Abstract
CD4+T cell epitopes plays a key role in anti-tuberculosis (TB) immunity, CD4+T cell epitopes suitable for the domestic population are lacking. Therefore, we predicted and identified novel CD4+T cell epitopes.
The bioinformatics software, namely, DNAStar (DNASTAR of the United States), SYFPEITHI (INTERFACTORS INSTITUT Für ZELL Biologie of Germany), RANKPEP, and NetMHC IIpan (National Cancer Institute, United States of America), were used to comprehensively predict the CD4+T cell immune epitope of Mycobacterium TB, and the predicted epitope polypeptide was synthesized by the standard Fmoc scheme. The proliferation of PBMC and CD4+T cells stimulated by peptides was preliminarily detected by the CCK8 method. Then, the candidate polypeptides screened out by the CCK8 method were verified again by the BrdU assay, and flow cytometry was performed to analyze further the extent of their stimulation on the proliferation of CD4+T cells. The changes in the secreted cytokines IFN-γ, TNF-α, IL-2, and IL-10 before and after the candidate polypeptide stimulation of CD4+T lymphocytes were detected by ELISA. The preliminary humoral immunity test was conducted by indirect ELISA to evaluate the serological diagnostic value of the CD4+T cell epitope polypeptide.
In this study, 5 novel candidate CD4+T cell epitope polypeptides with the amino acid sequences of LQGQWRGAAGTAAQA, PVTLAETGSTLLYPL, AAAWGGSGSEAYQGV, QFVYAGAMSGLLDPS, and KAALTRTASNMNAAA and others that have not been reported in the research were predicted. For convenience, the 5 candidates were successively named as P39, P50, P40, P185, and P62. P39, P62, and the mixed peptide P39+P62 could effectively induce the proliferation of CD4+T cells and increase the secretion of IFN-γ, TNF-α, and IL-2 from the CD4+T cells, while reducing the content of IL-10. The serological test showed that the sensitivity, specificity, and area under the receiver operating characteristic curve (AUC) of P39 were 75%, 67.71%, and 0.844, respectively. The sensitivity, specificity, and AUC of P62 were 91.66%, 46.87%, and 0.649, respectively. The sensitivity of the mixed peptide P39+P62 was 95.83%, the specificity was 97.91%, and the AUC was 0.793.
The P39 and P62 polypeptides were predicted and identified as potential CD4+T cell immune epitope polypeptides of M. TB. The polypeptide had better diagnosis effect, which provided potential candidate epitope polypeptides for the development of TB-specific diagnosis reagents and novel TB epitope vaccines.
Keywords: antigen, CD4+T cell epitope, identification, immunoinformatics
1. Introduction
Tuberculosis (TB) is a chronic infectious disease caused by Mycobacterium TB (MTB), and the leading cause of death from infectious diseases worldwide. China is one of the countries with a high burden of TB. In recent years, the emergence of drug-resistant TB, human immunodeficiency virus combined with MTB infection, and other issues have made the epidemic situation of TB more serious. Considering high morbidity and high mortality,[1] the traditional BCG vaccine cannot meet the needs of preventing TB, and the diagnostic technology based on traditional bacteriology and immunology cannot adapt to the current epidemic situation. It is urgent to develop novel TB vaccines, therapeutic drugs, and diagnostic methods for TB infection that are suitable for the Chinese population.
CD4+T cell-mediated antituberculosis immune protection, including the control of early TB infection, its spread to the lungs, as well as the helper CD8+T cells and natural killer cells, enhance the killing effect on MTB. The secreted cytokines IFN-γ, TNF-α, and IL-2 are the active ingredients that control the MTB infection and are important components for the body's antituberculosis response.[2] Epitopes are the material basis of immunogenicity. Studies have shown that protein antigens do not function through their complete molecules but reflect their specificity through their epitopes.[3] Since many immune reactions triggered by natural antigens cannot meet the needs of preventing TB infection and morbidity, in order to improve the protection of protein antigens, it is necessary to select and modify the epitopes, which requires epitope screening and identification. The research on epitopes is be of great significance to the diagnosis of antigens and design of epitope vaccines. CD4+T cell epitope is the basis of an antituberculosis T cell reaction and the key to the development and design of candidate vaccines, diagnostic reagents, and biomarkers for evaluating therapeutic efficacy.[4] Among the various protective antigens of MTB, CFP10, 38KDa, ESAT6, Ag85A, and TB27.4 have great advantages in the immune response of humans to MTB; these are markers for the serological detection of active TB and can stimulate the body to produce specific cellular immune responses, with multiple T cell epitope peptides, which are effective T cell antigens and show strong induction of IFN-γ. These are considered the most important protective antigens, the most widely studied antigens in the field of diagnosis of TB, but some of the genetic regions of the 5 antigenic proteins may be homologous with the genes outside the missing regions, leading to cross-reaction at the level of the immune response, thereby impairing the specificity of the experimental results. However, the epitope peptide derived from proteins is safe, eliminates other harmful components of the pathogens, can be obtained completely by artificial synthesis, and has a short research and development cycle. Multiple epitope peptides can also be used in series to enhance the immune effect and reduce immune escape. Therefore, it is necessary to use a protein-derived polypeptide or its truncated form to replace protein macromolecules clinically.[5]
With the rapid development of molecular biology, molecular immunology, bioinformatics, and other technologies, as well as a growing understanding of the binding characteristics of antigen epitopes and HLA-II molecules, many bioinformatics software technologies have emerged for efficient epitope prediction. The advantage of this method is that it can select the best set of peptide candidates for immunogenicity testing, eliminating the necessity for synthesizing a large number of overlapping peptides, and more importantly, avoiding the in vitro testing of the binding ability of each HLA allele.[6] Therefore, modern researchers in the field of identification of epitopes are becoming increasingly inclined to use bioinformatics software for predicting the binding of cell epitope peptides to the MHC molecules, synthesizing according to the results of the prediction, and finally, validating the synthetic epitope peptides in vitro or in vivo by traditional methods.[7] At present, the known epitope peptides of MTB are mainly identified for the European and American populations, and most of them are restricted by HLA-A∗0201. However, there is no suitable epitope peptide available for the people in Africa and Southeast Asia, where TB is the most prevalent. China is one of the countries with the heaviest burden of TB in the world, but there is little research on HLA-II allele restriction epitope peptides specifically targeting MTB infected people in China. The HLA-II allele frequency of the Chinese population is analyzed by the AFND website. Considering the common genes and the difficulty of blood source, we select HLA-DRB1∗0701 genotype to predict and identify CD4+T cell immune epitopes.
In this study, 4 bioinformatics software packages, namely, DNAStar (DNASTAR of the United States),[8] SYFPEITHI (INTERFACTORS INSTITUT Für ZELL Biologie of Germany),[9] RANKPEP,[10] and NetMHC IIpan (National Cancer Institute, United States of America),[11] were used to predict the restricted CD4+T cell epitopes of HLA-DRB1 ∗ 0701 for the 5 protective antigens of MTB. After excluding the epitope database and the epitopes published in the literature, the epitope polypeptides were synthesized. Finally, the synthesized polypeptides were immunologically verified, and clinical samples were used to evaluate the antigenicity of these epitope polypeptides to study their humoral immunity, cellular immunity, and their relationships, and to screen out the potential antigenic epitopes that could be used for the diagnosis of TB or for developing novel vaccines, which would provide some references for the future research on the antigenic epitopes of MTB as well as new ideas for further effective prevention and treatment of TB. It would be of great significance for the design and development of novel epitope vaccines suitable for the Chinese population.
2. Materials and methods
2.1. Selection of CD4+T cell epitopes within the proteins by bioinformatic prediction
The protein sequences of CFP10, 38KDa, ESAT6, Ag85A, and TB27.4 were obtained from the NCBI database, and the homology between the amino acid sequences and the human protein sequences was analyzed by EXPASY online software. The Protean template in the DNAStar (DNASTAR of the United States) software[12] was used to predict the secondary structure, hydrophobicity and hydrophilicity of the protein, and the possibility that a specific region is located on the surface of the protein. The plasticity of the protein was analyzed to predict the potential protein antigenic determinants, immune superiority auxiliary T lymphocyte antigenic sites, and potential T lymphocyte antigenic determinants with specific motifs. The restricted CD4+T cell epitopes of HLA-DRB1∗0701 were predicted and analyzed by SYFPEITHTI, RANKPEP, and NetMHC IIpan (National Cancer Institute, United States of America) software programs.[13] SYFPEITHTI concluded that the probability of the peptide with the first 2% score becoming an antigenic peptide was more than 80%. In order to improve the accuracy of epitope prediction,[14] 15 consecutive amino acid sequences with a score of more than 24 were selected for further analysis. In RANKPEP (Harvard Medical School of the United States) prediction, the results with red-labeled amino acid sequences were selected for further analysis,[15] while the polypeptides > 0.75 were selected in NetMHC IIpan (National Cancer Institute, United States of America)Server prediction.[16] In order to improve the accuracy of epitopes predicted by the bioinformatics software, the predicted results from the 3 bioinformatics software packages were analyzed comprehensively. The CD4+T cell epitopes with high scores in the 3 prediction methods were selected, and the known epitopes were removed. Combined with the results of homology between the antigen protein and the human protein, the secondary structure and the prediction results of protein hydrophilicity, hydrophobicity, flexibility and plasticity, surface possibility, and antigen epitope peptides were obtained, with significantly higher prediction scores selected as candidate CD4+T cell epitopes.[17]
2.2. Study subjects
The study was conducted in accordance with the ethical standards of the Declaration of Helsinki. This study did not infringe on the subjects’ right to know and privacy. All the subjects were aware of the experimental content and purpose before sampling and either signed or thumb printed the informed consent form to express their willingness to participate in the study. The study was approved by the Ethics Review Committee of the First Affiliated Hospital of Shihezi University under the symbol 2018–042–01.
