Supplemental Digital Content is available in the text
Keywords: CHRNA5/A3/B4 gene, lung cancer, meta-analysis, polymorphism, risk
Abstract
Background:
Genetic polymorphisms in the 15q25 region have been associated with the risk of lung cancer (LC). However, studies have yielded conflicting results.
Methods:
Searches were conducted in databases, including PubMed, EMbase, Web of Science, CNKI, and Wanfang, for case-control studies up to August 1, 2019. After retrieving eligible studies and data extraction, we calculated pooled odds ratios with 95% confidence intervals. In the meta-analysis, we included 32 publications with a total of 52,795 patients with LC and 97,493 control cases to evaluate the polymorphisms in the CHRNA5/A3/B4 gene cluster in the 15q25 region.
Results:
Data of the meta-analysis showed a significantly increased risk of LC in the presence of genetic polymorphisms (rs1051730, rs16969968, rs8034191). In the smoking subgroup, the CHRNA3 rs1051730 polymorphism was found to contribute to LC risk using following 5 models: the allelic model, the homozygous model, the heterozygous model, the dominant model, and the recessive model. Thus, the rs1051730 polymorphism may modify LC susceptibility under the condition of smoking. Stratification studies for CHRNA5-rs8034191 showed that Caucasian groups with the wild-type genotype (C/C) may be susceptible to LC in all 5 models. No significant relationship between CHRNA3 rs6495309 or rs3743073 and LC susceptibility was found. However, Asians with the rs3743037 B-allele showed an obviously higher risk of LC susceptibility than the Caucasian population, observed via allelic, heterozygous, and dominant models.
Conclusions:
The 3 polymorphisms of rs1051730, rs16969968 and rs8034191 in the CHRNA5/A3/B4 gene cluster in the 15q25 region were associated with LC risk, which might be influenced by ethnicity and smoking status.
1. Introduction
For decades, lung cancer (LC) has been the leading cause of malignancy-related mortality worldwide, and is considered a severe public health problem.[1] Carcinogenesis is a multifactorial process. Environmental exposure, primarily to cigarette smoke, has been cited as a significant contributor to the development of LC.[2] More recently, substantial genome-wide association studies have revealed genetic variants that mediate LC progression, which has provided valuable insight into its genetic architecture.[3]
A cluster of 3 genes, CHRNA5, CHRNA3, and CHRNB4, on chromosome 15q25 encodes neuronal nicotinic acetylcholine receptor subunits (nAChRs), which are the initial physiological targets of nicotine. As a potential lung carcinogen, nicotine has been hypothesized to play a role in forming bulky polycyclic aromatic hydrocarbon-like DNA adducts that may result in the mutation of key genes such as TP53.[4]
Furthermore, some reports indicated that Ach released from cell lines of non-small cell LC or small cell LC cells binds to nAChRs in the source and neighboring cells, which have been implicated in the regulation of cellular processes such as proliferation, cell-cell interaction, and cell death.[5–8] Catassi et al reported that nAChRs build a part of an autocrine-proliferative network that facilitates the growth of neoplastic cells.[5] Schulle et al and Jull et al[6,7] also revealed the interaction of nicotine and nAChR promote cell proliferation in LC cells via serotonin-induced stimulation of the Raf-1/MAPK/c-myc pathway. In addition, nicotine has been shown to inhibit apoptosis by phosphorylation of Bcl-2 family members.[8]
Recently, both single nucleotide polymorphisms (SNPs) and haplotypes in the CHRNA5/A3/B4 Gene Cluster have been identified to be associated with the etiology of LC risk,[9,10] dependent or independent of smoking behavior. In a study published by Zienolddiny et al,[11] the association was shown to be statistically significant for rs1051730 (P = .017) and rs16969968 (P = .020), which was further validated by Vander Weele et al[12] and Jaworowska et al[13] However, in studies by Spitz et al and Schwartz et al,[14,15] there was no evidence that carriers with the rs1051730 polymorphism have susceptibility to LC no matter they were smoking or non-smoking. In 2013, a case-control study was carried out among 106 LC patients and 116 controls also reached a null conclusion on the CHRNA5 rs16969968 polymorphism, though the variant allele appeared slightly more common among these cases.[16] Jaworowska et al[12] observed the strongest connection between the rs8034191 polymorphism and the small cell LC subtype in both smokers and non-smokers. In another case-control study in Chinese population, Wang et al[17] reported that neither genotype nor allele frequencies of rs8034191 showed statistically differences between LC patients and controls. Moreover, rs3743073 has been shown to be significantly correlated to LC in recent studies.[18,19]
The results remain controversial and ambiguous, and no consensus has been reached as to the relative impact of the variants on the propensity to nicotine dependence or direct carcinogenesis. In light of this controversy, we performed an updated systematic meta-analysis to evaluate the contribution of genetic variations in the CHRNA5/A3/B4 gene cluster to LC susceptibility.
2. Materials and methods
2.1. Literature search
A comprehensive literature search was performed in PubMed, EMbase, Web of Science, CNKI, and Wanfang databases (up to August 1, 2019). The following keywords were used: (CHRNA3 or cholinergic receptor nicotinic alpha 3 subunit) and (polymorphism or mutation or variation or snp or genotype) and (carcinoma or cancer or neoplasm or adenocarcinoma or tumor or malignancy); (CHRNB4 or cholinergic receptor nicotinic beta 4 subunit) and (polymorphism or mutation or variation or SNP or genotype) and (carcinoma or cancer or neoplasm or adenocarcinoma or tumor or malignancy); (CHRNA5 or cholinergic receptor nicotinic alpha 5 subunit) and (polymorphism or mutation or variation or snp or genotype) and (carcinoma or cancer or neoplasm or adenocarcinoma or tumor or malignancy). literature languages were not restricted. Articles with large sample sizes were enrolled if the data or datasets were repeated. All anayses were based on previous published studies, thus no ethical approval and patient consent are required.
2.2. Inclusion and exclusion criteria
We selected publications that satisfied the following inclusion criteria:
-
(1)
case-control studies;
-
(2)
studies concentrating on genotype or allele frequencies;
-
(3)
studies with sufficient genotype data to calculate odds ratios (ORs) and 95% confidence intervals (95% CIs).
The exclusion criteria were as follows:
-
(1)
case-only studies, case reports, or reviews;
-
(2)
insufficient data for the CHRNA5/A3/B4 gene genotype;
-
(3)
studies that compared the CHRNA5/A3/B4 gene variants in precancerous lesions with other cancers.
2.3. Data extraction
The first author's name, year of publication, ethnicity, source of controls, smoking status, and the number of cases and controls in the CHRNA5/A3/B4 gene were extracted from the articles. Data extraction was independently performed by 2 investigators. Any discrepancies were adjudicated by discussion until a consensus was reached. We distinguished the controls of eligible case-control studies by denoting them as either population-based (PB) or hospital-based (HB). Ethnicity was categorized as “Caucasian,” “Asian,” or “Mixed.” All eligible case-control studies were defined as either PB or HB. Additionally, smoking status was classified into smokers (Y), non-smokers (N), and unclassified groups (mixed).
2.4. Statistical analysis
We assessed the strength of the relationship between CHRNA5/A3/B4 gene polymorphisms and LC susceptibility by ORs and 95% CI in allelic, homozygous, heterozygous, dominant, and recessive models. The P values in our study were adjusted through using the Bonferroni Correction to compensate for the increases induced by testing each individual hypothesis at a significant level of a/m (a = the desired overall alpha level, m = the number of the hypothesis). The Bonferroni correction rejects the null hypothesis at a P value less than a/m (PA = PZ ∗ 5< .05 was considered statistically significant).[20] The heterogeneity assumption was determined by the Chi-Squared based Q-test and I2 statistics. If P > .05 for the Q test or I2 < 50%, the OR of each study was calculated by using a fixed-effects model (Mantel–Haenszel method); otherwise, the random-effects (DerSimonian–Laird method) model was used.[21] With the χ2 test, we inspected the Hardy–Weinberg equilibrium (HWE) of control genotypes. We conducted the stratified analyses by ethnicity, control source, smoking status, or HWE status. We also performed the sensitivity analysis to evaluate the stability of pooled results by neglecting each study in turn and determining the effect on the pooled analyses. Publication bias was assessed with Begg funnel plot and Egger test, wherein P < .05 was considered statistically significant.[22] Moreover, the trim and fill algorithm trimmed off the asymmetric outlying part of the funnel and estimated the true center of the funnel, further providing effective and relatively powerful testing for evaluating the existence of publication bias.[23] We used Stata software (version 12.0, StataCorp LP, College Station, TX) to perform statistical analyses, and used the Power and Sample Size Calculation to evaluate the power of this study.
The false-positive report probability (FPRP) threshold was set as 0.2, and the prior probability of 0.1 was used to detect an OR of 1.50 risk effects for the significant associations. Once the FPRP value of positive association is less than 0.2, we would assert that the results were noteworthy. All statistical tests were 2-sided, and a P-value < .05 was considered statistically significant. SAS software (version 9.1, SAS Institute, Cary, NC) was used to analyze the FPRP value and statistical power.
2.5. Trial sequential analysis (TSA)
We adopted TSA to minimize random errors and to increase the robustness of results as a series of sparse data and reduplicative testing in meta-analysis. The information size would be estimated based on the assumption of a plausible relative risk of 10% with low risk bias.
Risks for a type I error
-
(a)
of 5% and a type II error
-
(b)
of 20% would be acquired.
With the estimated information size and risks for type I and type II errors, TSA monitoring boundaries were built. If the Z-curve cross TSA monitoring boundary before reaching the required information, we would confirm that the results obtained were significant with strong evidence, and further trials would become unnecessary. Otherwise, it is necessary to continue conducting trials.
3. Results
3.1. Main characteristics of the enrolled studies
A total of 32 publications that met the inclusion criteria were utilized in the quantitative synthesis (Table 1). For CHRNA3 gene polymorphisms (rs1051730, rs6495309, rs3743073), 28 case-control studies with 25,516 cases and 35,547 controls met our criteria. Thirteen of these studies investigated the association between the s1051730 polymorphism and LC susceptibility in a Caucasian population, 14 studies focused on an Asian population, and a single study centered on an African population. Eleven studies had HB controls, and the others were PB. All except for 2 of the studies were consistent with the HWE. For the CHRNA5 gene polymorphisms (rs16969968, rs8034191), 23 Caucasian, and 5 Asian-focused studies qualified, with a total of 27,636 cases and 62,372 controls. Among them 20 studies had HB controls, and 9 had PB controls. The genotype distributions of all control groups were in accord with the HWE. Two case-control studies deviated from the HWE. The quality of these enrolled case-control studies was evaluated by using the Newcastle–Ottawa Scale (Supplementary Table 1, http://links.lww.com/MD/F568). All the data will be available at https://pan.baidu.com/s/1etP0shr0izf2W6AMe94jKg (extraction code: ywez) publicly.
Table 1.
