Abstract
The Escherichia coli ATP-consuming chaperonin machinery, a complex between GroEL and GroES, has evolved to facilitate folding of substrate proteins (SPs) that cannot do so spontaneously. A series of kinetic experiments show that the SPs are encapsulated in the GroEL/ES nanocage for a short duration. If confinement of the SPs is the mechanism by which GroEL/ES facilitates folding, it follows that the assisted folding rate, relative to the bulk value, should always be enhanced. Here, we show that this is not the case for the folding of rhodanese in the presence of the full machinery of GroEL/ES and ATP. The assisted folding rate of rhodanese decreases. On the basis of our finding and those reported in other studies, we suggest that the ATP-consuming chaperonin machinery has evolved to optimize the product of the folding rate and the yield of the folded SPs on the biological time scale. Neither the rate nor the yield is separately maximized.
Graphical Abstract
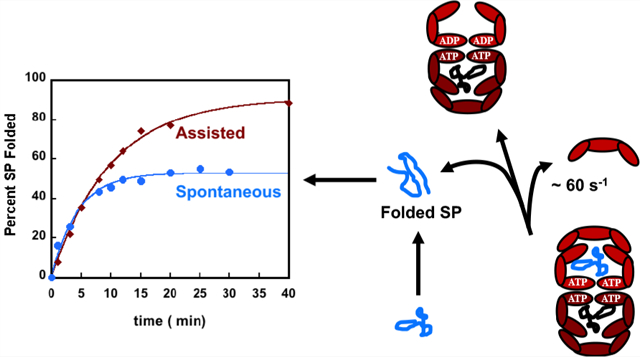
The bacterial chaperonin GroEL, a stochastic nanomachine powered by ATP binding and hydrolysis,1,2 assists the folding of substrate proteins (SPs) that are otherwise destined for aggregation.3,4 GroEL consists of two heptameric rings that are stacked in a back-to-back arrangement.5,6 Each ring has seven identical subunits, which are symmetrically arranged in the resting state (absence of ligands ATP or SP), termed the T state.6–8 Like other motors,9,10 GroEL undergoes a series of increasingly large scale conformational (allosteric) transitions upon binding of ATP, SP, and the co-chaperonin GroES.6–8 The roughly cylinder-shaped GroEL has a cavity.5 The volume of the cavity nearly doubles when both ATP and GroES are bound (termed the RI or RII state).7,11 The cavity volume increases roughly from 85000 Å3 in the T state to ~170000 Å3 in the RI or RII state.11 In the RII nucleotide state, the cavity can fully encapsulate a protein with a maximum of ~500 amino acid residues.12
How GroEL facilitates the folding of a large number of SPs that are unrelated by sequence or topology of the native state, as the machine executes a complex but well-defined catalytic cycle, has remained a topic of great interest for nearly 30 years. Three scenarios for the mechanism of GroEL/ES-assisted folding have been suggested. (i) The structures of GroEL and the complex between GroEL and GroES in the ADP-hydrolyzed state show the presence of a large cavity. From this observation, it is tempting to conclude that the cavity provides the encapsulated SP a protective passive chamber in which to fold, thus avoiding aberrant interactions with other non-native SPs.13–15 In such an Anfinsen cage, GroEL/ES functions in a passive manner. Although theoretically the assisted folding rate when the SP is in the cage for arbitrarily long times should increase relative to the bulk value, it is asserted that the rate is unchanged. (ii) In the active Anfinsen cage model,16,17 it is envisioned that efficient folding in the cage occurs because kinetic traps are “entropically” disrupted, and hence, the folding trajectories have unimpeded access to the folded state. There are two immediate consequences of the active cage model. (1) Confinement must enhance the folding rate relative to folding in the bulk, sometimes by a factor of ≥50.16,17 In contrast, theoretical arguments18,19 and simulations have shown that the maximum acceleration in the folding rate compared to spontaneous folding cannot exceed a factor of ~10. (2) The SP stability must also increase in the GroEL/ES cage. Because confinement of the SP in the cage decreases the entropy of the unfolded state, it follows that the native state stability increases. This argument has theoretical support20–22 and has been invoked to rationalize a number of experiments.23–26 More importantly, rate enhancement has been observed for certain proteins but is not universal as the active cage model asserts.16,17,27–29 We note parenthetically that a major weakness of the cage models is that they pay scant attention to the GroEL/ES catalytic cycle that occurs even in the absence of the SP, albeit at a much slower rate. (iii) The iterative annealing mechanism (IAM), the only theory that accounts for the coupling between the allosteric states visited by GroEL/ES during the reaction cycle and SP folding, predicts that it is the product of the folding rate and the yield of the native material (folded SP) that are maximized by repeated binding and release of the SP by GroEL/ES.1,12,30,31 Of relevance here is the implication that folding rates per se could be accelerated (modestly) or even retarded relative to the bulk. The IAM quantitatively explains the results of a large number of experiments,12 including observations that mutations in GroEL render it less efficient than the wild type.12 Recent experiments32,33 have also established that in the presence of SP the chaperonin machinery responds rapidly by processing folding in both chambers (GroEL/ES is a parallel processing machine), a discovery that is quite consistent with the IAM predictions. A consequence of the IAM is that the GroEL/ES machinery optimizes the product of the assisted folding rate (kF) and the yield of the native material on a biologically relevant time scale by driving the SP out of equilibrium.34 Neither kF nor the yield is separately maximized. We note parenthetically that the same optimization principle holds for RNA chaperones.
