Abstract
Canonical Wnt signaling plays an essential role in proper craniofacial morphogenesis, at least partially due to regulation of various aspects of cranial neural crest development. In an effort to gain insight into the etiology of craniofacial abnormalities resulting from Wnt signaling and/or cranial neural crest dysfunction, we sought to identify Wnt-responsive targets during chick cranial neural crest development. To this end, we leveraged overexpression of a canonical Wnt antagonist, Draxin, in conjunction with RNA-sequencing of cranial neural crest cells that have just activated their epithelial–mesenchymal transition (EMT) program. Through differential expression analysis, gene list functional annotation, hybridization chain reaction (HCR), and quantitative reverse transcription polymerase chain reaction (RT-qPCR), we validated a novel downstream target of canonical Wnt signaling in cranial neural crest – RHOB – and identified possible signaling pathway crosstalk underlying cranial neural crest migration. The results reveal novel putative targets of canonical Wnt signaling during cranial neural crest EMT and highlight important intersections across signaling pathways involved in craniofacial development.
Keywords: Draxin, Wnt, neural crest, EMT, craniofacial development
Introduction
The neural crest is a multipotent stem cell population in the vertebrate embryo that undergoes coordinated induction, specification, and epithelial–mesenchymal transition (EMT) events to migrate and ultimately differentiate into a wide range of cell types. The migratory pathways and derivatives formed by the neural crest are regionalized according to their axial level of origin, such that cells from a given axial level give rise to a characteristic array of progeny and follow distinct pathways from those arising at other axial levels (Gandhi and Bronner, 2018). The most anterior “cranial” neural crest population underlies much of the development of the face (Cordero et al., 2011), and is the only neural crest population in vivo with the ability to differentiate into facial skeleton, contributing to the upper and lower jaw, and bones of the neck (Noden, 1975; Le Douarin, 1982; Simoes-Costa and Bronner, 2015). Importantly, perturbation of various stages of cranial neural crest development results in a myriad of craniofacial malformations (Vega-Lopez et al., 2018).
Many facets of cranial neural crest development are regulated by Wnt signaling (Wu et al., 2003; Yanfeng et al., 2003; Steventon et al., 2009; Milet and Monsoro-Burq, 2012; Simoes-Costa et al., 2015; Rabadán et al., 2016; Hutchins and Bronner, 2018, 2019; Gandhi et al., 2020). Furthermore, Wnt signaling is critical for normal facial patterning; mutations in Wnt pathway components or dysregulation of canonical Wnt signaling output result in defects in craniofacial morphogenesis (Huelsken et al., 2000; Chiquet et al., 2008; Reid et al., 2011; He and Chen, 2012; Kurosaka et al., 2014). Thus, identification of canonical Wnt targets during cranial neural crest development would greatly enhance understanding the etiology of craniofacial abnormalities resulting from Wnt signaling or cranial neural crest dysfunction.
Here, we took advantage of a canonical Wnt signaling inhibitor, Draxin, to identify Wnt-responsive targets during chick cranial neural crest development. As Draxin overexpression impedes cranial neural crest EMT in a β-catenin-dependent mechanism (Hutchins and Bronner, 2018, 2019), here we utilize Draxin overexpression together with RNA-sequencing (RNA-seq) on sorted populations of cranial neural crest cells to identify novel downstream targets of canonical Wnt signaling during cranial neural crest EMT.
Materials and Methods
Embryo Electroporation and Expression Constructs
Electroporations were performed at Hamburger-Hamilton stage HH4 (Hamburger and Hamilton, 1951), using commercially available fertile chicken (Gallus gallus) eggs (Sunstate Ranch, Sylmar, CA, United States), as previously described (Hutchins and Bronner, 2018). The cranial neural crest-specific enhancer NC1.1m3:GFP (Simoes-Costa et al., 2012), Draxin overexpression (Hutchins and Bronner, 2018), BRE::GFP BMP reporter (Le Dreau et al., 2012), NC1-Δ90βcat canonical Wnt signaling activation (Hutchins and Bronner, 2018), and control expression (Betancur et al., 2010b) constructs were described previously.
Tissue Dissociation and FACS
Following electroporation, embryos were incubated at 37°C until HH9+. We then dissected embryonic heads anterior to the otic vesicle in Ringer’s solution, washed tissue with sterile PBS (Corning cellgro #21-031-CV), then incubated tissue in Accumax (Innovative Cell Technologies, Inc. #AM-105) at 37°C for 15 min, with trituration every 5 min. Following dissociation, cells were washed with Hanks’ Balanced Salt Solution (Thermo Fisher Scientific #88284), filtered through a 20 μM nylon net mesh filter (Millipore Product #NY2004700), and resuspended in Hanks’ supplemented with 0.25% bovine serum albumin and 5% RQ1 DNase (Promega #M6101). GFP + cells were then collected using fluorescence activated cell sorting (FACS) at the Caltech Flow Cytometry Cell Sorting Facility.
Library Preparation and Sequencing
We used 1500 GFP+ cranial neural crest cells per replicate to prepare libraries. cDNA libraries were prepared using the Takara Bio SMART-Seq v4 Ultra Low Input cDNA kit, according to manufacturer instructions. RNA-Seq was performed at the Caltech Millard and Muriel Jacobs Genetics and Genomics Laboratory at 35 million reads on two biological replicates for both the control cranial and Draxin overexpression cranial neural crest cells. Sequencing libraries were built according to Illumina Standard Protocols and SR50 sequencing was performed in a HiSeq Illumina machine by the Caltech Millard and Muriel Jacobs Genetics and Genomics Laboratory. Sequence reads were aligned to the G. gallus genome (galgal6) with Bowtie2 (Langmead and Salzberg, 2012), transcript counts were calculated with HTSeq-Count (Anders et al., 2015), and differential expression analysis was performed with DESeq2 (Love et al., 2014). Gene lists were analyzed for functional annotation using PANTHER (Mi et al., 2019) and DAVID (Huang da et al., 2009a,b).
