Abstract
Background:
Severe asthma exacerbations are a major cause of asthma morbidity and increased healthcare costs. Several studies have shown racial and ethnic differences in asthma exacerbation rates. We aimed to identify genetic variants associated with severe exacerbations in two high-risk populations for asthma.
Methods:
A genome-wide association study of asthma in children and youth with severe exacerbations was performed in 1283 exacerbators and 2027 controls without asthma of Latino ancestry. Independent suggestive variants (P ≤ 5 × 10−6) were selected for replication in 448 African Americans exacerbators and 595 controls. Case-only analyses were performed comparing the exacerbators with additional 898 Latinos and 524 African Americans asthma patients without exacerbations, while adjusting by treatment category as a proxy of asthma severity. We analyzed the functionality of associated variants with in silico methods and by correlating genotypes with methylation levels in whole blood in a subset of 473 Latinos.
Results:
We identified two genome-wide significant associations for susceptibility to asthma with severe exacerbations, including a novel locus located at chromosome 2p21 (rs4952375, odds ratio = 1.39, P = 3.8 × 10−8), which was also associated with asthma exacerbations in a case-only analysis (odds ratio = 1.25, P = 1.95 × 10−3). This polymorphism is an expression quantitative trait locus of the long intergenic non-protein coding RNA 1913 (LINC01913) in lung tissues (P = 1.3 × 10−7) and influences methylation levels of the protein kinase domain-containing cytoplasmic (PKDCC) gene in whole-blood cells (P = 9.8 × 10−5).
Conclusion:
We identified a novel susceptibility locus for severe asthma exacerbations in Hispanic/Latino and African American youths with functional effects in gene expression and methylation status of neighboring genes.
Keywords: African American, asthma, ethnic differences, exacerbations, gene expression, genome-wide association study, Hispanic, Latino, methylation, single nucleotide polymorphism, susceptibility
1 |. INTRODUCTION
Asthma is a chronic inflammatory disease influenced by both genetic and environmental factors. Despite treatment with controller medications, some patients experience severe asthma exacerbations, which are defined as episodes requiring emergency care, hospitalizations, or the use of systemic corticosteroids to prevent a serious or fatal out-come.1 These can be triggered by environmental influences, such as viral infections, fungal allergens, or air pollution.2 Genetic factors also predispose patients to higher susceptibility to asthma exacerbations.2 In children, these exacerbations are associated with lower quality of life,3 a progressive loss of lung function,4 and an increase in their parents’ work absence.5 Therefore, asthma exacerbations represent a major component of the economic burden of asthma.
Among pediatric populations in the United States, asthma prevalence and asthma-related emergency department or urgent care center visits are highest in African American and Puerto Rican populations.6 Moreover, African ancestry has been associated with a higher risk of developing asthma among African Americans7 and Puerto Ricans,8 as well as with an increased risk of exacerbations among African Americans.9,10 These data point to a genetic component potentially contributing to the disparities in asthma exacerbations.
The genetic determinants of severe asthma exacerbations have been studied mostly in European populations, focusing on genes previously associated with asthma susceptibility through candidate gene association studies.11 The only four genome-wide association studies (GWAS) focused on asthma exacerbations performed to date suggest that loci for exacerbations differ from those of asthma susceptibility.12–15 One GWAS analyzed extreme phenotypes by comparing exacerbators and children without asthma and revealed a novel locus at the cadherin-related family member 3 (CDHR3) gene, not associated in any GWAS focused on asthma susceptibility.12 The other three GWAS of asthma exacerbations performed case-only analyses, comparing asthma patients with and without exacerbations. One study identified several single nucleotide polymorphisms (SNPs) in alpha-T-cat-enin (CTNNA3) significantly associated with asthma exacerbations at genome-wide significant level.13 Two studies flagged CMTR114 and APOBEC3B-APOBEC3C15 as suggestively associated with asthma exacerbations despite treatment with inhaled corticosteroids.
We hypothesized that a GWAS comparing asthma patients with severe exacerbations with non-asthmatic controls could reveal novel specific association signals in two high-risk populations for asthma. We aimed to identify genetic variants associated with severe asthma exacerbations in Hispanics/Latinos and African Americans.
2 |. METHODS
2.1 |. Study populations
The Genes-Environments and Admixture in Latino Americans (GALA II) and the Study of African Americans, Asthma, Genes, and Environments (SAGE) are two case-control studies of pediatric asthma in minority populations of the United States.16 Both studies were approved by the Human Research Protection Program Institutional Review Board of the University of California, San Francisco. Participants/parents provided written assent/consent, respectively. Asthma was defined by physician diagnosis, use of controller or rescue medication, and report of symptoms in the 2 years preceding enrollment. Controls were excluded if they reported asthma, rhinitis, hay fever, allergy, or eczema, wheezing, shortness of breath, or use of medication for allergies. Asthma exacerbations were defined as the presence of any of the following asthma-related events in the 12 months before enrollment: administration of oral corticosteroids (OCS), seeking emergency care, and hospitalizations.1 A total of 1283 exacerbators and 2027 controls from GALA II were analyzed in the discovery stage. In the replication stage, 448 exacerbators and 595 controls from SAGE were included. Additionally, asthma patients without exacerbations (898 from GALA II and 524 from SAGE) were used as controls in case-only analyses.
2.2 |. Genotyping and imputation
Genotyping was conducted with the Axiom LAT1 array and with the Axiom LAT1 Array Plus HLA (Affymetrix), and the SNPs and individuals resulting from the intersection of the two arrays were subjected to quality control. The Haplotype Reference Consortium (HRC)17 r1.1 was used as imputation reference panel. Quality control and imputation are detailed in the Supporting Information.
