Abstract
Translating ribosomes slow down or completely stall when they encounter obstacles on mRNAs. Such events can lead to ribosomes colliding with each other and forming complexes of two (disome), three (trisome) or more ribosomes. While these events can activate surveillance pathways, it has been unclear if collisions are common on endogenous mRNAs and whether they are usually detected by these cellular pathways. Recent genome-wide surveys of collisions revealed widespread distribution of disomes and trisomes across endogenous mRNAs in eukaryotic cells. Several studies further hinted that the recognition of collisions and response to them by multiple surveillance pathways depend on the context and duration of the ribosome stalling. This review considers recent efforts in the identification of endogenous ribosome collisions and cellular pathways dedicated to sense their severity. We further discuss the potential role of collided ribosomes in modulating co-translational events and contributing to cellular homeostasis.
Introduction
Many kinds of impediments to translation can interrupt the movement of the ribosome, such as unfavorable codons, problematic amino acid sequences, downstream secondary structures or damaged mRNAs (Chandrasekaran et al. 2019; Doma and Parker 2006; Gamble et al. 2016; Letzring et al. 2013; Simms et al. 2014). Unresolved ribosome stalling has been linked to proteotoxicity and neurodegeneration (Choe et al. 2016; Chu et al. 2009; Nedialkova and Leidel 2015; Yonashiro et al. 2016), suggesting that severely slowed ribosomes must be detected and removed from the mRNA (“rescued”) to maintain cellular homeostasis. For this purpose, cells have evolved surveillance mechanisms that are responsible for recognizing and resolving aberrant ribosome stalling events, such as the Ribosome-associated Quality Control (RQC) system in eukaryotes (reviewed in (Brandman and Hegde 2016; Joazeiro 2019)). This recognition has been shown to occur via specific detection of disomes or trisomes, which form due to prolonged ribosome stalling that signals a potentially detrimental translation slowdown (Ikeuchi et al. 2019; Juszkiewicz et al. 2018; Matsuo et al. 2020; Simms et al. 2017). Ribosome collisions are detected by an E3 ubiquitin ligase, Hel2(yeast)/ZNF598(mammals) (Ikeuchi et al. 2019; Juszkiewicz et al. 2018), which ubiquitinates specific ribosomal proteins (uS3, uS10 in yeast; eS10, uS10 in mammals) in the 40S subunit of the leading ribosome in a collision complex (Garzia et al. 2017; Juszkiewicz and Hegde 2017; Juszkiewicz et al. 2020b; Matsuo et al. 2017; Sundaramoorthy et al. 2017). Ubiquitination by Hel2/ZNF598 elicits the activity of the RQC trigger (RQT) complex in yeast (Matsuo et al. 2017; Matsuo et al. 2020) or ASC-1 complex in mammals (Hashimoto et al. 2020; Juszkiewicz et al. 2020b); this splits the leading stalled ribosome into subunits and triggers myriad events that lead to degradation of the nascent peptide and recycling of ribosomal subunits (Bengtson and Joazeiro 2010; Brandman et al. 2012; Kostova et al. 2017; Meyer et al. 2020; Osuna et al. 2017; Sitron and Brandman 2019). Recognition of collisions can also induce mRNA degradation through multiple endonucleolytic cleavage events, also referred to as No-go Decay (NGD) (D’Orazio et al. 2019; Doma and Parker 2006; Glover et al. 2020; Guydosh and Green 2017; Guydosh et al. 2017; Ikeuchi et al. 2019; Navickas et al. 2020; Simms et al. 2018). Ribosomes may, in turn, stall at these cleavage sites.
One challenge for the RQC system is to differentiate the stalled ribosomes on defective mRNAs that need to be rescued from the occasional, or sometimes even functional (Collart and Weiss 2019), ribosome pauses on non-aberrant transcripts. Most RQC studies have been based on severe stall-inducing constructs containing 12–20 consecutive Lys or Arg, an inherently problematic sequence for the ribosome, inserted between two reporter genes to measure the level of translation readthrough of the stalling site (Brandman et al. 2012; Dimitrova et al. 2009; Letzring et al. 2013). Although this experimental system has been useful for identifying numerous components of the RQC pathway and for revealing crucial insights into the mechanism of RQC, a reporter cannot fully capture the diversity and degree of endogenous ribosome stalling events that occur in the cell. Therefore, answers to several questions have remained largely unknown: (1) Where and how frequently do endogenous collisions occur in the cell? (2) What is the cellular response to endogenous collisions? (3) Are all collisions detrimental or can some of them have functional roles? In this review, we present the results of recent studies that begin to answer these questions.
Ribosome collisions are widespread on endogenous mRNAs
One suitable technique to systematically identify ribosome stalling events in the cell is ribosome profiling (also known as Ribo-seq), which is deep-sequencing of ribosome-protected mRNA fragments and can provide a snapshot of transcriptome-wide ribosome positions (Brar and Weissman 2015; Ingolia et al. 2009; Ingolia et al. 2019). However, the traditional ribosome profiling approach is based on isolating mRNA fragments that are protected by single ribosomes (monosomes). To obtain the distribution of collisions, ribosome profiling methodology was revised by isolating the fragments that are specifically protected by disomes (Guydosh and Green 2014) or trisomes (Meydan and Guydosh 2020) (denoted as disome or trisome profiling). Recently, profiling of ribosome collisions has successfully revealed the cellular landscape of translational pauses in various model eukaryotic systems.