Hybridization Chain Reaction
Embryos to be processed for hybridization chain reaction (HCR) were fixed in 4% paraformaldehyde 1 h at room temperature, then dehydrated in graded methanol washes and stored at least one overnight at −20°C. HCR was performed as previously described (Gandhi et al., 2020), with custom probes designed and ordered through Molecular Technologies.
Image Acquisition and Analysis
Confocal images were acquired with an upright Zeiss LSM 880 at the Caltech Biological Imaging Facility. Images were minimally processed for brightness/contrast and pseudocolored using Fiji (Schindelin et al., 2012) and Adobe Photoshop 2020. Relative fluorescence intensity was determined in Fiji. For each whole mount image, the line tool was used to draw an ROI surrounding the area of neural crest indicated by positive HCR fluorescence for neural crest marker TFAP2β. Integrated density measurements were quantified for ROIs on the control electroporated (left) and experimental electroporated (right) sides from the same embryo. Relative fluorescence intensity was then calculated by dividing the integrated density measurements for the experimental versus the control side of the same embryo. Statistical analyses were performed using Prism (8; GraphPad Software). P values are defined in the text, and significance was established with P < 0.05. P values were calculated for embryos using one-tailed paired t-tests with integrated density measurements for control versus experimental sides, and for qRT-PCR using two-tailed one sample t-tests for ΔΔCT values.
Quantitative Reverse Transcription PCR (RT-qPCR)
RNA was extracted from sorted cells (Draxin overexpression) and dissected HH9+ embryonic half heads (NC1-Δ90βcat canonical Wnt signaling activation) using the RNAqueous-Micro Total RNA Isolation Kit (Invitrogen), according to manufacturer instructions. Following RNA isolation in elution buffer, cDNA was reverse transcribed using the SuperScriptIII First-Strand Synthesis System (Invitrogen) with oligo-dT priming. Quantitative PCR (qPCR) was performed using gene-specific primers with FastStart Universal SYBR Green Master Mix with Rox (Roche) and cDNA (diluted 1:10) on a QuantStudio 3 Real-Time PCR System (Applied Biosystems) in triplicate. We determined ΔCT with normalization against 18S ribosomal RNA (ΔCT = Target CT − 18S CT) for Draxin, SNAI2, and RHOB for samples, then calculated ΔΔCT values (ΔΔCT = Average Control ΔCT − Perturbation ΔCT) for each target and replicate. The gene-specific primers used for qPCR were: Draxin-F 5′-CTACGCTGTTATGCCAAATTCC; Draxin-R 5′-GAATGATCCCTGCTCTCCATT; SNAI2-F 5′-GCA ACAAGACCTATTCCACTTTC; SNAI2-R 5′-GTACTTG CAGCTGAACGATTTC; RHOB-F 5′-CGTGATCCTCATGT GCTTCT; RHOB-R 5′-TGCGCAGGTCTTTCTTGT; 18S-F 5′-CCATGATTAAGAGGGACGGC; 18S-R 5′-TGGCAAA TGCTTTCGCTTT.
Results
Identification of Draxin-Responsive Genes in Migrating Cranial Neural Crest
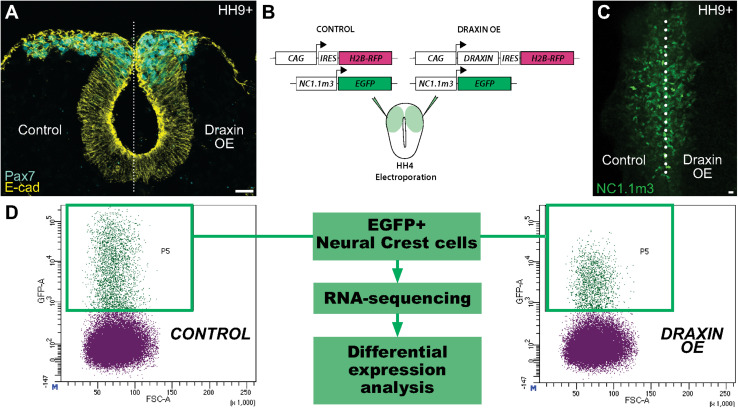
We have previously shown that the secreted protein Draxin functions as a potent inhibitor of cranial neural crest cell migration during EMT (Figure 1A; Hutchins and Bronner, 2018, 2019). Its effects on neural crest are elicited extracellularly via β-catenin-dependent Wnt signaling inhibition, precisely at the early stages of cranial neural crest EMT at HH9+ (Hutchins and Bronner, 2018). To parse the cranial-specific targets of Draxin underlying its effect on neural crest EMT and uncover potential novel targets of canonical Wnt signaling, we performed RNA-sequencing (RNA-seq) on sorted chick cranial neural crest cells, with and without Draxin-mediated Wnt inhibition. To this end, we co-electroporated the FoxD3 NC1.1m3 enhancer, which drives GFP expression specifically in the cranial neural crest population (Simoes-Costa et al., 2012), with either a Draxin overexpression construct containing an internal ribosomal entry site (IRES) driving H2B-RFP (Hutchins and Bronner, 2018) or the same construct without the Draxin coding region as a control (Figure 1B). Embryos were subsequently developed to the onset of cranial neural crest EMT (HH9+), by which point Draxin-mediated effects on EMT are evident (Figure 1C; Hutchins and Bronner, 2018, 2019), to identify EMT-related genes sensitive to canonical Wnt inhibition. From heads dissected anterior to the otic vesicle, we isolated 1500 GFP+ cranial neural crest cells per replicate by FACS, then performed cDNA library preparation and sequencing (Figure 1D).
FIGURE 1.
