the major group).36,52 The LMW-GS can also be classified based on the first N-terminal amino acid residue into m-type (Methionine), s-type (Serine) and i-type (Isoleucine) subclasses.4,52
The HMW-GS and LMW-GS genes are located on the Glu-A1, B1, and D1 loci (long arm) and Glu-A3, Glu-B3, and Glu-D3 loci (short arm) of the homoeologous group 1 chromosomes, respectively (Figure 1).52,54 The Glu-1 loci have multiple alleles and each of these loci includes two genes related to x- and y-type subunits.54 The genes encoding LMW-GS are more complex and each Glu-3 loci contain several genes and each gene has two or more alleles.55,56 Genomic studies revealed that Glu-3 loci are linked to Gli-1 loci, and Gli-1 loci encode LMW-GS in addition to γ - and ω-gliadin genes.4 Accordingly, the C and D subunits of LMW-GS are very similar in sequence to α-/γ- and ω-gliadins, respectively.36 LMW-GS and HMW-GS contribute peptides that are immunogenic in CD patients who carry the less common HLA-DQ2.2 and DQ8 genotypes.57
Gluten Protein and the Pathogenesis of Various GRDs
Celiac Disease (CD)
Studies reported that CD has a prevalence of approximately 1–3% in the general population worldwide especially in Western societies.58,59 CD is a gluten-induced immune-mediated inflammatory disorder of the small intestine caused by an intolerance to dietary gluten. CD is limited to genetically predisposed individuals who carry HLA-DQ2.5, HLA-DQ8, HLA-DQ2.2 and/or rarely HLA-DQ7 haplotypes located on the short arm of chromosome 6.60–64 The results of several Genome-wide association studies (GWAS), for example, most recently in a prospective study of 6010 children that carried HLA genotypes associated with increased risk of type-1 diabetes and CD, have also reported the role of non-HLA genes in CD presentation.65 Other immunologic and environmental factors are also involved in the development of CD.66 For instance, Caminero et al57 demonstrated that opportunistic bacterial pathogens (such as P. aeruginosa) in duodenal biopsies from active CD patients could increase mucosal injury caused by immunogenic gluten-derived peptides in a mouse model through protease production and protease-activated receptor-2 (PAR-2) signaling.57
In general, the most immunogenic wheat gluten peptides in CD are derived from α-gliadins (Figure 2).47,67 Some repetitive sequences include two or more overlapping immunodominant epitopes that bind to HLA-DQ2.5, while others include single copies of epitopes that bind HLA-DQ8 or HLA-DQ2.2 and stimulate effector memory CD4+ T cells.68 According to standardized nomenclature, the most immunogenic fragment of α-gliadin for patients positive for HLA-DQ2.5 encompasses multiple copies of the overlapping DQ2.5-glia-α1a, DQ2.5-glia-α1b, and DQ2.5-glia-α2 epitopes.68,69 Wheat α-gliadin also includes the subdominant DQ2.5-glia-α3 epitope, DQ8-glia-α1, DQ2.2-glia-α1 and the DQ2.2-glia-α2 epitopes, which are relevant in patients who are positive for HLA-DQ8, and/or HLA-DQ2.2.40,68,70,71 Ozuna et al46 studied six distinct types of α-gliadins in diploid and polyploid wheats through next-generation sequencing and Sanger sequencing. They found that α-gliadin sequences differed significantly in their frequencies and in the existence and abundance of CD immunogenic peptides. Their findings may help reduce the risk of CD incidence by the breeding/selection of wheat with low stimulatory properties.46 Wheat ω-gliadin, however, includes two overlapping immunodominant epitopes, DQ2.5-glia-ω1 and DQ2.5-glia-ω2, that resemble DQ2.5-glia-α1a and DQ2.5-glia-α2, but stimulate a distinct population of CD4+ T cells and appear to be responsible for many cross-reactive CD4+ T cells activated by wheat, barley and rye.47,72
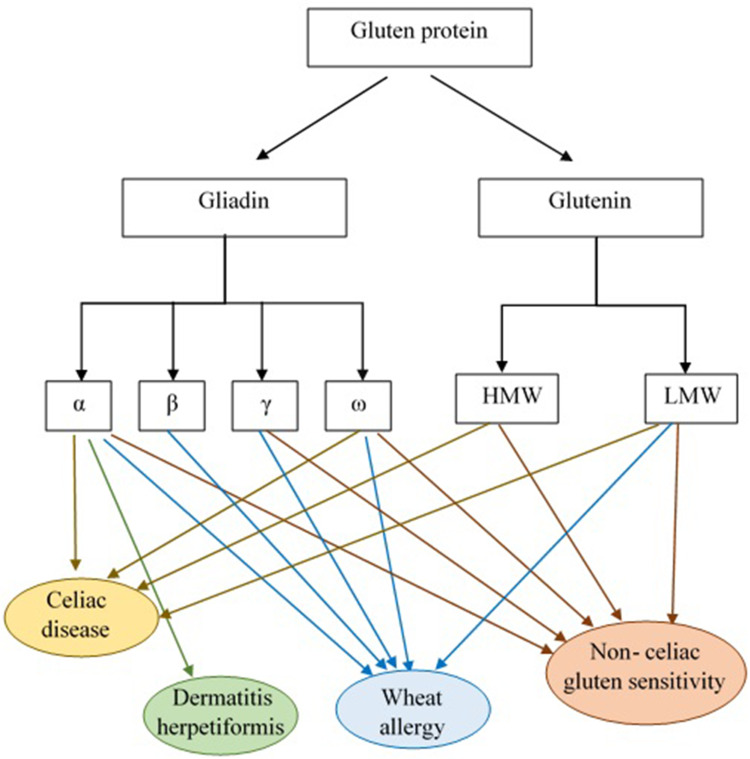
Figure 2.
Gluten protein components and the role of its subgroups in GRDs pathogenesis. According to the results of studies α and γ-gliadins and glutenin are considered to be CD pathogenic responses eliciting factors. The α-gliadin fraction is also reported as DH immunological response triggering agent. Allergic reactivity to the α-/β-, γ- and ω- gliadin fractions and LMW-GS was observed in WA patients. Moreover, patients with NCGS revealed high levels of IgG antibodies against α-, γ- and ω-gliadin and glutenin.
Abbreviations: HMW, high molecular weight; LMW, low molecular weight.