A total of 144 eligible subjects, which included TB patients and healthy volunteers, were recruited in this experimental study. All subjects with a history of chronic diseases (chronic obstructive pulmonary disease, asthma, lung cancer, and hypertension) and HIV infection, tumor, long-term use of hormones, organ transplantation, and immunocompromised condition were excluded from the study.
The inclusion criteria were[18]:
-
(1)
Patients with TB were diagnosed with clinical symptoms, signs, and chest imaging, or the sputum culture and sputum smear results were positive.
-
(2)
Healthy volunteers, including no clinical symptoms of TB, no TB contact history and previous history of TB, normal imaging examination, and negative serum TB antibody test result.
The basic situation and clinical characteristics of the selected subjects are shown in Table (Table 1). Serum and blood samples were collected, and the HLA-DRB1∗0701 positive TB patients were screened out using PCR-SBT for HLA typing.
Table 1.
Basic information and the clinical characteristics of the selected subjects.
| Characteristics | TB | Healthy control |
| N (Female, Male) | 48 (13,35) | 96 (50,46) |
| Average Age (range) | 56 (24,79) | 39 (20,59) |
| Diabetes | 2 | 0 |
| BCG Status | ||
| Vaccinated | 37 | 89 |
| Unvaccinated | 6 | 5 |
| Unknown | 5 | 2 |
| History of antituberculosis therapy | 30 | 0 |
| Sputum smear (+) | 12 | 0 |
| Tuberculin skin testing | ||
| Negative | Not done | 96 |
| Positive | 48 | Not done |
TB = tuberculosis
2.3. Synthesis, purification, identification, dissolution, and preservation of peptides
In this study, peptides were synthesized using the standard Fmoc protocol. The synthesized epitope peptides were purified by high performance liquid chromatography (HPLC), analyzed for purity, and qualitatively identified by mass spectrometry (MS). The purified peptides were directly mixed with the PBS solution in the required concentration, along with a small amount of DMSO if necessary. The peptide solution was kept in the refrigerator at–80 degrees to avoid repeated freezing and thawing.
2.4. Determination of the optimal peptide dilution concentration to stimulate cell proliferation
In this study, we chose to establish the appropriate concentration of polypeptide stimulation to reduce the cost of using the polypeptide and achieve a certain effect of stimulating cell proliferation. In this study, 1 verified epitope peptide of CD4+T cells was diluted with PBS into 5 different polypeptide concentrations: 2.5 μg/mL, 5 μg/mL, 10 μg/mL, 20 μg/mL, and 40 μg/mL, according to the reference of single polypeptide stimulation concentration in the domestic and international literature,[19] to stimulate PBMCs of the HLA-DRB1∗0701 positive TB patients. Then, 5 μL CCK8 reagent (Cat.No.ab228554, Abcam) was added. All the steps were performed according to the manufacturer's instructions. The optimal working concentration of the polypeptide for cell stimulation was determined by the stimulation index (SI); SI= optical density (OD)450 nm value of epitope peptide well/OD450 nm value of non-epitope peptide well. SI≥2 was positive, and the antigenic peptide with SI≥2 was considered to be able to induce lymphocyte proliferation.[20]
2.5. Preliminary detection of the proliferation levels of peripheral blood mononuclear cells and CD4+T cells stimulated by the candidate peptides
The peripheral blood mononuclear cells and CD4+T cells of the HLA-DRB1∗0701 positive sample of TB patients were added into a 96-well plate, and 100 μL of cell suspension with a cell concentration of 2.5 × 106/mL was added to each well. An experimental group and a blank control group were set up. The experimental group was added with 5 μL of candidate peptide at the optimal working concentration. No candidate peptide was added to the blank control well. Six patient samples were tested for each group. After 5 days of culture in the incubator, 5 μL CCK8 reagent (Cat.No.ab228554, Abcam) was added in all the steps according to the manufacturer's instructions. The mean and standard deviation of the experimental group and blank control group were calculated. The SI value was used to study the effect of candidate peptide on the proliferation and activation of peripheral blood mononuclear cells and CD4+T cells, and the final predicted candidate peptide was verified.
2.6. The preliminarily verified candidate peptides were subjected to BrdU detection to analyze cell proliferation and flow cytometry to analyze the phenotype of cell proliferation
A volume of 100 μL of the cells not stimulated by the candidate polypeptide and 100 μL of the cells stimulated by the candidate polypeptide were grown in 96-well cell culture plates using the BrdU cell proliferation assay kit (Cat.No.#11647229001, Roche), according to the manufacturer's instructions. Six patient samples were tested for each group. Three re-wells were set for each sample, the OD value of the cell culture well was measured at the wavelength of 450 nm by ELISA reader, and the average value was taken as the test result.
After the cells stimulated by the candidate peptides were cultured for 5 days, 5 μL FITC-labeled murine anti-human CD4 (Cat. No.#317408, Biolegend) and 5 μL PE-labeled murine anti-human CD3 (Cat. No.#300308, Biolegend) were added to the experimental tubes, which were then incubated on ice in the dark for 30 minutes. Then, 2 mL PBS washing solution containing 2% BSA was added, followed by centrifugation at 2000 rpm for 5 minutes, discarding the supernatant, and 2 washes. Then the precipitate was re-suspended with 350 μL PBS and subjected to flow cytometry to observe the changes in the CD4+T cells before and after the stimulation with the candidate peptide.
2.7. ELISA to detect secreted cytokine levels before and after stimulation with candidate peptides
The CD4+T cell suspensions with cell concentration of 1 × 106/mL (100uL each) were spread on 96-well culture plates. 10 μL of the candidate peptides were added to the experimental group and not to the blank group. Three parallel wells were used in each group. The cell suspension was collected at 2000 rpm for 5 minutes after 5 days of culture in strict accordance with Human IFN-γ ELISA Kit (Cat. No.ab231036, Abcam), Human TNF-α ELISA Kit (Cat. No.ab183218, Abcam), Human IL-2 ELISA Kit (Cat. No.ab222639, Abcam), and Human IL-10 ELISA Kit (Cat. No.ab134742, Abcam) instructions of the experimental steps for the quantitative detection of IFN-γ, TNF-α, IL-2, and IL-10 expression.
2.8. The candidate polypeptide was used as a coating antigen to determine the optimal condition for detecting serum immunoglobulin G (IgG) antibody by indirect ELISA
Antibody titrations were performed by indirect ELISA.[21] The candidate peptides were diluted with a carbonate buffer solution and then incubated overnight at 4°C in 96-well plates (Cat. No.#44-2404-21, Invitrogen) with 100 μg/mL per will. The plates were washed 5 times with PBST times and then incubated with 5% skimmed milk powder at 37°C for 2 hours. The plates were again washed 5 times with PBST, the second antibody was added [Goat anti-Human IgG/ Horseradish Peroxidase ] (Cat. No. SE101, Solarbio) and incubated at 37°C for 1 hour. The plates were washed again with PBST 5 times, single component TMB color developing solution was added and left at 37°C for 10 minutes. The reaction was stopped by the addition of 50 μL of 2 mol/L H2SO4 to each well. The plate was read at 450 nm on a multiwell plate reader (Molecular Devices, SpectraMax, Silicon Valley of the United States). The OD value was measured at 450 nm, and positive (P)/negative (N) [P/N] value = (positive well OD value − blank well OD value)/(negative well OD value − blank well OD value) was calculated.
