Abstract
The current COVID-19 pandemic has led to a devastating impact across the world. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (the virus causing COVID-19) is known to use the receptor-binding domain (RBD) at viral surface spike (S) protein to interact with the angiotensin-converting enzyme 2 (ACE2) receptor expressed on many human cell types. The RBD-ACE2 interaction is a crucial step to mediate the host cell entry of SARS-CoV-2. Recent studies indicate that the ACE2 interaction with the SARS-CoV-2 S protein has a higher affinity than its binding with the structurally identical S protein of SARS-CoV-1, the virus causing the 2002–2004 SARS outbreak. However, the biophysical mechanism behind such binding affinity difference is unclear. This study utilizes combined single-molecule force spectroscopy and steered molecular dynamics (SMD) simulation approaches to quantify the specific interactions between SARS-CoV-2 or SARS-CoV-1 RBD and ACE2. Depending on the loading rates, the unbinding forces between SARS-CoV-2 RBD and ACE2 range from 70 to 105 pN and are 30–40% higher than those of SARS-CoV-1 RBD and ACE2 under similar loading rates. SMD results indicate that SARS-CoV-2 RBD interacts with the N-linked glycan on Asn90 of ACE2. This interaction is mostly absent in the SARS-CoV-1 RBD-ACE2 complex. During the SMD simulations, the extra RBD-N-glycan interaction contributes to a greater force and prolonged interaction lifetime. The observation is confirmed by our experimental force spectroscopy study. After removing N-linked glycans on ACE2, its mechanical binding strength with SARS-CoV-2 RBD decreases to a similar level of the SARS-CoV-1 RBD-ACE2 interaction. Together, the study uncovers the mechanism behind the difference in ACE2 binding between SARS-CoV-2 and SARS-CoV-1 and could help develop new strategies to block SARS-CoV-2 entry.
Significance
This study utilizes combined single-molecule force spectroscopy and steered molecular dynamics simulation approaches to quantify the specific interactions between SARS-CoV-2 or SARS-CoV-1 receptor-binding domain and human ACE2. The study reveals the mechanism behind the difference in ACE2 binding between SARS-CoV-2 and SARS-CoV-1 and could help develop new strategies to block SARS-CoV-2 entry.
Introduction
Coronavirus disease 2019 (COVID-19) is a highly contagious infectious disease caused by severe acute respiratory syndrome CoV 2 (SARS-CoV-2) (1). First reported in Wuhan, China in December 2019, COVID-19 has rapidly spread to the entire world and become a devastating pandemic. As of February 2021, COVID-19 has infected over 100 million individuals and caused over two million deaths.
Coronaviruses (CoVs) are enveloped, positive-sense RNA viruses that belong to the family Coronaviridae (2). They are classified into four genera (α, β, γ, and δ). Both SARS-CoV-2 and SARS-CoV-1 (which caused the 2002–2004 outbreak) belong to the β-CoV genus. The genomes of SARS-CoV-2 and SARS-CoV-1 share 76% sequence identity (3). Both genomes encode four structural proteins: spike (S), envelope, membrane, and nucleocapsid. The membrane protein maintains the viral lipid membrane integrity. The envelope protein facilitates the virus’s assembly and release, and the nucleocapsid protein encapsulates and protects the viral genome (4).
The S protein (∼150 kDa) is a heavily N-linked glycosylated homotrimer projecting 20 nm from the CoV’s surface (2). The trimeric S glycoprotein is a class I fusion protein and mediates attachment to the host receptor. The S1 portion contains the large receptor-binding domain (RBD), and the S2 portion forms the S molecule’s stalk. The atomic structures of the SARS-CoV-2 S protein in a trimeric form (5), as well as the RBD-receptor complex, have been determined (6). These structures are similar to the previously reported structures of SARS-CoV-1 S protein (7, 8, 9), indicating that the two proteins might function similarly.
Angiotensin-converting enzyme 2 (ACE2) is a known receptor for SARS-CoV-1 and SARS-CoV-2 S proteins. The primary physiological function of ACE2 is to hydrolyze angiotensin II (a vasoconstrictor) into angiotensin-(1–7) (a vasodilator) and thereby to lower blood pressure (10,11). ACE2 is a type I transmembrane protein expressed in various organs, including the lungs, heart, kidneys, and intestine (12,13). Recent structural studies show that ACE2 is a homodimer with each monomer consisting of an N-terminal peptidase domain, a C-terminal collectrin-like domain, a single-pass transmembrane region, and a short cytoplasmic region (9). The RBD-binding region on ACE2 is located in its N-terminal peptidase domain with major contact regions located in the α1 and α2 helixes, as well as the linker between β3 and β4 strands (9).
The binding interactions between ACE2 and CoV S proteins have been widely studied recently. Although there are variations among different binding assays reported, most reports show a higher binding affinity between ACE2 and SARS-CoV-2 S than the binding between ACE2 and SARS-CoV-1 S (14,15). However, the mechanism behind such a difference is still unclear. Little is known about the biomechanical strength of ACE2-S interaction that drives viral adhesion and helps withstand the force exerted during viral entry.
In this work, using atomic force microscopy (AFM)-based single-molecule force spectroscopy, a method in which a single bond rupture (i.e., interaction) between two molecules can be measured directly, we have quantified the mechanical strengths between ACE2 and SARS-CoV-1 RBD (RBDCoV1) or SARS-CoV-2 RBD (RBDCoV2). As AFM can measure forces in the pico-Newton (pN) range, it is possible to detect intermolecular forces and allow for weak interactions between tip-bound ligands and surface-bound receptor molecules to be quantified in terms of their affinities and rate constants (16). Furthermore, AFM has been recently adopted by us and others to study the interactions between viruses and host cells (17, 18, 19, 20). We also used all-atom steered molecular dynamics (SMD) simulations to pull the RBDCoV1-ACE2 or RBDCoV2-ACE2 complexes with or without N-glycans. Both AFM and SMD confirmed a stronger force/energy associated with the dissociation of the RBDCoV2-ACE2 complex. This enhanced mechanical strength stems from an additional interaction of RBDCoV2 with an N-linked glycan of ACE2 Asn90.