Gluten proteins are highly resistant to human digestive proteases (due to their high content of proline) and do not fully degrade during gastric and pancreatic digestion.34,69 Two peptides that have attracted most attention and remain intact in the digestive process are the 33-mer (p55–87) and the 25-mer (p31–55) located in the α-gliadins encoded by the Gli-D2 locus on chromosome 6D.73,74 Of these two peptides, the 33-mer is the most digestion-resistant peptide with high immunogenic properties (contains DQ2.5-glia-α1a, DQ2.5-glia-α1b, and DQ2.5-glia-α2 epitopes).75,76 During or after absorption of partially digested gliadin peptides into the lamina propria, specific glutamine residues of the 33-mer peptide are susceptible to pH-dependent transamidation (covalent cross-linking to free amines, for example, lysine residues in other proteins) or direct deamidation to glutamate through the action of extracellular tissue transglutaminase (tTG) expressed in inflamed host tissues.77–79 Transamidation abolishes the immunotoxicity of gluten epitopes,80 but direct deamidation enhances their affinity for HLA-DQ2.5 and is essential for their immunogenicity.80–82 tTG-affected peptides are efficiently presented to CD4+ T-cells by the HLA-DQ molecules implicated in CD susceptibility. This results in gluten-specific CD4+ T cell activation with the secretion of pro-inflammatory cytokines like interleukin-2 (IL-2), IL-21 and interferon-gamma (IFNγ), antigen-non-specific activation of local cytotoxic CD8+ T cells,83,84 and enterocyte injury and apoptosis, which ultimately contribute to the characteristic mucosal lesions and local inflammation associated with active CD. Gluten-stimulated CD4+ T cells also provide help for gliadin and tTG-specific B cells, and support antibody production by specific plasma cells.84–87 Glutenin peptides are also implicated in T-cell responses.87 In contrast, the in vitro effects of the α-gliadin 25-mer peptide include induction of IL-15 production from enterocytes and dendritic cells, and innate immune activation.88 IL-15 promotes induction of inflammatory Th1 cell responses and also activation of cytotoxic CD8+ IELs leads to the development of the intestinal lesions.89,90 The contribution of IL-15 to mucosal injury facilitated by induction of CD4+ T-cell immunity to gluten has been supported by a recently reported HLA-DQ8-expressing mouse model with overproduction of IL-15 in the gut epithelium and lamina propria, which develop gluten-dependent small intestinal villous atrophy mimicking human CD.91
Tye-din et al47 in their follow-up study on HLA-DQ2.5+ CD patients screened for T cell–stimulatory gluten peptides in blood following a brief oral challenge with wheat, barley, and rye. They showed that α-gliadin 33-mer epitopes (DQ2-α-I and DQ2-α-II) are immunodominant only after the wheat challenge, while ω-gliadin/C-hordein–derived sequences encompassing DQ2-ω1/ω2 were the dominant T cell–stimulatory peptides in response to consumption of any of these cereals. Hence, they considered ω-gliadin/C-hordein–derived peptides as common T cell–stimulatory peptides in HLA-DQ2.5–associated CD patients.47
Nutrient malabsorption results from mucosal injury marked by villous blunting, crypt hyperplasia, increased intraepithelial lymphocytes (IELs) infiltrate, and immune-mediated enteropathy along with.92 Gluten-induced systemic inflammation in CD is characterized by the presence of intestinal and/or extra-intestinal manifestations or it can even be completely asymptomatic.59
Dermatitis herpetiformis (DH) is one of the extra-intestinal presentations of CD that is accompanied by the development of papulovesicular pruritic skin rash on the extensor aspects of the limbs, sacral region, and buttocks.59,93 As reported in previous studies, topical or intradermal use of gluten protein does not lead to DH formation and the incidence of this disorder is related to intestinal contact with gluten.93 In fact, anti-tTG antibodies, made in response to gluten consumption, interact with the epidermal transglutaminase (ETG) enzyme and cause DH symptoms.59 Allardyce and Shearman94 in their study reported that cellular immune reactivity to the α-gliadin fraction was also observed in DH patients. Moreover, Huff et al95 reported high levels of alpha-gliadin-specific antibodies in patients with DH (Figure 2). Therefore, more studies are required to evaluate the exact pathogenic fractions of gluten in DH.
Wheat Allergy (WA)
Wheat is one of the most common allergens and wheat allergy (WA) results from immunological adverse reactions to wheat ingredients, including water-soluble (albumin and globulin) and insoluble (glutenin and gliadin) proteins.56,96–99 In fact, skin contact, inhalation or ingestion of wheat can lead to the occurrence of these allergic reactions.97 Wheat-dependent exercise-induced anaphylaxis (WDEIA), where symptoms result from the ingestion of wheat in combination with physical exercise, and baker’s asthma, that caused by inhalation of wheat flour, are classified as two commonly WA.97,100 With a higher rate of reports in pediatrics, the prevalence of this disorder is reported to be between 0.5% and 1% in the world.59,98
Scientific reports have considered wheat ω-5 gliadin (fast ω-gliadin, Tri a 19), encoded by the Gli-1 locus on chromosome 1B, as the major allergen part of gluten protein for various types of WA especially WDEIA (also known as ω5-gliadin allergy) (Figure 2).101,102 Morita et al103 reported that fast ω-gliadin is the main allergen for Japanese WDEIA patients. Moreover, results of the study conducted by Palosuo et al104 showed that γ-70 and γ-35 secalins in rye and γ-3 hordein in barley that have structural homology and cross-reactivity with ω-5 gliadin could bind to IgE antibodies and elicit symptoms in WDEIA patients. Since WDEIA diagnosis is significantly delayed, Kennard et al105 recommended the use of ω-5 gliadin-specific IgE testing for patients with unexplained anaphylaxis.104 Moreover, according to Sandiford et al,106 α- and fast ω-gliadin are also associated with baker’s asthma allergic reactions (Figure 2).105 Positive IgE responses to several other wheat grain proteins, such as α/β- (Tri a 21), γ-gliadins, low molecular weight (LMW) glutenin, and α-amylase/trypsin inhibitors (ATIs) are also reported (Figure 2).98 For instance, Baar et al,107 using a molecular discovery approach, found that Tri a 36, which belongs to the LMW-GSs (GluB3-23), is a wheat food allergen with IgE-reactive sequences.56,107
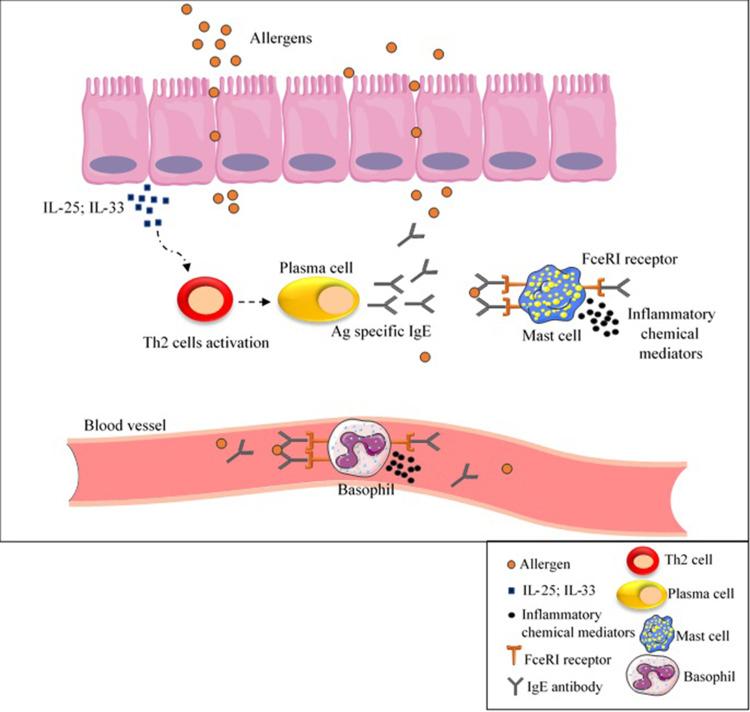
Following contact with allergens, secretion of IL-25 or IL-33 from epithelial cells leads to T helper type 2 (Th2) cell response activation and subsequently IgE antibodies production by B-cells.108 This secreted IgE antibody bound to FceRI receptor on mast cells and basophils as well as to specific epitopes in wheat allergens resulting in the release of inflammatory chemical mediators such as histamine and platelet activator factor (PAF) (Figure 3).25 As a result, allergic responses (such as itching, eczema, rhinitis, nausea) can be life-threatening and cause anaphylactic shock in some cases.25
Figure 3.
IgE- mediated wheat allergy. As a result of contact with allergens, IL-25 or IL-33 are secreted from epithelial cells, cause Th2 cell response activation and subsequently IgE antibodies production by B-cells. Inflammatory chemical mediators are released as a result of IgE antibody binding to FceRI receptor on mast cells and basophils as well as to specific epitopes in wheat allergens, causing allergic reactions.
Abbreviations: Ag, antigen; IgE, immunoglobulin E; IL, interleukin; Th2, T helper type 2.
