Abstract
With growing evidence on the therapeutic efficacy and safety of herbal drugs, there has been a substantial increase in their application in the lung cancer treatment. Meanwhile, their action mechanisms at the system level have not been comprehensively uncovered. To this end, we employed a network pharmacology methodology to elucidate the systematic action mechanisms of FDY2004, an anticancer herbal drug composed of Moutan Radicis Cortex, Persicae Semen, and Rhei Radix et Rhizoma, in lung cancer treatment. By evaluating the pharmacokinetic properties of the chemical compounds present in FDY2004 using herbal medicine-associated databases, we identified its 29 active chemical components interacting with 141 lung cancer-associated therapeutic targets in humans. The functional enrichment analysis of the lung cancer-related targets of FDY2004 revealed the enriched Gene Ontology terms, involving the regulation of cell proliferation and growth, cell survival and death, and oxidative stress responses. Moreover, we identified key FDY2004-targeted oncogenic and tumor-suppressive pathways associated with lung cancer, including the phosphatidylinositol 3-kinase-Akt, mitogen-activated protein kinase, tumor necrosis factor, Ras, focal adhesion, and hypoxia-inducible factor-1 signaling pathways. Overall, our study provides novel evidence and basis for research on the comprehensive anticancer mechanisms of herbal medicines in lung cancer treatment.
1. Introduction
Despite the advances in anticancer therapies, lung cancer (LC) remains the most common reason for cancer mortality, which accounts for 18.4% of global cancer deaths [1]. Accumulating evidence and increasing understanding of LC pathology have led to the development of effective anticancer therapies such as chemotherapy, targeted therapy, and cancer immunotherapy that can prolong the survival of patients with LC; however, these therapies may frequently and inevitably accompany therapeutic resistance and toxic adverse effects [2, 3]. Therefore, there has been a substantial increase in the application of herbal drugs in cancer treatment owing to their potent anticancer effects and less toxicities [4–6]. It has been shown that the administration of herbal drugs can enhance the efficacy and attenuate the adverse effects of anticancer therapies, alleviate cancer symptoms, and improve the survival and clinical outcomes of patients with cancer [6–8].
FDY2004 is a herbal drug composed of three herbal medicines, namely, Moutan Radicis Cortex (MRC), Persicae Semen (PS), and Rhei Radix et Rhizoma (RRR) (Supplementary Table S1) [9]. This herbal drug may exert potent antiproliferative effect against LC cells (Supplementary Table S1) [9]; however, its system-level anticancer mechanisms in LC treatment remain to be elucidated.
With advances in scientific and analytical technologies, various convergence research methodologies such as network pharmacology have emerged [5, 10–12]. They have been used to investigate complex multiple compound-multiple target pharmacological mechanisms of herbal drugs [5, 10–12]. Network pharmacology is used to explore the mechanisms underlying various diseases and action mechanisms of therapeutic herbal drugs [5, 10–12]. It involves the interactions among biological elements, such as genes, proteins, and metabolites, and integrates pharmacology, medicine, and network biology [5, 10–12]. This research strategy has been proven beneficial in understanding the multiple compound-multiple target mechanisms of herbal drugs via the following: (i) investigation of their active chemical components and disease-associated therapeutic targets and (ii) analysis of their therapeutic functions to uncover the polypharmacological mechanisms of herbal medicines [5, 10–12]. By employing a network pharmacology methodology, we uncovered the anti-LC mechanisms of FDY2004.
2. Materials and Methods
2.1. Investigation of the Active Chemical Components of FDY2004
We retrieved the chemical components of FDY2004 from the Traditional Chinese Medicine Systems Pharmacology (TCMSP) and Anticancer Herbs Database of Systems Pharmacology (CancerHSP) databases [13, 14]. We then used the pharmacokinetic characteristics obtained from the aforementioned databases [13, 14], including oral bioavailability, Caco-2 permeability, and drug-likeness, to determine the active chemical components of FDY2004. Oral bioavailability is a measure of an orally administered drug's capacity to be transported to the general circulation and sites of drug action [13, 15]. Chemical compounds whose oral bioavailability is higher than 30% are regarded to possess effective absorption abilities [13, 15]. Caco-2 permeability is an indicator of the diffusion ability of a chemical compound in the intestinal epithelium, assessed using Caco-2 human intestinal cells [13]. Compounds with a Caco-2 permeability of ≥−0.4 are considered to have effective permeability in the intestines [16, 17]. Drug-likeness is an index used to investigate the druggability of a chemical component with respect to its biochemical and physical properties using Tanimoto coefficients [13, 18]. The average drug-likeness of available drugs is 0.18; therefore, chemical compounds whose drug-likeness is higher than this average value are regarded to have druggable potential in network pharmacology analysis [13, 18]. Consequently, in this study, chemical components that meet the following criteria were determined to be bioactive: oral bioavailability ≥ 30%, drug-likeness ≥ 0.18, and Caco-2 permeability ≥ −0.4 [11, 13].
2.2. Identification of the Targets of Active Chemical Components
We retrieved the simplified molecular-input line-entry system (SMILES) notation for individual chemical components from the PubChem database [19]. By importing the SMILES information into diverse in silico tools, involving the SwissTargetPrediction [20], Search Tool for Interactions of Chemicals 5 [21], PharmMapper [22], and Similarity Ensemble Approach [23], the human targets of FDY2004 were obtained. The LC-associated human targets were searched using “Lung Neoplasms” (Medical Subject Headings Unique ID: D008175) as a keyword in the following comprehensive genomic databases: Comparative Toxicogenomics Database [24], Therapeutic Target Database [25], Human Genome Epidemiology Navigator [26], GeneCards [27], DisGeNET [28], DrugBank [29], Online Mendelian Inheritance in Man [30], and Pharmacogenomics Knowledge Base [31].
2.3. Construction of Herbal Drug-Associated Networks
Herbal medicine-active chemical component (H-C), active chemical component-target (C-T), and target-pathway (T-P) interaction networks were built by connecting the herbal components of FDY2004 with their active chemical components, the components with their interacting targets, and the targets with their relevant enriched pathways, respectively. A protein-protein interaction (PPI) network was built based on the interaction data for the targets obtained from the STRING database (interaction confidence score ≥ 0.7) [32]. Network visualization and analysis were conducted using Cytoscape [33]. In the network pharmacology analysis, individual constituents relevant to a herbal drug, including its herbal medicines, chemical components, targets, and pathways, are represented as nodes [34]. Their interactions are represented as links (or edges) [34]. The degree of nodes is defined as the number of their links, and nodes with a relatively high degree are called hubs [34].
2.4. Survival Analysis
The correlation between the expression status of FDY2004 targets and the survival of patients with LC was analyzed using Kaplan–Meier plotter [35].
2.5. Functional Enrichment Analysis
Functional enrichment of Gene Ontology (GO) terms and pathways for “Homo sapiens” by FDY2004 targets was analyzed using g:Profiler [36] and Kyoto Encyclopedia of Genes and Genomes [37].
2.6. Molecular Docking Analysis
We obtained the structures of the chemical components and their targets from PubChem [19] and RCSB Protein Data Bank [38], respectively. Then, we imported the collected structural information into Autodock Vina [39] and analyzed the docking scores of individual chemical component-target pairs. As reported previously, we considered that a chemical component-target pair might have a high binding affinity if it has a docking score of less than −5.0 [40, 41].
3. Results
3.1. Active Chemical Components of FDY2004
From the TCMSP and CancerHSP [13, 14], we obtained detailed information on the chemical components of FDY2004 (Supplementary Table S2). The chemical components that satisfied the following criteria were considered bioactive: oral bioavailability ≥ 30%, drug-likeness ≥ 0.18, and Caco-2 permeability ≥ −0.4 [11, 13]. We also considered numerous compounds as active compounds because of their high amounts and potent activity, although they did not meet the aforementioned criteria [42–56]. Thus, 35 bioactive chemical compounds of FDY2004 were identified (Supplementary Table S3).
3.2. Targets of the Active Chemical Components of FDY2004
We obtained 212 human molecular targets for the 29 bioactive chemical components of FDY2004 (see Materials and Methods) (Supplementary Table S4). The information on the LC-associated human genes and proteins was retrieved from various genomic databases (see Materials and Methods), and 141 of all the 212 FDY2004 targets were considered LC-related targets.
3.3. Network Pharmacological Identification of the Action Mechanisms of FDY2004
By integrating the comprehensive data of FDY2004, including its herbal and chemical components and their LC-related targets, we built a herbal medicine-active chemical component-target (H-C-T) network representing the polypharmacological features of the herbal drug (Figure 1). The network consisted of 173 nodes (3 herbal medicines, 29 bioactive chemical components, and 141 targets) and 304 links (Figure 1 and Supplementary Table S4). In this network, quercetin, kaempferol, gallic acid, emodin, campesterol, (+)-epicatechin, (+)-catechin, and (−)-catechin had the highest number of interacting targets (Figure 1), demonstrating that they may be the key pharmacological compounds underlying the anti-LC effects of FDY2004. It is noteworthy that 96 of the 141 LC-related genes/proteins were common therapeutic targets of two or more bioactive chemical components of FDY2004 (Figure 1), implying the polypharmacological and coordinated action mechanisms of FDY2004.
Figure 1.

The herbal medicine-active chemical component-target network of FDY2004. Green hexagon nodes, herbal medicines; red rectangle nodes, active chemical compounds; blue eclipse nodes, lung cancer-related targets.
To understand the interactions among the LC-related targets of FDY2004, a PPI network with 114 nodes and 304 edges was generated, where the targets served as nodes and their interactions represented edges (Figure 2). We then searched for nodes with a relatively high degree (i.e., hubs) [57, 58]. They are reported to have key roles in the pharmacological activities of drugs and serve as potential therapeutic targets [57, 58]. As reported previously, hubs were defined as nodes with a degree higher than or equal to twice the average degree of all nodes in a PPI network [11]. The nodes TP53, PIK3R1, HSP90AA1, AKT1, VEGFA, EGFR, JUN, PTK2, TNF, ESR1, NFKB1, and RAC1 were identified as hubs with high degree (Figure 2), demonstrating that these targets may be important for the exertion of anti-LC pharmacological effects of FDY2004. The expression status of these hub targets was further shown to be significantly related to the survival of patients with LC (Figure 3), implying their potential clinical significance and prognostic role.
Figure 2.

The protein-protein interaction network for lung cancer-related targets of FDY2004. Pink nodes, hub targets.
Figure 3.

Survival analysis of lung cancer-related hub targets of FDY2004. Kaplan–Meier curves for overall survival of the patients with lung cancer with respect to the expression of the indicated targets.
3.4. Functional Enrichment Analysis of FDY2004-Associated Targets and Pathways
To explore the molecular mechanisms of FDY004 in LC treatment based on the biological functions of its targets, we carried out the GO enrichment analysis. The GO terms involved in the various biological functions, including cell proliferation and growth, cell survival and death, and oxidative stress responses, were enriched by the LC-related targets of FDY2004 (Supplementary Figure S1), indicating the anticancer molecular characteristics of its pharmacological activity.