Characteristics of the enrolled studies.
| Case | Control | |||||||||||||
| SNP | Gene location | First Author | Yr | Ethnicity | Source of Control | Cancer Type | Smoking Statue | HWE | Common | Heterozygous | Rare | Common | Heterozygous | Rare |
| rs1051730 | 15q25.1 G > A | Takashi et al | 2011 | Asian | HB | LC | mixed | Y | 349 | 25 | 0 | 314 | 10 | 0 |
| Sakoda et al | 2011 | Cacausian | PB | LC | Y (s) | Y | 255 | 373 | 117 | 625 | 690 | 160 | ||
| Ren et al | 2013 | Asian | PB | LC | Y (s) | Y | 127 | 12 | 0 | 124 | 6 | 0 | ||
| Ren et al | 2013 | Asian | PB | LC | N (ns) | Y | 61 | 10 | 0 | 66 | 4 | 0 | ||
| Pérez-Morales et al | 2018 | Cacausian | PB | LC | Y (s) | Y | 45 | 26 | 3 | 138 | 51 | 3 | ||
| Yang et al | 2012 | Asian | HB | LC | mixed | Y | 1007 | 49 | 0 | 1025 | 36 | 0 | ||
| Christopher et al | 2008 | Cacausian | PB | LC | mixed | Y | 687 | 848 | 295 | 445 | 418 | 93 | ||
| Christopher et al | 2008 | Cacausian | PB | LC | Y (s) | Y | 683 | 871 | 301 | 767 | 771 | 193 | ||
| Schwartz et al | 2009 | Cacausian | PB | LC | mixed | Y | 207 | 280 | 95 | 344 | 379 | 121 | ||
| Schwartz et al | 2009 | African American | PB | LC | mixed | Y | 279 | 96 | 10 | 353 | 74 | 5 | ||
| Liu et al | 2008 | Cacausian | HB | LC | Y (s) | Y | 73 | 84 | 37 | 105 | 98 | 15 | ||
| Shiraishi et al | 2009 | Asian | HB | LC | N (ns) | Y | 248 | 16 | 1 | 560 | 15 | 0 | ||
| Shiraishi et al | 2009 | Asian | HB | LC | Y (s) | N | 922 | 61 | 2 | 350 | 10 | 1 | ||
| Zienolddiny et al | 2009 | Cacausian | PB | LC | Y (s) | Y | 110 | 184 | 58 | 174 | 195 | 56 | ||
| Kaur-Knudsen et al | 2010 | Cacausian | PB | LC | Y (s) | N | 112 | 146 | 50 | 4440 | 4181 | 1086 | ||
| VanderWeele et al | 2012 | Cacausian | HB | LC | mixed | Y | 2529 | 3198 | 1135 | 2902 | 3075 | 784 | ||
| Spitz et al | 2008 | Cacausian | PB | LC | N (ns) | Y | 294 | 198 | 55 | 317 | 281 | 55 | ||
| Spitz et al | 2008 | Cacausian | PB | LC | Y (s) | Y | 685 | 869 | 300 | 764 | 770 | 193 | ||
| rs6495309 | 15q25.1 T > C | Yang et al | 2012 | Asian | HB | LC | mixed | Y | 262 | 735 | 562 | 398 | 794 | 485 |
| Sakoda et al | 2011 | Caucasian | PB | LC | Y (s) | Y | 510 | 208 | 28 | 921 | 496 | 60 | ||
| Sun et al | 2018 | Asian | HB | LC | Y (s) | Y | 60 | 88 | 39 | 24 | 44 | 19 | ||
| Sun et al | 2018 | Asian | HB | LC | N (ns) | Y | 25 | 60 | 22 | 54 | 113 | 52 | ||
| Wu et al | 2009 | Asian | PB | LC | mixed | Y | 490 | 1578 | 920 | 622 | 1425 | 832 | ||
| Du et al | 2011 | Asian | HB | LC | Y (s) | Y | 8 | 32 | 20 | 22 | 28 | 10 | ||
| rs3743073 | 15q25.1 C > A | Shen et al | 2012 | Asian | PB | LC | mixed | Y | 124 | 258 | 218 | 186 | 291 | 123 |
| Tekpli et al | 2012 | Caucasians | HB | LC | Y (s) | Y | 132 | 146 | 31 | 136 | 147 | 51 | ||
| Niu et al | 2010 | Asian | PB | LC | N (ns) | Y | 38 | 118 | 56 | 133 | 246 | 106 | ||
| Niu et al | 2010 | Asian | PB | LC | Y (s) | Y | 62 | 123 | 85 | 33 | 37 | 17 | ||
| rs16969968 | 15q25.1 G > A | Sakoda et al | 2011 | Cacausian | PB | LC | Y (s) | Y | 258 | 370 | 115 | 624 | 689 | 163 |
| Gabrielsen et al | 2013 | Cacausian | PB | LC | Y (s) | Y | 125 | 189 | 69 | 12386 | 12685 | 3298 | ||
| Pérez-Morales et al | 2018 | Cacausian | PB | LC | Y (s) | Y | 45 | 27 | 2 | 136 | 52 | 4 | ||
| Falvella et al | 2009 | Cacausian | PB | LC | mixed | Y | 128 | 226 | 113 | 267 | 348 | 124 | ||
| Zienolddiny et al | 2009 | Cacausian | PB | LC | Y (s) | Y | 112 | 186 | 59 | 174 | 194 | 58 | ||
| Ji et al | 2015 | Cacausian | PB | LC | mixed | Y | 514 | 904 | 396 | 750 | 917 | 286 | ||
| Lips et al | 2009 | Cacausian | PB | LC | Y (s) | Y | 1183 | 1560 | 563 | 2470 | 2493 | 633 | ||
| Lips et al | 2009 | Cacausian | PB | LC | N (ns) | Y | 133 | 155 | 54 | 1432 | 1454 | 387 | ||
| Jaworowska et al | 2011 | Cacausian | HB | LC | mixed | Y | 280 | 433 | 129 | 373 | 369 | 99 | ||
| Ito et al | 2012 | Cacausian | HB | LC | mixed | Y | 678 | 37 | 1 | 681 | 34 | 1 | ||
| Weissfeld et al | 2016 | Cacausian | HB | LC | Y (s) | Y | 276 | 378 | 124 | 471 | 545 | 149 | ||
| Islam et sl. | 2013 | Asian | PB | LC | mixed | Y | 58 | 43 | 5 | 72 | 40 | 4 | ||
| Young et al | 2011 | Cacausian | PB | LC | Y (s) | Y | 81 | 69 | 18 | 225 | 205 | 45 | ||
| rs8034191 | 15q25.1 T > C | Christopher et al | 2008 | Cacausian | PB | LC | mixed | Y | 670 | 858 | 303 | 448 | 415 | 97 |
| Christopher et al | 2008 | Cacausian | PB | LC | Y (s) | Y | 685 | 864 | 302 | 762 | 775 | 191 | ||
| Schwartz et al | 2009 | Cacausian | PB | LC | mixed | Y | 185 | 264 | 90 | 326 | 367 | 116 | ||
| Schwartz et al | 2009 | African | PB | LC | mixed | Y | 231 | 119 | 10 | 300 | 106 | 15 | ||
| Shiraishi et al | 2009 | Asian | HB | LC | Y (s) | N | 241 | 17 | 1 | 559 | 15 | 1 | ||
| Shiraishi et al | 2009 | Asian | HB | LC | Y (s) | N | 919 | 64 | 2 | 346 | 13 | 2 | ||
| Liu et al | 2008 | Cacausian | HB | LC | Y (s) | Y | 71 | 77 | 46 | 109 | 81 | 18 | ||
| Zienolddiny et al | 2009 | Cacausian | PB | LC | Y (s) | Y | 117 | 178 | 57 | 176 | 187 | 61 | ||
| Jaworowska et al | 2011 | Cacausian | HB | LC | mixed | Y | 286 | 419 | 128 | 368 | 361 | 102 | ||
| Ito et al | 2012 | Cacausian | HB | LC | mixed | Y | 674 | 41 | 4 | 672 | 43 | 1 | ||
| VanderWeele et al | 2012 | Cacausian | HB | LC | mixed | Y | 2506 | 3243 | 1115 | 2897 | 3083 | 786 | ||
| Weissfeld et al | 2016 | Cacausian | HB | LC | Y (s) | Y | 270 | 374 | 134 | 469 | 546 | 151 | ||
| de Mello et al | 2012 | Cacausian | PB | LC | mixed | Y | 44 | 71 | 29 | 53 | 67 | 24 | ||
| Wang et al | 2012 | Asian | HB | LC | mixed | Y | 350 | 29 | 2 | 385 | 25 | 0 | ||
| Bae et al | 2012 | Asian | HB | LC | mixed | Y | 328 | 544 | 221 | 294 | 535 | 261 | ||
H-B = hospital-based, HWE = Hardy Weinberg equilibrium, LC = lung cancer, mixed = not mentioned, N (ns); sample without smoking, N = controls not conformed to HWE, P-B = population-based, SNP = single nucleotide polymorphism, Y (s) = sample with smoking, Y = controls conformed to HWE.
3.2. Quantitative synthesis
The main results of the meta-analysis of the CHRNA5/A3/B4 gene polymorphisms and the risk of lung neoplasm were listed in Table 2.
Table 2.
Results of the meta-analysis of single nucleotide polymorphisms in CHRNA5/A3/B4 gene and risk of lung neoplasm.
| SNP | Comparison | Subgroup | N | P H | P Z | P A | Random | Fixed |
| rs1051730 | A vs G | Overall | 18 | .005 | <.001 | <.001 | 1.321 (1.240–1.408) | 1.293 (1.251–1.336) |
| A vs G | Cacausian | 11 | .012 | <.001 | <.001 | 1.285 (1.210–1.364) | 1.281 (1.240–1.325) | |
| A vs G | Asian | 6 | .634 | <.001 | <.001 | 1.851 (1.410–2.429) | 1.855 (1.414–2.434) | |
| A vs G | HB | 6 | .04 | <.001 | <.001 | 1.585 (1.261–1.993) | 1.289 (1.229–1.352) | |
| A vs G | PB | 12 | .013 | <.001 | <.001 | 1.294 (1.200–1.396) | 1.296 (1.239–1.355) | |
| A vs G | N (ns) | 3 | .004 | .994 | 1.000 | 1.681 (0.709–3.986) | 0.999 (0.844–1.183) | |
| A vs G | Y (s) | 9 | .73 | <.001 | <.001 | 1.336 (1.267–1.408) | 1.336 (1.267–1.409) | |
| A vs G | mixed | 6 | .116 | <.001 | <.001 | 1.319 (1.206–1.443) | 1.286 (1.232–1.342) | |
| A vs G | N | 2 | .234 | <.001 | <.001 | 1.474 (1.089–1.995) | 1.411 (1.205–1.652) | |
| A vs G | Y | 16 | .005 | <.001 | <.001 | 1.312 (1.227–1.404) | 1.288 (1.246–1.332) | |
| AA vs GG | Overall | 18 | .204 | <.001 | <.001 | 1.709 (1.549–1.886) | 1.700 (1.583–1.825) | |
| AA vs GG | Cacausian | 11 | .124 | <.001 | <.001 | 1.705 (1.535–1.894) | 1.697 (1.580–1.822) | |
| AA vs GG | Asian | 2 | .283 | .535 | 1.000 | 1.738 (0.216–13.991) | 1.809 (0.278–11.761) | |
| AA vs GG | HB | 4 | .114 | <.001 | <.001 | 2.149 (1.185–3.900) | 1.694 (1.527–1.880) | |
| AA vs GG | PB | 10 | .28 | <.001 | <.001 | 1.693 (1.512–1.896) | 1.705 (1.547–1.880) | |