Non-Celiac Gluten Sensitivity (NCGS)
Non-celiac gluten sensitivity (NCGS) or gluten sensitivity, which remains ill-defined, is a condition resulting from reactions to gluten-containing grains without IgE-mediated or T-cell-mediated responses.59,109–111 Recent studies found that different non-gluten components of wheat flour such as ATIs, Fermentable Oligosaccharides, Disaccharides, Monosaccharides, and Polyols (FODMAPs) causing irritable bowel syndrome might also contribute to NCGS.59,112
The precise pathogenesis of NCGS is still obscure; nevertheless, it has been reported that activation of the innate immune system could have a role in some patients with this condition.113 In this regard, a decrease in T helper cell numbers and a reduction in regulatory T cell clones expansion and their cytokines production have been reported in mucosal biopsy specimens of NCGS patients.114,115 Some early studies reported that HLA-DQ2 and/or -DQ8 genotypes could be over-expressed in NCGS, although these molecules are found in only around 50% of NCGS patients, which is not dissimilar to the general population.56 NCGS is a widespread disorder with an estimated prevalence of 0.5% to 13%, which is more frequent in adult females.116
Given the lack of a definitive test to diagnose NCGS, which in some cases may be confused with CD or WA, several research studies focus on finding NCGS-specific serum biomarkers.117,118 Tye-Din et al119 in their study on CD and self-reported gluten sensitive (SR-GS) patients found that gluten challenge significantly increased IL-2, IL-8 and IL-10 serum levels in CD but not SR-GS patients (both groups had completely eliminated gluten from their diet before participating in the challenge). They concluded that cytokine assessment after acute gluten challenge could be used for distinguishing CD from SR-GS.119 Uhde et al120 also reported a significant increase in anti-gliadin IgG1 and IgG3, and IgG2 and IgG4 subclasses in CD and NCGS patients, respectively. There was also a correlation between the IgG4 and IgG3 antibodies and serum concentration of Fatty acid-binding protein 2 (FABP2), which is an intestinal cell damage marker. They proposed that these components might be additional biomarkers to differentiate CD and NCGS.120 The only presented case report by Vojdani and Perlmutter of a patient with NCGS and autoimmunity revealed high levels of IgG antibodies against α-gliadin 33 and 17 mer, γ- and ω-gliadin and glutenin (Figure 2).121
The chronic symptoms ascribed to NCGS are similar to those of untreated CD with a wide range of intestinal and extra-intestinal presentation as highlighted in Salerno expert criteria.4,117 In contrast, the double-blind sham-controlled gluten challenge that is low in FODMAPs does not generally induce measurable symptoms in patients self-reporting NCGS, whereas patients with treated CD typically experience acute upper gastrointestinal symptoms and show elevations in serum IL-2 within 2 hours.122,123
Wheat Genome Editing
Currently removing gluten from the diet is the only remedy to improve the symptoms of people prone to GRDs.124 However, the addition of gluten to numerous food products and the high cost of gluten-free foodstuffs have made it difficult to strictly adherence to this diet.15,27,125 As a result, several genetic studies have focused on breeding wheat with low immunological properties and preserved baking quality through biotechnological approaches. Numerous scientists believe that gene editing would be a definitive solution for GRDs; however, due to the variety of causal agents, it is not so easy to solve them all in this way126–128 Biotechnological approaches are used for precise and organized modifying of specific genomic sequences through their different functions such as gene replacement, targeted gene knock-out and knock-in, etc.129,130 Vasil et al131 were the first group to successfully produce transgenic wheat plants in 1992 through the Bar gene transferring by biolistic particle bombardment method. The Bar gene encodes phosphinothricin acetyltransferase (PAT) enzyme, which is the cause of herbicide tolerance of plants.131 RNA interference (RNAi) and CRISPR/Cas9 are the two recent biotechnology methods used in this regard.126
RNA interference (RNAi) is a post-transcriptional process present in almost all eukaryotic organisms and regulates the expression of protein-coding genes in a sequence-specific manner, which is capable of engineering novel phenotypes.132,133 In fact, RNAi suppresses protein synthesis by using short double-stranded RNA (dsRNA) complementary to target mRNA and degrading that (silencing of the gene).134 It has been shown that this method is very efficient in regulating gene expression in numerous plant systems.135 Gil-Humanes et al136 used this method to produce breads with up to 97% lower gliadin content (near gliadin-free). The results of their study showed that these reduced-gliadin breads had lower immunotoxicity compared to wild types, while physically no difference was observed between them. Additionally, the removal of gliadin leads to an increase in the number of lysine amino acids (due to the increase in glutenin content which contains more lysine residues) that increases the nutritional value of these breads.136 Altenbach et al137 suppressed the expression of ω-5 gliadins (as an important food allergen) in the US wheat cv Butte 86 using RNA interference technique. The results of their study showed that removing ω-5 gliadins from wheat did not affect flour functionality and had no effect on the expression of other grain proteins. Conversely, the removal of this part improved the dough properties and increased protein stability, indicates the negative role of ω-5 gliadins in flour quality.137 In comparison, Altenbach et al138 in their recent study used the same method to silence a subset of alpha-gliadin genes (containing CD epitopes) of wheat flour from the US spring wheat cultivar Butte 86. Analysing reactivities of IgG and IgA antibodies from patients with CD showed a significantly reduced immunoreactivity of the flour. However, their results showed a decrease in functional properties and dough strength in the transgenic lines. They proposed that the simultaneous removal of alpha and omega gliadins from wheat could be a more efficient approach in this regard.138 In a comprehensive study, Barro et al139 reported the effectiveness of seven RNAi containing plasmids with the ability to target α-, γ-, ω-gliadins, and LMW glutenin subunits in breeding non-toxic wheat variants without any CD epitopes.139 Targeted gene knockdown by RNAi is a fast, low-cost and easy-to-perform method, however, while effective, it provides only transitory inhibition of gene function and may also have unpredictable effects on target genes leads to limited use of this method.5,140
There are some editing genome tools based on the effect of site-specific DNA-binding domain and the use of engineered nucleases, that can identify and edit a particular DNA sequence, including zinc-finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs).5 The clustered regularly interspersed short palindromic repeats (CRISPR)/Cas system, especially CRISPR-associated protein 9 (CRISPR/Cas9), is a widely used prokaryotic nuclease-based target gene precise editing tool, which known as an effective alternative to ZFNs and TALENs.5,141,142 CRISPR/Cas9, identified as the most popular genetic engineering technique, causes genome modifications by delivering to plant cells and expressing there.129,142,143 In this method, the single guide RNA (sgRNA) directs the caspase to the target DNA sites and helps breeding low immunogenic epitopes containing plants with greater specificity.144 The first successful use of CRISPR-Cas9 system was to knock out TaMLO gene (Mildew-resistance locus O) in wheat protoplasts which leads to improved disease resistance that brings the importance of the CRISPR/Cas9 system to promote important traits.145 Sánchez-León et al,146 using two single guide RNAs (sgAlpha-1 and sgAlpha-2) targeted coding sequence for α-gliadin genes, show that CRISPR/Cas9 technology could be used for providing wheat lines with reduced immunoreactivity.145 Jouanin et al147 in their pilot study reported the efficacy of CRISPR/Cas9, using six sgRNA sequences, in mutating α- and γ-gliadin gene copies and preventing them from triggering the human immune system.147
The Court of Justice of the European Union (CJEU) in July 2018 considered any crop with altered genetic material caused by new plant breeding techniques (unnatural changes) as genetically modified organisms (GMOs), which are subjects within the scope of EU law and can be used. This judgment, although supported by some, has also provoked criticism, which led to the formation of the new European Commission that may result in the EU’s GMO legislation change.148
Conclusion
As lifelong adherence to a gluten-free diet remains a challenge for GRDs patients, it seems that increasing attention to the immunogenetic properties of gluten constituents is an essential element in improving the condition of patients. In this regard, technologies have been designed that can reduce the immunogenicity properties of gluten by promising a genomic editing approach. Although these techniques mostly work precisely on the target gene, it is important to note that changes in the expression of one gene how affect the expression of other genes? It can be said that one of the reasons for not including these manipulated products in patients’ diets is the lack of a clear answer to this question. Therefore, it is suggested that future studies in this regard consider all genome-editing results (either genetic or metabolic changes) to ensure the created product compatibility with food safety conditions.
Funding Statement
There is no funding to report.