Disome profiling in yeast showed that collisions are widespread on endogenous mRNAs (Guydosh and Green 2014; Meydan and Guydosh 2020), predicted to be as high as 20% of all translating ribosomes (Diament et al. 2018). This prediction is in line with the disome percentage (~10%) in mouse liver cells, as revealed by a quantitative comparison of monosome and disome profiling data by using spike-in RNA oligos (Arpat et al. 2020). Although in vitro experiments and theoretical modeling showed that the level of ribosome loading of an mRNA (translation efficiency or “TE”) is correlated with the presence of traffic jams or “stochastic” collisions (Juszkiewicz et al. 2018; Park and Subramaniam 2019; Simms et al. 2017), disome profiling data suggest that this may not be globally true for all transcripts (Arpat et al. 2020; Diament et al. 2018; Meydan and Guydosh 2020). Several reasons could explain the lack of correlation between TE and ribosome collisions. For instance, collisions on mRNAs could create a negative feedback loop to prevent further loading of ribosomes or transcripts with high TE could be evolutionarily tuned to avoid stochastic collisions. Alternatively, specific sequences or co-translational events, rather than chance encounters, might be the major driver of ribosome collisions and these cases may involve other features to prevent their detection. Consistent with the idea of collisions at specific sites, disomes in yeast, zebrafish, mouse and human cells were mostly enriched on Pro-, Asp-, and Gly-containing motifs as well as Lys- and Arg-rich polybasic tracts (Arpat et al. 2020; Han et al. 2020; Meydan and Guydosh 2020), demonstrating that endogenous collisions frequently occur on sequences known to slow down the ribosome.
Multiple surveillance pathways detect ribosome collisions
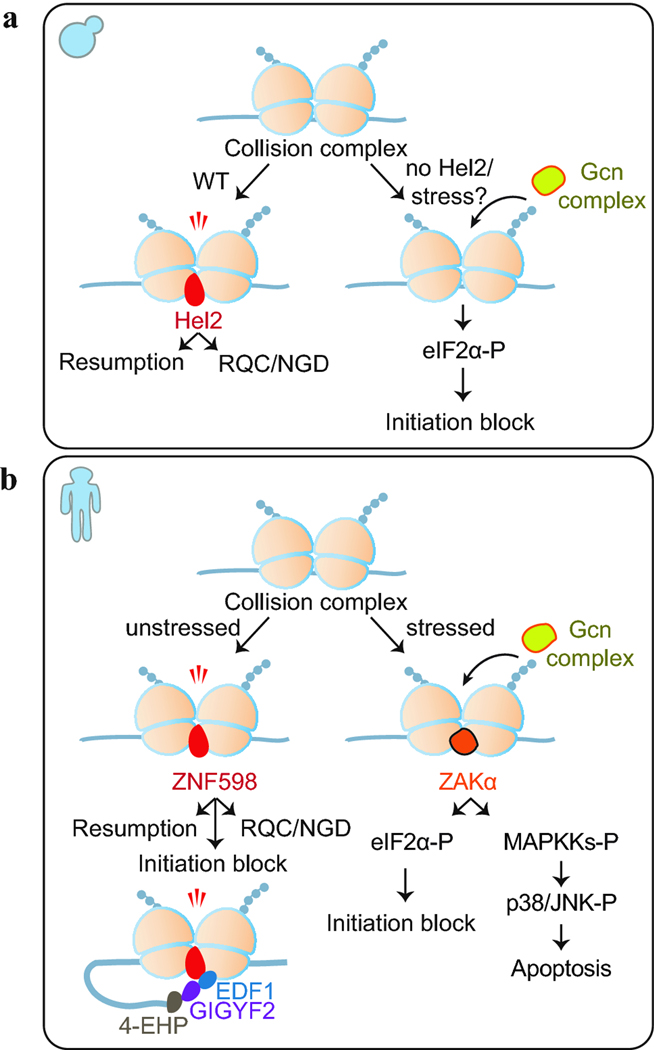
One of the keys to understanding how the cell responds to and distinguishes between different types of collisions is to identify the specific sensors of disomes. Recent studies have examined the role of many potential sensors, particularly Hel2/ZNF598, since it is tasked with triggering downstream steps of RQC via ubiquitination of the ribosome. The direct effect of Hel2 activity on the stability of collisions, however, has been unclear. At least two non-mutually exclusive models are possible: (1) Since Hel2/ZNF598 is needed to promote the rescue of the stalled ribosome in a collision complex, the lack of Hel2 should result in further accumulation of collided ribosomes. Indeed, in vitro experiments using an endogenous yeast RQC target (SDD1) and ribosome profiling of an optimized stalling reporter in yeast showed that either lack of Hel2 or deletion of RQT factors result in increased queuing upstream of stall sites (D’Orazio et al. 2019; Matsuo et al. 2020). Reporter studies in a mammalian cell-free system were also consistent with this model (Juszkiewicz et al. 2020b). (2) Hel2/ZNF598 enhances collision complexes to prevent any translation of the downstream, problematic sequence in favor of a slow rescue process. This activity may also prevent other sensors from recognizing the collision and triggering other surveillance pathways that may not be warranted. In this scenario, loss of Hel2/ZNF598 would lead to reduced pausing and decreased disome formation. Consistent with this second case, reporter experiments have demonstrated that lack of Hel2/ZNF598 causes readthrough of the stalling site (Juszkiewicz and Hegde 2017; Letzring et al. 2013; Matsuo et al. 2017; Sundaramoorthy et al. 2017). Similarly, disome profiling in yeast showed that loss of Hel2 globally decreased formation of the strongest disomes (Meydan and Guydosh 2020). One way to reconcile the observations comes down to the specific kinetics of the translational slowdown: severe ribosome arrest events may not require ubiquitination to block downstream translation but minor collisions on endogenous substrates may require ubiquitination to halt the resumption of translation (Juszkiewicz et al. 2020b). Ubiquitination by Hel2 could therefore act as a temporary translational checkpoint to decide the fate of a minor collision: either resuming translation or recruiting other proteins for triggering downstream RQC/NGD pathways (Fig. 1A). Consistent with the idea that the kinetics of stall events guide their fate come from RNA-seq experiments in the presence and absence of Hel2 showing very little change in levels of disome-associated mRNAs (Meydan and Guydosh 2020; Park and Subramaniam 2019). Therefore, endogenous collisions appear to not lead to much NGD and could be handled differently than the prolonged, detrimental stalls that are typically observed with the stalling reporters.
