Abstract
The third pandemic of coronavirus infection, called COVID-19 disease, was first detected in November 2019th. Various determinants of disease progression such as age, sex, virus mutations, comorbidity, lifestyle, host immune response, and genetic background variation have caused clinical variability of COVID-19. The causative agent of COVID-19 is an enveloped coronavirus named severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) that invades host cells using an endocytic pathway. The SARS-CoV-2 spike protein is the main viral protein that contributes to the fusion of the virus particle to the host cell through angiotensin-converting enzyme 2 (ACE2). The highly conserved expression of ACE2 is found in various animals, which indicates its pivotal physiological function. The ACE2 has a crucial role in vascular, renal, and myocardial physiology. Genetic factors contributing to the outcome of SARS-CoV-2 infection are unknown; however, variants in the specific sites of ACE2 gene could be regarded as a main genetic risk factor for COVID-19. Given that ACE2 is the main site for virus landing on host cells, the effect of amino acid sequences of ACE2 on host susceptibility to COVID-19 seems reasonable. It would likely have a substantial role in the occurrence of a wide range of clinical symptoms. Several ACE2 variants can affect the protein stability, influencing the interaction between spike protein and ACE2 through imposing conformational changes while some other variants are known to cause a decrease or an increase in the ligand-receptor affinity. The other variations are located at the proteolytic cleavage site, which can influence virus infection; because soluble ACE2 can act as a decoy receptor for virus and decrease virus intake by cell surface ACE2. Notably, polymorphisms of regulatory and non-coding regions such as promoter in ACE2, can play crucial role in different expression levels of ACE2 among different individuals. Many studies should be performed to investigate the involvement of ACE2 polymorphism with susceptibility to COVID-19. Herein, we discuss some reported associations between variants of ACE2 and COVID-19 in details. In addition, the mode of action of ACE2 and its role in SARS-CoV-2 infection are highlighted which is followed by addressing the effects of several ACE2 variants on its protein stability, viral tropism or ligand-receptor affinity, secondary and tertiary structure or protein conformation, proteolytic cleavage site, and finally inter-individual clinical variability in COVID-19. The polymorphisms of regulatory regions of ACE2 and their effect on expression levels of ACE2 are also provided in this review. Such studies can improve the prediction of the affinity of mutant ACE2 variations with spike protein, and help the biopharmaceutical industry to design effective approaches for recombinant hACE2 therapy and vaccination of COVID-19 disease.
Keywords: SARS-CoV-2, COVID-19, Pathogenicity, Pandemic Disease, ACE2
Graphical Abstract
1. COVID-19 disease
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) is a new coronavirus which was firstly reported on 2019. This virus causes Coronavirus Disease 2019 (COVID-19) The virus has infected about 80 million cases in various countries contributing to approximate 2 million deaths during one year (Chan et al., 2020; Huang et al., 2020; Organization., W.H, 2020; Zhu et al., 2020).
Prior to this outbreak, in 2003 and 2012, MERS-CoV and SARS-CoV, which caused acute respiratory disease, have spread in China and Saudi Arabia, respectively (Rabaan et al., 2020). Analyzing the entire length of the virus genome showed that the SARS-CoV-2 genome is about 70–80% and 50% similar to SARS-CoV and MERS-CoV, respectively (Chen et al., 2020b). Although SARS and MERS have far higher mortality rates than the SARS-CoV-2, this novel coronavirus spreads more easily among people, leading to an incomparable number of infections (Rabaan et al., 2020).
The results of different studies on the severity of the infection with SARS-CoV-2 reveals that about 81% of cases are mild, ~14% severe, and ~ 5% critical (Nikpouraghdam et al., 2020; Novel, 2020; Zhang et al., 2020b). Moderate cases suffering from COVID-19 usually show fever, dry cough, and headache (Zhong et al., 2003), but in the severe cases, it can affect multiple organs, including the cardiovascular system, lungs, gut, kidney, liver, and central nervous system (Chen et al., 2020b; Zou et al., 2020). The incubation period was estimated to range from 3 to 10 days (Poutanen et al., 2003). Comorbidities such as diabetes, chronic kidney diseases, chronic respiratory diseases, cardiovascular diseases, hypertension, and cancer affect the severity of the infection leading to an increased risk of death (Nikpouraghdam et al., 2020; Zhang et al., 2020b). In the early stage, case fatality rate (CFR), had been estimated 11% (11 of 99 confirmed cases) (Xu et al., 2020a), and nearly one month later 2.3% (1023 of 44,672 confirmed cases) (Zhang et al., 2020b). WHO Situation Report–134 on June 02, 2020, illustrates that the global CFR is 6.07 (WHO, 2020). CFR is different in countries depending on the quality of the healthcare system and the situation of the outbreak. The minimum amount of CFR has been reported by the Korea Centers for Disease Control and Prevention, which was 0.9% total (Choe, 2020). The group of patients more than 80 years old had the highest mortality rate that was 19.27% (Nikpouraghdam et al., 2020), 14.8% (Zhang et al., 2020b), 8.5% (Choe, 2020).
2. SARS-CoV-2
Some scientists suggested that bats are natural hosts of SARS-CoV-2 because a comparison of SARS-CoV-2 sequences and bat SARS coronavirus (SARS-CoV-RaTG13) revealed 96% similarity. Pangolins, camels and snakes are another potential intermediate hosts (Bravo et al., 2014; Nikpouraghdam et al., 2020; Xu et al., 2020a).
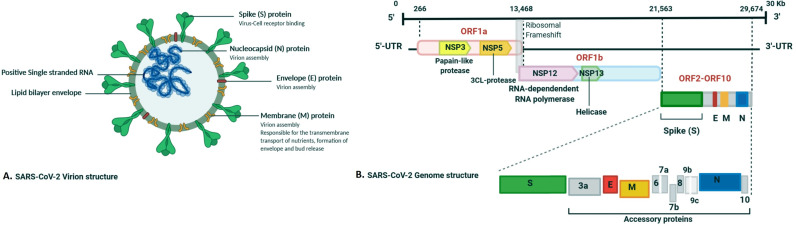
SARS-CoV-2, as a member of the beta coronaviruses family, is composed of positive-stranded RNA as genetic material, capsid proteins, and an outer envelope. The viral envelope consists of the membrane (M), spike glycoprotein (S), nucleocapsid (N), envelope protein (E), and hemagglutinin esterase (HE) as structural proteins. The spike protein is cleaved by furin-like protease into S1 and S2 functional domains (Fig. 1 ) (De Groot et al., 1987; Spaan et al., 1988). The coronavirus enters the host cell by endocytosis or direct merger of the viral envelope with the cell membrane (Magrone et al., 2020).
Fig. 1.
(A) Schematic illustration of SARS-CoV-2 structure and its major structural proteins (B) the Schematic representation of the genomic organization of SARS-CoV-2. The distinct genomic locations, including ORFs, are represented. SARS-CoV-2 is comprised of four structural proteins, containing the E and M proteins responsible for the viral envelope, the N protein binding to the viral genome, and the spike protein capable of binding to the hACE2 receptor. Besides, translation of ORF 1a and ORF 1b to non-structural proteins (nsp) and accessory proteins, containing 3a, 6, 7a, 7b, 8, 9b, 9c, and 10 are indicated. Kb, kilobase pair, 3′-UTR 5′-UTR, untranslated regions at the 3′ and 5′ regions, respectively.
SARS-CoV basic reproductive number (R0) has been estimated 2.7 by Riley et al. (Riley et al., 2003) and about three by Lipsitch et al. (Lipsitch et al., 2003). In many studies, the range of R0 has been estimated between 2.2 and 3.8 (Xu et al., 2020b),(Li et al., 2020). Another study using the exponential growth model method indicated the mean R0 range from 2.24 to 3.58 (Wu et al., 2020).
Although genetic evidence suggests that SARS-CoV-2 may have originated in animals, it is not yet clear exactly where the virus came from and what its primary origin was. Addressing this highly controversial issue requires further research around the world.
2.1. Cytokine storm in COVID-19
According to the previous investigations, increasing numbers of pro-inflammatory cytokines including interleukin 1B, IL-6, IL-12, and Interferon γ and chemokines, including CCL2 and CXCL10 are correlated to inflammation of lungs and extensive pulmonary involvement in MERS-CoV and SARS-CoV infected patients (Channappanavar and Perlman, 2017). Similar to other coronavirus pathogenesis, in COVID-19 patients, increased levels of chemokines and pro-inflammatory cytokines, and higher activation of T-helper-1 (Th1) cells induced by elevated levels of CCL2, CXCL10, IFN γ, and IL-1B are also observed. Prominently, cytokine storm plays a crucial role in the promotion of a more severe clinical state. Rapid replication of SARS-CoV-2 induced activation and then differentiation of T helper cell to Th1 cells which is responsible for releasing pro-inflammatory cytokines such as IL-6, GM-CSF, and IFN-γ. Accordingly, the clinical observations in patients admitted to intensive care unit (ICU) with SARS-CoV-2 demonstrated a higher concentration of TNFα, CCL2, and CXCL10 than other cases with less severe conditions (Coperchini et al., 2020). It should be noted that, in SARS-CoV infections, higher secretion of IL-10 and IL-4 as Th2-immune-oriented cytokines has been observed, which plays a prominent role in the inflammatory response suppression (Zhang et al., 2020a). In SARS cases, acute respiratory distress syndrome (ARDS) is the eventual consequence of cytokine storm. In this situation, massive amounts of chemokines (CCL3, CCL3, CCL, CXCL8, CXCL9, CXCL10) and pro-inflammatory cytokines (TNFα, IFNγ, IFNα, IL-12, IL-1β, IL-6, IL-33, IL-18, TGFβ) are released by immune effector cells, and subsequent precipitation drive to the excessive systemic inflammatory response (Cameron et al., 2008; Channappanavar and Perlman, 2017; Huang et al., 2020; Williams and Chambers, 2014).
The ARDS seen with SARS-CoV-2 infection is a cytokine release syndrome (CRS), which is a disorder induced by cytokine storms. Indeed, ARDS is a severe immune-pathological lung condition causing serious hypoxemia in SARS-CoV, MERS-CoV, and SARS-CoV-2 infections (Huang et al., 2020). Recent studies revealed that ARDS accounts for a significant number of deaths in COVID-19 patients, defined by severe hypoxemia and bilateral lung infiltrates (Zou et al., 2020). Increased lung permeability and the enhancement of the high protein content of edema fluid in the airway of the lungs lead to inadequate respiration, which is the main consequence of ARDS inflammatory injury (Bhatia et al., 2012). Overall, ARDS contributes to the cytokine storm that is a life-threatening condition characterized by a severe systemic inflammatory response induced by massive amounts of chemokines and pro-inflammatory cytokines.
Upon entry of SARS-CoV-2 into respiratory epithelial cells, the production of inflammatory cytokines combined with a weak interferon response induces an immune response mediated by membrane-bound immune receptors (e.g., Fc and Toll-like receptors). Ultimately, the infiltration of macrophages and neutrophils into lung tissue leads to a cytokine storm. One suggested mechanism for cytokine storms is that SARS-CoV-2 infection in the respiratory system can activate NF-κB via pattern recognition receptors (PPR). STAT3 is required for full activation of the NF-κB pathway. Both NF-κB and STAT3 are capable of activating the IL-6 amplifier (IL-6 Amp), which is a mechanism for the hyper-activation of NF-κB by STAT3. It in turn induces a variety of proinflammatory cytokines, including IL-6, TNFα, and chemokines (Hu et al., 2021).
Ultimately, the cytokine storm causes ARDS and organs failure, and final death in at least the most severe COVID-19 cases (Huang et al., 2020).
2.2. Innate immune evasion of coronavirus
Get the main antivirus response, innate immune cells as professional guards recognize the invasion of the virus by binding immunogenic antigens such as RNA coronavirus. Innate immunity is initiated by a molecular pattern related to pathogens, evolutionary conservation of special molecular structures for pathogens acting as a ligand for PRR. Ligand binding triggers signaling paths that include transcription factors including NF-κB, IRF3, and AP-1, which synergistically promotes type I interferon (IFN-I) production. Interferon-stimulated genes are an important component of innate antivirus defense, acting to limit virus entries and limit viral replication after the virus enters the host cell. The expression of the interferon stimulated gene was driven mainly by the activation of JAK/STAT pathway STAT mediated by IFNAR, which resulted in the binding of homodimer STAT1 and STAT1/2 heterodimer to the ISGS promoter area (Taefehshokr et al., 2020). Overproduction of IFN-I increases the penetration of neutrophils, macrophages, and inflammatory factors into the lung, which is then followed by cytokine storm syndrome, as well. In this situation, the numbers of Th1, Th17, M1 macrophage, NK cells and their secreted inflammatory cytokines increase (Rahmani-Kukia et al., 2020). An increase in the content of activated immune cells at the site of infection and their released inflammatory cytokines triggers cytokine storm. Subsequently, serious pathological complications induced in the patients' lung can worsen the illness from COVID-19 and even terminate the life. On the other hand, the virus provides ways to escape the immune system.
Generally, coronaviruses have multiple strategies to avoid the immune cells leading them to better survival and infection of the host cells (Guo et al., 2020c). The coronaviruses can form double-membrane vesicles (DMV) at the extracellular space as an avoidance strategy. The formation of DMV shields dsRNA, produced as an intermediate of viral replication, from recognition by intracellular cytosolic PRRs, including RIG-I and MDA5 (Kikkert, 2020). As another avoiding immune strategy, they can suppress interferons with the aid of their eight specific viral proteins (Guo et al., 2020c). The lack of a 5` cap in the genome of RNA viruses could facilitate the recognition of the viral RNA. However, the virus mimicry strategy of the host capping combats with immune system activation. The virus recruits two nonstructural proteins (nsp), nsp 14 and nsp 16, leading to cap formation and modification of viral RNA, respectively, following its similarity to host cell RNA and evading PRRs recognition (Chen et al., 2009; Daffis et al., 2010; Totura and Baric, 2012). Moreover, nsp3 is another nonstructural coronavirus protein capable of immune response prevention. The virus also encodes two functional proteins, including PLpro, to cleave nsps and macro domains. Studies in mice concluded that according to the loss of deubiquitinating enzyme activity in MERS-CoV, another possible role of PLpro is to contribute coronavirus to evade immune response through antagonizing the IFN response (Knaap et al., 2019; Nelemans and Kikkert, 2019). Besides, other protein accessories can be utilized by SARS-CoV as an alternative to the immune evasion strategy. An instance of antagonizing the IFN signaling pathways by the gene located on ORF3b of this virus leads to the inhibition of effector cell activation cascade, which is essential for inhibiting the virus replication (Freundt et al., 2009). In contrast, the results of SARS-CoV-2 investigations indicate that the SARS-CoV-2 is more susceptible to IFN-I pretreatment and shows STAT1 phosphorylation and higher ISG proteins in the context of IFN-I induction. Considering the homology of interferon antagonists among viral proteins of SARS-CoV and SARS-CoV-2, the absence of ORF3b and dramatic difference to ORF6 suggesting the differences in IFN-I response susceptibility between the members of coronaviruses (Lokugamage et al., 2020). Overall, the production of IFN-I plays an important role in the secretion of antiviral proteins, promoting phagocytosis by macrophages, and facilitating the protection of infected cells. Coronavirus accessory proteins can interfere with the activation of these pathways during virus replication. This interference facilitates the persistence of the virus and causes the innate immune response to be dysregulated(García-Salido, 2020).