2.3 |. Statistical analysis
The association between 9 737 707 SNPs with minor allele frequency (MAF) ≥1% and imputation quality score R2 ≥ 0.3, and asthma with severe exacerbations was tested through logistic regression models using EPACTS 3.2.6,18 with correction for age, sex, and the first two genotype principal components (PCs), which explained most genetic variation (Figure S1). Conditional analyses were performed with GCTA-COJO v1.92.219 using the default options to identify independent variants (r2 ≤ 0.9) within 10 Mb distance from the lead SNP at each locus with suggestive level of significance (P ≤ 5 × 10−6). These variants were examined for replication in SAGE, and shared variants were meta-analyzed with METASOFT.20 Random or fixed-effects models were chosen based on the heterogeneity estimated by Cochrane’s Q test. Genome-wide significance was declared with P ≤ 5 × 10−8 after the meta-analysis across both stages.
To refine the SNPs whose effect was driven by severe asthma exacerbations or asthma, we conducted a case-only analysis by comparing exacerbators and non-exacerbators with correction for age, sex, two PCs, and disease severity defined based on treatment category (Table 1). For the region associated with asthma exacerbations in this analysis, fine mapping was conducted within 1 Mb flanking the lead SNP in Latinos using three imputation panels: HRC, 1000 Genomes Project (1KGP) Phase III, and the Consortium on Asthma among African-Ancestry Populations in the Americas (CAAPA). For each SNP, imputed dosages were selected based on the highest R2.
TABLE 1.
Clinical and demographic characteristics of the subjects included in the case-control and case-only analyses
| GALA II | SAGE | |||||
|---|---|---|---|---|---|---|
| Asthma with exacerbations (n = 1283) | Asthma without exacerbations (n = 898) | Control (n = 2027) | Asthma with exacerbations (n = 448) | Asthma without exacerbations (n = 524) | Control (n = 595) | |
| Gender (% female) | 582 (45.4)a | 403 (44.9) | 1137 (56.1) | 214 (47.8)a | 263 (50.2) | 340 (57.1) |
| Mean age in years ± SD | 12.2 ± 3.1a,b | 13.4 ± 3.5 | 13.9 ± 3.6 | 13.3 ± 3.6a,b | 14.4 ± 3.6 | 15.6 ± 3.8 |
| African ancestry (%) | 18.9 ± 13.2a,b | 12.6 ± 11.9 | 14.2 ± 12.0a | 83.6 ± 18.1 | 81.5 ± 21.5 | 81.7 ± 18.1 |
| Native American ancestry (%) | 22.9 ± 22.6b | 37.4 ± 25.5 | 33.9 ± 27.8 | NA | NA | NA |
| Asthma exacerbations in the last 12 mo, n (%) | ||||||
| Acute asthma care | 1092 (85.1) | NA | NA | 231 (51.6) | NA | NA |
| Oral corticosteroid use | 712 (55.5) | NA | NA | 337 (75.2) | NA | NA |
| Hospitalizations | 192 (15.0) | NA | NA | 88 (19.6) | NA | NA |
| Asthma control, n (%)c | ||||||
| Well controlled | 210 (16.4)b | 431 (48.0) | NA | 60 (15.7)b | 182 (36.8) | NA |
| Partially controlled | 533 (41.5)b | 282 (31.4) | NA | 112 (29.4) | 150 (30.4) | NA |
| Poorly controlled | 540 (42.1)b | 185 (20.6) | NA | 209 (54.9)b | 162 (32.8) | NA |
| Treatment category n (%)d | ||||||
| Step 1 | 464 (36.4)b | 505 (56.4) | NA | 97 (21.7)b | 242 (46.5) | NA |
| Step 2 | 417 (32.7)b | 233 (26.1) | NA | 182 (40.6) | 210 (40.3) | NA |
| Step 3 | 394 (30.9)b | 157 (17.5) | NA | 169 (37.7)b | 69 (13.2) | NA |
| Eczema, n (%)e | 224 (17.7) | 134 (15.1) | NA | 188 (48.5)b | 159 (32.4) | NA |
| Hay fever/rhinitis, n (%)f | 564 (45.2)b | 314 (35.4) | NA | 148 (36.2) | 142 (28.9) | NA |
| FEV1 (%)g | 97.9 ± 15.7a,b | 103.6 ± 14.4 | 108.0 ± 12.6 | 91.2 ± 11.9 | 92.4 ± 11.6 | 96.1 ± 10.8 |
| FVC (%)g | 102.0 ± 16.0a,b | 107.3 ± 15.3 | 108.0 ± 13.6 | 96.5 ± 11.8 | 97.0 ± 10.9 | 98.4 ± 11.8 |
| FEV1/FVC (%)g | 96.4 ± 7.6a | 97.2 ± 7.2 | 99.7 ± 6.7 | 94.1 ± 8.3 | 95.3 ± 7.6 | 97.8 ± 7.4 |
Note: For continuous variables, the mean and standard deviation are displayed, and the Mann-Whitney-Wilcoxon test was applied for the comparison of cases vs controls. For categorical variables, the number and proportion of subjects in each category are shown and a chi-square test was applied for the comparison of cases vs controls.
Abbreviations: FEV1, forced expiratory volume in the first second; FVC, forced vital capacity; NA, not available/applicable; SD, standard deviation.
P < .01 for the comparison between asthma cases with exacerbations and controls.
P < .01 for the comparison between asthma cases with exacerbations and asthma cases without exacerbations.