Figure 1:
Multiple surveillance pathways recognize ribosome collisions in yeast (A) and in mammalian (B) cells. We note it has been suggested that Mbf1/EDF1 broader roles at additional points in these pathways.
Disome profiling combined with biochemical experiments in yeast also suggested that disomes can be detected by other sensors (Meydan and Guydosh 2020). The lack of Hel2 led to activation of Gcn4, a master regulator of the integrated stress response (ISR) in yeast, which is turned on via phosphorylation of eIF2α (eIF2α-P) by the Gcn2 kinase (Dever et al. 1992; Hinnebusch 2005). Gcn2 per se, or as a part of the Gcn complex (consisting of Gcn2 and its partners, Gcn1 and Gcn20), was suggested to be activated in response ribosome stalling (Garcia-Barrio et al. 2000; Harding et al. 2019; Inglis et al. 2019; Ishimura et al. 2016; Visweswaraiah et al. 2012). In addition, Gcn1 and Gcn20 were found to be associated with nuclease resistant collisions via sucrose gradient fractionation experiments (Marton et al. 1997), hinting that Gcn proteins can readily sense collided ribosomes. According to this model, when Hel2 is knocked out or overwhelmed during amino acid starvation, the collision burden of the Gcn pathway increases, resulting in increased eIF2α-P, activation of the ISR and subsequent blockage of translational initiation (Meydan and Guydosh 2020) (Fig. 1A). Reduced initiation could also help explain the observation that formation of the strongest disomes is reduced under loss of Hel2. Future studies will address if the initiation block by ISR activation is evolved to act as a negative feedback loop to decrease disome formation under conditions where the availability of Hel2 could be limited, such as globally severe ribosome stalling.
A collision-mediated stress response has also been discovered in human cells. Biochemical experiments along with a selective disome profiling revealed that ZAKα, an isoform of mitogen-activated protein kinase kinase kinase (MAPKKK) ZAK protein, can detect collisions that are formed due to cellular stress (Wu et al. 2020). Strikingly, ZAKα activates the ISR by inducing eIF2α phosphorylation in response to less severe collisions that occur when cells are subjected to amino acid starvation or UV irradiation. eIF2α-P is then suggested to block initiation to decrease further disome formation (Fig. 1B). On the other hand, if the collisions are more severe, such as those that are formed due to treatment with translation elongation inhibitors, then ZAKα activates the Ribotoxic Stress Response (RSR) pathway, by inducing phosphorylation of p38 or c-Jun N-terminal kinase, which can lead to cell cycle arrest or apoptosis, respectively (Wu et al. 2020) (Fig. 1B). Deficiency in RSR activation has been linked to decreased life span of Caenorhabditis elegans, further supporting the physiological importance of the ZAK pathway (Vind et al. 2020). Interestingly, proximity proteomics analysis showed that GCN1/2/20 interact with ZAK and ZNF598 (Go et al. 2019; Wu et al. 2020). Similarly, in yeast, Gcn1/2/20 co-immunoprecipitate with Hel2 (Meydan and Guydosh 2020; Simms et al. 2017). It is yet to be investigated, however, if the proteins of these different pathways (ISR/RSR and RQC) can co-exist and interact on the same colliding ribosome or each pathway recognizes a distinct set of collision substrates.
In addition to the Gcn complex, other proteins can dynamically control collisions by modulating initiation on mRNAs with stalled ribosomes. For example, ZNF598 was shown to interact with 4EHP (also called as EIF4E2), an inhibitory cap binding protein, and its partner GIGYF1/2 (Morita et al. 2012; Tollenaere et al. 2019). A parallel study utilizing a growth based CRISPRi screen in human cells revealed that 4EHP and GIGYF2 cooperate with ZNF598 to suppress translation initiation on mRNAs with ribosome collisions (Fig. 1B) (Hickey et al. 2020). 4EHP is critical for healthy embryonic development in Drosophila (Cho et al. 2005), C. elegans (Dinkova et al. 2005) and mice (Morita et al. 2012), suggesting that 4EHP-GIGYF2-ZNF598-mediated translation repression may play an essential role in development. On the other hand, the yeast homologs of GIGYF2, Syh1 and Smy2, both lack the 4EHP binding domain, and do not induce inhibition of initiation (Hickey et al. 2020). Interestingly, however, Syh1 and Smy2 mediate decay of mRNAs with stalled ribosomes and therefore could be linked to NGD.