Here, we focus on the effect of GroES/EL-mediated folding rates (kF) of rhodanese, which has been extensively studied previously.35,36 In an important single-molecule experiment, Hofmann et al.29 showed that the kF of an encapsulated protein rhodanese (which is also the substrate of choice in this study) in a single-ring mutant (SR1) of GroEL decreases relative to the bulk due to potential interactions with the wall, as suggested using computations.18 Because SR1 is an artificial construct in which GroES does not dissociate from SR1 for ~300 min,12 it is unclear if a decrease in the folding rate is also observed in the full wild-type cycling system. In the full chaperonin machinery, the residence time of the SP in the expanded internal cage of GroEL is just a few seconds, and not 300 min.12 Nevertheless, we find using the cycling system consisting of GroES, GroEL, and ATP that the folding rate of rhodanese is retarded relative to its value in the bulk. Surprisingly, the extent of retardation is similar to that found in the SR1 mutant.
RESULTS AND DISCUSSION
To investigate the role of the wild-type GroEL/ES machine in the rhodanese folding pathway, we compared the kinetics of spontaneous and assisted folding. Under our experimental conditions, we were concerned that the spontaneous folding rate (kF) of rhodanese released from the GroES−GroEL cage before folding is complete could contribute to the observed rate of GroEL−GroES-assisted rhodanese folding. Thus, the GroEL−GroES-assisted reaction was initially performed with SR1 and ADP·AlFx, a transition state analogue of ATP. Previous experimental data have shown that the GroES-SR1-ADP·AlFx complex assisted in the folding of a number of substrates and this complex is stable and long-lived.37 In the SR1 mutant, the SPs are trapped inside the GroES-SR1-ADP·AlFx cage without the possibility of escaping from the cage. In this case, it has been argued that folding occurs as it would in the absence of chaperones.37 In other words, the SPs are forced to fold in the expanded GroEL cavity.
Figure 1A shows the dependence of the extent of rhodanese folding versus time in the presence of SR1 and ATP, GroES-SR1-ADP·AlFx, and spontaneous folding. It is clear that, under these conditions, rhodanese is a stringent substrate, which means that there is a higher probability that this SP would fold with the assistance of the chaperonin machinery than it would otherwise. Consequently, SR1 and ATP alone are not sufficient for rhodanese folding, and the intact machinery is required to rescue the SP. Moreover, the data for folding of rhodanese in the presence of ATP and SR1 demonstrate that spontaneous folding of rhodanese does not occur before the addition of GroES to the reaction mixture. SR1 captures the unfolded rhodanese and prevents folding in solution. In other words, the pseudo-first-order rate for SP capture is greater than the spontaneous folding rate, which is a generic kinetic requirement for all SPs. Thus, the rhodanese folded in the presence of SR1, GroES, and ADP·AlFx is folded completely inside the GroES-SR1-ADP·AlFx cage.
Figure 1.