Asthma control was available for 2181 and 875 asthma cases from GALA II and SAGE, respectively.
The severity medication regime was categorized into three levels based on the use of short beta-agonists (step 1), one inhaled corticosteroid, leukotriene inhibitor, or theophylline tablet (step 2), more than one or a combination of an inhaled corticosteroid, leukotriene inhibitor, or theophylline tablet or a combination of inhaled corticosteroids and long-beta-agonists (step 3). Medication data were available for 2170 and 969 asthma cases from GALA II and SAGE, respectively.
Eczema status was available for 2155 and 879 subjects from GALA II and SAGE, respectively.
Hay fever status was available for 2133 and 901 subjects from GALA II and SAGE, respectively.
FEV1, FVC, and FEV1/FVC were available for 2698, 2710, and 2662 GALA II subjects, respectively, and 1058, 1075, and 1056 SAGE subjects, respectively.
In secondary analyses, we evaluated the association of the most significant SNP with lung function measurements by regression models adjusted for the first two PCs and asthma status. Moreover, we performed stratified analyses to assess the potential confounding effect of atopic comorbidities and asthma control. See Supporting Information for more details.
2.4 |. Methylation quantitative trait loci (cis-meQTL) analysis
We performed a cis-methylation quantitative trait locus (meQTL) analysis in 473 GALA II whole-blood samples (236 asthma cases and 237 controls) profiled with the Infinium HumanMethylation450 BeadChip array (Illumina, Inc). We tested for association with the M-values within 500 kb of the most strongly associated SNP with correction for sex, asthma, exacerbations, African and Native American ancestries, and inferred white cell counts. A false discovery rate (FDR) of 5% was used to declare significance accounting for the multiple comparisons performed. Detailed procedures are provided in the Supporting Information.
2.5 |. In silico functional evaluation
Functional annotation was conducted with HaploReg v4.221 based on linkage disequilibrium (LD) data of admixed American populations from the 1KGP. Evidence of expression quantitative trait loci (eQTLs) was searched in the Genotype-Tissue Expression (GTEx)22 v8.
3 |. RESULTS
3.1 |. Characteristics of study populations
The demographics and clinical characteristics of participants included in the analysis are shown in Table 1. Among asthma cases, Latinos sought unexpected asthma care more often than African Americans, while hospitalizations and OCS use were more frequent in African Americans.
3.2 |. Discovery in Latinos from GALA II
The quantile-quantile plot showed slight genomic inflation (Figure S2A, λGC = 1.02), which was driven by the strong association signals in LD at the known asthma locus 17q12-q21 (Figure S2B). A total of 171 SNPs were suggestively associated with severe exacerbations (P ≤ 5 × 10−6) (Figure 1; Table S1). From these, conditional analyses identified 12 independent SNPs with imputation quality R2 values ranging from 0.91 to 1.00 (Table 2), of which rs12946510 at the GRB7/IKZF3 region showed the strongest association (odds ratio [OR] for T allele = 0.68, 95% confidence interval [CI]:0.60–0.76, P = 6.82 × 10−11).
FIGURE 1.

Manhattan plot of the GWAS for GALA II (represented as -log10 P-value on the y-axis) along the chromosomes (x-axis). The suggestive significance threshold for replication is indicated by the gray line (P = 5 × 10−6). GALA II, Genes-Environments and Admixture in Latino Americans; GWAS, genome-wide association studies
TABLE 2.
Association and meta-analysis results for the independent SNPs identified in Hispanics/Latinos followed up for replication in African Americans