Two parallel studies utilizing a quantitative proteomics approach of colliding ribosomes in human cells further identified EDF1 as a ZNF598-independent sensor for collisions (Juszkiewicz et al. 2020a; Sinha et al. 2020). Upon detecting collisions, EDF1 was proposed to block initiation by stabilizing 4EHP-GIGYF2-ZNF598 complex on the collided ribosomes (Fig. 1B). Mbf1 (yeast homolog of EDF1) and to some extent, EDF1, were shown to suppress frameshifting at stalling sites (Juszkiewicz et al. 2020a; Wang et al. 2018). Since collisions have been shown to stimulate frameshifting (Simms et al. 2019; Wang et al. 2018), one model posits that Mbf1/EDF1 blocks initiation, thereby limiting ribosome collisions and decreasing frameshifting. Alternatively, Mbf1/EDF1 could have a direct role on the ribosome in preventing slippage events that lead to frameshifting. Mbf1/EDF1 was also shown to have a role in stabilizing association of ZNF598 (Juszkiewicz et al. 2020a; Sinha et al. 2020) and allowing ribosome collisions to trigger activation of c-Jun, perhaps in coordination with RACK1 and ZAKα (Sinha et al. 2020).
In addition to aforementioned pathways, it is also intriguing to propose that other proteins can detect or regulate ribosome collisions. For example, disome profiling experiments showed that knockdown of EIF5A in human cells caused accumulation of some disomes at Pro-rich motifs, suggesting that EIF5A often alleviates ribosomes pausing at problematic amino acid sequences and can directly affect disome distribution (Han et al. 2020). Another factor that can potentially modulate formation of collisions is MKRN1, which is a poly(A) sensing protein and a ubiquitin ligase that was proposed to stall ribosomes to prevent them from translating premature poly(A) sequences (Hildebrandt et al. 2019). MKRN1-induced stalling was suggested to cause ribosomes to collide and thereby promote ZNF598 binding, hinting that MKRN1 may dynamically affect collision occupancy. Finally, FMRP, encoded by the FMR1 gene, was suggested to stall ribosomes on specific mRNAs (Darnell et al. 2011; Das Sharma et al. 2019; Shah et al. 2020). In addition, FMRP-induced stalling is thought to be important for neuronal health, since ribosome profiling demonstrated that ribosome pausing was abolished in brains of mice with Fragile-X syndrome, which is a neurodevelopmental disorder and occurs to due to genetic silencing of FMR1 (Das Sharma et al. 2019). If FMRP has a direct effect on translation elongation, it is intriguing to hypothesize that it can also influence the distribution of collisions. It is also worth noting that two autosomal paralogs of FMRP, FXR1 and FXR2, are present in mammalian cells and can form complexes with FMRP (Suardi and Haddad 2020). Curiously, both FMRP and FXR2 appear to interact with ZAK (Wu et al. 2020), suggesting that ribosome-mediated cross-talk between the two pathways could exist. It will be exciting to address whether FMRP, FXR1 or FXR2, have any role in modulating ribosome collisions, if they are linked to other surveillance pathways and if the collisions could be important for neuronal health.
Can collisions modulate functional events?
The presence of multiple surveillance pathways sensing and modulating collisions suggest that both the context and severity could play roles in determining the fate of a collision (Fig. 2). It is therefore intriguing to propose that the response may be tuned to allow for collisions that are beneficial or regulatory, and respond to those that are detrimental. For example, ribosome collisions appear to lead to mRNA degradation on aberrant transcripts such as premature poly(A) within a coding region (Garzia et al. 2017; Guydosh and Green 2017) and likely during conditions of stress (Guydosh et al. 2017; Simms et al. 2017; Wu et al. 2020). However, ribosome pausing is also known to modulate co-translational events (reviewed in (Collart and Weiss 2019)), such as nascent peptide folding, peptide targeting into membranes/organelles or assembly of the peptide emerging from the ribosome with chaperones and other proteins. In such cases, the formation of collided ribosomes that trigger a robust RQC response that leads to mRNA degradation would be seemingly detrimental.
Figure 2:
Potential schema for how cellular response is determined by the severity of the ribosome collision.
Several pieces of evidence suggest the presence of potentially functional collisions in the cell. For example, in mammalian cells, prominent disome peaks observed at known programmed translational pause sites, such as SEC61B and the AZIN1 upstream ORF, could be important for modulating co-translational membrane insertion of the nascent peptide and regulation of polyamine synthesis, respectively (Arpat et al. 2020; Han et al. 2020). In the latter case, the regulation has been proposed to rely on queues of ribosomes (perhaps mixed with 40S subunits) under conditions of eIF5A deficiency and can likely regulate initiation on uORFs globally (Eisenberg et al. 2020; Ivanov et al. 2018). Disomes were also found in the vicinity of signal peptide coding regions, suggesting that programmed ribosome queuing on these mRNAs can mediate co-translational targeting of the nascent protein into the secretory pathway. In addition, disome peaks in mRNA regions encoding unstructured segments of some proteins further points to the potential involvement of collisions in modulating co-translational folding (Arpat et al. 2020). In yeast, collisions were also found at a known stalling site in the CPA1 upstream ORF, which is known to regulate arginine metabolism. In addition, disomes that are observed around +1 programmed ribosomal frameshifting (PRF) sites in the OAZ1, EST3 and ABP140 genes may be important for modulating PRF efficiency (Meydan and Guydosh 2020; Olson and Dinman 2020), consistent with the hypothesis that disomes can regulate some PRF events in bacteria (Smith et al. 2019). Strikingly, prominent disome peaks were also detected in yeast genes coding for ubiquitin precursors, located ~120 nt downstream of the ubiquitin cleavage sites. This distance positions the ubiquitin nascent peptide with the cleavage site protruding out of the ribosomal exit tunnel (which spans ~30–40 amino acids) and may potentially allow co-translational processing of ubiquitin precursor into free ubiquitin monomers (Meydan and Guydosh 2020). In this case, it is plausible to speculate that ribosome collision can also act as a feedback control mechanism to regulate free ubiquitin homeostasis.