To investigate the pathway-level pharmacological properties of FDY2004 against LC, we conducted the pathway enrichment analysis (Figure 4 and Supplementary Figure S1). The following signaling pathways were found to be enriched by the LC-associated targets of FDY2004: “Pathways in cancer,” “PI3K-Akt signaling pathway,” “MAPK signaling pathway,” “TNF signaling pathway,” “Ras signaling pathway,” “Apoptosis,” “Focal adhesion,” “HIF-1 signaling pathway,” “Cellular senescence,” “EGFR tyrosine kinase inhibitor resistance,” “Estrogen signaling pathway,” “PD-L1 expression and PD-1 checkpoint pathway in cancer,” “Small cell lung cancer,” “Non-small cell lung cancer,” “Platinum drug resistance,” “ErbB signaling pathway,” “p53 signaling pathway,” and “VEGF signaling pathway” (Figure 4 and Supplementary Figure S1).
Figure 4.
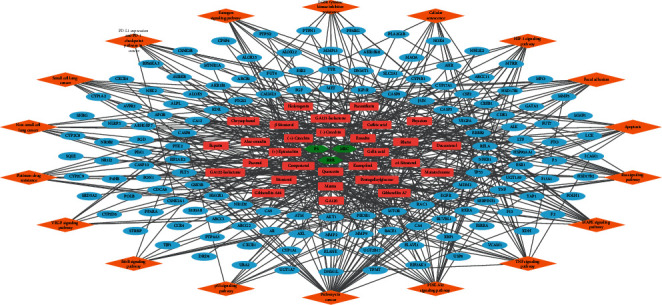
The herb-compound-target-pathway network of FDY2004. Green hexagon nodes, herbal medicines; red rectangle nodes, active chemical compounds; blue eclipse nodes, lung cancer-related targets; orange diamond nodes, signaling pathways.
Together, the results suggest the system-level mechanisms of FDY2004 against LC from the molecular- and pathway perspectives.
3.5. Molecular Docking of the FDY2004 Targets
To investigate the binding activities of compound-target interactions for FDY2004, we analyzed their molecular docking affinities (see Materials and Methods). In the docking analysis, 95.19% of the active compound-target pairs presented docking scores of ≤−5.0, implying the potential pharmacological binding activities of the herbal drug (Figure 5 and Supplementary Table S5).
Figure 5.
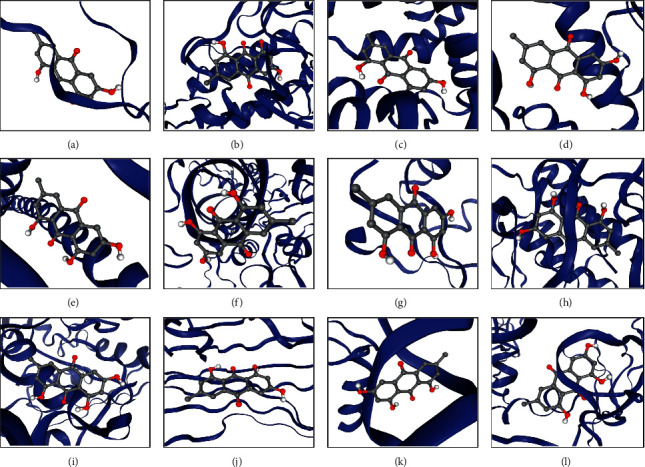
Molecular docking analysis for the active chemical compounds of FDY2004 and the targets. The analysis results of quercetin and the hub targets are shown as representatives. (a) Quercetin-AKT1 (score = −6.4). (b) Quercetin-EGFR (score = −7.9). (c) Quercetin-ESR1 (score = −6.5). (d) Quercetin-HSP90AA1 (score = −6.1). (e) Quercetin-JUN (score = −5.4). (f) Quercetin-NFKB1 (score = −6.6). (g) Quercetin-PI3KR1 (score = −6.3). (h) Quercetin-PTK2 (score = −6.1). (i) Quercetin-RAC1 (score = −9.3). (j) Quercetin-TNF (score = −6.5). (k) Quercetin-TP53 (score = −7.0). (l) Quercetin-VEGFA (score = −7.2).
4. Discussion
Although there has been increasing use of herbal drugs in LC treatment, their system-level anticancer mechanisms have not been comprehensively understood. Here, we employed a network pharmacological approach to uncover the therapeutic mechanisms of FDY2004 [9] in LC treatment from a system-level view. The network pharmacological investigation of FDY2004 revealed 29 active chemical components that interact with 141 lung cancer-associated therapeutic targets, mediating the anti-LC effects of the herbal drug. The GO enrichment analysis of the FDY2004 targets revealed the molecular action mechanisms of FDY2004, involving the regulation of cell proliferation and growth, cell survival and death, and oxidative stress responses. Furthermore, the key FDY2004-targeted oncogenic and tumor-suppressive pathways implicated in LC development and progression were the phosphatidylinositol 3-kinase (PI3K)-Akt, mitogen-activated protein kinase (MAPK), tumor necrosis factor (TNF), Ras, focal adhesion, and hypoxia-inducible factor (HIF)-1 signaling pathways.
The LC-related hub targets of FDY2004 were found to be closely associated with LC pathology and play a role as prognostic indicators for the survival and therapeutic sensitivity of patients with LC. The tumor suppressor TP53 is one of the most frequently mutated and malfunctioned genes in the pathological process of LC, and its genetic and functional status may serve as a predictor for the risk, survival, and therapeutic outcomes of LC [59–63]. PIK3R1 is involved in the regulation of LC cell growth [64]. The upregulation of HSP90AA1 correlates with the occurrence, progression, and clinical outcomes of LC, and its inhibition can repress the proliferation, survival, and metastasis of LC cells [65, 66]. The abnormal regulation of Akt1 (encoded by AKT1) and TNF-α (encoded by TNF) may enhance the growth, survival, proliferation, metastasis, epithelial-to-mesenchymal transition (EMT), and stemness capacity of LC cells, and they are potential targets that can alleviate chemotherapy and radiotherapy resistance [67–76]. Clinical studies have also reported that AKT1 and TNF may be prognostic determinants for patients' survival and treatment outcomes with LC [73, 77–80]. Vascular endothelial growth factor (VEGF)-A (encoded by VEGFA) enhances the metastasis and angiogenesis of LC cells and thereby contribute to the progression of LC, and its activation profile is related to a poor clinical prognosis and the survival of patients with LC [81–86]. Dysregulated expression of EGFR and its encoded receptor tyrosine kinase activity may lead to the induction of various cancerous cellular processes underlying the LC pathology, making it a key target of widely used antitumor agents against LC in clinical settings [87–89]. c-Jun (encoded by JUN) functions as a modulator of the growth, proliferation, and apoptosis of LC cells as well as a mediator of the pharmacological effects of cytotoxic drugs [90–92]. Pharmacological modulation of focal adhesion kinase (FAK; encoded by PTK2) and Ras-related C3 botulinum toxin substrate 1 (RAC1; encoded by RAC1) reduces the proliferation, migration, invasion, EMT, motility, angiogenesis, and stemness activity of LC cells, and this reverses chemotherapy and radiotherapy resistance [93–103]. The expression of estrogen receptor (ER)-α (encoded by ESR1) might be correlated with the survival and prognosis of patients with LC, and previous studies have reported its role as a therapeutic target in LC treatment [104, 105]. The polymorphisms of NFKB1 are associated with the risk of LC occurrence [106].
The signaling pathways targeted by FDY2004 are known to function as crucial regulators of LC development and progression, mediate treatment resistance to anticancer therapies, and play a role as therapeutic targets. The PI3K-Akt, MAPK, Ras, focal adhesion, HIF-1, and erythroblastic leukemia viral oncogene homolog (ErbB) signaling pathways coordinate diverse tumorigenic processes of cancer cells, involving cell proliferation and growth, survival and cell death, anoikis resistance, metastasis, EMT, self-renewal potential and stemness properties, and angiogenesis, of LC cells [101, 102, 107–125]. In addition, aberrant regulations of these signaling pathways may contribute to therapeutic resistance, which can be overcome by genetic and pharmacological interventions of their activities [101, 102, 107–125]. The TNF signaling pathway is a key inflammation mediator involved in the development, progression, metastasis, and recurrence of LC, and the pathway constituents have prognostic significance in the clinical outcome of patients with LC [74–76, 126, 127]. The estrogen pathway and its components may possess carcinogenic properties in LC and act as potential targets [128–131]. The programmed cell death protein 1 (PD-1)/programmed death-ligand 1 (PD-L1) pathway is involved in the regulation of tumor-related immune processes, and it is a key target of cancer immunotherapy, which attempts to suppress immune escape and enhance antitumor immunity for the durable regression of malignant tumors of LC [132–134]. The dysfunction of genes and proteins comprising the p53 pathway, one of the common carcinogenic causes, is associated with various cancerous behaviors of LC cells, such as uncontrolled proliferation, survival, and cell cycle progression [61, 63, 135–143]. The genetic and functional activities of the pathway components might be correlated with the survival and anticancer therapeutic sensitivity of patients with LC [61, 63, 135–137, 139, 141–143]. The VEGF pathway may induce the progression of LC tumors by activating malignant angiogenic, metastatistic, and proliferative programs of cancer cells, and it is the primary pharmacological target of antiangiogenic anticancer drugs [144–146]. Defects in the regulation of important cellular phenotypes such as apoptosis and cellular senescence are the major drivers of the development and progression of LC, and their proper regulation is the key mechanism of anticancer therapeutics [147–152]. Resistance to platinum-based chemotherapeutics and epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors is mediated by diverse oncogenic signaling mechanisms, and co-targeting the resistance-associated pathways may enhance the efficacy of LC treatment [153–157].