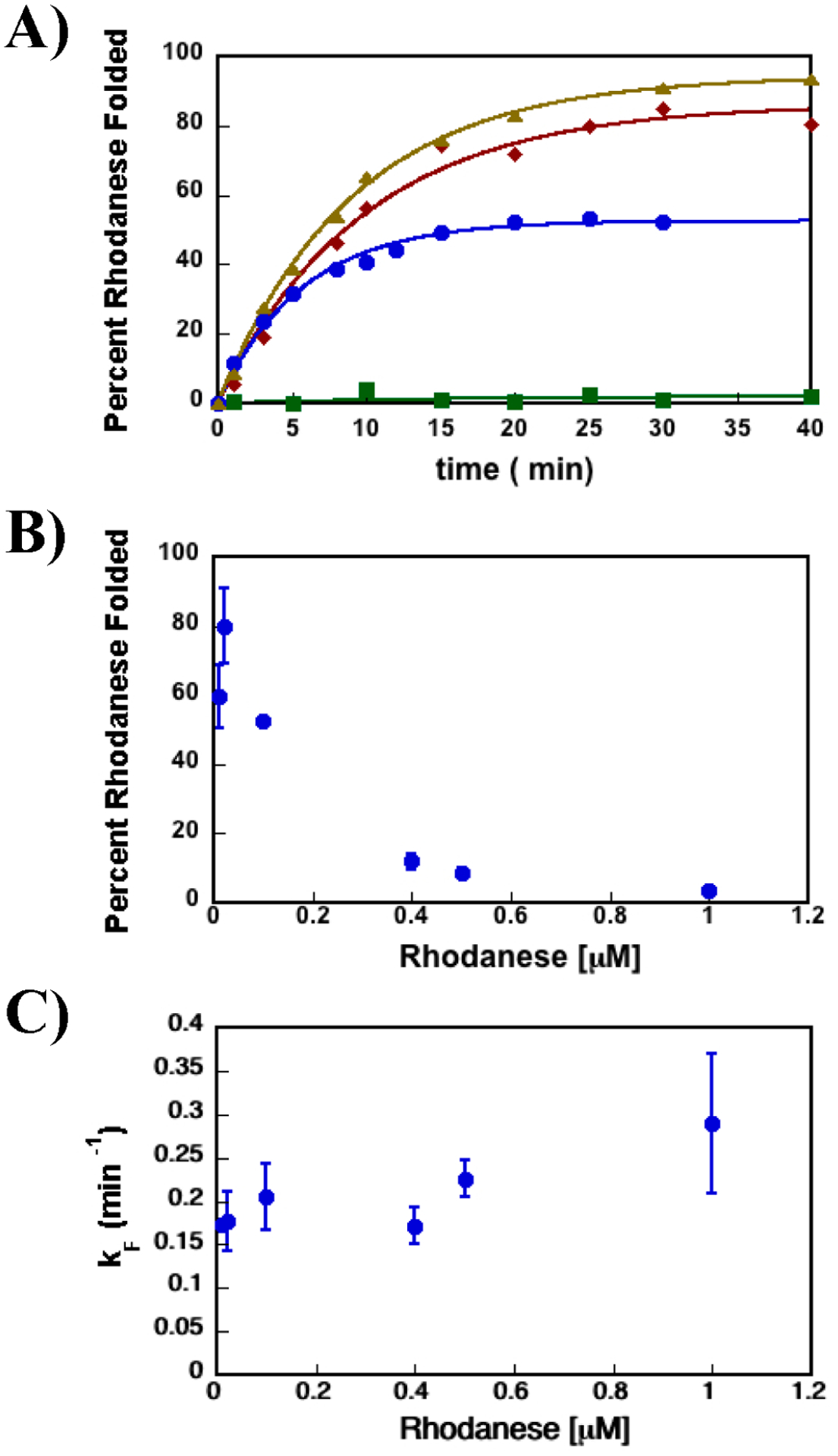
SR1-GroES cage that increases the yield of the folded SP while decreasing the SP’s rate of folding. (A) Rhodanese enzymatic activity was used to monitor the extent of rhodanese folded vs time. The data shown here are representative of the folding experiments. The rate constants’ average values and standard deviations for the kinetic folding experiments are listed in Table 1. The data for rhodanese folded in the presence of SR1-ADP·AlFx with GroES are shown as brown triangles, the data for rhodanese folded in the presence of SR1 D398A-GroES-ATP with GroES as red diamonds, and the data for rhodanese folded in the presence of SR1 and ATP with no GroES as green squares. Data for spontaneous folding of rhodanese are shown as blue circles. (B) Percentage of the rhodanese protein folded spontaneously vs rhodanese concentration. The extent of rhodanese folded spontaneously decreases as the concentration of rhodanese increases, consistent with earlier works showing rhodanese protein is prone to aggregation.13,38 (C) Dependence of spontaneous folding rate constant vs rhodanese concentration. The rate of rhodanese spontaneous folding is independent of its concentration. It should be emphasized that the pseudo-first-order rate of conversion of the aggregated product to native rhodanese is not significant. More importantly, this process is expected to have no effect on the folding rate of formation of native rhodanese from the unfolded protein. Previous work has shown that rhodanese aggregates are dead-end folding products, which do not convert to the native rhodanese over time.39
More importantly, Figure 1A also shows that the yield of the folded state in the secluded SR1-GroES-ADP·AlFx cage is larger than the fraction of spontaneously folded rhodanese. This is because the SR1-GroES-ADP·AlFx cage sequesters rhodanese for times that far exceed kF, thus preventing aggregation. In the bulk folding reaction, unfolded and misfolded rhodanese molecules are free to interact with each other and form higher-order aggregates. In addition, evidence that spontaneously folded rhodanese has a propensity to aggregate comes from the dependence of the fraction of rhodanese folded spontaneously as a function of rhodanese concentration. As the concentration of rhodanese increases, the fraction of folded rhodanese decreases because of a competing aggregation reaction that dominates as the concentration of the protein in solution is increased (Figure 1B). Of particular importance here is the finding that the observed rate of folding of rhodanese inside the SR1-GroES-ADP·AlFx cage is 2-fold lower than the spontaneous rhodanese folding rate29 (Table 1).