| rsID | Chr.: position (bp)a | A1/A2 | Closest gene | GALA II (n = 3310) | SAGE (n = 1043) | Meta-analysis (n = 4353) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Freq.b | OR (95% CI)b | P-valueb | Freq.b | OR (95% CI)b | P-valueb | OR (95% CI)b | P-valueb | Model | ||||
| rs4952375 | 2:42 016 863 | A/T | snoZ247/UNC01913 | 0.23 | 1.37(1.20–1.55) | 1.24 × 10−6 | 0.10 | 1.53 (1.12–2.08) | 7.43 × 10−3 | 1.39(1.23–1.56) | 3.79 × 10−8 | FE |
| rs17759529 | 2:162 851 013 | G/C | DPP4 | 0.16 | 0.67 (0.58–0.77) | 6.35 × 10−8 | 0.14 | 0.91 (0.69–1.18) | .467 | 0.76 (0.57–1.02) | 3.80 × 10−7 | RE2 |
| rs187089444 | 3:174 519 230 | A/C | NAALADL2 | 0.03 | 2.19 (1.60–2.99) | 8.70 × 10−7 | NA | NA | NA | NA | NA | NA |
| rs181042383 | 4:14 887 486 | A/G | UNC00504/CPEB2-DT | 0.04 | 1.95 (1.49–2.56) | 1.35 × 10−6 | NA | NA | NA | NA | NA | NA |
| rs78968521 | 4:152 214 673 | G/A | PRSS48 | 0.02 | 2.38(1.68–3.38) | 1.16 × 10−6 | 0.01 | 0.69 (0.31–1.54) | .360 | 1.35 (0.40–4.55) | 1.45 × 10−5 | RE2 |
| rs112599943 | 6:133 499 724 | C/T | EYA4 | 0.04 | 1.92 (1.47–2.50) | 1.55 × 10−6 | 0.08 | 1.40(1.00–1.95) | .047 | 1.70(1.38–2.09) | 6.00 × 10−7 | FE |
| rs76605301 | 10:12 227 306 | T/C | NUDT5 | 0.03 | 2.14 (1.56–2.94) | 2.87 × 10−6 | NA | NA | NA | NA | NA | NA |
| rs591709 | 11:93 520 315 | T/C | MED17 | 0.15 | 0.70 (0.60–0.81) | 2.39 × 10−6 | 0.20 | 1.28 (1.02–1.60) | .037 | 0.94 (0.52–1.70) | 9.65 × 10−6 | RE2 |
| rs11613333 | 12:116 880 378 | C/T | MIR4472–2 | 0.42 | 1.34(1.20–1.50) | 2.90 × 10−7 | 0.08 | 1.22 (0.87–1.71) | .243 | 1.33 (1.20–1.48) | 1.64 × 10−7 | FE |
| rs12946510 | 17:37 912 377 | T/C | GRB7/IKZF3 | 0.32 | 0.68 (0.60–0.76) | 6.82 × 10−11 | 0.21 | 0.85 (0.67–1.07) | .164 | 0.71 (0.64–0.79) | 1.12 × 10−10 | FE |
| rs113516984 | 17:45 964 442 | C/T | RP11–6N17.4/SP2 | 0.11 | 0.66 (0.56–0.79) | 4.64 × 10−6 | 0.07 | 0.67 (0.43–1.02) | .064 | 0.66(0.56–0.78) | 7.82 × 10−7 | FE |
| rs6132057 | 20:18 390 980 | G/A | DZANK1 | 0.15 | 0.69 (0.59–0.81) | 3.33 × 10−6 | 0.07 | 1.33 (0.93–1.91) | .117 | 0.94 (0.49–1.79) | 2.58 × 10−5 | RE2 |
Abbreviations: A1, effect allele; A2, non-effect allele; Chr., chromosome; CI, confidence interval; FE, fixed-effects model; Freq., effect allele frequency; NA, not available/applicable; OR, odds ratio; RE2, Han and Eskin’s random-effects models; rsID, reference SNP identifier.
Coordinates are referred to the GRCh37 reference genome.
Referred to the effect alleles (additive model).
3.3 |. Replication in African Americans from SAGE and meta-analysis
Of the 12 SNPs selected for replication, 3 SNPs were unsuitable for replication in African Americans due to their low frequency in this population (MAF <1%). From the remaining SNPs (Table 2), rs4952375 at the 2p21 region near LINC01913 replicated (OR for A allele = 1.53, 95% CI: 1.12–2.08, P = 7.43 × 10−3). A combined meta-analysis resulted in two genome-wide significant associations for rs12946510 (OR for C allele = 0.71, 95% CI: 0.64–0.79, P = 1.12 × 10−10) and rs4952375 (OR for A allele = 1.39, 95% CI: 1.23–1.56, P = 3.79 × 10−8) (Figure 2). We next assessed these two genome-wide significant associations in a case-only meta-analysis (n = 3139) including exacerbators and non-exacerbators (Table S2). The SNP rs4952375 was associated with increased risk of severe exacerbations among asthma patients (OR for A allele = 1.25, 95% CI: 1.09–1.44, P = 1.95 × 10−3). However, rs12946510 showed no association in the case-only analysis, suggesting that its effect is likely driven by asthma rather than severe exacerbations. The results of case-only analyses of the suggestive associations identified in the discovery are also provided in Table S2.
FIGURE 2.
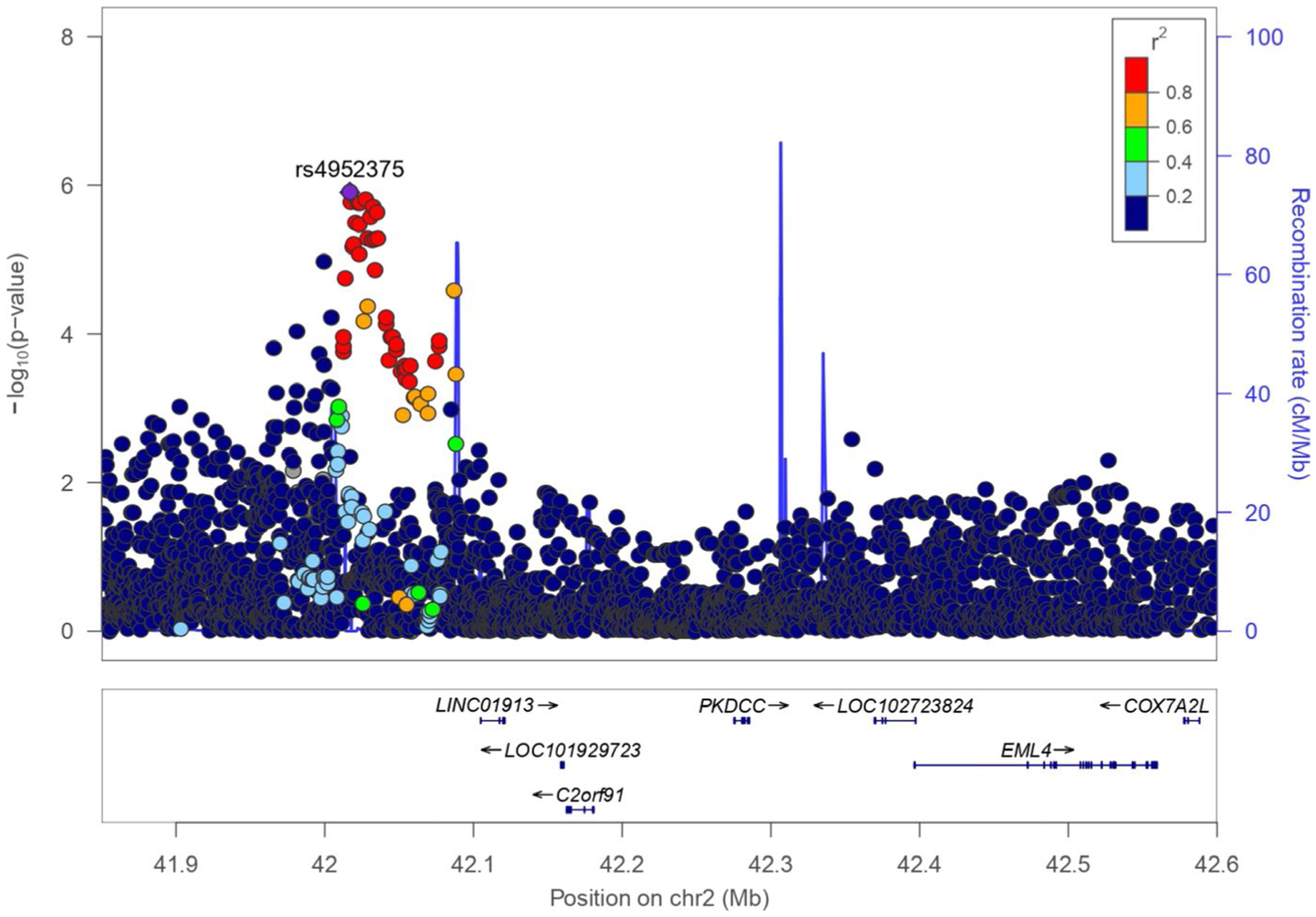