The active chemical components of FDY2004 have been reported to act as anticancer compounds in LC. Aloe-emodin induces DNA damage, autophagy, and death of LC cells by regulating reactive oxygen species (ROS) generation and signaling activities of the PI3K/Akt/mammalian target of rapamycin, MAPK, protein kinase C (PKC), and caspase pathways [158–162]. It also functions as a photosensitizer that enhances irradiation-induced anoikis in LC cells [158–162]. Caffeic acid has been shown to improve the cytotoxicity of chemotherapeutics in LC cells [163]. Catechins may suppress the growth and promote cell cycle arrest of LC cells by inactivating proliferation-inducing oncogenic kinases and cell cycle regulators [164, 165]. Chrysophanol regulates the activation of oxidative stress responses and relevant signaling pathways to reduce the proliferation, migration, invasion, and survival potential of LC cells [166, 167]. Daucosterol disturbs redox homeostasis and cell cycle processes to elicit growth arrest and death of LC cells [168, 169]. Emodin inhibits cell proliferation and migration and promotes EMT, autophagic cell death, and cell cycle arrest coordinated by chemokine, endoplasmic reticulum (ER) stress, ROS, p53, cell cycle, nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB), tribbles pseudokinase 3, and PKC signaling; it enhances the efficacy of anticancer drugs [159, 170–180]. The proapoptotic and chemosensitizing effects of gallic acid are mediated by the EGFR, PD-L1, ROS, NF-κB, caspase, janus kinase (JAK)-signal transducer and activator (STAT), and mitochondrial pathways [181–188]. Hederagenin exerts cytotoxic effects and further synergizes with chemotherapeutic agents in LC cells [189]. Kaempferol may block the growth, survival, EMT, and migration of LC cells and enhance anti-LC therapies [190–192]. Previous studies have reported the anticancer roles of mairin (betulinic acid) in inducing apoptosis, suppressing proliferation, and reversing drug resistance of LC cells [193–195]. The antiproliferative, antimetastatic, and cell cycle arrest activities of paeoniflorin are mediated by the modulation of the FAS pathways and macrophage activation [196, 197]. Paeonol represses the proliferation and bone metastasis of LC cells and also serves as a radiosensitizer by inhibiting the PI3K/Akt pathway to enhance their apoptosis [198, 199]. Physcion increases the pharmacological sensitivity of LC cells to cytotoxic drugs [200]. Rhein induces apoptosis while suppressing the proliferation of LC cells mediated by the modulation of the calcium, ER stress, and STAT3 pathways [201, 202]. Previous studies have reported the inhibitory roles of quercetin on the growth, survival, metastasis, and chemotherapy and radiotherapy resistance of LC cells via cancer pathways such as Akt, MAPKs, NF-κB, inflammation, and apoptotic caspase signaling [203–209]. β-Sitosterol inhibits cancerous autophagic, proliferative, survival, and cell cycle regulatory processes in LC cells [169, 210, 211]. These observations support the pharmacological mechanisms underlying the anti-LC effects of FDY2004.
Overall, our study presents novel and comprehensive insights into and evidence of the anti-LC effects of FDY2004. Further preclinical and clinical studies are warranted to investigate the action mechanisms of FDY2004 and evaluate the pharmacological effects of its combinatorial use with standard anticancer strategies such as chemotherapy, targeted therapy, cancer immunotherapy, and radiotherapy.
Data Availability
The data used to support the findings of this study are included within the article and Supplementary Materials file.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Supplementary Materials
Supplementary Figure S1: functional enrichment analysis for the lung cancer-related targets of FDY2004. Supplementary Table S1: general information and reports on evidence of biological activities of FDY2004 and its herbal constituents. Supplementary Table S2: chemical components of FDY2004. Supplementary Table S3: active chemical components of FDY2004. Supplementary Table S4: Targets of active chemical components of FDY2004. Supplementary Table S5: docking scores between active chemical components of FDY2004 and the lung cancer-associated targets.
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R. L., Torre L. A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer Journal for Clinicians. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Ishikawa A., Ohara G., Nakazawa K., et al. Chemotherapy-induced complications in patients with lung cancer: an evaluation by pharmacists. Molecular and Clinical Oncology. 2013;1(1):65–68. doi: 10.3892/mco.2012.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ricciardi S., Tomao S., de Marinis F. Toxicity of targeted therapy in non-small-cell lung cancer management. Clinical Lung Cancer. 2009;10(1):28–35. doi: 10.3816/clc.2009.n.004. [DOI] [PubMed] [Google Scholar]
- 4.Ohnishi S., Takeda H. Herbal medicines for the treatment of cancer chemotherapy-induced side effects. Frontiers in Pharmacology. 2015;6:p. 14. doi: 10.3389/fphar.2015.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poornima P., Kumar J. D., Zhao Q., Blunder M., Efferth T. Network pharmacology of cancer: from understanding of complex interactomes to the design of multi-target specific therapeutics from nature. Pharmacological Research. 2016;111:290–302. doi: 10.1016/j.phrs.2016.06.018. [DOI] [PubMed] [Google Scholar]
- 6.Yin S. Y., Wei W. C., Jian F. Y., Yang N. S. Therapeutic applications of herbal medicines for cancer patients. Evidence-Based Complementary and Alternative Medicine. 2013;2013:15. doi: 10.1155/2013/302426.302426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li S. G., Chen H. Y., Ou-Yang C. S., et al. The efficacy of Chinese herbal medicine as an adjunctive therapy for advanced non-small cell lung cancer: a systematic review and meta-analysis. PLoS One. 2013;8(2) doi: 10.1371/journal.pone.0057604.e57604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang X.-W., Liu W., Jiang H.-L., Mao B. Chinese herbal medicine for advanced non-small-cell lung cancer: a systematic review and meta-analysis. The American Journal of Chinese Medicine. 2018;46(5):923–952. doi: 10.1142/s0192415x18500490. [DOI] [PubMed] [Google Scholar]
- 9.Lee I.-H., Lee H.-S., Kang K., et al. Influence of decoction duration of FDY2004 on its physicochemical components and antioxidant and antiproliferative activities. Natural Product Communications. 2020;15(10) doi: 10.1177/1934578x20968437.1934578X2096843 [DOI] [Google Scholar]
- 10.Lee W. Y., Lee C. Y., Kim Y. S., Kim C. E. The methodological trends of traditional herbal medicine employing network pharmacology. Biomolecules. 2019;9(8) doi: 10.3390/biom9080362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee H. S., Lee I. H., Park S. I., Lee D. Y. Network pharmacology-based investigation of the system-level molecular mechanisms of the hematopoietic activity of Samul-Tang, a traditional Korean herbal formula. Evidence-based Complementary and Alternative Medicine. 2020;2020:17. doi: 10.1155/2020/9048089.9048089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang S. Q., Xu H. B., Zhang S. J., Li X. Y. Identification of the active compounds and significant pathways of artemisia annua in the treatment of non-small cell lung carcinoma based on network pharmacology. Medical Science Monitor. 2020;26 doi: 10.12659/MSM.923624.e923624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ru J., Li P., Wang J., et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. Journal of Cheminformatics. 2014;6(1):p. 13. doi: 10.1186/1758-2946-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tao W., Li B., Gao S., et al. CancerHSP: anticancer herbs database of systems pharmacology. Scientific Reports. 2015;5(1):p. 11481. doi: 10.1038/srep11481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang C. K., Craik D. J. Cyclic peptide oral bioavailability: lessons from the past. Biopolymers. 2016;106(6):901–909. doi: 10.1002/bip.22878. [DOI] [PubMed] [Google Scholar]
- 16.Li Y., Zhang J., Zhang L., et al. Systems pharmacology to decipher the combinational anti-migraine effects of Tianshu formula. Journal of Ethnopharmacology. 2015;174:45–56. doi: 10.1016/j.jep.2015.07.043. [DOI] [PubMed] [Google Scholar]
- 17.Zhang J., Li Y., Chen X., Pan Y., Zhang S., Wang Y. Systems pharmacology dissection of multi-scale mechanisms of action for herbal medicines in stroke treatment and prevention. PLoS One. 2014;9(8) doi: 10.1371/journal.pone.0102506.e102506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee A. Y., Park W., Kang T.-W., Cha M. H., Chun J. M. Network pharmacology-based prediction of active compounds and molecular targets in Yijin-Tang acting on hyperlipidaemia and atherosclerosis. Journal of Ethnopharmacology. 2018;221:151–159. doi: 10.1016/j.jep.2018.04.027. [DOI] [PubMed] [Google Scholar]
- 19.Kim S., Chen J., Cheng T., et al. PubChem 2019 update: improved access to chemical data. Nucleic Acids Research. 2019;47(D1):D1102–D1109. doi: 10.1093/nar/gky1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Daina A., Michielin O., Zoete V. Swiss target prediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Research. 2019;47(1):W357–W364. doi: 10.1093/nar/gkz382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Szklarczyk D., Santos A., von Mering C., Jensen L. J., Bork P., Kuhn M. Stitch 5: augmenting protein-chemical interaction networks with tissue and affinity data. Nucleic Acids Research. 2016;44(1):D380–D384. doi: 10.1093/nar/gkv1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang X., Shen Y., Wang S., et al. PharmMapper 2017 update: a web server for potential drug target identification with a comprehensive target pharmacophore database. Nucleic Acids Research. 2017;45(1):W356–W360. doi: 10.1093/nar/gkx374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keiser M. J., Roth B. L., Armbruster B. N., Ernsberger P., Irwin J. J., Shoichet B. K. Relating protein pharmacology by ligand chemistry. Nature Biotechnology. 2007;25(2):197–206. doi: 10.1038/nbt1284. [DOI] [PubMed] [Google Scholar]
