Genome-wide polygenic scores (GPS) integrate information from millions of sites of common DNA variation into a single metric of inherited susceptibility. When applied to coronary artery disease (CAD), we and others have noted three key observations: (i) GPSCAD identifies a subgroup of the population at substantially increased risk not reliably identified using traditional risk factors; (ii) GPSCAD can be quantified from birth, yet demonstrates similar or better discrimination power as clinical risk factors ascertained in middle age, and (iii) individuals with high GPSCAD derive the greatest risk reduction from lipid-lowering therapy.1
Despite growing evidence for integration of polygenic scores into routine clinical practice, the Eurocentric bias in genome-wide association studies – needed as input to polygenic score derivation – results in reduced performance of most polygenic scores in non-European ancestries.
To further quantify the transethnic portability of GPSCAD, we assembled a meta-analysis of 30,365 CAD cases and 482,063 controls across five ancestral groups derived from six studies. First, the UK Biobank, a cohort study of middle-aged individuals that included 22,250 cases and 441,490 controls. Second, two hospital-based biobanks in the United States, the Partners Biobank with 2,772 cases and 15,124 controls, and the BioMe biobank with 2,824 cases and 21,547 controls.2 Third, two ancestry-specific case-control studies, the Bangladesh Risk of Acute Vascular Events (BRAVE) study of early-onset myocardial infarction with 247 cases and 244 controls, and the TaiChi consortium in Taiwan with 288 cases and 457 controls.3,4 Fourth, 1,984 cases with early-onset myocardial infarction from the hospital-based VIRGO study and 3,201 controls free of CAD from the MESA prospective cohort study.5 CAD was defined as myocardial infarction and/or history of coronary revascularization in all studies, with some variability across studies based on study design or available data as detailed previously.1–5
Participants within each study provided informed consent to study investigators, with the meta-analysis approved by the Mass General Brigham institutional review board. UK Biobank data is made available to qualified researchers via application (http://www.ukbiobank.ac.uk/register-apply). Data for the VIRGO, MESA, TaiChi, and BRAVE studies have been uploaded to the database of Genotypes and Phenotypes (dbGaP) under accession numbers phs001259, phs001416, phs001487, and phs001398. Data from BioMe and Partners Biobank may be available via outreach to study investigators.
To quantify the relationship of GPSCAD with disease, we first computed the GPSCAD in each study participant based on either genotyping array and imputation (UK Biobank, Partners Biobank, and BioMe) or whole-genome sequencing (TaiChi, BRAVE, VIRGO/MESA). Sample and variant quality control was performed separately in each study as previously described.1,2,5 Ancestral subgroups were based on concordance between self-report and genetically-defined ancestry, excluding individuals with discordant self-report and genetically-determined ancestry.2,5 GPSCAD was calculated using the raw weights for ~6.6 million variants (available at: www.broadcvdi.org).1 We calculated the association of GPSCAD with CAD using a logistic regression model adjusted for the first four principal components of ancestry. We then performed a meta-analysis of the odds ratio per standard deviation increment of GPSCAD within ancestral groupings for each study (Figure).