Regional plot of association in the discovery phase for the novel association detected at 2p21. The statistical significance of association results (−log10 P-value) is represented for each SNP as a dot (left y-axis) and recombination rate (right y-axis) by chromosome position (x-axis). SNPs are colored to show their LD with the top hit based on the pairwise r2 values from the American admixed populations of the 1KGP. LD, linkage disequilibrium; SNP, single nucleotide polymorphism
3.4 |. Fine mapping of 2p21 and assessment of additional phenotypes
Since rs4952375 was associated with asthma exacerbations in the case-control and the case-only analyses and has not been identified by previous GWAS, we performed fine mapping with additional imputation reference panels. Despite increasing the number of variants analyzed from 4817 to 6906 (27.8% and 2.4% exclusive of 1KGP and CAAPA, respectively), no additional genome-wide or suggestively associated SNPs were found (Figure S3).
Further meta-analyses for asthma-related traits were conducted for the SNP rs4952375 (Table S3). We first evaluated whether the association found for asthma with exacerbations differed by asthma control status, stratifying the exacerbators as well controlled or poorly controlled and comparing them with the controls. Although the association was only significant among poorly controlled exacerbators, no heterogeneity was found for the effect of rs4952375 among the two groups when compared to the controls (Cochran’s Q P = 0.263 and P = 0.988 for GALA II and SAGE, respectively). Additionally, given that almost half of the asthma patients reported eczema or rhinitis, we assessed whether atopic comorbidities could be a confounder performing stratified analyses (Table S3). No heterogeneity for the effect of rs4952375 was found between exacerbators with these morbidities vs controls or exacerbators without them when compared to controls (Cochran’s Q P = 0.842 for both studies). Moreover, rs4952375 was not associated with predicted baseline forced expiratory volume in one second (FEV1), forced vital capacity (FVC), or the FEV1/FVC ratio, in an analysis corrected for asthma status and the top two PCs (Table S3).
3.5 |. Functional analysis of the associated variant
In a subset of GALA II participants with methylation data available, we identified rs4952375 as a cis-meQTL for the CpG site cg17892159, which lies near the protein kinase domain-containing cytoplasmic (PKDCC) gene. Carriers of the risk allele showed increased methylation levels for cg17892159 (P = 9.79 × 10−5; FDR = 1.3%) (Figure 3A).
FIGURE 3.

Functional analysis of the rs4952375. A, Boxplot of blood methylation levels at cg17892159 annotated to PKDCC by rs4952375 genotype in a subset of GALA II subjects (n = 473). B, Gene expression levels of LINC01913 in lung tissue by genotype at rs4952375 indicating that this SNP is an eQTL, based on data obtained from GTEx (n = 450). Abbreviations: eQTL, expression quantitative trait loci; GALA II, Genes-Environments and Admixture in Latino Americans; SNP, single nucleotide polymorphism
Additionally, rs4952375 is a lung eQTL for the uncharacterized long intergenic non-coding RNA (lncRNA) 1913 (LINC01913) in the GTEx database (P = 1.3 × 10−7). Lung LINC01913 expression increases when the risk allele is present (Figure 3B). Furthermore, this variant is in high LD (r2 ≥ 0.8) with 31 lung eQTLs for LINC01913 (Table S4).
3.6 |. Validation of associations identified by previous GWAS
We assessed previous associations reported for asthma exacerbations11 and moderate-to-severe asthma23 for validation in GALA II and SAGE. We replicated the association of GSDMB and IKZF3 at genome-wide significant level and 10 additional SNPs at nominal level with asthma with severe exacerbations (Table S5). However, only the SNP rs1837253 from TSLP was also nominally associated with severe exacerbations in a case-only analysis (Table S6).