- 24.Davis A. P., Grondin C. J., Johnson R. J., et al. The comparative toxicogenomics database: update 2019. Nucleic Acids Research. 2019;47(1):D948–D954. doi: 10.1093/nar/gky868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu F., Han B., Kumar P., et al. Update of TTD: therapeutic target database. Nucleic Acids Research. 2010;38(1):D787–D791. doi: 10.1093/nar/gkp1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu W., Gwinn M., Clyne M., Yesupriya A., Khoury M. J. A navigator for human genome epidemiology. Nature Genetics. 2008;40(2):124–125. doi: 10.1038/ng0208-124. [DOI] [PubMed] [Google Scholar]
- 27.Safran M., Dalah I., Alexander J., et al. GeneCards version 3: the human gene integrator. Database. 2010;2010 doi: 10.1093/database/baq020.baq020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Piñero J., Bravo À., Queralt-Rosinach N., et al. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Research. 2017;45(1):D833–D839. doi: 10.1093/nar/gkw943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wishart D. S., Feunang Y. D., Guo A. C., et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Research. 2018;46(1):D1074–D1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Amberger J. S., Bocchini C. A., Schiettecatte F., Scott A. F., Hamosh A. OMIM.org: online mendelian inheritance in man (OMIM), an online catalog of human genes and genetic disorders. Nucleic Acids Research. 2015;43(1):D789–D798. doi: 10.1093/nar/gku1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Whirl-Carrillo M., McDonagh E. M., Hebert J. M., et al. Pharmacogenomics knowledge for personalized medicine. Clinical Pharmacology & Therapeutics. 2012;92(4):414–417. doi: 10.1038/clpt.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Szklarczyk D., Gable A. L., Lyon D., et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Research. 2019;47(1):D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shannon P., Markiel A., Ozier O., et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Research. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barabási A.-L., Oltvai Z. N. Network biology: understanding the cell’s functional organization. Nature Reviews Genetics. 2004;5(2):101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 35.Nagy A., Lanczky A., Menyhart O., Gyorffy B. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Scientific Reports. 2018;8(1):p. 9227. doi: 10.1038/s41598-018-29514-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raudvere U., Kolberg L., Kuzmin I., et al. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update) Nucleic Acids Research. 2019;47(1):W191–W198. doi: 10.1093/nar/gkz369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Research. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Burley S. K., Berman H. M., Bhikadiya C., et al. RCSB protein data bank: biological macromolecular structures enabling research and education in fundamental biology, biomedicine, biotechnology and energy. Nucleic Acids Research. 2019;47(1):D464–D474. doi: 10.1093/nar/gky1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trott O., Olson A. J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhuang Z., Wen J., Zhang L., et al. Can network pharmacology identify the anti-virus and anti- inflammatory activities of Shuanghuanglian oral liquid used in Chinese medicine for respiratory tract infection? European Journal of Integrative Medicine. 2020;37:p. 101139. doi: 10.1016/j.eujim.2020.101139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang M., Yuan Y., Zhou W., et al. Network pharmacology analysis of Chaihu Lizhong Tang treating non-alcoholic fatty liver disease. Computational Biology and Chemistry. 2020;86:p. 107248. doi: 10.1016/j.compbiolchem.2020.107248. [DOI] [PubMed] [Google Scholar]
- 42.Behrendt P., Perin P., Menzel N., et al. Pentagalloylglucose, a highly bioavailable polyphenolic compound present in cortex moutan, efficiently blocks hepatitis C virus entry. Antiviral Research. 2017;147:19–28. doi: 10.1016/j.antiviral.2017.09.006. [DOI] [PubMed] [Google Scholar]
- 43.Fukuda T., Ito H., Mukainaka T., Tokuda H., Nishino H., Yoshida T. Anti-tumor promoting effect of glycosides from Prunus persica seeds. Biological & Pharmaceutical Bulletin. 2003;26(2):271–273. doi: 10.1248/bpb.26.271. [DOI] [PubMed] [Google Scholar]
- 44.He X.-Y., Wu L.-J., Wang W.-X., Xie P.-J., Chen Y.-H., Wang F. Amygdalin—a pharmacological and toxicological review. Journal of Ethnopharmacology. 2020;254:p. 112717. doi: 10.1016/j.jep.2020.112717. [DOI] [PubMed] [Google Scholar]
- 45.Saleem M., Asif J., Asif M., Saleem U. Amygdalin from apricot kernels induces apoptosis and causes cell cycle arrest in cancer cells: an updated review. Anti-cancer Agents in Medicinal Chemistry. 2018;18(12):1650–1655. doi: 10.2174/1871520618666180105161136. [DOI] [PubMed] [Google Scholar]
- 46.Tanaka R., Nitta A., Nagatsu A. Application of a quantitative 1H-NMR method for the determination of amygdalin in Persicae semen, Armeniacae semen, and Mume fructus. Journal of Natural Medicines. 2014;68(1):225–230. doi: 10.1007/s11418-013-0783-y. [DOI] [PubMed] [Google Scholar]
- 47.Wang G.-Y., Qi H.-Y., Shi Y.-P. Ultrasonic cell grinder extraction of anthraquinones from radix et rhizoma rhei and determination by ultra-performance liquid chromatography. Journal of Separation Science. 2010;33(12):1730–1738. doi: 10.1002/jssc.200900861. [DOI] [PubMed] [Google Scholar]
- 48.Wu M., Gu Z. Screening of bioactive compounds from moutan cortex and their anti-inflammatory activities in rat synoviocytes. Evidence-Based Complementary and Alternative Medicine. 2009;6(1):57–63. doi: 10.1093/ecam/nem066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang C., Li X., Rong J. Amygdalin isolated from Semen Persicae (Tao Ren) extracts induces the expression of follistatin in HepG2 and C2C12 cell lines. Chinese Medicine. 2014;9(1):p. 23. doi: 10.1186/1749-8546-9-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang C., Zhao J., Cheng Y., Li X., Rong J. Bioactivity-guided fractionation identifies amygdalin as a potent neurotrophic agent from herbal medicine Semen Persicae extract. BioMed Research International. 2014;2014:14. doi: 10.1155/2014/306857.306857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang M.-H., Feng L., Zhu M.-M., et al. The anti-inflammation effect of moutan cortex on advanced glycation end products-induced rat mesangial cells dysfunction and high-glucose-fat diet and streptozotocin-induced diabetic nephropathy rats. Journal of Ethnopharmacology. 2014;151(1):591–600. doi: 10.1016/j.jep.2013.11.015. [DOI] [PubMed] [Google Scholar]
- 52.Singh P., Joshi nee Pant G., Rawat M. Phytochemistry and biological activity perspectives of Rheum species. The Natural Products Journal. 2016;6(2):84–93. doi: 10.2174/2210315506666151208212726. [DOI] [Google Scholar]
- 53.Zhan H., Fang J., Wu H.-W., et al. Rapid determination of total content of five major anthraquinones in Rhei Radix et Rhizoma by NIR spectroscopy. Chinese Herbal Medicines. 2017;9(3):250–257. doi: 10.1016/s1674-6384(17)60101-1. [DOI] [Google Scholar]
- 54.Dehghan H., Salehi P., Amiri M. S. Bioassay-guided purification of α-amylase, α-glucosidase inhibitors and DPPH radical scavengers from roots of Rheum turkestanicum. Industrial Crops and Products. 2018;117:303–309. doi: 10.1016/j.indcrop.2018.02.086. [DOI] [Google Scholar]
- 55.Xie J. L., Zhang Z. Q., Liang S., et al. Simultaneous determination of the 11 contents in the combination extracts of Rhei Radix et Rhizoma and Moutan Cortex by HPLC wavelength switching method. China Journal of Chinese Materia Medica. 2013;33(1):103–107. [Google Scholar]
- 56.Zhu J. J., Wang Z. M., Feng W. H., Zhang Q. W., Medicines J. C. H. A quantitative method for simultaneous determination of four anthraquinones with one marker in Rhei Radix et Rhizoma. Chinese Herbal Medicines. 2012;4(2):157–163. [Google Scholar]
- 57.Cho D. Y., Kim Y. A., Przytycka T. M. Chapter 5: network biology approach to complex diseases. PLoS Computational Biology. 2012;8(12) doi: 10.1371/journal.pcbi.1002820.e1002820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jeong H., Mason S. P., Barabási A.-L., Oltvai Z. N. Lethality and centrality in protein networks. Nature. 2001;411(6833):41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- 59.Gibbons D. L., Byers L. A., Kurie J. M. Smoking, p53 mutation, and lung cancer. Molecular Cancer Research. 2014;12(1):3–13. doi: 10.1158/1541-7786.mcr-13-0539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gu J., Zhou Y., Huang L., et al. TP53 mutation is associated with a poor clinical outcome for non-small cell lung cancer: evidence from a meta-analysis. Molecular and Clinical Oncology. 2016;5(6):705–713. doi: 10.3892/mco.2016.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mogi A., Kuwano H. TP53 mutations in nonsmall cell lung cancer. Journal of Biomedicine and Biotechnology. 2011;2011:9. doi: 10.1155/2011/583929.583929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ahrendt S. A., Hu Y., Buta M., et al. p53 mutations and survival in stage I non-small-cell lung cancer: results of a prospective study. JNCI Journal of the National Cancer Institute. 2003;95(13):961–970. doi: 10.1093/jnci/95.13.961. [DOI] [PubMed] [Google Scholar]
- 63.Steels E., Paesmans M., Berghmans T., et al. Role of p53 as a prognostic factor for survival in lung cancer: a systematic review of the literature with a meta-analysis. European Respiratory Journal. 2001;18(4):705–719. doi: 10.1183/09031936.01.00062201. [DOI] [PubMed] [Google Scholar]
- 64.Tian F., Wang J., Ouyang T., et al. MiR-486-5p serves as a good biomarker in nonsmall cell lung cancer and suppresses cell growth with the involvement of a target PIK3R1. Frontiers in Genetics. 2019;10:p. 688. doi: 10.3389/fgene.2019.00688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu K., Kang M., Li J., Qin W., Wang R. Prognostic value of the mRNA expression of members of the HSP90 family in non-small cell lung cancer. Experimental and Therapeutic Medicine. 2019;17(4):2657–2665. doi: 10.3892/etm.2019.7228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rong B., Yang S. Molecular mechanism and targeted therapy of Hsp90 involved in lung cancer: new discoveries and developments (Review) International Journal of Oncology. 2018;52(2):321–336. doi: 10.3892/ijo.2017.4214. [DOI] [PubMed] [Google Scholar]