3. Angiotensin-converting enzyme (ACE)
The renin-angiotensin system (RAS) serves as the primary volume regulator in mammals and has an essential role in regulating body homeostasis, including blood pressure, electrolyte balance, and vascular tone (Rice et al., 2004; Verma et al., 2019). In the RAS, angiotensinogen is transformed into angiotensin (Ang) I by renin, followed by hydrolysis of Ang I to Ang II by angiotensin-converting enzyme (ACE) (Bernstein et al., 2018a). As a pivotal enzyme in the RAS, ACE is expressed in most of the tissues in the human body(Donoghue et al., 2000a). ACE is a zinc-dependent di-carboxypeptidase that catalyzes the alternation of Ang I to Ang II and Ang (1–9) to Ang (1–7) (Tikellis et al., 2003a). It is a membrane-bound enzyme and exists in secretory form (Bernstein et al., 2018b). ACE has two different isoforms, including somatic ACE and testicular ACE (tACE). Somatic ACE possesses two homologous domains for active sites, while tACE has just one (Fig. 2 ) (Guy et al., 2003a).
Fig. 2.
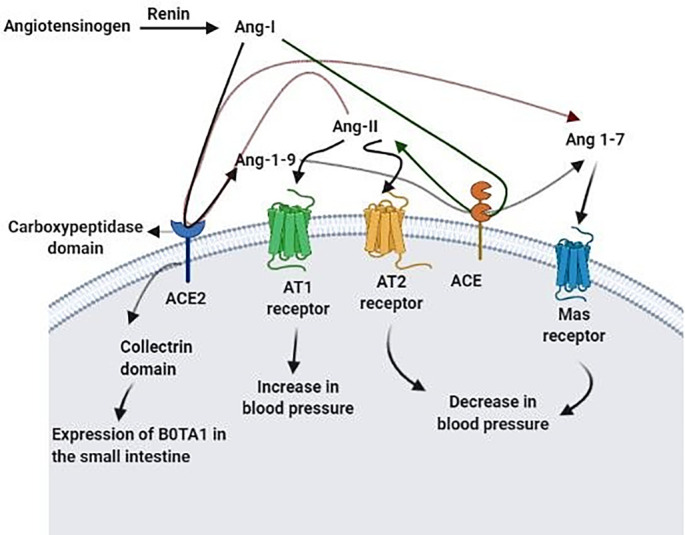
Renin hydrolyzes Angiotensin to Ang l. ACE2 carboxypeptidase domain catalyzes the conversion of Ang l to Ang 1–9 and Ang II to Ang 1–7. ACE, as another enzyme in the RAS transforms Ang l to Ang II and Ang 1–9 to Ang 1–7. Ang II signals through AT1 and AT2 receptors, which leads to an increase and decrease in blood pressure, respectively. Mas receptor is also a receptor for Ang 1–7 and causes a decrease in blood pressure. These altogether controls blood pressure homeostasis. ACE2 collectrin domain also plays a critical role in expressing the Hartnup amino acid transporter B0AT1 in the small intestine.
4. Angiotensin-Converting enzyme 2 (ACE2)
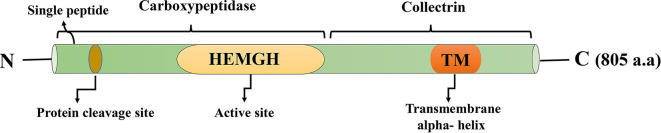
In 2000, ACE2 was discovered as the first homolog of ACE (Donoghue et al., 2000b; Tipnis et al., 2000), although some years later, ACE3, another homolog of ACE, was identified, which is expressed in sperm (Inoue et al., 2010). ACE2 was identified in a study conducted on the genes contributing to heart failure and also in independent research, reported as angiotensin-converting enzyme homolog (ACEH) (Tipnis et al., 2000). ACE2 consists of 805 amino acids, and its molecular weight is about 120 kDa (Alenina and Bader, 2019). ACE2 has one zinc-binding motif His-Glu-Xaa-Xaa-His as an active site (residues 374–378) (Tipnis et al., 2000), in which two zinc ions coordinate with histidine residues (Guy et al., 2003a). This enzyme is anchored in the membrane from the C-terminal, and 17 residues at the N-terminal act as a signal peptide (Fig. 3 ) (Tipnis et al., 2000).
Fig. 3.
ACE2, an enzyme with 805 amino acid residues, consists of two domains: the collectrin domain locates at the C-terminal and includes a transmembrane alpha-helix, the carboxypeptidase domain, is at the N-terminal and has one active site. There is also a signal peptide at the end of the N-terminal, which the protein cleavage site is located next to it.
It is believed that ACE, ACE2, neurolysin, and PfuCP have a common ancestral protein (Donoghue et al., 2000a) and were evolved from the same clan (Guy et al., 2003a). ACE2 is a bio-chimeric protein; the fusion between ACE gene and the collectrin gene during evolution resulted in the ACE2 gene (Turner and Hooper, 2002). In ACE2, the N-terminal domain sequence is 42% similar to the catalytic domains of endothelial ACE (Donoghue et al., 2000a), and its C-terminal sequence is 48% identical to collectrin (Alenina and Bader, 2019). Its global alignment also shows a 33% identity to human tACE (Donoghue et al., 2000a).
When ACE2 was characterized, its expression in endothelial cells of the heart, kidney, and testis was identified (Donoghue et al., 2000a; Tipnis et al., 2000). However, further studies showed that this enzyme also exists in the gastrointestinal tract, lungs, and lower brain (Alenina and Bader, 2019; Gu et al., 2005; Komatsu et al., 2002). Based on research, ACE2 has the highest expression levels in the small intestine, duodenum, gall bladder, kidney, and heart (Fagerberg et al., 2014). According to an investigation on the expression of ACE2 in 31 human tissues, the kidneys, testis, heart, small intestine, thyroid, and adipose tissue have significant levels of ACE2 expression, and the bladder, lungs, liver, large intestine, and adrenal gland express to a lesser extent (Chen et al., 2020b). ACE2 expression also occurs in the oral cavity mucosa and mainly exists in the tongue epithelial cells. Overall, ACE2 does not exhibit a tissue-specific expression pattern (Zhao et al., 2020).
4.1. ACE2 orthologues in other species
ACE2 expression is found in a wide variety of animals and highly conserved, especially in mammals. This conservation indicates the pivotal physiological function of ACE2. Some scientists have constructed the phylogenic tree of mammalian ACE2 using the Maximum Likelihood method (Luan et al., 2020). Species, which express ACE2, could be infected by SARS-CoV or SARS-CoV-2. Many different studies have supported this subject. Although receptor recognition by spike protein of the virus mainly determines the host range (Li, 2013), other steps in the infectious cycle of the virus, including genome replication, translation, virion assembly, and budding, must be done correctly for the host infection (Zhao et al., 2020). It has been reported that murine and rat ACE2 has less affinity for attachment to the S1 domain of SARS-CoV in comparison with human ACE2. SARS-CoV can infect some primates, cats, hamsters, bats, raccoon dogs, and ferrets (Li et al., 2004; Li et al., 2006). In a study on SARS-CoV-2 hosts, ACE2 of 14 mammalian species have been investigated, and ACE2 of human and rhesus monkey showed the highest receptor activity (Zhao et al., 2020). ACE2 in Bovidae, Cetacea, Cricetidae, and Primates species could recognize the spike protein of SARS-CoV-2, in contrast to turtle and snake ACE2 (Luan et al., 2020).
4.2. Enzymatic activity of ACE2
ACE2 is a type l transmembrane enzyme (Donoghue et al., 2000b), and its domain on the cell surface has carboxypeptidase activity (Donoghue et al., 2000b; Vickers et al., 2002) and hydrolyzes circulating peptides. Its ectodomain catalyzes the transformation of Ang l to Ang (1–9) (via cleavage of the C-terminal Leu) (Donoghue et al., 2000b) and Ang II to Ang 1–7 (Vickers et al., 2002). Ang l, Ang II, Ang (1–7), and Ang (1–9) are the RAS peptide hormones. Ang II is a vasoconstrictor, while Ang (1–7) is a vasodilator hormone. Ang II type (AT) 1 and AT2 receptors are two receptors for Ang II, which have different roles. Ang (1–7) signaling also occurs via the Mas receptor (Fournier et al., 2012). Attachment of Ang II to AT1 receptors leads to the increase of the blood pressure, and the stimulation of the Mas receptor by Ang (1–7) eventuates in nitric oxide release followed by prevention of neurogenic hypertension (Xia et al., 2013). ACE2 opposes the increase in blood pressure by transforming Ang II to Ang (1–7). It also turns Ang l, a substrate converted to Ang II by ACE, to Ang (1–9), decreasing Ang II levels. Furthermore, ACE hydrolyzes Ang (1–9) and other vasoactive peptides and produces Ang (1–7). These all lead to a reduction in blood pressure (Donoghue et al., 2000b; Rice et al., 2004; Tikellis et al., 2003b; Xia et al., 2013).
In the small intestine, the expression of B0AT1 on the surface of enterocytes depends on ACE2 (Camargo et al., 2009). Collectrin is necessary for the intake of amino acid by kidneys and the action of Hartnup amino acid transporter B0AT1, which is encoded by the SLC6A19 gene, in renal epithelial cells (Danilczyk et al., 2006). Similarly, the function of B0AT1 in the small intestine is controlled by the cytoplasmic domain of ACE2, a homologous of collectrin (Camargo et al., 2009).
The active site sequence and structure in ACE is almost conserved in ACE2, except one alternation in ACE2 active site residues (Guy et al., 2003b). Despite the high similarity in ACE and ACE2, they differ in substrate specificity. ACE acts as a peptidyl dipeptidase; however, ACE2 omits just an amino acid residue from its substrate's C-terminal. The differences between these two enzymes in ligand-binding pockets, which results in different substrate specificity, were studied (Guy et al., 2003b). Optimum pH for ACE2 activity is ~6.5, and like ACE, monovalent anions such as Cl− and F− improve its function up to ~10-fold (Vickers et al., 2002). Another study stated that pH 7.0 with 300 mM NaCl is the optimum condition for ACE2 activity, and Cl− rises Ang I hydrolysis but declines Ang II cleavage (Guy et al., 2003b). ACE2 has relatively high specificity and cannot cleave unrelated hormonal peptides (Donoghue et al., 2000b). In a study on the possible substrates of ACE2, elevens of the peptides were hydrolyzed, especially Ang II, APLN13 (the fragment of apelin consists of thirteen residues), dynorphin A (1–13) and des-Arg(9)-bradykinin with higher efficiency (Tipnis et al., 2000; Vickers et al., 2002). Neurotensin and kinesin are other substrates that ACE2 acts on them (Donoghue et al., 2000b). However, unlike ACE, it has not any effect on bradykinin (Donoghue et al., 2000b; Vickers et al., 2002). It also hydrolyzes Ang I, but ~400-fold less efficient in comparison with Ang II (Rice et al., 2004; Vickers et al., 2002). So it is suggested that its primary role is in the metabolism of Ang II to Ang (1–7) (Rice et al., 2004). Pro-Xaa (1–3)-Pro-hydrophobic was recognized as a consensus sequence in all substrates of ACE2, and the peptide bond between proline and hydrophobic residues as the cleavage site (Vickers et al., 2002). However, in another study, by analyzing ACE2 different substrates, Pro-Xaa-Pro-hydrophobic/basic was known as its protease specificity (Guy et al., 2003b). Although ACE2 and ACE have similar catalytic domains (Guy et al., 2003b), ACE inhibitors that block the conversion of Ang II to Ang I, have no effect on ACE2 activity (Donoghue et al., 2000b; Tipnis et al., 2000). ACE inhibitors, including lisinopril, enalaprilat, and captopril which bind in the S1′ and S2′ pockets of ACE, cannot bind to ACE2. This is probably because the S2′ pocket of the active site in ACE2 is smaller than that of ACE. The active site model was able to help design new ACE2 inhibitors, for example the presence of Pro-Phe in the concentration of 180 μM could inhibit ACE2 activity (Guy et al., 2003b). ACE2 cleaves the peptide bond between Pro and Phe residues at the Ang II C-terminal. Dipeptide Pro-Phe can itself be a substrate for this enzyme and also act as an inhibitor for ACE2 while hydrolyzing Ang II (Guy et al., 2003b).
Ethylenediamine-tetraacetic acid (EDTA), which inhibits metalloprotease by the chelation of the metal ion, is another inhibitor for ACE2 (Tipnis et al., 2000). In a study by Anguiano et al., circulating ACE2 and ACE activity were measured in EDTA-plasma; EDTA inhibited ACE2 and ACE activity. When zinc chloride was added to plasma samples to prevent the effect of EDTA chelation, enzymatic activity returned. The results showed that in the presence of EDTA, endogenous and exogenous ACE2 activities are inhibited (Anguiano et al., 2015).
4.3. Physiological roles of ACE2
ACE2 has a crucial role in vascular, renal, and myocardial physiology (Crackower et al., 2002; Donoghue et al., 2000b; Tikellis et al., 2003b). In ACE2 knocked out mice, heart dysfunction, memory, and cognition problems were observed (Guy et al., 2003b; Wang et al., 2016). ACE2 has a profound antihypertensive and sympatholytic activity in the hypothalamus (Alenina and Bader, 2019) and is necessary for protecting against metabolic and cardiovascular disorders like hypertension, diabetes, and Ang II-mediated vascular inflammation (Sahara et al., 2014; Verma et al., 2019). Its brain action helps the human body regulate the stress response, cognition, brain damage, and neurogenesis (Alenina and Bader, 2019). ACE2 modulates the RAS negatively (Imai et al., 2005), and its functions contradict ACE activity. Diabetes is also associated with a decrease in renal ACE2, which could be the target of treatment (Tikellis et al., 2003b). In acute lung injury, an increase in Ang ll resulting from ACE activity improves the damage through the AT1 receptor. ACE2 and the AT2 receptor could prevent lung failure, but they are downregulated (Imai et al., 2005). Despite ACE2 function in reducing the blood pressure, its expression decreases in neurogenic hypertension. A disintegrin and metalloproteinase 17 (ADAM17 or TNFα convertase) cleaves ACE2 ectodomain and releases it in a soluble form. The brain's RAS activation improves ADAM17 activity and reduces membrane-bound ACE2 in the brain, inhibiting its role in compensating hypertension, although ACE2 soluble form preserves its carboxypeptidase activity (Xia et al., 2013).