Abbreviations
ATIs, Alpha-amylase/Trypsin Inhibitors; CD, Celiac Disease; CRISPR, Clustered Regularly Interspersed Short Palindromic Repeats; DH, Dermatitis herpetiformis; ETG, Epidermal Transglutaminase; FABP2, Fatty Acid-Binding Protein 2; FODMAPs, Fermentable Oligosaccharides, Disaccharides, Monosaccharides, and Polyols; g, Gram; GWASs, Genome-Wide Association Studies; HMW, High Molecular Weight; IELs, Intraepithelial Lymphocytes; IgE, Immunoglobulin E; IL, Interleukin; kDa, Kilodalton; LMW, Low Molecular Weight; NCGS, Non-Celiac Gluten Sensitivity; P, Proline; PAF, Platelet Activator Factor; PAT, Phosphinothricin Acetyltransferase; PI, Isoelectric Point; Q, Qlutamine; RNAi, RNA interference; sgRNA, single guide RNA; SR-GS, Self-Reported Gluten Sensitive; TALENs, Transcription Activator-Like Effectors Nucleases; Th2, T helper type 2; tTG, Tissue Transglutaminase; WA, Wheat Allergy; WDEIA, Wheat-Dependent Exercise-Induced Anaphylaxis; ZFNs, Zinc-Finger Nucleases.
Author Contributions
All authors made a significant contribution to the work reported in all of the following areas: took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
Dr Robert P Anderson reports personal fees from consultancies for GSK (Wilmington, DE, USA; since 2019), Allero Therapeutics BV (Rotterdam, Netherlands; since 2020), Takeda Pharmaceutical Company Ltd/Millennium Pharmaceuticals (Cambridge MA USA; since 2019), Kanyos Bio Inc (Cambridge MA USA; since 2019), Atheneum Partners GmbH (Berlin, Germany; since 2019), being an ImmusanT Inc, CSO, employee until September 2019, principal and founder of Specific Pharma Consulting, and founder and part-owner of Novoviah Pharmaceuticals Pty Ltd, outside the submitted work; in addition, Dr Robert P Anderson has multiple patents licensed to ImmusanT (patents lodged from 2009 to 2019 relating diagnosis, and treatment of coeliac disease) and BTG International (patents lodged from 2009 to 2019 relating diagnosis, treatment, and modification of gluten for coeliac disease). The authors declare that they have no other potential conflicts of interest for this work.
References
- 1.Shiferaw B, Smale M, Braun H-J, et al. Crops that feed the world 10. Past successes and future challenges to the role played by wheat in global food security. Food Secur. 2013;5(3):291–317. doi: 10.1007/s12571-013-0263-y [DOI] [Google Scholar]
- 2.Utebayev M, Dashkevich S, Bome N, et al. Genetic diversity of gliadin-coding alleles in bread wheat (Triticum aestivum L.) from Northern Kazakhstan. PeerJ. 2019;7:e7082. doi: 10.7717/peerj.7082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bell G. The History of Wheat Cultivation. Wheat Breeding. Springer; 1987:31–49. [Google Scholar]
- 4.Wang D, Li F, Cao S, Zhang K. Genomic and functional genomics analyses of gluten proteins and prospect for simultaneous improvement of end-use and health-related traits in wheat. Theor Appl Genet. 2020;133(5):1521–1539. doi: 10.1007/s00122-020-03557-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liaqat N, Liaqat A, Ali M, et al. Chapter 24 - Wheat genomics and genome editing In: Ozturk M, Gul A, editors. Climate Change and Food Security with Emphasis on Wheat. Academic Press; 2020:331–346. [Google Scholar]
- 6.Rostami K, Malekzadeh R, Shahbazkhani B, et al. Coeliac disease in Middle Eastern countries: a challenge for the evolutionary history of this complex disorder? Dig Liver Dis. 2004;36(10):694–697. doi: 10.1016/j.dld.2004.05.010 [DOI] [PubMed] [Google Scholar]
- 7.Huo N, Zhu T, Altenbach S, et al. Dynamic evolution of α-gliadin prolamin gene family in homeologous genomes of hexaploid wheat. Sci Rep. 2018;8(1):5181. doi: 10.1038/s41598-018-23570-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Metakovsky E, Melnik V, Rodriguez-Quijano M, et al. A catalog of gliadin alleles: polymorphism of 20th-century common wheat germplasm. Crop J. 2018;6(6):628–641. doi: 10.1016/j.cj.2018.02.003 [DOI] [Google Scholar]
- 9.Altenbach SB, Kothari KM, Lieu D. Environmental conditions during wheat grain development alter temporal regulation of major gluten protein genes. Cereal Chem J. 2002;79(2):279–285. doi: 10.1094/CCHEM.2002.79.2.279 [DOI] [Google Scholar]
- 10.Shewry PR, Hey SJ. The contribution of wheat to human diet and health. Food Energy Secur. 2015;4(3):178–202. doi: 10.1002/fes3.64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shewry P. What is gluten-why is it special? Front Nutr. 2019;6:101. doi: 10.3389/fnut.2019.00101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Islam S, Yu Z, She M, et al. Wheat gluten protein and its impacts on wheat processing quality. Front Agric Sci Eng. 2019;6(3):279–287. doi: 10.15302/J-FASE-2019267 [DOI] [Google Scholar]
- 13.Kumar P, Yadava R, Gollen B, et al. Nutritional contents and medicinal properties of wheat: a review. Life Sci Med Res. 2011;22:1–10. [Google Scholar]
- 14.Wieser H. Chemistry of gluten proteins. Food Microbiol. 2007;24:115–119. doi: 10.1016/j.fm.2006.07.004 [DOI] [PubMed] [Google Scholar]
- 15.Jouanin A, Boyd L, Visser RGF, Smulders MJM. Development of wheat with hypoimmunogenic gluten obstructed by the gene editing policy in Europe. Front Plant Sci. 2018;9:1523. doi: 10.3389/fpls.2018.01523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Freeman HJ. Celiac Disease☆. Reference Module in Biomedical Sciences. Elsevier; 2017. [Google Scholar]
- 17.Pasha I, Saeed F, Sultan MT, et al. Wheat allergy and intolerance; recent updates and perspectives. Crit Rev Food Sci Nutr. 2016;56(1):13–24. doi: 10.1080/10408398.2012.659818 [DOI] [PubMed] [Google Scholar]
- 18.Barak S, Mudgil D, Khatkar BS. Influence of gliadin and glutenin fractions on rheological, pasting, and textural properties of dough. Int J Food Prop. 2014;17(7):1428–1438. doi: 10.1080/10942912.2012.717154 [DOI] [Google Scholar]
- 19.Biesiekierski JR. What is gluten? J Gastroenterol Hepatol. 2017;32(S1):78–81. doi: 10.1111/jgh.13703 [DOI] [PubMed] [Google Scholar]
- 20.Kajendran K, Chandrasekharan NV, Hettiarachchi CM, Sulochana Wijesundera WS. Molecular characterization and expression of α-gliadin genes from wheat cultivar Dacke in Bg 250 rice variety. GM Crops Food. 2019;10(2):102–114. doi: 10.1080/21645698.2019.1622990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tian N, Leffler DA, Kelly CP, et al. Despite sequence homologies to gluten, salivary proline-rich proteins do not elicit immune responses central to the pathogenesis of celiac disease. Am J Physiol Gastrointest Liver Physiol. 2015;309(11):G910–G917. doi: 10.1152/ajpgi.00157.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar J, Kumar M, Pandey R, Chauhan NS. Physiopathology and management of gluten-induced celiac disease. J Food Sci. 2017;82(2):270–277. doi: 10.1111/1750-3841.13612 [DOI] [PubMed] [Google Scholar]
- 23.Phiarais BPN, Arendt EK. 15 - Malting and brewing with gluten-free cereals In: Arendt EK, Dal Bello F, editors. Gluten-Free Cereal Products and Beverages. San Diego: Academic Press; 2008:347–372. [Google Scholar]
- 24.Sharma N, Bhatia S, Chunduri V, et al. Pathogenesis of celiac disease and other gluten related disorders in wheat and strategies for mitigating them. Front Nutr. 2020;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cabanillas B. Gluten-related disorders: celiac disease, wheat allergy, and nonceliac gluten sensitivity. Crit Rev Food Sci Nutr. 2020;60(15):2606–2621. doi: 10.1080/10408398.2019.1651689 [DOI] [PubMed] [Google Scholar]