- 67.Kanda R., Kawahara A., Watari K., et al. Erlotinib resistance in lung cancer cells mediated by integrin β1/src/akt-driven bypass signaling. Cancer Research. 2013;73(20):6243–6253. doi: 10.1158/0008-5472.can-12-4502. [DOI] [PubMed] [Google Scholar]
- 68.Lee M. W., Kim D. S., Lee J. H., et al. Roles of AKT1 and AKT2 in non-small cell lung cancer cell survival, growth, and migration. Cancer Science. 2011;102(10):1822–1828. doi: 10.1111/j.1349-7006.2011.02025.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lee M. W., Kim D. S., Min N. Y., Kim H. T. Akt1 inhibition by RNA interference sensitizes human non-small cell lung cancer cells to cisplatin. International Journal of Cancer. 2008;122(10):2380–2384. doi: 10.1002/ijc.23371. [DOI] [PubMed] [Google Scholar]
- 70.Liu L.-Z., Zhou X.-D., Qian G., Shi X., Fang J., Jiang B.-H. AKT1 amplification regulates cisplatin resistance in human lung cancer cells through the mammalian target of rapamycin/p70S6K1 pathway. Cancer Research. 2007;67(13):6325–6332. doi: 10.1158/0008-5472.can-06-4261. [DOI] [PubMed] [Google Scholar]
- 71.Rao G., Pierobon M., Kim I. K., et al. Inhibition of AKT1 signaling promotes invasion and metastasis of non-small cell lung cancer cells with K-RAS or EGFR mutations. Scientific Reports. 2017;7(1):p. 7066. doi: 10.1038/s41598-017-06128-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gong K., Guo G., Gerber D. E., et al. TNF-driven adaptive response mediates resistance to EGFR inhibition in lung cancer. Journal of Clinical Investigation. 2018;128(6):2500–2518. doi: 10.1172/jci96148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pal S., Yadav P., Sainis K. B., Shankar B. S. TNF-α and IGF-1 differentially modulate ionizing radiation responses of lung cancer cell lines. Cytokine. 2018;101:89–98. doi: 10.1016/j.cyto.2016.06.015. [DOI] [PubMed] [Google Scholar]
- 74.Shang G.-S., Liu L., Qin Y.-W. IL-6 and TNF-α promote metastasis of lung cancer by inducing epithelial-mesenchymal transition. Oncology Letters. 2017;13(6):4657–4660. doi: 10.3892/ol.2017.6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang S., Yan Y., Cheng Z., Hu Y., Liu T. Sotetsuflavone suppresses invasion and metastasis in non-small-cell lung cancer A549 cells by reversing EMT via the TNF-alpha/NF-κB and PI3K/AKT signaling pathway. Cell Death Discovery. 2018;4(1):p. 26. doi: 10.1038/s41420-018-0026-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang X., Chen W., Lin Y. Sensitization of TNF-induced cytotoxicity in lung cancer cells by concurrent suppression of the NF-κB and Akt pathways. Biochemical and Biophysical Research Communications. 2007;355(3):807–812. doi: 10.1016/j.bbrc.2007.02.030. [DOI] [PubMed] [Google Scholar]
- 77.Kim M. J., Kang H.-G., Lee S. Y., et al. AKT1 polymorphisms and survival of early stage non-small cell lung cancer. Journal of Surgical Oncology. 2012;105(2):167–174. doi: 10.1002/jso.22071. [DOI] [PubMed] [Google Scholar]
- 78.Zhang X., Fan J., Li Y., et al. Polymorphisms in epidermal growth factor receptor (EGFR) and AKT1 as possible predictors of clinical outcome in advanced non-small-cell lung cancer patients treated with EGFR tyrosine kinase inhibitors. Tumor Biology. 2016;37(1):1061–1069. doi: 10.1007/s13277-015-3893-1. [DOI] [PubMed] [Google Scholar]
- 79.Seifart C., Plagens A., Dempfle A., et al. TNF-α, TNF-β, IL-6, and IL-10 polymorphisms in patients with lung cancer. Disease Markers. 2005;21(3):157–165. doi: 10.1155/2005/707131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shih C.-M., Lee Y.-L., Chiou H.-L., et al. Association of TNF-α polymorphism with susceptibility to and severity of non-small cell lung cancer. Lung Cancer. 2006;52(1):15–20. doi: 10.1016/j.lungcan.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 81.Chen C. H., Lai J. M., Chou T. Y., et al. VEGFA upregulates FLJ10540 and modulates migration and invasion of lung cancer via PI3K/AKT pathway. PLoS One. 2009;4(4) doi: 10.1371/journal.pone.0005052.e5052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Han H., Silverman J. F., Santucci T. S., et al. Vascular endothelial growth factor expression in stage I non-small cell lung cancer correlates with neoangiogenesis and a poor prognosis. Annals of Surgical Oncology. 2001;8(1):72–79. doi: 10.1007/s10434-001-0072-y. [DOI] [PubMed] [Google Scholar]
- 83.Liu W., Xu J., Wang M., Wang Q., Bi Y., Han M. Tumor-derived vascular endothelial growth factor (VEGF)-a facilitates tumor metastasis through the VEGF-VEGFR1 signaling pathway. International Journal of Oncology. 2011;39(5):1213–1220. doi: 10.3892/ijo.2011.1138. [DOI] [PubMed] [Google Scholar]
- 84.Niu G., Chen X. Vascular endothelial growth factor as an anti-angiogenic target for cancer therapy. Current Drug Targets. 2010;11(8):1000–1017. doi: 10.2174/138945010791591395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.O’Byrne K. J., Koukourakis M. I., Giatromanolaki A., et al. Vascular endothelial growth factor, platelet-derived endothelial cell growth factor and angiogenesis in non-small-cell lung cancer. British Journal of Cancer. 2000;82(8):1427–1432. doi: 10.1054/bjoc.1999.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang S.-D., McCrudden C. M., Kwok H. F. Prognostic significance of combining VEGFA, FLT1 and KDR mRNA expression in lung cancer. Oncology Letters. 2015;10(3):1893–1901. doi: 10.3892/ol.2015.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gazdar A. F. Activating and resistance mutations of EGFR in non-small-cell lung cancer: role in clinical response to EGFR tyrosine kinase inhibitors. Oncogene. 2009;28(1):S24–S31. doi: 10.1038/onc.2009.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Morgillo F., Della Corte C. M., Fasano M., Ciardiello F. Mechanisms of resistance to EGFR-targeted drugs: lung cancer. ESMO Open. 2016;1(3) doi: 10.1136/esmoopen-2016-000060.e000060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang Z., Stiegler A. L., Boggon T. J., Kobayashi S., Halmos B. EGFR-mutated lung cancer: a paradigm of molecular oncology. Oncotarget. 2010;1(7):497–514. doi: 10.18632/oncotarget.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Levresse V., Marek L., Blumberg D., Heasley L. E. Regulation of platinum-compound cytotoxicity by the c-Jun N-terminal kinase and c-Jun signaling pathway in small-cell lung cancer cells. Molecular Pharmacology. 2002;62(3):689–697. doi: 10.1124/mol.62.3.689. [DOI] [PubMed] [Google Scholar]
- 91.Wang C., Liu E., Li W., Cui J., Li T. MiR-3188 inhibits non-small cell lung cancer cell proliferation through FOXO1-mediated mTOR-p-PI3K/AKT-c-JUN signaling pathway. Frontiers in Pharmacology. 2018;9:p. 1362. doi: 10.3389/fphar.2018.01362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yang Y., Ikezoe T., Saito T., Kobayashi M., Koeffler H. P., Taguchi H. Proteasome inhibitor PS-341 induces growth arrest and apoptosis of non-small cell lung cancer cells via the JNK/c-Jun/AP-1 signaling. Cancer Science. 2004;95(2):176–180. doi: 10.1111/j.1349-7006.2004.tb03200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Akunuru S., Palumbo J., Zhai Q. J., Zheng Y. Rac1 targeting suppresses human non-small cell lung adenocarcinoma cancer stem cell activity. PLoS One. 2011;6(2) doi: 10.1371/journal.pone.0016951.e16951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chen Q. Y., Xu L. Q., Jiao D. M., et al. Silencing of Rac1 modifies lung cancer cell migration, invasion and actin cytoskeleton rearrangements and enhances chemosensitivity to antitumor drugs. International Journal of Molecular Medicine. 2011;28(5):769–776. doi: 10.3892/ijmm.2011.775. [DOI] [PubMed] [Google Scholar]
- 95.Li Z., Guo C., Liu X., et al. TIPE2 suppresses angiogenesis and non-small cell lung cancer (NSCLC) invasiveness via inhibiting Rac1 activation and VEGF expression. Oncotarget. 2016;7(38):62224–62239. doi: 10.18632/oncotarget.11406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tan S., Yi P., Wang H., et al. RAC1 involves in the radioresistance by mediating epithelial-mesenchymal transition in lung cancer. Frontiers in Oncology. 2020;10:p. 649. doi: 10.3389/fonc.2020.01106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yuan K., Qian C., Zheng R. Prognostic significance of immunohistochemical Rac1 expression in survival in early operable non-small cell lung cancer. Medical Science Monitor: International Medical Journal of Experimental and Clinical Research. 2009;15(11):313–319. [PubMed] [Google Scholar]
- 98.Zhou Y., Liao Q., Han Y., et al. Rac1 overexpression is correlated with epithelial mesenchymal transition and predicts poor prognosis in non-small cell lung cancer. Journal of Cancer. 2016;7(14):2100–2109. doi: 10.7150/jca.16198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zou T., Mao X., Yin J., et al. Emerging roles ofRAC1in treating lung cancer patients. Clinical Genetics. 2017;91(4):520–528. doi: 10.1111/cge.12908. [DOI] [PubMed] [Google Scholar]
- 100.Cao Q., Mao Z.-D., Shi Y.-J., et al. MicroRNA-7 inhibits cell proliferation, migration and invasion in human non-small cell lung cancer cells by targeting FAK through ERK/MAPK signaling pathway. Oncotarget. 2016;7(47):77468–77481. doi: 10.18632/oncotarget.12684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ichihara E., Westover D., Meador C. B., et al. SFK/FAK signaling attenuates osimertinib efficacy in both drug-sensitive and drug-resistant models of EGFR-mutant lung cancer. Cancer Research. 2017;77(11):2990–3000. doi: 10.1158/0008-5472.can-16-2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lu H., Wang L., Gao W., et al. IGFBP2/FAK pathway is causally associated with dasatinib resistance in non-small cell lung cancer cells. Molecular Cancer Therapeutics. 2013;12(12):2864–2873. doi: 10.1158/1535-7163.mct-13-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tong X., Tanino R., Sun R., et al. Protein tyrosine kinase 2: a novel therapeutic target to overcome acquired EGFR-TKI resistance in non-small cell lung cancer. Respiratory Research. 2019;20(1):p. 270. doi: 10.1186/s12931-019-1244-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hsu L. H., Chu N. M., Kao S. H. Estrogen, estrogen receptor and lung cancer. International Journal of Molecular Sciences. 2017;18(8) doi: 10.3390/ijms18081713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kawai H., Ishii A., Washiya K., et al. Estrogen receptor α and β are prognostic factors in non-small cell lung cancer. Clinical Cancer Research. 2005;11(14):5084–5089. doi: 10.1158/1078-0432.ccr-05-0200. [DOI] [PubMed] [Google Scholar]