| AA vs GG | N (ns) | 2 | .265 | .589 | 1.000 | 1.321 (0.427–4.086) | 1.117 (0.748–1.668) | |
| AA vs GG | Y (s) | 8 | .634 | <.001 | <.001 | 1.791 (1.597–2.010) | 1.793 (1.598–2.011) | |
| AA vs GG | mixed | 4 | .153 | <.001 | <.001 | 1.690 (1.411–2.023) | 1.681 (1.532–1.845) | |
| AA vs GG | N | 2 | .479 | <.001 | <.001 | 1.794 (1.281–2.512) | 1.790 (1.277–2.510) | |
| AA vs GG | Y | 12 | .131 | <.001 | <.001 | 1.706 (1.527–1.905) | 1.696 (1.577–1.824) | |
| AG vs GG | Overall | 18 | .007 | <.001 | <.001 | 1.299 (1.189–1.420) | 1.246 (1.189–1.307) | |
| AG vs GG | Cacausian | 11 | .026 | <.001 | <.001 | 1.234 (1.135–1.341) | 1.223 (1.165–1.284) | |
| AG vs GG | Asian | 6 | .662 | <.001 | <.001 | 1.873 (1.415–2.478) | 1.889 (1.429–2.497) | |
| AG vs GG | HB | 6 | .073 | .002 | .050 | 1.478 (1.154–1.892) | 1.225 (1.142–1.313) | |
| AG vs GG | PB | 12 | .012 | <.001 | <.001 | 1.281 (1.149–1.428) | 1.266 (1.186–1.350) | |
| AG vs GG | N (ns) | 3 | .002 | .373 | 1.000 | 1.547 (0.593–4.039) | 0.890 (0.712–1.112) | |
| AG vs GG | Y (s) | 9 | .747 | <.001 | <.001 | 1.316 (1.217–1.422) | 1.319 (1.220–1.425) | |
| AG vs GG | mixed | 6 | .238 | <.001 | <.001 | 1.278 (1.157–1.413) | 1.234 (1.160–1.313) | |
| AG vs GG | N | 2 | .163 | .001 | .025 | 1.619 (1.016–2.579) | 1.492 (1.182–1.882) | |
| AG vs GG | Y | 16 | .009 | <.001 | <.001 | 1.280 (1.168–1.402) | 1.237 (1.178–1.298) | |
| AG+AA vs GG | Overall | 18 | .006 | <.001 | <.001 | 1.381 (1.269–1.503) | 1.336 (1.277–1.397) | |
| AG+AA vs GG | Cacausian | 11 | .009 | <.001 | <.001 | 1.326 (1.218–1.443) | 1.316 (1.257–1.378) | |
| AG+AA vs GG | Asian | 6 | .647 | <.001 | <.001 | 1.876 (1.421–2.477) | 1.886 (1.430–2.487) | |
| AG+AA vs GG | HB | 6 | .124 | <.001 | <.001 | 1.548 (1.245–1.925) | 1.319 (1.235–1.409) | |
| AG+AA vs GG | PB | 12 | .006 | <.001 | <.001 | 1.358 (1.220–1.512) | 1.350 (1.270–1.435) | |
| AG+AA vs GG | N (ns) | 3 | .002 | .317 | 1.000 | 1.618 (0.630–4.157) | 1.336 (1.277–1.397) | |
| AG+AA vs GG | Y (s) | 9 | .894 | <.001 | <.001 | 1.411 (1.310–1.519) | 1.413 (1.313–1.521) | |
| AG+AA vs GG | mixed | 6 | .281 | <.001 | <.001 | 1.354 (1.240–1.479) | 1.324 (1.249–1.405) | |
| AG+AA vs GG | N | 2 | .272 | <.001 | <.001 | 1.583 (1.180–2.122) | 1.554 (1.247–1.938) | |
| AG+AA vs GG | Y | 16 | .006 | <.001 | <.001 | 1.365 (1.249–1.491) | 1.327 (1.267–1.389) | |
| AA vs AG+GG | Overall | 18 | .371 | <.001 | <.001 | 1.519 (1.409–1.639) | 1.521 (1.424–1.625) | |
| AA vs AG+GG | Cacausian | 11 | .262 | <.001 | <.001 | 1.516 (1.393–1.649) | 1.519 (1.421–1.623) | |
| AA vs AG+GG | Asian | 2 | .283 | .555 | 1.000 | 1.677 (0.208–13.518) | 1.758 (0.271–11.425) | |
| AA vs AG+GG | HB | 4 | .096 | .026 | .650 | 1.974 (1.086–3.585) | 1.540 (1.398–1.696) | |
| AA vs AG+GG | PB | 10 | .577 | <.001 | <.001 | 1.501 (1.371–1.644) | 1.505 (1.374–1.648) | |
| AA vs AG+GG | N (ns) | 2 | .307 | .256 | 1.000 | 1.288 (0.706–2.350) | 1.252 (0.850–1.846) | |
| AA vs AG+GG | Y (s) | 8 | .479 | <.001 | <.001 | 1.552 (1.395–1.726) | 1.555 (1.398–1.730) | |
| AA vs AG+GG | mixed | 4 | .151 | <.001 | <.001 | 1.507 (1.273–1.784) | 1.514 (1.389–1.651) | |
| AA vs AG+GG | N | 2 | .548 | .008 | .200 | 1.520 (1.118–2.066) | 1.518 (1.116–2.065) | |
| AA vs AG+GG | Y | 12 | .251 | <.001 | <.001 | 1.520 (1.390–1.663) | 1.521 (1.422–1.628) | |
| rs6495309 | T vs C | Overall | 6 | 0 | .268 | 1.000 | 1.110 (0.923–1.334) | 1.153 (1.094–1.216) |
| T vs C | Asian | 5 | .004 | .025 | .625 | 1.198 (1.023–1.404) | 1.206 (1.139–1.276) | |
| T vs C | HB | 4 | .008 | .178 | 1.000 | 1.224 (0.912–1.643) | 1.288 (1.177–1.409) | |
| T vs C | PB | 2 | 0 | .898 | 1.000 | 0.978 (0.696–1.374) | 1.086 (1.017–1.160) | |
| T vs C | Y(s) | 3 | .001 | .658 | 1.000 | 1.119 (0.681–1.838) | 0.893 (0.777–1.027) | |
| T vs C | mixed | 2 | .023 | .003 | .075 | 1.235 (1.074–1.419) | 1.214 (1.145–1.287) | |
| TT vs CC | Overall | 6 | .002 | .066 | 1.000 | 1.325 (0.982–1.789) | 1.450 (1.297–1.622) | |
| TT vs CC | Asian | 5 | .01 | .013 | .325 | 1.462 (1.082–1.976) | 1.502 (1.338–1.687) | |
| TT vs CC | HB | 4 | .01 | .182 | 1.000 | 1.501 (0.827–2.726) | 1.655 (1.382–1.982) | |
| TT vs CC | PB | 2 | .039 | .597 | 1.000 | 1.142 (0.699–1.865) | 1.334 (1.157–1.539) | |
| TT vs CC | Y(s) | 3 | .007 | .499 | 1.000 | 1.382 (0.542–3.524) | 1.047 (0.734–1.495) | |
| TT vs CC | mixed | 2 | .074 | <.001 | <.001 | 1.557 (1.248–1.943) | 1.526 (1.354–1.721) | |
| TC vs CC | Overall | 6 | 0 | .290 | 1.000 | 1.179 (0.869–1.601) | 1.203 (1.097–1.320) | |
| TC vs CC | Asian | 5 | .16 | <.001 | <.001 | 1.358 (1.135–1.624) | 1.385 (1.246–1.541) | |
| TC vs CC | HB | 4 | .091 | .165 | 1.000 | 1.303 (0.897–1.894) | 1.357 (1.149–1.601) | |
| TC vs CC | PB | 2 | 0 | .909 | 1.000 | 1.036 (0.565–1.899) | 1.139 (1.019–1.274) | |
| TC vs CC | Y(s) | 3 | .017 | .845 | 1.000 | 1.066 (0.563–2.018) | 0.806 (0.673–0.965) | |
| TC vs CC | mixed | 2 | .997 | .019 | .475 | 1.406 (1.258–1.571) | 1.406 (1.258–1.571) | |
| TC+TT vs CC | Overall | 6 | 0 | .224 | 1.000 | 1.219 (0.886–1.678) | 1.240 (1.136–1.354) | |
| TC+TT vs CC | Asian | 5 | .038 | .004 | .100 | 1.386 (1.110–1.732) | 1.428 (1.292–1.579) | |
| TC+TT vs CC | HB | 4 | .018 | .161 | 1.000 | 1.378 (0.880–2.158) | 1.462 (1.251–1.708) | |
| TC+TT vs CC | PB | 2 | 0 | .893 | 1.000 | 1.042 (0.575–1.886) | 1.147 (1.031–1.276) | |
| TC+TT vs CC | Y(s) | 3 | .004 | .680 | 1.000 | 1.161 (0.571–2.361) | 0.823 (0.693–0.978) | |
| TC+TT vs CC | mixed | 2 | .408 | <.001 | <.001 | 1.453 (1.308–1.613) | 1.453 (1.308–1.614) | |
| TT vs TC+CC | Overall | 6 | .031 | .145 | 1.000 | 1.155 (0.952–1.402) | 1.176 (1.080–1.280) | |
| TT vs TC+ CC | Asian | 5 | .025 | .106 | 1.000 | 1.192 (0.964–1.474) | 1.186 (1.088–1.293) | |
| TT vs TC+CC | HB | 4 | .103 | <.001 | <.001 | 1.236 (0.869–1.757) | 1.338 (1.167–1.535) | |
| TT vs TC+CC | PB | 2 | .472 | .147 | 1.000 | 1.084 (0.972–1.208) | 1.084 (0.972–1.208) | |
| TT vs TC+CC | Y(s) | 3 | .119 | .622 | 1.000 | 1.173 (0.692–1.989) | 1.087 (0.779–1.518) | |
| TT vs TC+CC | mixed | 2 | .013 | .085 | 1.000 | 1.225 (0.973–1.543) | 1.193 (1.091–1.304) | |
| rs3743073 | A vs C | Overall | 4 | 0 | .081 | 1.000 | 1.341 (0.965–1.865) | 1.375 (1.234–1.532) |
| A vs C | Asian | 3 | .201 | <.001 | <.001 | 1.565 (1.323–1.852) | 1.580 (1.397–1.788) | |
| A vs C | PB | 3 | .201 | <.001 | <.001 | 1.565 (1.323–1.852) | 1.580 (1.397–1.788) | |
| A vs C | Y(s) | 2 | .001 | .608 | 1.000 | 1.197 (0.602–2.378) | 1.059 (0.876–1.279) | |
| AA vs CC | Overall | 4 | 0 | .122 | 1.000 | 1.692 (0.869–3.296) | 1.846 (1.486–2.292) | |
| AA vs CC | Asian | 3 | .449 | <.001 | <.001 | 2.420 (1.891–3.097) | 2.419 (1.891–3.095) | |
| AA vs CC | PB | 3 | .449 | <.001 | <.001 | 2.420 (1.891–3.097) | 2.419 (1.891–3.095) | |
| AA vs CC | Y (s) | 2 | .001 | .742 | 1.000 | 1.269 (0.307–5.236) | 1.069 (0.725–1.577) | |
| AC vs CC | Overall | 4 | .206 | .002 | .050 | 1.347 (1.067–1.700) | 1.325 (1.106–1.587) | |
| AC vs CC | Asian | 3 | .526 | <.001 | <.001 | 1.476 (1.188–1.832) | 1.477 (1.190–1.833) | |
| AC vs CC | PB | 3 | .526 | <.001 | <.001 | 1.476 (1.188–1.832) | 1.477 (1.190–1.833) | |
| AC vs CC | Y (s) | 2 | .099 | .357 | 1.000 | 1.282 (0.756–2.174) | 1.177 (0.886–1.565) | |
| AC+AA vs CC | Overall | 4 | .007 | .026 | .650 | 1.507 (1.050–2.163) | 1.461 (1.234–1.731) | |
| AC+AA vs CC | Asian | 3 | .835 | <.001 | <.001 | 1.772 (1.447–2.170) | 1.769 (1.444–2.167) | |
| AC+AA vs CC | PB | 3 | .835 | <.001 | <.001 | 1.772 (1.447–2.170) | 1.769 (1.444–2.167) | |
| AC+AA vs CC | Y (s) | 2 | .01 | .363 | 1.000 | 1.337 (0.611–2.924) | 1.133 (0.866–1.481) | |
| AA vs AC+CC | Overall | 4 | 0 | .274 | 1.000 | 1.363 (0.783–2.373) | 1.573 (1.313–1.884) | |
| AA vs AC+CC | Asian | 3 | .063 | .003 | .075 | 1.767 (1.219–2.563) | 1.866 (1.530–2.275) | |