Materials and methods
Protein constructs
Immortalized human embryonic kidney (HEK) 293T cells purchased from American Type Culture Collection (ATCC; Manassas, VA) were cultured in DMEM medium (American Type Culture Collection) and supplemented with 4 mM L-glutamine, 4500 mg/L glucose, 1 mM sodium pyruvate, 1500 mg/L sodium bicarbonate, 1% penicillin-streptomycin, and 10% fetal bovine serum. RBD proteins were expressed as previously described with some modifications (21). Briefly, genes encoding RBDCoV1 (residues 318–510) and RBDCoV2 (residues 331–524) proteins containing a C-terminal Fc tag were amplified by PCR using codon-optimized SARS-CoV-1 (GenBank accession: AY278488.2) or SARS-CoV-2 S (GenBank accession: QHD43416.1) plasmid and inserted into pFUSE-hIgG1-Fc expression vector (Invitrogen, Carlsbad, CA). The recombinant RBD plasmids were transiently transfected into HEK293T cells through the calcium phosphate transfection method. The RBD proteins were expressed in the culture supernatants and purified by protein A affinity chromatography following the manuscript’s instructions (GE Healthcare, Chicago, IL). The purity of the protein was >90%, as determined by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE).
The ACE2 protein was purchased from ACRObiosystems (catalog AC2-H52H8; Newark, DE). It is a recombinant N-terminal His-tagged protein consisting of Gln18-Ser740 of human ACE2 (GenBank accession: AF291820.1), expressed from HEK293 cells. According to the manufacturer, the protein had a purity of ∼95%, as determined by SDS-PAGE. It has been shown to bind the SARS-CoV-2 S protein with high affinity (22). The Middle East respiratory syndrome CoV (MERS-CoV) RBD was from Sino Biological (catalog 40071-V08B1; Wayne, PA). It is a recombinant C-terminal His-tagged protein consisting of Glu367-Tyr606 of MERS-CoV S protein (GenBank accession: AFS88936.1) (23). The purity of the protein was >90%, as determined by SDS-PAGE.
Cantilever preparation/coverslip preparation
To functionalize AFM cantilevers (MLCT-BIO-DC; Bruker Nano, Billerica, MA) with RBD, the cantilever was first silanized with (3-aminopropyl)-triethoxysilane. RBDCoV1, RBDCoV2, or MERS-CoV (RBDMERS-CoV, as a negative control) at 1 μM were immobilized onto the silanized cantilever using a heterobifunctional polyethylene glycol (PEG) cross-linker, Acetal-PEG-NHS (2000 MW; Creative PEGworks, Durham, NC), according to the detailed protocol developed by Dr. Hermann J. Gruber of Johannes Kepler University (https://www.jku.at/fileadmin/gruppen/216/03_AFM_tip_aminofunctionalization_2016_05_06.pdf (19,24)). A 5000-MW Acetal-PEG-NHS (Creative PEGworks) was also used as a control study. Soluble recombinant ACE2 (1 μM) was attached to the (3-aminopropyl)-triethoxysilane-silanized glass coverslips (NovaScan, Chicago, IL) using the same cross-linking approach. Functionalized cantilevers and glass surfaces were stored in phosphate-buffered saline (3 × 5 min) and used for the AFM experiment within 8 h.
Single-molecule force measurements
All single-molecule force measurements were conducted using a custom-designed AFM apparatus (25,26) used previously to quantify viral protein-receptor interactions (27,28). AFM measurements were collected at cantilever retraction speeds ranging from 0.19 to 7.5 μm/s to achieve the desired loading rate (5000–20,000 pN/s). All measurements were conducted at 25°C in phosphate-buffered saline (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 1.8 mM KH2PO4 (pH 7.4)).
Calibration of the cantilevers (lever C of MLCT) was done by first measuring the inverse optical lever sensitivity via recording force curves on a hard glass surface, followed by obtaining the spring constant of the cantilever via thermally induced fluctuations (29). The spring constants (9.6 ± 2.8 pN/nm, mean ± SD) of the calibrated cantilevers agreed with the values specified by the manufacturer (10 pN/nm). All the unbinding rupture forces have been corrected for viscous drag force (30,31), which was obtained by multiplying the tip movement velocity by the viscous drag coefficient. The viscous drag coefficient was measured by moving the cantilever at varying velocities near the substrate (30,32) and is 4 pN⋅s/μm for the lever C of MLCT.
To enable measurement of a single-molecular interaction, the contact between the cantilever tip and the substrate was minimized by reducing both the contact duration (<100 ms) and the contact force (100–200 pN). The brief contact duration was chosen to ensure that, for the majority of contacts (70% or greater), no adhesion (rupture force) was observed between the AFM tip and surface. Assuming the adhesion bond formation obeyed Poisson statistics, an adhesion frequency of ∼30% in the force measurements implies that among the observed unbinding events, the probabilities of forming a single, double, and triple adhesion bonds between AFM tip and surface were 83, 16, and 1, respectively (33). Therefore, our experimental condition ensured a >83% probability that the adhesion event was mediated by a single bond (34). Loading rates of the rupture forces were determined from each unbinding force curve by conducting a linear fit to the force-time curve shortly (the last 50 data points) before rupture (35).
Statistical analysis
300–500 force curves were typically recorded for each pulling speed, which yielded 60–100 unbinding forces with an adhesion frequency of 25–30%. At least four independent cantilevers were used to conduct the measurement of each ligand-receptor pair. Curve fitting was performed using IGOR Pro or Origin software by minimizing the chi-square statistic for the optimal fit. Unless otherwise stated, the data are reported as the mean and the standard error of the estimate. Statistical analyses between groups were performed using an unpaired t-test or ANOVA using the Prism software, with a p-value less than 0.05 considered statistically significant.
SMD simulation
All SMD simulations were performed using NAMD (36). The CHARMM36(m) (37,38) force field was used for protein and carbohydrates. PDB: 2AJF (7) from Protein Data Bank (PDB) was used for an RBDCoV1-ACE2 complex structure and PDB: 6VW1 (6) for an RBDCoV2-ACE2 complex structure. We used a TIP3P water model (39), and K+ and Cl− ions with a concentration of 0.15 M were added to neutralize the system. All simulation systems and parameters were set up through CHARMM-GUI Solution Builder (40,41). The analysis was done with CHARMM (42) and visualization through VMD (43).
PDB: 6VW1 has five N-linked glycans in ACE2 (Asn53, Asn90, Asn103, Asn322, and Asn549) and one N-glycan in RBDCoV2 (Asn343), and PDB: 2AJF has four N-linked glycans in ACE2 (Asn53, Asn90, Asn322, and Asn549) and one N-glycan in RBDCoV1 (Asn330). Similar to other crystal or cryo-electron microscopy structures, all N-glycan structures in both PDB structures are incomplete as they are truncated in the experiment or not observable because of low resolution and high structural flexibility. Because we did not know the glycoforms of the ACE2 glycosylation sites at the time of this study, we used the N-glycan core pentasaccharide (a minimal structure of all N-glycans: Fig. S1) in all N-glycosylation sites, including Asn103 of ACE2 in PDB: 2AJF. Glycan Reader & Modeler (44, 45, 46) in CHARMM-GUI was used to model N-glycan core pentasaccharide in all glycosylation sites using the templates from Glycan Fragment Database (47). To compare the receptor-binding affinity between RBDCoV1 and RBDCoV2 and to explore the influences of N-glycans on binding affinity, we made four systems: SCoV1+G (RBDCoV1-ACE2 with N-glycans), SCoV1−G (RBDCoV1-ACE2 without N-glycans), SCoV2+G (RBDCoV2-ACE2 with N-glycans), and SCoV2−G (RBDCoV2-ACE2 without N-glycans).