Dietary tryptophan uptake almost occurs through the transport pathway mediated by B0AT1 and ACE2 in the small intestine. ACE2 inactivation causes a decrease in B0AT1 expression. Because the absorption of the neutral amino acids is regulated by B0AT, this would result in low concentrations of Thr, Val, Tyr, and essential amino acid Trp in the serum. ACE2 deficiency leads to deterioration of local Trp hemostasis that is the cause of reducing regenerative responses and so susceptibility to intestinal damage. So inactivation of ACE2 can lead to colitis and intestinal injury. It also alters the intestinal microbiome (Hashimoto et al., 2012). Trp is a precursor of serotonin, so its deficiency reduces the concentration of serotonin in the blood and the brain (Klempin et al., 2018).
4.4. ACE2 as a receptor for pathologic viruses
Enveloped viruses almost enter the host cells by attaching to the cell surface proteins. ACE2, as a cell surface protein, acts as a receptor for some coronaviruses, including SARS-CoV (Li et al., 2003), HCoV-NL63 (Hofmann et al., 2005), and the recently emerged novel coronavirus SARS-CoV-2 (Lu et al., 2020).
The spike proteins of a coronavirus consist of two domains: S1 domain, the N-terminal domain that attaches to the receptor and S2 domain, the membrane fusion domain (Gallagher, 2001). As an enveloped virus, SARS-CoV enters the target cell by fusing the host cell membrane (Simmons et al., 2004). Indeed, direct membrane fusion and receptor-mediated endocytosis are two possible mechanisms for virus entry (Peiris et al., 2004b). As mentioned earlier, ACE2 is the receptor for the virus and attaches the S1 domain of SARS-CoV with excellent specificity and affinity (affinity of 1.7 nM) (Sui et al., 2004). Spike protein is the only protein of a virus that contributes to the fusion of the virus particle with the host cell, and it is not cleaved after the fusion (Li et al., 2003). Nevertheless, the enzymatic activity of ACE2 does not play a role in virus entry (Spiga et al., 2003), and after attachment to S1 domain, its catalytic activity is not affected (Kuhn et al., 2004). A small region of the S1 domain, residues 318 to 510, attaches to ACE2, and two amino acid Glu and Asp at the position of 452 and 454, respectively, play a critical role in this attachment (Wong et al., 2004). Different approaches for preventing the infection with this virus have been studied, like using soluble ACE2, an antibody against ACE2, or the ACE2-binding domain of the virus spike protein (Sui et al., 2004). Some researchers have identified CD209L, another human cellular glycoprotein, as a second receptor for SARS-CoV, which associates with spike protein less efficiently in comparison with ACE2 (Jeffers et al., 2004).
Human coronavirus NL63 (HCoV-NL63) is the fourth identified human coronavirus, which, unlike SARS-CoV, causes mild respiratory disease in infants or immune-deficient adults with severe respiratory tract infections (Van Der Hoek et al., 2004). At the time of discovery, it was not an emerging virus and existed before, but it had not been identified (Van Der Hoek et al., 2006). HCoV-NL63 infection is observed globally but mostly in winters (Arden et al., 2005). This virus stands under group 1 coronaviruses in the classification, and its genome has a 65% sequence identity to HCoV-229E, another human coronavirus (Van Der Hoek et al., 2004). Due to this similarity, it was expected that HCoV-NL63 occupies the same receptor for entering the cell as HCoV-229E, which employs CD13 (Yeager et al., 1992). However, the lack of homology in critical regions of the S protein changes the HCoV-NL63 receptor to ACE2. Like SARS-CoV, the S2 domain binds ACE2, but in HCoV-NL63, with less affinity. So, SARS-CoV and HCoV-NL63 have the same target cells, although HCoV-NL63 is a weaker pathogen. It could be because of the lack of pathogenicity factors or lower affinity to ACE2 in this virus. The spike proteins of these two viruses are just 14% similar, and it could be concluded that they have different ways of interacting with ACE2 (Hofmann et al., 2005). A similar receptor for these two viruses could lead to recombination between them and the evolution of more pathogenic variants in the future (Van Der Hoek et al., 2006). ACE2 also contributes to HCoV-NL63 replication in the cell lines (Hofmann et al., 2005). The recently emerged novel coronavirus SARS-CoV-2 also enters the cell via ACE2 (Lu et al., 2020). The viruses which employ ACE2 as a receptor are summarized in Table 1 .
Table 1.
Viruses that employ ACE2 as a receptor.
| Virus | Group Of Coronavirus | Year Of Emergence | Disease | Symptoms | References |
|---|---|---|---|---|---|
| SARS-CoV | Group 2 | 2003 | SARS | fever, dry cough, dyspnea, headache, hypoxemia | Li et al. (2003) |
| HCoV-NL63 | Group 1 | Much before discovery | Mild/moderate respiratory disease | fever, cough, sore throat, rhinitis | Hofmann et al. (2005) |
| SARS-CoV-2 | Group 2 | 2019 | COVID-19 | cough, fever, myalgia, fatigue, dyspnea, pneumonia | Coperchini et al. (2020) |
Recently a study demonstrated that the comparison of ACE2 WES data between the cohorts of 131 patients and 258 controls allowed identifying higher allelic variability statistically in control group compared to patients in the Italian population (Benetti et al., 2020b). This opens the possibility that some people can be less or more susceptible to SARS-CoV-2 infection than others.
5. The pathogenic mechanism of SARS-CoV-2 in human
Clinical presentations of COVID-19 patients are familiar to symptoms of other coronavirus infections, including SARS-CoV and MERS-CoV (Huang et al., 2020; Peiris et al., 2004a). According to the poor understanding of the new coronavirus pathogenesis, more investigations need to be elucidated. However, the similar pathogenesis of MERS-coronavirus and SARS-coronavirus can help understand SARS-CoV-2 infections better. The pathogenesis of coronavirus begins with the binding to host airway epithelial cells using ACE2 receptors, leading to viral entry and its replication (Liu et al., 2010; Mason, 2020). The viral entry to host cells is mediated by direct membrane fusion between the host cell membrane receptor (ACE2) and the virus (spike protein), leading to the release of the viral ssRNA genome into the host cell. Following the viral entry, ACE2 is down-regulated, causing excessive activity of ACE/Ang II and increasing the permeability of lung vascular and consequent lung damage (Magrone et al., 2020). It has been reported that coronaviruses show dual entry pathways including, early and late pathways. Subsequent to the binding of the virus to the cellular receptor, it can be accomplished by plasma membranes or endosomes. The presence of trypsin and TMPRSS2 as membrane-bound exogenous proteases leads to the entry of viruses through early fusion pathways or by the endocytosis pathway. On the other hand, the investigations on the MERS-CoV have been reported that the cleavage of spike protein at S1/S2 by furin during its biosynthesis and the presence of aforementioned membrane bound exogenous proteases mediates the early entry pathway. Likewise, spike protein cleavage at the S1/S2 site leading to endocytosis of the virus. In both entry pathways, the low acidic pH within the endosomes activates another cleavage site at the spike protein, leading to the fusion pathways and releasing of the virus genome into the host cell cytoplasm. Indeed, current studies indicate that the entry pathways of SARS-CoV-2 are accomplished by both endosomal cathepsin L or membrane-bound TMPRSS2 and direct fusion of virus membrane with host cell membrane without formation of endosome. Moreover, calcium and cholesterol are other influencing factors on viral entry (Straus et al., 2020).
While SARS-CoV-2 enters the cell, the positive sense single-stranded RNA genome of the virus is entered into the host cell cytoplasm. Both ORF1a and ORF1ab are readily translated into pp1a and pp1ab polyproteins, respectively, which are then readily undergo the proteolytic cleavage process by the transcription complex (RTC) proteases. During the replication, RTC contributes to the synthesis of full length (−) RNA copies of the viral genome, which is utilized as templates for the full-length synthesis of positive RNA genomes. At the time of transcription, a set of sub-genomic RNAs (sgRNAs) is synthesized using fragmented transcription (Perlman and Netland, 2009). Despite the presence of several open reading frames (ORFs) in these sgRNAs, the closest ORF to the 5′ end will be exclusively translated. Afterward, the recently translated envelope glycoprotein is then inserted into the ER and Golgi membranes, respectively, and the viral nucleocapsid is constituted from the assembling of nucleocapsid protein and new synthesized genomic RNA. Afterward, new virions start budding into the lumen of ER-Golgi intermediate compartments. The vesicles carrying the virions fuse to the cytoplasmic membrane, and the newly synthesized viruses can be released by exocytosis and infect other surrounding host cells (Alanagreh et al., 2020).
Studies suggest that the high transmissibility and higher infectivity of SARS-CoV-2 are related to its characteristics, including the longer spike protein and the higher affinity of receptor-binding domain (RBD) to ACE2. In addition, the existence of the furin-like cleavage site facilitates the priming of the viral spike protein (Wrapp et al., 2020).
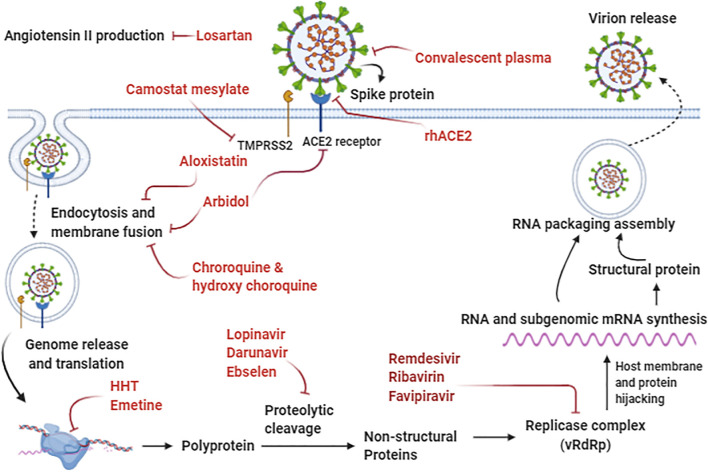
The roles of some biochemical inhibitors on SARS-CoV-2 entry and infection are presented in Fig. 4 . At the beginning of viral entry into the cell, Losartan, as an Ang II receptor blocker, can alleviate the vasocontraction induced by the viral infection. Indeed, Camostat mesylate can inhibit the entry of the viral spike protein into the host cell via inhibition of TMPRSS2. Furthermore, rhACE2 functionally binds to the virus and sequesters circulating viruses, and contributes to the prevention of further spike protein interactions with ACE2 on the host cell. Moreover, convalescent plasma can contribute to passive immunization by neutralizing antibodies against the virus. In the cytoplasm, E64d, the cysteine proteinase inhibitor, can inhibit the viral RNA synthesis and protein processing. As a potent antiviral drug, Arbidol can inhibit viral entry through the induction of structural rigidity in the virus glycoprotein leading to disruption of membrane attachment and counteract clathrin-dependent trafficking. Moreover, Chloroquine and Hydroxychloroquine prevent viral entry and endocytosis, and also play the role of potential immunomodulatory agents. Homoharringtonine (HHT) and Emetine as protein synthesis inhibitors can prevent viral replication. The anti-retroviral drugs, including Lopinavir and darunavir, act as a potential protease inhibitor, probably can inhibit the 3C-like protease 3CLpro of SARS-CoV-2. Additionally, Ebselen and N3 can effectively inhibit the viral main protease (Mpro), leading to viral replication inhibition. Nucleoside analogs, including Remdesivir, Favipiravir, and Ribavirin, act as interfering with the viral RNA-dependent RNA polymerase (RdRp), consequently leading to the termination of viral protein synthesis.
Fig. 4.
Cell entry and viral replication mechanisms of SARS-CoV-2 and potential drugs targeting different virus life cycle stages. Abbreviations: ACE2; Angiotensin-Converting Enzyme 2, HHT; Homoharringtonine, rhACE2; Recombinant Human Angiotensin-converting Enzyme 2, TMPRSS2; Transmembrane protease serine 2, vRdRp; viral RNA-dependent RNA polymerase.
Following the entry of SARS-CoV-2 into the cells, the specific viral antigen will be processed and presented to the antigen-presenting cells (APC), thereby stimulating the body's antiviral immunity to both CD4-positive T cells, and CD8-positive T cells. Viral Antigenic peptides can be detected by the virus-specific CTL, mediated by their presentation by MHC (major histocompatibility complex) on the surface of the infected cells. As a result, a comprehensive perception of the SARS-CoV-2 antigen presentation will help us understanding COVID-19 pathogenesis better. Regretfully, there is not still sufficient report about it, and the previous investigation on MERS-CoV and SARS-CoV can be beneficial to get some information. The previously published reports about SARS-CoV indicates the importance of both MHCI and MHCII molecules in its antigen presentation ( Liu et al., 2010 ). Moreover, it has been reported that numerous HLA polymorphisms are associated with the SARS-CoV susceptibility including HLA-Cw*0801, HLA-DR B1*1202, HLA-B*4601, and HLA-B*0703 (Chen et al., 2006), whilst the HLA-A*0201, HLA-DR030, and HLA-Cw1502 related to the prevention from being infected by SARS-CoV (Wang et al., 2011). It has been also demonstrated that in MERS-CoV infected patients, MHC II molecules, including HLA-DQB1*02:0 and HLA-DRB1*11:01, are correlated with the higher MERS-coronavirus infection susceptibility (Hajeer et al., 2016). Furthermore, the association of Mannose-binding lectin (MBL) gene polymorphisms with antigen polymorphisms shows correlations with a higher rate of SARS-CoV infections (Tu et al., 2015). Aforementioned studies will contribute to beneficial clues for SARS-CoV-2 prevention, treatment, and pathogenesis. Indeed, various clinical trials evaluate the effectiveness of different therapeutic strategies to treat individuals with COVID-19 (Table 2 ).
Table 2.