Table 1.
Kinetic Parameters for Spontaneous and GroEL-Assisted Rhodanese Protein Folding
| kFa (min−1) | |
|---|---|
| spontaneous | 0.21 ± 0.04 |
| SR1-ADP·AlFx | 0.09 ± 0.04 |
| SR1 D398A | 0.1 ± 0.01 |
| wild-type GroEL | 0.09 ± 0.015 |
Rhodanese folding rates. The values represent the average folding rate constants from at least two independent data sets, and the errors are the standard deviations from these averages.
The decreased rate of folding inside the GroES-SR1-ADP·AlFx cage could result from interactions of the rhodanese protein with the SR1-GroES cage formed in the presence of ADP·AlFx, and not as a consequence of rhodanese−GroEL interactions. To explore this possibility, we took advantage of the SR1 D398A construct, which forms a stable folding active chamber37 when bound to GroES. The rate of rhodanese folded inside the SR1 D398A-GroES-ATP chamber, as measured by the rhodanese enzyme assay, is identical within experimental error to the rate of rhodanese folded inside the GroES-SR1-ADP·AlFx cage and twice as slow as the rate of spontaneously folded rhodanese (Figure 1A and Table 1). Hence, the secluded SR1-GroES chamber slows the folding of the rhodanese protein, as reported previously.29 Rhodanese is a monomeric protein, and the rhodanese aggregates are dead-end folding products that do not convert to native rhodanese.39 Therefore, the folding rate should be independent of the protein concentration, and our experimental results completely agree with that conclusion (Figure 1C).
The slower rate of folding inside the stable GroES-SR1 cage could be a consequence of the inability of the SR1 construct to progress through the allosteric states of the GroEL ATP-driven reaction cycle. To rule out this possibility, we investigated rhodanese folding using the full wild-type machinery, GroELGroES-ATP. Figure 2 shows the GroEL-GoES-ATP-assisted rhodanese folding versus the reaction time. The observed rate of GroES-GroEL-ATP-assisted rhodanese folding is very similar to the observed rates inside the SR1-GroES SR1-ADP·AlFx and SR1 D398A-GroES-ATP complexes (Table 1). Thus, the full GroEL/ES chaperonin system, while protecting the SP from aggregation, also decreases the rhodanese substrate overall rate of folding.
Figure 2.
