Supplemental digital content is available in the text.
Key Words: BIOLOGICAL AGING, METHYLATION, QUANTITATIVE GENETICS, SMOKING
ABSTRACT
Purpose
Greater leisure-time physical activity (LTPA) associates with healthier lives, but knowledge regarding occupational physical activity (OPA) is more inconsistent. DNA methylation (DNAm) patterns capture age-related changes in different tissues. We aimed to assess how LTPA and OPA are associated with three DNAm-based epigenetic age estimates, namely, DNAm age, PhenoAge, and GrimAge.
Methods
The participants were young adult (21–25 yr, n = 285) and older (55–74 yr, n = 235) twin pairs, including 16 pairs with documented long-term LTPA discordance. Genome-wide DNAm from blood samples was used to compute DNAm age, PhenoAge, and GrimAge Age acceleration (Acc), which describes the difference between chronological and epigenetic ages. Physical activity was assessed with sport, leisure-time, and work indices based on the Baecke Questionnaire. Genetic and environmental variance components of epigenetic age Acc were estimated by quantitative genetic modeling.
Results
Epigenetic age Acc was highly heritable in young adult and older twin pairs (~60%). Sport index was associated with slower and OPA with faster DNAm GrimAge Acc after adjusting the model for sex. Genetic factors and nonshared environmental factors in common with sport index explained 1.5%–2.7% and 1.9%–3.5%, respectively, of the variation in GrimAge Acc. The corresponding proportions considering OPA were 0.4%–1.8% and 0.7%–1.8%, respectively. However, these proportions were minor (<0.5%) after adjusting the model for smoking status.
Conclusions
LTPA associates with slower and OPA with faster epigenetic aging. However, adjusting the models for smoking status, which may reflect the accumulation of unhealthy lifestyle habits, attenuated the associations.
The health benefits of leisure-time physical activity (LTPA) are well documented. High LTPA is associated with a low risk of several diseases, such as cardiovascular disease and metabolic syndrome (1,2), and with low risk of premature death in a dose-dependent manner (3). The benefits of occupational physical activity (OPA) are more controversial. There is some evidence that OPA may even adversely affect health outcomes, such as risk of all-cause mortality, cardiovascular disease, and long-term sickness absence, whereas LTPA associates with low risk (1,4,5). This contrasting association of OPA with health outcomes is referred to as “the physical activity health paradox” (6).
Several possible reasons for the paradox have been proposed (7). LTPA and OPA may produce divergent physiological responses, as LTPA typically differs in terms of intensity, duration, and movement type from the monotonous OPA. The different physiological demands of LTPA and OPA are accompanied by other environmental and psychological factors regulating the response to physical activity (PA). Therefore, these two forms of activities may produce an inverse impact on levels of inflammation and autonomic imbalance (6). However, there are also many selection issues and third variables (confounders such as genetic and social factors as well as other health habits) possibly explaining some of the difference in the associations (8), but these relationships are as yet poorly understood.
Epigenetics refers to DNA or chromatin modifications that regulate gene expression without altering the underlying DNA sequence itself. DNA methylation (DNAm; attachment of a methyl group to C-5 of cytosine base in the context of CpG dinucleotide in a DNA strand) is one type of epigenetic modification. Many studies have provided evidence of age-related hypomethylation or hypermethylation within specific CpG sites or islands (9). These findings have laid the ground for the development of epigenetic biomarkers of aging, also known as epigenetic clocks. Horvath’s DNAm age was the first widely used epigenetic estimate for chronological age (10). It has been argued that DNAm age may exclude CpGs whose methylation patterns may reflect a deviation of the biological age from the chronological age (11). Therefore, novel DNAm-based biomarkers for aging that capture CpGs associated with the functional stage along with the chronological age have been developed in recent years. DNAm PhenoAge was developed using “phenotypic age measure” instead of chronological age as a reference in the biological age prediction (11). DNAm GrimAge is a combination of DNAm-based surrogate biomarkers for health-related plasma proteins and smoking pack-years as well as sex and chronological age, predicting best mortality risk (12).
Epigenetic age acceleration measures the discrepancy between chronological age and epigenetic age. According to a systematic review and meta-analysis, there was no consistent association between LTPA and epigenetic aging assessed with Horvath’s DNAm age (13). The first results considering the novel epigenetic age estimates seem to be more promising. LTPA has been shown to be correlated negatively with both DNAm PhenoAge and GrimAge acceleration (12,14). However, there is no conclusive evidence on this topic, as the novel epigenetic clocks were published very recently. To our knowledge, no studies are investigating the association between OPA and epigenetic aging.
Here, we investigated the relative contributions of the genetic and environmental factors predicting epigenetic aging measured by DNAm age, PhenoAge, and GrimAge estimates in young adulthood and older age. We further assessed cross-sectional associations of LTPA and OPA with epigenetic aging, as well as the genetic and environmental factors explaining the association. Finally, we studied long-term effects of LTPA on epigenetic aging by comparing co-twins that differed for LTPA for over three decades at least.
MATERIALS AND METHODS
The participants were members of the FinnTwin12 study (born in 1983–1987) and an older cohort (born before 1958) of The Finnish Twin Cohort (FTC) (15,16). Both cohorts included monozygotic (MZ) and dizygotic (DZ) twin pairs. Data on health-related behavior were collected with questionnaires and interviews. More detailed information on data collection is available in Supplemental Text Appendix (Text, Supplemental Digital Content 1, additional information on material and methods, http://links.lww.com/MSS/C106). Blood samples for DNA analyses were collected during in-person clinical studies after a written informed consent form was signed. A total of 1295 twins of the FinnTwin12 study and 447 of the older cohort were examined and measured. The same-sex twin pairs in which both had information on DNAm in a young adult (age range, 21–25 yr; 163 MZ and 122 DZ pairs) and an older cohort (age range, 55–72 yr; 140 MZ and 78 DZ pairs) were included in this study. The FTC data collection has been approved by the ethics committees of the University of Helsinki and Helsinki University Central Hospital.
PA-discordant twin pairs
The PA-discordant twin pairs (TWINACTIVE) initiated from the older cohort of the FTC (15,17). The comprehensive selection process that included multiple measurements of PA since 1975 has been described in detail by Leskinen et al. (18,19). LTPA was examined with standardized repeated questions, which were quantified as metabolic equivalent (MET; intensity × duration × frequency) and expressed as a sum score of leisure-time MET hours per day. Twin pairs whose difference in the volume of PA were >3 MET·h·d−1 were invited to the retrospective follow-up interviews on leisure activity (covering the years from 1980 to 2005 in 5-yr intervals), which were carried out during the years 2005–2007 (19). Of the 5663 originally healthy same-sex twin pairs, 16 twin pairs (age range, 50–74 yr; 7 MZ and 9 DZ pairs, total 5 female pairs) participated in the TWINACTIVE study. During the 30+ yr before the clinical examination and DNA sample collection, the mean intrapair difference in LTPA was 8.8 MET·h·d−1. The participants representing pairs with the longest and highest discordance in LTPA were comprehensively selected from the FTC. The ethics committee of the Central Finland Health Care District has approved the TWINACTIVE study.