Given the observation of collisions at functional sites, it is important to address how the surveillance pathways respond to them. In some cases, disome formation and downstream RQC events are beneficial, as was recently shown for the elimination of the protein encoded by the XBP1u (unspliced) mRNA (Han et al. 2020). Another option is that recognition of a “benign” collision could mediate ribosome pausing without committing the mRNA or peptide to degradation. For example, in yeast, ubiquitination by Hel2 is required to maintain the strongest sites of disome/trisome formation (Meydan and Guydosh 2020) and this enhanced pausing could be beneficial for a functional stalling event. In this case, de-ubiquitinases could potentially remove the ubiquitin mark before the collisions are irreversibly targeted to downstream pathways or recognized by other sensors such as the Gcn complex (Garshott et al. 2020; Jung et al. 2017; Matsuo et al. 2017; Meyer et al. 2020). In addition, some tissues or cell types may purposely suppress the activity of surveillance pathways when needed, e.g. ZNF598 is downregulated in reticulocytes, possibly to avoid degradation of hemoglobin mRNA with unusually high ribosome traffic (Juszkiewicz et al. 2018). Alternatively, problematic sequences may be favored within the extreme 5’ region of the transcript where the close proximity of the start codon would physically prevent another ribosome from loading onto the message and forming a disome. Interestingly, some endogenous stalling events, such as those that occur on RQC1 and SDD1 mRNAs are thought to trigger RQC (Brandman et al. 2012; Matsuo et al. 2020). Perhaps the surveillance systems are not entirely failproof and some “innocent” collisions, if they persist long enough, can be sacrificed at the cost of keeping the cell safe.
Concluding remarks
We look forward to structures of colliding ribosomes bound to various surveillance factors since they will likely illuminate mechanisms of collision detection and could possibly identify novel proteins bound to a collision complex. It will be very interesting to find out if different collision sensors can co-exist on the ribosome and how the surveillance pathways are connected with each other.
We also envision that monosome/disome/trisome profiling tools, in combination with other techniques, can illuminate how translation efficiency is affected by ribosome collisions. Beyond its importance for understanding gene expression, this information is crucial for synthetic biology studies, such as those that are dedicated to optimizing in vivo translation of mRNA vaccines and therapeutics.
Finally, it is important to emphasize that the absence of surveillance factors results in dysregulation of protein homeostasis, disease phenotypes or developmental defects. We therefore look forward to studies of ribosome collisions and surveillance pathways in various model organisms, where the role of translational pausing in different tissues, developmental stages and disease phenotypes can be explored.
Acknowledgments
Funding
This work was funded by the Intramural Research Program of the NIH, the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) (DK075132 to N.R.G.) and the Postdoctoral Research Associate Training Program (PRAT) at the National Institute of General Medical Sciences (NIGMS) (1FI2GM137845 to S.M.).
Footnotes
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
References
- Arpat AB, Liechti A, De Matos M, Dreos R, Janich P, Gatfield D (2020) Transcriptome-wide sites of collided ribosomes reveal principles of translational pausing Genome Res 30:985–999 doi: 10.1101/gr.257741.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtson MH, Joazeiro CA (2010) Role of a ribosome-associated E3 ubiquitin ligase in protein quality control Nature 467:470–473 doi: 10.1038/nature09371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandman O, Hegde RS (2016) Ribosome-associated protein quality control Nat Struct Mol Biol 23:7–15 doi: 10.1038/nsmb.3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandman O et al. (2012) A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress Cell 151:1042–1054 doi: 10.1016/j.cell.2012.10.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brar GA, Weissman JS (2015) Ribosome profiling reveals the what, when, where and how of protein synthesis Nat Rev Mol Cell Biol 16:651–664 doi: 10.1038/nrm4069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekaran V et al. (2019) Mechanism of ribosome stalling during translation of a poly(A) tail Nat Struct Mol Biol 26:1132–1140 doi: 10.1038/s41594-0190331-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho PF, Poulin F, Cho-Park YA, Cho-Park IB, Chicoine JD, Lasko P, Sonenberg N (2005) A new paradigm for translational control: inhibition via 5’−3’ mRNA tethering by Bicoid and the eIF4E cognate 4EHP Cell 121:411–423 doi: 10.1016/j.cell.2005.02.024 [DOI] [PubMed] [Google Scholar]
- Choe YJ, Park SH, Hassemer T, Korner R, Vincenz-Donnelly L, Hayer-Hartl M, Hartl FU (2016) Failure of RQC machinery causes protein aggregation and proteotoxic stress Nature 531:191–195 doi: 10.1038/nature16973 [DOI] [PubMed] [Google Scholar]