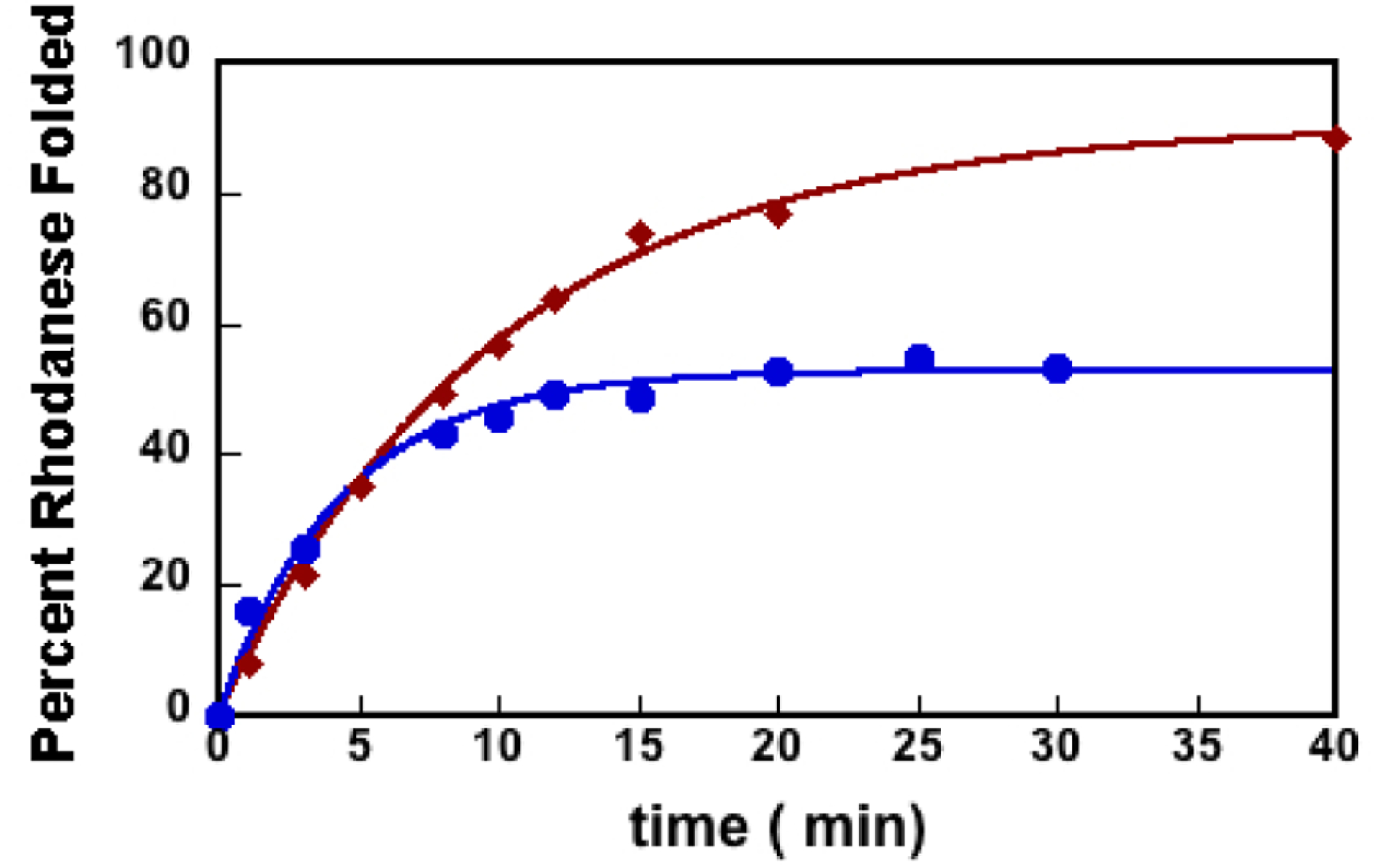
GroEL-GroES-ATP complex increases the SP folding yield while decreasing its folding rate. The extent of GroEL-ES-ATP-assisted rhodanese folding vs reaction time is shown as red diamonds. The extent of spontaneous rhodanese folding vs reaction time is shown as blue circles. These data are representative of kinetic folding experiments as measured by the rhodanese enzymatic assay. The average rate constants and standard deviations for the representative data depicted here are listed in Table 1.
Taken together, the results of the experiments show that GroEL/ES does decrease the folding rate of the SP, contradicting the often-stated assertion that an active chaperonin always increases the rate of SP folding. Although folding in a cavity could accelerate the rate of folding of SPs, it is neither necessary for GroEL function nor valid universally.