- 106.Yin J., Wang H., Vogel U., Wang C., Hou W., Ma Y. Association and interaction of NFKB1 rs28362491 insertion/deletion ATTG polymorphism and PPP1R13L and CD3EAP related to lung cancer risk in a Chinese population. Tumor Biology. 2016;37(4):5467–5473. doi: 10.1007/s13277-015-4373-3. [DOI] [PubMed] [Google Scholar]
- 107.Carelli S., Zadra G., Vaira V., et al. Up-regulation of focal adhesion kinase in non-small cell lung cancer. Lung Cancer. 2006;53(3):263–271. doi: 10.1016/j.lungcan.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 108.Giatromanolaki A., Koukourakis M. I., Sivridis E., et al. Relation of hypoxia inducible factor 1α and 2α in operable non-small cell lung cancer to angiogenic/molecular profile of tumours and survival. British Journal of Cancer. 2001;85(6):881–890. doi: 10.1054/bjoc.2001.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hung J.-J., Yang M.-H., Hsu H.-S., Hsu W.-H., Liu J.-S., Wu K.-J. Prognostic significance of hypoxia-inducible factor-1 , TWIST1 and Snail expression in resectable non-small cell lung cancer. Thorax. 2009;64(12):1082–1089. doi: 10.1136/thx.2009.115691. [DOI] [PubMed] [Google Scholar]
- 110.Jiang S., Wang R., Yan H., Jin L., Dou X., Chen D. MicroRNA-21 modulates radiation resistance through upregulation of hypoxia-inducible factor-1α-promoted glycolysis in non-small cell lung cancer cells. Molecular Medicine Reports. 2016;13(5):4101–4107. doi: 10.3892/mmr.2016.5010. [DOI] [PubMed] [Google Scholar]
- 111.Kim W.-Y., Oh S. H., Woo J.-K., Hong W. K., Lee H.-Y. Targeting heat shock protein 90 overrides the resistance of lung cancer cells by blocking radiation-induced stabilization of hypoxia-inducible factor-1α. Cancer Research. 2009;69(4):1624–1632. doi: 10.1158/0008-5472.can-08-0505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lee S.-H., Koo K. H., Park J.-W., et al. HIF-1 is induced via EGFR activation and mediates resistance to anoikis-like cell death under lipid rafts/caveolae-disrupting stress. Carcinogenesis. 2009;30(12):1997–2004. doi: 10.1093/carcin/bgp233. [DOI] [PubMed] [Google Scholar]
- 113.Liu W., Liang Y., Chan Q., Jiang L., Dong J. CX3CL1 promotes lung cancer cell migration and invasion via the Src/focal adhesion kinase signaling pathway. Oncology Reports. 2019;41(3):1911–1917. doi: 10.3892/or.2019.6957. [DOI] [PubMed] [Google Scholar]
- 114.Liu Y.-L., Yu J.-M., Song X.-R., Wang X.-W., Xing L.-G., Gao B.-B. Regulation of the chemokine receptor CXCR4 and metastasis by hypoxia-inducible factor in non small cell lung cancer cell lines. Cancer Biology and Therapy. 2006;5(10):1320–1326. doi: 10.4161/cbt.5.10.3162. [DOI] [PubMed] [Google Scholar]
- 115.Park S., Ha S. Y., Cho H. Y., et al. Prognostic implications of hypoxia-inducible factor-1α in epidermal growth factor receptor-negative non-small cell lung cancer. Lung Cancer. 2011;72(1):100–107. doi: 10.1016/j.lungcan.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 116.Ren W., Mi D., Yang K., et al. The expression of hypoxia-inducible factor-1α and its clinical significance in lung cancer: a systematic review and meta-analysis. Swiss Medical Weekly. 2013;143 doi: 10.4414/smw.2013.13855.w13855 [DOI] [PubMed] [Google Scholar]
- 117.Rothschild S. Targeted therapies in non-small cell lung cancer-beyond EGFR and ALK. Cancers. 2015;7(2):930–949. doi: 10.3390/cancers7020816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Swinson D. E. B., Jones J. L., Cox G., Richardson D., Harris A. L., O’Byrne K. J. Hypoxia-inducible factor-1α in non small cell lung cancer: relation to growth factor, protease and apoptosis pathways. International Journal of Cancer. 2004;111(1):43–50. doi: 10.1002/ijc.20052. [DOI] [PubMed] [Google Scholar]
- 119.Takasaki C., Kobayashi M., Ishibashi H., Akashi T., Okubo K. Expression of hypoxia-inducible factor-1α affects tumor proliferation and antiapoptosis in surgically resected lung cancer. Molecular and Clinical Oncology. 2016;5(2):295–300. doi: 10.3892/mco.2016.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Volm M., Koomagi R. Hypoxia-inducible factor (HIF-1) and its relationship to apoptosis and proliferation in lung cancer. Anticancer Research. 2000;20(3A):1527–1533. [PubMed] [Google Scholar]
- 121.Wang M., Han J., Marcar L., et al. Radiation resistance in KRAS-mutated lung cancer is enabled by stem-like properties mediated by an osteopontin-EGFR pathway. Cancer Research. 2017;77(8):2018–2028. doi: 10.1158/0008-5472.can-16-0808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wu D.-W., Wu T.-C., Wu J.-Y., et al. Phosphorylation of paxillin confers cisplatin resistance in non-small cell lung cancer via activating ERK-mediated Bcl-2 expression. Oncogene. 2014;33(35):4385–4395. doi: 10.1038/onc.2013.389. [DOI] [PubMed] [Google Scholar]
- 123.Xu L., Nilsson M. B., Saintigny P., et al. Epidermal growth factor receptor regulates MET levels and invasiveness through hypoxia-inducible factor-1α in non-small cell lung cancer cells. Oncogene. 2010;29(18):2616–2627. doi: 10.1038/onc.2010.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhang H., Shao H., Golubovskaya V. M., et al. Efficacy of focal adhesion kinase inhibition in non-small cell lung cancer with oncogenically activated MAPK pathways. British Journal of Cancer. 2016;115(2):203–211. doi: 10.1038/bjc.2016.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhu H., Zhang S. Hypoxia inducible factor-1α/vascular endothelial growth factor signaling activation correlates with response to radiotherapy and its inhibition reduces hypoxia-induced angiogenesis in lung cancer. Journal of Cellular Biochemistry. 2018;119(9):7707–7718. doi: 10.1002/jcb.27120. [DOI] [PubMed] [Google Scholar]
- 126.Hildebrandt M. A., Roth J. A., Vaporciyan A. A., et al. Genetic variation in the TNF/TRAF2/ASK1/p38 kinase signaling pathway as markers for postoperative pulmonary complications in lung cancer patients. Scientific Reports. 2015;5(1):p. 12068. doi: 10.1038/srep12068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.McGrath E. E. The tumor necrosis factor-related apoptosis-inducing ligand and lung cancer: still following the right TRAIL? Journal of Thoracic Oncology. 2011;6(6):983–987. doi: 10.1097/jto.0b013e318217b6c8. [DOI] [PubMed] [Google Scholar]
- 128.Burns T. F., Stabile L. P. Targeting the estrogen pathway for the treatment and prevention of lung cancer. Lung Cancer Management. 2014;3(1):43–52. doi: 10.2217/lmt.13.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hamilton D. H., Griner L. M., Keller J. M., et al. Targeting estrogen receptor signaling with fulvestrant enhances immune and chemotherapy-mediated cytotoxicity of human lung cancer. Clinical Cancer Research. 2016;22(24):6204–6216. doi: 10.1158/1078-0432.ccr-15-3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Márquez-Garbán D. C., Chen H.-W., Goodglick L., Fishbein M. C., Pietras R. J. Targeting aromatase and estrogen signaling in human non-small cell lung cancer. Annals of the New York Academy of Sciences. 2009;1155(1):194–205. doi: 10.1111/j.1749-6632.2009.04116.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Tang H., Liao Y., Zhang C., et al. Fulvestrant-mediated inhibition of estrogen receptor signaling slows lung cancer progression. Oncology Research Featuring Preclinical and Clinical Cancer Therapeutics. 2014;22(1):13–20. doi: 10.3727/096504014x14077751730315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.He J., Hu Y., Hu M., Li B. Development of PD-1/PD-L1 pathway in tumor immune microenvironment and treatment for non-small cell lung cancer. Scientific Reports. 2015;5(1):p. 13110. doi: 10.1038/srep13110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ji M., Liu Y., Li Q., et al. PD-1/PD-L1 pathway in non-small-cell lung cancer and its relation with EGFR mutation. ournal of Translational Medicine. 2015;13(1):p. 5. doi: 10.1186/s12967-014-0373-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Santarpia M., Aguilar A., Chaib I., et al. Non-small-cell lung cancer signaling pathways, metabolism, and PD-1/PD-L1 antibodies. Cancers. 2020;12(6) doi: 10.3390/cancers12061475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Breen L., Heenan M., Amberger-Murphy V., Clynes M. Investigation of the role of p53 in chemotherapy resistance of lung cancer cell lines. Anticancer Research. 2007;27(3A):1361–1364. [PubMed] [Google Scholar]
- 136.Campling B. G., El-Deiry W. S. Clinical implications of p53 mutations in lung cancer. Methods in Molecular Medicine. 2003;75:53–77. doi: 10.1385/1-59259-324-0:53. [DOI] [PubMed] [Google Scholar]
- 137.Fernández-Aroca D. M., Roche O., Sabater S., et al. P53 pathway is a major determinant in the radiosensitizing effect of Palbociclib: implication in cancer therapy. Cancer Letters. 2019;451:23–33. doi: 10.1016/j.canlet.2019.02.049. [DOI] [PubMed] [Google Scholar]
- 138.Lai S.-L., Perng R.-P., Hwang J. p53 gene status modulates the chemosensitivity of non-small cell lung cancer cells. Journal of Biomedical Science. 2000;7(1):64–70. doi: 10.1007/bf02255920. [DOI] [PubMed] [Google Scholar]
- 139.Liu L., Wu C., Wang Y., et al. Combined effect of genetic polymorphisms in P53, P73, and MDM2 on non-small cell lung cancer survival. Journal of Thoracic Oncology. 2011;6(11):1793–1800. doi: 10.1097/jto.0b013e3182272273. [DOI] [PubMed] [Google Scholar]
- 140.Mirzayans R., Andrais B., Scott A., Murray D. New insights into p53 signaling and cancer cell response to DNA damage: implications for cancer therapy. Journal of Biomedicine and Biotechnology. 2012;2012:16. doi: 10.1155/2012/170325.170325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Robles A. I., Linke S. P., Harris C. C. The p53 network in lung carcinogenesis. Oncogene. 2002;21(45):6898–6907. doi: 10.1038/sj.onc.1205563. [DOI] [PubMed] [Google Scholar]