PA
PA was assessed by the Baecke Questionnaire (20). It includes four questions on sports activity and leisure-time activity excluding sports and eight questions on work-related PA on a 5-point scale. Activities were scored as 1, 3, or 5 according to how physically demanding they are. A sport index, a nonsport leisure-time (leisure) index and a work index, respectively, were yielded from mean scores of each section as described earlier by Baecke et al. (20) and Mustelin et al. (21) for the FinnTwin12 study.
Confounding variables
Body mass index was calculated as the ratio of measured body weight (in kilograms) to height squared (in meters per squared). Sex, education years, and smoking status (never, former, and current smoker (includes regular and occasional use)) and alcohol use (in grams per day) were assessed through interviews.
Epigenetic Age Estimates
DNAm
Generating and normalizing the DNAm data have been described in Supplementary Text Appendix (Text, Supplemental Digital Content 1, preprocessing the DNAm data, http://links.lww.com/MSS/C106). Genome-wide DNAm from blood samples was determined on Illumina 450K and EPIC BeadChips, and the epigenetic age estimates DNAm age (10), DNAm PhenoAge (11), and DNAm GrimAge (12) were calculated by an online calculator (https://dnamage.genetics.ucla.edu/new; Text, Supplemental Digital Content 1, additional information on epigenetic age estimates, http://links.lww.com/MSS/C106). Epigenetic age acceleration (Acc), which describes the difference between chronological age and epigenetic age estimate, was calculated as the residuals from a linear regression model of epigenetic age estimate on chronological age.
The components of DNAm GrimAge (adjusted for age) were obtained using the calculator as well, including DNAm-based smoking pack-years and the surrogate biomarkers for plasma proteins (DNAm-based plasma proteins): DNAm adrenomedullin, DNAm β2-microglobulin, DNAm cystatin-C, DNAm growth differentiation factor 15, DNAm leptin, DNAm plasminogen activator inhibitor 1, and DNAm tissue inhibitor metalloproteinases 1.
Statistical Methods
Descriptive statistics were calculated using IBM SPSS statistics, and further modeling was performed by using Mplus statistical package (version 8.2) (22).
Quantitative genetic modeling
Quantitative genetic modeling was conducted using a structural equation framework. First, intraclass correlation coefficients (ICCs) and correlations between epigenetic age Acc measures and LTPA (sport index and leisure index), as well as OPA (work index) were studied. Second, univariate modeling was carried out to study the magnitude of genetic and environmental factors affecting epigenetic age Acc measures and PA indices in young adult and older twin pairs as described in Supplemental Text Appendix (Text, Supplemental Digital Content 1, modeling procedure of the univariate models, http://links.lww.com/MSS/C106) (23). The univariate models were adjusted for sex, education, and smoking status. Third, structural equation modeling was used to study the associations of PA indices on epigenetic aging. Shared genetic and environmental effects between epigenetic age Acc and PA, as well as the genetic and environmental factors unique to epigenetic age Acc were studied. For these purposes, Cholesky’s decomposition was applied to PA indices and epigenetic age Acc measures after controlling for covariates (Fig. 1). The latent variables representing PA (CH1) and the residual of epigenetic age Acc after the effect of PA has been taken into account (CH2) were specified (24). The model was initially adjusted for sex only. The variation in both the latent variables CH1 and CH2 was partitioned in the genetic and environmental components as described in Supplementary Text Appendix (Text, Supplemental Digital Content 1, modeling procedure of the bivariate models, http://links.lww.com/MSS/C106).
FIGURE 1.
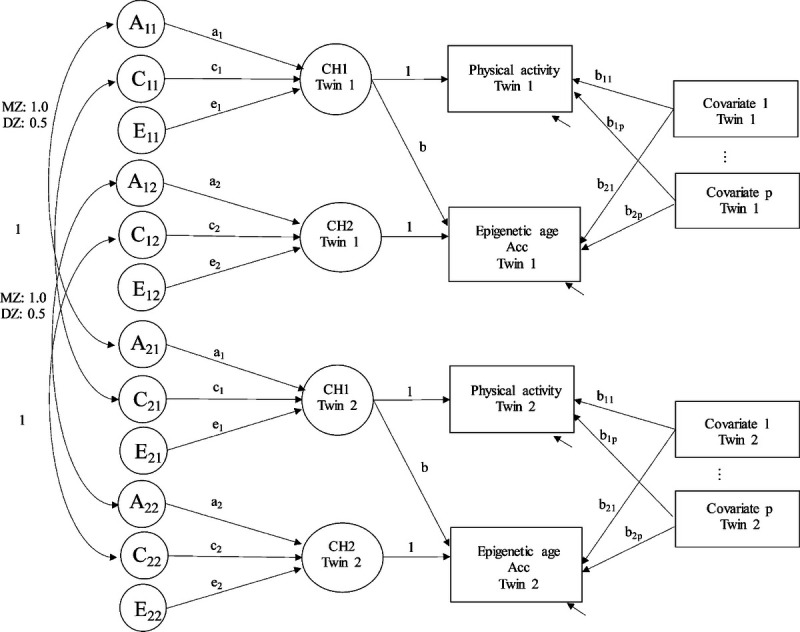
The path diagram of the structural equation model. Circles denote latent variables; rectangles denote observed variables. CH1 represents PA, and CH2 represents the residual of the epigenetic age Acc after the effect of PA has been taken into account. A, additive genetic; C, shared environmental; E, nonshared environmental components.
The potential confounding variables, including sex, education, body mass index, smoking status, and alcohol use, were added sequentially to the model. At each stage, the regression coefficients between PA and epigenetic age Acc (b) were studied. In addition, the parameters of the model were used to calculate the relative proportions in the total variation of epigenetic age Acc [( + (] explained by the genetic and environmental effects in common with PA (, and , respectively) as well as the unique genetic and environmental factors of epigenetic age Acc (, , and , respectively).
Discordant twin-pair analysis
The mean within-pair differences in epigenetic age estimates between active and inactive co-twins were calculated. Standardized mean difference (SMD; within-pair difference/SD of the variable among the members of the pairs) was used to evaluate effect size. The within-pair differences were regressed on zygosity to study whether there were differences in the effect of LTPA on epigenetic aging between MZ and DZ twins. The study design controls for chronological age and sex, as the twins are of the same sex and age.
RESULTS
Among the young adults, DZ twins had a slightly higher body mass index and leisure PA index compared with MZ twins (Table 1). The mean age of the participants was 22.4 yr, whereas the mean of the different epigenetic age estimates ranged from 15.0 to 28.6 yr, depending on the estimate utilized. In the older cohort, DZ twins were slightly older compared with MZ twins (mean, 62.9 vs 62.0 yr), and there were also differences in the means of the epigenetic age estimates. The mean of the different epigenetic age estimates ranged from 54.7 to 62.0 yr.
TABLE 1.