For the SMD simulations, the protein complex structures were initially aligned along the x axis in a cubic water box with an initial size of 171 Å for SCoV1 ± G and 172 Å for SCoV2 ± G; a total number of atoms is ∼470,000. The pulling forces were applied to the center of mass (COM) of each protein (i.e., RBD and ACE2). In the pulling process, the spring constant was set to 5 kcal/mol/Å2 and its moving speed to 0.5 Å/ns in the opposite directions along the x axis. Gentle restrains with a force constant of 5 kcal/mol/Å2 were applied to each protein’s COM to restrict their movement along with the y/z directions during the pulling process. The SMD simulations stopped at 30 ns when two proteins were detached from each other. 9 independent simulations for each system were performed for better statistics.
The van der Waals interactions were smoothly switched off over 10–12 Å by a force-based switching function (48). The electrostatic interactions were calculated by the particle-mesh Ewald method with a mesh size of 1 Å for fast Fourier transformation and sixth-order B-spline interpolation. SHAKE algorithm was used to constrain bond lengths involving hydrogen atom (49), and the simulation time step was set to 2 fs. We first relaxed the system in NVT (constant particle number, volume, and temperature) ensemble at 303.15 K with harmonic restraints to all solute atoms. The constant temperature was controlled by Langevin dynamics with a damping frequency of 50 fs−1. 100–120 ps NPT (constant particle number, pressure, and temperature) simulation was then applied to adjust the solvent density. The Langevin piston method was used to control the pressure. A dihedral restraint with a force constant of 1 kcal/mol/rad2 was applied to carbohydrates to keep the carbohydrate chair conformation during these equilibration steps. To perform the SMD simulation, a COLVARS method was used (50), and the COMs of two proteins were calculated first and used as the external forces’ initial positions. The effective spring potential (whose negative derivative is used to represent external forces) acting on the COM of each protein was calculated using the following equation: , where k is the spring constant, v is the moving speed of the spring potentials, R(t) is the current position of the selected protein COM, and n is the unit vector along the protein COMs. As a result of this spring potential, the spring-connected protein would move following the energy well, so that two proteins are pulled apart.
Results
RBD of SARS-CoV-2 S protein binds ACE2 stronger compared to SARS-CoV-1
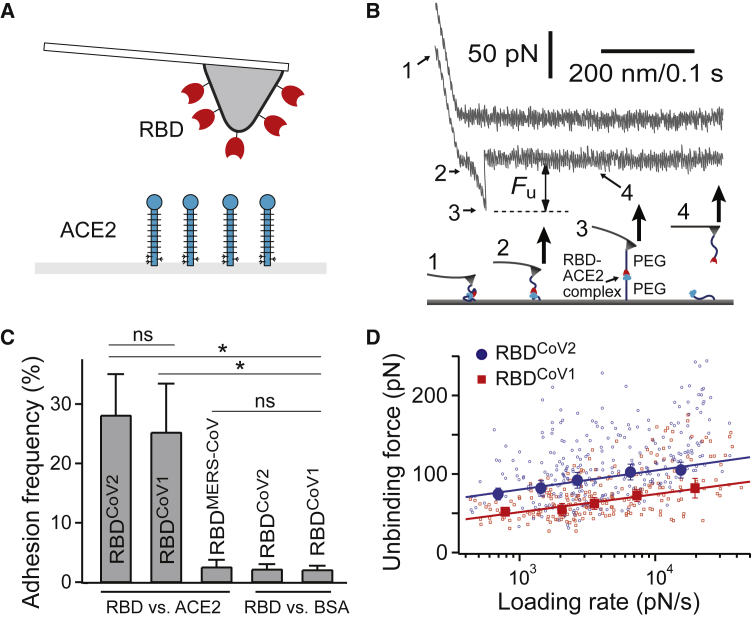
First, we characterized the mechanical interaction between the RBD proteins and ACE2 using AFM. We have attached RBDCoV1, RBDCoV2, or RBDMERS-CoV (negative control) to a microcantilever, the force probe, via an established protocol using PEG coupling chemistry (19,20). The cantilever-bound RBD was brought to interact with surface-immobilized soluble ACE2 via AFM force scans (Fig. 1 A).
Figure 1.
Single-molecule studies of CoV RBD-ACE2 interactions. (A) Shown is a schematic of the experimental system. The microcantilever is functionalized with RBD. Soluble human ACE2 is immobilized on the opposing surface using established protocols. The molecules are not drawn to scale. (B) The upper panel shows two sample AFM force-piezo displacement/time traces of the RBDCoV2-ACE2 interaction. The first (upper) trace had no interaction, and the second (lower) trace shows the rupture force of the protein-protein complex. Fu is the unbinding force. The loading rate was measured by conducting a linear fit to the force-time curve shortly (the last 50 data points) before rupture (35). The lower panel illustrates the four stages of stretching and rupturing a single RBD-ACE2 complex using the AFM. (C) Interaction specificity was demonstrated by the adhesion frequency measurement for different interacting pairs (N = 6, 10, 6, 5, and 5 for the RBDCoV2-ACE2, RBDCoV1-ACE2, RBDMERS-CoV-ACE2, RBDCoV2-BSA, and RBDCoV1-BSA groups, respectively). Contact force, contact time, and retraction speed for all the interacting AFM tips and surfaces were set at 150 pN, 0.1 s, and 3.7 μm/s, respectively. Error bars are SD. ∗: significant differences (p < 0.001) against each control group were determined by unpaired t-tests. ns: statistically insignificant was determined by unpaired t-tests. (D) The dynamic force spectra (i.e., the plot of most probable unbinding force (Fu∗)) is shown as a function of loading rate (rf) of the RBD-ACE2 interactions. The data are fitted to the single-barrier Bell-Evans model (Eq. 3) to extract the off-rate k0 (51). The bars denote half bin-widths of the unbinding force histograms (shown in Fig. S4), representing the force determination error. Individual data points of RBDCoV2-ACE2 (N = 305) and RBDCoV1-ACE2 (N = 245) unbinding forces were plotted as scatter plots, using smaller symbols and the same color scheme. To see this figure in color, go online.