The ongoing clinical trials being tested on COVID-19 subjects identified at Clinicaltrials (https://clinicaltrials.gov) using the listed keywords: SARS-CoV-2, 2019-nCoV, COVID-19, SARS-Coronavirus-2 along with the name of each administered drug.
| Drug | Mechanism of Action | Medical Condition | Sponsor | Trial Phase | Ref. |
|---|---|---|---|---|---|
| Arbidol (Umifenovir) | Blocking SARS-Coronavirus-2 binding to receptor and prevention of intracellular vesicle trafficking | 380 participants with COVID-19 pneumonia | Jieming QU, Ruijin Hospital | 4 | (Wang et al., 2020) |
| Bromhexine Hydrochloride | Inhibiting the viral entry by interfering with TMPRSS2 | 140 participants with Increased Risk of SARS-CoV-2 Infection | Federal State Budgetary Institution | 4 | (Habtemariam et al., 2020) |
| Camostat Mesilate | TMPRSS2 inhibitor | 580 participants with COVID-19 | University of Aarhus | 1 and 2 | (Uno, 2020) |
| Chloroquine | Preventing the Endosomal maturation | Preventive treatment for COVID-19 with 10,000 participants | University of Oxford | N/A | (Duan et al., 2020) |
| Danoprevir - Ritonavir | HCV NS3 protease inhibitor | 11 participants with COVID-19 | The Ninth Hospital of Nanchang | 4 | (Chen et al., 2020a) |
| Darunavir-Cobicistat | Inhibitor of HIV protease / blocking Darunavir metabolism by the enzyme CYP3A | 30 participants with COVID-19 pneumonia | Shanghai Public Health Clinical Center | 3 | (De Meyer et al., 2020) |
| Favipiravir | Inhibitor of RNA-dependent RNA polymerase | 210 participants with COVID-19 | Peking University First Hospital | N/A | (Shannon et al., 2020) |
| Hydroxychloroquine | Less-toxic derivative of chloroquine, Inhibition of Endosome maturation | 30 participants with COVID-19 pneumonia | Shanghai Public Health Clinical Center | 3 | (Chowdhury et al., 2020) |
| Lopinavir-Ritonavir | Acting as a protease inhibitor, interfering with the action of 3CLpro, inhibiting the viral replication and viral release from host cells | 1220 participants in a preventive treatment study for COVID-19 | St. Paul's Hospital | 3 | (Dorward and Gbinigie, 2020) |
| Nitric Oxide Gas | Blocking the synthesis of viral protein and RNA | 240 participants with COVID-19 | Massachusetts General Hospital | 2 | (Gianni et al., 2020) |
| Interferon Α2β | Initiating the JAK-STAT signaling cascades | 328 participants infected by 2019-nCoV | Tongji Hospital | 1 | (Mantlo et al., 2020) |
| Remdesivir | Inhibition of SARS-CoV-2 RNA synthesis | 452 participants with COVID-19 | Capital Medical University | 3 | (Saha et al., 2020) |
| Baricitinib | JAK/STAT inhibitor | 80 participants with COVID-19 | University of Colorado, Denver | 2 and 3 | (Lo Caputo et al., 2020) |
| Bevacizumab | Monoclonal antibody against VEGF | 130 participants with COVID-19 pneumonia | Assistance Publique - Hôpitaux de Paris | 2 | (Tu et al., 2020) |
| Clazakizumab | Humanized monoclonal anti-IL-6 antibody | 60 participants with COVID-19 | Cedars-Sinai Medical Center | 2 | (Vaidya et al., 2020) |
| Colchicine | Blockage of the NLRP3 inflammasome assembly | 6000 adult and older participants with COVID-19 | Montreal Heart Institute | 3 | (Shah, 2020) |
| Convalescent Plasma | Plasma with the specific antibody | 100 participants with COVID-19 | Thomas Jefferson University | 2 | (Rojas et al., 2020) |
| Eculizumab | Humanized anti-C5 monoclonal Ab | 120 participants with COVID-19 | Alexion Pharmaceuticals | 2 | (Diurno et al., 2020) |
| Fingolimod | Sphingosine-1-phosphate receptor regulator | 30 individuals with COVID-19 | First Affiliated Hospital of Wenzhou Medical University | 2 | (Foerch et al., 2020) |
| IVIG | Block FcR activation | 50 participants with COVID-19 pneumonia | Dow University of Health Sciences | 1 and 2 | (Xie et al., 2020) |
| Kineret | Human Interleukin-1(IL-1) receptor Antagonist | 240 individuals with acute pneumonia caused by 2019-nCoV | University Hospital, Tours | 3 | (Huet et al., 2020) |
| Methylprednisolone | Inhibition of several cytokines (e.g. IL-6, IL-1, IL-2, IFN-δ and TNF-β) gene expression | 80 individuals with COVID-19 | Peking Union Medical College Hospital | 2 and 3 | (Xie et al., 2020) |
| Naproxen | Antiviral function against COX-2 of influenza A virus nucleoprotein | 584 participants with COVID-19 | Assistance Publique - Hôpitaux de Paris |
3 | (Alizadeh-Navaei et al., 2020) |
| Pirfenidone | inhibition of IL-1β and IL-4 | 294 individuals with severe COVID-1 pneumonia | Huilan Zhang | 3 | (George et al., 2020) |
| Ruxolitinib | Inhibitor of JAK-1 and 2 | 100 participants with COVID-19 | Marcelo Iastrebner |
2 | (Mukherjee et al.) |
| Sarilumab | Monoclonal recombinant human anti-IL6 receptor antibody |
120 participants with COVID-19 | Westyn Branch Elliman | 2 | (Gremese et al., 2020) |
| Siltuximab | chimeric monoclonal Anti-IL-6 antibody |
220 participants with COVID-19 | A.O. Ospedale Papa Giovanni XXIII |
4 | (Palanques-Pastor et al., 2020) |
| Stem Cells Therapy | Anti-inflammation and immunoregulation– restoring immune balance and homeostasis and induction of immunological tolerance in autoimmune T cells | ARDS caused by SARS-Coronavirus-2 with 24 participants | University of Miami | 1 and 2 | (Golchin et al., 2020) |
| Thalidomide | anti-angiogenesis, anti-fibrotic, anti-inflammatory, and immune regulatory effect | 100 individuals with COVID-19 pneumonia | First Affiliated Hospital of Wenzhou Medical University |
2 | (Zhao et al., 2020) |
| Tocilizumab | Monoclonal humanized Recombinant anti-interleukin-6 antibody | 100 participants with SARS-CoV-2 | University Hospital Inselspital, Berne |
2 | (Luo et al., 2020) |
| Vitamin C | Antioxidant properties | 20 participants with COVID-19 | Hunter Holmes Mcguire Veteran Affairs Medical Center |
1 and 2 | (Erol, 2020) |
| Rhace2 | Blocking the Angiotensin II receptor | 2000 participants with COVID-19 | Apeiron Biologics |
2 | (Guo et al., 2020a) |
| Carrimycin | Macrolide antibiotic | 520 participants with 2019-nCoV | Beijing YouAn Hospital |
4 | (Cortegiani et al., 2020) |
| Heparin | Anticoagulant | 3000 individuals with COVID-19 pneumonia | University of Manitoba |
2 and 3 | (Liu et al., 2020) |
| Losartan | Blocking the Angiotensin II receptor | 200 participants with 2019-nCoV | University of Minnesota |
2 | (Gurwitz, 2020) |
SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; TMPRSS2, transmembrane serine protease 2; HCV, hepatitis C virus; HIV, Human immunodeficiency viruses; 3CL pro; 3-chymotrypsin-like protease; VEGF, vascular endothelial growth factor; IL-6, Interleukin 6; NLRP3, NLR family pyrin domain containing 3; FcR, Fc Receptor; TNFα, tumor necrosis factor α; COX-2, cyclooxygenase-2; RHACE, Recombinant human angiotensin-converting enzyme.
6. The role of ACE2 variants in SARS-CoV-2 infection and severity of COVID-19 disease
Clinical variability of COVID-19 is because of various known and unknown determinants of disease progression such as age, sex, host immune response, virus mutations, comorbidity, lifestyle, and host genetic background (Qiu et al., 2020). Genetic factors contributing to the outcome of SARS-CoV-2 infection are unknown; however, variants in the specific sites of the ACE2 gene could be regarded as a main genetic risk factor for COVID-19 (Hou et al., 2020b). Due to the central role of ACE2 as the primary cellular receptor for COVID-19, many researches have investigated the association of ACE2 variants and COVID-19 susceptibility. The relationship between genetic variations in ACE2 and their effects on ACE2 structure and function, which may also affect the binding of the virus and subsequent infection severity, has been described previously (Guo et al., 2020c).
By bioinformatics and docking studies, the interacting residues of ACE2 at the interface between ACE2 and spike protein have been identified. Some residues have been suggested to map to the interface site of viral ligand and human receptors, such as residues 19, 35, 38, 206, 211,219, 341, 353, 378, 468, and 547 (Benetti et al., 2020a; Lippi et al., 2020a). In another in silico study using molecular docking, six ACE2 missense variants, Ile21Thr, Ala25Thr, Lys26Arg, Glu37Lys, Thr55Ala, and Glu75Gly with higher affinity to spike protein and 11 variants, Ile21Val, Glu23Lys, Lys26Asp, Thr27Ala, Glu35Lys, Ser43Arg, Tyr50Phe, Asn51Asp, Asn58His, Lys68Asp, Met82Ile with lower affinity were computationally predicted (Calcagnile et al., 2021).
Missense mutations in ACE2 may perturb the receptor–ligand interactions. Some important missense mutations and their effects on the function and structure of ACE2 are demonstrated in Table 3 .
Table 3.
List of potential missense mutations of ACE2 from gnomAD and their possible alteration in SARS-CoV-2 infection.
| Wild aa | Mutant aa | Position | Function of wild residue | Effect of the mutation |
|---|---|---|---|---|
| His | Arg | 378 | By forming hydrogen bond and π-interaction stabilize the structure of the catalytic center and metal atom | This mutation weakens ACE2 peptidase activity by break the chelation network to Zn atoms. |
| Ser | Pro | 19 | Locating at the beginning of helix Ser19-Ile54 - forming hydrogen bonds and hydrophilic interactions to stabilize the helical structure. | This mutation would destabilize the helix structure since Proline has poor helix-forming tendencies. |
| Gly | Arg | 211 | Locating at the turn point of a loop and stabilizing the secondary structure of ACE2 by its strong hydrophobic interaction | It would weaken the hydrophobic interaction as arginine is not suitable for the loop turning and because of its hydrophilic feature. |
| Asp | Gly | 206 | This asparagine is on a helix to stabilize secondary structures by a hydrogen bond | This mutation influences on the ACE2 inhibitor binding site and disturbs the binding site of the catalytic zinc atom. |
| Arg | Cys/His | 219 | This arginine has a strong salt-bridge, hydrogen bond, and charge interaction to stabilize protein | Its mutation disrupts the strong interactions and destabilizes the protein structure. |
| Lys | Arg | 341 | Stabilizing the loop structure by a strong hydrogen bond | Weakening hydrogen bond and so destabilizing the loop structure |
| Ile | Val | 468 | This isoleucine located at a loop to stabilize two helical structures by hydrophobic and π-stacking interaction | Because valine has a shorter side-chain and weaker hydrophobic interaction so this mutation destabilizes the protein structure. |
| Ser | Cys | 547 | This serin stabilizes local helix by hydrogen bonds |
This mutation weakens the hydrogen bond so destabilizes the helical structure. |
Replaced amino acids can lead to various outcomes depend to replaced amino acids that is more impact potential variant would be appearing when the exchanged amino acids have different specifications; for instance, in p. Ser19Pro and p. Arg206Gly mutations, a polar amino acid (Ser or Arg) replaced with a nonpolar amino acid (Pro or Gly) or inversely, in p. Gly211Arg mutation, a nonpolar amino acid (Gly) replaced with a polar amino acid (Arg) (Guo et al., 2020b). As mentioned in Benetti et al. study, some of the ACE2 variants have direct effect on SARS-CoV-2 spike protein binding such as p. Leu351Val and p. Pro389His which interfere with the internalization process of virus to the host cell in Italian population.
A study reported ACE2 variants in two populations, Caucasians and Asians, and their possible effect on SARS-CoV-2 infection Rojas et al. (2020). Their results showed that two mutations have different frequencies in these two races, affecting ACE2 binding to spike protein. In another study on ACE2 variants in different ethnicities, Lys26Arg mutation in Ashkenazi Jews reduced ACE2 affinity, in contrast to mutations in other populations, including East Asian (Ile468Val), South Asian (Arg219Cys), African and African American (Lys341Arg), European (Asp206Gly), European and South Asian (Gly211Arg) (Ali et al., 2020). In an independent study, ACE2 genotypes in Africa and Eastern Mediterranean population were considered protective against COVID-19 (Karahalil and Elkama, 2020). In a cohort of Russian patients infected by COVID-19, three rare variants (rs146598386, rs755766792, rs73195521) may affect SARS-CoV-2 infection outcome were found (Shikov et al., 2020).
Hussain et al. examined multiple ACE2 gene variants and the binding efficiencies of various encoded proteins to SARS-CoV-2 spike protein, each varying in the polymorphisms of the RBD binding sequence (Hussain et al., 2020). Exchange of amino acids leads to alteration in chemical characteristics, peptidase activity of ACE2, ligand-receptor affinity, stability, conformational changes, hydrogen bonds and hydrophilic interactions, secondary and tertiary structures of ACE2, and subsequently, its interaction with RBD of spike protein and behavior with other involved molecules in ACE2 functions.
Although most ACE2 gene variants showed a high percentage of structural homologies, remarkable differences were also determined in central binding residues' spatial orientation. Two particular ACE2 alleles, including rs143936383 and rs73635825, demonstrate a relatively reduced binding affinity for the viral spike protein, which likely indicate a reduced likelihood of SARS-CoV-2 attachment and possible lowered susceptibility to infection. In a subsequent study, Stawiski and colleagues assessed the probability that several ACE2 variants, particularly those variants that estimated to interact with SARS-CoV-2 spike proteins, might be correlated to different host-virus interactions, leading to the alteration of pathogenicity and the SARS-CoV-2 virulence (Stawiski et al., 2020a). In brief, the authors performed a broader genomic analysis to construct a map involving synthetic mutants of ACE2, using samples from over 400 populations and 290,000 cases. Remarkably, they reported a list of ACE2 variants from existing genomic datasets utilizing structural modeling to characterize and identify ACE2 variants that were predicted to be protective against viral entry since they exhibited a lower binding affinity to SARS-CoV-2 spike protein such as S19P, I21V, E23K, K26R, T27A, N64K, T92I, Q102P, H378R. While ACE2 gene variants including K31R, N33I, H34R, E35K, E37K, D38V, Y50F, N51S, M62V, K68E, F72V, Y83H, G326E, G352V, D355N, Q388L, D509Y were predicted an enhanced binding propensity to the viral binding based on the interactions with the spike glycoprotein. Another study reported that some of ACE2 gene variants were not capable of biding to the spike protein, while other variants showed higher binding affinity (Procko, 2020). Renieri et al. mined whole-exome sequencing data obtained from about 7000 Italian individuals to identify 33 ACE2 gene variations, explaining various SARS-CoV-2 spike protein binding, affinity, internalization, and processing (Benetti et al., 2020a). Particularly, variant N720D was closely located to TMPRSS2 cleavage-dependent viral intake site. However, several mutations (P389H, L351V, W69C) were predicted to modulate conformational changes that modify interactions with the RBD of the spike protein due to their location in the proximity of sequences is fundamental for SARS-CoV-2 spike protein binding. In part, this finding elucidated the greater rate of case fatality in the Italian population than in china (Lippi et al., 2020c). Significantly, other particular ACE2 gene variants were identified in mentioned investigation, some of which are probably able to establish conformational variation in the RBD binding sequence, which can alter the SARS-CoV-2 spike protein binding affinity. Consequently, it is possible that some of the ACE2 variants can affect the stability of ACE2, and imposing conformational changes can impact its interaction with RBD of spike protein and decrease or increase the ligand-receptor affinity.