Descriptive statistics of the study variables by zygosity among young adult (n = 570) and older (n = 470) twin individuals.
| Young Adult (21–25 yr) | Older (55–72 yr) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| MZ Twins (n = 326) | DZ Twins (n = 244) | MZ Twins (n = 294) | DZ Twins (n = 176) | |||||||
| Mean (SD) | n | Mean (SD) | n | Pa | Mean (SD) | n | Mean (SD) | n | Pa | |
| Women, n (%) | 208 (63.8) | 142 (58.2) | 0.174 | 188 (63.9) | 94 (53.4) | 0.024 | ||||
| Chronological age, yr | 22.4 (0.7) | 326 | 22.4 (0.7) | 244 | 0.672 | 62.0 (4.1) | 294 | 62.9 (4.3) | 176 | 0.036 |
| DNAm age, yr | 25.0 (3.5) | 326 | 24.4 (3.4) | 244 | 0.057 | 60.2 (6.2) | 294 | 62.1 (7.6) | 176 | 0.004 |
| DNAm PhenoAge, yr | 15.0 (4.6) | 326 | 15.3 (4.8) | 244 | 0.496 | 54.7 (6.5) | 294 | 56.0 (8.1) | 176 | 0.045 |
| DNAm GrimAge, yr | 28.6 (2.9) | 326 | 28.6 (3.4) | 244 | 0.843 | 59.8 (5.3) | 294 | 60.8 (5.5) | 176 | 0.061 |
| Body mass index, kg·m−2 | 23.1 (3.8) | 326 | 23.7 (4.2) | 244 | 0.028 | 27.1 (4.7) | 294 | 27.2 (4.8) | 176 | 0.727 |
| Education, yr | 16.7 (3.5) | 325 | 16.6 (3.6) | 244 | 0.429 | 11.9 (3.8) | 252 | 11.4 (3.5) | 96 | 0.266 |
| Smoking status | 325 | 244 | 292 | 176 | ||||||
| Never smokers, n (%) | 182 (56.0) | 116 (47.5) | 148 (50.8) | 88 (50.0) | ||||||
| Former smokers, n (%) | 37 (11.4) | 30 (12.3) | 97 (33.3) | 63 (35.8) | ||||||
| Current smokers, n (%) | 106 (32.6) | 98 (40.2) | 0.121 | 47 (15.9) | 25 (14.2) | 0.787 | ||||
| Alcohol use, g·d−1 | 12.4 (16.7) | 326 | 13.4 (16.7) | 244 | 0.460 | 6.9 (11.2) | 278 | 8.8 (11.6) | 158 | 0.080 |
| LTPA | ||||||||||
| Sport index | 2.9 (0.8) | 251 | 3.0 (0.8) | 233 | 0.358 | 3.2 (0.9) | 241 | 3.1 (0.8) | 156 | 0.421 |
| Nonsport leisure index | 2.9 (0.6) | 253 | 3.1 (0.6) | 234 | 0.023 | 2.9 (0.7) | 241 | 2.8 (0.7) | 156 | 0.715 |
| OPA | ||||||||||
| Work index | 2.7 (0.6) | 256 | 2.8 (0.6) | 234 | 0.315 | 2.4 (1.0) | 234 | 2.3 (1.0) | 150 | 0.935 |
| Out of working life | 322 | 243 | 241 | 156 | ||||||
| Retired or unemployed, n (%) | 49 (15.2) | 26 (10.7) | 105 (43.6) | 76 (50) | 0.209 | |||||
aP value for the difference between the groups from independent-samples t-test or χ2 test.
Heritability
In both cohorts, ICCs for epigenetic age Acc measures were consistently higher in MZ twins than in DZ twins, suggesting the influence of a genetic component (Table 2). According to univariate modeling, additive genetic factors accounted for 69% of the variation in DNAm age Acc, 64% of the variation in DNAm PhenoAge Acc, and 62% of the variation in DNAm GrimAge Acc in young adult twin pairs (Text, Supplemental Digital Content 1, http://links.lww.com/MSS/C106; Table, Supplemental Digital Content 2, the estimation results of the univariate model for epigenetic aging among young adult twin pairs, http://links.lww.com/MSS/C107). Correspondingly, nonshared environmental factors accounted for the remainder (31%–38%) of the variation in epigenetic aging. In older twin pairs, additive genetic factors explained 61% of the variation in DNAm age Acc, 60% of the variation in DNAm PhenoAge Acc, and 58% of the variation in DNAm GrimAge Acc (Text, Supplemental Digital Content 1, http://links.lww.com/MSS/C106; Table, Supplemental Digital Content 3, the estimation results of the univariate model for epigenetic aging among older twin pairs, http://links.lww.com/MSS/C108), whereas unique environmental factors accounted for the remaining variance. The proportions of the variation in epigenetic age Acc measures explained by the genetic factors did not differ between young adult and older twin pairs (Wald test, P = 0.076–0.220). The results considering PA are presented in the supplementary material (Text, Supplemental Digital Content 1, http://links.lww.com/MSS/C106; Tables, Supplemental Digital Contents 4–5, the estimation results of the univariate model for PA indices among young adult and older twin pairs, respectively, http://links.lww.com/MSS/C109 and http://links.lww.com/MSS/C110).
TABLE 2.
The ICCs of epigenetic age Acc measures and PA indices by zygosity, as well as the correlation coefficients between the measures among young adult (n = 570) and older (n = 470) twins individuals.
| ICCs and Their 95% CI | Correlation Coefficients and Their 95% CIa,b | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MZ | DZ | 1 | 2 | 3 | 4 | 5 | ||||||||
| Young adult twins | ||||||||||||||
| 1. DNAm age Acc | 0.70 | 0.61 to 0.79 | 0.49 | 0.35 to 0.64 | ||||||||||
| 2. DNAm PhenoAge Acc | 0.67 | 0.58 to 0.75 | 0.18 | 0.02 to 0.34 | 0.35 | 0.28 to 0.43 | ||||||||
| 3. DNAm GrimAge Acc | 0.67 | 0.57 to 0.76 | 0.42 | 0.22 to 0.62 | 0.08 | −0.01 to 0.17 | 0.34 | 0.25 to 0.42 | ||||||
| 4. Sport index | 0.61 | 0.48 to 0.75 | 0.32 | 0.15 to 0.50 | 0.09 | −0.02 to 0.19 | −0.09 | −0.19 to 0.01 | −0.21 | −0.30 to −0.12 | ||||
| 5. Leisure index | 0.43 | 0.27 to 0.58 | 0.34 | 0.17 to 0.51 | 0.05 | −0.05 to 0.15 | 0.03 | −0.07 to 0.13 | −0.12 | −0.21 to −0.02 | 0.28 | 0.19 to 0.37 | ||
| 6. Work index | 0.51 | 0.37 to 0.65 | 0.26 | 0.07 to 0.46 | −0.01 | −0.12 to 0.10 | 0.00 | −0.10 to 0.10 | 0.26 | 0.16 to 0.35 | −0.06 | −0.17 to 0.04 | −0.01 | −0.11 to 0.10 |
| Older twins | ||||||||||||||
| 1. DNAm age Acc | 0.62 | 0.51 to 0.73 | 0.28 | 0.15 to 0.41 | ||||||||||
| 2. DNAm PhenoAge Acc | 0.62 | 0.52 to 0.72 | 0.32 | 0.13 to 0.52 | 0.34 | 0.23 to 0.45 | ||||||||
| 3. DNAm GrimAge Acc | 0.64 | 0.54 to 0.75 | 0.40 | 0.19 to 0.62 | 0.21 | 0.11 to 0.31 | 0.40 | 0.32 to 0.48 | ||||||
| 4. Sport index | 0.32 | 0.15 to 0.50 | 0.21 | −0.04 to 0.46 | −0.03 | −0.13 to 0.07 | −0.06 | −0.19 to 0.06 | −0.20 | −0.31 to −0.08 | ||||
| 5. Leisure index | 0.27 | 0.10 to 0.44 | 0.26 | 0.03 to 0.48 | −0.12 | −0.21 to −0.03 | 0.00 | −0.13 to 0.12 | −0.21 | −0.31 to −0.12 | 0.45 | 0.36 to 0.53 | ||
| 6. Work index | 0.33 | 0.14 to 0.51 | 0.34 | 0.12 to 0.55 | 0.09 | −0.02 to 0.20 | 0.07 | −0.04 to 0.17 | 0.13 | 0.03 to 0.23 | −0.13 | −0.23 to −0.03 | −0.02 | −0.13 to 0.09 |
aCIs were corrected for nested sampling.