The epitope polypeptide coating concentration, serum dilution ratio, and enzyme-labeled secondary anti-dilution ratio corresponding to the P/N maximum (P/N value ≥2.1 was taken as effective data) were taken as the optimal conditions for the indirect ELISA detection of serum IgG antibody.[22]
2.9. Indirect ELISA was used to test the clinical samples and serological evaluation was conducted for the candidate peptides
In order to evaluate the efficacy of CD4+T epitope peptides in the detection of TB and to screen out the CD4+T epitope peptides with diagnostic potential, the indirect ELISA established under the optimal conditions of IgG antibody test was used to detect 48 sera of TB patients and 96 serum samples of healthy people. The absorbance was measured at 450 nm, and the serological evaluation of the candidate peptides was conducted to calculate the sensitivity, specificity, and receiver operating characteristic (ROC) curve of each polypeptide in the detection of TB. The ROC curve reflected the relationship between sensitivity and specificity, with the false positive rate (1-specificity) in the horizontal coordinate and the true positive rate (sensitivity) in the vertical coordinate. The ROC curve was analyzed and drawn using the R software (MathSoft of New Zealand) to calculate the maximum area under the ROC curve (AUC) for evaluation of the diagnostic energy efficiency of the polypeptide. If AUC≈1, the method was the ideal test index. If 0.7<AUC≤ 0.9, the experimental accuracy was high, and it had reliable diagnostic value. If 0.5<AUC≤0.7, the experimental accuracy was low, and it had a certain diagnostic value. If AUC< 0.5, the diagnostic experiment was worthless. Youden index is a measure of the effectiveness of biomarkers. Youden index= sensitivity+specificity−1. The larger the value of the index, the better the authenticity. The cut-off value with the highest sum of sensitivity and specificity was considered to be the optimal cut-off value. According to the cut-off value of each epitope polypeptide, it was determined that the serum was negative and positive, that is, the OD value of the serum to be tested≥the cut-off value was considered positive, while the OD value of the sample to be tested < the cut-off value was considered negative.[23]
2.10. Statistical analysis
Statistical analysis of the data was performed using the SPSS 20.0 statistical software (IBM in the United State). All measurement data were expressed as mean±standard deviation (mean±SD). Statistical correlation of data was checked for significance by ANOVA and the Student–Newman–Keμls (SNK) Test. The mean comparison between the 2 samples was an independent sample test. At P< .05, the difference was significant.
3. Results
3.1. Selection of CD4+T cell epitopes within the proteins by bioinformatics prediction
The results of BLAST analysis showed that the amino acid sequence at positions 58 to 100 of the CFP10 protein had the highest similarity with HCG, which was 28.1% identical. The 100 to 360 amino acid sequence of the 38KDa protein has the highest similarity to the DING protein P38, and its homology was 41.2%. The amino acid sequence at positions 60 to 89 of the ESAT6 protein had the highest similarity with the regulation-related protein of mTOR (raptor), which was 29.6% identical. The amino acid sequence from 69 to 127 of Ag85A protein has the highest similarity with human protein TULP3, and its homology was 33.82%. The TB27.4 protein had no amino acid sequence similarity to the human protein, and its homology was 0.
The structures of the 5 antigenic proteins were analyzed by using the Protean template in the DNAStar (DNASTAR of the United States) software package. The results showed that the protein skeletons of the 5 proteins contained more flexible regions, which were consistent with the distribution of regions with a high antigenic index. Besides, they had strong hydrophilicity, similar to the regions with surface possibility, and the probability of these regions being exposed to the cell surface was higher, so that they have a high probability of distortion and folding,[24] suggesting that the 5 proteins are relatively flexible, may have strong antigenicity, and are easy to be chimeric with antibodies. As antigenic epitopes, they have a high probability of being exposed to the cell surface, and it is preliminarily predicted that the 5 proteins have more CD4+T cell immune epitopes.
After a specific analysis, the CD4+T cell epitope of CFP10 protein was found to be mainly located at amino acid residues 15 to 22, 32 to 44, 57 to 60, and 70 to 76. The CD4+T cell epitope of the 38KDa protein was mainly located at amino acid residues 49 to 68, 154 to 164, 202 to 260, 141 to 146, and 297 to 305. The CD4+T cell epitope of ESAT6 protein was mainly located at amino acid residues 17 to 31, 34 to 43, 46 to 55, and 64 to 81. It is predicted that the CD4+T cell epitopes of Ag85A protein are mainly located at amino acid residues 7 to 42, 161 to 195, and 269 to 301. The CD4+T cell epitopes of TB27.4 protein are mainly located at amino acids 38 to 82, 100 to 134, 141 to 151, 167 to 181, and 236 to 268.
The 3 bioinformatics software SYFPEITHTI, RANKPEP, and Net MHC II Pan were independently predicted and analyzed to obtain candidate peptides with high scores and were combined with the antigen protein and human protein homology analysis, secondary structure and protein hydrophilicity, hydrophobicity, flexible plasticity, and surface possibility prediction results for a comprehensive analysis to obtain the candidate peptides with significantly high comprehensive scores. After the known epitopes were removed, the 5 candidate CD4+T cell epitope polypeptides were finally predicted, with the basic information shown in Table (Table 2).
Table 2.
The basic information of the predicted CD4+T epitopes.
| Epitope peptides sequence | Source of protein | Amino acid position | Naming | Formula | Theoretical PI | Aliphatic index | Instability index |
| LQGQWRGAAGTAAQA | CFP10 | 39–53 | P39 | C63H100N22O20 | 9.75 | 59.33 | 14.09 |
| PVTLAETGSTLLYPL | 38KDa | 50–64 | P50 | C73H119N15O23 | 4.00 | 130 | 19.08 |
| AAAWGGSGSEAYQGV | ESAT6 | 40–54 | P40 | C61H87N17O22 | 4.00 | 46 | 23.49 |
| QFVYAGAMSGLLDPS | Ag85A | 185–199 | P185 | C70H106N16O22S1 | 3.80 | 84.67 | 51.54 |
| KAALTRTASNMNAAA | TB27.4 | 62–76 | P62 | C60H107N21O21S1 | 11.00 | 66 | 9.33 |
3.2. Results of RP-HPLC and MS analysis of the synthetic candidate epitope peptides
Five candidate peptides P39, P50, P40, P185, and P62, were successfully synthesized. Peptide purity was > 90%, as detected by HPLC. The peptide purities of P39, P50, P40, P185, and P62 were 96.54%, 96.27%, 92.61%, 91.79%, and 96.99%, respectively. The results of MS analysis to identify the synthesized candidate peptides P39, P50, P40, P185, and P62 showed that the measured molecular masses of synthesized candidate peptides were 1485.63, 1574.85, 1410.47, 1555.78, and 1490.71, respectively, consistent with the theoretical values.
The obtained peptides met the experimental requirements and were the desired candidate peptides. The experimental results confirmed that the obtained epitope polypeptide was a high-purity epitope polypeptide and provided a basis for further immune verification experiments with these CD4+T dominant epitopes.
3.3. Optimal peptide dilution concentration to stimulate cell proliferation
The PBMCs of HLA-DRB1∗0701 positive TB patients were stimulated with 5 different concentrations of CD4+T cell epitope peptides. When the stimulation concentration is 40 μg/mL peptide, the SI value is the highest, and the cell proliferation effect is the most suitable. Therefore, we determined that the optimal working concentration of the peptide is 40 μg/mL (Fig. 1).
Figure 1.
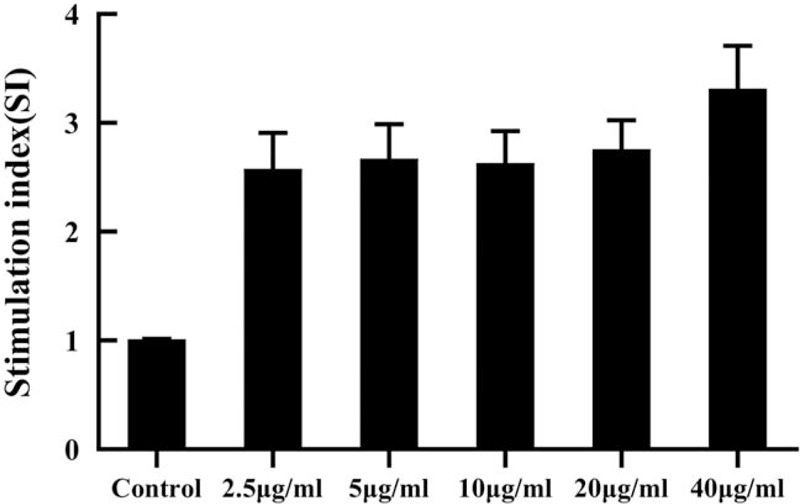
Effects of different concentrations of CD4+T cell epitope peptide on lymphocyte proliferation. Notes: the results were expressed as a stimulation index (SI= OD450 nm value of epitope peptide group /OD450 nm value of non-epitope peptide group). SI ≥ 2 was considered positive.