All single-molecule force measurements were conducted using a custom-built AFM designed for operation in the force spectroscopy mode (25, 26, 27,52,53). Using a piezoelectric translator, the functionalized cantilever was lowered onto an ACE2-functionalized surface to allow possible binding between RBD and ACE2 to occur. After a brief contact, the cantilever was retracted from the surface. Any binding interaction between tip and substrate would lead to an adhesive pull-off force determined from the cantilever’s deflection via a position-sensitive two-segment photodiode. Fig. 1 B shows two typical pulling traces. The first (upper) trace represents a majority (∼70%) of all the pulling curves, indicating no interaction (i.e., no adhesive force) between the AFM tip and sample surface. The second (lower) trace, representing ∼30% of the pulling curves in our single-molecule assay, shows the unbinding (i.e., pull-off) force of the tip-substrate interaction.
Interaction specificity was demonstrated by the adhesion frequency measurement under the same measurement conditions. Fig. 1 C shows a significant decrease in adhesion when either RBDCoV1, RBDCoV2, or ACE2 was absent, confirming that the vast majority of the recorded unbinding forces stemmed from specific interactions. RBDMERS-CoV was also used here as negative control proteins, as RBDMERS-CoV does not bind ACE2 and its known receptor is dipeptidyl peptidase-4 (54). Indeed, cantilever tips functionalized with RBDMERS-CoV yielded almost no adhesion (∼2% adhesion frequency) on an ACE2-functionalized surface. Fig. S2 contains additional pulling traces, showing an RBDCoV2-functionalized AFM tip having no adhesion on a bovine serum albumin (BSA) substrate but displaying adhesion (2 out of 10) on an ACE2 substrate.
To validate the PEG linker used in this study, we analyzed the force-extension property of the unbinding curves using the worm-like chain (WLC) model (Fig. S3; (55,56)). In this work, the molecular tether being stretch consists of the RBD-ACE2 complex in the middle, flanked by two pieces of PEG linkers of 2000 MW (PEG2000) (Fig. 1 B, lower panel). Fitting the force-extension curve of RBDCoV2-ACE2 interaction to the WLC model yielded a contour length of 45 ± 11 nm (average ± SD), consistent with the predicted 37-nm contour length of the molecular tether (Fig. S3). The WLC fit also yielded a persistence length of 0.52 ± 0.15 nm (average ± SD), which is consistent with the persistence length of PEG linker or polypeptide (∼0.4 nm) (57). Moreover, we conducted a control study using a longer PEG linker of 5000 MW, PEG5000. Using the RBDCoV2-ACE2 system, we found that the contour length for the PEG5000 experimental group increased by 2.2-fold to 99 ± 14 nm, whereas the persistence length remained at a similar range at 0.48 ± 0.09 nm. Together, the data shown in Fig. S3 validate the PEG linker used in the study and suggests that the unbinding events indeed occur between the proteins coupled to the two PEG linkers’ termini.
The biophysical properties of RBD-ACE2 interactions were studied through a dynamic force spectrum (DFS), the plot of the most probable unbinding force as a function of the loading rate. The unbinding forces of each RBD-ACE2 interaction were first grouped into five groups by their loading rates. The distribution of forces within the same group was analyzed by histograms (see Fig. S4). The most probable unbinding forces were then recorded as the center of the tallest bin of each histogram. Fig. 1 D shows that the unbinding force of both RBD-ACE2 complexes increased linearly with the logarithm of the loading rate. However, the unbinding forces of RBDCoV2-ACE2 are stronger, ranging from 70 to 105 pN over a loading rate of 500–20,000 pN/s, whereas the RBDCoV1-ACE2 unbinding forces are 30–40% lower under similar loading rates.
A more detailed analysis of the biophysical properties of RBD-ACE2 interactions was conducted by fitting the acquired DFS data to the Bell-Evans model (58). According to this model, a pulling force (F) distorts the intermolecular potential of a receptor-ligand complex, leading to a lowering of the activation energy and an increase in the dissociation rate k(f) as follows:
| (1) |
where k0 is the dissociation rate constant in the absence of a pulling force, γ is the position of the transition state, T is the absolute temperature, and kB is the Boltzmann constant. If the applied force increases linearly with a loading rate (RF) (such as in the case of our AFM unbinding experiment), the probability for protein-protein unbinding as a function of the force f is given by:
| (2) |
The most probable unbinding force F∗ at a given loading rate RF can then be written as:
| (3) |
Hence, as predicted by the model, the most probable unbinding force F∗ is a linear function of the logarithm of the loading rate. Experimentally, F∗ was determined from the unbinding force histograms. Fitting the DFS of RBDCoV2-ACE2 interaction to the Bell-Evans model (Eq. 3) yielded a k0 of 0.047 s−1 and a γ (i.e., activation barrier width) of 0.39 nm. The best-fit parameters for RBDCoV2-ACE2 and RBDCoV1-ACE2 interactions are summarized in Table 1. Clearly, compared to RBDCoV1, RBDCoV2 binds ACE2 with a 12-fold smaller k0 and a similar γ, indicating that the RBDCoV2-ACE2 interaction is stronger.
Table 1.
Bell-Evans model parameters of RBD-ACE2 interactions
| Receptor-ligand pairs | k0 (s−1) | γ (nm) |
|---|---|---|
| RBDCoV2 versus ACE2 | 0.047 ± 0.034 | 0.39 ± 0.05 |
| RBDCoV1 versus ACE2 | 0.57 ± 0.39 | 0.41 ± 0.05 |
| RBDCoV2 versus dgACE2 | 0.82 ± 0.72 | 0.42 ± 0.06 |
| RBDCoV1 versus dgACE2 | 3.4 ± 2.2 | 0.37 ± 0.06 |
Uncertainties are the standard errors of the fits. dgACE2 represents deglycosylated ACE2 treated with PNGase F.