bSignificant correlations at the level 0.05 are presented in bold.
Bivariate twin models
Sport index and leisure index were associated with slower and work index with faster DNAm GrimAge Acc in both young adult and older twins as shown by correlation coefficients in Table 2. Epigenetic age Acc and PA indices were not consistently correlated, when DNAm age and DNAm PhenoAge estimates were used to assess epigenetic age. Therefore, no further modeling was considered for these measures. Additional information on the model selection is available in the supplemental material (Text, Supplemental Digital Content 1, http://links.lww.com/MSS/C106; Table, Supplemental Digital Content 6, the model fit of the bivariate models, http://links.lww.com/MSS/C111).
In young adult twin pairs, sport index was associated with slower DNAm GrimAge acceleration after all the adjustments, but in an older cohort, the association was no longer significant after controlling for smoking status (Table 3). In both cohorts, leisure index was associated with slower and work index with accelerated epigenetic aging, but these associations attenuated after controlling for covariates, especially for smoking status.
TABLE 3.
The estimation results of the structural equation modeling: the standardized linear regression coefficients of DNAm GrimAge Acc on PA indices among young adult (MZ, n = 163; DZ:, n = 122) and older (MZ, n = 147; DZ, n = 88) twin pairs.
| Sport Index | Leisure Index | Work Indexa | |||||||
|---|---|---|---|---|---|---|---|---|---|
| B | 95% CI | P | B | 95% CI | P | B | 95% CI | P | |
| Young adult twin pairs | |||||||||
| PA index + sex | −0.212 | −0.288 to −0.136 | <0.001 | −0.113 | −0.192 to −0.033 | 0.006 | 0.189 | 0.105 to 0.273 | <0.001 |
| + Education years | −0.172 | −0.244 to −0.099 | <0.001 | −0.089 | −0.167 to −0.012 | 0.024 | 0.103 | 0.027 to 0.179 | 0.008 |
| + Body mass index | −0.161 | −0.236 to −0.087 | <0.001 | −0.085 | −0.163 to −0.007 | 0.108 | 0.106 | 0.029 to 0.182 | 0.007 |
| + Smoking status | −0.085 | −0.153 to −0.017 | 0.015 | −0.029 | −0.104 to 0.046 | 0.451 | 0.051 | −0.020 to 0.122 | 0.157 |
| + Alcohol use | −0.082 | −0.151 to −0.014 | 0.019 | −0.029 | −0.104 to 0.046 | 0.451 | 0.052 | −0.018 to 0.123 | 0.147 |
| Older twin pairs | |||||||||
| PA index + sex | −0.212 | −0.298 to −0.126 | <0.001 | −0.169 | −0.248 to −0.090 | <0.001 | 0.098 | 0.014 to 0.182 | 0.023 |
| + Education years | −0.211 | −0.296 to −0.126 | <0.001 | −0.165 | −0.244 to −0.086 | <0.001 | 0.078 | −0.005 to 0.160 | 0.066 |
| + Body mass index | −0.198 | −0.283 to −0.113 | <0.001 | −0.153 | −0.232 to −0.075 | <0.001 | 0.077 | −0.006 to 0.161 | 0.070 |
| + Smoking status | −0.053 | −0.127 to 0.020 | 0.154 | −0.048 | −0.112 to 0.017 | 0.145 | 0.028 | −0.038 to 0.094 | 0.410 |
| + Alcohol use | −0.054 | −0.126 to 0.017 | 0.138 | −0.053 | −0.118 to 0.012 | 0.108 | 0.029 | −0.037 to 0.095 | 0.388 |
aThe model was additionally adjusted for indicator of being out of working life.
+, The model was additionally adjusted for the following variables; B, standardized regression coefficient.
Significant associations at the level 0.05 are presented in bold.
In young twin pairs, genetic and environmental factors in common with sport index explained 2.7% and 1.9%, respectively, of the variation in DNAm GrimAge Acc, after adjusting the model for sex (Table 4). The corresponding proportions were 0.6% and 0.7% for leisure index and 1.8% and 1.7% for work index, respectively. In older twin pairs, genetic and environmental factors in common with sport index explained 1.5% and 3.5%, respectively, of the variation in DNAm GrimAge Acc (Table 5). The corresponding proportions for leisure index were 0.8% and 2.4% and for OPA 0.4% and 0.7%, respectively. In both cohorts, the proportions of the variation in DNAm GrimAge Acc explained by genetic and environmental factors in common with PA indices were minor (<0.5%) after including smoking status in the models.
TABLE 4.
The proportion of the variation in DNAm GrimAge Acc explained by genetic and environmental effects in young adult twin pairs.