The molecular origin in the decrease in the folding rate of the SP is hard to quantify precisely. It should be noted that the observed rate decrease, by a factor of slightly more than 2, is not large but is clearly outside the experimental errors (see Table 1). Our finding is consistent with previous experiments,29 which reported a factor of 2–8 decrease in the rate of rhodanese folding in the SR1 mutant, depending on the temperature. One explanation, favored in the earlier work,29 is that the favorable interaction of the SP with exposed residues29 in the interior wall of the expanded GroEL cavity18,19 could increase the folding barrier. This would also imply that certain SPs are thermodynamically destabilized in the GroEL/ES cage40 not by interactions between the SP and the walls of the GroEL. Destabilization could occur because of alterations due to solvent-mediated interactions between hydrophobic residues in the SP in a confined environment. Previous atomic detailed simulations have suggested this mechanism as a possibility.41 Regardless of the molecular mechanism, which is important to decipher, it is clear that the GroEL/ES machinery has not evolved to enhance the folding rates of proteins but to maximize the yield, YN, of the native material on biological time scales.12,13,29,42 More precisely, using theory with validation by experiments, Chakrabarty et al.34 established that it is the product, kFYN, that is maximized. The chaperonin machinery has not evolved to maximize kF or YN separately. In the example presented here, kF decreases, but this is compensated by an increase in YN. Optimization of kFYN is a general feature of protein and RNA chaperones. In the case of RNA chaperones, as well, YN decreases but is compensated by an increase in kF in such a way that kFYN is maximized. Such an optimization is possible if chaperones drive the substrates out of equilibrium because folding of the misfolded substrate protein due to equilibrium fluctuations is possible only on time scales that far exceed biologically relevant times. The potential biological advantage of maximizing kFYN could be rationalized using the following argument. The chaperonin machinery processes a number of SPs, which must occur on time scales that are shorter than the cell doubling time. If the kF is maximized at the expense of YN, then the functions of the processed SP may be compromised because of the paucity of folded proteins. Similarly, if accumulation of sufficient native SP takes a very long time, then various processes in the cell cycle, requiring exquisite timing, might be disrupted. The compromise is to maximize kFYN so that functionally competent proteins are produced on reasonable time scales, which should be considerably shorter than the cell doubling time. Moreover, it is unlikely that the GroEL/ES machine, which facilitates SPs that are unrelated by sequence or structure of the folded states, has evolved to maximize the folding rates of all of the SPs in the cavity.
MATERIALS AND METHODS
Protein Preparation.
Bos taurus (B. taurus) (bovine) rhodanese bearing a C-terminal six-His tag was purified under denaturing conditions, as previously described, and stored as a lyophilized powder.43 GroEL, GroES, SR1, and SR1 D398A were purified as native proteins.44
Rhodanese Folding.
Rhodanese was unfolded for 30 min at 24 °C in 8 M urea, 20 mM DTT, and 50 mM Tris (pH 7.5). Unfolded rhodanese was diluted to a concentration of 0.1 μM in folding buffer [10 mM DTT, 50 mM KCl, 10 mM MgCl2, 50 mM Tris (pH 7.5), and 50 mM Na2S2O3] with 0.2 μM GroEL. The formation of the rhodanese−GroEL binary complex was allowed to proceed for 5 min at 24 °C. Subsequently, GroES was added to the reaction mixture to a concentration of 0.4 μM. The folding reaction was initiated by the addition of ATP to a final concentration of 5 mM. For the SR1-ADP·AlFx-assisted folding reaction, the folding buffer also contained 5 mM ADP and 30 mM KF, and the folding was initiated by the addition of KAl(SO4)2 to a final concentration of 3 mM.37 The spontaneous rhodanese folding was initiated by diluting the unfolded rhodanese to a specific concentration in folding buffer. The extent of correctly folded SP was measured by monitoring the absorbance at 460 nm of the complex formed among thiocyanate, one of the rhodanese reaction products, and ferric ion.45
We used A(1 − e−kFt), where A is the amplitude, t is the reaction time, and kF is the folding rate constant, to fit the extent of folded SP versus t.
ACKNOWLEDGMENTS
The authors thank Ben Schuler and Hagen Hoffmann for useful comments. E.K. performed the experiments shown in this paper in Art Horwich’s laboratory at Yale University (New Haven, CT). She thanks Art Horwich and his group members for the helpful discussions.
Funding
This work was supported in part by a National Institutes of Health NRSA fellowship (F32GM079981 to E.K.) and National Institute of General Medical Sciences Grant R01-GM131062 to E.K. D.T. is grateful to the National Science Foundation (CHE 19-00093) and the Collie-Welch Chair (F-0019) administered through the Welch Foundation.
Footnotes
Accession Codes
B. taurus (bovine) rhodanese, UniProt entry P00586; Escherichia coli (E.coli) GroEL, UniProt entry P0A6F5; E. coli GroES, UniProt entry P0A6F9.
Complete contact information is available at: https://pubs.acs.org/10.1021/acs.biochem.0c00903
The authors declare no competing financial interest.
Contributor Information
Eda Koculi, Department of Biology, Johns Hopkins University, Baltimore, Maryland 21218, United States;.