4 |. DISCUSSION
To our knowledge, this is the first GWAS of asthma with severe exacerbations in Latino and African American children and youth. We performed a two-stage GWAS and identified a novel locus for susceptibility to asthma with exacerbations that reached genome-wide significance in the combined meta-analysis. The strongest association at chromosome 2p21 is located at rs4952375, which was also associated with severe asthma exacerbations in a case-only meta-analysis of Latinos and African Americans. The sentinel SNP rs4952375 and its variants in LD (r2 ≥ 0.8) are located nearby LINC01913. Specifically, rs4952375 is located 87.8 kb away from the 5’UTR of LINC01913. In fact, the SNP rs4952375 and their proxies in LD regulate gene expression of the long non-coding RNA LINC01913 in the lung. We found that the A allele of rs4952375 is associated with a higher risk of asthma exacerbations and increased gene expression of LINC01913 in the lung. Long non-protein coding RNAs have been previously implicated in asthma phenotypes and other lung diseases as they interact with microRNAs and transcription factors involved in different biologic processes.24 Our results suggest that lncRNAs could also contribute to the pathogenesis of asthma exacerbations, although the mechanism involving LINC01913 in asthma remains unknown.
Our findings indicate that rs4952375 is also a meQTL for a CpG site annotated to PKDCC, a gene located 258 kb away from the associated marker. Genetic variation from PKDCC or its neighbor gene encoding for the echinoderm microtubule-associated protein-like 4 (EML4) has been associated with lung function measurements25 and atopy.26 However, those polymorphisms were not associated with severe asthma exacerbations in our study and are not in LD with rs4952375 (Table S7), suggesting that they are independent association signals. Likewise, rs4952375 was not associated with lung function measurements in our study and did not show differences between severe exacerbators with or without atopic comorbidities.
PKDCC is a tyrosine-protein kinase that regulates cell adhesion through signal transduction,27 including phosphorylation of MMP1,28 which is involved in airway remodeling and whose activity is increased during exacerbations in relation to the exacerbation severity.29 Based on the biologic processes ontology described in the database Open Targets,30 PKDCC is involved in cell differentiation, lung alveolus development, multicellular organism growth, and bone mineralization. Furthermore, mice deficient in Pkdcc show lung abnormalities.30 Our results suggest that the risk allele for asthma exacerbations is associated with higher methylation levels of PKDCC, which would imply that its gene expression could be downregulated in individuals with asthma exacerbations.
Hispanics/Latinos are descendants of the admixture of European, African, and Native American populations, with large variability in the ancestral proportions depending on the historical events that each particular subgroup has undergone.31 On the other hand, African Americans derive from the admixture of European and African populations, with large proportions of African ancestry, with mean values ranging from 73% to 93%.31 The high asthma prevalence among African-admixed individuals, particularly in developed countries,6–10 suggests a shared genetic basis that may be a risk factor for asthma under environmental influences that are absent in non-Westernized societies.8 In this work, we leveraged this specific genetic background to identify a novel locus for severe asthma exacerbations. Given the different ancestral composition, the risk allele of rs4952375 was more frequent in Latinos than in African Americans (22.7% vs 10.1%), in agreement with 1KGP (Figure S4). However, the association had consistent effects in both populations.
We also attempted to validate previous SNPs associated with asthma exacerbations or moderate-to-severe asthma in Latinos and African Americans. Although we found replication of several associations, no evidence of association was detected with severe asthma exacerbations in a case-only analysis, except for one SNP, suggesting that the effect of those variants is due to asthma susceptibility rather than exacerbations. Therefore, our findings suggest that the genetic determinants of severe exacerbations may differ from those involved in moderate-to-severe asthma. Alternatively, the lack of replication could be due to the differences in the characteristics of the patients analyzed in the current study compared to Shrine et al,23 in terms of ancestry, age, and study design. Of note, the SNPs rs6967330, located in the human rhinovirus C receptor CDHR3 gene and previously associated with asthma with severe exacerbations,12 did not replicate in our study, which could reflect different pathogenic mechanisms between the two studies, which differed in age, ethnicity, and definition of exacerbations.
We acknowledge some limitations in this study. First, reporting of asthma exacerbations was retrospective in both studies, which could introduce some imprecision into the phenotypes analyzed. Second, the fact that several SNPs are in strong LD in the region complicates the identification of the causal variant(s). Third, asthma is a complex disease resulting from the interaction between genetic and environmental exposures, which could be critical for the development of exacerbations, and those exposures were not addressed in our study. Fourth, limited functional data were available within the samples analyzed. However, data available on public databases (GTEx22 and OpenTargets30) provided evidence about the functional role of the associated variant regulating gene expression and the importance of the PKDCC in lung development based on knockout mice models.
On the other hand, this study has several strengths. Foremost, this is the first GWAS of asthma with severe exacerbations specifically focused on minority racial/ethnic groups at high risk of asthma morbidity and underrepresented in biomedical studies. Second, we showed that examining a specific asthma phenotype revealed genetic variants that have not been identified by previous GWAS of asthma susceptibility on the same populations. Third, we performed an in-depth fine mapping of the region of interest using a combination of three different panels and assessed the association with several clinical subtypes.
In summary, we conducted a GWAS of asthma with severe exacerbations in Hispanic/Latino and African American children and youth. Our study identified a novel genome-wide significant association for severe asthma exacerbations on chromosome 2p21. The associated variant is a lung eQTL for the LINC01913 gene and is also a whole-blood meQTL of a CpG site annotated to PKDCC. Additional functional follow-up is required to confirm the role of this locus in asthma exacerbations.
Supplementary Material
Key Message.
The first genomic analysis comparing asthma cases with severe exacerbations against controls performed in Hispanic/Latino and African American revealed a novel genome-wide significant susceptibility locus. The associat-edvariant demonstrated to have functional consequences in methylation and gene expression in blood and lung tissue, respectively. Our results provide two different gene targets of interest for asthma exacerbations whose specific role should be further explored in future functional analysis.