Experimental design for identification of Draxin-responsive targets in cranial neural crest EMT. (A) Cross-section through embryo electroporated with a Draxin overexpression construct on the right side of the embryo. Immunostaining for E-cadherin (E-cad) and the neural crest marker Pax7 at HH9+ highlights the deleterious effects of Draxin overexpression on cranial neural crest EMT and migration away from the midline (dotted line), compared to the contralateral control side. (B–D) Experimental design to isolate cranial neural crest cells with or without Draxin overexpression. Gastrula stage chick embryos were co-electroporated with a neural crest-specific enhancer driving EGFP expression in cranial neural crest cells (NC1.1m3) and either a Draxin overexpression or control construct (B). NC1.1m3 enhancer expression revealed EGFP + cranial neural crest cells were responsive to Draxin overexpression and exhibited EMT defects (C). Fluorescence activated cell sorting (FACS) was used to isolate EGFP+ cranial neural crest cells with and without Draxin overexpression that were subsequently processed for RNA-sequencing and differential expression analysis (D). Scale bar, 20 μm. HH, Hamburger-Hamilton stage; OE, overexpression; IRES, internal ribosomal entry site.
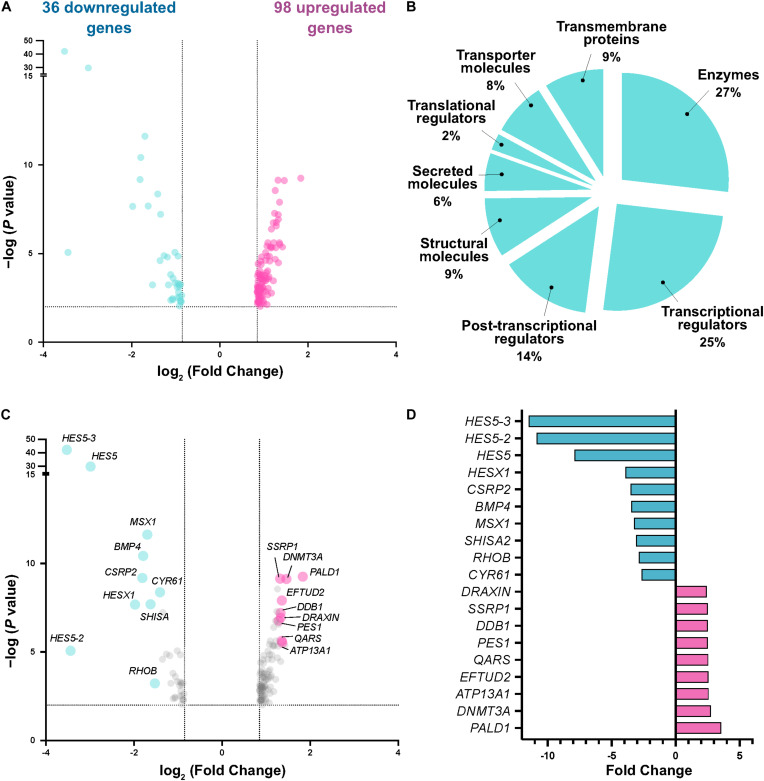
Differential expression analysis initially revealed 284 differentially expressed genes with ≥1.8 fold change and FDR < 0.01. For subsequent functional analysis, we filtered the gene lists to exclude lowly expressed genes (average normalized count values < 1000), resulting in a filtered list of 134 differentially expressed genes (36 downregulated, 98 upregulated) (Figure 2A). Using PANTHER analysis (Mi et al., 2019) to probe molecular functions of these gene targets, we observed enrichment of factors highly associated with transcriptional regulation, enzymatic reactions (including kinases) and secreted proteins indicative of targets associated with intracellular signaling pathways, and structural molecules (such as cytoskeletal and extracellular matrix proteins) indicative of cell migration-associated targets (Figure 2B). Among the most highly changed genes, we found significant enrichment of Draxin, as expected due to its experimental overexpression. Interestingly, we also detected significant downregulation of the Notch pathway effector HES5 (and related genes), and BMP4 (as well as its downstream target MSX1) (Figures 2C,D), suggesting potential signaling pathway crosstalk between Draxin, canonical Wnt signaling, and other pathways with critical roles in neural crest development. Given that Draxin has been shown to intersect with additional signaling pathways in other contexts (Ahmed et al., 2011; Hossain et al., 2013; Meli et al., 2015), further studies are needed to parse direct and indirect effects relevant to craniofacial morphogenesis and neural crest EMT.
FIGURE 2.
Transcriptome analysis reveals differentially expressed genes responsive to Draxin-mediated canonical Wnt inhibition. (A) Volcano plot following differential expression analysis and filtering. Dotted lines indicate significance and fold change cutoffs for downstream analysis. (B) Molecular functions for the 134 differentially expressed genes. (C,D) Most enriched and depleted genes in cranial neural crest with Draxin overexpression.