- 26.Lammers KM, Herrera MG, Dodero VI. Translational chemistry meets gluten-related disorders. ChemistryOpen. 2018;7(3):217–232. doi: 10.1002/open.201700197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rostami Nejad M, Karkhane M, Marzban A, et al. Gluten related disorders. Gastroenterol Hepatol Bed Bench. 2012;5(Suppl 1):S1–S7. [PMC free article] [PubMed] [Google Scholar]
- 28.Czerwińska K, Czerwiński G, Poniewierka E, et al. Spectrum of gluten-related disorders: celiac disease, wheat allergy, baker’s asthma and non-celiac gluten sensitivity. World Sci News. 2018;100:154–164. [Google Scholar]
- 29.Tanner G, Juhász A, Florides CG, et al. Preparation and characterization of avenin-enriched oat protein by chill precipitation for feeding trials in celiac disease. Front Nutr. 2019;6(162). doi: 10.3389/fnut.2019.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spector Cohen I, Day AS, Shaoul R. To be oats or not to be? An update on the ongoing debate on oats for patients with celiac disease. Front Pediatr. 2019;7:384. doi: 10.3389/fped.2019.00384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hollén E, Holmgren Peterson K, Sundqvist T, et al. Coeliac children on a gluten-free diet with or without oats display equal anti-avenin antibody titres. Scand J Gastroenterol. 2006;41(1):42–47. doi: 10.1080/00365520510023945 [DOI] [PubMed] [Google Scholar]
- 32.Haboubi NY, Taylor S, Jones S. Coeliac disease and oats: a systematic review. Postgrad Med J. 2006;82(972):672–678. doi: 10.1136/pgmj.2006.045443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hardy MY, Tye-Din JA, Stewart JA, et al. Ingestion of oats and barley in patients with celiac disease mobilizes cross-reactive T cells activated by avenin peptides and immuno-dominant hordein peptides. J Autoimmun. 2015;56:56–65. [DOI] [PubMed] [Google Scholar]
- 34.Balakireva AV, Zamyatnin AA. Properties of gluten intolerance: gluten structure, evolution, pathogenicity and detoxification capabilities. Nutrients. 2016;8(10):644. doi: 10.3390/nu8100644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Békés F, Gianibelli MC, Wrigley CW. The gluten proteins of the wheat grain in relation to flour quality In: Wrigley C, Corke H, Seetharaman K, Faubion J, editors. Encyclopedia of Food Grains (Second Edition). Oxford: Academic Press; 2016:375–383. [Google Scholar]
- 36.Shewry PR, Halford NG, Lafiandra D. Genetics of wheat gluten proteins In: Hall JC, Dunlap JC, Friedmann T, editors. Advances in Genetics. Vol. 49 Academic Press; 2003:111–184. [DOI] [PubMed] [Google Scholar]
- 37.Zaefizadeh M, Jamaati-e-Somarin S, Ojaghi J, et al. Genetic diversity for gliadin patterns of durum wheat landraces in the Northwest of Iran and Azerbaijan. Pesquisa Agropecuária Brasileira. 2010;45(12):1425–1432. doi: 10.1590/S0100-204X2010001200013 [DOI] [Google Scholar]
- 38.Wang D-W, Li D, Wang J, et al. Genome-wide analysis of complex wheat gliadins, the dominant carriers of celiac disease epitopes. Sci Rep. 2017;7(1):44609. doi: 10.1038/srep44609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Žilić S. Wheat Gluten: Composition and Health Effects. Nova Science Publishers, lnc. 2013:71–86. [Google Scholar]
- 40.Dubois B, Bertin P, Mingeot D. Molecular diversity of α-gliadin expressed genes in genetically contrasted spelt (Triticum aestivum ssp. spelta) accessions and comparison with bread wheat (T. aestivum ssp. aestivum) and related diploid Triticum and Aegilops species. Mol Breed. 2016;36(11):152. doi: 10.1007/s11032-016-0569-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gianibelli M, Larroque O, Macritchie F, Wrigley C. Biochemical, genetic, and molecular characterization of wheat endosperm proteins. Cereal Chem Rev. 2001;78(6):635–646. doi: 10.1094/CCHEM.2001.78.6.635 [DOI] [Google Scholar]
- 42.Li Y, Xin R, Zhang D, Li S. Molecular characterization of α-gliadin genes from common wheat cultivar Zhengmai 004 and their role in quality and celiac disease. Crop J. 2014;2(1):10–21. doi: 10.1016/j.cj.2013.11.003 [DOI] [Google Scholar]
- 43.Harberd N, Bartels D, Thompson R. Analysis of the gliadin multigene loci in bread wheat using nullisomic-tetrasomic lines. Mol Gen Genet. 1985;198(2):234–242. doi: 10.1007/BF00383001 [DOI] [Google Scholar]
- 44.Anderson O, Litts J, Greene F. The α-gliadin gene family. I. Characterization of ten new wheat α-gliadin genomic clones, evidence for limited sequence conservation of flanking DNA, and southern analysis of the gene family. Theor Appl Genet. 1997;95(1–2):50–58. doi: 10.1007/s001220050531 [DOI] [Google Scholar]
- 45.Okita T, Cheesbrough V, Reeves CD. Evolution and heterogeneity of the alpha-/beta-type and gamma-type gliadin DNA sequences. J Biol Chem. 1985;260(13):8203–8213. doi: 10.1016/S0021-9258(17)39582-0 [DOI] [PubMed] [Google Scholar]
- 46.Ozuna CV, Iehisa JC, Giménez MJ, et al. Diversification of the celiac disease α-gliadin complex in wheat: a 33-mer peptide with six overlapping epitopes, evolved following polyploidization. Plant J. 2015;82(5):794–805. doi: 10.1111/tpj.12851 [DOI] [PubMed] [Google Scholar]
- 47.Tye-Din JA, Stewart JA, Dromey JA, et al. Comprehensive, quantitative mapping of T cell epitopes in gluten in celiac disease. Sci Transl Med. 2010;2(41):41ra51. doi: 10.1126/scitranslmed.3001012 [DOI] [PubMed] [Google Scholar]
- 48.Rasheed F, Plivelic TS, Kuktaite R, et al. Unraveling the structural puzzle of the giant glutenin polymer-an interplay between protein polymerization, nanomorphology, and functional properties in bioplastic films. ACS Omega. 2018;3(5):5584–5592. doi: 10.1021/acsomega.7b02081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fu BX, Sapirstein HD. Procedure for isolating monomeric proteins and polymeric glutenin of wheat flour. Cereal Chem J. 1996;73(1):143–152. [Google Scholar]
- 50.Dangi P, Khatkar BS. Extraction and purification of low molecular weight glutenin subunits using size exclusion chromatography. J Food Sci Technol. 2019;56(2):951–956. doi: 10.1007/s13197-018-03560-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Y, Zhen S, Luo N, et al. Low molecular weight glutenin subunit gene Glu-B3h confers superior dough strength and breadmaking quality in wheat (Triticum aestivum L.). Sci Rep. 2016;6(1):27182. doi: 10.1038/srep27182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu L, Ikeda TM, Branlard G, et al. Comparison of low molecular weight glutenin subunits identified by SDS-PAGE, 2-DE, MALDI-TOF-MS and PCR in common wheat. BMC Plant Biol. 2010;10:124. doi: 10.1186/1471-2229-10-124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang Z, Huang L, Wu B, et al. Characterization of an integrated active Glu-1Ay allele in common wheat from wild emmer and its potential role in flour improvement. Int J Mol Sci. 2018;19(4):923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Anjum FM, Khan MR, Din A, et al. Wheat gluten: high molecular weight glutenin subunits—structure, genetics, and relation to dough elasticity. J Food Sci. 2007;72(3):R56–R63. doi: 10.1111/j.1750-3841.2007.00292.x [DOI] [PubMed] [Google Scholar]
- 55.Wang Z, Li Y, Yang Y, et al. New insight into the function of wheat glutenin proteins as investigated with two series of genetic mutants. Sci Rep. 2017;7(1):3428. doi: 10.1038/s41598-017-03393-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang D, Zhang K, Dong L, et al. Molecular genetic and genomic analysis of wheat milling and end-use traits in China: progress and perspectives. Crop J. 2018;6(1):68–81. doi: 10.1016/j.cj.2017.10.001 [DOI] [Google Scholar]