ACKNOWLEDGEMENTS
The GALA II and SAGE study collaborators include Shannon Thyne, UCSF; Harold J. Farber, Texas Children’s Hospital; Denise Serebrisky, Jacobi Medical Center; Rajesh Kumar, Lurie Children’s Hospital of Chicago; Emerita Brigino-Buenaventura, Kaiser Permanente; Michael A. LeNoir, Bay Area Pediatrics; Kelley Meade, UCSF Benioff Children’s Hospital, Oakland; William Rodriguez-Cintron, VA Hospital, Puerto Rico; Pedro C. Avila, Northwestern University; Jose R. Rodríguez-Santana, Centro de Neumología Pediátrica; Luisa N. Borrell, City University of New York; Adam Davis, UCSF Benioff Children’s Hospital, Oakland; Saunak Sen, University of Tennessee; and Fred Lurmann, Sonoma Technologies, Inc; Michael A. LeNoir, Bay Area Pediatrics; Kelley Meade, UCSF Benioff Children’s Hospital, Oakland.
The authors would like to thank the families and patients for their participation and thank the numerous healthcare providers and community clinics for their support and participation. In particular, the authors thank Sandra Salazar, the study coordinator, and the recruiters who obtained the data: Duanny Alva, Gaby Ayala-Rodriguez, Lisa Caine, Elizabeth Castellanos, Jaime Colon, Denise DeJesus, Blanca Lopez, Brenda Lopez, Louis Martos, Vivian Medina, Juana Olivo, Mario Peralta, Esther Pomares, Jihan Quraishi, Johanna Rodriguez, Shahdad Saeedi, Dean Soto, and Ana Taveras.
Funding information
This study was funded by the Spanish Ministry of of Science, Innovation, and Universities, the State Research Agency, and the European Regional Development Funds from the European Union (MICIU/AEI/FEDER, UE, SAF2017-83417R). EH-L was funded by a fellowship (PRE2018-083837) from the Spanish Ministry of Science, Innovation, and Universities. MP-Y was supported by the Ramón y Cajal Program from the Spanish Ministry of Economy, Industry, and Competitiveness (RYC-2015-17205). KLK was supported by the National Heart, Lung, and Blood Institute (NHLBI) (R01HL135156-S1), the UCSF Bakar Computational Health Sciences Institute, the Gordon and Betty Moore Foundation (GBMF3834), and the Alfred P. Sloan Foundation grant 2013-10-27 to UC Berkeley through the Moore-Sloan Data Sciences Environment initiative at the Berkeley Institute for Data Science (BIDS). Financial support to conduct part of this work onsite at Universidad de La Laguna (ULL) was supported by ULL Campus America 2019 Initiative.
The Genes-Environments and Admixture in Latino Americans (GALA II) Study and the Study of African Americans, Asthma, Genes and Environments (SAGE) were supported by the National Heart, Lung, and Blood Institute (NHLBI) of the National Institutes of Health (NIH) (R01HL117004 and X01HL134589). Additional study enrollment was supported by the Sandler Family Foundation, the American Asthma Foundation, the Amos Medical Faculty Development Program from the Robert Wood Johnson Foundation, the Harry Wm. and Diana V. Hind Distinguished Professorship in Pharmaceutical Sciences II, and the National Institute of Environmental Health Sciences (NIEHS) (R01ES015794). The generation, cleaning, quality control, and analysis of GALA II and SAGE data was funded by the NHLBI (R01HL128439, R01HL135156, R01HL141992, and R01HL141845); the NIEHS (R21ES24844); the National Institute on Minority Health and Health Disparities (NIMHD) (P60MD006902, R01MD010443, and R56MD013312); the National Institute of General Medical Sciences (NIGMS) (RL5GM118984); the Tobacco-Related Disease Research Program (24RT-0025 and 27IR-0030); and the National Human Genome Research Institute (NHGRI) (U01HG009080) to EGB.
Footnotes
CONFLIC TS OF INTEREST
EH-L reports a fellowship from the Spanish Ministry of Science, Innovation, and Universities. KLK reports grants from National Heart, Lung, and Blood Institute (NHLBI), the Gordon and Betty Moore Foundation to UC Berkeley, the UCSF Bakar Computational Health Sciences Institute, and the Alfred P. Sloan Foundation to UC Berkeley. KLK also reports non-financial support from Universidad de La Laguna Campus America 2019 Initiative. MP-Y and FLD report grants from Spanish Ministry of Science, Innovation, and Universities, the State Research Agency, and the European Regional Development Funds from the European Union (MICIU/AEI/FEDER, UE) and MP-Y reports grants from the Spanish Ministry of Economy, Industry, and Competitiveness. EGB reports grants from the National Institutes of Health, the Tobacco-Related Disease Research Program, the Sandler Family Foundation, the American Asthma Foundation, the Amos Medical Faculty Development Program from the Robert Wood Johnson Foundation and from the Harry Wm. and Diana V. Hind Distinguished Professorship in Pharmaceutical Sciences II. The rest of authors declare no conflict of interest.
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section.