3.4. Candidate peptides stimulate the proliferation of peripheral blood mononuclear cells and CD4+T cells
The results of SI values in Figure (Fig. 2) show that the P39, P50, P40, P185, and P62 groups could effectively stimulate the proliferation of peripheral blood mononuclear cells (SI≥2), and the 5 candidate epitopes P39, P50, P40, P185, and P62 could be screened out. The CD4+T cells of the same population were used as feeder cells to observe the proliferation of CD4+T cells. The results showed that the proliferation of CD4+T cells in P39 and P62 groups was similar to that of the peripheral blood mononuclear cells, and there was no significant difference in proliferation degree (P > .05). It was concluded that P39 and P62 mainly induced the proliferation of CD4+T cells. According to the experimental results, the candidate peptides P39 and P62 can stimulate the proliferation of CD4+T cells of the HLA-DRB1∗0701-positive TB patients with good immunogenicity and are effective HLA-DRB1∗0701-restricted CD4+T epitopes of MTB.
Figure 2.
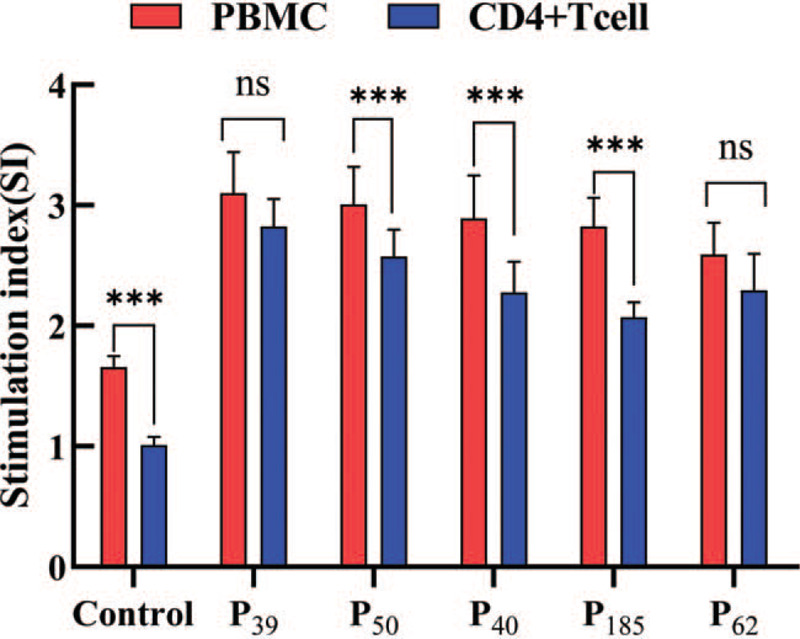
Comparison of CD4+T cell proliferation and peripheral blood lymphocytes proliferation. Notes:∗P < .05 vs Control group, ∗∗P < .01 vs Control group, ∗∗∗P < .001 vs Control group. nsP > .05 vs Control group.
3.5. The candidate polypeptides screened by the CCK8 method were analyzed for the cell proliferation effect again by the BrdU method and flow cytometry
The candidate polypeptides preliminarily screened by the CCK8 method stimulated CD4+T cell proliferation in vitro in the patients with HLA-DRB1∗0701-positive TB. The OD values of P39, P62, and the mixed peptide P39+P62 for the stimulation of CD4+T cell proliferation are shown in Table (Table 3). The results showed that the differences were significant between the control group and the P39, P62 and the mixed peptide P39+P62 groups (P < .01), indicating that P39, P62, and the mixed peptide P39+P62 could all stimulate CD4+T cell proliferation.
Table 3.
Comparison of cell phenotypes and cytokines before and after CD4+T cell stimulation by candidate epitope peptides (mean ± SD).
| Group | OD value (mean ± SD) |
| Control | 0.21 ± 0. 01 |
| P39 | 0.44 ± 0 .15∗∗∗ |
| P62 | 0.51 ± 0. 05∗∗∗ |
| P39+P62 | 0.52 ± 0 .06∗∗∗ |
According to the normality and homogeneity of variance, the experimental data were analyzed by single factor and multilevel variance analysis, and the Student–Newman–KeμLs test was used for pairwise analysis and comparison of data between groups. When P < .05, there was a significant difference.
∗P < .05 vs Control group.
∗∗P < .01 vs Control group.
P < .001 vs Control group.
Using flow cytometry to analyze the cell phenotype, P39, P62, and the mixed peptide P39+P62 stimulated the lymphocytes in the patients with HLA-DRB1∗0701-positive TB, and the proportion of CD4+T lymphocytes in the blank control without synthetic peptides stimulation was 36.40% ± 0.91% (Fig. 3A). The percentages of CD4+T lymphocytes stimulated by P39, P62, and the mixed peptide P39+P62 were 38.17% ± 1.00%(Fig. 3B), 41.96% ± 1.66% (Fig. 3C), and 44.57% ± 1.18% (Fig. 3D), respectively, which were significantly higher than those in the blank control group (P < .05). It was inferred that P39, P62, and the mixed peptide P39+P62 could stimulate the proliferation of CD4+T lymphocytes (Fig. 3 is the flow diagram of one of the experiments). Previous studies showed that P39 and P62 had good immunogenicity and were effective HLA-DRB1∗0701-restricted CD4+T epitopes of MTB.
Figure 3.
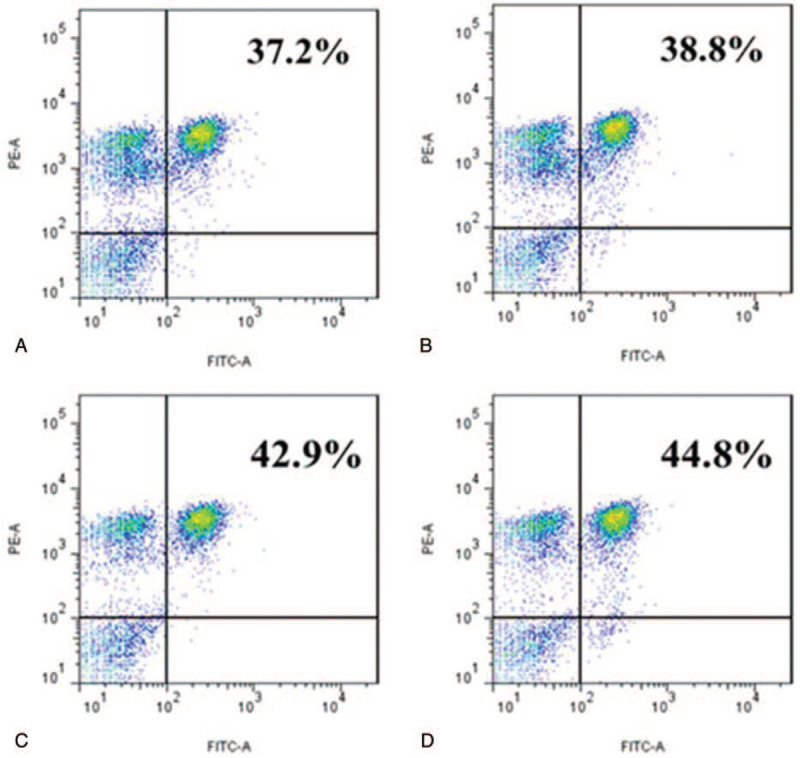
Phenotype analysis of the lymphocytes stimulated by CD4+T cell epitope peptides. Notes: The negative control (Fig. 3A 3Aof the proliferation assay. (The mean of 3 separate experiments is 36.40% ± 0.91%); the percent of CD4+T cells after stimulation by P39 (Fig. 3B), P62 (Fig. 3C), and P39 + P62 (Fig. 3D). (The mean of 3 separate experiments is 38.17% ± 1.00%, 41.96% ± 1.66%, and 44.57% ± 1.18% respectively); B, C, D vs A: P < .05Values represent mean ± SD from 3 independent experiments. This figure illustrates 1 of the 3 independent experiments.
3.6. Analysis of cytokine content before and after epitope polypeptide stimulation
The results are shown in Table (Table 4). The ELISA results showed that the levels of IFN-γ, TNF-α, and IL-2 were increased in the HLA-DRB1∗0701-positive serum samples from the TB patients stimulated with P39, P62, and mixed peptide P39+P62, while the levels of IL-10 were decreased and were significant compared to their levels before stimulation. The levels of the 4 cytokines remained in a stable range. The results indicated that P39, P62, and mixed peptide P39+P62 enhanced the secretion of IFN-γ, TNF-α, and IL-2 in HLA-DRB1∗0701-positive TB patients, while the secretion of IL-10 was down regulated. Therefore, P39 and P62 could induce the mixed cellular immune response of CD4+Th1 and CD4+Th2 in HLA-DRB1∗0701-positive TB patients, but CD4+Th1 was the major cellular immune response.
Table 4.