| AA vs AC+CC | Taqman | 2 | .275 | .020 | .500 | 1.454 (1.019–2.075) | 1.448 (1.060–1.979) | |
| AA vs AC+CC | PB | 3 | .063 | .003 | .075 | 1.767 (1.219–2.563) | 1.866 (1.530–2.275) | |
| AA vs AC+CC | Y (s) | 2 | .004 | .908 | 1.000 | 1.067 (0.357–3.191) | 0.982 (0.689–1.400) | |
| rs16969968 | A vs G | Overall | 13 | .408 | <.001 | <.001 | 1.331 (1.281–1.384) | 1.333 (1.285–1.383) |
| A vs G | Cacausian | 12 | .334 | <.001 | <.001 | 1.328 (1.274–1.385) | 1.333 (1.285–1.384) | |
| A vs G | HB | 3 | .284 | <.001 | <.001 | 1.259 (1.125–1.410) | 1.261 (1.148–1.385) | |
| A vs G | PB | 10 | .493 | <.001 | <.001 | 1.347 (1.294–1.402) | 1.347 (1.294–1.401) | |
| A vs G | Y (s) | 7 | .273 | <.001 | <.001 | 1.305 (1.232–1.383) | 1.317 (1.258–1.378) | |
| A vs G | mixed | 5 | .813 | <.001 | <.001 | 1.393 (1.302–1.491) | 1.393 (1.302–1.491) | |
| AA vs GG | Overall | 13 | .692 | <.001 | <.001 | 1.784 (1.650–1.928) | 1.782 (1.649–1.926) | |
| AA vs GG | Cacausian | 12 | .617 | <.001 | <.001 | 1.784 (1.650–1.929) | 1.783 (1.649–1.927) | |
| AA vs GG | HB | 3 | .607 | <.001 | <.001 | 1.554 (1.265–1.909) | 1.555 (1.266–1.909) | |
| AA vs GG | PB | 10 | .724 | <.001 | <.001 | 1.825 (1.678–1.986) | 1.824 (1.677–1.985) | |
| AA vs GG | Y (s) | 7 | .448 | <.001 | <.001 | 1.749 (1.588–1.926) | 1.743 (1.583–1.920) | |
| AA vs GG | mixed | 5 | .909 | <.001 | <.001 | 1.922 (1.665–2.218) | 1.922 (1.665–2.218) | |
| AG vs GG | Overall | 13 | .478 | <.001 | <.001 | 1.334 (1.262–1.410) | 1.334 (1.262–1.410) | |
| AG vs GG | Cacausian | 12 | .398 | <.001 | <.001 | 1.335 (1.259–1.416) | 1.334 (1.262–1.411) | |
| AG vs GG | HB | 3 | .117 | <.001 | <.001 | 1.310 (1.050–1.635) | 1.327 (1.157–1.523) | |
| AG vs GG | PB | 10 | .598 | <.001 | <.001 | 1.336 (1.257–1.419) | 1.336 (1.257–1.419) | |
| AG vs GG | Y (s) | 7 | .438 | <.001 | <.001 | 1.310 (1.224–1.402) | 1.310 (1.224–1.402) | |
| AG vs GG | mixed | 5 | .71 | <.001 | <.001 | 1.433 (1.291–1.590) | 1.433 (1.291–1.590) | |
| AG+AA vs GG | Overall | 13 | .298 | <.001 | <.001 | 1.422 (1.337–1.512) | 1.426 (1.353–1.502) | |
| AG+AA vs GG | Cacausian | 12 | .235 | <.001 | <.001 | 1.421 (1.331–1.516) | 1.426 (1.353–1.504) | |
| AG+AA vs GG | HB | 3 | .108 | <.001 | <.001 | 1.348 (1.086–1.673) | 1.369 (1.202–1.561) | |
| AG+AA vs GG | PB | 10 | .416 | <.001 | <.001 | 1.436 (1.353–1.524) | 1.437 (1.357–1.521) | |
| AG+AA vs GG | Y (s) | 7 | .314 | <.001 | <.001 | 1.392 (1.288–1.505) | 1.398 (1.311–1.491) | |
| AG+AA vs GG | mixed | 5 | .63 | <.001 | <.001 | 1.537 (1.393–1.697) | 1.537 (1.392–1.696) | |
| AA vs AG+GG | Overall | 13 | .882 | <.001 | <.001 | 1.515 (1.412–1.627) | 1.514 (1.410–1.625) | |
| AA vs AG+GG | Cacausian | 12 | .833 | <.001 | <.001 | 1.516 (1.412–1.627) | 1.514 (1.410–1.626) | |
| AA vs AG+GG | HB | 3 | .952 | <.001 | <.001 | 1.320 (1.092–1.595) | 1.320 (1.092–1.595) | |
| AA vs AG+GG | PB | 10 | .898 | <.001 | <.001 | 1.550 (1.436–1.673) | 1.549 (1.435–1.672) | |
| AA vs AG+GG | Y (s) | 7 | .586 | <.001 | <.001 | 1.507 (1.379–1.647) | 1.503 (1.375–1.643) | |
| AA vs AG+GG | mixed | 5 | .855 | <.001 | <.001 | 1.555 (1.368–1.767) | 1.555 (1.368–1.767) | |
| rs8034191 | C vs T | Overall | 15 | .000 | <.001 | <.001 | 1.273 (1.166–1.390) | 1.251 (1.210–1.294) |
| C vs T | Cacausian | 10 | .107 | <.001 | <.001 | 1.300 (1.229–1.375) | 1.288 (1.243–1.334) | |
| C vs T | Asian | 4 | .002 | .190 | 1.000 | 1.385 (0.851–2.255) | 0.943 (0.844–1.054) | |
| C vs T | HB | 6 | .000 | .001 | .025 | 1.294 (1.111–1.508) | 1.227 (1.178–1.277) | |
| C vs T | PB | 9 | .502 | <.001 | <.001 | 1.308 (1.231–1.391) | 1.309 (1.232–1.391) | |
| C vs T | Y (s) | 5 | .064 | <.001 | <.001 | 1.347 (1.187–1.529) | 1.309 (1.221–1.404) | |
| C vs T | mixed | 9 | .000 | .002 | .050 | 1.208 (1.072–1.361) | 1.231 (1.185–1.279) | |
| C vs T | N | 2 | .216 | .007 | .175 | 1.871 (1.108–3.159) | 1.796 (1.173–2.751) | |
| C vs T | Y | 13 | .000 | <.001 | <.001 | 1.254 (1.149–1.369) | 1.248 (1.207–1.291) | |
| CC vs TT | Overall | 15 | .000 | <.001 | <.001 | 1.529 (1.254–1.863) | 1.553 (1.446–1.669) | |
| CC vs TT | Cacausian | 10 | .135 | <.001 | <.001 | 1.698 (1.511–1.908) | 1.683 (1.559–1.816) | |
| CC vs TT | Asian | 4 | .434 | .031 | .775 | 0.766 (0.606–0.970) | 0.772 (0.611–0.976) | |
| CC vs TT | HB | 6 | .000 | .014 | .350 | 1.551 (1.095–2.199) | 1.498 (1.375–1.632) | |
| CC vs TT | PB | 9 | .169 | <.001 | <.001 | 1.623 (1.349–1.952) | 1.692 (1.482–1.931) | |
| CC vs TT | Y (s) | 5 | .032 | <.001 | <.001 | 1.752 (1.296–2.369) | 1.717 (1.479–1.994) | |
| CC vs TT | mixed | 9 | .000 | .016 | .400 | 1.409 (1.065–1.865) | 1.507 (1.388–1.636) | |
| CC vs TT | N | 2 | .294 | .677 | 1.000 | 0.710 (0.130–3.881) | 0.710 (0.142–3.542) | |
| CC vs TT | Y | 13 | .000 | <.001 | <.001 | 1.546 (1.267–1.887) | 1.556 (1.448–1.672) | |
| CT vs TT | Overall | 15 | .043 | <.001 | <.001 | 1.271 (1.170–1.380) | 1.244 (1.184–1.307) | |
| CT vs TT | Cacausian | 10 | .569 | <.001 | <.001 | 1.257 (1.193–1.324) | 1.257 (1.193–1.324) | |
| CT vs TT | Asian | 4 | .007 | <.001 | <.001 | 1.437 (0.880–2.347) | 1.067 (0.900–1.266) | |
| CT vs TT | HB | 6 | .010 | .002 | .050 | 1.247 (1.081–1.437) | 1.213 (1.143–1.288) | |
| CT vs TT | PB | 9 | .875 | <.001 | <.001 | 1.315 (1.204–1.437) | 1.315 (1.204–1.437) | |
| CT vs TT | Y (s) | 5 | .563 | <.001 | <.001 | 1.272 (1.147–1.411) | 1.275 (1.150–1.414) | |
| CT vs TT | mixed | 9 | .034 | <.001 | <.001 | 1.239 (1.110–1.383) | 1.230 (1.162–1.301) | |
| CT vs TT | N | 2 | .461 | .002 | .050 | 2.149 (1.353–3.412) | 2.108 (1.316–3.375) | |
| CT vs TT | Y | 13 | .108 | <.001 | <.001 | 1.248 (1.158–1.345) | 1.236 (1.176–1.299) | |
| CT+CC vs TT | Overall | 15 | .002 | <.001 | <.001 | 1.336 (1.215–1.469) | 1.313 (1.253–1.376) | |
| CT+CC vs TT | Cacausian | 10 | .373 | <.001 | <.001 | 1.352 (1.278–1.430) | 1.344 (1.279–1.412) | |
| CT+CC vs TT | Asian | 4 | .003 | .185 | 1.000 | 1.409 (0.848–2.340) | 1.011 (0.860–1.189) | |
| CT+CC vs TT | HB | 6 | .000 | .001 | .025 | 1.323 (1.123–1.559) | 1.280 (1.210–1.354) | |
| CT+CC vs TT | PB | 9 | .865 | <.001 | <.001 | 1.391 (1.279–1.513) | 1.391 (1.279–1.513) | |
| CT+CC vs TT | Y (s) | 5 | .421 | <.001 | <.001 | 1.367 (1.240–1.507) | 1.368 (1.241–1.508) | |
| CT+CC vs TT | mixed | 9 | .001 | <.001 | <.001 | 1.274 (1.118–1.452) | 1.292 (1.225–1.363) | |
| CT+CC vs TT | N | 2 | .319 | .004 | .100 | 1.995 (1.283–3.102) | 1.953 (1.246–3.062) | |
| CT+CC vs TT | Y | 13 | .002 | <.001 | <.001 | 1.313 (1.196–1.442) | 1.307 (1.247–1.370) | |
| CC vs CT+TT | Overall | 15 | .000 | <.001 | <.001 | 1.352 (1.139–1.605) | 1.382 (1.293–1.476) | |
| CC vs CT+TT | Cacausian | 10 | .089 | <.001 | <.001 | 1.475 (1.315–1.655) | 1.482 (1.381–1.591) | |
| CC vs CT+TT | Asian | 4 | .449 | .043 | 1.000 | 0.809 (0.662–0.989) | 0.813 (0.666–0.994) | |
| CC vs CT+TT | HB | 6 | .000 | <.001 | <.001 | 1.389 (1.034–1.866) | 1.348 (1.246–1.458) | |
| CC vs CT+TT | PB | 9 | .129 | <.001 | <.001 | 1.395 (1.164–1.671) | 1.466 (1.297–1.658) | |
| CC vs CT+TT | Y (s) | 5 | .025 | <.001 | <.001 | 1.532 (1.150–2.042) | 1.516 (1.320–1.741) | |
| CC vs CT+TT | mixed | 9 | .000 | <.001 | <.001 | 1.256 (0.993–1.590) | 1.344 (1.246–1.449) | |
| CC vs CT+TT | N | 2 | .297 | .651 | 1.000 | 0.683 (0.126–3.693) | 0.690 (0.138–3.443) | |
| CC vs CT+TT | Y | 13 | .000 | <.001 | <.001 | 1.363 (1.147–1.619) | 1.383 (1.295–1.478) |
Heterogeneity was considered to be significant when the P-value was less than .1. If there was no significant heterogeneity, a fixed effect model (Der-Simonian Laird) was used to evaluate the point estimates and 95% CI; otherwise, a random effects model (Der-Simonian Laird) was used. And the PZ was calculated based on the actual model adopted.