- Chu J et al. (2009) A mouse forward genetics screen identifies LISTERIN as an E3 ubiquitin ligase involved in neurodegeneration Proc Natl Acad Sci U S A 106:2097–2103 doi: 10.1073/pnas.0812819106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collart MA, Weiss B (2019) Ribosome pausing, a dangerous necessity for co-translational events Nucleic Acids Res doi: 10.1093/nar/gkz763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Orazio KN, Wu CC, Sinha N, Loll-Krippleber R, Brown GW, Green R (2019) The endonuclease Cue2 cleaves mRNAs at stalled ribosomes during No Go Decay Elife 8 doi: 10.7554/eLife.49117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC et al. (2011) FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism Cell 146:247–261 doi: 10.1016/j.cell.2011.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das Sharma S et al. (2019) Widespread Alterations in Translation Elongation in the Brain of Juvenile Fmr1 Knockout Mice Cell Rep 26:3313–3322 e3315 doi: 10.1016/j.celrep.2019.02.086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever TE, Feng L, Wek RC, Cigan AM, Donahue TF, Hinnebusch AG (1992) Phosphorylation of initiation factor 2 alpha by protein kinase GCN2 mediates gene-specific translational control of GCN4 in yeast Cell 68:585–596 doi: 10.1016/0092-8674(92)90193-g [DOI] [PubMed] [Google Scholar]
- Diament A, Feldman A, Schochet E, Kupiec M, Arava Y, Tuller T (2018) The extent of ribosome queuing in budding yeast PLoS Comput Biol 14:e1005951 doi: 10.1371/journal.pcbi.1005951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova LN, Kuroha K, Tatematsu T, Inada T (2009) Nascent peptide-dependent translation arrest leads to Not4p-mediated protein degradation by the proteasome J Biol Chem 284:10343–10352 doi: 10.1074/jbc.M808840200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinkova TD, Keiper BD, Korneeva NL, Aamodt EJ, Rhoads RE (2005) Translation of a small subset of Caenorhabditis elegans mRNAs is dependent on a specific eukaryotic translation initiation factor 4E isoform Mol Cell Biol 25:100–113 doi: 10.1128/MCB.25.1.100-113.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doma MK, Parker R (2006) Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation Nature 440:561–564 doi: 10.1038/nature04530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg AR et al. (2020) Translation Initiation Site Profiling Reveals Widespread Synthesis of Non-AUG-Initiated Protein Isoforms in Yeast Cell Syst 11:145–160 e145 doi: 10.1016/j.cels.2020.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble CE, Brule CE, Dean KM, Fields S, Grayhack EJ (2016) Adjacent Codons Act in Concert to Modulate Translation Efficiency in Yeast Cell 166:679–690 doi: 10.1016/j.cell.2016.05.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Barrio M, Dong J, Ufano S, Hinnebusch AG (2000) Association of GCN1-GCN20 regulatory complex with the N-terminus of eIF2alpha kinase GCN2 is required for GCN2 activation EMBO J 19:1887–1899 doi: 10.1093/emboj/19.8.1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garshott DM, Sundaramoorthy E, Leonard M, Bennett EJ (2020) Distinct regulatory ribosomal ubiquitylation events are reversible and hierarchically organized Elife 9 doi: 10.7554/eLife.54023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzia A et al. (2017) The E3 ubiquitin ligase and RNA-binding protein ZNF598 orchestrates ribosome quality control of premature polyadenylated mRNAs Nat Commun 8:16056 doi: 10.1038/ncomms16056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover ML, Burroughs AM, Monem PC, Egelhofer TA, Pule MN, Aravind L, Arribere JA (2020) NONU-1 Encodes a Conserved Endonuclease Required for mRNA Translation Surveillance Cell Rep 30:4321–4331 e4324 doi: 10.1016/j.celrep.2020.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Go CD et al. (2019) A proximity biotinylation map of a human cell bioRxiv:796391 doi: 10.1101/796391 [DOI] [Google Scholar]
- Guydosh NR, Green R (2014) Dom34 rescues ribosomes in 3’ untranslated regions Cell 156:950–962 doi: 10.1016/j.cell.2014.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guydosh NR, Green R (2017) Translation of poly(A) tails leads to precise mRNA cleavage RNA 23:749–761 doi: 10.1261/rna.060418.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guydosh NR, Kimmig P, Walter P, Green R (2017) Regulated Ire1-dependent mRNA decay requires no-go mRNA degradation to maintain endoplasmic reticulum homeostasis in S. pombe Elife 6 doi: 10.7554/eLife.29216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han P et al. (2020) Genome-wide Survey of Ribosome Collision Cell Rep 31:107610 doi: 10.1016/j.celrep.2020.107610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding HP, Ordonez A, Allen F, Parts L, Inglis AJ, Williams RL, Ron D (2019) The ribosomal P-stalk couples amino acid starvation to GCN2 activation in mammalian cells Elife 8 doi: 10.7554/eLife.50149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto S, Sugiyama T, Yamazaki R, Nobuta R, Inada T (2020) Identification of a novel trigger complex that facilitates ribosome-associated quality control in mammalian cells Sci Rep 10:3422 doi: 10.1038/s41598-020-60241-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickey KL et al. (2020) GIGYF2 and 4EHP Inhibit Translation Initiation of Defective Messenger RNAs to Assist Ribosome-Associated Quality Control Mol Cell doi: 10.1016/j.molcel.2020.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt A et al. (2019) The RNA-binding ubiquitin ligase MKRN1 functions in ribosome-associated quality control of poly(A) translation Genome Biol 20:216 doi: 10.1186/s13059-019-1814-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG (2005) Translational regulation of GCN4 and the general amino acid control of yeast Annu Rev Microbiol 59:407–450 doi: 10.1146/annurev.micro.59.031805.133833 [DOI] [PubMed] [Google Scholar]