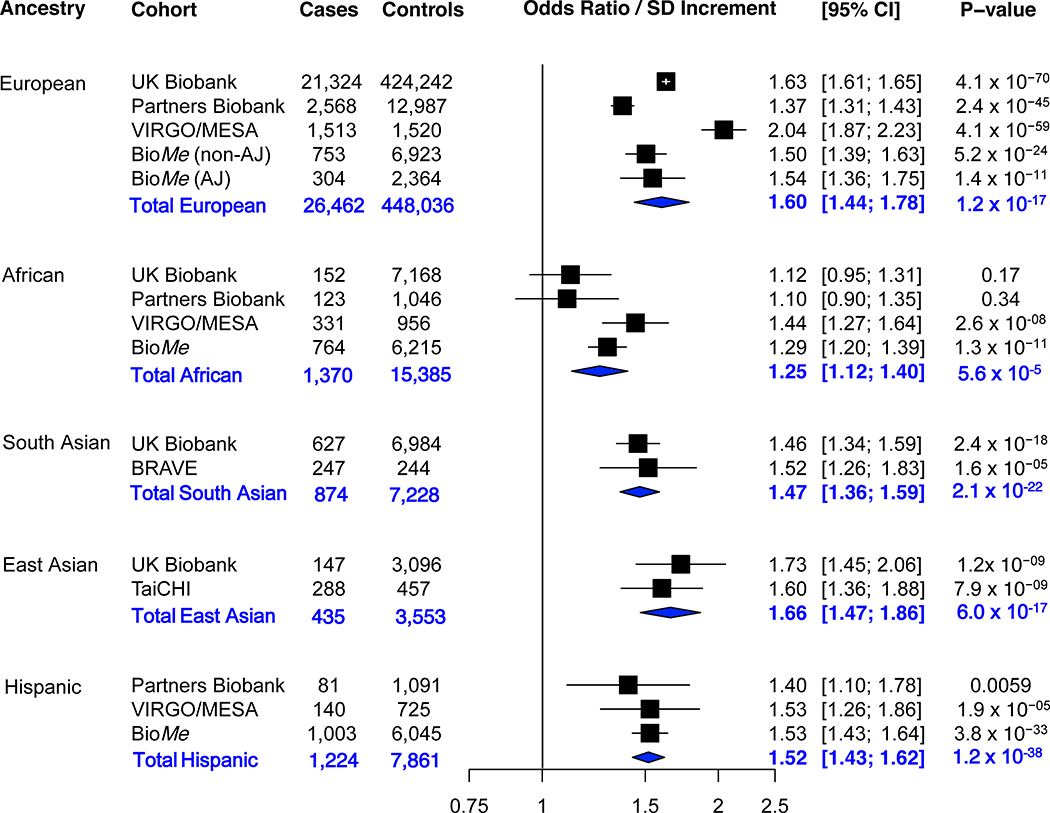
Figure.
Association of a genome-wide polygenic score with coronary artery disease in five ancestry groups. In each cohort, the polygenic score is residualized to the first four principal components of ancestry (to adjust for the effect of ancestry on the score). In each ancestry of each study, the odds ratio per standard deviation is calculated using a logistic regression model adjusted for the first four principal components of ancestry (to adjust for the effect of ancestry on disease). Sensitivity analyses adjusting for sex and the first four principal components of ancestry, or age, sex, and the first four principal components of ancestry demonstrated similar results. A random-effects meta-analysis of odds ratio per standard deviation increment was performed. The p-value for subgroup differences was 0.005 (Cochran-Mantel-Haenszel test). In pairwise ancestry group comparisons, the effect estimate in African ancestry group was significantly lower than each of the other ancestry groups, European (p=0.002), South Asian (p=0.02), East Asian (0.001), and Hispanic (0.003). None of the other ancestry group pairwise comparisons were statistically significant (p>0.05). The random effects model results for each ancestry group showed an I2 of 95.4%, 63.7%, 0%, 0%, and 0% for European, African, South Asian, East Asian, and Hispanic, respectively. Abbreviations: AJ Ashkenazi Jewish, CI confidence interval, SD standard deviation.
GPSCAD was robustly associated with CAD within each ancestral group, but significant variability in effect size was noted (p-value=0.005). Consistent with prior reports, we observe an odds ratio per standard deviation (ORSD) of 1.60 (1.44–1.78) in individuals of European ancestry. Similar effect sizes were noted in East Asian (ORSD = 1.66 [1.47–1.86]), South Asian (ORSD =1.47 [1.36–1.59]), and Hispanic (ORSD =1.52 [1.43–1.62]) ancestries. Among individuals of African ancestry, the GPSCAD remained associated, but effect size was significantly reduced (ORSD =1.25 [1.12–1.40]).
The transethnic variability – most notable for a decreased effect size in individuals of African ancestry – is likely attributed to four factors: (i) use of non- causal variants that – due to variable linkage disequilibrium patterns – may not correlate with the causal signal across ancestries; (ii) allele frequency differences or population-specific risk variants that have not been optimally modeled; (iii) varying genetic architectures, as can occur with gene-gene or gene-environment interactions; (iv) different heritabilities, as can occur with population-specific environmental exposures. More diverse genome-wide association studies remain a top priority for the scientific community. In parallel, ongoing efforts seek to improve performance of polygenic scores in individuals of African ancestry with existing data, such as improving fine mapping to identify causal variants, optimizing methods that combine data from different ancestries, and including other sources of information (risk factors or other ‘omics data).