Comparison of cell phenotype and cytokine before and after the stimulation of CD4+T cell with candidate epitope peptide.
| Group | Control | P39 | P62 | P39+P62 |
| CD4+T% | 36.40 ± 0.91 | 38.17 ± 1.00∗∗∗ | 41.96 ± 1.66∗∗∗ | 44.57 ± 1.18∗∗∗ |
| IFN-γ(pg/mL) | 52.29 ± 7.52 | 86.24 ± 4.91∗∗∗ | 91.54 ± 1.86∗∗∗ | 102.72 ± 5.19∗∗∗ |
| TNF-α(pg/mL) | 64.42 ± 9.44 | 83.65 ± 7.30∗∗∗ | 88.24 ± 5.22∗∗∗ | 99.53 ± 4.47∗∗∗ |
| IL-2 (pg/mL) | 2.96 ± 0.14 | 4.94 ± 0.27∗∗∗ | 5.03 ± 0.36∗∗∗ | 6.11 ± 0.67∗∗∗ |
| IL-10 (pg/mL) | 9.91 ± 0.68 | 9.76 ± 0.53 | 8.98 ± 0.78 | 5.87 ± 0.29∗∗∗ |
According to the normality and homogeneity of variance, the experimental data were analyzed by single factor and multilevel variance analysis, and the Student–Newman–KeμLs test was used for pairwise analysis and comparison of data between groups. When P < .05, there was a significant difference.
∗P < .05 vs Control group.
∗∗P < .01 vs Control group.
P < .001 vs Control group.
Based on the results of the lymphocyte proliferation test, CD4+T cell proliferation analysis, and flow cytometric analysis, the epitope peptides P39 and P62 are the HLA-DRB1∗0701-restricted CD4+T epitopes of MTB immunity. In this study, the cytokines secreted by CD4+T lymphocytes stimulated by epitope peptides were detected by ELISA. The results showed that P39 and P62 could stimulate the CD4+Th0 cells to differentiate into CD4+Th1 cells.
3.7. Establishment of indirect ELISA for detection of TB antibody
The candidate epitope polypeptide was used as the coating antigen, and positive (P)/negative (N) [P/N] value = (positive well OD value − blank well OD value)/(negative well OD value − blank well OD value) was calculated. The optimal condition for detecting serum IgG antibody by indirect ELISA was established based on the coating concentration of the epitope polypeptide, the serum dilution ratio, and the enzyme-labeled secondary antibody dilution ratio corresponding to the maximum P/N. The experimental results are shown in Table (Table 5).
Table 5.
The optimal conditions for the detection of serum immunoglobulin G antibodies by different polypeptides in the establishment of indirect ELISA.
| Group | Polypeptide coating concentration | Serum dilution ratio | IgG secondary antibody dilution ratio | P/N value |
| P39 | 10 μg/mL | 1:500 | 1:40000 | 5.58 |
| P62 | 0.25 μg/mL | 1:100 | 1:20000 | 8.11 |
| P39+P62 | 10 μg/mL | 1:50 | 1:10000 | 21.71 |
positive (P)/ negative (N) [P/N] value = (positive well OD value − blank well OD value)/ (negative well OD value − blank well OD value); When P/N value ≥ 2.1, diagnosis is meaningful. When the P/N value was maximum, the corresponding coating concentration, serum concentration and secondary antibody concentration were the optimal conditions.
IgG = immunoglobulin G.
3.8. Analysis and evaluation of candidate epitope peptides in the detection of TB
The results of indirect Elisa showed that the specific IgG antibody levels of candidate peptides P39, P62, and mixed peptide P39+P62 in active TB group were significantly higher than those in the healthy control group (P < .001). In the preliminary test, 36 positive samples of P39 were detected in the active TB patients, while 31 positive samples were detected in a healthy population, with a sensitivity of 75%, a specificity of 67.71%, a Youden index of 0.667, and a cut-off value of 0.259. ROC analysis revealed that the AUC under the curve was 0.844 (Fig. 4). The sensitivity and specificity of P62 were 91.66% and 46.87%, respectively, in 44 positive samples from the active TB patients and 51 positive samples from the healthy controls. The Youden index was 0.458, the cut-off value was 0.238, and the area under the curve was 0.649, as determined by the ROC analysis and shown in Figure (Fig. 5). The 2 epitope peptides combined with P39 +P62 detected 46 positive samples in the active TB population and 2 positive samples in the healthy population, with a sensitivity of 95.83%, a specificity of 97.91%, a Youden index of 0.625, and a cut-off of 0.184. Using the ROC analysis, the AUC was found to be 0.793 (Fig. 6)
Figure 4.
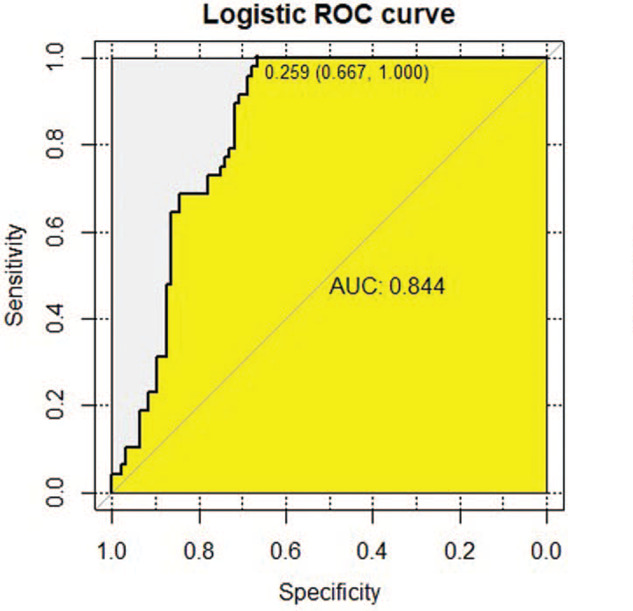
The receiver operating characteristic (ROC) curve analysis of immunoglobulin G antibody response for the Candidate Peptide P39. Notes: ROC, receiver operating characteristics; area under the curve (AUC) Diagnostic basis of ROC curve: When the area of the curve was between 0.5 to 1, the closer to 1, that is, the larger the area, the higher the diagnostic accuracy would be. When 0.5 < AUC≤0.7, there is a lower accuracy; When 0.7 < AUC≤0.9, there is a certain accuracy; When the AUC > 0.9, the diagnosis has a higher accuracy. The sensitivity of P39 was 75%, the specificity was 67.71%, the Yoden Index was 0.667, the cut-off value was 0.259, and the AUC value was 0.844.
Figure 5.
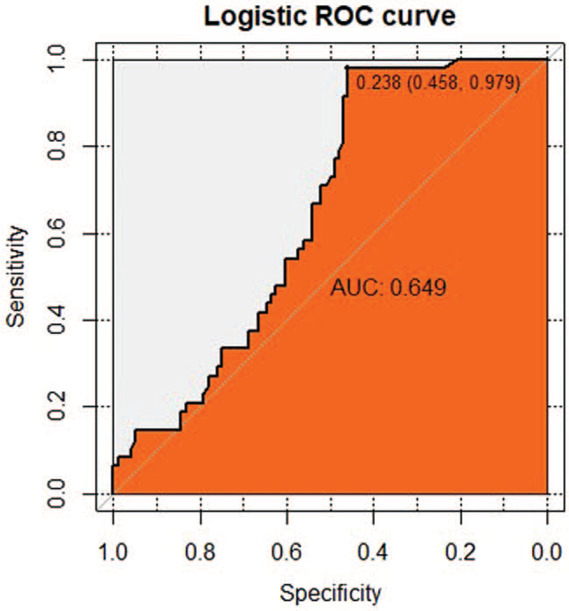
The receiver operating characteristic (ROC) curve analysis of immunoglobulin G antibody response for the Candidate Peptide P62. Notes: ROC, receiver operating characteristics; area under the curve (AUC), area under the curve. Diagnostic basis of ROC curve: When the area of the curve was between 0.5 to 1, the closer to 1, that is, the larger the area, the higher the diagnostic accuracy would be. When 0.5 < AUC≤0.7, there is a lower accuracy; When 0.7 < AUC≤0.9, there is a certain accuracy; When the AUC> 0.9, the diagnosis has higher accuracy. The sensitivity of P62 was 91.66%, the specificity was 46.87%, the Yoden Index was 0.458, the cut-off value was 0.238, and the AUC value was 0.649.
Figure 6.