SMD identifies an additional interaction of SARS-CoV-2 S RBD with an N-linked glycan of ACE2 Asn90
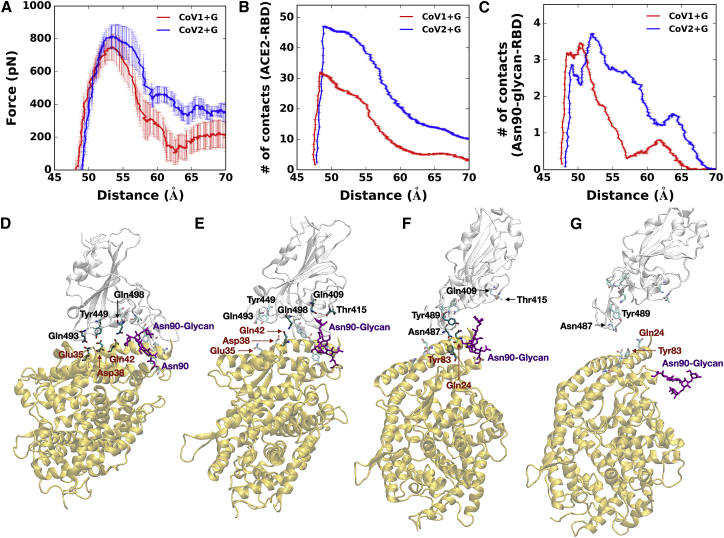
To gain molecular insight into the receptor-binding affinity between RBDCoV1 and RBDCoV2 and to explore influences of N-glycans on binding affinity, we performed SMD simulations on the following four systems: SCoV1+G (RBDCoV1-ACE2 with N-glycans), SCoV1−G (RBDCoV1-ACE2 without N-glycans), SCoV2+G (RBDCoV2-ACE2 with N-glycans), and SCoV2−G (RBDCoV2-ACE2 without N-glycans). To compare RBDCoV2-ACE2 interactions with RBDCoV1-ACE2 interactions, pulling force analysis was performed as a function of distance (DRBD-ACE2) between the COMs of RBD and ACE2. Also, to investigate how many residues between RBD and ACE2 interact as a function of DRBD-ACE2, the number of contacts analysis was performed. A contact was counted if any heavy atom of RBD was within 4.5 Å from any heavy atom of ACE2.
As shown in Fig. 2 A, the overall force profile of SCoV2+G shows higher forces than SCoV1+G because of greater numbers of RBDCoV2-ACE2 contacts compared to RBDCoV1-ACE2 (Fig. 2 B); see Fig. S5 for the force profiles of individual replicas. Initially, SCoV2+G has more contacts than SCoV1+G, and the difference in the number of contacts between SCoV2+G and SCoV1+G is ∼20 at DRBD-ACE2 of 52 Å (Fig. 2 B). The difference decreases to ∼17 starting from 55 Å and to 9 at 65 Å, in which ACE2 Asn90-glycan maintains its interactions with RBDCoV2, whereas such interactions are lost in SCoV1 +G (Fig. 2 C). Note that the force profile in SCoV2+G has a plateau around 60 Å and a small peak around 66 Å, which are attributed to the interactions between ACE2 Asn90-glycan and RBDCoV2 from 55 to 65 Å (Fig. 2 C). Because of relatively negligible interactions between ACE2 Asn90-glycan and RBDCoV1, the plateau is not observed around 60 Å in SCoV1+G. Fig. S6 shows representative snapshots of SCoV1+G during the pulling simulation at the initial state (Fig. S6 A) and DRBD-ACE2 of 57 Å (Fig. S6 B). It shows that Asn90-glycan of SCoV1+G barely interacts with RBDCoV1, whereas Asn90-glycan of SCoV2+G still contacts with RBDCoV2 at the same distance (Fig. 2 E; Fig. S6 B). This result indicates that the interaction between ACE2 Asn90-glycan and RBDCoV2 somewhat blocks the direct contact between RBDCoV2 and ACE2 at 55 Å < DRBD-ACE2 < 65 Å, suggesting that ACE2 Asn90-glycan can hinder the association of RBDCoV2 to ACE2 more than RBDCoV1 but makes RBDCoV2-ACE2 dissociation harder than RBDCoV1-ACE2.
Figure 2.
SMD simulation results. (A) Shown are the average force profiles of SCoV1+G (red) and SCoV2+G (blue) as a function of distance (DRBD-ACE2) between the centers of mass of RBD and ACE2. (B) Shown are the average numbers of contacts between RBD and ACE2 in SCoV1+G (red) and SCoV2+G (blue). (C) Shown are the average numbers of contacts between RBD and ACE2 Asn90-glycan in SCoV1+G (red) and SCoV2+G (blue). In (A–C), the average data are obtained based on 9 independent SMD simulations for each system, and error bars represent the SD with 68% confident intervals. (D–G) Shown are representative snapshots of SMD simulations of SCoV2+G at DRBD-ACE2 of (D) 49 Å, (E) 57 Å, (F) 65 Å, and (G) 70 Å. Key interacting residues are depicted as the solid sticks, and residues losing their interactions are shown as the transparent sticks. The black residue names are for RBDCoV2 and brown ones for ACE2. The RBDCoV2 and ACE2 are shown by transparent light gray and yellow, respectively. Asn90-glycan is colored in purple. To see this figure in color, go online.
Using SCoV2+G as an example, the overall RBD and ACE2 dissociation during the pulling simulation can be divided into three states: state I (<55 Å, Fig. 2 D), state II (56∼70 Å, Fig. 2, E and F), and state III (>70 Å, Fig. 2 G). In state I, RBDCoV2-ACE2 has a number of interactions. As DRBD-ACE2 increases to 56 Å (state II), RBDCoV2 and ACE2 start to lose some of its polar interactions (RBDCoV2-ACE2: Gln493-Glu35 and Tyr449-Asp38), but the interaction between Gln498 and Glu42 is intact. Note that ACE2 Asn90-glycan has polar interactions with Gln409 and Thr415 (Fig. 2 E). At 65 Å (Fig. 2 F), Asn487 and Try489 of the RBDCoV2 loop can still interact with ACE2 Tyr83 and Gln24 because of the flexibility of the loop, and Asn487 can also contact Gln24 from time to time. At this period, Asn90-glycan loses its contacts with RBDCoV2. In state III, RBDCoV2 and ACE2 are fully detached with no close interactions (Fig. 2 G). Although the average forces show a subtle difference inbetween SCoV1+G and SCoV1−G when RBDCoV1 and ACE2 start to detach at DRBD-ACE2 = 56 Å (Fig. S7 A), SCoV2+G has higher forces over 56 Å to 70 Å than SCoV2−G (Fig. S7 B). RBDCoV2 shows slightly higher forces than RBDCoV1, even with no glycans (Fig. S7 C).
Removal of ACE2 N-linked glycans leads to a decrease in unbinding forces
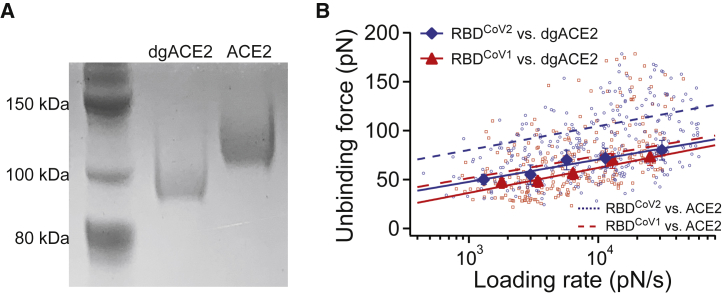
In light of the SMD results, we tested the effect of ACE2 N-linked glycan on the mechanical strength of RBD-ACE2 interactions. To remove the ACE2 N-linked glycans, surface-immobilized ACE2 was incubated with PNGase F (New England BioLabs, Ipswich, MA) for 1 h at 37°C. The effect of PNGase F treatment was analyzed by SDS-PAGE (Fig. 3 A). After 1 h of treatment, the molecular weight of ACE2 was visibly reduced from ∼115 to 95 kDa. Assuming each N-linked glycosylation adds 2.5 kDa of molecular mass, the result is consistent with seven N-glycosylation sites on ACE2.