Besides, some variations of ACE2 are located at its proteolytic cleavage site (C terminal domain of ACE2), which can influence virus infection (Benetti et al., 2020a); because it was suggested that soluble ACE2 could act as a decoy receptor for virus and decrease virus intake by cell surface ACE2.
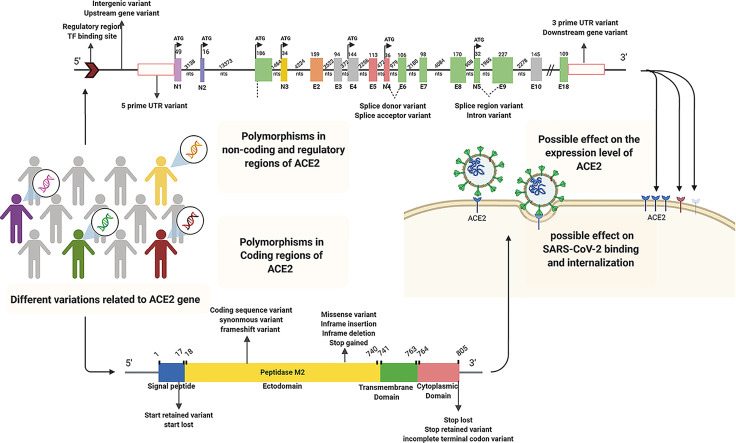
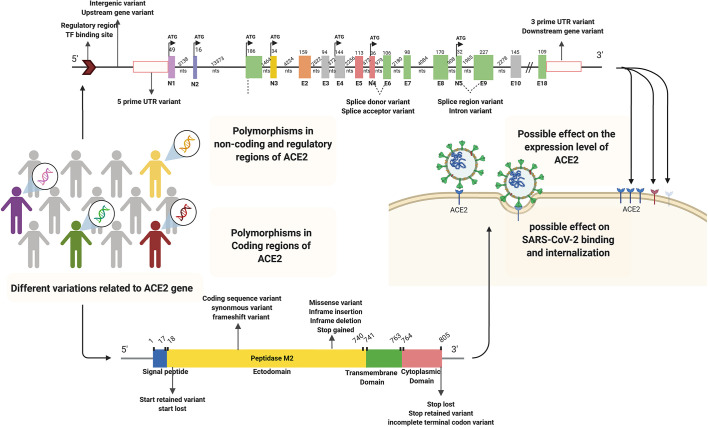
Notably, ACE2 level of expression, which differs among people, could correlate with susceptibility to the infection (Devaux et al., 2020). Polymorphisms of regulatory and non-coding regions such as promoters or 3’-UTR in ACE2 can have a role in the different expression levels of ACE2 among individuals (Chaudhary, 2020; Lippi et al., 2020b). Association between some mutation of ACE2 (affecting its expression) with the SARS-CoV intake was also strengthened the possible effect of ACE2 variant on SARS-CoV-2 entrance (Lippi et al., 2020a). In one study, 6 SNPs (rs4240157, rs6632680, rs1548474, rs4830965, rs1476524, and rs2048683) out of 61 evaluated ones in the interionic sequence of ACE2 showed significant association with the expression level of ACE2 and hospitalization in COVID-19 patients (Wooster et al., 2020). Therefore, polymorphisms of both coding and non-coding regions of ACE2 have influencing potential on virus susceptibility and COVID-19 severity (Fig. 5 ).
Fig. 5.
Schematic representation of different ACE2 gene polymorphisms in regulatory, non-coding, and coding regions with possible effects on SARS-CoV-2 binding, internalization, and ACE2 expression levels, respectively. The human ACE2 gene includes 5′UTR, 3′UTR, intronic, 18 exogenic regions (E1-E18), and five novel exogenic regions (N1-N5) participating in various alternative splicing. The polymorphisms contribute to ACE2 variations. Solid and dashed lines illustrate introns and possible splicing patterns, respectively. Abbreviations: ACE2; Angiotensin I converting enzyme 2, E; Exon, N; Novel exon, NTS; nucleotides, TF; Transcription factor, UTR; Untranslated Region.
Several studies investigated coding variants of ACE2 in multiple global databases. They estimated an increase or decrease of affinity between ligand and receptor by calculating the changes in the binding energy of those variants. As shown in Table 4 , the predicted function for some of the variants of ACE2, as mentioned above, was in contrast with the results of Gibson et al., which worked on the same database (Gibson et al., 2020).
Table 4.
Reported variants of ACE2 and their possible functional roles.
| Cohort/Database | Variants in ACE2 Protein (accession number) | Possible Effect | Ref. |
|---|---|---|---|
| Italian Cohort | V506A (rs775181355) | Destabilizing effect on spike protein and ACE2 interaction | (Benetti et al., 2020a) |
| N720D (rs41303171), K26R (rs4646116), and G211R (rs148771870) | Effect on the interaction between spike protein and ACE2 | ||
| GnomAD database (Canadian group) | E37K (rs146676783), T27A (rs781255386), K329G (rs143936283), and K26E (rs1299103394) | Increase the binding affinity between S protein and ACE2 | (Gibson et al., 2020) |
| N720D (rs41303171), S43R (rs1447927937), G326E (rs759579097), M82I (rs766996587), K26R (rs4646116) | Decrease the binding affinity between S protein and ACE2 | ||
| GnomAD database (UK group) | G326E (rs759579097) | Enhance ACE2 binding with spike protein | (MacGowan and Barton, 2020) |
| E37K (rs146676783), G352V (rs370610075), and D355N (rs961360700) | Weaken ACE2 binding with spike protein | ||
| Other large genomic datasets | S19P (rs73635825), I21V (rs778030746), E23K (rs756231991), K26R (rs4646116), T27A (rs781255386), N64K (rs1199100713), T92I (rs763395248), Q102P (rs1395878099), H378R (rs142984500) | Enhanced susceptibility to viral attachment | (Procko, 2020; Stawiski et al., 2020b) |
| K31R (rs758278442), N33I, H34R, E35K (rs1348114695), E37K (rs146676783), D38V, Y50F (rs1192192618), N51S (rs1569243690), M62V (rs1325542104), K68E (rs755691167), F72V (rs1256007252), Y83H (rs759134032), G326E (rs759579097), G352V (rs370610075), D355 N (rs961360700), Q388L (rs751572714), D509Y | Decrease attachment propensity to spike protein |
On the other hand, an evaluation of exome sequence data in the UK biobank in which some subjects were infected by SARS-CoV-2, showed that none of the variants of ACE2 and TMPRSS2 determine COVID-19 severity (Curtis, 2020). However, MacGowan and Barton from UK analyzed the variants in gnomAD for ACE2 and predicted some variants of ACE2 modify the structure and function of ACE2 and affect its binding affinity to spike protein (MacGowan and Barton, 2020). The impact of the missense variant depends on the functional and structural context of mutating residue, besides the physicochemical characteristics of the mutant residue. In Fig. 6 , according to gnomAD database, possible interactions between the ACE2 affinity variants and ACE2 expression polymorphism are highlighted.
Fig. 6.
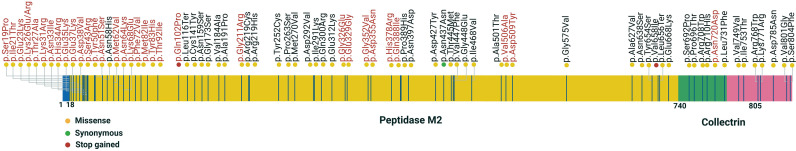
Schematic representation of the distribution of gnomAD (v2.1.1, v3) missense, synonymous, and stop gained variants in the coding sequence of ACE2. Altogether, 338 ACE2 coding variants in gnomAD are reported, of which 241 numbers of them are missense and 88 synonymous variants. In the illustration, 44 deleterious coding variants of the ACE2 gene identified in the gnomAD databases and 33 coding variants (red) mentioned in Table 4, possibly affecting the host-virus interactions (Hou et al., 2020a). Yellow, green and red dots indicate the missense, synonymous, and stop gained variants. Synonymous variants do not change the protein sequence and subsequently are less likely to have a functional effect. Generally, the influence of missense variants depends on the functional and structural context of the residues that have been mutated and the mutant's physicochemical features. Indeed, missense variants are determined in all the main functional domains in ACE2, involving the peptidase M2, collectrin domains. Synonymous variants, including frameshifts and stop, are also reported in gnomAD for ACE2. These mutation types can influence the expression levels and the protein structure, but their influence is less associated with the residue context than missense ones (Cao et al., 2020).
Altogether the variant could potentially affect ACE2 expression and function, which can contribute to SARS-CoV-2 spread among populations around the worldwide. Indeed, this study will be useful to identify patients at high risk of complications of the underlying disease, which may need monitoring and treatment strategies.
7. Conclusion
The increasing data available from SARS-CoV-2 infections highlighting the potential correlation to be made between particular gene loci and variable susceptibility to COVID-19. The immune system components react to SARS-CoV-2 mainly related to the inter-individual variation in COVID-19 severity, whereas genes linked with the binding affinity to the ACE2 cell receptor and the viral entry at the early stages of infection, considerably determine the variable COVID-19 severity and different susceptibility to SARS-CoV-2 infection. At this time, it is not clear whether the structure and function modulation of ACE2 in different variants could have a role in the probability of the infection or not. There are contradictory results for the impact of ACE2 variants in COVID-19 severity. Obviously, variants and mutations of spike protein and mainly its RBD have influenced the interaction between ACE2 and SARS-CoV-2.
There can be regarded as two possible scenarios for the effect of ACE2 variants in the results of SARS-CoV-2 infection. In the first scenario, multiple variants of ACE2 may have a synergistic effect on disease progression, and in the second, one/multiple rare variants may have a major impact on infection outcome. In order to confirmation of the findings, many studies need to be performed. Such studies could benefit the biopharmaceutical industry to design effective approaches for therapy and vaccination of COVID-19 disease.
Funding
The authors received no specific funding for this work.
Declaration of Competing Interest
The authors declare no conflicts of interest.