H-B = hospital-based, HWE = Hardy Weinberg equilibrium, LC = lung cancer, mixed = not mentioned, N (ns) = sample without smoking, P (Adjust) = multiple testing P value according to Bonferroni Correction (P value less than .05 / 5 models was considered as statistically significant, which was marked with bold font in the table), P-B = Population-based, PH = P value of Q test for heterogeneity test, PZ = means statistically significant, SNP = single nucleotide polymorphism, Y (s) = sample with from smoking.
3.2.1. CHRNA3 rs1051730
Overall analysis of the rs1051730 polymorphism showed there is significant LC risks in the allelic, homozygous, heterozygous, dominant, and recessive models (PA < .0001 in all 5 models). However, there were no significant differences of the ORs when the sources of control and HWE status were match or not match.
In stratification analysis by ethnicity, it was found that in the homozygous and recessive models, the Caucasian population was at higher risk of developing LC (AA vs GG: OR = 1.697, 95%CI = 1.580–1.822, PA < .0001, Fig. 1; AA vs AG+GG: OR = 1.519, 95%CI = 1.421–1.623, PA < .001). Ethnicity exhibited no influence on the results of analyses in other 3 models.
Figure 1.
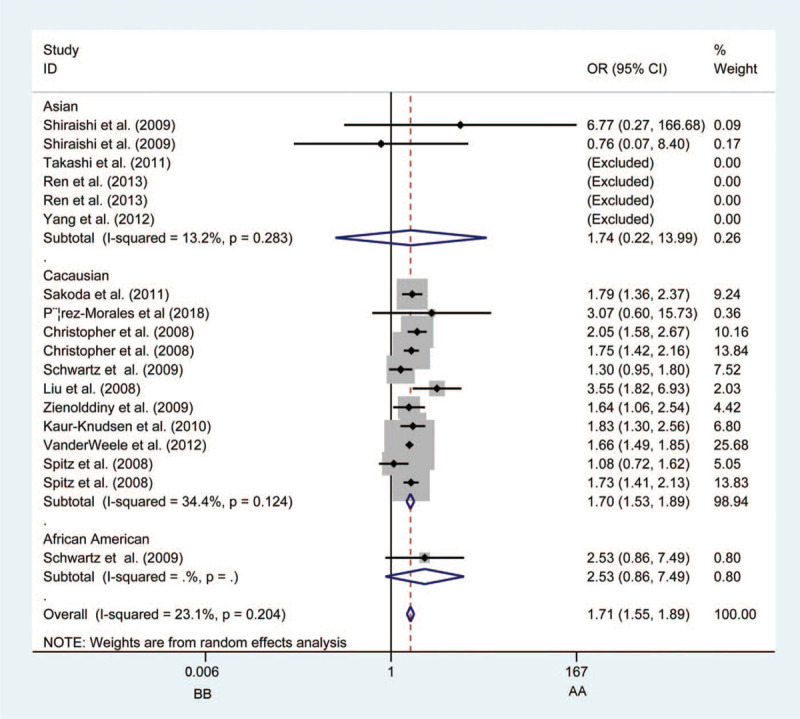
Forest plots of the association between CHRNA3 rs1051730 polymorphism and the risk of lung cancer in Caucasian population (BB vs AA). Each square indicates a study, and the area of squares is proportional to the weight of the study. The diamond represents the summary odds ratio and 95% confidence interval. CI = confidence interval, OR = odds ratio.
Consistent with previous studies, the rs1051730 polymorphism was more significant in smokers within all 5 models. When compared with the non-smokers, the results were as follows: in A vs G model (OR = 1.336, 95%CI = 1.267–1.409, P < .0001, Fig. 2); in AA vs GG model (OR = 1.809, 95%CI = 0.278–11.761, P < .0001); in AG vs GG model (OR = 1.319, 95%CI = 1.220–1.425, P < .0001); in AG + AA vs GG model (OR = 1.413, 95%CI = 1.313–1.521, P < .0001), and in AA vs AG + GG model (OR = 1.555, 95%CI = 1.398–1.730, P < .0001).
Figure 2.
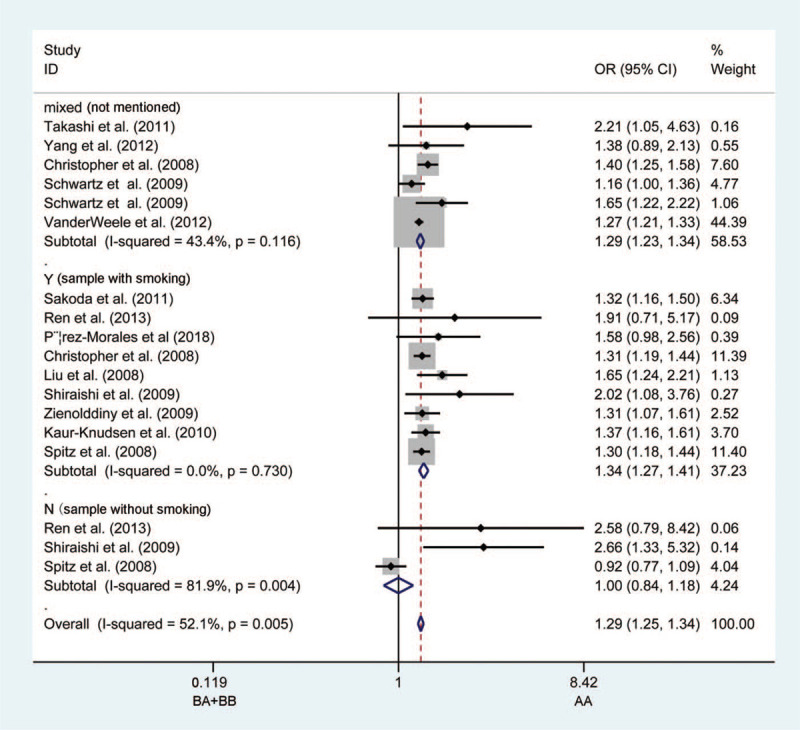
Forest plots of the association between CHRNA3 rs1051730 polymorphism and the risk of lung cancer in the non-smoking population (BA + BB vs AA). Each square indicates a study, and the area of squares is proportional to the weight of the study. The diamond represents the summary odds ratio and 95% confidence interval. CI = confidence interval, OR = odds ratio.
3.2.2. CHRNA3 rs6495309
No link was observed between the rs6495309 polymorphism and LC risk in the overall analysis. However, we did observe that the rs6495309 polymorphism was associated with LC susceptibility in an Asian population upon heterozygous comparison (TC vs CC: OR = 1.385, 95%CI = 1.246–1.541, P < .001, Fig. 3). Stratified by the source of the controls, HB groups with “B” variants had an increased OR of being diagnosed with LC in recessive models (TT vs TC + CC: OR = 1.338, 95%CI = 1.167–1.535, P < .0001), indicating there is a considerable heterogeneity based on the source of the controls.
Figure 3.
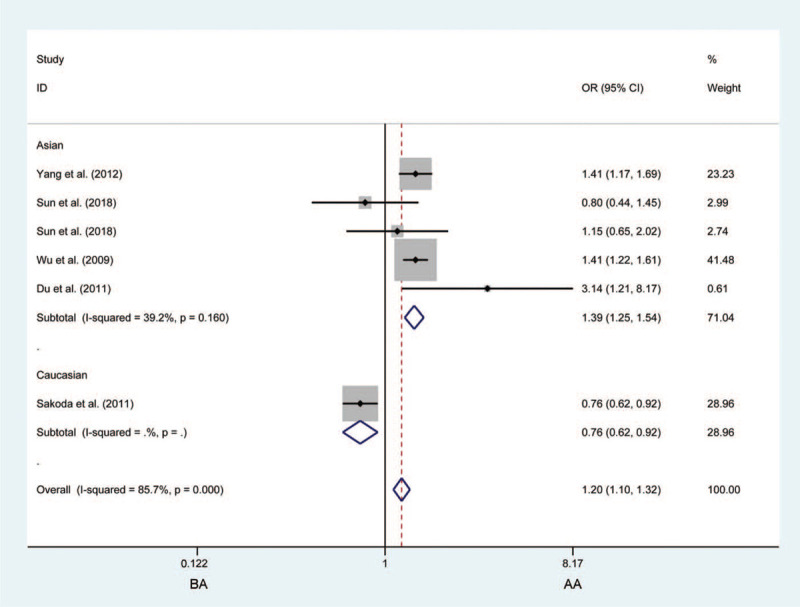
Forest plots of the association between CHRNA3 rs6495309 polymorphism and the risk of lung cancer in the Asian population (BA vs AA). Each square indicates a study, and the area of squares is proportional to the weight of the study. The diamond represents the summary odds ratio and 95% confidence interval. CI = confidence interval, OR = odds ratio.
3.2.3. CHRNA3 rs3743073
Analysis of the rs3743073 polymorphism revealed no remarkable effect on LC susceptibility. When ethnicity was taken into account, it was observed that the rs3743037 B-allele increases the risk of LC significantly in Asian via allelic contrast (A vs C: OR = 1.580, 95%CI: 1.397–1.788, P < .0001, Fig. 4), heterozygous contrast (AC vs CC: OR = 1.477, 95%CI = 1.190–1.833, P < .0001), and dominant contrast (AC + CC vs CC: OR = 1.769, 95%CI = 1.444–2.167, P < .0001). When the source of the control subgroup considered, the risk in PB groups with the B allele of developing LC was higher than that in HB groups under allelic contrast (A vs C: OR = 1.580, 95%CI = 1.397–1.788, P < .0001), heterozygous contrast (AC vs CC: OR = 1.477, 95%CI = 1.190–1.833, P < .0001), and dominant contrast (AC + CC vs CC: OR = 1.769, 95%CI = 1.444–2.167, P < .0001).
Figure 4.
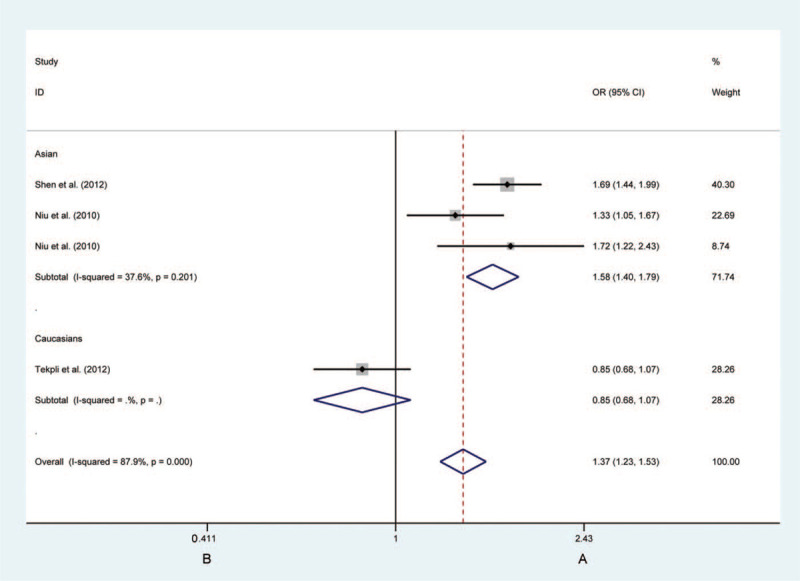
Forest plots of the association between CHRNA3 rs3743073 polymorphism and the risk of lung cancer in the Asian individuals (B vs A). Each square indicates a study, and the area of squares is proportional to the weight of the study. The diamond represents the summary odds ratio and 95% confidence interval. CI = confidence interval, OR = odds ratio.
3.2.4. CHRNA5 rs16969968
For the rs16969968 polymorphism, the pooled analysis demonstrated a significant link with LC risk in all 5 models, as displayed in Table 3. The ORs of the 16969968-C allele were obviously elevated in LC cases in the stratified analysis with matched or none matched ethnicity, source of controls, and smoking status.
Table 3.