- 57.Caminero A, McCarville JL, Galipeau HJ, et al. Duodenal bacterial proteolytic activity determines sensitivity to dietary antigen through protease-activated receptor-2. Nat Commun. 2019;10(1):1198. doi: 10.1038/s41467-019-09037-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dubé C, Rostom A, Sy R, et al. The prevalence of celiac disease in average-risk and at-risk Western European populations: a systematic review. Gastroenterology. 2005;128(4 Suppl 1):S57–S67. doi: 10.1053/j.gastro.2005.02.014 [DOI] [PubMed] [Google Scholar]
- 59.Taraghikhah N, Ashtari S, Asri N, et al. An updated overview of spectrum of gluten-related disorders: clinical and diagnostic aspects. BMC Gastroenterol. 2020;20(1):258. doi: 10.1186/s12876-020-01390-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yoosuf S, Makharia GK. Evolving therapy for celiac disease. Front Pediatr. 2019;7:193. doi: 10.3389/fped.2019.00193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Asri N, Rostami-Nejad M. The facts of celiac disease; a comprehensive review. Int J Celiac Dis. 2019;7(2):48–52. [Google Scholar]
- 62.Asri N, Rostami-Nejad M, Barzegar M, et al. Suppressive mechanisms induced by tregs in celiac disease. Iran Biomed J. 2020;24(3):140–147. doi: 10.29252/ibj.24.3.140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kårhus LL, Thuesen BH, Skaaby T, et al. The distribution of HLA DQ2 and DQ8 haplotypes and their association with health indicators in a general Danish population. United European Gastroenterol J. 2018;6(6):866–878. doi: 10.1177/2050640618765506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tye-Din JA, Daveson AJM, Goldstein KE, et al. Patient factors influencing acute gluten reactions and cytokine release in treated coeliac disease. BMC Med. 2020;18(1):362. doi: 10.1186/s12916-020-01828-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sharma A, Liu X, Hadley D, et al. Identification of non-HLA genes associated with celiac disease and Country-specific differences in a large, international pediatric Cohort. PLoS One. 2016;11:e0152476. doi: 10.1371/journal.pone.0152476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sarno M, Discepolo V, Troncone R, Auricchio R. Risk factors for celiac disease. Ital J Pediatr. 2015;41:57. doi: 10.1186/s13052-015-0166-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Henderson KN, Tye-Din JA, Reid HH, et al. A structural and immunological basis for the role of human leukocyte antigen DQ8 in celiac disease. Immunity. 2007;27(1):23–34. doi: 10.1016/j.immuni.2007.05.015 [DOI] [PubMed] [Google Scholar]
- 68.Sollid LM, Tye-Din JA, Qiao SW, et al. Update 2020: nomenclature and listing of celiac disease-relevant gluten epitopes recognized by CD4(+) T cells. Immunogenetics. 2020;72(1–2):85–88. doi: 10.1007/s00251-019-01141-w [DOI] [PubMed] [Google Scholar]
- 69.Shan L, Molberg Ø, Parrot I, et al. Structural basis for gluten intolerance in celiac sprue. Science (New York, NY). 2002;297(5590):2275–2279. doi: 10.1126/science.1074129 [DOI] [PubMed] [Google Scholar]
- 70.Koning F. Adverse effects of wheat gluten. Ann Nutr Metab. 2015;67(Suppl. 2):7–14. doi: 10.1159/000440989 [DOI] [PubMed] [Google Scholar]
- 71.Sollid LM, Qiao S-W, Anderson RP, et al. Nomenclature and listing of celiac disease relevant gluten T-cell epitopes restricted by HLA-DQ molecules. Immunogenetics. 2012;64(6):455–460. doi: 10.1007/s00251-012-0599-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dahal-Koirala S, Risnes LF, Christophersen A, et al. TCR sequencing of single cells reactive to DQ2.5-glia-α2 and DQ2.5-glia-ω2 reveals clonal expansion and epitope-specific V-gene usage. Mucosal Immunol. 2016;9(3):587–596. doi: 10.1038/mi.2015.147 [DOI] [PubMed] [Google Scholar]
- 73.Cebolla Á, Moreno M, Coto L, Sousa C. Gluten Immunogenic peptides as standard for the evaluation of potential harmful prolamin content in food and human specimen. Nutrients. 2018;10(12):1927. doi: 10.3390/nu10121927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li D, Jin H, Zhang K, et al. Analysis of the Gli-D2 locus identifies a genetic target for simultaneously improving the breadmaking and health-related traits of common wheat. Plant J. 2018;95(3):414–426. doi: 10.1111/tpj.13956 [DOI] [PubMed] [Google Scholar]
- 75.Schalk K, Lang C, Wieser H, Koehler P, Scherf KA. Quantitation of the immunodominant 33-mer peptide from α-gliadin in wheat flours by liquid chromatography tandem mass spectrometry. Sci Rep. 2017;7(1):45092. doi: 10.1038/srep45092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhou L, Kooy-Winkelaar YMC, Cordfunke RA, et al. Abrogation of immunogenic properties of gliadin peptides through transamidation by microbial transglutaminase Is Acyl-acceptor dependent. J Agric Food Chem. 2017;65(34):7542–7552. doi: 10.1021/acs.jafc.7b02557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bonciani D, Verdelli A, Bonciolini V, et al. Dermatitis herpetiformis: from the genetics to the development of skin lesions. Clin Dev Immunol. 2012;2012:239691. doi: 10.1155/2012/239691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Skovbjerg H, Koch C, Anthonsen D, Sjöström H. Deamidation and cross-linking of gliadin peptides by transglutaminases and the relation to celiac disease. Biochim Biophys Acta Mol Basis Dis. 2004;1690(3):220–230. doi: 10.1016/j.bbadis.2004.06.009 [DOI] [PubMed] [Google Scholar]
- 79.Quarsten H, Molberg O, Fugger L, McAdam SN, Sollid LM. HLA binding and T cell recognition of a tissue transglutaminase-modified gliadin epitope. Eur J Immunol. 1999;29(8):2506–2514. doi: [DOI] [PubMed] [Google Scholar]
- 80.Anderson RP, Degano P, Godkin AJ, Jewell DP, Hill AV. In vivo antigen challenge in celiac disease identifies a single transglutaminase-modified peptide as the dominant A-gliadin T-cell epitope. Nat Med. 2000;6(3):337–342. doi: 10.1038/73200 [DOI] [PubMed] [Google Scholar]
- 81.Molberg O, McAdam SN, Körner R, et al. Tissue transglutaminase selectively modifies gliadin peptides that are recognized by gut-derived T cells in celiac disease. Nat Med. 1998;4(6):713–717. doi: 10.1038/nm0698-713 [DOI] [PubMed] [Google Scholar]
- 82.Arentz-Hansen H, Körner R, Molberg O, et al. The intestinal T cell response to alpha-gliadin in adult celiac disease is focused on a single deamidated glutamine targeted by tissue transglutaminase. J Exp Med. 2000;191(4):603–612. doi: 10.1084/jem.191.4.603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Goel G, Tye-Din JA, Qiao S-W, et al. Cytokine release and gastrointestinal symptoms after gluten challenge in celiac disease. Sci Adv. 2019;5(8):eaaw7756. doi: 10.1126/sciadv.aaw7756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kooy-Winkelaar YMC, Bouwer D, Janssen GMC, et al. CD4 T-cell cytokines synergize to induce proliferation of malignant and nonmalignant innate intraepithelial lymphocytes. Proc Natl Acad Sci. 2017;114(6):E980–E989. doi: 10.1073/pnas.1620036114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ciccocioppo R, Di Sabatino A, Corazza GR. The immune recognition of gluten in coeliac disease. Clin Exp Immunol. 2005;140(3):408–416. doi: 10.1111/j.1365-2249.2005.02783.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gianfrani C, Auricchio S, Troncone R. Adaptive and innate immune responses in celiac disease. Immunol Lett. 2005;99(2):141–145. doi: 10.1016/j.imlet.2005.02.017 [DOI] [PubMed] [Google Scholar]