Biological Pathway Analysis of Draxin-Responsive Genes in Cranial Neural Crest
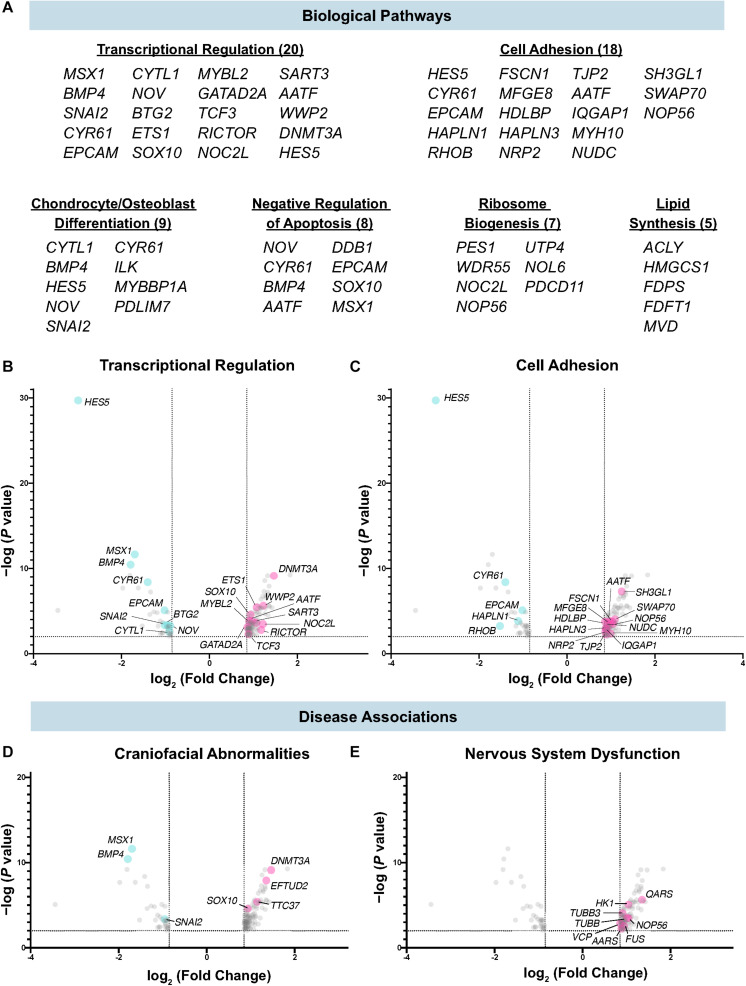
To better understand the molecular processes in which Draxin, and by extension canonical Wnt signaling, function during cranial neural crest EMT, we performed functional annotation for the dataset using the Database for Annotation, Visualization and Integrated Discovery (DAVID) (Huang da et al., 2009a,b). Consistent with established roles of canonical Wnt signaling and Draxin-mediated inhibition during cranial neural crest EMT, we observed enrichment of genes associated with transcriptional regulation, cell adhesion, and lipid synthesis, which we have recently shown is important for cell signaling during cranial neural crest EMT (Piacentino et al., 2020). In addition, we found numerous genes associated with bone/cartilage formation (e.g., CYTL1, ILK, NOV), a critical function of cranial neural crest, and genes involved in ribosome biogenesis (e.g., NOP56, PES1, NOC2L), which has implications for craniofacial development (Ross and Zarbalis, 2014) (Figure 3A).
FIGURE 3.
Biological pathways and diseases associated with Draxin-responsive transcriptome dataset. (A–C) Biological pathway analysis identified enrichment of targets associated with transcriptional regulation, cell adhesion, chondrocyte/osteoblast differentiation, negative regulation of apoptosis, ribosome biogenesis, and lipid synthesis. (D,E) Functional annotation identified genes highly correlated with craniofacial abnormalities and nervous system dysfunction.
Among the targets associated with transcriptional regulation, we detected significant downregulation of SNAI2, which has been shown to be a direct target of canonical Wnt signaling, and subsequently Draxin (LaBonne and Bronner-Fraser, 1998; Monsoro-Burq et al., 2005; Wu et al., 2005; Hutchins and Bronner, 2018) (Figure 3B). In addition, we observed significant downregulation of genes that mediate cell adhesion and EMT (Figure 3C), including RHOB, shown to be required for neural crest delamination in the trunk (Liu and Jessell, 1998), and EPCAM, which has been shown to participate in cancer cell EMT (Wang et al., 2018). We also observed significant correlations for disease-associated genes, including those involved in craniofacial (Figure 3D) and nervous system dysfunction (Figure 3E). This was expected given the role of the cranial neural crest in craniofacial development, and established roles for Draxin in nervous system development (Islam et al., 2009; Tawarayama et al., 2018).
Functional Validation of a Novel Immediate Early Canonical Wnt Target
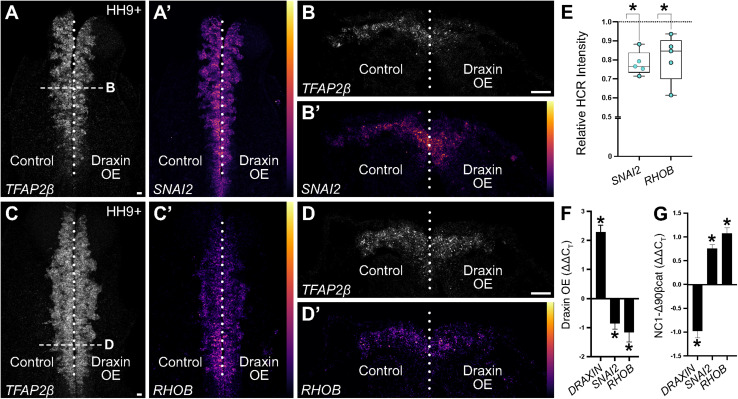
To validate Draxin-responsive targets from our dataset, we performed quantitative HCR on embryos bilaterally electroporated with the Draxin overexpression construct on the right side of embryos and the control construct on the left side (as in Figure 1B). To establish the area of neural crest migration from which to measure target fluorescence intensities, we visualized expression of TFAP2β, a neural crest marker and non-target of Draxin. We focused on early HH9 + embryos, corresponding to the beginning of cranial neural crest EMT and initial stages of migration, in order to probe immediate early gene changes. As a result, modest defects were evident in the distance cranial neural crest cells migrated away from the midline (Figures 4A–D), consistent with a Draxin overexpression phenotype, albeit to a lesser extent than later stage HH9+ embryos in which migration has progressed more laterally (Figures 1A,C; Hutchins and Bronner, 2018, 2019). We measured SNAI2 and RHOB fluorescence intensities for Draxin overexpression versus control sides of individual embryos, and found significant downregulation of gene expression (Figure 4E; 78.0 ± 2.8% of the control side for SNAI2 and 81.0 ± 5.5% of the control side for RHOB; P ≤ 0.01, one-tailed paired t-test), consistent with predicted trends based on our transcriptomic analyses. This is consistent with our previously published work indicating that Draxin acts upstream of Snail2 protein expression (Hutchins and Bronner, 2018). We further validated the effects of Draxin overexpression on SNAI2 and RHOB using quantitative reverse transcription PCR (RT-qPCR) with sorted cells collected alongside sequenced cells from Figure 1; consistent with the HCR data (Figures 4A–E), we detected significant downregulation of both SNAI2 and RHOB with Draxin overexpression (Figure 4F).