Figure 3.
Effect of ACE2 N-glycans on RBD-ACE2 interaction. (A) Deglycosylation (dgACE2) was performed by treatment with PNGase F for 1 h at 37°C. Deglycosylation was confirmed via SDS-PAGE stained with EZBlue. (B) The dynamic force spectra of the RBD-dgACE2 interactions are shown. Solid lines are linear fits to Eq. 3 for the interactions. The dashed line is the linear fits for N-glycosylated ACE2 interactions taken from Fig. 1. The bars denote half bin-widths of the unbinding force histograms (see Fig. S4), representing the errors of force determination. Individual data points of RBDCoV2-dgACE2 (N = 346) and RBDCoV1-dgACE2 (N = 260) unbinding forces were plotted as scatter plots, using smaller symbols and the same color scheme. To see this figure in color, go online.
Next, AFM unbinding experiments were performed between tip-immobilized RBD and surface-immobilized, PNGase F-treated ACE2. As shown in Fig. 3 B, N-linked glycan removal resulted in a significant decrease of the unbinding forces between RBDCoV2 and ACE2, from 70–105 to 50–70 pN. The unbinding forces between RBDCoV1 and ACE2 also decreased but to a lesser extent. The DFS of RBDCoV2 and RBDCoV1 are almost overlapped with each other. This trend is also similar to the SMD results, showing that the force profiles of SCoV1−G and SCoV2−G are within the error bars (Fig. S7 C). The Bell-Evans model fit confirmed that after N-glycan removal, the k0 of RBDCoV2-ACE2 interaction increases by 17-fold (from 0.047 to 0.82 s−1), and the k0 of RBDCoV1-ACE2 interaction increases by only sixfold.
Discussion
Interactions between the viral protein and host receptors require direct physical contact between viral and host cell membranes. Unlike interactions in solution (three-dimensional (3D)), which have at least one interacting molecular species in the fluid phase, the interactions between receptors and ligands anchored on two opposing membranes (two-dimensional (2D)) are constrained in molecular movement or transport and are under common tensile force. Hence, the 2D reaction kinetics may be different from 3D kinetics (59,60). To study the mechanism underlying virus-cell interaction, it is necessary to probe the interaction between anchored molecules using 2D binding assays such as the single-molecule AFM used in this study. Using this method, we found that the dissociate rate for RBDCoV2-ACE2 and RBDCoV1-ACE2 bonds (or interactions) are significantly different. The one order magnitude slower dissociation rate could partially account for the greater infectivity of SARS-CoV-2.
In a recent study, Shang et al. used surface plasmon resonance, a technique detecting the 3D binding kinetics of bulk molecules, to measure the binding kinetics of RBD-ACE2 interactions. They reported the dissociate rate for RBDCoV2-ACE2 and RBDCoV1-ACE2 interactions to be 0.008 and 0.04 s−1, respectively (6). These two values are qualitatively agreed with our AFM results, i.e., k0 values of 0.047 ± 0.034 and 0.57 ± 0.39 s−1 for RBDCoV2-ACE2 and RBDCoV1-ACE2 interactions, respectively, suggesting that our 2D binding assay is overall agreeable with the 3D bulk assay. Also, RBDCoV2-ACE2 interaction was studied recently using AFM-based force spectroscopy by Yang et al., who reported a k0 value of 0.009 ± 0.006 s−1 (61). This k0 value is also agreeable (within fitting error) with the k0 value reported in Table 1. In addition, the unbinding forces of RBDCoV2-ACE2 interactions shown in Fig. 1 D are relatively larger than those reported by Yang et al. This difference could be due to the different ACE2 variants being studied; whereas Yang et al. studied a Rhesus ACE2 protein, a human ACE2 was used in the current study.
Using the k0 values from Table 1, we can estimate the activation energy differences among different RBD-ACE2 complexes. According to the traditional transition state theory, the dissociation rate of a reaction can be described as koff = α⋅Exp(−ΔG∗/kBT), where kB, T, and ΔG∗ are the Boltzmann constant, absolute temperature, and activation energy, respectively, and α is a prefactor. Assuming all the RBD-ACE2 interactions have the same α value, estimates of the difference between the transition state energies can be calculated as ΔG12 = ΔG∗2−ΔG∗1 = −kBT ln(k1/k2), where k1 and k2 are the k0 values of two RBD-ACE2 interactions used for comparison, respectively. Using this equation, the activation energy for RBDCoV1-ACE2 dissociation is estimated to be 2.5 kBT lower than that of the RBDCoV2-ACE2 interaction. After deglycosylation of ACE2, the activation energy are lower by 2.9 kBT (RBDCoV2) and 1.8 kBT (RBDCoV1), compared with the unbinding of glycosylated ACE2, suggesting that ACE2 deglycosylation has a greater effect on RBDCoV2 binding than on the RBDCoV1.
SMD simulations provide molecular-level insight into RBDCoV-ACE2 interactions and help us to interpret the AFM data. The SMD simulations manifest that RBDCoV2 interacts stronger with ACE2 than RBDCoV1 because the former has more direct contacts with ACE2 than the latter. In particular, ACE2 Asn90-glycan appears to have an important role in having stronger interactions with RBDCoV2 than RBDCoV1 by retaining contacts with residues of RBDCoV2, Gln409, and Thr415, even when the original contacts of RBDCoV2-ACE2 start to lose (Fig. 2 E). This additional interaction implies that ACE2 Asn90-glycan can have effects on the association and dissociation of RBDCoV2-ACE2. In other words, ACE2 Asn90-glycan could hinder the association of RBDCoV2 with ACE2 more than RBDCoV1 but make RBDCoV2-ACE2 dissociation harder than RBDCoV1-ACE2. In addition, based on the SMD simulations, we propose a three-step dissociation mechanism of RBDCoV2-ACE2 complex.
It should be noted that the current models utilize only RBD out of trimeric SARS-CoV-2 S protein and the N-glycan core structure for all N-glycans. Having a fully glycosylated SARS-CoV-2 S protein and ACE2 models would provide further insight into the RBD-ACE2 interactions. With a recently modeled fully glycosylated SARS-CoV-2 S protein model (62) and recently determined glycosylation patterns of ACE2 (63), we plan to study the RBD-ACE2 interactions in a more realistic model.