REFERENCES
- 1.Reddel HK, Taylor DR, Bateman ED, et al. Asthma control and exacerbations - standardizing endpoints for clinical asthma trials and clinical practice. Am J Respir Crit Care Med. 2009;180:59–99. [DOI] [PubMed] [Google Scholar]
- 2.Cook J, Saglani S. Pathogenesis and prevention strategies of severe asthma exacerbations in children. Curr Opin Pulm Med. 2016;22:25–31. [DOI] [PubMed] [Google Scholar]
- 3.Chipps BE, Haselkorn T, Rosén K, Mink DR, Trzaskoma BL, Luskin AT. Asthma exacerbations and triggers in children in TENOR: impact on quality of life. J Allergy Clin Immunol Pract. 2018;6:169–176. e2. [DOI] [PubMed] [Google Scholar]
- 4.Calhoun WJ, Haselkorn T, Miller DP, Omachi TA. Asthma exacerbations and lung function in patients with severe or difficult-to-treat asthma. J Allergy Clin Immunol. 2015;136:1125–1127.e4. [DOI] [PubMed] [Google Scholar]
- 5.Sullivan PW, Ghushchyan V, Navaratnam P, et al. The national burden of poorly controlled asthma, school absence and parental work loss among school-aged children in the United States. J Asthma. 2018;55:659–667. [DOI] [PubMed] [Google Scholar]
- 6.Oraka E, Iqbal S, Flanders WD, Brinker K, Garbe P. Racial and ethnic disparities in current asthma and emergency department visits: findings from the national health interview survey, 2001–2010. J Asthma. 2013;50:488–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flores C, Ma S-F, Pino-Yanes M, et al. African ancestry is associated with asthma risk in African Americans. PLoS One. 2012;7:e26807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pino-Yanes M, Thakur N, Gignoux CR, et al. Genetic ancestry influences asthma susceptibility and lung function among Latinos. J Allergy Clin Immunol. 2015;135:228–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rumpel JA, Ahmedani BK, Peterson EL, et al. Genetic ancestry and its association with asthma exacerbations among African American subjects with asthma. J Allergy Clin Immunol. 2012;130:1302–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grossman NL, Ortega VE, King TS, et al. Exacerbation-prone asthma in the context of race and ancestry in Asthma Clinical Research Network trials. J Allergy Clin Immunol. 2019;144:1524–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herrera-Luis E, Hernandez-Pacheco N, Vijverberg SJ, Flores C, Pino-Yanes M. Role of genomics in asthma exacerbations. Curr Opin Pulm Med. 2019;25:101–112. [DOI] [PubMed] [Google Scholar]
- 12.Bønnelykke K, Sleiman P, Nielsen K, et al. A genome-wide association study identifies CDHR3 as a susceptibility locus for early childhood asthma with severe exacerbations. Nat Genet. 2014;46:51–55. [DOI] [PubMed] [Google Scholar]
- 13.McGeachie MJ, Wu AC, Tse SM, et al. CTNNA3 and SEMA3D: promising loci for asthma exacerbation identified through multiple genome-wide association studies. J Allergy Clin Immunol. 2015;136:1503–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dahlin A, Denny J, Roden DM, et al. CMTR1 is associated with increased asthma exacerbations in patients taking inhaled corticosteroids. Immun Inflamm Dis. 2015;3:350–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hernandez-Pacheco N, Farzan N, Francis B, et al. Genome-wide association study of inhaled corticosteroid response in admixed children with asthma. Clin Exp Allergy. 2019;49:789–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nishimura KK, Galanter JM, Roth LA, et al. Early-life air pollution and asthma risk in minority children. The GALA II and SAGE II studies. Am J Respir Crit Care Med. 2013;188:309–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCarthy S, Das S, Kretzschmar W, et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet. 2016;48:1279–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang HM. EPACTS (Efficient and Parallelizable Association Container Toolbox). Date last updated: February 19, 2019. https://genome.sph.umich.edu/wiki/EPACTS. Accessed June 4, 2019.
- 19.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han B, Eskin E. Random-effects model aimed at discovering associations in meta-analysis of genome-wide association studies. Am J Hum Genet. 2011;88:586–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ward LD, Kellis M. HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 2016;44:D877–D881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lonsdale J, Thomas J, Salvatore M, et al. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shrine N, Portelli MA, John C, et al. Moderate-to-severe asthma in individuals of European ancestry: a genome-wide association study. Lancet Respir Med. 2019;7:20–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Z, Ji N, Chen Z, et al. Next generation sequencing for long non-coding RNAs profile for CD4+ T cells in the mouse model of acute asthma. Front Genet. 2019;10:545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kichaev G, Bhatia G, Loh P-R, et al. Leveraging polygenic functional enrichment to improve GWAS power. Am J Hum Genet. 2019;104:65–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Castro-Giner F, Bustamante M, Ramon González J, et al. A pooling-based genome-wide analysis identifies new potential candidate genes for atopy in the European Community Respiratory Health Survey (ECRHS). BMC Med Genet. 2009;10:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maddala R, Skiba NP, Rao PV. Vertebrate lonesome kinase regulated extracellular matrix protein phosphorylation, cell shape, and adhesion in trabecular meshwork cells. J Cell Physiol. 2017;232:2447–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bordoli M, Yum J, Breitkopf S, et al. A secreted tyrosine kinase acts in the extracellular environment. Cell. 2014;158:1033–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Naveed S-u-N, Clements D, Jackson DJ, et al. Matrix metalloprotein-ase-1 activation contributes to airway smooth muscle growth and asthma severity. Am J Respir Crit Care Med. 2017;195:1000–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koscielny G, An P, Carvalho-Silva D, et al. Open Targets: a platform for therapeutic target identification and validation. Nucleic Acids Res. 2017;45:D985–D994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bryc K, Durand EY, Macpherson JM, Reich D, Mountain JL. The genetic ancestry of African Americans, Latinos, and European Americans across the United States. Am J Hum Genet. 2015;96:37–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