- Ikeuchi K et al. (2019) Collided ribosomes form a unique structural interface to induce Hel2-driven quality control pathways EMBO J 38 doi: 10.15252/embj.2018100276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglis AJ, Masson GR, Shao S, Perisic O, McLaughlin SH, Hegde RS, Williams RL (2019) Activation of GCN2 by the ribosomal P-stalk Proc Natl Acad Sci U S A 116:4946–4954 doi: 10.1073/pnas.1813352116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS (2009) Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling Science 324:218–223 doi: 10.1126/science.1168978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Hussmann JA, Weissman JS (2019) Ribosome Profiling: Global Views of Translation Cold Spring Harb Perspect Biol 11 doi: 10.1101/cshperspect.a032698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimura R, Nagy G, Dotu I, Chuang JH, Ackerman SL (2016) Activation of GCN2 kinase by ribosome stalling links translation elongation with translation initiation Elife 5 doi: 10.7554/eLife.14295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov IP et al. (2018) Polyamine Control of Translation Elongation Regulates Start Site Selection on Antizyme Inhibitor mRNA via Ribosome Queuing Mol Cell 70:254–264 e256 doi: 10.1016/j.molcel.2018.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joazeiro CAP (2019) Mechanisms and functions of ribosome-associated protein quality control Nat Rev Mol Cell Biol 20:368–383 doi: 10.1038/s41580-019-0118-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung Y, Kim HD, Yang HW, Kim HJ, Jang CY, Kim J (2017) Modulating cellular balance of Rps3 mono-ubiquitination by both Hel2 E3 ligase and Ubp3 deubiquitinase regulates protein quality control Exp Mol Med 49:e390 doi: 10.1038/emm.2017.128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juszkiewicz S, Chandrasekaran V, Lin Z, Kraatz S, Ramakrishnan V, Hegde RS (2018) ZNF598 Is a Quality Control Sensor of Collided Ribosomes Mol Cell 72:469–481 e467 doi: 10.1016/j.molcel.2018.08.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juszkiewicz S, Hegde RS (2017) Initiation of Quality Control during Poly(A) Translation Requires Site-Specific Ribosome Ubiquitination Mol Cell 65:743–750 e744 doi: 10.1016/j.molcel.2016.11.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juszkiewicz S, Slodkowicz G, Lin Z, Freire-Pritchett P, Peak-Chew SY, Hegde RS (2020a) Ribosome collisions trigger cis-acting feedback inhibition of translation initiation Elife 9 doi: 10.7554/eLife.60038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juszkiewicz S, Speldewinde SH, Wan L, Svejstrup JQ, Hegde RS (2020b) The ASC-1 Complex Disassembles Collided Ribosomes Mol Cell doi: 10.1016/j.molcel.2020.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostova KK, Hickey KL, Osuna BA, Hussmann JA, Frost A, Weinberg DE, Weissman JS (2017) CAT-tailing as a fail-safe mechanism for efficient degradation of stalled nascent polypeptides Science 357:414–417 doi: 10.1126/science.aam7787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letzring DP, Wolf AS, Brule CE, Grayhack EJ (2013) Translation of CGA codon repeats in yeast involves quality control components and ribosomal protein L1 RNA 19:1208–1217 doi: 10.1261/rna.039446.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marton MJ, Vazquez de Aldana CR, Qiu H, Chakraburtty K, Hinnebusch AG (1997) Evidence that GCN1 and GCN20, translational regulators of GCN4, function on elongating ribosomes in activation of eIF2alpha kinase GCN2 Mol Cell Biol 17:4474–4489 doi: 10.1128/mcb.17.8.4474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo Y et al. (2017) Ubiquitination of stalled ribosome triggers ribosome-associated quality control Nat Commun 8:159 doi: 10.1038/s41467-017-00188-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo Y et al. (2020) RQT complex dissociates ribosomes collided on endogenous RQC substrate SDD1 Nat Struct Mol Biol doi: 10.1038/s41594-020-0393-9 [DOI] [PubMed] [Google Scholar]
- Meydan S, Guydosh NR (2020) Disome and Trisome Profiling Reveal Genome-wide Targets of Ribosome Quality Control Mol Cell doi: 10.1016/j.molcel.2020.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer C, Garzia A, Morozov P, Molina H, Tuschl T (2020) The G3BP1-Family-USP10 Deubiquitinase Complex Rescues Ubiquitinated 40S Subunits of Ribosomes Stalled in Translation from Lysosomal Degradation Mol Cell 77:1193–1205 e1195 doi: 10.1016/j.molcel.2019.12.024 [DOI] [PubMed] [Google Scholar]