References
- Alanagreh L.a., Alzoughool F., Atoum M. The human coronavirus disease COVID-19: Its origin, characteristics, and insights into potential drugs and its mechanisms. Pathogens. 2020;9:331. doi: 10.3390/pathogens9050331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alenina N., Bader M. ACE2 in brain physiology and pathophysiology: evidence from transgenic animal models. Neurochem. Res. 2019;44:1323–1329. doi: 10.1007/s11064-018-2679-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali F., Elserafy M., Alkordi M.H., Amin M. ACE2 coding variants in different populations and their potential impact on SARS-CoV-2 binding affinity. Biochem. Biophys. Rep. 2020;24 doi: 10.1016/j.bbrep.2020.100798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alizadeh-Navaei R., Lagzian M., Valadan R., Saeedi M., Roozbeh F., Hedayatizadeh-Omran A., Amanlou M. 2020. Repurposing Naproxen as a Potential Antiviral Agent Against SARS-CoV-2. [DOI] [PubMed] [Google Scholar]
- Anguiano L., Riera M., Pascual J., Valdivielso J.M., Barrios C., Betriu A., Mojal S., Fernández E., Soler M.J., study, o.b.o.t.i.f.t.N, Castro E., María V., Molí T., Soria M., Regidor A., José M., Jaume A., Esther P., Coloma A., Rubio B., Rodríguez B., Sara B.-G., Jordi B.S., Josep B.A., Romero C., Juan B., Salomé M.C., Jesús C.V., Pilar C.A., Jordi C.B., Aleix C.A., Elisabet M.J., Jesús C.P., Secundino C.G., Saray L.P., Lourdes C.M., Isabel C., Ma Teresa, C.J, Marta C.I., Fernando, d.Á, Covadonga H.O., Gabriel, d.A.d.l.F, Ma Dolores, d.P.y.P, Rafael D.-T.I., Marta D., Verónica D., Sara E.T., Ma José, F.R, Ma Loreto, F.R, Guillermina F., Antonio G.S., Cesar G.C., Herrera G., Antonio L., Mercedes G.M., Luis G.S., Maria A., Luis G.J., Emma H.L., Luis L.J., Antonio L.C., Álvarez M., Pedro J., Nàdia M.A., Jesús M.G., Alberto M.C., María M.V., Isabel M., Iñigo M.E., Silvia M.L.H., Ricardo M.M., Antonia M.V., Ana Beatriz M.D., González N., Juan F., Javier N., Agustín C., Enrique N.F., Alberto O., Beatriz F., Vicente P., Miguel P.F., Ana P.D., Celestino P.H., Ma Dolores P.G., Mario P.V., Carmina P.M., Maite R.G., Esther R., Pilar R., Mercedes S.L., Puerto M., Isabel A., Tomero S., Antonio J., Emilio S.J., Ramon S.L., Ramon S., Maria S., Oreto P., Fernando S., Daniel T., Fernando T.M., Carrasco U., Javier J., Ildefonso V.C., Ma Merce, V.d.P, Ruiz V., Rafael C., study, o.b.o.t.i.f.t.N Circulating angiotensin-converting enzyme 2 activity in patients with chronic kidney disease without previous history of cardiovascular disease. Nephrol. Dial. Transplant. 2015;30:1176–1185. doi: 10.1093/ndt/gfv025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arden K.E., Nissen M.D., Sloots T.P., Mackay I.M. New human coronavirus, HCoV-NL63, associated with severe lower respiratory tract disease in Australia. J. Med. Virol. 2005;75:455–462. doi: 10.1002/jmv.20288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benetti E., Tita R., Spiga O., Ciolfi A., Birolo G., Bruselles A., Doddato G., Giliberti A., Marconi C., Musacchia F., Pippucci T., Torella A., Trezza A., Valentino F., Baldassarri M., Brusco A., Asselata R., Bruttini M., Furini S., Seri M., Nigro V., Matullao G., Tartaglia M., Mari F., Renieri A., Pinto M. ACE2 gene variants may underlie interindividual variability and susceptibility to COVID-19 in the Italian population. Eur. J. Hum. Genet. 2020;28:1602–1614. doi: 10.1038/s41431-020-0691-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benetti E., Tita R., Spiga O., Ciolfi A., Birolo G., Bruselles A., Doddato G., Giliberti A., Marconi C., Musacchia F., Pippucci T., Torella A., Trezza A., Valentino F., Baldassarri M., Brusco A., Asselta R., Bruttini M., Furini S., Seri M., Nigro V., Matullo G., Tartaglia M., Mari F., Frullanti E., Fallerini C., Daga S., Croci S., Amitrano S., Fava F., Montagnani F., Di Sarno L., Tommasi A., Palmieri M., Emiliozzi A., Fabbiani M., Rossetti B., Zanelli G., Bergantini L., D’Alessandro M., Cameli P., Bennet D., Anedda F., Marcantonio S., Scolletta S., Franchi F., Mazzei M.A., Conticini E., Cantarini L., Frediani B., Tacconi D., Feri M., Scala R., Spargi G., Corridi M., Nencioni C., Caldarelli G.P., Spagnesi M., Piacentini P., Bandini M., Desanctis E., Canaccini A., Spertilli C., Donati A., Guidelli L., Croci L., Verzuri A., Anemoli V., Ognibene A., Vaghi M., D’Arminio Monforte A., Merlini E., Mondelli M.U., Mantovani S., Ludovisi S., Girardis M., Venturelli S., Sita M., Cossarizza A., Antinori A., Vergori A., Rusconi S., Siano M., Gabrieli A., Riva A., Francisci D., Schiaroli E., Scotton P.G., Andretta F., Panese S., Scaggiante R., Parisi S.G., Castelli F., Quiros-Roldan M.E., Magro P., Minardi C., Castelli D., Polesini I., Della Monica M., Piscopo C., Capasso M., Russo R., Andolfo I., Iolascon A., Carella M., Castori M., Merla G., Aucella F., Raggi P., Marciano C., Perna R., Bassetti M., Di Biagio A., Sanguinetti M., Masucci L., Gabbi C., Valente S., Guerrini S., Meloni I., Mencarelli M.A., Rizzo C.L., Bargagli E., Mandalà M., Giorli A., Salerni L., Fiorentino G., Zucchi P., Parravicini P., Menatti E., Baratti S., Trotta T., Giannattasio F., Coiro G., Lena F., Coviello D.A., Mussini C., Renieri A., Pinto A.M., Study G.-C.M. ACE2 gene variants may underlie interindividual variability and susceptibility to COVID-19 in the Italian population. Eur. J. Hum. Genet. 2020;28:1602–1614. doi: 10.1038/s41431-020-0691-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein K.E., Khan Z., Giani J.F., Cao D.-Y., Bernstein E.A., Shen X.Z. Angiotensin-converting enzyme in innate and adaptive immunity. Nat. Rev. Nephrol. 2018;14:325. doi: 10.1038/nrneph.2018.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein K.E., Khan Z., Giani J.F., Cao D.Y., Bernstein E.A., Shen X.Z. Angiotensin-converting enzyme in innate and adaptive immunity. Nat. Rev. Nephrol. 2018;14:325–336. doi: 10.1038/nrneph.2018.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia M., Zemans R.L., Jeyaseelan S. Role of chemokines in the pathogenesis of acute lung injury. Am. J. Respir. Cell Mol. Biol. 2012;46:566–572. doi: 10.1165/rcmb.2011-0392TR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo D., Solano C., Giménez E., Remigia M.J., Corrales I., Amat P., Navarro D. Effect of the IL28B Rs12979860 C/T polymorphism on the incidence and features of active cytomegalovirus infection in allogeneic stem cell transplant patients. J. Med. Virol. 2014;86:838–844. doi: 10.1002/jmv.23865. [DOI] [PubMed] [Google Scholar]
- Calcagnile M., Forgez P., Iannelli A., Bucci C., Alifano M., Alifano P. Molecular docking simulation reveals ACE2 polymorphisms that may increase the affinity of ACE2 with the SARS-CoV-2 Spike protein. Biochimie. 2021;180:143–148. doi: 10.1016/j.biochi.2020.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camargo S.M.R., Singer D., Makrides V., Huggel K., Pos K.M., Wagner C.A., Kuba K., Danilczyk U., Skovby F., Kleta R., Penninger J.M., Verrey F. Tissue-Specific Amino Acid Transporter Partners ACE2 and Collectrin Differentially Interact With Hartnup Mutations. Gastroenterology 136. 2009;e873:872–882. doi: 10.1053/j.gastro.2008.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron M.J., Bermejo-Martin J.F., Danesh A., Muller M.P., Kelvin D.J. Human immunopathogenesis of severe acute respiratory syndrome (SARS) Virus Res. 2008;133:13–19. doi: 10.1016/j.virusres.2007.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Li L., Feng Z., Wan S., Huang P., Sun X., Wen F., Huang X., Ning G., Wang W. Comparative genetic analysis of the novel coronavirus (2019-nCoV/SARS-CoV-2) receptor ACE2 in different populations. Cell Discov. 2020;6:1–4. doi: 10.1038/s41421-020-0147-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F.-W., Kok K.-H., Zhu Z., Chu H., To, K.K.-W, Yuan S., Yuen K.-Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microb. Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Channappanavar R., Perlman S. Springer; 2017. Pathogenic Human Coronavirus Infections: Causes and Consequences of Cytokine Storm and Immunopathology, Seminars in Immunopathology; pp. 529–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary M. COVID-19 susceptibility: potential of ACE2 polymorphisms. Egypt. J. Med. Human Genet. 2020;21:1–8. doi: 10.1186/s43042-020-00099-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.-M.A., Liang S.-Y., Shih Y.-P., Chen C.-Y., Lee Y.-M., Chang L., Jung S.-Y., Ho M.-S., Liang K.-Y., Chen H.-Y. Epidemiological and genetic correlates of severe acute respiratory syndrome coronavirus infection in the hospital with the highest nosocomial infection rate in Taiwan in 2003. J. Clin. Microbiol. 2006;44:359–365. doi: 10.1128/JCM.44.2.359-365.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Cai H., Xiang N., Tien P., Ahola T., Guo D. Functional screen reveals SARS coronavirus nonstructural protein nsp14 as a novel cap N7 methyltransferase. Proc. Natl. Acad. Sci. 2009;106:3484–3489. doi: 10.1073/pnas.0808790106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Zhang Z., Wang L., Huang Z., Gong F., Li X., Chen Y. 2020. First clinical study using HCV protease inhibitor danoprevir to treat naive and experienced COVID-19 patients. MedRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe Y.J. 2020. Coronavirus disease-19: The First 7,755 Cases in the Republic of Korea. medRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury M.S., Rathod J., Gernsheimer J. A Rapid systematic review of clinical trials utilizing chloroquine and hydroxychloroquine as a treatment for COVID-19. Acad. Emerg. Med. 2020;27:493–504. doi: 10.1111/acem.14005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coperchini F., Chiovato L., Croce L., Magri F., Rotondi M. The Cytokine storm in COVID-19: an overview of the involvement of the chemokine/chemokine-receptor system. Cytokine Growth Factor Rev. 2020;53:25–32. doi: 10.1016/j.cytogfr.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortegiani A., Ingoglia G., Ippolito M., Giarratano A., Einav S. A systematic review on the efficacy and safety of chloroquine for the treatment of COVID-19. J. Crit. Care. 2020;57:279–283. doi: 10.1016/j.jcrc.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crackower M.A., Sarao R., Oliveira-dos-Santos A.J., Da Costa J., Zhang L. Angiotensin-converting enzyme 2 is an essential regulator of heart function. Nature. 2002;417:822–828. doi: 10.1038/nature00786. [DOI] [PubMed] [Google Scholar]
- Curtis D. 2020. Variants in ACE2 and TMPRSS2 genes are not major determinants of COVID-19 severity in UK Biobank subjects. medRxiv preprint. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daffis S., Szretter K.J., Schriewer J., Li J., Youn S., Errett J., Lin T.-Y., Schneller S., Zust R., Dong H. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature. 2010;468:452–456. doi: 10.1038/nature09489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilczyk U., Sarao R., Remy C., Benabbas C., Stange G., Richter A., Arya S., Pospisilik J.A., Singer D., Camargo S.M.R., Makrides V., Ramadan T., Verrey F., Wagner C.A., Penninger J.M. Essential role for collectrin in renal amino acid transport. Nature. 2006;444:1088–1091. doi: 10.1038/nature05475. [DOI] [PubMed] [Google Scholar]
- De Groot R., Luytjes W., Horzinek M., Van der Zeijst B., Spaan W., Lenstra J. Evidence for a coiled-coil structure in the spike proteins of coronaviruses. J. Mol. Biol. 1987;196:963–966. doi: 10.1016/0022-2836(87)90422-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Meyer S., Bojkova D., Cinatl J., Van Damme E., Meng C.B., Van Loock M., Woodfall B., Ciesek S. Lack of antiviral activity of darunavir against SARS-CoV-2. Int. J. Infect. Dis. 2020;97:7–10. doi: 10.1016/j.ijid.2020.05.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaux C.A., Rolain J.M., Raoult D. ACE2 receptor polymorphism: Susceptibility to SARS-CoV-2, hypertension, multi-organ failure, and COVID-19 disease outcome. J. Microbiol. Immunol. Infect. 2020;53:425–435. doi: 10.1016/j.jmii.2020.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diurno F., Numis F., Porta G., Cirillo F., Maddaluno S., Ragozzino A., De Negri P., Di Gennaro C., Pagano A., Allegorico E. Eculizumab treatment in patients with COVID-19: preliminary results from real life ASL Napoli 2 Nord experience. Eur. Rev. Med. Pharmacol. Sci. 2020;24:4040–4047. doi: 10.26355/eurrev_202004_20875. [DOI] [PubMed] [Google Scholar]
- Donoghue M., Hsieh F., Baronas E., Godbout K., Gosselin M., Stagliano N., Donovan M., Woolf B., Robison K., Jeyaseelan R. A novel angiotensin-converting enzyme–related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1-9. Circ. Res. 2000;87:e1–e9. doi: 10.1161/01.res.87.5.e1. [DOI] [PubMed] [Google Scholar]
- Donoghue M., Hsieh F., Baronas E., Godbout K., Gosselin M., Stagliano N., Donovan M., Woolf B., Robison K., Jeyaseelan R., Breitbart R.E., Acton S. A novel angiotensin-converting enzyme-related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1-9. Circ. Res. 2000;87:E1–E9. doi: 10.1161/01.res.87.5.e1. [DOI] [PubMed] [Google Scholar]
- Dorward J., Gbinigie K. 2020. Lopinavir/ritonavir: A rapid review of effectiveness in COVID-19. [Google Scholar]
- Duan Y., Liu Q., Zhao S., Huang F., Ren L., Liu L., Zhou Y. The trial of chloroquine in the treatment of Corona virus disease 2019 (COVID-19) and its research progress in forensic toxicology. Fa yi xue za zhi. 2020;36 doi: 10.12116/j.issn.1004-5619.2020.02.002. [DOI] [PubMed] [Google Scholar]
- Erol A. 2020. High-dose intravenous vitamin C treatment for COVID-19. [Google Scholar]
- Fagerberg L., Hallstrom B.M., Oksvold P., Kampf C., Djureinovic D., Odeberg J., Habuka M., Tahmasebpoor S., Danielsson A., Edlund K., Asplund A., Sjostedt E., Lundberg E., Szigyarto C.A.K., Skogs M., Ottosson Takanen J., Berling H., Tegel H., Mulder J., Nilsson P., Schwenk J.M., Lindskog C., Danielsson F., Mardinoglu A., Sivertsson A., Von Feilitzen K., Forsberg M., Zwahlen M., Olsson I., Navani S., Huss M., Nielsen J., Ponten F., Uhlen M. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell. Proteomics. 2014;13:397–406. doi: 10.1074/mcp.M113.035600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foerch C., Friedauer L., Bauer B., Wolf T., Adam E.H. Severe COVID-19 infection in a patient with multiple sclerosis treated with fingolimod. Mult. Sclerosis Related Disord. 2020;102180 doi: 10.1016/j.msard.2020.102180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier D., Luft F.C., Bader M., Ganten D., Andrade-Navarro M.A. Emergence and evolution of the renin-angiotensin-aldosterone system. J. Mol. Med. 2012;90:495–508. doi: 10.1007/s00109-012-0894-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freundt E.C., Yu L., Park E., Lenardo M.J., Xu X.-N. Molecular determinants for subcellular localization of the severe acute respiratory syndrome coronavirus open reading frame 3b protein. J. Virol. 2009;83:6631–6640. doi: 10.1128/JVI.00367-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher T.M. Murine coronavirus spike glycoprotein: Receptor binding and membrane fusion activities. Adv. Exp. Med. Biol. 2001:183–192. [PubMed] [Google Scholar]