False-positive report probability values for associations between the risk of lung cancer and CHRNA5/A3/B4 gene.
| Prior probability | ||||||||||
| Genotype | Comparison | Supgroup | Crude OR (95%) | P-value | Stastical power | 0.250 | 0.100 | 0.010 | 0.001 | 0.0001 |
| rs1051730 | A vs G | Overall | 1.321 (1.240–1.408) | <.001 | 0.856 | 0.000 | 0.000 | 0.001 | 0.006 | 0.061 |
| A vs G | Cacausian | 1.285 (1.210–1.364) | <.001 | 0.926 | 0.000 | 0.000 | 0.001 | 0.006 | 0.057 | |
| A vs G | Asian | 1.855 (1.414–2.434) | <.001 | 0.053 | 0.000 | 0.001 | 0.010 | 0.095 | 0.513 | |
| A vs G | HB | 1.585 (1.261–1.993) | <.001 | 0.142 | 0.000 | 0.000 | 0.004 | 0.038 | 0.283 | |
| A vs G | PB | 1.294 (1.200–1.396) | <.001 | 1.000 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 | |
| A vs G | Y (s) | 1.336 (1.267–1.409) | <.001 | 1.000 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 | |
| A vs G | mixed | 1.286 (1.232–1.342) | <.001 | 1.000 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 | |
| A vs G | N | 1.411 (1.205–1.652) | <.001 | 0.691 | 0.000 | 0.000 | 0.001 | 0.008 | 0.075 | |
| A vs G | Y | 1.312 (1.227–1.404) | <.001 | 1.000 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 | |
| AA vs GG | Overall | 1.700 (1.583–1.825) | <.001 | 1.000 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 | |
| AA vs GG | Cacausian | 1.697 (1.580–1.822) | <.001 | 1.000 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 | |
| AA vs GG | HB | 1.694 (1.527–1.880) | <.001 | 1.000 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 | |
| AA vs GG | PB | 1.705 (1.547–1.880) | <.001 | 1.000 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 | |
| AA vs GG | Y (s) | 1.793 (1.598–2.011) | <.001 | 1.000 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 | |
| AA vs GG | mixed | 1.681 (1.532–1.845) | <.001 | 1.000 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 | |
| AA vs GG | N | 1.790 (1.277–2.510) | <.001 | 0.014 | 0.001 | 0.004 | 0.037 | 0.282 | 0.797 | |
| AA vs GG | Y | 1.696 (1.577–1.824) | <.001 | 1.000 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 | |
| AG vs GG | Cacausian | 1.234 (1.135–1.341) | <.001 | 1.000 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 | |
| AG vs GG | Asian | 1.889 (1.429–2.497) | <.001 | 0.045 | 0.000 | 0.001 | 0.012 | 0.110 | 0.553 | |
| AG vs GG | PB | 1.281 (1.149–1.428) | <.001 | 0.997 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 | |
| AG vs GG | Y (s) | 1.319 (1.220–1.425) | <.001 | 1.000 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 | |
| AG vs GG | mixed | 1.234 (1.160–1.313) | <.001 | 1.000 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 | |
| AG vs GG | N | 1.492 (1.182–1.882) | <.001 | 0.132 | 0.000 | 0.000 | 0.004 | 0.041 | 0.298 | |
| AG vs GG | Y | 1.280 (1.168–1.402) | <.001 | 1.000 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 | |
| AG+AA vs GG | Overall | 1.381 (1.269–1.503) | <.001 | 1.000 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 | |
| AG+AA vs GG | Cacausian | 1.326 (1.218–1.443) | <.001 | 1.000 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 | |
| AG+AA vs GG | Asian | 1.886 (1.430–2.487) | <.001 | 0.048 | 0.000 | 0.001 | 0.011 | 0.105 | 0.540 | |
| AG+AA vs GG | HB | 1.319 (1.235–1.409) | <.001 | 1.000 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 | |
| AG+AA vs GG | PB | 1.358 (1.220–1.512) | <.001 | 1.000 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 | |
| AG+AA vs GG | Y (s) | 1.413 (1.313–1.521) | <.001 | 1.000 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 | |
| AG+AA vs GG | mixed | 1.324 (1.249–1.405) | <.001 | 1.000 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 | |
| AG+AA vs GG | N | 1.554 (1.247–1.938) | <.001 | 0.173 | 0.000 | 0.000 | 0.003 | 0.031 | 0.244 | |
| AG+AA vs GG | Y | 1.365 (1.249–1.491) | <.001 | 1.000 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 | |
| AA vs AG+GG | Overall | 1.521 (1.424–1.625) | <.001 | 1.000 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 | |
| AA vs AG+GG | Cacausian | 1.519 (1.421–1.623) | <.001 | 1.000 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 | |
| AA vs AG+GG | PB | 1.501 (1.371–1.644) | <.001 | 1.000 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 | |
| AA vs AG+GG | Y (s) | 1.552 (1.395–1.726) | <.001 | 0.998 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 | |
| AA vs AG+GG | mixed | 1.507 (1.273–1.784) | <.001 | 0.567 | 0.000 | 0.000 | 0.001 | 0.010 | 0.090 | |
| AA vs AG+GG | Y | 1.520 (1.390–1.663) | <.001 | 1.000 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 | |
| rs6495309 | TT vs CC | mixed | 1.526 (1.354–1.721) | <.001 | 0.981 | 0.000 | 0.000 | 0.001 | 0.006 | 0.054 |
| TC vs CC | Asian | 1.358 (1.135–1.624) | <.001 | 0.461 | 0.000 | 0.000 | 0.001 | 0.012 | 0.108 | |
| TC+TT vs CC | mixed | 1.453 (1.308–1.613) | <.001 | 0.999 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 | |
| TT vs TC+CC | HB | 1.338 (1.167–1.535) | <.001 | 0.893 | 0.000 | 0.000 | 0.001 | 0.006 | 0.059 | |
| rs3743073 | A vs C | Asian | 2.419 (1.891–3.095) | <.001 | 0.094 | 0.000 | 0.001 | 0.006 | 0.056 | 0.373 |
| A vs C | PB | 2.419 (1.891–3.095) | <.001 | 0.094 | 0.000 | 0.001 | 0.006 | 0.056 | 0.373 | |
| AA vs CC | Asian | 2.419 (1.891–3.095) | <.001 | 0.094 | 0.000 | 0.001 | 0.006 | 0.056 | 0.373 | |
| AA vs CC | PB | 2.419 (1.891–3.095) | <.001 | 0.094 | 0.000 | 0.001 | 0.006 | 0.056 | 0.373 | |
| AC vs CC | Asian | 1.477 (1.190–1.833) | <.001 | 0.195 | 0.000 | 0.000 | 0.003 | 0.028 | 0.223 | |
| AC vs CC | PB | 1.477 (1.190–1.833) | <.001 | 0.195 | 0.000 | 0.000 | 0.003 | 0.028 | 0.223 | |
| AC+AA vs CC | Asian | 1.769 (1.444–2.167) | <.001 | 0.266 | 0.000 | 0.000 | 0.002 | 0.021 | 0.174 | |
| AC+AA vs CC | PB | 1.769 (1.444–2.167) | <.001 | 0.266 | 0.000 | 0.000 | 0.002 | 0.021 | 0.174 | |
| rs16969968 | A vs G | Overall | 1.333 (1.285–1.383) | <.001 | 1.000 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 |
| A vs G | Cacausian | 1.333 (1.285–1.384) | <.001 | 1.000 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 | |
| A vs G | HB | 1.261 (1.148–1.385) | <.001 | 1.000 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 | |
| A vs G | PB | 1.347 (1.294–1.401) | <.001 | 1.000 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 | |
| A vs G | Y (s) | 1.317 (1.258–1.378) | <.001 | 1.000 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 | |
| A vs G | mixed | 1.393 (1.302–1.491) | <.001 | 1.000 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 | |
| AA vs GG | Overall | 1.782 (1.649–1.926) | <.001 | 1.000 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 | |
| AA vs GG | Cacausian | 1.783 (1.649–1.927) | <.001 | 1.000 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 | |
| AA vs GG | HB | 1.555 (1.266–1.909) | <.001 | 0.253 | 0.000 | 0.000 | 0.002 | 0.022 | 0.181 | |
| AA vs GG | PB | 1.824 (1.677–1.985) | <.001 | 1.000 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 | |
| AA vs GG | Y (s) | 1.743 (1.583–1.920) | <.001 | 1.000 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 | |
| AA vs GG | mixed | 1.922 (1.665–2.218) | <.001 | 0.843 | 0.000 | 0.000 | 0.001 | 0.007 | 0.062 | |
| AG vs GG | Overall | 1.334 (1.262–1.410) | <.001 | 1.000 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 | |
| AG vs GG | Cacausian | 1.334 (1.262–1.411) | <.001 | 1.000 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 | |
| AG vs GG | HB | 1.327 (1.157–1.523) | <.001 | 0.890 | 0.000 | 0.000 | 0.001 | 0.006 | 0.059 | |
| AG vs GG | PB | 1.336 (1.257–1.419) | <.001 | 1.000 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 | |
| AG vs GG | Y (s) | 1.310 (1.224–1.402) | <.001 | 1.000 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 | |
| AG vs GG | mixed | 1.433 (1.291–1.590) | <.001 | 0.999 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 | |
| AG+AA vs GG | Overall | 1.426 (1.353–1.502) | <.001 | 1.000 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 | |
| AG+AA vs GG | Cacausian | 1.426 (1.353–1.504) | <.001 | 1.000 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 | |
| AG+AA vs GG | HB | 1.369 (1.202–1.561) | <.001 | 0.953 | 0.000 | 0.000 | 0.001 | 0.006 | 0.057 | |
| AG+AA vs GG | PB | 1.437 (1.357–1.521) | <.001 | 1.000 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 | |
| AG+AA vs GG | Y (s) | 1.398 (1.311–1.491) | <.001 | 1.000 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 | |
| AG+AA vs GG | mixed | 1.537 (1.392–1.696) | <.001 | 1.000 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 | |
| AA vs AG+GG | Overall | 1.514 (1.410–1.625) | <.001 | 1.000 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 | |
| AA vs AG+GG | Cacausian | 1.514 (1.410–1.626) | <.001 | 1.000 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 | |
| AA vs AG+GG | HB | 1.320 (1.092–1.595) | <.001 | 0.366 | 0.000 | 0.000 | 0.002 | 0.015 | 0.133 | |
| AA vs AG+GG | PB | 1.549 (1.435–1.672) | <.001 | 1.000 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 | |
| AA vs AG+GG | Y (s) | 1.503 (1.375–1.643) | <.001 | 1.000 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 | |
| AA vs AG+GG | mixed | 1.555 (1.368–1.767) | <.001 | 0.953 | 0.000 | 0.000 | 0.001 | 0.006 | 0.055 | |
| rs8034191 | C vs T | Overall | 1.273 (1.166–1.390) | <.001 | 1.000 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 |
| C vs T | Cacausian | 1.288 (1.243–1.334) | <.001 | 1.000 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 | |
| C vs T | HB | 1.294 (1.111–1.508) | .025 | 0.743 | 0.000 | 0.000 | 0.001 | 0.007 | 0.070 | |
| C vs T | PB | 1.309 (1.232–1.391) | <.001 | 1.000 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 | |
| B vs A | Y (s) | 1.347 (1.187–1.529) | <.001 | 0.958 | 0.000 | 0.000 | 0.001 | 0.006 | 0.055 | |
| B vs A | mixed | 1.208 (1.072–1.361) | .050 | 0.983 | 0.000 | 0.000 | 0.001 | 0.006 | 0.054 | |
| B vs A | Y | 1.254 (1.149–1.369) | <.001 | 1.000 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 | |
| BB vs AA | Overall | 1.529 (1.254–1.863) | <.001 | 0.302 | 0.000 | 0.000 | 0.002 | 0.018 | 0.156 | |
| BB vs AA | Cacausian | 1.683 (1.559–1.816) | <.001 | 1.000 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 | |
| BB vs AA | PB | 1.692 (1.482–1.931) | <.001 | 0.930 | 0.000 | 0.000 | 0.001 | 0.006 | 0.057 | |
| BB vs AA | Y (s) | 1.752 (1.296–2.369) | <.001 | 0.028 | 0.001 | 0.002 | 0.019 | 0.165 | 0.665 | |
| BB vs AA | Y | 1.546 (1.267–1.887) | <.001 | 0.290 | 0.000 | 0.000 | 0.002 | 0.019 | 0.162 | |
| BA vs AA | Overall | 1.271 (1.170–1.380) | <.001 | 1.000 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 | |
| BA vs AA | Cacausian | 1.257 (1.193–1.324) | <.001 | 1.000 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 | |
| BA vs AA | PB | 1.315 (1.204–1.437) | <.001 | 1.000 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 | |
| BA vs AA | Y (s) | 1.275 (1.150–1.414) | <.001 | 0.999 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 | |
| BA vs AA | mixed | 1.239 (1.110–1.383) | <.001 | 0.996 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 | |
| BA vs AA | Y | 1.236 (1.176–1.299) | <.001 | 1.000 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 | |
| BA+BB vs AA | Overall | 1.336 (1.215–1.469) | <.001 | 1.000 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 | |
| BA+BB vs AA | Cacausian | 1.344 (1.279–1.412) | <.001 | 1.000 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 | |
| BA+BB vs AA | PB | 1.391 (1.279–1.513) | <.001 | 1.000 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 | |
| BA+BB vs AA | Y (s) | 1.368 (1.241–1.508) | <.001 | 1.000 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 | |
| BA+BB vs AA | mixed | 1.274 (1.118–1.452) | <.001 | 0.938 | 0.000 | 0.000 | 0.001 | 0.006 | 0.056 | |
| BA+BB vs AA | Y | 1.313 (1.196–1.442) | <.001 | 1.000 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 | |
| BB vs BA+AA | Overall | 1.352 (1.139–1.605) | <.001 | 0.537 | 0.000 | 0.000 | 0.001 | 0.010 | 0.094 | |
| BB vs BA+AA | Cacausian | 1.475 (1.315–1.655) | <.001 | 0.991 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 | |
| BB vs BA+AA | HB | 1.389 (1.034–1.866) | <.001 | 0.032 | 0.001 | 0.002 | 0.017 | 0.148 | 0.635 | |
| BB vs BA+AA | PB | 1.466 (1.297–1.658) | <.001 | 0.972 | 0.000 | 0.000 | 0.001 | 0.006 | 0.054 | |
| BB vs BA+AA | Y (s) | 1.532 (1.150–2.042) | <.001 | 0.038 | 0.000 | 0.001 | 0.014 | 0.129 | 0.569 | |
| BB vs BA+AA | mixed | 1.344 (1.246–1.449) | <.001 | 1.000 | 0.000 | 0.000 | 0.001 | 0.006 | 0.053 | |
| BB vs BA+AA | Y | 1.363 (1.147–1.619) | <.001 | 0.530 | 0.000 | 0.000 | 0.001 | 0.010 | 0.095 | |
H-B = Hospital-based, P-B = Population-based, Y (s) = sample with from smoking.