| Sport Index | Leisure Index | Work Indexa | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Estimate | 95% CI | P | Estimate | 95% CI | P | Estimate | 95% CI | P | ||
| A1 | PA index + sex | 0.027 | 0.006 to 0.048 | 0.011 | 0.006 | −0.002 to 0.014 | 0.167 | 0.018 | 0.001 to 0.036 | 0.039 |
| + Education | 0.018 | 0.002 to 0.034 | 0.029 | 0.004 | −0.003 to 0.010 | 0.253 | 0.003 | −0.002 to 0.008 | 0.232 | |
| + Body mass index | 0.016 | 0.000 to 0.032 | 0.044 | 0.003 | −0.003 to 0.010 | 0.278 | 0.003 | −0.002 to 0.009 | 0.225 | |
| + Smoking status | 0.006 | −0.004 to 0.015 | 0.234 | 0.001 | −0.002 to 0.003 | 0.702 | 0.001 | −0.002 to 0.004 | 0.499 | |
| + Alcohol use | 0.005 | −0.004 to 0.015 | 0.249 | 0.001 | −0.002 to 0.003 | 0.703 | 0.002 | −0.002 to 0.004 | 0.487 | |
| A2 | PA index +sex | 0.680 | 0.597 to 0.762 | <0.001 | 0.706 | 0.629 to 0.782 | <0.001 | 0.661 | 0.576 to 0.747 | <0.001 |
| + Education | 0.689 | 0.606 to 0.771 | <0.001 | 0.706 | 0.627 to 0.784 | <0.001 | 0.689 | 0.606 to 0.772 | <0.001 | |
| + Body mass index | 0.685 | 0.602 to 0.769 | <0.001 | 0.701 | 0.622 to 0.780 | <0.001 | 0.689 | 0.606 to 0.771 | <0.001 | |
| + Smoking status | 0.617 | 0.516 to 0.718 | <0.001 | 0.620 | 0.520 to 0.721 | <0.001 | 0.617 | 0.515 to 0.718 | <0.001 | |
| + Alcohol use | 0.619 | 0.515 to 0.718 | <0.001 | 0.618 | 0.518 to 0.719 | <0.001 | 0.615 | 0.513 to 0.717 | <0.001 | |
| E1 | PA index + sex | 0.019 | 0.005 to 0.033 | 0.008 | 0.007 | −0.003 to 0.017 | 0.178 | 0.018 | 0.002 to 0.034 | 0.030 |
| + Education | 0.014 | 0.002 to 0.025 | 0.023 | 0.005 | −0.004 to 0.013 | 0.277 | 0.008 | −0.004 to 0.021 | 0.183 | |
| + Body mass index | 0.012 | 0.001 to 0.023 | 0.038 | 0.004 | −0.004 to 0.013 | 0.301 | 0.009 | −0.004 to 0.021 | 0.176 | |
| + Smoking status | 0.004 | −0.003 to 0.011 | 0.220 | 0.001 | −0.003 to 0.004 | 0.709 | 0.003 | −0.005 to 0.010 | 0.478 | |
| + Alcohol use | 0.004 | −0.003 to 0.011 | 0.235 | 0.001 | −0.003 to 0.004 | 0.710 | 0.003 | −0.005 to 0.010 | 0.467 | |
| E2 | PA index + sex | 0.275 | 0.200 to 0.349 | <0.001 | 0.281 | 0.203 to 0.360 | <0.001 | 0.302 | 0.223 to 0.382 | <0.001 |
| + Education | 0.280 | 0.204 to 0.356 | <0.001 | 0.286 | 0.206 to 0.367 | <0.001 | 0.299 | 0.220 to 0.379 | <0.001 | |
| + Body mass index | 0.287 | 0.210 to 0.364 | <0.001 | 0.291 | 0.210 to 0.372 | <0.001 | 0.299 | 0.220 to 0.379 | <0.001 | |
| + Smoking status | 0.373 | 0.274 to 0.471 | <0.001 | 0.378 | 0.277 to 0.480 | <0.001 | 0.380 | 0.279 to 0.480 | <0.001 | |
| + Alcohol use | 0.371 | 0.275 to 0.473 | <0.001 | 0.381 | 0.279 to 0.482 | <0.001 | 0.382 | 0.281 to 0.481 | <0.001 | |
aThe model was additionally adjusted for indicator of being out of working life.
+, The model was additionally adjusted for the following variables; A1, genetic factors of DNAm GrimAge Acc in common with PA index; A2, unique genetic factors of DNAm GrimAge Acc; E1, environmental factors of DNAm GrimAge Acc in common with PA index; E2, unique environmental factors of DNAm GrimAge Acc.
Significant associations at the level 0.05 are presented in bold.
TABLE 5.
The proportion of the variation in DNAm GrimAge Acc explained by genetic and environmental effects in older twin pairs.
| Sport Index | Leisure Index | Work Indexa | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Estimate | 95% CI | P | Estimate | 95% CI | P | Estimate | 95% CI | P | ||
| A1 | PA index + sex | 0.015 | 0.001 to 0.03 | 0.034 | 0.008 | −0.001 to 0.018 | 0.074 | 0.004 | −0.003 to 0.011 | 0.256 |
| + Education | 0.015 | 0.001 to 0.03 | 0.035 | 0.008 | −0.001 to 0.017 | 0.079 | 0.002 | −0.002 to 0.006 | 0.380 | |
| + Body mass index | 0.014 | 0.000 to 0.027 | 0.047 | 0.007 | −0.001 to 0.015 | 0.099 | 0.002 | −0.002 to 0.006 | 0.381 | |
| + Smoking status | 0.002 | −0.003 to 0.007 | 0.455 | 0.001 | −0.002 to 0.005 | 0.471 | 0.000 | −0.002 to 0.003 | 0.683 | |
| + Alcohol use | 0.002 | −0.003 to 0.008 | 0.437 | 0.002 | −0.002 to 0.003 | 0.426 | 0.001 | −0.002 to 0.003 | 0.669 | |
| A2 | PA index + sex | 0.627 | 0.516 to 0.738 | <0.001 | 0.623 | 0.509 to 0.738 | <0.001 | 0.618 | 0.493 to 0.743 | <0.001 |
| + Education | 0.627 | 0.517 to 0.738 | <0.001 | 0.624 | 0.509 to 0.738 | <0.001 | 0.623 | 0.499 to 0.748 | <0.001 | |
| + Body mass index | 0.622 | 0.508 to 0.737 | <0.001 | 0.617 | 0.497 to 0.737 | <0.001 | 0.612 | 0.480 to 0.744 | <0.001 | |
| + Smoking status | 0.584 | 0.489 to 0.680 | <0.001 | 0.583 | 0.486 to 0.680 | <0.001 | 0.579 | 0.481 to 0.678 | <0.001 | |
| + Alcohol use | 0.597 | 0.505 to 0.690 | <0.001 | 0.598 | 0.524 to 0.723 | <0.001 | 0.595 | 0.501 to 0.690 | <0.001 | |
| E1 | PA index + sex | 0.035 | 0.005 to 0.065 | 0.023 | 0.024 | 0.001 to 0.046 | 0.041 | 0.007 | −0.005 to 0.018 | 0.267 |
| + Education | 0.035 | 0.005 to 0.065 | 0.022 | 0.023 | 0.000 to 0.045 | 0.045 | 0.005 | −0.006 to 0.016 | 0.361 | |
| + Body mass index | 0.031 | 0.003 to 0.059 | 0.032 | 0.020 | −0.001 to 0.041 | 0.062 | 0.005 | −0.006 to 0.016 | 0.370 | |
| + Smoking status | 0.004 | −0.007 to 0.015 | 0.487 | 0.004 | −0.006 to 0.013 | 0.467 | 0.001 | −0.004 to 0.007 | 0.678 | |
| + Alcohol use | 0.004 | −0.007 to 0.015 | 0.470 | 0.004 | −0.003 to 0.004 | 0.423 | 0.001 | −0.005 to 0.007 | 0.664 | |
| E2 | PA index + sex | 0.323 | 0.221 to 0.424 | <0.001 | 0.345 | 0.235 to 0.455 | <0.001 | 0.371 | 0.246 to 0.496 | <0.001 |
| + Education | 0.322 | 0.222 to 0.422 | <0.001 | 0.346 | 0.235 to 0.456 | <0.001 | 0.370 | 0.245 to 0.495 | <0.001 | |
| + Body mass index | 0.333 | 0.228 to 0.438 | <0.001 | 0.356 | 0.240 to 0.473 | <0.001 | 0.381 | 0.248 to 0.514 | <0.001 | |
| + Smoking status | 0.410 | 0.316 to 0.503 | <0.001 | 0.412 | 0.316 to 0.508 | <0.001 | 0.419 | 0.322 to 0.516 | <0.001 | |
| + Alcohol use | 0.396 | 0.305 to 0.488 | <0.001 | 0.396 | 0.275 to 0.476 | <0.001 | 0.403 | 0.310 to 0.496 | <0.001 | |
aThe model was additionally adjusted for indicator of being out of working life.