In conclusion, the study shows that the biomechanical parameters are important for RBDCoV1 and RBDCoV2 to attach to host cells. Our results suggest important viral-host cell interaction through ACE2 Asn90-glycan. Nonetheless, the potential effect of ACE2 Asn90-glycan on the transmission of COVID-19 remains to be further investigated.
Author contributions
W.C. designed the study, performed experiments, and analyzed results. C.D. and S.K. designed the study, performed simulations, analyzed results, and wrote the manuscript. D.H. and W.T. performed experiments. L.D. analyzed results and wrote the manuscript. X.F.Z. and W.I. designed the study, analyzed results, and wrote the manuscript.
Acknowledgments
This work was supported in part by a National Institutes of Health grant AI133634, an National Science Foundation grant 1804117 (to X.F.Z.), NIH GM126140, National Science Foundation DBI-1707207, and MCB-1810695 (to W.I.), NIH grant AI157975 (to L.D.), and an internal grant from Lehigh University to X.F.Z, and W.I.
Editor: Thomas Perkins.
Footnotes
Wenpeng Cao, Chuqiao Dong, and Seonghan Kim contributed equally to this study.
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2021.02.007.
Contributor Information
Wonpil Im, Email: woi216@lehigh.edu.
X. Frank Zhang, Email: xiz310@lehigh.edu.
Supporting material
References
- 1.WHO . 2020. Coronavirus.https://www.who.int/health-topics/coronavirus#tab=tab_1 [Google Scholar]
- 2.Heald-Sargent T., Gallagher T. Ready, set, fuse! The coronavirus spike protein and acquisition of fusion competence. Viruses. 2012;4:557–580. doi: 10.3390/v4040557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andersen K.G., Rambaut A., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bar-On Y.M., Flamholz A., Milo R. SARS-CoV-2 (COVID-19) by the numbers. eLife. 2020;9:e57309. doi: 10.7554/eLife.57309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wrapp D., Wang N., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shang J., Ye G., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li F., Li W., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 8.Song W., Gui M., Xiang Y. Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2. PLoS Pathog. 2018;14:e1007236. doi: 10.1371/journal.ppat.1007236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan R., Zhang Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keidar S., Kaplan M., Gamliel-Lazarovich A. ACE2 of the heart: from angiotensin I to angiotensin (1-7) Cardiovasc. Res. 2007;73:463–469. doi: 10.1016/j.cardiores.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 11.Tikellis C., Thomas M.C. Angiotensin-converting enzyme 2 (ACE2) is a key modulator of the renin angiotensin system in health and disease. Int. J. Pept. 2012;2012:256294. doi: 10.1155/2012/256294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jia H.P., Look D.C., McCray P.B., Jr. ACE2 receptor expression and severe acute respiratory syndrome coronavirus infection depend on differentiation of human airway epithelia. J. Virol. 2005;79:14614–14621. doi: 10.1128/JVI.79.23.14614-14621.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li M.-Y., Li L., Wang X.-S. Expression of the SARS-CoV-2 cell receptor gene ACE2 in a wide variety of human tissues. Infect. Dis. Poverty. 2020;9:45. doi: 10.1186/s40249-020-00662-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Q., Zhang Y., Qi J. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181:894–904.e9. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoffmann M., Kleine-Weber H., Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hinterdorfer P., Dufrêne Y.F. Detection and localization of single molecular recognition events using atomic force microscopy. Nat. Methods. 2006;3:347–355. doi: 10.1038/nmeth871. [DOI] [PubMed] [Google Scholar]
- 17.Delguste M., Koehler M., Alsteens D. Probing single virus binding sites on living mammalian cells using AFM. Methods Mol. Biol. 2018;1814:483–514. doi: 10.1007/978-1-4939-8591-3_29. [DOI] [PubMed] [Google Scholar]
- 18.Dobrowsky T.M., Rabi S.A., Wirtz D. Adhesion and fusion efficiencies of human immunodeficiency virus type 1 (HIV-1) surface proteins. Sci. Rep. 2013;3:3014. doi: 10.1038/srep03014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rankl C., Kienberger F., Hinterdorfer P. Multiple receptors involved in human rhinovirus attachment to live cells. Proc. Natl. Acad. Sci. USA. 2008;105:17778–17783. doi: 10.1073/pnas.0806451105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sieben C., Kappel C., Herrmann A. Influenza virus binds its host cell using multiple dynamic interactions. Proc. Natl. Acad. Sci. USA. 2012;109:13626–13631. doi: 10.1073/pnas.1120265109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tai W., He L., Du L. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell. Mol. Immunol. 2020;17:613–620. doi: 10.1038/s41423-020-0400-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ravichandran S., Coyle E.M., Khurana S. Antibody signature induced by SARS-CoV-2 spike protein immunogens in rabbits. Sci. Transl. Med. 2020;12:eabc3539. doi: 10.1126/scitranslmed.abc3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Steiner D.J., Cognetti J.S., Miller B.L. Array-based analysis of SARS-CoV-2, other coronaviruses, and influenza antibodies in convalescent COVID-19 patients. Biosens. Bioelectron. 2020;169:112643. doi: 10.1016/j.bios.2020.112643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wildling L., Unterauer B., Gruber H.J. Linking of sensor molecules with amino groups to amino-functionalized AFM tips. Bioconjug. Chem. 2011;22:1239–1248. doi: 10.1021/bc200099t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang X., Wojcikiewicz E., Moy V.T. Force spectroscopy of the leukocyte function-associated antigen-1/intercellular adhesion molecule-1 interaction. Biophys. J. 2002;83:2270–2279. doi: 10.1016/S0006-3495(02)73987-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang X.H., Chen A., Moy V.T. In: Protein-Protein Interactions: A Molecular Cloning Manual. Golemis E.A., editor. Cold Spring Harbor Laboratory Press; 2002. Probing ligand-receptor interactions with atomic force microscopy; pp. 241–254. [Google Scholar]
- 27.Dragovich M.A., Fortoul N., Zhang X.F. Biomechanical characterization of TIM protein-mediated Ebola virus-host cell adhesion. Sci. Rep. 2019;9:267. doi: 10.1038/s41598-018-36449-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bondu V., Wu C., Buranda T. Low-affinity binding in cis to P2Y2R mediates force-dependent integrin activation during hantavirus infection. Mol. Biol. Cell. 2017;28:2887–2903. doi: 10.1091/mbc.E17-01-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hutter J.L., Bechhoefer J. Calibration of atomic-force microscope tips. Rev. Sci. Instrum. 1993;64:1868–1873. [Google Scholar]