3.2.5. CHRNA5 rs8034191
In the overall analyses of the rs8034191 polymorphism, we identified that this independent locus may be associated with risk for LC (P < .0001). When subgroup analysis was conducted based on ethnicity, source of control, smoking status, and HWE status, the ORs of the 5 models remained significant for both smoking and non-smoking patients (P < .0001). With respect to the stratification analysis by ethnicity, Caucasian groups were related to an elevated risk of LC in allelic (C vs T: OR = 1.288, 95%CI = 1.243–1.334, P < .0001, Fig. 5), homozygous (CC vs TT: OR = 1.683, 95%CI = 1.559–1.816, P < .0001), heterozygous (CT vs TT: OR = 1.257, 95%CI = 1.193–1.324, P < .0001), dominant (CT + CC vs TT: OR = 1.344, 95%CI = 1.279–1.412, P < .0001), and recessive models (CC vs CT + TT: OR = 1.482 95%CI = 1.381–1.591, P < .0001) when compared with Asian groups. In the stratified analysis of HWE status, the deviation of the rs8034191 genotype frequency may occur in the allelic (C vs T: OR = 1.796, 95%CI = 1.173–2.751, P < .05), homozygous (CC vs TT: OR = 0.710, 95%CI = 0.142–3.542, P < .05), dominant (CT + CC vs TT: OR = 1.953, 95%CI = 1.246–3.062, P < .05), and recessive models (CC vs CT + TT: OR = 0.690, 95%CI = 0.138–3.443, P < .05).
Figure 5.
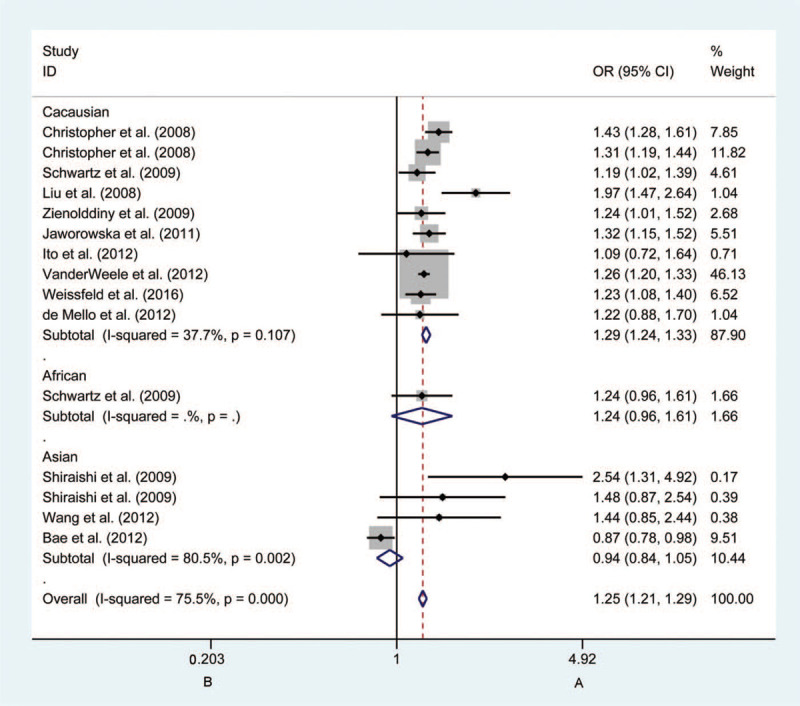
Forest plots of the association between CHRNA5 rs8034191 polymorphism and the risk of lung cancer in the Caucasian group (B vs A). Each square indicates a study, and the area of squares is proportional to the weight of the study. The diamond represents the summary odds ratio and 95% confidence interval. CI = confidence interval, OR = odds ratio.
3.3. Sensitivity analysis and publication bias
We repeated the meta-analysis and omitted each study one by one to examine the effects of all eligible studies. The results showed that there was no material alteration in the corresponding pooled ORs for CHRNA3 rs1051730, CHRNA3 rs6495309, CHRNA3 rs3743073, CHRNA5 rs8034191, or CHRNA5 rs16969968 polymorphisms (Supplemental Table 2 and Supplementary Figs. 1–5, http://links.lww.com/MD/F568). We further performed Egger tests that proposed a marked association between the 5 polymorphisms and LC risk. The results demonstrated no obvious publication bias for CHRNA3 rs6495309, CHRNA3 rs3743073, CHRNA5 rs8034191, or CHRNA5 rs16969968 polymorphisms (Supplemental Table 3 and Supplementary Figs. 6–10, http://links.lww.com/MD/F568). For CHRNA3 rs1051730, it was observed that a publication bias existed in the overall analysis (P > |t| = .008), hospital-based control (P > |t| = .012), and smoking status analysis (P > |t| = .002). After adjusting with the trim and fill method, the relative symmetrical figure appeared, indicating no publication bias for CHRNA3 rs1051730.
3.4. FPRP results
The FPRP values of significant results at different prior probability levels are summarized in Table 3. When the prior probability was set as 0.2, the association of rs1051730 SNP with an increasing risk of LC in overall, Caucasian, smoking groups were still noteworthy (FPRP <= 0.001 in the 5 comparisons), and the statistical power were more than 0.8. While for the association between rs6495309, rs3743073, and LC in the Asian group, we observed a lower statistical power of 0.461 and 0.195 in the heterozygous group, suggesting possible bias in the findings due to the limited reduced sample size of the Asian group, which requires further validation in larger studies. Positive associations (rs6495309, rs3743073) among the allelic, homozygous, and dominant comparisons were observed in the overall analysis, subgroups of Caucasian population, and ever-smokers significantly associated with LC risk, were considered noteworthy because their probability of being a false-positive was < 20%.
Rs6495309 and rs3743073 polymorphisms were associated with LC susceptibility in overall population, Caucasians, and people with ever-smokers in all genetic models excepting recessive model. In addition, the false positive probability of these results is less than 20%. For rs8034191polymorphism, we found that the risk effect of rs8034191 genotypes was increased in the subgroups of Caucasian population and ever-smokers from the allelic, homozygous, heterozygous, and dominant comparisons.
3.5. TSA analysis
Taken the data of the allelic model for the TSA analysis, the required information size for rs1051073 polymorphism was estimated as 29,018 (Supplementary Fig. 11, http://links.lww.com/MD/F568). The cumulative z-curve crossed the z = 1.96 and the trial-monitoring boundary with the required information size, confirming that the rs-1051073 polymorphism is significantly associated with increased LC risk among Caucasian and smoking populations. Similar results were also obtained for rs8034191 and rs16969968 (data not shown). As for the significant finding of the rs6495309 and rs3743073 polymorphisms in the Asian population, the heterozygous model was selected to perform the TSA. The cumulative z-curve crossed both the traditional threshold and the TSA threshold, indicating that although the cumulative amount of information did not meet the expected value, a positive conclusion might be reached in advance (Supplementary Figs. 12–13, http://links.lww.com/MD/F568).
4. Discussion
Lung cancer is the leading cause of cancer-related death worldwide, accounting for 13% of all cases and 23% of all cancer-related deaths globally.[24] As the etiology mechanism of LC is unknown, differences in LC morbidity exposed to the same risk, such as smoking and carcinogen exposure, have yet to be sufficiently illustrated.[25] Recently, scientists have worked to elaborate on the functional role of genetic factors, such as CHRNA5-CHRNA3-CHRNB4 polymorphisms, on LC susceptibility.[25] Alterations in the expression of nicotine receptor protein have been demonstrated in many studies. It was reported that CHRNA3 and CHRNA5 mRNA levels are regulated in lung adenocarcinoma[26] which may be one of the reasons for LC recurrence. CHRNA3 expression was downregulated whereas CHRNA5 expression was upregulated in tissues of lung adenocarcinoma compared with those in control lung tissue.[27]
In the current study, we found a strong association between the rs1051730 polymorphism and LC risk, consistent with previous studies by Ji et al and Gu et al.[28,29] Located on CHRNA3, SNP rs1051730 was reported to be related to diseases including LC and COPD, tobacco consumption through nicotine dependence, and exposure to a cytotoxic and genotoxic microenvironment.[30,31] The involved mechanisms include regulating cell apoptosis and increasing cellular proliferation. We further calculated the total OR and 95% CI in smoking and non-smoking patients, which yielded a significant difference in smokers (P < .0001) vs non-smokers (P > .05). It validates the presence of the polymorphism depending on nicotine self-administration among smoking patients.
Studies have shown that rs6495309, located in the CHRNA3 gene promoter region, inhibits gene transcription of CHRNA3 by affecting the binding ability of transcription factor Oct-1, thus promoting cell apoptosis and LC progression.[24] However, no contribution of the SNP rs6495309 to LC susceptibility was observed in the overall analysis. When the comparison was carried out among the heterozygous models, the Asian population showed elevated susceptibility to LC. While analyzing the FPRP for positive associations, we observed lower statistical power in the heterozygous group, suggesting possible bias in the findings due to the reduced sample size of the Asian group, thus it requires further validation in larger studies.