- 87.van de Wal Y, Kooy YM, van Veelen P, et al. Glutenin is involved in the gluten-driven mucosal T cell response. Eur J Immunol. 1999;29(10):3133–3139. doi: [DOI] [PubMed] [Google Scholar]
- 88.Barone MV, Troncone R, Auricchio S. Gliadin peptides as triggers of the proliferative and stress/innate immune response of the celiac small intestinal mucosa. Int J Mol Sci. 2014;15(11):20518–20537. doi: 10.3390/ijms151120518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Abadie V, Jabri B. IL-15: a central regulator of celiac disease immunopathology. Immunol Rev. 2014;260(1):221–234. doi: 10.1111/imr.12191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hardy MY, Tye-Din JA. Coeliac disease: a unique model for investigating broken tolerance in autoimmunity. Clin Transl Immunol. 2016;5(11):e112. doi: 10.1038/cti.2016.58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Abadie V, Kim SM, Lejeune T, et al. IL-15, gluten and HLA-DQ8 drive tissue destruction in coeliac disease. Nature. 2020;578(7796):600–604. doi: 10.1038/s41586-020-2003-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dickson BC, Streutker CJ, Chetty R. Coeliac disease: an update for pathologists. J Clin Pathol. 2006;59(10):1008–1016. doi: 10.1136/jcp.2005.035345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Clarindo MV, Possebon AT, Soligo EM, et al. Dermatitis herpetiformis: pathophysiology, clinical presentation, diagnosis and treatment. An Bras Dermatol. 2014;89(6):865–877. doi: 10.1590/abd1806-4841.20142966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Allardyce RA, Shearman DJ. Leukocyte reactivity to alpha-gliadin in dermatitis herpetiformis and adult coeliac disease. Int Arch Allergy Appl Immunol. 1975;48(3):395–400. doi: 10.1159/000231324 [DOI] [PubMed] [Google Scholar]
- 95.Clark Huff J, Weston WL, Zirker DK. Wheat protein antibodies in dermatitis herpetiformis. J Invest Dermatol. 1979;73(6):570–574. doi: 10.1111/1523-1747.ep12541611 [DOI] [PubMed] [Google Scholar]
- 96.Catassi C, Fasano A. 1 - Celiac disease In: Arendt EK, Dal Bello F, editors. Gluten-Free Cereal Products and Beverages. San Diego: Academic Press; 2008:1. [Google Scholar]
- 97.Cianferoni A. Wheat allergy: diagnosis and management. J Asthma Allergy. 2016;9:13. doi: 10.2147/JAA.S81550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ricci G, Andreozzi L, Cipriani F, et al. Wheat allergy in children: a comprehensive update. Medicina. 2019;55(7):400. doi: 10.3390/medicina55070400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Koehler P, Wieser H, Konitzer K. Chapter 1 - Celiac Disease—A Complex Disorder In: Koehler P, Wieser H, Konitzer K, editors. Celiac Disease and Gluten. Boston: Academic Press; 2014:1–96. [Google Scholar]
- 100.Scherf K, Brockow K, Biedermann T, et al. Wheat-dependent exercise-induced anaphylaxis. Clin Exp Allergy. 2015;46. [DOI] [PubMed] [Google Scholar]
- 101.De Santis MA, Giuliani MM, Giuzio L, et al. Differences in gluten protein composition between old and modern durum wheat genotypes in relation to 20th century breeding in Italy. Eur J Agron. 2017;87:19–29. doi: 10.1016/j.eja.2017.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hofmann S, Fischer J, Eriksson C, et al. IgE detection to α/β/γ-gliadin and its clinical relevance in wheat-dependent exercise-induced anaphylaxis. Allergy. 2012;67:1457–1460. doi: 10.1111/all.12020 [DOI] [PubMed] [Google Scholar]
- 103.Morita E, Matsuo H, Mihara S, et al. Fast ω-gliadin is a major allergen in wheat-dependent exercise-induced anaphylaxis. J Dermatol Sci. 2003;33(2):99–104. doi: 10.1016/s0923-1811(03)00156-7 [DOI] [PubMed] [Google Scholar]
- 104.Palosuo K, Alenius H, Varjonen E, Kalkkinen N, Reunala T. Rye γ-70 and γ-35 secalins and barley γ-3 hordein cross-react with ω-5 gliadin, a major allergen in wheat-dependent, exercise-induced anaphylaxis. Clin Exp Allergy. 2001;31(3):466–473. doi: 10.1046/j.1365-2222.2001.01023.x [DOI] [PubMed] [Google Scholar]
- 105.Kennard L, Thomas I, Rutkowski K, et al. A multicenter evaluation of diagnosis and management of omega-5 gliadin allergy (Also known as wheat-dependent exercise-induced anaphylaxis) in 132 adults. J Allergy Clin Immunol Pract. 2018;6(6):1892–1897. doi: 10.1016/j.jaip.2018.02.013 [DOI] [PubMed] [Google Scholar]
- 106.Sandiford CP, Tatham AS, Fido R, et al. Identification of the major water/salt insoluble wheat proteins involved in cereal hypersensitivity. Clin Exp Allergy. 1997;27(10):1120–1129. doi: 10.1111/j.1365-2222.1997.tb01148.x [DOI] [PubMed] [Google Scholar]
- 107.Baar A, Pahr S, Constantin C, et al. Molecular and immunological characterization of Tri a 36, a low molecular weight glutenin, as a novel major wheat food allergen. J Immunol. 2012;189:3018–3025. doi: 10.4049/jimmunol.1200438 [DOI] [PubMed] [Google Scholar]
- 108.Galli SJ, Tsai M. IgE and mast cells in allergic disease. Nat Med. 2012;18(5):693–704. doi: 10.1038/nm.2755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Volta U, Caio G, Tovoli F, De Giorgio R. Non-celiac gluten sensitivity: questions still to be answered despite increasing awareness. Cell Mol Immunol. 2013;10(5):383–392. doi: 10.1038/cmi.2013.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rostami K, Hogg-Kollars S. A patient’s journey. non-coeliac gluten sensitivity. BMJ. 2012;345:e7982. doi: 10.1136/bmj.e7982 [DOI] [PubMed] [Google Scholar]
- 111.Rostami-Nejad M, Lahmi F, Zali M. Non-celiac Gluten Sensitivity. J Army Univ Med Sci. 2013;11:243–251. [Google Scholar]
- 112.Catassi C, Alaedini A, Bojarski C, et al. The overlapping area of non-celiac gluten sensitivity (NCGS) and wheat-sensitive irritable bowel syndrome (IBS): an update. Nutrients. 2017;9(11):1268. doi: 10.3390/nu9111268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ierardi E, Losurdo G, Piscitelli D, et al. Biological markers for non-celiac gluten sensitivity: a question awaiting for a convincing answer. Gastroenterol Hepatol Bed Bench. 2018;11(3):203–208. [PMC free article] [PubMed] [Google Scholar]
- 114.Losurdo G, Piscitelli D, Pezzuto F, et al. T helper lymphocyte and mast cell immunohistochemical pattern in nonceliac gluten sensitivity. Gastroenterol Res Pract. 2017;2017:5023680. doi: 10.1155/2017/5023680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sapone A, Lammers KM, Casolaro V, et al. Divergence of gut permeability and mucosal immune gene expression in two gluten-associated conditions: celiac disease and gluten sensitivity. BMC Med. 2011;9(1):23. doi: 10.1186/1741-7015-9-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Barbaro MR, Cremon C, Stanghellini V, Barbara G. Recent advances in understanding non-celiac gluten sensitivity. F1000Res. 2018;7:F1000Faculty Rev–1631. doi: 10.12688/f1000research.15849.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Catassi C, Elli L, Bonaz B, et al. Diagnosis of non-celiac gluten sensitivity (NCGS): the Salerno experts’ criteria. Nutrients. 2015;7(6):4966–4977. doi: 10.3390/nu7064966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ortiz C, Valenzuela R, Lucero AY. [Celiac disease, non celiac gluten sensitivity and wheat allergy: comparison of 3 different diseases triggered by the same food]. [Spanish]. Rev Chil Pediatr. 2017;88(3):417–423. doi: 10.4067/S0370-41062017000300017 [DOI] [PubMed] [Google Scholar]