+, The model was additionally adjusted for the following variables; A1, genetic factors of DNAm GrimAge Acc in common with PA index; A2, unique genetic factors of DNAm GrimAge Acc; E1, environmental factors of DNAm GrimAge Acc in common with PA index; E2, unique environmental factors of DNAm GrimAge Acc.
Significant proportions at the level 0.05 are presented in bold.
The associations of the DNAm-based surrogates (adjusted for age) included in DNAm GrimAge with DNAm GrimAge Acc, as well as with PA indices, were studied using correlation coefficients (Table, Supplemental Digital Content 7, the correlations among young adult and older individual twins, http://links.lww.com/MSS/C112). Sport index in young adults and PA indices in older cohort were associated with lower levels of several DNAm-based plasma proteins, whereas inconsistent associations of work index were observed in both cohorts. In both cohorts, sport index was negatively associated with DNAm-based smoking pack-years (r = −0.20 to −0.16), whereas the opposite association of work index was observed (r = 0.11 to 0.30). DNAm-based smoking pack-years and DNAm GrimAge Acc were highly correlated (r > 0.80).
Twin pairs discordant for LTPA for 32 yr: the TWINACTIVE sample
The mean (SD) age of the participants was 60.4 (6.2) yr, whereas the means of the epigenetic age estimates DNAm age, PhenoAge, and GrimAge were 56.5 (4.8), 46.8 (5.8), and 59.7 (5.7) yr, respectively. There was no difference in Horvath’s DNAm age between the active and inactive co-twins among the LTPA discordant twin pairs, as we have reported previously (25). The two newer epigenetic age estimates, however, differed between active and inactive co-twins. Active twins were on average 3.27 yr (95% confidence interval (CI), 1.34 to 5.20 yr; SMD, 0.56 yr) younger compared with their inactive co-twins in DNAm PhenoAge and 2.08 yr (95% CI, 0.75 to 3.41 yr; SMD, 0.37 yr) younger in DNAm GrimAge.
Mean within-pair difference for DNAm PhenoAge was among MZ pairs 1.80 (95% CI, −1.40 to 4.96; SMD, 0.30) and among DZ pairs 4.42 (95% CI, 2.33 to 6.51; SMD, 0.77; Figure, Supplemental Digital Content 8, the epigenetic age estimates in the LTPA discordant MZ and DZ twin pairs, http://links.lww.com/MSS/C113). In DNAm GrimAge, active MZ twins were on average 1.97 yr (95% CI, −0.04 to 4.00 yr; SMD, 0.40 yr) and DZ twins 2.27 (95% CI, 0.40 to 3.94 yr; SMD, 0.36 yr) younger compared with their inactive co-twins. Among the small number of twin pairs, the difference in the association between LTPA and DNAm PhenoAge or DNAm GrimAge between MZ and DZ twin pairs was not significant (P = 0.091 and P = 0.887, respectively).
DISCUSSION
In this study, we showed that the heritability estimates of different epigenetic clocks (namely DNAm age, PhenoAge, and GrimAge) are very similar. Genetic factors accounted for about 60% of the variation in epigenetic age acceleration, whereas nonshared environmental factors explained the remainder. Models with no genetic effects showed poorer fit to the data than did the models with genetic effects. Our twin models did not require the inclusion of shared environmental effects as genetic models with (ACE) or without (AE) shared environmental effects fit the data adequately. The observed heritability estimates of the epigenetic age acceleration based on the newer epigenetic age estimates were considerably higher than those reported in previous studies, possibly because of the methodological differences in constructing epigenetic age estimates and the age differences of the target cohorts. Lu and colleagues reported low to moderate estimates of heritability for DNAm GrimAge- and DNAm PhenoAge-based epigenetic age acceleration (30% and 11%, respectively) (12). However, in other studies, moderate to high heritability estimates have been reported for the latter one (33%–51%) (11,26). The AE model was more parsimonious and therefore used as the basis for the bivariate models to explore the common genetic and environmental factors underlying both epigenetic aging and PA.
Our findings revealed that the associations between PA and epigenetic aging depended on the utilized epigenetic age estimate and the form of PA. The results supported the existence of the PA paradox: high-intensity LTPA (sport index) was related to slower epigenetic aging and OPA (work index) to faster epigenetic aging when the newest epigenetic age estimate DNAm GrimAge was used. However, the associations mainly attenuated after controlling for smoking status. Associations between other epigenetic clocks (DNAm age and PhenoAge acceleration) and PA were very minor or nonexistent. Only a few previous studies have reported on the associations between PA and epigenetic aging with the novel epigenetic clocks. Stevenson and colleagues (14) showed a cross-sectional negative association of DNAm PhenoAge acceleration with LTPA at the age of 70 yr, but the analysis was controlled only for age and childhood cognitive ability. Zhao and colleagues (27) did not observe significant associations between LTPA and DNAm PhenoAge or DNAm GrimAge acceleration in older African Americans.
In addition to the cross-sectional associations, we provided evidence for beneficial association of long-term LTPA using a discordant twin pair design. Twin pairs discordant for LTPA for 32 yr differed in epigenetic aging measured with DNAm PhenoAge and DNAm GrimAge, although this was not seen when measured with DNAm age. Active twin pairs were epigenetically 2 to 3 yr younger on average compared with their inactive co-twins, when the genetic factors were controlled for partially (DZ pairs) or fully (MZ pairs). There were no differences in the effects between MZ and DZ twin pairs, but the mean within-pair differences were not significant in MZ pairs.
All the utilized epigenetic aging acceleration measures have been shown to predict mortality and morbidity risk, but DNAm GrimAge acceleration stands out in the prediction accuracy (12,28,29). Previous studies have shown that DNAm GrimAge may capture the stimulus of a variety of health- and lifestyle-related factors (12,27). In our study, LTPA was most consistently associated with DNAm GrimAge acceleration. DNAm GrimAge is a composite biomarker based on seven DNAm surrogates for plasma markers and smoking pack-years, which strongly predict time to death (12). Whereas CpGs for the other clocks were selected based on their association with a single reference, DNAm GrimAge was developed in two stages. First, CpGs for DNAm surrogates were selected based on their associations with the corresponding plasma protein levels and self-reported smoking pack-years. Second, DNAm-based surrogates for DNAm GrimAge estimator were selected based on their ability to predict mortality risk. This approach may have efficiently captured the CpGs associated with diverse health-related factors.