CHRNA rs3743073 has been investigated as a functional genetic variation site specific to Chinese individuals[19] and can be a prognostic indicator of non-small cell LC in the Chinese population.[31] This was verified in current studies. The rs16969968 SNP leads to a D to N substitution at position 398 of the CHRNA5 protein, which is a region highly conserved within species.[32] In a genome-wide association study, Sacconers et al initially demonstrated that rs16969968 in CHRNA5 is related to nicotine dependence,[33] and this finding was subsequently supported by other studies.[34,35] We found that a G to T substitution in rs16969968 of the CHRNA3 gene on chromosome 15q25 was significantly concerned with an increased risk of LC, regardless of the source of control, smoking status, or HWE status. The rs1051730 polymorphism is located in gene CHRNA3, and is in tight linkage disequilibrium with rs16969968. As it reportedly increases LC susceptibility and nicotine dependence, further studies of this polymorphism need to be conducted.[28]
Collectively, our data provided evidence that, although the nicotinic acetylcholine receptor may play a role in smoking behavior, the variation at 15q5.4 defined by rs8034191 directly contributes to LC susceptibility, and the LC risk was more strongly associated in the Caucasian population than in the Asian population. The stratified analysis provided evidence that the HWE status was an important factor for determining bias in the allelic, heterozygous, dominant, and recessive model results.
Our synthesis approach has shown some advantages. Firstly, a comprehensive analysis allows for larger sample size, enhancing the statistical validity, and reliability of the conclusions. Secondly, we performed various subgroup analyses based upon ethnicity, control sources, smoking status, and HWE status. It was done to provide heterogeneity of origin. In addition, the Bonferroni correction was adopted to adjust P values for a more precise estimation. Lastly, the FPRP and TSA were performed to evaluate the significant findings and validate statistical power. All these analyses help to minimize random errors and increase the robustness of conclusions.
Several limitations exist in the current work. First, some heterogeneity exists among studies because of the differences in ethnicities, sources of controls, smoking status, and HWE status. Second, only published studies were included in this meta-analysis, and publication bias may exist. Additionally, linkage disequilibrium is present in different CHRNA5-CHRNA3-CHRNB4 SNPs, and relevant haplotype analysis needs to be performed. Finally, as there are associations between genes and the environment, our findings should be applied to larger sample size studies with diverse covariates (including age, family history, environmental factors, lifestyle), as well as to further in-depth functional studies.
In conclusion, by analyzing and summarizing published studies, we are able to provide ideas and references for the relationships between the 5 SNPs in the CHRNA5-CHRNA3-CHRNB4 cluster and LC. Given the discordance in the subgroup, further studies with a larger sample size are still required.
Acknowledgments
The authors are thankful to the participants of the study.
Author contributions
Xingxu Yi, Wanzhen Li, Xueran Chen, and Fang Ye contributed to the conceptualization and design of the study, data collection, or analysis and interpretation of data. Xingxu Yi and Yiyuan Wang wrote the manuscript and were involved in the critical revision. Jingxian Chen, Gengyun Sun participated in the design of the research and approved the final version to be submitted.
Conceptualization: Xingxu Yi, Wanzhen Li.
Data curation: Xingxu Yi, Wanzhen Li.
Formal analysis: Xingxu Yi, Gengyun Sun, Jingxian Chen.
Investigation: Wanzhen Li, Yiyuan Wang.
Methodology: Wanzhen Li, Jingxian Chen.
Project administration: Xueran Chen.
Resources: Xueran Chen.
Software: Wanzhen Li, Yiyuan Wang.
Supervision: Gengyun Sun, Jingxian Chen.
Validation: Wanzhen Li, Fang Ye.
Visualization: Fang Ye.
Writing – original draft: Wanzhen Li.
Writing – review & editing: Gengyun Sun, Jingxian Chen.
Glossary
Abbreviations: CI = confidence interval, FPRP = false-positive report probability, HB = hospital-based, HWE = Hardy–Weinberg equilibrium, LC = lung cancer, nAChRs = nicotinic acetylcholine receptor subunits, OR = odds ratio, PB = population based, SCLC = small cell lung cancer, SNP = single nucleotide polymorphism, TSA = trial sequential analysis.
References
- [1].Herbst M. Fact Sheet on Lung Cancer. Cancer Asso South Africa 2015. [Google Scholar]
- [2].Obtel M, Nejjari C, Tachfouti N, et al. Estimating attributable fraction of lung cancer linked to smoking in Morocco. East Mediterr Health J 2016;21:871–7. [PubMed] [Google Scholar]
- [3].Bierut LJ, Stitzel JA, Wang JC, et al. Variants in nicotinic receptors and risk for nicotine dependence. Am J Psychiatry 2008;165:1163–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Le Calvez F, Mukeria A, Hunt JD, et al. TP53 and KRAS mutation load and types in lung cancers in relation to tobacco smoke: distinct patterns in never, former, and current smokers. Cancer Res 2005;65:5076–83. [DOI] [PubMed] [Google Scholar]
- [5].Catassi A, Servent D, Paleari L, et al. Multiple roles of nicotine on cell proliferation and inhibition of apoptosis: implications on lung carcinogenesis. Mutat Res 2008;659:221–31. [DOI] [PubMed] [Google Scholar]
- [6].Schuller HM, Orloff M. Tobacco-specific carcinogenic nitrosamines. Ligands for nicotinic acetylcholine receptors in human lung cancer cells. Biochem Pharmacol 1998;55:1377–84. [DOI] [PubMed] [Google Scholar]
- [7].Jull BA, Plummer HK, 3rd, Schuller HM. Nicotinic receptor-mediated activation by the tobacco-specific nitrosamine NNK of a Raf-1/MAP kinase pathway, resulting in phosphorylation of c-myc in human small cell lung carcinoma cells and pulmonary neuroendocrine cells. J Cancer Res Clin Oncol 2001;127:707–17. [DOI] [PubMed] [Google Scholar]
- [8].Jin Z, Gao F, Flagg T, et al. Nicotine induces multi-site phosphorylation of bad in association with suppression of apoptosis. J Biol Chem 2004;279:23837–44. [DOI] [PubMed] [Google Scholar]
- [9].Niu XM, Lu S. Acetylcholine receptor pathway in lung cancer: New twists to an old story. World J Clin Oncol 2014;5:667–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Pillai SG, Ge D, Zhu G, et al. A genome-wide association study in chronic obstructive pulmonary disease (COPD): identification of two major susceptibility loci. PLoS Genet 2009;5:e1000421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Zienolddiny S, Skaug V, Landvik NE, et al. The TERT-CLPTM1L lung cancer susceptibility variant associates with higher DNA adduct formation in the lung. Carcinogenesis 2009;30:1368–71. [DOI] [PubMed] [Google Scholar]
- [12].VanderWeele TJ, Asomaning K, Tchetgen Tchetgen EJ, et al. Genetic variants on 15q25.1, smoking, and lung cancer: an assessment of mediation and interaction. Am J Epidemiol 2012;175:1013–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Jaworowska E, Trubicka J, Lener MR, et al. Smoking related cancers and loci at chromosomes 15q25, 5p15, 6p22.1 and 6p21.33 in the Polish population. PLoS One 2011;6:e25057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Spitz MR, Amos CI, Dong Q, et al. The CHRNA5-A3 region on chromosome 15q24-25.1 is a risk factor both for nicotine dependence and for lung cancer. J Natl Cancer Inst 2008;100:1552–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Schwartz AG, Cote ML, Wenzlaff AS, et al. Racial differences in the association between SNPs on 15q25.1, smoking behavior, and risk of non-small cell lung cancer. J Thorac Oncol 2009;4:1195–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Islam MS, Ahmed MU, Sayeed MS, et al. Lung cancer risk in relation to nicotinic acetylcholine receptor, CYP2A6 and CYP1A1 genotypes in the Bangladeshi population. Clin Chim Acta 2013;416:11–9. [DOI] [PubMed] [Google Scholar]
- [17].Wang H, Zhao Y, Ma J, et al. The genetic variant rs401681C/T is associated with the risk of non-small cell lung cancer in a Chinese mainland population. Genet Mol Res 2013;12:67–73. [DOI] [PubMed] [Google Scholar]
- [18].Tekpli X, Landvik NE, Skaug V, et al. Functional effect of polymorphisms in 15q25 locus on CHRNA5 mRNA, bulky DNA adducts and TP53 mutations. Int J Cancer 2013;132:1811–20. [DOI] [PubMed] [Google Scholar]
- [19].Shen B, Shi M-Q, Zheng M-Q, et al. Correlation between polymorphisms of nicotine acetylcholine acceptor subunit CHRNA3 and lung cancer susceptibility. Mol Med Rep 2012;6:1389–92. [DOI] [PubMed] [Google Scholar]
- [20].Bonferroni C. Teoria Statistica Delle Classi e Calcolo Delle Probabilit?? Pubblicazioni del R Istituto Superiore di Scienze Economiche e Commerciali di Firenze 1935;8: [Google Scholar]
- [21].Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 1959;22:719–48. [PubMed] [Google Scholar]
- [22].Seagroatt V, Stratton I. Bias in meta-analysis detected by a simple, graphical test. Test had 10% false positive rate. BMJ (Clinical research ed ) 1998;316:470. [PMC free article] [PubMed] [Google Scholar]
- [23].Duval S, Tweedie R. Trim and fill: a simple funnel-plot-based method of testing and adjusting for publication bias in meta-analysis. Biometrics 2000;56:455–63. [DOI] [PubMed] [Google Scholar]
- [24].Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin 2011;61:69–90. [DOI] [PubMed] [Google Scholar]
- [25].Sun Y, Li J, Zheng C, et al. Study on polymorphisms in CHRNA5/CHRNA3/CHRNB4 gene cluster and the associated with the risk of non-small cell lung cancer. Oncotarget 2018;9:2435–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Zhang L, Zhang M, Wang H, et al. Comprehensive review of genetic association studies and meta-analysis on polymorphisms in microRNAs and urological neoplasms risk. Sci Rep 2018;8:3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Falvella FS, Galvan A, Frullanti E, et al. Transcription deregulation at the 15q25 locus in association with lung adenocarcinoma risk. Clin Cancer Res 2009;15:1837–42. [DOI] [PubMed] [Google Scholar]
- [28].Ji X, Bossé Y, Landi MT, et al. Identification of susceptibility pathways for the role of chromosome 15q25.1 in modifying lung cancer risk. Nat Commun 2018;9:3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Gu M, Dong X, Zhang X, et al. Strong association between two polymorphisms on 15q25.1 and lung cancer risk: a meta-analysis. PLoS One 2012;7:e37970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Paliwal A, Vaissière T, Krais A, et al. Aberrant DNA methylation links cancer susceptibility locus 15q25.1 to apoptotic regulation and lung cancer. Cancer Res 2010;70:2779–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Pérez Morales R, González Zamora A, González Delgado MF, et al. CHRNA3 rs1051730 and CHRNA5 rs16969968 polymorphisms are associated with heavy smoking, lung cancer, and chronic obstructive pulmonary disease in a Mexican population. Ann Hum Genet 2018;82:415–24. [DOI] [PubMed] [Google Scholar]
- [32].Lips EH, Gaborieau V, McKay JD, et al. Association between a 15q25 gene variant, smoking quantity and tobacco-related cancers among 17 000 individuals. Int J Epidemiol 2010;39:563–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Saccone SF, Hinrichs AL, Saccone NL, et al. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum Mol Genet 2007;16:36–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Hung RJ, McKay JD, Gaborieau V, et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature 2008;452:633–7. [DOI] [PubMed] [Google Scholar]
- [35].Xu ZW, Wang GN, Dong ZZ, et al. CHRNA5 rs16969968 polymorphism association with risk of lung cancer--evidence from 17,962 lung cancer cases and 77,216 control subjects. Asian Pac J Cancer Prev 2015;16:6685–90. [DOI] [PubMed] [Google Scholar]