D. Thirumalai, Department of Chemistry, The University of Texas at Austin, Austin, Texas 78712, United States;.
REFERENCES
- (1).Thirumalai D, and Lorimer GH (2001) Chaperonin-mediated protein folding. Annu. Rev. Biophys. Biomol. Struct 30, 245–269. [DOI] [PubMed] [Google Scholar]
- (2).Horwich AL, Farr GW, and Fenton WA (2006) GroELGroES-mediated protein folding. Chem. Rev 106, 1917–1930. [DOI] [PubMed] [Google Scholar]
- (3).Horwich AL, Low KB, Fenton WA, Hirshfield IN, and Furtak K (1993) Folding in vivo of bacterial cytoplasmic proteins: role of GroEL. Cell 74, 909–917. [DOI] [PubMed] [Google Scholar]
- (4).Chapman E, Farr GW, Usaite R, Furtak K, Fenton WA, Chaudhuri TK, Hondorp ER, Matthews RG, Wolf SG, Yates JR, Pypaert M, and Horwich AL (2006) Global aggregation of newly translated proteins in an Escherichia coli strain deficient of the chaperonin GroEL. Proc. Natl. Acad. Sci. U. S. A 103, 15800–15805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Braig K, Otwinowski Z, Hegde R, Boisvert DC, Joachimiak A, Horwich AL, and Sigler PB (1994) The crystal structure of the bacterial chaperonin GroEL at 2.8 A. Nature 371, 578–586. [DOI] [PubMed] [Google Scholar]
- (6).Sigler PB, Xu Z, Rye HS, Burston SG, Fenton WA, and Horwich AL (1998) Structure and function in GroEL-mediated protein folding. Annu. Rev. Biochem 67, 581–608. [DOI] [PubMed] [Google Scholar]
- (7).Horovitz A, Fridmann Y, Kafri G, and Yifrach O (2001) Review: allostery in chaperonins. J. Struct. Biol 135, 104–114. [DOI] [PubMed] [Google Scholar]
- (8).Thirumalai D, and Hyeon C (2018) Signalling networks and dynamics of allosteric transitions in bacterial chaperonin GroEL: implications for iterative annealing of misfolded proteins. Philos. Trans. R. Soc., B 373, 20170182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Svoboda K, and Block SM (1994) Force and velocity measured for single kinesin molecules. Cell 77, 773–784. [DOI] [PubMed] [Google Scholar]
- (10).Schliwa M, and Woehlke G (2003) Molecular motors. Nature 422, 759–765. [DOI] [PubMed] [Google Scholar]
- (11).Xu Z, Horwich AL, and Sigler PB (1997) The crystal structure of the asymmetric GroEL-GroES-(ADP)7 chaperonin complex. Nature 388, 741–750. [DOI] [PubMed] [Google Scholar]
- (12).Thirumalai D, Lorimer GH, and Hyeon C (2020) Iterative annealing mechanism explains the functions of the GroEL and RNA chaperones. Protein Sci. 29, 360–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Ellis RJ (2001) Molecular chaperones: inside and outside the Anfinsen cage. Curr. Biol 11, R1038–1040. [DOI] [PubMed] [Google Scholar]
- (14).Ellis RJ (1996) Revisiting the Anfinsen cage. Folding Des. 1, R9–R15. [PubMed] [Google Scholar]
- (15).Horwich AL, and Fenton WA (2009) Chaperonin-mediated protein folding: using a central cavity to kinetically assist polypeptide chain folding. Q. Rev. Biophys 42, 83–116. [DOI] [PubMed] [Google Scholar]
- (16).Hayer-Hartl M, Bracher A, and Hartl FU (2016) The GroEL-GroES Chaperonin Machine: A Nano-Cage for Protein Folding. Trends Biochem. Sci 41, 62–76. [DOI] [PubMed] [Google Scholar]
- (17).Lin Z, and Rye HS (2006) GroEL-mediated protein folding: making the impossible, possible. Crit. Rev. Biochem. Mol. Biol 41, 211–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Betancourt MR, and Thirumalai D (1999) Exploring the kinetic requirements for enhancement of protein folding rates in the GroEL cavity. J. Mol. Biol 287, 627–644. [DOI] [PubMed] [Google Scholar]
- (19).Baumketner A, Jewett A, and Shea JE (2003) Effects of confinement in chaperonin assisted protein folding: rate enhancement by decreasing the roughness of the folding energy landscape. J. Mol. Biol 332, 701–713. [DOI] [PubMed] [Google Scholar]
- (20).Cheung MS, and Thirumalai D (2006) Nanopore-protein interactions dramatically alter stability and yield of the native state in restricted spaces. J. Mol. Biol 357, 632–643. [DOI] [PubMed] [Google Scholar]