- García-Salido A. Narrative review of the immune response against coronavirus: an overview, applicability for SARS-COV-2, and therapeutic implications. Anales de Pediatría (English Edition) 2020;93 doi: 10.1016/j.anpede.2020.04.006. 60. e61-60. e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George P.M., Wells A.U., Jenkins R.G. Pulmonary fibrosis and COVID-19: the potential role for antifibrotic therapy. Lancet Respir. Med. 2020;8:807–815. doi: 10.1016/S2213-2600(20)30225-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianni S., Fakhr B.S., Morais C.C.A., Di Fenza R., Larson G., Pinciroli R., Houle T., Mueller A.L., Bellavia A., Kacmarek R. 2020. Nitric oxide gas inhalation to prevent COVID-2019 in healthcare providers. medRxiv. [Google Scholar]
- Gibson W.T., Evans D.M., Jones J.A., Jones S.J. ACE 2 Coding Variants: A Potential X-linked Risk Factor 1 for COVID-19 Disease. bioRxiv preprint. 2020;2020 2004.2005.026633. [Google Scholar]
- Golchin A., Seyedjafari E., Ardeshirylajimi A. Mesenchymal stem cell therapy for COVID-19: present or future. Stem Cell Rev. Rep. 2020:1–7. doi: 10.1007/s12015-020-09973-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gremese E., Cingolani A., Bosello S.L., Alivernini S., Tolusso B., Perniola S., Landi F., Pompili M., Murri R., Santoliquido A. 2020. Sarilumab use in severe SARS-CoV-2 pneumonia. medRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J., Gong E., Zhang B., Zheng J., Gao Z., Zhong Y., Zou W., Zhan J., Wang S., Xie Z., Zhuang H., Wu B., Zhong H., Shao H., Fang W., Gao D., Pei F., Li X., He Z., Xu D., Shi X., Anderson V.M., Leong A.S.Y. Multiple organ infection and the pathogenesis of SARS. J. Exp. Med. 2005;202:415–424. doi: 10.1084/jem.20050828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J., Huang Z., Lin L., Lv J. Coronavirus disease 2019 (covid-19) and cardiovascular disease: a viewpoint on the potential influence of angiotensin-converting enzyme inhibitors/angiotensin receptor blockers on onset and severity of severe acute respiratory syndrome coronavirus 2 infection. J. Am. Heart Assoc. 2020;9 doi: 10.1161/JAHA.120.016219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X., Chen Z., Xia Y., Lin W., Li H. 2020. Investigation of the genetic variation in ACE2 on the structural recognition by the novel coronavirus (SARS-CoV-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X., Chen Z., Xia Y., Lin W., Li H. Investigation of the genetic variation in ACE2 on the structural recognition by the novel coronavirus (SARS-CoV-2) J. Transl. Med. 2020;18:1–9. doi: 10.1186/s12967-020-02486-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurwitz D. Angiotensin receptor blockers as tentative SARS-CoV-2 therapeutics. Drug Dev. Res. 2020;81:537–540. doi: 10.1002/ddr.21656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy J., Jackson R., Acharya K., Sturrock E., Hooper N., Turner A. Angiotensin-converting enzyme-2 (ACE2): comparative modelling of the active site, substrate specificity and chloride sensitivity. Biochemistry. 2003;42:13185. doi: 10.1021/bi035268s. [DOI] [PubMed] [Google Scholar]
- Guy J.L., Jackson R.M., Acharya K.R., Sturrock E.D., Hooper N.M., Turner A.J. Angiotensin-converting enzyme-2 (ACE2): comparative modeling of the active site, specificity requirements, and chloride dependence. Biochemistry. 2003;42:13185–13192. doi: 10.1021/bi035268s. [DOI] [PubMed] [Google Scholar]
- Habtemariam S., Nabavi S.F., Ghavami S., Cismaru C.A., Neagoe I.B., Nabavi S.M. Possible use of the mucolytic drug, bromhexine hydrochloride, as a prophylactic agent against SARS-CoV-2 infection based on its action on the Transmembrane Serine Protease 2. Pharmacol. Res. 2020;157:104853. doi: 10.1016/j.phrs.2020.104853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajeer A.H., Balkhy H., Johani S., Yousef M.Z., Arabi Y. Association of human leukocyte antigen class II alleles with severe Middle East respiratory syndrome-coronavirus infection. Ann. Thorac. Med. 2016;11:211. doi: 10.4103/1817-1737.185756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto T., Perlot T., Rehman A., Trichereau J., Ishiguro H., Paolino M., Sigl V., Hanada T., Hanada R., Lipinski S., Wild B., Camargo S.M.R., Singer D., Richter A., Kuba K., Fukamizu A., Schreiber S., Clevers H., Verrey F., Rosenstiel P., Penninger J.M. ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature. 2012;487:477–481. doi: 10.1038/nature11228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann H., Pyrc K., Van Der Hoek L., Geier M., Berkhout B., Pöhlmann S. Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc. Natl. Acad. Sci. U. S. A. 2005;102:7988–7993. doi: 10.1073/pnas.0409465102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y., Zhao J., Martin W., Kallianpur A., Chung M.K., Jehi L., Sharifi N., Erzurum S., Eng C., Cheng F. New insights into genetic susceptibility of COVID-19: an ACE2 and TMPRSS2 polymorphism analysis. BMC Med. 2020;18:1–8. doi: 10.1186/s12916-020-01673-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y., Zhao J., Martin W., Kallianpur A., Chung M.K., Jehi L., Sharifi N., Erzurum S., Eng C., Cheng F. New insights into genetic susceptibility of COVID-19: an ACE2 and TMPRSS2 polymorphism analysis. BMC Med. 2020;18:216. doi: 10.1186/s12916-020-01673-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B., Huang S., Yin L. The cytokine storm and COVID-19. J. Med. Virol. 2021;93:250–256. doi: 10.1002/jmv.26232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huet T., Beaussier H., Voisin O., Jouveshomme S., Dauriat G., Lazareth I., Sacco E., Naccache J.-M., Bézie Y., Laplanche S. Anakinra for severe forms of COVID-19: a cohort study. Lancet Rheumatol. 2020;2:e393–e400. doi: 10.1016/S2665-9913(20)30164-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain M., Jabeen N., Amanullah A., Baig A.A., Aziz B., Shabbir S., Raza F. 2020. Structural basis of SARS-CoV-2 spike protein priming by TMPRSS2. BioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai Y., Kuba K., Rao S., Huan Y., Guo F., Guan B., Yang P., Sarao R., Wada T., Leong-Poi H., Crackower M.A., Fukamizu A., Hui C.C., Hein L., Uhlig S., Slutsky A.S., Jiang C., Penninger J.M. Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature. 2005;436:112–116. doi: 10.1038/nature03712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue N., Kasahara T., Ikawa M., Okabe M. Identification and disruption of sperm-specific angiotensin converting enzyme-3 (ACE3) in mouse. PLoS One. 2010;5 doi: 10.1371/journal.pone.0010301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffers S.A., Tusell S.M., Gillim-Ross L., Hemmila E.M., Achenbach J.E., Babcock G.J., Thomas W.D., Jr., Thackray L.B., Young M.D., Mason R.J., Ambrosino D.M., Wentworth D.E., DeMartini J.C., Holmes K.V. CD209L (L-SIGN) is a receptor for severe acute respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. U. S. A. 2004;101:15748–15753. doi: 10.1073/pnas.0403812101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karahalil B., Elkama A. The impact of ACE2 gene polymorphism in the development of COVID-19 disease. Gazi Med. J. 2020;31 [Google Scholar]
- Kikkert M. Innate Immune Evasion by Human Respiratory RNA Viruses. J. Innate Immun. 2020;12:4–20. doi: 10.1159/000503030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klempin F., Mosienko V., Matthes S., Villela D.C., Todiras M., Penninger J.M., Bader M., Santos R.A.S., Alenina N. Depletion of angiotensin-converting enzyme 2 reduces brain serotonin and impairs the running-induced neurogenic response. Cell. Mol. Life Sci. 2018;75:3625–3634. doi: 10.1007/s00018-018-2815-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knaap R.C., Fernández-Delgado R., Dalebout T.J., Oreshkova N., Bredenbeek P.J., Enjuanes L., Sola I., Snijder E.J., Kikkert M. 2019. The deubiquitinating activity of Middle East respiratory syndrome coronavirus papain-like protease delays the innate immune response and enhances virulence in a mouse model. bioRxiv; p. 751578. [Google Scholar]
- Komatsu T., Suzuki Y., Imai J., Sugano S., Hida M., Tanigami A., Muroi S., Yamada Y., Hanaoka K. Molecular cloning, mRNA expression and chromosomal localization of mouse angiotensin-converting enzyme-related carboxypeptidase (mACE2) Mitochondrial DNA. 2002;13:217–220. doi: 10.1080/1042517021000021608. [DOI] [PubMed] [Google Scholar]
- Kuhn J.H., Li W., Choe H., Farzan M. Angiotensin-converting enzyme 2: a functional receptor for SARS coronavirus. Cell. Mol. Life Sci. 2004;61:2738–2743. doi: 10.1007/s00018-004-4242-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. Receptor recognition and cross-species infections of SARS coronavirus. Antivir. Res. 2013;100:246–254. doi: 10.1016/j.antiviral.2013.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasllieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greeneugh T.C., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Greenough T.C., Moore M.J., Vasilieva N., Somasundaran M., Sullivan J.L., Farzan M., Choe H. Efficient replication of severe acute respiratory syndrome coronavirus in mouse cells is limited by murine angiotensin-converting enzyme 2. J. Virol. 2004;78:11429–11433. doi: 10.1128/JVI.78.20.11429-11433.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Wong S.K., Li F., Kuhn J.H., Huang I.C., Choe H., Farzan M. Animal origins of the severe acute respiratory syndrome coronavirus: insight from ACE2-S-protein interactions. J. Virol. 2006;80:4211–4219. doi: 10.1128/JVI.80.9.4211-4219.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Zhao R., Zheng S., Chen X., Wang J., Sheng X., Zhou J., Cai H., Fang Q., Yu F. Lack of vertical transmission of severe acute respiratory syndrome coronavirus 2, China. Emerg. Infect. Dis. 2020;26:1335. doi: 10.3201/eid2606.200287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippi G., Lavie C.J., Henry B.M., Sanchis-Gomar F. Do genetic polymorphisms in angiotensin converting enzyme 2 (ACE2) gene play a role in coronavirus disease 2019 (COVID-19)? Clin. Chem. Lab. Med. 2020;58:1415–1422. doi: 10.1515/cclm-2020-0727. [DOI] [PubMed] [Google Scholar]
- Lippi G., Lavie C.J., Henry B.M., Sanchis-Gomar F. Do genetic polymorphisms in angiotensin converting enzyme 2 (ACE2) gene play a role in coronavirus disease 2019 (COVID-19)? Clin. Chem. Lab. Med. (CCLM) 2020;1 doi: 10.1515/cclm-2020-0727. [DOI] [PubMed] [Google Scholar]
- Lippi G., Mattiuzzi C., Sanchis-Gomar F., Henry B.M. Clinical and demographic characteristics of patients dying from COVID-19 in Italy vs China. J. Med. Virol. 2020;92:1759–1760. doi: 10.1002/jmv.25860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipsitch M., Cohen T., Cooper B., Robins J.M., Ma S., James L., Gopalakrishna G., Chew S.K., Tan C.C., Samore M.H. Transmission dynamics and control of severe acute respiratory syndrome. science. 2003;300:1966–1970. doi: 10.1126/science.1086616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Wu P., Gao F., Qi J., Kawana-Tachikawa A., Xie J., Vavricka C.J., Iwamoto A., Li T., Gao G.F. Novel immunodominant peptide presentation strategy: a featured HLA-A* 2402-restricted cytotoxic T-lymphocyte epitope stabilized by intrachain hydrogen bonds from severe acute respiratory syndrome coronavirus nucleocapsid protein. J. Virol. 2010;84:11849–11857. doi: 10.1128/JVI.01464-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Li J., Arnold K., Pawlinski R., Key N.S. Using heparin molecules to manage COVID-2019. Res Pract Thromb Haemost. 2020;4:518–523. doi: 10.1002/rth2.12353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Caputo S., Corso G., Clerici M., Santantonio T.A. Baricitinib: a chance to treat COVID-19? J. Med. Virol. 2020;92:2343–2344. doi: 10.1002/jmv.26033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokugamage K.G., Hage A., Schindewolf C., Rajsbaum R., Menachery V.D. 2020. SARS-CoV-2 sensitive to type I interferon pretreatment. BioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan J., Jin X., Lu Y., Zhang L. SARS-CoV-2 spike protein favors ACE2 from Bovidae and Cricetidae. J. Med. Virol. 2020;92:1649–1656. doi: 10.1002/jmv.25817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo P., Liu Y., Qiu L., Liu X., Liu D., Li J. Tocilizumab treatment in COVID-19: A single center experience. J. Med. Virol. 2020;92:814–818. doi: 10.1002/jmv.25801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGowan S., Barton G.J. Missense variants in ACE2 are predicted to encourage and inhibit interaction with SARS-CoV-2 Spike and contribute to genetic risk in COVID-19. bioRxiv. 2020 2020.2005.2003.074781. [Google Scholar]
- Magrone T., Magrone M., Jirillo E. Focus on receptors for coronaviruses with special reference to angiotensin-converting enzyme 2 as a potential drug target-a perspective. Endocr Metab Immune Disord Drug Targets (Formerly Current Drug Targets-Immune, Endocrine & Metabolic Disorders) 2020;20:807–811. doi: 10.2174/1871530320666200427112902. [DOI] [PubMed] [Google Scholar]
- Mantlo E., Bukreyeva N., Maruyama J., Paessler S., Huang C. Antiviral activities of type I interferons to SARS-CoV-2 infection. Antivir. Res. 2020;104811 doi: 10.1016/j.antiviral.2020.104811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason R.J. Pathogenesis of COVID-19 from a cell biology perspective. Eur Respiratory Soc. 2020;55 doi: 10.1183/13993003.00607-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelemans T., Kikkert M. Viral innate immune evasion and the pathogenesis of emerging RNA virus infections. Viruses. 2019;11:961. doi: 10.3390/v11100961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikpouraghdam M., Farahani A.J., Alishiri G., Heydari S., Ebrahimnia M., Samadinia H., Sepandi M., Jafari N.J., Izadi M., Qazvini A. Epidemiological characteristics of coronavirus disease 2019 (COVID-19) patients in IRAN: a single center study. J. Clin. Virol. 2020;127:104378. doi: 10.1016/j.jcv.2020.104378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novel C.P.E.R.E. The epidemiological characteristics of an outbreak of 2019 novel coronavirus diseases (COVID-19) in China. Zhonghua liu xing bing xue za zhi= Zhonghua liuxingbingxue zazhi. 2020;41:145. doi: 10.3760/cma.j.issn.0254-6450.2020.02.003. [DOI] [PubMed] [Google Scholar]
- Organization., W.H . 2020. Coronavirus disease 2019 (COVID-19) Situation Report-63. [Google Scholar]