- 30.Janovjak H., Struckmeier J., Müller D.J. Hydrodynamic effects in fast AFM single-molecule force measurements. Eur. Biophys. J. 2005;34:91–96. doi: 10.1007/s00249-004-0430-3. [DOI] [PubMed] [Google Scholar]
- 31.Alcaraz J., Buscemi L., Navajas D. Correction of microrheological measurements of soft samples with atomic force microscopy for the hydrodynamic drag on the cantilever. Langmuir. 2002;18:716–721. [Google Scholar]
- 32.Franz C.M., Taubenberger A., Muller D.J. Studying integrin-mediated cell adhesion at the single-molecule level using AFM force spectroscopy. Sci. STKE. 2007;2007:pl5. doi: 10.1126/stke.4062007pl5. [DOI] [PubMed] [Google Scholar]
- 33.Chesla S.E., Selvaraj P., Zhu C. Measuring two-dimensional receptor-ligand binding kinetics by micropipette. Biophys. J. 1998;75:1553–1572. doi: 10.1016/S0006-3495(98)74074-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Evans E. Probing the relation between force--lifetime--and chemistry in single molecular bonds. Annu. Rev. Biophys. Biomol. Struct. 2001;30:105–128. doi: 10.1146/annurev.biophys.30.1.105. [DOI] [PubMed] [Google Scholar]
- 35.Sluysmans D., Devaux F., Duwez A.-S. Dynamic force spectroscopy of synthetic oligorotaxane foldamers. Proc. Natl. Acad. Sci. USA. 2018;115:9362–9366. doi: 10.1073/pnas.1712790115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Phillips J.C., Braun R., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hatcher E., Guvench O., Mackerell A.D. CHARMM additive all-atom force field for aldopentofuranoses, methyl-aldopentofuranosides, and fructofuranose. J. Phys. Chem. B. 2009;113:12466–12476. doi: 10.1021/jp905496e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang J., Rauscher S., MacKerell A.D., Jr. CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat. Methods. 2017;14:71–73. doi: 10.1038/nmeth.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jorgensen W.L., Chandrasekhar J., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 40.Jo S., Kim T., Im W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 41.Lee J., Cheng X., Im W. CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J. Chem. Theory Comput. 2016;12:405–413. doi: 10.1021/acs.jctc.5b00935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brooks B.R., Bruccoleri R.E., Karplus M. CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 1983;4:187–217. [Google Scholar]
- 43.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38, 27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 44.Jo S., Song K.C., Im W. Glycan Reader: automated sugar identification and simulation preparation for carbohydrates and glycoproteins. J. Comput. Chem. 2011;32:3135–3141. doi: 10.1002/jcc.21886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park S.J., Lee J., Im W. Glycan Reader is improved to recognize most sugar types and chemical modifications in the Protein Data Bank. Bioinformatics. 2017;33:3051–3057. doi: 10.1093/bioinformatics/btx358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Park S.J., Lee J., Im W. CHARMM-GUI Glycan Modeler for modeling and simulation of carbohydrates and glycoconjugates. Glycobiology. 2019;29:320–331. doi: 10.1093/glycob/cwz003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jo S., Im W. Glycan fragment database: a database of PDB-based glycan 3D structures. Nucleic Acids Res. 2013;41:D470–D474. doi: 10.1093/nar/gks987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dion M., Rydberg H., Lundqvist B.I. van der Waals density functional for general geometries. Phys. Rev. Lett. 2004;92:246401. doi: 10.1103/PhysRevLett.92.246401. [DOI] [PubMed] [Google Scholar]
- 49.Ryckaert J.-P., Ciccotti G., Berendsen H.J.C. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- 50.Bernardi R., Bhandarkar M., Zhu F. 2018. NAMD user’s guide, Version 2.13.http://www.ks.uiuc.edu/Research/namd/2.13/ug/ [Google Scholar]
- 51.Zhang X., Rico F., Moy V. In: Handbook of Single-Molecule Biophysics. Hinterdorfer P., van Oijen A., editors. Springer Press; 2009. Atomic force microscopy of protein-protein interactions; pp. 555–570. [Google Scholar]
- 52.Zhang X., Craig S.E., Moy V.T. Molecular basis for the dynamic strength of the integrin alpha4beta1/VCAM-1 interaction. Biophys. J. 2004;87:3470–3478. doi: 10.1529/biophysj.104.045690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang S., Wu C., Chen J. Integrin α4β7 switches its ligand specificity via distinct conformer-specific activation. J. Cell Biol. 2018;217:2799–2812. doi: 10.1083/jcb.201710022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Raj V.S., Mou H., Haagmans B.L. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Milstein J.N., Meiners J.-C. In: Encyclopedia of Biophysics. Roberts G.C.K., editor. Springer; 2013. Worm-like chain (WLC) model; pp. 2757–2760. [Google Scholar]
- 56.Tong Z., Mikheikin A., Lyubchenko Y.L. Novel polymer linkers for single molecule AFM force spectroscopy. Methods. 2013;60:161–168. doi: 10.1016/j.ymeth.2013.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kienberger F., Pastushenko V.P., Hinterdorfer P. Static and dynamical properties of single poly(ethylene glycol) molecules investigated by force spectroscopy. Single Mol. 2000;1:123–128. [Google Scholar]
- 58.Evans E., Ritchie K. Dynamic strength of molecular adhesion bonds. Biophys. J. 1997;72:1541–1555. doi: 10.1016/S0006-3495(97)78802-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dustin M.L., Bromley S.K., Zhu C. Identification of self through two-dimensional chemistry and synapses. Annu. Rev. Cell Dev. Biol. 2001;17:133–157. doi: 10.1146/annurev.cellbio.17.1.133. [DOI] [PubMed] [Google Scholar]
- 60.Dustin M.L., Zhu C. T cells like a firm molecular handshake. Proc. Natl. Acad. Sci. USA. 2006;103:4335–4336. doi: 10.1073/pnas.0600899103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang J., Petitjean S.J.L., Alsteens D. Molecular interaction and inhibition of SARS-CoV-2 binding to the ACE2 receptor. Nat. Commun. 2020;11:4541. doi: 10.1038/s41467-020-18319-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Woo H., Park S.-J., Im W. Developing a fully glycosylated full-length SARS-CoV-2 spike protein model in a viral membrane. J. Phys. Chem. B. 2020;124:7128–7137. doi: 10.1021/acs.jpcb.0c04553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shajahan A., Archer-Hartmann S., Azadi P. Comprehensive characterization of N- and O- glycosylation of SARS-CoV-2 human receptor angiotensin converting enzyme 2. Glycobiology. 2020 doi: 10.1093/glycob/cwaa101. Published online October 29, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.