FIGURE 4.
Hybridization chain reaction (HCR) validation of identified neural crest EMT genes. (A–D) Representative images of quantitative HCR for neural crest marker TFAP2β and targets SNAI2 and RHOB in embryos bilaterally electroporated with a control (left) or Draxin overexpression (right) construct. SNAI2 and RHOB are pseudocolored based on fluorescence intensities as indicated by color scale in panels (A′–D′). (E) Integrated density measurements revealed significant downregulation of SNAI2 and RHOB (78.0 ± 2.8% of the control side for SNAI2 and 81.0 ± 5.5% of the control side for RHOB; P ≤ 0.01, one-tailed paired t-test) with Draxin overexpression. Scale bar, 20 μm. (F) ΔΔCT values from quantitative reverse transcription PCR (RT-qPCR) for DRAXIN, SNAI2, and RHOB (normalized against 18S, comparing control versus overexpression cells) from HH9+ sorted cranial neural crest cells co-electroporated with NC1.1m3 fluorescent GFP reporter with or without Draxin overexpression (DraxinOE). As expected, DRAXIN was significantly upregulated whereas SNAI2 and RHOB were significantly downregulated in DraxinOE cranial neural crest cells. *P < 0.05, two-tailed one sample t-test. (G) ΔΔCT values from quantitative reverse transcription PCR (RT-qPCR) for DRAXIN, SNAI2, and RHOB (normalized against 18S, comparing control versus Wnt-activated cells) from dissected HH9+ cranial tissue from embryos co-electroporated with NC1.1m3 fluorescent RFP reporter with or without stabilized β-catenin expression (NC1-Δ90βcat). In contrast to Wnt inhibition via DraxinOE (F), SNAI2 and RHOB were significantly upregulated with Wnt activation via NC1-Δ90βcat, whereas DRAXIN was significantly downregulated. *P ≤ 0.02, two-tailed one sample t-test.
Given that RHOB has been previously shown to be a BMP-responsive target (Liu and Jessell, 1998) and insensitive to Wnt signaling (Taneyhill and Bronner-Fraser, 2005) in trunk neural crest, we next sought to determine whether the reduction in RHOB we observed in cranial neural crest was due to direct effects from Wnt signaling, or indirect effects through downregulation of BMP. We have previously shown that canonical Wnt signaling is active in cranial neural crest at the onset of EMT using a fluorescent reporter (Hutchins and Bronner, 2018), while BMP signaling is active in the presumptive cranial neural crest at earlier stages during their induction (Piacentino and Bronner, 2018); here we employed a similar approach to investigate the timing of BMP signaling activation in cranial neural crest at the onset of EMT. Electroporation of a fluorescent BMP reporter (BRE::GFP) revealed a lack of active BMP signaling in cranial neural crest cells that have undergone EMT and commenced migration at HH9+ (Supplementary Figure 1). Thus, it is unlikely that the reduction in RHOB we observed was due to suppressive effects on BMP signaling by Draxin. To more fully examine whether RHOB downregulation was due to direct effects from Wnt signaling, we performed RT-qPCR on embryos with and without canonical Wnt signaling activation during cranial neural crest EMT. Here, we specifically activated canonical Wnt signaling in specified cranial neural crest by driving expression of a stabilized form of β-catenin under the control of a neural crest-specific enhancer (NC1-Δ90βcat). Importantly, we observed upregulation of SNAI2, an established direct target of canonical Wnt signaling (LaBonne and Bronner-Fraser, 1998; Monsoro-Burq et al., 2005; Wu et al., 2005), as well as upregulation of RHOB, suggesting a direct link with Wnt signaling. Interestingly, we also observed concomitant downregulation of endogenous DRAXIN, suggesting the possibility of a negative feedback loop with respect to DRAXIN expression (Figure 4G).
Taken together, our data identify a novel target of Draxin and canonical Wnt signaling during cranial neural crest EMT (RHOB), and suggest that Draxin downregulation, and subsequent activation of Wnt signaling, is essential for crosstalk and feedback of signaling pathways that alter cranial neural crest transcriptional activation, and ultimately EMT.
Discussion
Using transcriptome profiling of Draxin-responsive targets, we identified likely gene targets of canonical Wnt signaling during cranial neural crest EMT. Consistent with our previously published work examining protein expression (Hutchins and Bronner, 2018), we verified transcript downregulation of canonical Wnt target SNAI2 in response to Draxin overexpression. Furthermore, we also identified and validated a novel target – RHOB. RhoB is BMP-responsive in trunk neural crest and is necessary for delamination (Liu and Jessell, 1998); its misexpression has been associated with defects in laminin organization within the basement membrane (Perez-Alcala et al., 2004). Interestingly, we have previously demonstrated that Draxin, via regulation of canonical Wnt signaling, also is involved in regulating laminin organization and remodeling of the basement membrane during cranial neural crest development (Hutchins and Bronner, 2019). Furthermore, we also observed downregulation of the BMP ligand BMP4, suggesting that BMP signaling may act downstream of Wnt signaling during or immediately after cranial neural crest EMT. In trunk, it has been shown that neural crest delamination is regulated by BMP, and that canonical Wnt signaling is controlled by BMP signaling through BMP-responsive expression of the Wnt1 ligand (Burstyn-Cohen et al., 2004). This is particularly interesting in light of our observations from a GFP reporter construct that BMP signaling is inactive in cranial neural crest at the onset of EMT (Supplementary Figure 1). Interestingly, Draxin has also been shown to inhibit neural crest migration in the trunk (Su et al., 2009; Zhang et al., 2017). Thus, whether RHOB expression is differentially regulated based on axial level (i.e., in response to BMP signaling in trunk neural crest versus in response to Wnt signaling in cranial neural crest) or based on signaling pathway crosstalk (which may also be dependent on axial level) remains to be explored.