This study should be interpreted in the context of potential limitations. Within each ancestral group, studies were somewhat variable in their design and does not consider non-genetic risk factors. Additionally, we quantify relative risk associated with GPSCAD using odds ratios, but epidemiological differences in CAD risk across ancestral groups influence estimates of absolute risk. For example, despite a lower relative risk related to a polygenic score, a greater number of individuals of Africa ancestry may benefit from reclassification in the context of higher absolute event rates.
Supplementary Material
Acknowledgments:
This research has been conducted using the UK Biobank resource under application number 7089.
Sources of Funding: Dr. Fahed is supported by T32HL007208 from the National Heart, Lung, and Blood Institute. Dr. Do is supported by R35GM124836 from the National Institute of General Medical Sciences and R01HL139865 from the National Heart, Lung, and Blood Institute. Dr. Ellinor is supported by grant 14CVD01 Fondation Leducq, grants 1RO1HL092577, R01HL128914, K24HL105780 from the National Heart, Lung, and Blood Institute, and grant 18SFRN34110082 from the American Heart Association. Dr. Khera reports funding support from an institutional grant from the Broad Institute of MIT and Harvard (BroadIgnite), grant 1K08HG010155 and 5UM1HG008895 from the National Human Genome Research Institute, a Hassenfeld Scholar Award from Massachusetts General Hospital, a Merkin Institute Fellowship from the Broad Institute of MIT and Harvard, and a sponsored research agreement from IBM Research. Molecular data for the Trans-Omics in Precision Medicine (TOPMed) program was supported by the National Heart, Lung and Blood Institute (NHLBI). General study coordination was provided by the TOPMed Data Coordinating Center (3R01HL-12393-02S1). MESA and the MESA SHARe project are conducted and supported by the National Heart, Lung, and Blood Institute (NHLBI) in collaboration with MESA investigators. Support for MESA is provided by contracts 75N92020D00001, HHSN268201500003I, N01-HC-95159, 75N92020D00005, N01-HC-95160, 75N92020D00002, N01-HC-95161, 75N92020D00003, N01-HC-95162, 75N92020D00006, N01-HC-95163, 75N92020D00004, N01-HC-95164, 75N92020D00007, N01-HC-95165, N01-HC-95166, N01-HC-95167, N01-HC-95168, N01-HC-95169, UL1-TR-000040, UL1-TR-001079, UL1-TR-001420; also supported in part by the National Center for Advancing Translational Sciences, CTSI grant UL1TR001881, and the National Institute of Diabetes and Digestive and Kidney Disease Diabetes Research Center (DRC) grant DK063491 to the Southern California Diabetes Endocrinology Research Center. The VIRGO study was supported by grant R01 HL081153-01A1K from the National Heart, Lung, and Blood Institute. Whole-genome sequencing of the VIRGO cohort was funded by grant 5UM1HG008895-02 from the National Human Genome Research Institute’s Center for Common Disease Genomics.