- 142.Tsao M.-S., Aviel-Ronen S., Ding K., et al. Prognostic and predictive importance of p53 and RAS for adjuvant chemotherapy in non-small-cell lung cancer. Journal of Clinical Oncology. 2007;25(33):5240–5247. doi: 10.1200/jco.2007.12.6953. [DOI] [PubMed] [Google Scholar]
- 143.Viktorsson K., De Petris L., Lewensohn R. The role of p53 in treatment responses of lung cancer. Biochemical and Biophysical Research Communications. 2005;331(3):868–880. doi: 10.1016/j.bbrc.2005.03.192. [DOI] [PubMed] [Google Scholar]
- 144.Aita M., Fasola G., Defferrari C., et al. Targeting the VEGF pathway: antiangiogenic strategies in the treatment of non-small cell lung cancer. Critical Reviews in Oncology/Hematology. 2008;68(3):183–196. doi: 10.1016/j.critrevonc.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 145.Alevizakos M., Kaltsas S., Syrigos K. N. The VEGF pathway in lung cancer. Cancer Chemotherapy and Pharmacology. 2013;72(6):1169–1181. doi: 10.1007/s00280-013-2298-3. [DOI] [PubMed] [Google Scholar]
- 146.Korpanty G., Smyth E., Sullivan L. A., Brekken R. A., Carney D. N. Antiangiogenic therapy in lung cancer: focus on vascular endothelial growth factor pathway. Experimental Biology and Medicine. 2010;235(1):3–9. doi: 10.1258/ebm.2009.009191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Gewirtz D. A., Holt S. E., Elmore L. W. Accelerated senescence: an emerging role in tumor cell response to chemotherapy and radiation. Biochemical Pharmacology. 2008;76(8):947–957. doi: 10.1016/j.bcp.2008.06.024. [DOI] [PubMed] [Google Scholar]
- 148.Salim H., Akbar N. S., Zong D., et al. miRNA-214 modulates radiotherapy response of non-small cell lung cancer cells through regulation of p38MAPK, apoptosis and senescence. British Journal of Cancer. 2012;107(8):1361–1373. doi: 10.1038/bjc.2012.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Shivapurkar N., Reddy J., Chaudhary P. M., Gazdar A. F. Apoptosis and lung cancer: a review. Journal of Cellular Biochemistry. 2003;88(5):885–898. doi: 10.1002/jcb.10440. [DOI] [PubMed] [Google Scholar]
- 150.Chen J., Huang X., Tao C., et al. Artemether attenuates the progression of non-small cell lung cancer by inducing apoptosis, cell cycle arrest and promoting cellular senescence. Biological and Pharmaceutical Bulletin. 2019;42(10):1720–1725. doi: 10.1248/bpb.b19-00391. [DOI] [PubMed] [Google Scholar]
- 151.Luo H., Wang L., Schulte B. A., Yang A., Tang S., Wang G. Y. Resveratrol enhances ionizing radiation-induced premature senescence in lung cancer cells. International Journal of Oncology. 2013;43(6):1999–2006. doi: 10.3892/ijo.2013.2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Luo H., Yount C., Lang H., et al. Activation of p53 with Nutlin-3a radiosensitizes lung cancer cells via enhancing radiation-induced premature senescence. Lung Cancer. 2013;81(2):167–173. doi: 10.1016/j.lungcan.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Chen J., Emara N., Solomides C., Parekh H., Simpkins H. Resistance to platinum-based chemotherapy in lung cancer cell lines. Cancer Chemotherapy and Pharmacology. 2010;66(6):1103–1111. doi: 10.1007/s00280-010-1268-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kobayashi S., Boggon T. J., Dayaram T., et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. New England Journal of Medicine. 2005;352(8):786–792. doi: 10.1056/nejmoa044238. [DOI] [PubMed] [Google Scholar]
- 155.Leonetti A., Sharma S., Minari R., Perego P., Giovannetti E., Tiseo M. Resistance mechanisms to osimertinib in EGFR-mutated non-small cell lung cancer. British Journal of Cancer. 2019;121(9):725–737. doi: 10.1038/s41416-019-0573-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Martin L. P., Hamilton T. C., Schilder R. J. Platinum resistance: the role of DNA repair pathways. Clinical Cancer Research. 2008;14(5):1291–1295. doi: 10.1158/1078-0432.ccr-07-2238. [DOI] [PubMed] [Google Scholar]
- 157.Tartarone A., Lazzari C., Lerose R., et al. Mechanisms of resistance to EGFR tyrosine kinase inhibitors gefitinib/erlotinib and to ALK inhibitor crizotinib. Lung Cancer. 2013;81(3):328–336. doi: 10.1016/j.lungcan.2013.05.020. [DOI] [PubMed] [Google Scholar]
- 158.Chang W. T., You B. J., Yang W. H., Wu C. Y., Bau D. T., Lee H. Z. Protein kinase C delta-mediated cytoskeleton remodeling is involved in aloe-emodin-induced photokilling of human lung cancer cells. Anticancer Research. 2012;32(9):3707–3713. [PubMed] [Google Scholar]
- 159.Lee H.-Z. Protein kinase C involvement in aloe-emodin- and emodin-induced apoptosis in lung carcinoma cell. British Journal of Pharmacology. 2001;134(5):1093–1103. doi: 10.1038/sj.bjp.0704342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Lee H., Lin C., Yang W., Leung W., Chang S. Aloe-emodin induced DNA damage through generation of reactive oxygen species in human lung carcinoma cells. Cancer Letters. 2006;239(1):55–63. doi: 10.1016/j.canlet.2005.07.036. [DOI] [PubMed] [Google Scholar]
- 161.Lee H. Z., Yang W. H., Hour M. J., et al. Photodynamic activity of aloe-emodin induces resensitization of lung cancer cells to anoikis. European Journal of Pharmacology. 2010;648(1–3):50–58. doi: 10.1016/j.ejphar.2010.08.033. [DOI] [PubMed] [Google Scholar]
- 162.Shen F., Ge C., Yuan P. Aloe-emodin induces autophagy and apoptotic cell death in non-small cell lung cancer cells via Akt/mTOR and MAPK signaling. European Journal of Pharmacology. 2020;886:p. 173550. doi: 10.1016/j.ejphar.2020.173550. [DOI] [PubMed] [Google Scholar]
- 163.Min J., Shen H., Xi W., et al. Synergistic anticancer activity of combined use of caffeic acid with paclitaxel enhances apoptosis of non-small-cell lung cancer H1299 cells in vivo and in vitro. Cellular Physiology and Biochemistry. 2018;48(4):1433–1442. doi: 10.1159/000492253. [DOI] [PubMed] [Google Scholar]
- 164.Saha A., Kuzuhara T., Echigo N., Suganuma M., Fujiki H. New role of (−)-Epicatechin in enhancing the induction of growth inhibition and apoptosis in human lung cancer cells by curcumin. Cancer Prevention Research. 2010;3(8):953–962. doi: 10.1158/1940-6207.capr-09-0247. [DOI] [PubMed] [Google Scholar]
- 165.Sun H., Yin M., Hao D., Shen Y. Anti-cancer activity of catechin against A549 lung carcinoma cells by induction of cyclin kinase inhibitor p21 and suppression of cyclin E1 and P-AKT. Applied Sciences. 2020;10(6):p. 2065. doi: 10.3390/app10062065. [DOI] [Google Scholar]
- 166.Ni C.-H., Yu C.-S., Lu H.-F., et al. Chrysophanol-induced cell death (necrosis) in human lung cancer A549 cells is mediated through increasing reactive oxygen species and decreasing the level of mitochondrial membrane potential. Environmental Toxicology. 2014;29(7):740–749. doi: 10.1002/tox.21801. [DOI] [PubMed] [Google Scholar]
- 167.Zhang J., Wang Q., Wang Q., et al. Chrysophanol exhibits anti-cancer activities in lung cancer cell through regulating ROS/HIF-1a/VEGF signaling pathway. Naunyn-Schmiedeberg’s Archives of Pharmacology. 2020;393(3):469–480. doi: 10.1007/s00210-019-01746-8. [DOI] [PubMed] [Google Scholar]
- 168.Rajavel T., Banu Priya G., Suryanarayanan V., Singh S. K., Pandima Devi K. Daucosterol disturbs redox homeostasis and elicits oxidative-stress mediated apoptosis in A549 cells via targeting thioredoxin reductase by a p53 dependent mechanism. European Journal of Pharmacology. 2019;855:112–123. doi: 10.1016/j.ejphar.2019.04.051. [DOI] [PubMed] [Google Scholar]
- 169.Rajavel T., Mohankumar R., Archunan G., Ruckmani K., Devi K. P. Beta sitosterol and daucosterol (phytosterols identified in Grewia tiliaefolia) perturbs cell cycle and induces apoptotic cell death in A549 cells. Scientific Reports. 2017;7(1):p. 3418. doi: 10.1038/s41598-017-03511-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Chen S., Zhang Z., Zhang J. Emodin enhances antitumor effect of paclitaxel on human non-small-cell lung cancer cells in vitro and in vivo. Drug Design, Development and Therapy. 2019;13:1145–1153. doi: 10.2147/dddt.s196319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Haque E., Kamil M., Irfan S., et al. Blocking mutation independent p53 aggregation by emodin modulates autophagic cell death pathway in lung cancer. The International Journal of Biochemistry and Cell Biology. 2018;96:90–95. doi: 10.1016/j.biocel.2018.01.014. [DOI] [PubMed] [Google Scholar]
- 172.Lai J. M., Chang J. T., Wen C. L., Hsu S. L. Emodin induces a reactive oxygen species-dependent and ATM-p53-Bax mediated cytotoxicity in lung cancer cells. European Journal of Pharmacology. 2009;623(1–3):1–9. doi: 10.1016/j.ejphar.2009.08.031. [DOI] [PubMed] [Google Scholar]
- 173.Ok S., Kim S.-M., Kim C., et al. Emodin inhibits invasion and migration of prostate and lung cancer cells by downregulating the expression of chemokine receptor CXCR4. Immunopharmacology and Immunotoxicology. 2012;34(5):768–778. doi: 10.3109/08923973.2012.654494. [DOI] [PubMed] [Google Scholar]
- 174.Su J., Yan Y., Qu J., Xue X., Liu Z., Cai H. Emodin induces apoptosis of lung cancer cells through ER stress and the TRIB3/NF-κB pathway. Oncology Reports. 2017;37(3):1565–1572. doi: 10.3892/or.2017.5428. [DOI] [PubMed] [Google Scholar]
- 175.Su Y.-J., Tsai M.-S., Kuo Y.-H., et al. Role of Rad51 down-regulation and extracellular signal-regulated kinases 1 and 2 inactivation in emodin and mitomycin C-induced synergistic cytotoxicity in human non-small-cell lung cancer cells. Molecular Pharmacology. 2010;77(4):633–643. doi: 10.1124/mol.109.061887. [DOI] [PubMed] [Google Scholar]
- 176.Su Y.-T., Chang H.-L., Shyue S.-K., Hsu S.-L. Emodin induces apoptosis in human lung adenocarcinoma cells through a reactive oxygen species-dependent mitochondrial signaling pathway. Biochemical Pharmacology. 2005;70(2):229–241. doi: 10.1016/j.bcp.2005.04.026. [DOI] [PubMed] [Google Scholar]
- 177.Wang X., Li L., Guan R., Zhu D., Song N., Shen L. Emodin inhibits ATP-induced proliferation and migration by suppressing P2Y receptors in human lung adenocarcinoma cells. Cellular Physiology and Biochemistry. 2017;44(4):1337–1351. doi: 10.1159/000485495. [DOI] [PubMed] [Google Scholar]
- 178.Ying Y., Qingwu L., Mingming X., Zhenju S., Chaoyang T., Zhengang T. Emodin: one main ingredient of shufeng jiedu capsule reverses chemoresistance of lung cancer cells through inhibition of EMT. Cellular Physiology and Biochemistry. 2017;42(3):1063–1072. doi: 10.1159/000478754. [DOI] [PubMed] [Google Scholar]