In searching our datasets for neural crest-specific factors, we also noted modest upregulation of SOX10 and ETS1, genes associated with neural crest EMT (Tahtakran and Selleck, 2003; Theveneau et al., 2007; Simoes-Costa and Bronner, 2015), which seemed contradictory to the antagonistic role of Draxin in modulating cranial neural crest EMT. ETS1 expression is restricted to the cranial population of neural crest and is itself activated via cMYB (Betancur et al., 2010a); together with Sox9, Ets1 and cMYB function as activating gene regulatory inputs into a SOX10E2 enhancer (Betancur et al., 2010b), regulating SOX10 expression in cranial neural crest. Interestingly, in other contexts, canonical Wnt signaling has been shown to trigger degradation of cMYB protein (Kanei-Ishii et al., 2004); given that Draxin is endogenously expressed at HH9, it is possible that its normal inhibitory effects on canonical Wnt signaling may be necessary to reduce degradation of cMYB to activate endogenous levels of ETS1 and SOX10, which initiate expression prior to the onset of EMT. Thus, we postulate here that during early cranial neural crest migration, excess cMYB protein is stabilized via exogenous Draxin-mediated inhibition of canonical Wnt signaling; as a result, this may trigger upregulation of ETS1 and SOX10 gene expression. However, despite upregulation of factors positively associated with EMT, downregulation of SNAI2 alone is sufficient to impede cranial neural crest migration (Nieto et al., 1994; Hutchins and Bronner, 2019).
Taken together, our data identify novel targets of canonical Wnt signaling during cranial neural crest EMT, and highlight potential avenues of intersection for signaling pathways involved in craniofacial development. The results raise the intriguing possibility that the sequence and magnitude of signaling and gene expression crosstalk during cranial neural crest development may help precisely regulate craniofacial morphogenesis.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA673315.
Ethics Statement
Ethical review and approval was not required for the animal study because our study uses chicken embryos at E1-2. These are not considered vertebrate embryos until E10, and thus we do not require ethics committee approvals or protocols, as they are not considered vertebrates at the stages we work.
Author Contributions
EH, MP, and MB conceived the project and conducted the experimental design and data interpretation. EH and MP performed the cell dissociations, library preparations, and RNA-seq analyses. EH performed the functional annotation, hybridization chain reaction experiments, imaging, quantitation, and statistical analyses. EH and MB wrote the manuscript with editing by MP. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank A. Collazo and G. Spigolon for imaging assistance at the Caltech Biological Imaging Facility; P. Cannon and R. Diamond of the Caltech Flow Cytometry Cell Sorting Facility for cell sorting assistance; I. Antoshechkin of the Caltech Millard and Muriel Jacobs Genetics and Genomics Laboratory for sequencing of our RNA-seq libraries; and M. Martik and S. Gandhi for assistance with data processing.
Footnotes
Funding. This work was supported by the National Institutes of Health (R01DE027538 and R01DE027568 to MB, K99DE028592 to EH, and K99DE029240 to MP).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2021.624037/full#supplementary-material
BMP-sensitive reporter expression is absent from delaminating and migratory cranial neural crest during EMT. (A) Schematic of cross section of cranial region at HH9+ examined in panel (B). Locations of premigratory (pNC) and migratory crest cells (mNC) are indicated. (B) Following electroporation of a BMP fluorescent reporter construct (BRE:GFP), cross sections stained for a neural crest marker (Snail2, magenta) and membrane label (WGA, white) revealed BMP reporter activity (green) in pNC (white dotted line), but not in delaminating or mNC (yellow dotted line). WGA, wheat germ agglutinin; pNC, premigratory neural crest; mNC, migratory neural crest. Scale bar, 10 μm.