- 119.Tye-Din JA, Skodje GI, Sarna VK, et al. Cytokine release after gluten ingestion differentiates coeliac disease from self-reported gluten sensitivity. United European Gastroenterol J. 2020;8(1):108–118. doi: 10.1177/2050640619874173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Uhde M, Caio G, De Giorgio R, et al. Subclass profile of IgG antibody response to gluten differentiates nonceliac gluten sensitivity from celiac disease. Gastroenterology. 2020;159(5):1965–1967.e2. doi: 10.1053/j.gastro.2020.07.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Vojdani A, Perlmutter D. Differentiation between celiac disease, nonceliac gluten sensitivity, and their overlapping with Crohn’s disease: a case series. Case Rep Immunol. 2013;2013:248482. doi: 10.1155/2013/248482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Skodje GI, Sarna VK, Minelle IH, et al. Fructan, rather than gluten, induces symptoms in patients with self-reported non-celiac gluten sensitivity. Gastroenterology. 2018;154(3):529–539.e522. doi: 10.1053/j.gastro.2017.10.040 [DOI] [PubMed] [Google Scholar]
- 123.Daveson AJM, Tye-Din JA, Goel G, et al. Masked bolus gluten challenge low in FODMAPs implicates nausea and vomiting as key symptoms associated with immune activation in treated coeliac disease. Aliment Pharmacol Ther. 2020;51(2):244–252. doi: 10.1111/apt.15551 [DOI] [PubMed] [Google Scholar]
- 124.El Khoury D, Balfour-Ducharme S, Joye IJ. A review on the gluten-free diet: technological and nutritional challenges. Nutrients. 2018;10(10):1410. doi: 10.3390/nu10101410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Rostami K, Bold J, Parr A, Johnson MW. Gluten-free diet indications, safety, quality, labels, and challenges. Nutrients. 2017;9(8):846. doi: 10.3390/nu9080846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Jouanin A, Gilissen LJWJ, Schaart JG, et al. CRISPR/Cas9 gene editing of gluten in wheat to reduce gluten content and exposure-reviewing methods to screen for coeliac safety. Front Nutr. 2020;7:51. doi: 10.3389/fnut.2020.00051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Jouanin A, Schaart JG, Boyd LA, et al. Outlook for coeliac disease patients: towards bread wheat with hypoimmunogenic gluten by gene editing of α- and γ-gliadin gene families. BMC Plant Biol. 2019;19(1):333. doi: 10.1186/s12870-019-1889-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Borisjuk N, Kishchenko O, Eliby S, et al. Genetic modification for wheat improvement: from transgenesis to genome editing. Biomed Res Int. 2019;2019:6216304. doi: 10.1155/2019/6216304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Li H, Yang Y, Hong W, Huang M, Wu M, Zhao X. Applications of genome editing technology in the targeted therapy of human diseases: mechanisms, advances and prospects. Signal Transduct Target Ther. 2020;5(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wang K, Riaz B, Ye X. Wheat genome editing expedited by efficient transformation techniques: progress and perspectives. Crop J. 2018;6(1):22–31. doi: 10.1016/j.cj.2017.09.009 [DOI] [Google Scholar]
- 131.Vasil V, Castillo AM, Fromm ME, Vasil IK. Herbicide resistant fertile transgenic wheat plants obtained by microprojectile bombardment of regenerable embryogenic callus. Bio/Technology. 1992;10(6):667–674. [Google Scholar]
- 132.Agrawal N, Dasaradhi PVN, Mohmmed A, et al. RNA interference: biology, mechanism, and applications. Microbiol Mol Biol Rev. 2003;67(4):657–685. doi: 10.1128/MMBR.67.4.657-685.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Tenea G, Burlibasa L. RNAi Towards Functional Genomics Studies. InTech Publisher. 2012:67–94. [Google Scholar]
- 134.Ansari WA, Chandanshive SU, Bhatt V, et al. Genome editing in cereals: approaches, applications and challenges. Int J Mol Sci. 2020;21(11):4040. doi: 10.3390/ijms21114040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Travella S, Klimm TE, Keller B. RNA interference-based gene Silencing as an efficient tool for functional genomics in hexaploid bread wheat. Plant Physiol. 2006;142(1):6–20. doi: 10.1104/pp.106.084517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Gil-Humanes J, Pistón F, Altamirano-Fortoul R, et al. Reduced-gliadin wheat bread: an alternative to the gluten-free diet for consumers suffering gluten-related pathologies. PLoS One. 2014;9(3):e90898. doi: 10.1371/journal.pone.0090898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Altenbach SB, Tanaka CK, Seabourn BW. Silencing of omega-5 gliadins in transgenic wheat eliminates a major source of environmental variability and improves dough mixing properties of flour. BMC Plant Biol. 2014;14(1):393. doi: 10.1186/s12870-014-0393-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Altenbach SB, Chang H-C, Rowe MH, et al. Reducing the immunogenic potential of wheat flour: silencing of alpha gliadin genes in a U.S. wheat cultivar. Front Plant Sci. 2020;11(20). doi: 10.3389/fpls.2020.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Barro F, Iehisa J, Gimenez M, et al. Targeting of prolamins by RNAi in bread wheat: effectiveness of seven silencing-fragment combinations for obtaining lines devoid of coeliac disease epitopes from highly immunogenic gliadins. Plant Biotechnol J. 2015;14. doi: 10.1111/pbi.12455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ervin E-H, Pook M, Teino I, et al. Targeted gene silencing in human embryonic stem cells using cell-penetrating peptide PepFect 14. Stem Cell Res Ther. 2019;10(1):43. doi: 10.1186/s13287-019-1144-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Gaj T, Gersbach CA, Barbas CF 3rd ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31(7):397–405. doi: 10.1016/j.tibtech.2013.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Deshpande K, Vyas A, Balakrishnan A, Vyas D. Clustered regularly interspaced short palindromic Repeats/Cas9 genetic engineering: robotic genetic surgery. Am J Robot Surg. 2015;2(1):49–52. doi: 10.1166/ajrs.2015.1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Campenhout CV, Cabochette P, Veillard A-C, et al. Guidelines for optimized gene knockout using CRISPR/Cas9. BioTechniques. 2019;66(6):295–302. doi: 10.2144/btn-2018-0187 [DOI] [PubMed] [Google Scholar]
- 144.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157(6):1262–1278. doi: 10.1016/j.cell.2014.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Shan Q, Wang Y, Li J, et al. Targeted genome modification of crop plants using a CRISPR-Cas system. Nat Biotechnol. 2013;31(8):686–688. doi: 10.1038/nbt.2650 [DOI] [PubMed] [Google Scholar]
- 146.Sánchez-León S, Gil-Humanes J, Ozuna CV, et al. Low-gluten, nontransgenic wheat engineered with CRISPR/Cas9. Plant Biotechnol J. 2018;16(4):902–910. doi: 10.1111/pbi.12837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Jouanin A, Borm T, Boyd LA, et al. Development of the GlutEnSeq capture system for sequencing gluten gene families in hexaploid bread wheat with deletions or mutations induced by γ-irradiation or CRISPR/Cas9. J Cereal Sci. 2019;88:157–166. doi: 10.1016/j.jcs.2019.04.008 [DOI] [Google Scholar]
- 148.Purnhagen K, Wesseler J. EU regulation of new plant breeding technologies and their possible economic implications for the EU and beyond. Appl Econ Perspect Policy. 2020:1–17. [Google Scholar]