- 179.Zhang L., Hung M. C. Sensitization of HER-2/neu-overexpressing non-small cell lung cancer cells to chemotherapeutic drugs by tyrosine kinase inhibitor emodin. Oncogene. 1996;12(3):571–576. [PubMed] [Google Scholar]
- 180.Lee C., Cheng M., Chen T., Yang Y., Chung M., Chiu H. Cell cycle regulation of emodin through p53, p21, and cyclin D pathway in human lung adenocarcinoma A549 cells. Planta Medica. 2008;74(9):p. PA124. doi: 10.1055/s-0028-1084122. [DOI] [Google Scholar]
- 181.Choi K.-C., Lee Y.-H., Jung M. G., et al. Gallic acid suppresses lipopolysaccharide-induced nuclear factor-κb signaling by preventing RelA acetylation in A549 lung cancer cells. Molecular Cancer Research. 2009;7(12):2011–2021. doi: 10.1158/1541-7786.mcr-09-0239. [DOI] [PubMed] [Google Scholar]
- 182.Ji B.-C., Hsu W.-H., Yang J.-S., et al. Gallic acid induces apoptosis via caspase-3 and mitochondrion-dependent pathways in vitro and suppresses lung xenograft tumor growth in vivo. Journal of Agricultural and Food Chemistry. 2009;57(16):7596–7604. doi: 10.1021/jf901308p. [DOI] [PubMed] [Google Scholar]
- 183.Kang D. Y., Sp N., Jo E. S., et al. The inhibitory mechanisms of tumor PD-L1 expression by natural bioactive gallic acid in non-small-cell lung cancer (NSCLC) cells. Cancers. 2020;12(3) doi: 10.3390/cancers12030727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Nam B., Rho J. K., Shin D.-M., Son J. Gallic acid induces apoptosis in EGFR-mutant non-small cell lung cancers by accelerating EGFR turnover. Bioorganic & Medicinal Chemistry Letters. 2016;26(19):4571–4575. doi: 10.1016/j.bmcl.2016.08.083. [DOI] [PubMed] [Google Scholar]
- 185.Phan A. N. H., Hua T. N. M., Kim M.-K., et al. Gallic acid inhibition of Src-Stat3 signaling overcomes acquired resistance to EGF receptor tyrosine kinase inhibitors in advanced non-small cell lung cancer. Oncotarget. 2016;7(34):54702–54713. doi: 10.18632/oncotarget.10581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Wang R., Ma L., Weng D., Yao J., Liu X., Jin F. Gallic acid induces apoptosis and enhances the anticancer effects of cisplatin in human small cell lung cancer H446 cell line via the ROS-dependent mitochondrial apoptotic pathway. Oncology Reports. 2016;35(5):3075–3083. doi: 10.3892/or.2016.4690. [DOI] [PubMed] [Google Scholar]
- 187.You B. R., Park W. H. Gallic acid-induced lung cancer cell death is related to glutathione depletion as well as reactive oxygen species increase. Toxicology in Vitro. 2010;24(5):1356–1362. doi: 10.1016/j.tiv.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 188.Zhang T., Ma L., Wu P., et al. Gallic acid has anticancer activity and enhances the anticancer effects of cisplatin in nonsmall cell lung cancer A549 cells via the JAK/STAT3 signaling pathway. Oncology Reports. 2019;41(3):1779–1788. doi: 10.3892/or.2019.6976. [DOI] [PubMed] [Google Scholar]
- 189.Wang K., Liu X., Liu Q., et al. Hederagenin potentiated cisplatin- and paclitaxel-mediated cytotoxicity by impairing autophagy in lung cancer cells. Cell Death and Disease. 2020;11(8):p. 611. doi: 10.1038/s41419-020-02880-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Nguyen T. T. T., Tran E., Ong C. K., et al. Kaempferol-induced growth inhibition and apoptosis in A549 lung cancer cells is mediated by activation of MEK-MAPK. Journal of Cellular Physiology. 2003;197(1):110–121. doi: 10.1002/jcp.10340. [DOI] [PubMed] [Google Scholar]
- 191.Jo E., Park S. J., Choi Y. S., Jeon W.-K., Kim B.-C. Kaempferol suppresses transforming growth factor-β1-induced epithelial-to-mesenchymal transition and migration of A549 lung cancer cells by inhibiting akt1-mediated phosphorylation of Smad3 at threonine-179. Neoplasia. 2015;17(7):525–537. doi: 10.1016/j.neo.2015.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Kuo W.-T., Tsai Y.-C., Wu H.-C., et al. Radiosensitization of non-small cell lung cancer by kaempferol. Oncology Reports. 2015;34(5):2351–2356. doi: 10.3892/or.2015.4204. [DOI] [PubMed] [Google Scholar]
- 193.Ko J.-L., Lin C.-H., Chen H.-C., et al. Effects and mechanisms of betulinic acid on improving EGFR TKI-resistance of lung cancer cells. Environmental Toxicology. 2018;33(11):1153–1159. doi: 10.1002/tox.22621. [DOI] [PubMed] [Google Scholar]
- 194.Kutkowska J., Strzadala L., Rapak A. Synergistic activity of sorafenib and betulinic acid against clonogenic activity of non‐small cell lung cancer cells. Cancer Science. 2017;108(11):2265–2272. doi: 10.1111/cas.13386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Zhan X. K., Li J. L., Zhang S., Xing P. Y., Xia M. F. Betulinic acid exerts potent antitumor effects on paclitaxel-resistant human lung carcinoma cells (H460) via G2/M phase cell cycle arrest and induction of mitochondrial apoptosis. Oncology Letters. 2018;16(3):3628–3634. doi: 10.3892/ol.2018.9097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Hung J. Y., Yang C. J., Tsai Y. M., Huang H. W., Huang M. S. Antiproliferative activity of paeoniflorin is through cell cycle arrest and the Fas/Fas ligand-mediated apoptotic pathway in human non-small cell lung cancer A549 cells. Clinical and Experimental Pharmacology and Physiology. 2008;35(2):141–147. doi: 10.1111/j.1440-1681.2008.04935.x. [DOI] [PubMed] [Google Scholar]
- 197.Wu Q., Chen G.-L., Li Y.-J., Chen Y., Lin F.-Z. Paeoniflorin inhibits macrophage-mediated lung cancer metastasis. Chinese Journal of Natural Medicines. 2015;13(12):925–932. doi: 10.1016/s1875-5364(15)30098-4. [DOI] [PubMed] [Google Scholar]
- 198.Tian Y., Chen C., Zhang Y., Zhang Z., Xie H. Paeonol inhibits migration, invasion and bone adhesion of small cell lung cancer cells. Current Signal Transduction Therapy. 2015;10(2):126–130. doi: 10.2174/1574362410666150625191130. [DOI] [Google Scholar]
- 199.Lei Y., Li H.-X., Jin W.-S., et al. The radiosensitizing effect of Paeonol on lung adenocarcinoma by augmentation of radiation-induced apoptosis and inhibition of the PI3K/Akt pathway. International Journal of Radiation Biology. 2013;89(12):1079–1086. doi: 10.3109/09553002.2013.825058. [DOI] [PubMed] [Google Scholar]
- 200.Savenkova D., Havrysh К., Skripova V., et al. Physcion enhances sensitivity of pancreatic adenocarcinoma and lung carcinoma cell lines to cisplatin. BioNanoScience. 2020;10(3):549–553. doi: 10.1007/s12668-020-00740-2. [DOI] [Google Scholar]
- 201.Hsia T. C., Yang J. S., Chen G. W., et al. The roles of endoplasmic reticulum stress and Ca2+ on rhein-induced apoptosis in A-549 human lung cancer cells. Anticancer Research. 2009;29(1):309–318. [PubMed] [Google Scholar]
- 202.Yang L., Li J., Xu L., et al. Rhein shows potent efficacy against non-small-cell lung cancer through inhibiting the STAT3 pathway. Cancer Management and Research. 2019;11:1167–1176. doi: 10.2147/cmar.s171517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Chang J.-H., Lai S.-L., Chen W.-S., et al. Quercetin suppresses the metastatic ability of lung cancer through inhibiting Snail-dependent Akt activation and Snail-independent ADAM9 expression pathways. Biochimica et Biophysica Acta (BBA)—Molecular Cell Research. 2017;1864(10):1746–1758. doi: 10.1016/j.bbamcr.2017.06.017. [DOI] [PubMed] [Google Scholar]
- 204.Klimaszewska-Wisniewska A., Halas-Wisniewska M., Izdebska M., Gagat M., Grzanka A., Grzanka D. Antiproliferative and antimetastatic action of quercetin on A549 non-small cell lung cancer cells through its effect on the cytoskeleton. Acta Histochem. 2017;119(2):99–112. doi: 10.1016/j.acthis.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 205.Lee S. H., Lee E. J., Min K. H., et al. Quercetin enhances chemosensitivity to gemcitabine in lung cancer cells by inhibiting heat shock protein 70 expression. Clinical Lung Cancer. 2015;16(6):e235–e243. doi: 10.1016/j.cllc.2015.05.006. [DOI] [PubMed] [Google Scholar]
- 206.Nguyen T. T., Tran E., Nguyen T. H., Do P. T., Huynh T. H., Huynh H. The role of activated MEK-ERK pathway in quercetin-induced growth inhibition and apoptosis in A549 lung cancer cells. Carcinogenesis. 2004;25(5):647–659. doi: 10.1093/carcin/bgh052. [DOI] [PubMed] [Google Scholar]
- 207.Wang Q., Chen Y., Lu H., et al. Quercetin radiosensitizes non‐small cell lung cancer cells through the regulation of miR‐16‐5p/WEE1 axis. IUBMB Life. 2020;72(5):1012–1022. doi: 10.1002/iub.2242. [DOI] [PubMed] [Google Scholar]
- 208.Wu T.-C., Chan S.-T., Chang C.-N., Yu P.-S., Chuang C.-H., Yeh S.-L. Quercetin and chrysin inhibit nickel-induced invasion and migration by downregulation of TLR4/NF-κB signaling in A549 cells. Chemico-Biological Interactions. 2018;292:101–109. doi: 10.1016/j.cbi.2018.07.010. [DOI] [PubMed] [Google Scholar]
- 209.Lim E. J., Heo J., Kim Y.-H. Quercetin induces cell death by caspase-dependent and p38 MAPK pathway in EGFR mutant lung cancer cells. Kosin Medical Journal. 2016;31(1):30–40. doi: 10.7180/kmj.2016.31.1.30. [DOI] [Google Scholar]
- 210.Wang X., Li M., Hu M., Wei P., Zhu W. BAMBI overexpression together with β-sitosterol ameliorates NSCLC via inhibiting autophagy and inactivating TGF-β/Smad2/3 pathway. Oncology Reports. 2017;37(5):3046–3054. doi: 10.3892/or.2017.5508. [DOI] [PubMed] [Google Scholar]
- 211.Lee J., Kim S., Ju J. Effects of white sesame seed extract and β-sitosterol on growth, migration, and adhesion of H1299 human lung cancer cells. Journal of the Korean Society of Food Science and Nutrition. 2015;44(9):1279–1285. doi: 10.3746/jkfn.2015.44.9.1279. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1: functional enrichment analysis for the lung cancer-related targets of FDY2004. Supplementary Table S1: general information and reports on evidence of biological activities of FDY2004 and its herbal constituents. Supplementary Table S2: chemical components of FDY2004. Supplementary Table S3: active chemical components of FDY2004. Supplementary Table S4: Targets of active chemical components of FDY2004. Supplementary Table S5: docking scores between active chemical components of FDY2004 and the lung cancer-associated targets.
Data Availability Statement
The data used to support the findings of this study are included within the article and Supplementary Materials file.