In our study, both genetic and nonshared environmental factors common to PA and the DNAm GrimAge acceleration explained the observed associations, but these influences attenuated after controlling the model for smoking status. Therefore, the observed opposite associations of LTPA and OPA on DNAm GrimAge acceleration may reflect an accumulation of unhealthy lifestyle habits among individuals in the lower socioeconomic class performing physically demanding work (30). Both genetic and environmental factors regulate smoking (31). Moreover, smoking has been shown to predict lower LTPA, also independently of genetic factors (32). Smoking is one of the most detrimental lifestyle factors and has been seen not only to increase the risk for multiple diseases (33) and mortality (34) but also to accelerate cellular aging (35). Interestingly, DNAm-based smoking pack-years (a component of DNAm GrimAge) has been shown to predict mortality risk better than original self-reported measure (12) and fully mediate the effects of self-reported smoking on biological aging (36). In line with this, we observed a very high correlation between DNAm smoking pack-years and DNAm GrimAge acceleration. Moreover, the opposite association of LTPA and OPA with the components of GrimAge was most evident in the case of DNAm smoking pack-years. These findings may support the recently stated arguments that the PA paradox is probably partly explained by an insufficient adjustment for smoking (8,37).
We observed that the lower values in several DNAm-based surrogate biomarkers included in the GrimAge estimator were correlated with higher levels of LTPA. LTPA promotes changes in multiple mechanistic and regulatory pathways that underlie the exercise-induced adaptations in metabolic profile, fitness, and body fat and muscle distribution. Lack of these LTPA-induced adaptations may increase the risk of cardiovascular and metabolic diseases at the population level, as different metabolic profiles have also been found among LTPA-discordant twin pairs (38), although differences in life expectancy have not been observed (39). Our study suggests that benefits of the LTPA may also be seen in epigenetic aging based on DNAm levels in blood, but its role is minor. The effect size was about half or less of the magnitude of the previously reported effect of LTPA on certain other health-related traits such as body fat, liver fat, and artery structure (38) known to be associated cardiovascular and other inactivity-related diseases. LTPA induces adaptations also directly in muscle tissue, which plays an important role in age-related decline in physical functioning. Future studies utilizing recently published epigenetic clock for human skeletal muscle (40) may show whether LTPA has a more substantial effect on epigenetic aging of muscle tissue.
Strengths and limitations
To our knowledge, this is the first study investigating the association of both LTPA and OPA with epigenetic aging. Our study utilizes novel epigenetic clocks that were published very recently. Twin design and the use of quantitative genetic modeling enabled us to study the genetic and environmental effects on epigenetic aging. In addition, we were able to investigate the effects of long-term LTPA on epigenetic aging after controlling for genetic factors by comparing co-twins of pairs discordant for LTPA for 30+ yr.
We acknowledge that our results are based on self-reported measure of PA, and potential recall bias and the effect of social desirability cannot be excluded. Baecke questionnaire has been shown to be valid and reliable tool to assess high-intensity LTPA, but all the light-intensity activities may not be properly measured (41). Activities such as gardening and household, which are increasingly important determinants of physical functioning with age (42), are not directly assessed by the questionnaire. In addition, the sample size of the LTPA discordant twins is limited, and therefore, statistical power to detect small effects may be insufficient. It should be noted that recent studies have shown that biological aging may be distinct stages rather than a continuous process, and aging progression may not be linear throughout the studied age ranges (43,44).
CONCLUSIONS
We show that LTPA associates with slower epigenetic aging, whereas OPA associates with accelerated epigenetic aging. The observed associations are explained by both common genetic and environmental factors. Importantly, adjusting the models for smoking status, which may reflect the accumulation of unhealthy lifestyle habits, attenuated the negative association of LTPA and the positive association of OPA with epigenetic aging.
Supplementary Material
Acknowledgments
The Gerontology Research Center is a joint effort between the University of Jyväskylä and the University of Tampere. This work was supported by the Academy of Finland (260001 to E. S.; 213506, 308248, and 312073 to J. K.; 251316 and 297908 to M. O.), EC FP5 GenomEUtwin (J. K.), National Institutes of Health/National Heart, Lung, and Blood Institute (grant HL104125), EC MC ITN Project EPITRAIN (J. K. and M. O.) project, and the University of Helsinki Research Funds to M. O., Sigrid Juselius Foundation to M. O. and J. K., and Yrjö Jahnsson Foundation (6868) and Juho Vainio Foundation to E. S. The TWINACTIVE study was supported by the Finnish Ministry of Education and Culture (U. M. K.), Academy of Finland (U. M. K.), and Juho Vainio Foundation (U. M. K.).
The authors declare that there are no conflicts of interest. The results of the present study do not constitute endorsement by the American College of Sports Medicine. The authors declare that the results of the study are presented clearly, honestly, and without fabrication, falsification, or inappropriate data manipulation.
Footnotes
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal’s Web site (www.acsm-msse.org).
Contributor Information
ASKO TOLVANEN, Email: asko.j.tolvanen@jyu.fi.
SAILALITHA BOLLEPALLI, Email: sailalitha.bollepalli@helsinki.fi.
TUIJA LESKINEN, Email: tuija.leskinen@utu.fi.
URHO M. KUJALA, Email: urho.m.kujala@jyu.fi.
JAAKKO KAPRIO, Email: jaakko.kaprio@helsinki.fi.
MIINA OLLIKAINEN, Email: miina.ollikainen@helsinki.fi.
ELINA SILLANPÄÄ, Email: elina.sillanpaa@jyu.fi.
REFERENCES
- 1.Li J, Loerbroks A, Angerer P. Physical activity and risk of cardiovascular disease: what does the new epidemiological evidence show? Curr Opin Cardiol. 2013;28(5):575–83. [DOI] [PubMed] [Google Scholar]
- 2.Myers J, Kokkinos P, Nyelin E. Physical activity, cardiorespiratory fitness, and the metabolic syndrome. Nutrients. 2019;11(7):1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Löllgen H, Böckenhoff A, Knapp G. Physical activity and all-cause mortality: an updated meta-analysis with different intensity categories. Int J Sports Med. 2009;30(3):213–24. [DOI] [PubMed] [Google Scholar]
- 4.Coenen P Huysmans MA Holtermann A, et al. Do highly physically active workers die early? A systematic review with meta-analysis of data from 193 696 participants. Br J Sports Med. 2018;52(20):1320–6. [DOI] [PubMed] [Google Scholar]
- 5.Holtermann A, Hansen JV, Burr H, Søgaard K, Sjøgaard G. The health paradox of occupational and leisure-time physical activity. Br J Sports Med. 2011;46(4):291–5. [DOI] [PubMed] [Google Scholar]
- 6.Hallman DM, Jørgensen MB, Holtermann A. On the health paradox of occupational and leisure-time physical activity using objective measurements: effects on autonomic imbalance. PLoS One. 2017;12(5):e0177042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holtermann A, Krause N, van der Beek AJ, Straker L. The physical activity paradox: six reasons why occupational physical activity (OPA) does not confer the cardiovascular health benefits that leisure time physical activity does. Br J Sports Med. 2018;52(3):149–50. [DOI] [PubMed] [Google Scholar]