- Palanques-Pastor T., López-Briz E., Andrés J.L.P. Involvement of interleukin 6 in SARS-CoV-2 infection: siltuximab as a therapeutic option against COVID-19. Eur. J. Hosp. Pharm. 2020;27:297–298. doi: 10.1136/ejhpharm-2020-002322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris J., Guan Y., Yuen K. Severe acute respiratory syndrome. Nat. Med. 2004;10:S88–S97. doi: 10.1038/nm1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris J.S.M., Guan Y., Yuen K.Y. Severe acute respiratory syndrome. Nat. Med. 2004;10:S88–S97. doi: 10.1038/nm1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman S., Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat. Rev. Microbiol. 2009;7:439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poutanen S.M., Low D.E., Henry B., Finkelstein S., Rose D., Green K., Tellier R., Draker R., Adachi D., Ayers M. Identification of severe acute respiratory syndrome in Canada. N. Engl. J. Med. 2003;348:1995–2005. doi: 10.1056/NEJMoa030634. [DOI] [PubMed] [Google Scholar]
- Procko E. The sequence of human ACE2 is suboptimal for binding the S spike protein of SARS coronavirus 2. BioRxiv. 2020 [Google Scholar]
- Qiu H., Wu J., Hong L., Luo Y., Song Q., Chen D. Clinical and epidemiological features of 36 children with coronavirus disease 2019 (COVID-19) in Zhejiang, China: an observational cohort study. Lancet Infect. Dis. 2020;20:689–696. doi: 10.1016/S1473-3099(20)30198-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabaan A.A., Al-Ahmed S.H., Haque S., Sah R., Tiwari R., Malik Y.S., Dhama K., Yatoo M.I., Bonilla-Aldana D.K., Rodriguez-Morales A.J. SARS-CoV-2, SARS-CoV, and MERS-CoV: a comparative overview. Infez Med. 2020;28:174–184. [PubMed] [Google Scholar]
- Rahmani-Kukia N., Abbasi A., Pakravan N., Hassan Z.M. Measurement of oxidized albumin: An opportunity for diagnoses or treatment of COVID-19. Bioorg. Chem. 2020;105 doi: 10.1016/j.bioorg.2020.104429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice G.I., Thomas D.A., Grant P.J., Turner A.J., Hooper N.M. Evaluation of angiotensin-converting enzyme (ACE), its homologue ACE2 and neprilysin in angiotensin peptide metabolism. Biochem. J. 2004;383:45–51. doi: 10.1042/BJ20040634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley S., Fraser C., Donnelly C.A., Ghani A.C., Abu-Raddad L.J., Hedley A.J., Leung G.M., Ho L.-M., Lam T.-H., Thach T.Q. Transmission dynamics of the etiological agent of SARS in Hong Kong: impact of public health interventions. Science. 2003;300:1961–1966. doi: 10.1126/science.1086478. [DOI] [PubMed] [Google Scholar]
- Rojas M., Rodríguez Y., Monsalve D.M., Acosta-Ampudia Y., Camacho B., Gallo J.E., Rojas-Villarraga A., Ramírez-Santana C., Díaz-Coronado J.C., Manrique R. Convalescent plasma in Covid-19: Possible mechanisms of action. Autoimmun. Rev. 2020;102554 doi: 10.1016/j.autrev.2020.102554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha A., Sharma A.R., Bhattacharya M., Sharma G., Lee S.-S., Chakraborty C. Probable molecular mechanism of remdesivir for the treatment of COVID-19: need to know more. Arch. Med. Res. 2020;51:585–586. doi: 10.1016/j.arcmed.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahara M., Ikutomi M., Morita T., Minami Y., Nakajima T., Hirata Y., Nagai R., Sata M. Deletion of angiotensin-converti enzyme 2 promotes the development of atherosclerosis and arterial neointima formation. Cardiovasc. Res. 2014;101:236–246. doi: 10.1093/cvr/cvt245. [DOI] [PubMed] [Google Scholar]
- Shah A. Novel Coronavirus-induced NLRP3 inflammasome activation: a potential drug target in the treatment of COVID-19. Front. Immunol. 2020:11. doi: 10.3389/fimmu.2020.01021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon A., Selisko B., Le N., Huchting J., Touret F., Piorkowski G., Fattorini V., Ferron F., Decroly E., Meier C. 2020. Favipiravir strikes the SARS-CoV-2 at its Achilles heel, the RNA polymerase. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikov A.E., Barbitoff Y.A., Glotov A.S., Danilova M.M., Tonyan Z.N., Nasykhova Y.A., Mikhailova A.A., Bespalova O.N., Kalinin R.S., Mirzorustamova A.M. Analysis of the spectrum of ACE2 variation suggests a possible influence of rare and common variants on susceptibility to COVID-19 and severity of outcome. Front. Genet. 2020;11 doi: 10.3389/fgene.2020.551220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons G., Reeves J.D., Rennekamp A.J., Amberg S.M., Piefer A.J., Bates P. Characterization of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) spike glycoprotein-mediated viral entry. Proc. Natl. Acad. Sci. U. S. A. 2004;101:4240–4245. doi: 10.1073/pnas.0306446101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaan W., Cavanagh D., Horzinek M. Coronaviruses: structure and genome expression. J. Gen. Virol. 1988;69:2939–2952. doi: 10.1099/0022-1317-69-12-2939. [DOI] [PubMed] [Google Scholar]
- Spiga O., Bernini A., Ciutti A., Chiellini S., Menciassi N., Finetti F., Causarono V., Anselmi F., Prischi F., Niccolai N. Molecular modelling of S1 and S2 subunits of SARS coronavirus spike glycoprotein. Biochem. Biophys. Res. Commun. 2003;310:78–83. doi: 10.1016/j.bbrc.2003.08.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stawiski E.W., Diwanji D., Suryamohan K., Gupta R., Fellouse F.A., Sathirapongsasuti F., Liu J., Jiang Y.-P., Ratan A., Mis M. 2020. Human ACE2 receptor polymorphisms predict SARS-CoV-2 susceptibility. BioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stawiski E.W., Diwanji D., Suryamohan K., Gupta R., Fellouse F.A., Sathirapongsasuti J.F., Liu J., Jiang Y.P., Ratan A., Mis M., Santhosh D., Somasekar S., Mohan S., Phalke S., Kuriakose B., Antony A., Junutula R.J., Schuster S.C., Jura N., Seshagiri S. bioRxiv 024752. 2020. Human ACE2 receptor polymorphisms predict SARS-CoV-2 susceptibility. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straus M.R., Tang T., Lai A.L., Flegel A., Bidon M., Freed J.H., Daniel S., Whittaker G.R. Ca2+ ions promote fusion of Middle East respiratory syndrome coronavirus with host cells and increase infectivity. J. Virol. 2020;94 doi: 10.1128/JVI.00426-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui J., Li W., Murakami A., Tamin A., Matthews L.J., Wong S.K., Moore M.J., Tallarico A.S.C., Olurinde M., Choe H., Anderson L.J., Bellini W.J., Farzan M., Marasco W.A. Potent neutralization of severe acute respiratory syndrome (SARS) coronavirus by a human mAb to S1 protein that blocks receptor association. Proc. Natl. Acad. Sci. U. S. A. 2004;101:2536–2541. doi: 10.1073/pnas.0307140101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taefehshokr N., Taefehshokr S., Hemmat N., Heit B. Covid-19: perspectives on innate immune evasion. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.580641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikellis C., Johnston C.I., Forbes J.M., Burns W.C., Burrell L.M., Risvanis J., Cooper M.E. Characterization of renal angiotensin-converting enzyme 2 in diabetic nephropathy. Hypertension. 2003;41:392–397. doi: 10.1161/01.HYP.0000060689.38912.CB. [DOI] [PubMed] [Google Scholar]
- Tikellis C., Johnston C.I., Forbes J.M., Burns W.C., Burrell L.M., Risvanis J., Cooper M.E. Characterization of renal angiotensin - converting enzyme 2 in diabetic nephropathy. Hypertension. 2003;41:392–397. doi: 10.1161/01.HYP.0000060689.38912.CB. [DOI] [PubMed] [Google Scholar]
- Tipnis S.R., Hooper N.M., Hyde R., Karran E., Christie G., Turner A.J. A human homolog of angiotensin-converting enzyme: cloning and functional expression as a captopril-insensitive carboxypeptidase. J. Biol. Chem. 2000;275:33238–33243. doi: 10.1074/jbc.M002615200. [DOI] [PubMed] [Google Scholar]
- Totura A.L., Baric R.S. SARS coronavirus pathogenesis: host innate immune responses and viral antagonism of interferon. Curr. Opin. Virol. 2012;2:264–275. doi: 10.1016/j.coviro.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu X., Chong W.P., Zhai Y., Zhang H., Zhang F., Wang S., Liu W., Wei M., Siu N.H.O., Yang H. Functional polymorphisms of the CCL2 and MBL genes cumulatively increase susceptibility to severe acute respiratory syndrome coronavirus infection. J. Inf. Secur. 2015;71:101–109. doi: 10.1016/j.jinf.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Y.-F., Chien C.-S., Yarmishyn A.A., Lin Y.-Y., Luo Y.-H., Lin Y.-T., Lai W.-Y., Yang D.-M., Chou S.-J., Yang Y.-P. A review of SARS-CoV-2 and the ongoing clinical trials. Int. J. Mol. Sci. 2020;21:2657. doi: 10.3390/ijms21072657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner A.J., Hooper N.M. The angiotensin–converting enzyme gene family: genomics and pharmacology. Trends Pharmacol. Sci. 2002;23:177–183. doi: 10.1016/s0165-6147(00)01994-5. [DOI] [PubMed] [Google Scholar]
- Uno Y. Camostat mesilate therapy for COVID-19. Intern. Emerg. Med. 2020:1–2. doi: 10.1007/s11739-020-02345-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaidya G., Czer L.S., Kobashigawa J., Kittleson M., Patel J., Chang D., Kransdorf E., Shikhare A., Tran H., Vo A. Elsevier; 2020. Successful treatment of Severe COVID-19 Pneumonia with Clazakizumab in a Heart Transplant Recipient: Case Report, Transplantation Proceedings. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Hoek L., Pyrc K., Jebbink M.F., Vermeulen-Oost W., Berkhout R.J.M., Wolthers K.C., Wertheim-Van Dillen P.M.E., Kaandorp J., Spaargaren J., Berkhout B. Identification of a new human coronavirus. Nat. Med. 2004;10:368–373. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Hoek L., Pyrc K., Berkhout B. Human coronavirus NL63, a new respiratory virus. FEMS Microbiol. Rev. 2006;30:760–773. doi: 10.1111/j.1574-6976.2006.00032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma A., Xu K., Du T., Zhu P., Liang Z., Liao S., Zhang J., Raizada M.K., Grant M.B., Li Q. Expression of Human ACE2 in Lactobacillus and Beneficial Effects in Diabetic Retinopathy in Mice. Mol. Ther. - Methods Clin. Dev. 2019;14:161–170. doi: 10.1016/j.omtm.2019.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickers C., Hales P., Kaushik V., Dick L., Gavin J., Tang J., Godbout K., Parsons T., Baronas E., Hsieh F., Acton S., Patane M., Nichols A., Tummino P. Hydrolysis of biological peptides by human angiotensin-converting enzyme-related carboxypeptidase. J. Biol. Chem. 2002;277:14838–14843. doi: 10.1074/jbc.M200581200. [DOI] [PubMed] [Google Scholar]
- Wang S.-F., Chen K.-H., Chen M., Li W.-Y., Chen Y.-J., Tsao C.-H., Yen M.-y., Huang J.C., Chen Y.-M.A. Human-leukocyte antigen class I Cw 1502 and class II DR 0301 genotypes are associated with resistance to severe acute respiratory syndrome (SARS) infection. Viral Immunol. 2011;24:421–426. doi: 10.1089/vim.2011.0024. [DOI] [PubMed] [Google Scholar]
- Wang X.L., Iwanami J., Min L.J., Tsukuda K., Nakaoka H., Bai H.Y., Shan B.S., Kan-No H., Kukida M., Chisaka T., Yamauchi T., Higaki A., Mogi M., Horiuchi M. Deficiency of angiotensin-converting enzyme 2 causes deterioration of cognitive function. npj Aging Mech. Dis. 2016;2 doi: 10.1038/npjamd.2016.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Cao R., Zhang H., Liu J., Xu M., Hu H., Li Y., Zhao L., Li W., Sun X. The anti-influenza virus drug, arbidol is an efficient inhibitor of SARS-CoV-2 in vitro. Cell Discov. 2020;6:1–5. doi: 10.1038/s41421-020-0169-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . 2020. Coronavirus disease (COVID-19) Situation Report–134. [Google Scholar]
- Williams A.E., Chambers R.C. The mercurial nature of neutrophils: still an enigma in ARDS? Am. J. Phys. Lung Cell. Mol. Phys. 2014;306:L217–L230. doi: 10.1152/ajplung.00311.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S.K., Li W., Moore M.J., Choe H., Farzan M. A 193-amino acid fragment of the SARS Coronavirus S protein efficiently binds angiotensin-converting enzyme 2. J. Biol. Chem. 2004;279:3197–3201. doi: 10.1074/jbc.C300520200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooster L., Nicholson C.J., Sigurslid H.H., Lino Cardenas C.L., Malhotra R. 2020. Polymorphisms in the ACE2 Locus Associate with Severity of COVID-19 Infection. medRxiv preprint. [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P., Meng J., Zhu Z., Zhang Z., Wang J. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27:325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia H., Sriramula S., Chhabra K.H., Lazartigues E. Brain angiotensin-converting enzyme type 2 shedding contributes to the development of neurogenic hypertension. Circ. Res. 2013;113:1087–1096. doi: 10.1161/CIRCRESAHA.113.301811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y., Cao S., Dong H., Li Q., Chen E., Zhang W., Yang L., Fu S., Wang R. Effect of regular intravenous immunoglobulin therapy on prognosis of severe pneumonia in patients with COVID-19. J. Inf. Secur. 2020;81:318–356. doi: 10.1016/j.jinf.2020.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Chen P., Wang J., Feng J., Zhou H., Li X., Zhong W., Hao P. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci. China Life Sci. 2020;63:457–460. doi: 10.1007/s11427-020-1637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Shi L., Wang Y., Zhang J., Huang L., Zhang C., Liu S., Zhao P., Liu H., Zhu L. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020;8:420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeager C.L., Ashmun R.A., Williams R.K., Cardellichio C.B., Shapiro L.H., Look A.T., Holmes K.V. Human aminopeptidase N is a receptor for human coronavirus 229E. Nature. 1992;357:420–422. doi: 10.1038/357420a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Wu Z., Li J.-W., Zhao H., Wang G.-Q. The cytokine release syndrome (CRS) of severe COVID-19 and Interleukin-6 receptor (IL-6R) antagonist Tocilizumab may be the key to reduce the mortality. Int. J. Antimicrob. Agents. 2020;105954 doi: 10.1016/j.ijantimicag.2020.105954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Wu Q., Zhang Z. Probable pangolin origin of SARS-CoV-2 associated with the COVID-19 outbreak. Curr. Biol. 2020;30:1578. doi: 10.1016/j.cub.2020.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Chen D., Szabla R., Zheng M., Li G., Du P., Zheng S., Li X., Song C., Li R., Guo J.-T., Junop M., Zeng H., Lin H. Broad and differential animal ACE2 receptor usage by SARS-CoV-2. J. Virol. JVI. 2020;94 doi: 10.1128/JVI.00940-20. 00940–00920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong N., Zheng B., Li Y., Poon L., Xie Z., Chan K., Li P., Tan S., Chang Q., Xie J. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People’s Republic of China, in February, 2003. Lancet. 2003;362:1353–1358. doi: 10.1016/S0140-6736(03)14630-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L., Ruan F., Huang M., Liang L., Huang H., Hong Z., Yu J., Kang M., Song Y., Xia J. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N. Engl. J. Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]