References
- Ahmed G., Shinmyo Y., Ohta K., Islam S. M., Hossain M., Naser I. B., et al. (2011). Draxin inhibits axonal outgrowth through the netrin receptor DCC. J. Neurosci. 31 14018–14023. 10.1523/JNEUROSCI.0943-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Pyl P. T., Huber W. (2015). HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31 166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancur P., Bronner-Fraser M., Sauka-Spengler T. (2010a). Assembling neural crest regulatory circuits into a gene regulatory network. Annu. Rev. Cell Dev. Biol. 26 581–603. 10.1146/annurev.cellbio.042308.113245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancur P., Bronner-Fraser M., Sauka-Spengler T. (2010b). Genomic code for Sox10 activation reveals a key regulatory enhancer for cranial neural crest. Proc. Natl. Acad. Sci. U.S.A. 107 3570–3575. 10.1073/pnas.0906596107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burstyn-Cohen T., Stanleigh J., Sela-Donenfeld D., Kalcheim C. (2004). Canonical Wnt activity regulates trunk neural crest delamination linking BMP/noggin signaling with G1/S transition. Development 131 5327–5339. 10.1242/dev.01424 [DOI] [PubMed] [Google Scholar]
- Chiquet B. T., Blanton S. H., Burt A., Ma D., Stal S., Mulliken J. B., et al. (2008). Variation in WNT genes is associated with non-syndromic cleft lip with or without cleft palate. Hum. Mol. Genet. 17 2212–2218. 10.1093/hmg/ddn121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordero D. R., Brugmann S., Chu Y., Bajpai R., Jame M., Helms J. A. (2011). Cranial neural crest cells on the move: their roles in craniofacial development. Am. J. Med. Genet. A 155A 270–279. 10.1002/ajmg.a.33702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi S., Bronner M. E. (2018). Insights into neural crest development from studies of avian embryos. Int. J. Dev. Biol. 62 183–194. 10.1387/ijdb.180038sg [DOI] [PubMed] [Google Scholar]
- Gandhi S., Hutchins E. J., Maruszko K., Park J. H., Thomson M., Bronner M. E. (2020). Bimodal function of chromatin remodeler Hmga1 in neural crest induction and Wnt-dependent emigration. Elife 9:e57779. 10.7554/eLife.57779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamburger V., Hamilton H. L. (1951). A series of normal stages in the development of the chick embryo. J. Morphol. 88 49–92. [PubMed] [Google Scholar]
- He F., Chen Y. (2012). Wnt signaling in lip and palate development. Front. Oral Biol. 16:81–90. 10.1159/000337619 [DOI] [PubMed] [Google Scholar]
- Hossain M., Ahmed G., Naser I. B., Shinmyo Y., Ito A., Riyadh M. A., et al. (2013). The combinatorial guidance activities of draxin and Tsukushi are essential for forebrain commissure formation. Dev. Biol. 374 58–70. 10.1016/j.ydbio.2012.11.029 [DOI] [PubMed] [Google Scholar]
- Huang da W., Sherman B. T., Lempicki R. A. (2009a). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4 44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Huang da W., Sherman B. T., Zheng X., Yang J., Imamichi T., Stephens R., et al. (2009b). Extracting biological meaning from large gene lists with DAVID. Curr. Protoc. Bioinformatics Chapter 13:Unit 13.11. 10.1002/0471250953.bi1311s27 [DOI] [PubMed] [Google Scholar]
- Huelsken J., Vogel R., Brinkmann V., Erdmann B., Birchmeier C., Birchmeier W. (2000). Requirement for beta-catenin in anterior-posterior axis formation in mice. J. Cell Biol. 148 567–578. 10.1083/jcb.148.3.567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins E. J., Bronner M. E. (2018). Draxin acts as a molecular rheostat of canonical Wnt signaling to control cranial neural crest EMT. J. Cell Biol. 217 3683–3697. 10.1083/jcb.201709149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins E. J., Bronner M. E. (2019). Draxin alters laminin organization during basement membrane remodeling to control cranial neural crest EMT. Dev. Biol. 446 151–158. 10.1016/j.ydbio.2018.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam S. M., Shinmyo Y., Okafuji T., Su Y., Naser I. B., Ahmed G., et al. (2009). Draxin, a repulsive guidance protein for spinal cord and forebrain commissures. Science 323 388–393. 10.1126/science.1165187 [DOI] [PubMed] [Google Scholar]
- Kanei-Ishii C., Ninomiya-Tsuji J., Tanikawa J., Nomura T., Ishitani T., Kishida S., et al. (2004). Wnt-1 signal induces phosphorylation and degradation of c-Myb protein via TAK1, HIPK2, and NLK. Genes Dev. 18 816–829. 10.1101/gad.1170604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosaka H., Iulianella A., Williams T., Trainor P. A. (2014). Disrupting hedgehog and WNT signaling interactions promotes cleft lip pathogenesis. J. Clin. Invest. 124 1660–1671. 10.1172/JCI72688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaBonne C., Bronner-Fraser M. (1998). Neural crest induction in Xenopus: evidence for a two-signal model. Development 125 2403–2414. [DOI] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Douarin N. (1982). The Neural Crest. Cambridge: Cambridge University Press. [Google Scholar]
- Le Dreau G., Garcia-Campmany L., Rabadan M. A., Ferronha T., Tozer S., Briscoe J., et al. (2012). Canonical BMP7 activity is required for the generation of discrete neuronal populations in the dorsal spinal cord. Development 139 259–268. 10.1242/dev.074948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. P., Jessell T. M. (1998). A role for rhoB in the delamination of neural crest cells from the dorsal neural tube. Development 125 5055–5067. [DOI] [PubMed] [Google Scholar]
- Love M. I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meli R., Weisova P., Propst F. (2015). Repulsive axon guidance by Draxin is mediated by protein Kinase B (Akt), glycogen synthase kinase-3beta (GSK-3beta) and microtubule-associated protein 1B. PLoS One 10:e0119524. 10.1371/journal.pone.0119524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H., Muruganujan A., Ebert D., Huang X., Thomas P. D. (2019). PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 47 D419–D426. 10.1093/nar/gky1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milet C., Monsoro-Burq A. H. (2012). Neural crest induction at the neural plate border in vertebrates. Dev. Biol. 366 22–33. 10.1016/j.ydbio.2012.01.013 [DOI] [PubMed] [Google Scholar]
- Monsoro-Burq A. H., Wang E., Harland R. (2005). Msx1 and Pax3 cooperate to mediate FGF8 and WNT signals during Xenopus neural crest induction. Dev. Cell 8 167–178. 10.1016/j.devcel.2004.12.017 [DOI] [PubMed] [Google Scholar]