- 8.Kujala UM. Is physical activity a cause of longevity? It is not as straightforward as some would believe. A critical analysis. Br J Sports Med. 2018;52(14):914–8. [DOI] [PubMed] [Google Scholar]
- 9.Christensen BC Houseman EA Marsit CJ, et al. Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CPG island context. PLoS Genet. 2009;5(8):e1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14:R115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levine ME Lu AT Quach A, et al. An epigenetic biomarker of aging for lifespan and healthspan. Aging (Albany NY). 2018;10(4):573–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu AT Quach A Wilson JG, et al. DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging (Albany NY). 2019;11(2):303–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ryan J, Wrigglesworth J, Loong J, Fransquet PD, Woods RL. A systematic review and meta-analysis of environmental, lifestyle, and health factors associated with DNA methylation age. J Gerontol A Biol Sci Med Sci. 2020;75(3):481–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stevenson AJ McCartney DL Hillary RF, et al. Childhood intelligence attenuates the association between biological ageing and health outcomes in later life. Transl Psychiatry. 2019;9:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaprio J. The Finnish Twin Cohort study: an update. Twin Res Hum Genet. 2013;16(1):157–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaprio J Bollepalli S Buchwald J, et al. The older Finnish Twin Cohort: 45 years of follow-up. Twin Res Hum Genet. 2019;22(4):240–54. [DOI] [PubMed] [Google Scholar]
- 17.Kaprio J, Koskenvuo M. Genetic and environmental factors in complex diseases: the older Finnish Twin Cohort. Twin Res. 2002;5(5):358–65. [DOI] [PubMed] [Google Scholar]
- 18.Leskinen T, Sipilä S, Kaprio J, Kainulainen H, Alen M, Kujala UM. Physically active vs. inactive lifestyle, muscle properties, and glucose homeostasis in middle-aged and older twins. Age (Omaha). 2013;35(5):1917–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leskinen T Waller K Mutikainen S, et al. Effects of 32-year leisure time physical activity discordance in twin pairs on health (TWINACTIVE study): aims, design and results for physical fitness. Twin Res Hum Genet. 2009;12(1):108–17. [DOI] [PubMed] [Google Scholar]
- 20.Baecke JAH, Burema J, Fritjters JER. A short questionnaire for the measurement of habitual physical activity in epidemiological studies. Am J Clin Nutr. 1982;96(5):936–42. [DOI] [PubMed] [Google Scholar]
- 21.Mustelin L, Joutsi J, Latvala A, Pietiläinen KH, Rissanen A, Kaprio J. Genetic influences on physical activity in young adults: a twin study. Med Sci Sports Exerc. 2012;44(7):1293–301. [DOI] [PubMed] [Google Scholar]
- 22.Muthén LK, Muthén BO. Mplus User’s Guide [internet]. 8th ed. Los Angeles, CA: Muthén & Muthén; Available from: https://www.statmodel.com. [Google Scholar]
- 23.Neale MC, Cardon LR. Methodology for Genetic Studies of Twins and Families. Dordrecht, the Netherlands: Kluver Academic Publisher; 1992. [Google Scholar]
- 24.de Jong PF. Hierarchical regression analysis in structural equation modeling. Struct Equ Model. 1999;6(2):198–211. [Google Scholar]
- 25.Sillanpää E Ollikainen M Kaprio J, et al. Leisure-time physical activity and DNA methylation age—a twin study. Clin Epigenetics. 2019;11(1):12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jylhävä J Hjelmborg J Soerensen M, et al. Longitudinal changes in the genetic and environmental influences on the epigenetic clocks across old age: evidence from two twin cohorts. EBioMedicine. 2019;40:710–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao W Ammous F Ratli S, et al. Education and lifestyle factors are associated with DNA methylation clocks in older African Americans. Int J Environ Res Public Health. 2019;16(17):3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hillary RF Stevenson AJ McCartney DL, et al. Epigenetic clocks predict prevalence and incidence of leading causes of death and disease burden. bioRxiv [Internet]. 2020 [cited 2020 May 2]. Available from: 10.1101/2020.01.31.928648. [DOI] [Google Scholar]
- 29.Li X Ploner A Wang Y, et al. Longitudinal trajectories, correlations and mortality associations of nine biological ages across 20-years follow-up. Elife. 2020;9:e51507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thebault JL, Ringa V, Panjo H, Bloy G, Falcoff H, Rigal L. Accumulation of unhealthy behaviors: marked social inequalities in men and women. Prev Med Rep. 2018;12:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vink JM, Boomsma DI. Interplay between heritability of smoking and environmental conditions? A comparison of two birth cohorts. BMC Public Health. 2011;11(1):316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Waller K Vähä-Ypyä H Törmäkangas T, et al. Long-term leisure-time physical activity and other health habits as predictors of objectively monitored late-life physical activity—a 40-year twin study. Sci Rep. 2018;8(1):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alberg AJ, Shopland DR, Cummings KM. The 2014 Surgeon General’s report: commemorating the 50th Anniversary of the 1964 Report of the Advisory Committee to the US Surgeon General and updating the evidence on the health consequences of cigarette smoking. Am J Epidemiol. 2014;179(4):403–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kujala UM, Kaprio J, Koskenvuo M. Modifiable risk factors as predictors of all-cause mortality: the roles of genetics and childhood environment. Am J Epidemiol. 2002;156(11):985–93. [DOI] [PubMed] [Google Scholar]
- 35.Valdes AM Andrew T Gardner JP, et al. Obesity, cigarette smoking, and telomere length in women. Lancet. 2005;366(9486):662–4. [DOI] [PubMed] [Google Scholar]
- 36.Lei M, Gibbons FX, Simons RL, Philibert RA, Beach SRH. The effect of tobacco smoking differs across indices of DNA methylation-based aging in an African American sample: DNA methylation-based indices of smoking capture these effects. Genes (Basel). 2020;11(3):311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shephard RJ. Is there a ‘recent occupational paradox’ where highly active physically active workers die early? or are there failures in some study methods? Br J Sports Med. 2019;53(24):1557–9. [DOI] [PubMed] [Google Scholar]
- 38.Leskinen T, Kujala UM. Health-related findings among twin pairs discordant for leisure-time physical activity for 32 years: the TWINACTIVE study synopsis. Twin Res Hum Genet. 2015;18(3):266–72. [DOI] [PubMed] [Google Scholar]
- 39.Karvinen S Waller K Silvennoinen M, et al. Physical activity in adulthood: genes and mortality. Sci Rep. 2015;5:18259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Voisin S Harvey NR Haupt LM, et al. An epigenetic clock for human skeletal muscle. J Cachexia Sarcopenia Muscle. 2020;11(4):887–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Richardson MT, Ainsworth BE, Wu HC, Jacobs DR, Leon AS. Ability of the Atherosclerosis Risk in Communities (ARIC)/Baecke Questionnaire to assess leisure-time physical activity. Int J Epidemiol. 1995;24(4):685–93. [DOI] [PubMed] [Google Scholar]
- 42.Peeters G, Van Gellecum YR, Van Uffelen JGZ, Burton NW, Brown WJ. Contribution of house and garden work to the association between physical activity and well-being in young, mid-aged and older women. Br J Sports Med. 2014;48(12):996–1001. [DOI] [PubMed] [Google Scholar]
- 43.Holzscheck N Söhle J Kristof B, et al. Multi-omics network analysis reveals distinct stages in the human aging progression in epidermal tissue. Aging (Albany NY). 2020;12(12):12393–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Timmons JA Volmar CH Crossland H, et al. Longevity-related molecular pathways are subject to midlife “switch” in humans. Aging Cell. 2019;18(4):e12970. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