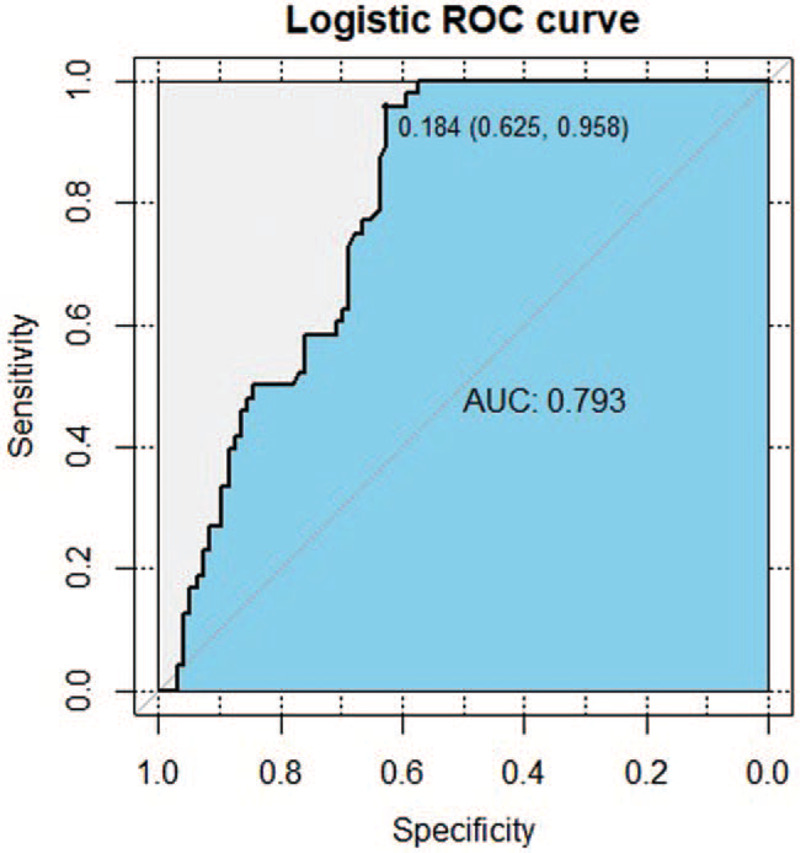
The receiver operating characteristic (ROC) curve analysis of immunoglobulin G antibody response for Mixed Peptide P39+ P62. Notes: ROC, receiver operating characteristics; area under the curve (AUC), area under the curve. Diagnostic basis of ROC curve: When the area of the curve was between 0.5 to 1, the closer to 1, that is, the larger the area, the higher the diagnostic accuracy would be. When 0.5 < AUC≤0.7, there is a lower accuracy; When 0.7 < AUC≤0.9, there is a certain accuracy; When the AUC> 0.9, the diagnosis has higher accuracy. The sensitivity of Mixed Peptide P39+ P62was 95.83%, the specificity was 97.91%, the Yoden Index was 0.625, the cut-off value was 0.184, and the AUC value was 0.793.
4. Discussion
According to the immune protection mechanism of antituberculosis infection, the immune response induced by antigen protein is mainly based on the epitopes; CD4+T cell epitopes are usually composed of 9 to 25 amino acid residues and play a key role in the process of stimulating the CD4+T cell immune response.[25] The vaccine design based on CD4+T cell epitopes can selectively remove the antigenic part without any toxic effects, directly stimulate the body to produce a stronger and more-specific immune response that is different from the immune response to a natural infection, as well as reduce the side effects of the vaccine which may overcome the problems of low immune protective effects of BCG and the shortcomings of subunit vaccines.[26] The candidate antigens of epitope vaccines are highly immunogenic epitope peptides and antigenic fragments. Obtaining the ideal epitope with the potential immune protective effect is the basis for developing multi-epitope vaccines. Understanding the amino acid sequence of CD4+T cell epitopes would contribute to the design and development of epitope vaccines and diagnostic reagents and increase the understanding of the immune mechanism involved.[27] In this study, novel CD4+T cell epitopes were screened and identified against 5 protective antigens CFP10, 38KDa, ESAT6, Ag85A, and TB27.4 to provide a reference for the construction of epitope vaccines and diagnostic reagents, as well as to lay a foundation for the exploration of CD4+T cell epitope-mediated immune response mechanism in vivo.
With the completion of whole-genome sequencing of MTB H37Rv, each protective antigen may be retrieved from the genome sequence, marking a new stage in the design of TB vaccine candidates, diagnostic reagents, and biomarkers to evaluate their efficacy.[28] Bioinformatics technology can be used to predict and screen epitopes on a large scale in multiple antigen proteins, verify the affinity between the epitopes and the HLA molecules quickly, accurately, and on a large scale, find the immune-dominant epitopes further efficiently, enhance the purpose of experiments, and improve the predictability and success rate of the experimental results.[29] Although no prediction software can achieve 100% accuracy, and there may be peptides with high prediction scores that do not truly stimulate lymphocyte proliferation, this method can nonetheless greatly reduce the blindness of epitope finding and reduce the experimental cost.[30] In general, for the selection of antigenic epitopes of amino acid sequences of the antigenic proteins, in addition to using appropriate computer protein analysis software, it is also necessary to comprehensively consider the hydrophilicity, hydrophobicity, plasticity, and surface possibility of the selected sequence fragments, and then comprehensively score the peptides in the protein antigens using epitope prediction software with different algorithms, so that the specificity, accuracy, and applicability of epitope prediction are greatly improved. Sabarth et al screened 15 peptides with strong binding affinity to the MHC-II molecules by software prediction of the epitope peptides, of which 6 peptides have been experimentally confirmed. These studies suggest that bioinformatics-based epitope prediction is an effective approach to identify the dominant CD4+T epitopes associated with TB.[31] In this study, SYFPEITHTI, RANKPEP, and NetMHC II pan software (National Cancer Institute, United States of America) packages were used, which are of great value in many application fields.[32] These software packages predict the binding affinity of peptides to the MHC-II alleles and have been applied to design novel vaccines and develop disease diagnostic markers. These methods can improve the accuracy of epitope prediction by integrating prediction. On the basis of predicting multiple CD4+T cell epitopes using these 4 bioinformatics software packages, according to the score values of different epitopes, the homology with human proteins, amino acid sequence repeatability, comparative analysis, and preliminary screening with published epitope sequences was performed to remove the CD4+T epitopes confirmed by the studies and perform a comprehensive all-round analysis. Restrictive epitopes of HLA-DRB1∗0701 were predicted from CFP10, 38KDa, ESAT6, Ag85A, and TB27.4 through comprehensive analysis. However, the prediction of cell epitopes should consider not only their strong binding but also their immunogenicity. Therefore, in this study, the reliability and effect of these predicted epitopes were experimentally verified.
Cellular immunity in TB patients is suppressed, and CD4+T cells play an important role in the immune response against TB. Therefore, antigen epitopes that can stimulate CD4+T cell proliferation and activation are needed to induce the suppression of cellular immunity in TB patients. The ability of the epitopes to stimulate cell proliferation was used as an evaluation criterion for screening the cell epitopes.[33] In this study, we evaluated the immune effects of 5 predicted CD4+T candidate peptides in terms of stimulating cell proliferation and phenotypic changes before and after the PBMC stimulation. In this way, the research cost and workload can be greatly reduced, the proliferation of PBMC and CD4+T cells from HLA-DRB1∗ 0701 positive samples of patients with TB can be analyzed, and the immune effect of the candidate peptides can be evaluated. P39 and P62 could stimulate the proliferation of HLA-DRB1∗ 0701-positive PBMC and CD4+T cells in patients with TB. By comparing the proliferation of PBMC and CD4+T cells, it was concluded that the proliferation cells stimulated by P39 and P62 were mainly CD4+T cells, which indicated that P39 and P62 were potential candidates CD4+T cell epitopes. According to the change in the CD4+T cell ratio before and after stimulation by the candidate peptides, P39 and P62 had good immunogenicity and were effective HLA- DRB1∗0701-restricted CD4+T epitopes of MTB Stimulation with both P39 and P62 showed that the combined effect of P39 and P62 was stronger than that of a single epitope peptide (P < .05), which indicated that P39 and P62 could mediate mutual synergistic stimulation.
Humoral immunity plays an important role during an antituberculosis response. IFN-γ is an important cytokine of MTB and an important evaluation criterion for evaluating the immune efficacy of a TB vaccine. Screening antigenic epitopes that can enhance the secretion of IFN-γ has become the focus of the research related to MTB.[34] TNF-α can induce the activation of macrophages and has a certain immunomodulatory effect, which has a certain reference value for the detection of TB.[35] IL-2 can promote T-cell activation and proliferation and can induce the production of IFN-γ, IL-6, and other cytokines. The lack of IL-2 in the human body can lead to abnormal immune response and disease progression[36]; IL-10 can reduce the body's inflammatory response and inhibit the action of pro-inflammatory factors. Studies have shown that IL-10 is significantly increased in the serum of patients with pulmonary TB, whereas after the TB patients are treated, the IL-10 levels decrease. If the secretion of IL-10 in patients with TB can be reduced, the ability of the human body to resist/fight TB would improve.[37] In this study, P39, P62, and their combination P39+P62 could not only induce the secretion of Th1-type cytokines such as IFN-γ, TNF-α, and IL-2 but could also reduce the secretion of IL-10, indicating that these 2 candidate peptides along with their combination have potential in the development of novel TB epitope vaccines.