- Nieto M. A., Sargent M. G., Wilkinson D. G., Cooke J. (1994). Control of cell behavior during vertebrate development by Slug, a zinc finger gene. Science 264 835–839. [DOI] [PubMed] [Google Scholar]
- Noden D. M. (1975). An analysis of migratory behavior of avian cephalic neural crest cells. Dev. Biol. 42 106–130. [DOI] [PubMed] [Google Scholar]
- Perez-Alcala S., Nieto M. A., Barbas J. A. (2004). LSox5 regulates RhoB expression in the neural tube and promotes generation of the neural crest. Development 131 4455–4465. 10.1242/dev.01329 [DOI] [PubMed] [Google Scholar]
- Piacentino M. L., Bronner M. E. (2018). Intracellular attenuation of BMP signaling via CKIP-1/Smurf1 is essential during neural crest induction. PLoS Biol. 16:e2004425. 10.1371/journal.pbio.2004425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piacentino M. L., Hutchins E. J., Andrews C. J., Bronner M. E. (2020). Temporal changes in plasma membrane lipid content induce endocytosis to regulate developmental epithelial-to-mesenchymal transition. bioRxiv [Preprint] 10.1101/2020.10.18.344523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabadán M. A., Herrera A., Fanlo L., Usieto S., Carmona-Fontaine C., Barriga E. H., et al. (2016). Delamination of neural crest cells requires transient and reversible Wnt inhibition mediated by Dact1/2. Development 143 2194–2205. 10.1242/dev.134981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid B. S., Yang H., Melvin V. S., Taketo M. M., Williams T. (2011). Ectodermal Wnt/beta-catenin signaling shapes the mouse face. Dev. Biol. 349 261–269. 10.1016/j.ydbio.2010.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross A. P., Zarbalis K. S. (2014). The emerging roles of ribosome biogenesis in craniofacial development. Front. Physiol. 5:26. 10.3389/fphys.2014.00026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9 676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoes-Costa M. S., McKeown S. J., Tan-Cabugao J., Sauka-Spengler T., Bronner M. E. (2012). Dynamic and differential regulation of stem cell factor FoxD3 in the neural crest is Encrypted in the genome. PLoS Genet. 8:e1003142. 10.1371/journal.pgen.1003142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoes-Costa M., Bronner M. E. (2015). Establishing neural crest identity: a gene regulatory recipe. Development 142 242–257. 10.1242/dev.105445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoes-Costa M., Stone M., Bronner M. E. (2015). Axud1 Integrates Wnt signaling and transcriptional inputs to drive neural crest formation. Dev. Cell 34 544–554. 10.1016/j.devcel.2015.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steventon B., Araya C., Linker C., Kuriyama S., Mayor R. (2009). Differential requirements of BMP and Wnt signalling during gastrulation and neurulation define two steps in neural crest induction. Development 136 771–779. 10.1242/dev.029017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y., Naser I. B., Islam S. M., Zhang S., Ahmed G., Chen S., et al. (2009). Draxin, an axon guidance protein, affects chick trunk neural crest migration. Dev. Growth Differ. 51 787–796. 10.1111/j.1440-169X.2009.01137.x [DOI] [PubMed] [Google Scholar]
- Tahtakran S. A., Selleck M. A. (2003). Ets-1 expression is associated with cranial neural crest migration and vasculogenesis in the chick embryo. Gene Expr. Patterns 3 455–458. [DOI] [PubMed] [Google Scholar]
- Taneyhill L. A., Bronner-Fraser M. (2005). Dynamic alterations in gene expression after Wnt-mediated induction of avian neural crest. Mol. Biol. Cell 16 5283–5293. 10.1091/mbc.e05-03-0210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tawarayama H., Yamada H., Amin R., Morita-Fujimura Y., Cooper H. M., Shinmyo Y., et al. (2018). Draxin regulates hippocampal neurogenesis in the postnatal dentate gyrus by inhibiting DCC-induced apoptosis. Sci. Rep. 8:840. 10.1038/s41598-018-19346-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theveneau E., Duband J. L., Altabef M. (2007). Ets-1 confers cranial features on neural crest delamination. PLoS One 2:e1142. 10.1371/journal.pone.0001142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega-Lopez G. A., Cerrizuela S., Tribulo C., Aybar M. J. (2018). Neurocristopathies: new insights 150 years after the neural crest discovery. Dev. Biol. 444 S110–S143. 10.1016/j.ydbio.2018.05.013 [DOI] [PubMed] [Google Scholar]
- Wang M. H., Sun R., Zhou X. M., Zhang M. Y., Lu J. B., Yang Y., et al. (2018). Epithelial cell adhesion molecule overexpression regulates epithelial-mesenchymal transition, stemness and metastasis of nasopharyngeal carcinoma cells via the PTEN/AKT/mTOR pathway. Cell Death Dis. 9:2. 10.1038/s41419-017-0013-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Saint-Jeannet J. P., Klein P. S. (2003). Wnt-frizzled signaling in neural crest formation. Trends Neurosci. 26 40–45. [DOI] [PubMed] [Google Scholar]
- Wu J., Yang J., Klein P. S. (2005). Neural crest induction by the canonical Wnt pathway can be dissociated from anterior-posterior neural patterning in Xenopus. Dev. Biol. 279 220–232. 10.1016/j.ydbio.2004.12.016 [DOI] [PubMed] [Google Scholar]
- Yanfeng W., Saint-Jeannet J. P., Klein P. S. (2003). Wnt-frizzled signaling in the induction and differentiation of the neural crest. Bioessays 25 317–325. 10.1002/bies.10255 [DOI] [PubMed] [Google Scholar]
- Zhang S., Su Y., Gao J., Zhang C., Tanaka H. (2017). A potential inhibitory function of draxin in regulating mouse trunk neural crest migration. In Vitro Cell. Dev. Biol. Anim. 53 43–53. 10.1007/s11626-016-0079-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
BMP-sensitive reporter expression is absent from delaminating and migratory cranial neural crest during EMT. (A) Schematic of cross section of cranial region at HH9+ examined in panel (B). Locations of premigratory (pNC) and migratory crest cells (mNC) are indicated. (B) Following electroporation of a BMP fluorescent reporter construct (BRE:GFP), cross sections stained for a neural crest marker (Snail2, magenta) and membrane label (WGA, white) revealed BMP reporter activity (green) in pNC (white dotted line), but not in delaminating or mNC (yellow dotted line). WGA, wheat germ agglutinin; pNC, premigratory neural crest; mNC, migratory neural crest. Scale bar, 10 μm.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA673315.