Disclosures: Dr. Fahed is a consultant and owns shares in Goodpath. Dr. Krumholz works under contract with the Centers for Medicare & Medicaid Services to support quality measurement programs; was a recipient of a research grant, through Yale, from Medtronic and the U.S. Food and Drug Administration to develop methods for post-market surveillance of medical devices; was a recipient of a research grant from Johnson & Johnson, through Yale University, to support clinical trial data sharing; was a recipient of a research agreement, through Yale University, from the Shenzhen Center for Health Information for work to advance intelligent disease prevention and health promotion; collaborates with the National Center for Cardiovascular Diseases in Beijing; receives payment from the Arnold & Porter Law Firm for work related to the Sanofi clopidogrel litigation, from the Martin Baughman Law Firm for work related to the Cook Celect IVC filter litigation, and from the Siegfried and Jensen Law Firm for work related to Vioxx litigation; chairs a Cardiac Scientific Advisory Board for UnitedHealth; was a member of the IBM Watson Health Life Sciences Board; is a member of the Advisory Board for Element Science, the Healthcare Advisory Board for Facebook, and the Physician Advisory Board for Aetna; and is the co-founder of HugoHealth, a personal health information platform, and co-founder of Refactor Health, an enterprise healthcare AI-augmented data management company. He is a Venture Partner at F-Prime.Dr. Do has received research support from AstraZeneca and Goldfinch Bio. Dr. Ellinor is supported by a grant from Bayer AG to the Broad Institute focused on the genetics and therapeutics of cardiovascular diseases, and has served on advisory boards or consulted for Bayer AG, Quest Diagnostics, MyoKardia and Novartis. Dr. Kathiresan is an employee of Verve Therapeutics, and holds equity in Verve Therapeutics, Maze Therapeutics, Catabasis, and San Therapeutics. He is a member of the scientific advisory boards for Regeneron Genetics Center and Corvidia Therapeutics; he has served as a consultant for Acceleron, Eli Lilly, Novartis, Merck, Novo Nordisk, Novo Ventures, Ionis, Alnylam, Aegerion, Haug Partners, Noble Insights, Leerink Partners, Bayer Healthcare, Illumina, Color Genomics, MedGenome, Quest, and Medscape; he reports patents related to a method of identifying and treating a person having a predisposition to or afflicted with cardiometabolic disease (20180010185) and a genetics risk predictor (20190017119). Dr. Khera has served as a consultant to Sanofi, Medicines Company, Maze Pharmaceuticals, Navitor Pharmaceuticals, Verve Therapeutics, Amgen, and Color Genomics; received speaking fees from Illumina, the Novartis Institute for Biomedical Research; received sponsored research agreements from the Novartis Institute for Biomedical Research, and reports a patent related to a genetic risk predictor (20190017119).
Nonstandard Abbreviations and Acronyms
- GPSCAD
Genome-wide polygenic score for coronary artery disease
- CAD
Coronary artery disease
- ORSD
Odds ratio per standard deviation increment
- BRAVE
Bangladesh Risk of Acute Vascular Events study
- MESA
Multi-Ethnic Study of Atherosclerosis
- VIRGO
Variation In Recovery: role of Gender on Outcomes of young acute myocardial infarction patients study
- BioMe
BioMe biobank of Icahn School of Medicine at Mount Sinai
- SNP
Single nucleotide polymorphism
- AJ
Ashkenazi Jewish
References:
- 1.Khera AV, Chaffin M, Aragam KG, Haas ME, Roselli C, Choi SH, Natarajan P, Lander ES, Lubitz SA, Ellinor PT, et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet. 2018;50:1219–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aragam KG, Dobbyn A, Judy R, Chaffin M, Chaudhary K, Hindy G, Cagan A, Finneran P, Weng L-C, Loos RJF, et al. Limitations of Contemporary Guidelines for Managing Patients at High Genetic Risk of Coronary Artery Disease. J Am Coll Cardiol. 2020;75:2769–2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chowdhury R, Alam DS, Fakir II, Adnan SD, Naheed A, Tasmin I, Monower MM, Hossain F, Hossain FM, Rahman MM, et al. The Bangladesh Risk of Acute Vascular Events (BRAVE) Study: objectives and design. Eur J Epidemiol. 2015;30:577–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Assimes TL, Lee I-T, Juang J-M, Guo X, Wang T-D, Kim ET, Lee W-J, Absher D, Chiu Y-F, Hsu C-C, et al. Genetics of Coronary Artery Disease in Taiwan: A Cardiometabochip Study by the Taichi Consortium. PLoS ONE. 2016;11:e0138014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khera AV, Chaffin M, Zekavat SM, Collins RL, Roselli C, Natarajan P, Lichtman JH, D’Onofrio G, Mattera J, Dreyer R, et al. Whole-Genome Sequencing to Characterize Monogenic and Polygenic Contributions in Patients Hospitalized With Early-Onset Myocardial Infarction. Circulation. 2019;139:1593–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.