This study has explored and verified that P39 and P62 are potential HLA-DRB1∗0701-restricted MTB CD4+T epitope candidates with certain immunogenicity. This study explored the ELISA conditions of P39, P62, and their combination. Firstly, the optimal coating concentration, serum dilution, and goat anti-human IgG labeled with horseradish peroxidase were determined by chessboard titration. Then, the IgG levels in the sera of TB patients and healthy volunteers were detected, and the Youden index, cut-off value, sensitivity, specificity, and ROC curve of the candidate peptides were analyzed. Youden index is a method to evaluate the authenticity of the experiment.[38] The resulting value indicated that the experimental results were better and more reliable. The ROC curve visually showed the relationship between sensitivity and specificity and reflected the effectiveness of the diagnostic method.[39] In terms of evaluating the application value of P39, P62, and P39+P62 combination peptide in TB serological detection and humoral immune diagnosis and determining the most reasonable reaction system, the experimental protocol was found to be reasonable, and the results were credible. Studies have shown that the sensitivity and specificity of CFP10 are 67.9% to 74.3% and 91.5% to 92.8%, respectively. The sensitivity of TB27.4 is 84% to 88%, while the specificity of these 2 proteins in detecting TB is 91% to 95%.[40] Baassi L et al studied the 23-amino acid peptide of TB27.4 protein and reported the sensitivity and specificity of 65% and 100%, respectively, for the protein.[41] In this study, the sensitivity of P39 epitope peptide was 75%, the specificity was 67.71%, and the cut-off was 0.259; the sensitivity of the P62 epitope peptide was 91.66%, its specificity was 46.87%, and the cut-off was 0.238; and the sensitivity of the P39+P62 combined epitope peptide was 95.83%, its specificity was 97.91%, and the cut-off was 0.184. The result is relatively ideal, and the specificities of P39 and P62 are relatively low. The biggest problem in the serological diagnosis is the low specificity of the selected antigen, which may be due to the low immunogenicity of the peptide or several interfering substances in the serum.[42] The Youden index of P62 was less than 0.5, which indicates that the authenticity of the results of ELISA as a diagnostic tool is low, and it is, therefore, not recommended for serological detection. The Youden indices of P39 and P39+P62 combined epitope peptide were 0.667 and 0.625, respectively. The AUC values of P39, P62, and P39+P62 combined polypeptide were 0.844, 0.649, and 0.793, respectively, indicating that the diagnostic accuracy of P62 ELISA is low. The P39 and P39+P62 combined epitope peptide had high diagnostic efficiency, that is, there were differences in the antibody levels against the mixture of P39 and P39+P62 combined peptide between the TB patients and the healthy volunteers, which has certain accuracy when used as a diagnostic tool for ELISA diagnosis. Therefore, P39 and the P39+P62 combined polypeptide have certain authenticity and accuracy in TB diagnosis and have a potential diagnostic value, so they can be considered for use as immunological diagnostic reagents for the auxiliary diagnosis of TB. The combination of P39+P62 has better diagnostic energy efficiency in detecting the TB-specific antibody IgG, and this can provide a theoretical basis for the rapid serological diagnosis of TB.
In this study, the sensitivity and specificity of these 2 CD4+T cell epitope peptides were lower than those of the identified intact antigens CFP10 and TB27.4. The following speculations were made. Firstly, compared to the antigen, a single peptide containing CD4+T cell epitope is relatively simple, so its sensitivity and specificity are lower. Secondly, each HLA-DRB1∗0701 -positive TB patient is in a different disease stage, and his/her immune response state is also different.[43] The immune recognition of different CD4+T cell epitope peptides in different patients varied in a random manner. In addition, these epitope peptides were confirmed to be CD4+T cell epitopes in this study, but it was not proven whether they are also B cell epitopes. Thus, it was expected that no relatively strong humoral immune response would be elicited in humans.
There are a few limitations of this study. The predicted CD4+T epitopes were restricted by HLA-DRB1∗0701. However, in the real epitope peptide recognition, these epitopes cannot be excluded from being restricted by the other MHCs, which may be one of the reasons for the low positivity rate of the epitope peptides screened in this study in the TB patients. This requires experimental verification at a later date. Although P39 and P62 were confirmed as potential candidate CD4+T cell epitopes in the in vitro experiments, we still need to verify whether the candidate peptides are protective CD4+T cell epitopes by injecting the candidate peptides into mice. In this study, when evaluating the application value of the CD4+T cell epitope peptides in TB diagnosis, the sample size was insufficient, and all subjects were part of a case-control study. The evidence-based level was low and is required to be further validated by a large-sample prospective cohort study. A large number of clinical samples need to be included for a more comprehensive study in the future. In subsequent research, we would expand the sample size for the joint screening of TB and latent TB patients to further evaluate the clinical application value of the epitope peptides. Further clinical trials are needed to evaluate the immune response of the candidate peptides in patients with latent TB and TB.
In this study, the CD4+T cell epitopes of MTB were predicted and identified, and 2 novel MTB CD4+T cell epitopes P39 and P62 were discovered, which would provide some reference for the study of MTB antigenic epitopes, serve as potential candidate epitope polypeptides for the design and development of an antituberculosis epitope vaccine based on the Chinese population, and contribute to the study of MTB.
Acknowledgments
We would like to thank the key laboratory of Xinjiang endemic and ethnic diseases.
Author contributions
Conceptualization: Jing Liu, Xue Feng Chen, Ju Wang, Wei Cheng, Chunjun Zhang, Wanjiang Zhang.
Data curation: Jie Zhang, Jiangtao Dong, Hui Zhang, Jing Liu, Xuefeng Chen, Ju Wang.
Formal analysis: Fang Wu, Jiangtao Dong.
Investigation: Jing Liu, Xuefeng Chen, Ju Wang, Fang Wu, Jie Zhang, Hui Zhang, Le Zhang, Wanjiang Zhang.
Methodology: Jing Liu, Xuefeng Chen, Ju Wang, Fang Wu, Jie Zhang, Jiangtao Dong, Hui Zhang, Xiaoling Liu, Na Hu, Jiangdong Wu, Wei Cheng, Chunjun Zhang, Wanjiang Zhang.
Project administration: Chunjun Zhang.
Resources: Le Zhang, Chunjun Zhang, Xiaoling Liu, Na Hu.
Software: Xiaoling Liu, Na Hu, Jiangdong Wu, Le Zhang.
Supervision: Wei Cheng.
Validation: Fang Wu.
Writing – original draft: Jing Liu, Wanjiang Zhang, Xuefeng Chen, Ju Wang.
Writing – review & editing: Jing Liu, Wanjiang Zhang, Wei Cheng, Chunjun Zhang.
Glossary
Abbreviations: AUC = area under the receiver operating characteristic curve, HPLC = high performance liquid chromatography, IgG = immunoglobulin G, MS = mass spectrometry MTB = Mycobacterium tuberculosis, OD = Optical density, ROC = receiver operating characteristic, SI = stimulation index, TB = Tuberculosis.
