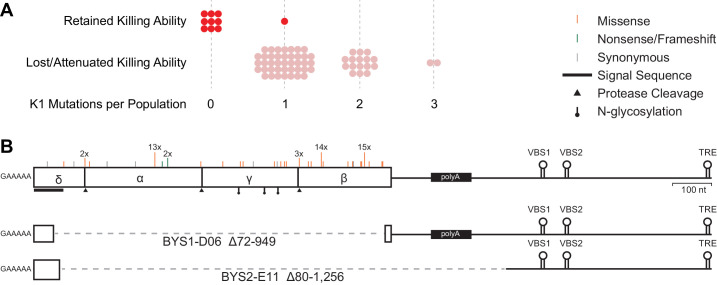
Figure 3. Loss of killer phenotype correlates with the presence of mutations in the K1 toxin gene.
(A) Number of mutations in the K1 gene in yeast populations that retain or lose killing ability. Each data point represents a single yeast population. (B) Observed spectrum of point mutations across the K1 toxin in 67 evolved yeast populations. Mutations were detected in a single population unless otherwise noted. Large internal deletion variants from two yeast populations (BYS1-D06 and BYS2-E11). The deletions span the region indicated by the dashed gray line. VBS: viral binding site. TRE: terminal recognition element.