- Morita M et al. (2012) A novel 4EHP-GIGYF2 translational repressor complex is essential for mammalian development Mol Cell Biol 32:3585–3593 doi: 10.1128/MCB.00455-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navickas A, Chamois S, Saint-Fort R, Henri J, Torchet C, Benard L (2020) No-Go Decay mRNA cleavage in the ribosome exit tunnel produces 5’-OH ends phosphorylated by Trl1 Nat Commun 11:122 doi: 10.1038/s41467-019-13991-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedialkova DD, Leidel SA (2015) Optimization of Codon Translation Rates via tRNA Modifications Maintains Proteome Integrity Cell 161:1606–1618 doi: 10.1016/j.cell.2015.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson AN, Dinman JD (2020) Two Ribosomes Are Better Than One...Sometimes Mol Cell 79:541–543 doi: 10.1016/j.molcel.2020.07.022 [DOI] [PubMed] [Google Scholar]
- Osuna BA, Howard CJ, Kc S, Frost A, Weinberg DE (2017) In vitro analysis of RQC activities provides insights into the mechanism and function of CAT tailing Elife 6 doi: 10.7554/eLife.27949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H, Subramaniam AR (2019) Inverted translational control of eukaryotic gene expression by ribosome collisions PLoS Biol 17:e3000396 doi: 10.1371/journal.pbio.3000396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah S, Molinaro G, Liu B, Wang R, Huber KM, Richter JD (2020) FMRP Control of Ribosome Translocation Promotes Chromatin Modifications and Alternative Splicing of Neuronal Genes Linked to Autism Cell Rep 30:4459–4472 e4456 doi: 10.1016/j.celrep.2020.02.076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simms CL, Hudson BH, Mosior JW, Rangwala AS, Zaher HS (2014) An active role for the ribosome in determining the fate of oxidized mRNA Cell Rep 9:1256–1264 doi: 10.1016/j.celrep.2014.10.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simms CL, Kim KQ, Yan LL, Qiu J, Zaher HS (2018) Interactions between the mRNA and Rps3/uS3 at the entry tunnel of the ribosomal small subunit are important for no-go decay PLoS Genet 14:e1007818 doi: 10.1371/journal.pgen.1007818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simms CL, Yan LL, Qiu JK, Zaher HS (2019) Ribosome Collisions Result in +1 Frameshifting in the Absence of No-Go Decay Cell Rep 28:1679–1689 e1674 doi: 10.1016/j.celrep.2019.07.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simms CL, Yan LL, Zaher HS (2017) Ribosome Collision Is Critical for Quality Control during No-Go Decay Mol Cell 68:361–373 e365 doi: 10.1016/j.molcel.2017.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha NK et al. (2020) EDF1 coordinates cellular responses to ribosome collisions Elife 9 doi: 10.7554/eLife.58828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitron CS, Brandman O (2019) CAT tails drive degradation of stalled polypeptides on and off the ribosome Nat Struct Mol Biol 26:450–459 doi: 10.1038/s41594-019-0230-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AM, Costello MS, Kettring AH, Wingo RJ, Moore SD (2019) Ribosome collisions alter frameshifting at translational reprogramming motifs in bacterial mRNAs Proc Natl Acad Sci U S A 116:21769–21779 doi: 10.1073/pnas.1910613116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suardi GAM, Haddad LA (2020) FMRP ribonucleoprotein complexes and RNA homeostasis Adv Genet 105:95–136 doi: 10.1016/bs.adgen.2020.01.001 [DOI] [PubMed] [Google Scholar]
- Sundaramoorthy E, Leonard M, Mak R, Liao J, Fulzele A, Bennett EJ (2017) ZNF598 and RACK1 Regulate Mammalian Ribosome-Associated Quality Control Function by Mediating Regulatory 40S Ribosomal Ubiquitylation Mol Cell 65:751–760 e754 doi: 10.1016/j.molcel.2016.12.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollenaere MAX et al. (2019) GIGYF1/2-Driven Cooperation between ZNF598 and TTP in Posttranscriptional Regulation of Inflammatory Signaling Cell Rep 26:3511–3521 e3514 doi: 10.1016/j.celrep.2019.03.006 [DOI] [PubMed] [Google Scholar]
- Vind AC et al. (2020) ZAKalpha Recognizes Stalled Ribosomes through Partially Redundant Sensor Domains Mol Cell 78:700–713 e707 doi: 10.1016/j.molcel.2020.03.021 [DOI] [PubMed] [Google Scholar]
- Visweswaraiah J, Lee SJ, Hinnebusch AG, Sattlegger E (2012) Overexpression of eukaryotic translation elongation factor 3 impairs Gcn2 protein activation J Biol Chem 287:37757–37768 doi: 10.1074/jbc.M112.368266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zhou J, Yang Q, Grayhack EJ (2018) Multi-protein bridging factor 1(Mbf1), Rps3 and Asc1 prevent stalled ribosomes from frameshifting Elife 7 doi: 10.7554/eLife.39637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CC, Peterson A, Zinshteyn B, Regot S, Green R (2020) Ribosome Collisions Trigger General Stress Responses to Regulate Cell Fate Cell 182:404–416 e414 doi: 10.1016/j.cell.2020.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonashiro R et al. (2016) The Rqc2/Tae2 subunit of the ribosome-associated quality control (RQC) complex marks ribosome-stalled nascent polypeptide chains for aggregation Elife 5:e11794 doi: 10.7554/eLife.11794 [DOI] [PMC free article] [PubMed] [Google Scholar]