References
- [1].Floyd K, Glaziou P, Zumla A, et al. The global tuberculosis epidemic and progress in care, prevention, and research: an overview in year 3 of the End TB era. Lancet Respir Med 2018;6:299–314. [DOI] [PubMed] [Google Scholar]
- [2].Prezzemolo T, Guggino G, La Manna MP, et al. Functional signatures of human CD4 and CD8 T cell responses to Mycobacterium tuberculosis. front. Immunol 2014;5:18–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Wang S, Li J, Chen X, et al. Analyzing the effect of peptide-HLA-binding ability on the immunogenicity of potential CD8+ and CD4+ T cell epitopes in a large dataset. Immunol Res 2016;64:908–18. [DOI] [PubMed] [Google Scholar]
- [4].Lindestam CA, Lewinsohn D, Sette A. Antigens for CD4 and CD8 T cells in tuberculosis. Cold Spring Harb Perspect Med 2014;4:19–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Shi C, Zhang H, Zhang T, et al. New alternative vaccine component against MMycobacterium tuberculosis–heat shock protein 16.3 or its T-cell epitope. Scand J Immunol 2009;70:465–74. [DOI] [PubMed] [Google Scholar]
- [6].Soria-Guerra RE, Nieto-Gomez R, Govea-Alonso DO, et al. An overview of bioinformatics tools for epitope prediction: implications on vaccine development. J Biomed Inform 2015;53:405–14. [DOI] [PubMed] [Google Scholar]
- [7].Raoufi E, Hemmati M, Eftekhari S, et al. Epitope prediction by novel immunoinformatics approach: a state-of-the-art review. Int J Pept Res Ther 2019;26:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Burland TG. DNASTAR's Lasergene sequence analysis software. Methods Mol Biol 2000;132:71–91. [DOI] [PubMed] [Google Scholar]
- [9].Schuler MM, Nastke MD, Stevanovikć S. SYFPEITHI: database for searching and T-cell epitope prediction. Methods Mol Biol 2007;409:75–93. [DOI] [PubMed] [Google Scholar]
- [10].Reche PA, Glutting JP, Zhang H, et al. Enhancement to the RANKPEP resource for the prediction of peptide binding to MHC molecules using profiles. Immunogenetics 2004;56:405–19. [DOI] [PubMed] [Google Scholar]
- [11].Lundegaard C, Lund O, Nielsen M. Prediction of epitopes using neural network based methods. J Immunol Methods 2011;374:26–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Zhou J, Wang W, Song P, et al. Structural predication and antigenic analysis of Toxoplasma gondii ROP20. Acta Parasitol 2018;63:244–51. [DOI] [PubMed] [Google Scholar]
- [13].Ma X, Zhou X, Zhu Y, et al. The prediction of T- and B-combined epitope and tertiary structure of the Eg95 antigen of Echinococcus granulosus. Exp Ther Med 2013;6:657–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Akya A, Farasat A, Ghadiri K, et al. Identification of HLA-I restricted epitopes in six vaccine candidates of Leishmania tropica using immunoinformatics and molecular dynamics simulation approaches. Infect Genet Evol 2019;75:92–109. [DOI] [PubMed] [Google Scholar]
- [15].León Y, Zapata L, Salas-Burgos A, et al. In silico design of a vaccine candidate based on autotransporters and HSP against the causal agent of shigellosis, Shigella flexneri. Mol Immunol 2020;121:47–58. [DOI] [PubMed] [Google Scholar]
- [16].Khanna D, Rana PS. Ensemble technique for prediction of T-cell Mycobacterium tuberculosis epitopes. Interdiscip Sci 2019;11:611–27. [DOI] [PubMed] [Google Scholar]
- [17].Zhao JW, Yan M, Shi G, et al. In silico identification of cytotoxic T lymphocyte epitopes encoded by RD5 region of Mycobacterium tuberculosis. J Infect Dev Ctries 2017;11:806–10. [DOI] [PubMed] [Google Scholar]
- [18].Wilson D, Badri M, Maartens G. Performance of serum C-reactive protein as a screening test for smear-negative tuberculosis in an ambulatory high HIV prevalence population. PLoS One 2011;6:13–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Jurcevic S, Hills A, Pasvol G, et al. T cell responses to a mixture of Mycobacterium tuberculosis peptides with complementary HLA-DR binding profiles. Clin Exp Immunol 1996;105:416–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Sun J, Tang T, Duan J, et al. Biocompatibility of bacterial magnetosomes: acute toxicity, immunotoxicity and cytotoxicity. Nanotoxicology 2010;4:271–83. [DOI] [PubMed] [Google Scholar]
- [21].Chen Y, Giri BR, Li X, et al. Preliminary evaluation of the diagnostic potential of Schistosoma japonicum extracellular vesicle proteins for Schistosomiasis japonica. Acta Trop 2020;201:21–38. [DOI] [PubMed] [Google Scholar]
- [22].Arroyo L, Marín D, Franken KL, et al. Potential of DosR and Rpf antigens from Mycobacterium tuberculosis to discriminate between latent and active tuberculosis in a tuberculosis endemic population of Medellin Colombia. BMC Infect Dis 2018;18:26–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].La Manna MP, Orlando V, Li Donni P, et al. Identification of plasma biomarkers for discrimination between tuberculosis infection/disease and pulmonary non tuberculosis disease. PLoS One 2018;13:33–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Kuo C-J, Gao J, Huang J-W, et al. Functional and structural investigations of fibronectin-binding protein Apa from Mycobacterium tuberculosis. Biochim Biophys Acta 2019;1863:1351–9. [DOI] [PubMed] [Google Scholar]
- [25].Rosa DS, Ribeiro SP, Cunha-Neto E. CD4+ T cell epitope discovery and rational vaccine design. Arch Immunol Ther Exp (Warsz ) 2010;58:121–30. [DOI] [PubMed] [Google Scholar]
- [26].Abdulla F, Adhikari UK, Uddin MK. Exploring T & B-cell epitopes and designing multi-epitope subunit vaccine targeting integration step of HIV-1 lifecycle using immunoinformatics approach. Microb Pathog 2019;137:31–48. [DOI] [PubMed] [Google Scholar]
- [27].Adhikari UK, Rahman MM. Overlapping CD8+ and CD4+ T-cell epitopes identification for the progression of epitope-based peptide vaccine from nucleocapsid and glycoprotein of emerging Rift valley fever virus using immunoinformatics approach. Infect Genet Evol 2017;56:75–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Panigada M, Sturniolo T, Besozzi G, et al. Identification of a promiscuous T-cell epitope in Mycobacterium tuberculosis Mce proteins. Infect Immun 2002;70:79–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Li Y, Li ZJ, Han WY, et al. Identification and characterization of Th cell epitopes in MrkD adhesin of Klebsiella pneumoniae. Microb Pathog 2010;49:8–13. [DOI] [PubMed] [Google Scholar]
- [30].Konstantinou GN. T-Cell epitope prediction. Methods Mol Biol 2017;1592:211–22. [DOI] [PubMed] [Google Scholar]
- [31].Sabarth N, Chamberlain L, Brett S, et al. Induction of homologous rather than heterologous antigen-specific CD4 T cell responses is critical for functional CD8 T cell responses in mice transgenic for a foreign antigen. J Immunol 2010;185:4590–601. [DOI] [PubMed] [Google Scholar]
- [32].Li Z, Zhang F, Zhang C, et al. Immunoinformatics prediction of OMP2b and BCSP31 for designing multi-epitope vaccine against Brucella. Mol Immunol 2019;114:651–60. [DOI] [PubMed] [Google Scholar]
- [33].Oliveira FM, Coelho IE, Lopes MD, et al. The use of reverse vaccinology and molecular modeling associated with cell proliferation stimulation approach to select promiscuous epitopes from Schistosoma mansoni. Appl Biochem Biotechnol 2016;179:1023–40. [DOI] [PubMed] [Google Scholar]
- [34].Bobadilla K, Sada E, Jaime ME, et al. Human phagosome processing of Mycobacterium tuberculosis antigens is modulated by interferon-γ and interleukin-10. Immunology 2013;138:34–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Godfrey MS, Friedman LN. Tuberculosis and biologic therapies: anti-tumor necrosis factor-α and beyond. Clin Chest Med 2019;40:721–39. [DOI] [PubMed] [Google Scholar]
- [36].Della Bella C, Spinicci M, Grassi A, et al. Novel M. tuberculosis specific IL-2 ELISpot assay discriminates adult patients with active or latent tuberculosis. PLoS One 2018;13:26–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Dai Y-C, Wang W-D, Zhang J-A, et al. MTB driven B cells producing IL-35 and secreting high level of IL-10 in the patients with active pulmonary tuberculosis. Mol Immunol 2019;112:175–81. [DOI] [PubMed] [Google Scholar]
- [38].Martínez-Camblor P, Pardo-Fernández JC. The Youden index in the generalized receiver operating characteristic curve context. Int J Biostat 2019;46:58–70. [DOI] [PubMed] [Google Scholar]
- [39].Dahiya B, Khan A, Mor P, et al. Detection of Mycobacterium tuberculosis lipoarabinomannan and CFP-10 (Rv3874) from urinary extracellular vesicles of tuberculosis patients by immuno-PCR. Pathog Dis 2019;77:5–17. [DOI] [PubMed] [Google Scholar]
- [40].Mukherjee P, Dutta M, Datta P, et al. The RD1-encoded antigen Rv3872 of Mycobacterium tuberculosis as a potential candidate for serodiagnosis of tuberculosis. Clin Microbiol Infect 2007;13:146–52. [DOI] [PubMed] [Google Scholar]
- [41].Baassi L, Sadki K, Seghrouchni F, et al. Evaluation of a multi-antigen test based on B-cell epitope peptides for the serodiagnosis of pulmonary tuberculosis. Int J Tuberc Lung Dis 2009;13:848–54. [PubMed] [Google Scholar]
- [42].Arlehamn CSL, Paul S, Mele F, et al. Immunological consequences of intragenus conservation of Mycobacterium tuberculosis T-cell epitopes. Proc Natl Acad Sci 2015;112:147–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Jain S, Baranwal M. Conserved peptide vaccine candidates containing multiple Ebola nucleoprotein epitopes display interactions with diverse HLA molecules. Med Microbiol Immunol 2019;208:227–38. [DOI] [PubMed] [Google Scholar]