- (21).Thirumalai D, Klimov DK, and Lorimer GH (2003) Caging helps proteins fold. Proc. Natl. Acad. Sci. U. S. A 100, 11195–11197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Takagi F, Koga N, and Takada S (2003) How protein thermodynamics and folding mechanisms are altered by the chaperonin cage: molecular simulations. Proc. Natl. Acad. Sci. U. S.A 100, 11367–11372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Eggers DK, and Valentine JS (2001) Crowding and hydration effects on protein conformation: a study with sol-gel encapsulated proteins. J. Mol. Biol 314, 911–922. [DOI] [PubMed] [Google Scholar]
- (24).Ravindra R, Zhao S, Gies H, and Winter R (2004) Protein encapsulation in mesoporous silicate: the effects of confinement on protein stability, hydration, and volumetric properties. J. Am. Chem. Soc 126, 12224–12225. [DOI] [PubMed] [Google Scholar]
- (25).Campanini B, Bologna S, Cannone F, Chirico G, Mozzarelli A, and Bettati S (2005) Unfolding of Green Fluorescent Protein mut2 in wet nanoporous silica gels. Protein Sci. 14, 1125–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Bolis D, Politou AS, Kelly G, Pastore A, and Temussi PA (2004) Protein stability in nanocages: a novel approach for influencing protein stability by molecular confinement. J. Mol. Biol 336, 203–212. [DOI] [PubMed] [Google Scholar]
- (27).Jewett AI, and Shea JE (2010) Reconciling theories of chaperonin accelerated folding with experimental evidence. Cell. Mol. Life Sci 67, 255–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Weaver J, Jiang M, Roth A, Puchalla J, Zhang J, and Rye HS (2017) GroEL actively stimulates folding of the endogenous substrate protein PepQ. Nat. Commun 8, 15934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Hofmann H, Hillger F, Pfeil SH, Hoffmann A, Streich D, Haenni D, Nettels D, Lipman EA, and Schuler B (2010) Single-molecule spectroscopy of protein folding in a chaperonin cage. Proc. Natl. Acad. Sci. U. S. A 107, 11793–11798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Tehver R, and Thirumalai D (2008) Kinetic model for the coupling between allosteric transitions in GroEL and substrate protein folding and aggregation. J. Mol. Biol 377, 1279–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Todd MJ, Lorimer GH, and Thirumalai D (1996) Chaperonin-facilitated protein folding: optimization of rate and yield by an iterative annealing mechanism. Proc. Natl. Acad. Sci. U. S. A 93, 4030–4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Yang D, Ye X, and Lorimer GH (2013) Symmetric GroEL:GroES2 complexes are the protein-folding functional form of the chaperonin nanomachine. Proc. Natl. Acad. Sci. U. S. A 110, E4298–4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Fei X, Ye X, LaRonde NA, and Lorimer GH (2014) Formation and structures of GroEL:GroES2 chaperonin footballs, the protein-folding functional form. Proc. Natl. Acad. Sci. U. S. A 111, 12775–12780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Chakrabarti S, Hyeon C, Ye X, Lorimer GH, and Thirumalai D (2017) Molecular chaperones maximize the native state yield on biological times by driving substrates out of equilibrium. Proc. Natl. Acad. Sci. U. S. A 114, E10919–E10927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Mendoza JA, Rogers E, Lorimer GH, and Horowitz PM (1991) Chaperonins facilitate the in vitro folding of monomeric mitochondrial rhodanese. J. Biol. Chem 266, 13044–13049. [PubMed] [Google Scholar]
- (36).Mendoza JA, Rogers E, Lorimer GH, and Horowitz PM (1991) Unassisted refolding of urea unfolded rhodanese. J. Biol. Chem 266, 13587–13591. [PubMed] [Google Scholar]
- (37).Chaudhry C, Farr GW, Todd MJ, Rye HS, Brunger AT, Adams PD, Horwich AL, and Sigler PB (2003) Role of the gamma-phosphate of ATP in triggering protein folding by GroEL-GroES: function, structure and energetics. EMBO J. 22, 4877–4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Weber F, and Hayer-Hartl M (2000) Prevention of rhodanese aggregation by the chaperonin GroEL. Methods Mol. Biol 140, 111–115. [DOI] [PubMed] [Google Scholar]
- (39).Bhattacharyya AM, and Horowitz PM (2001) The aggregation state of rhodanese during folding influences the ability of GroEL to assist reactivation. J. Biol. Chem 276, 28739–28743. [DOI] [PubMed] [Google Scholar]
- (40).Korobko I, Mazal H, Haran G, and Horovitz A (2020) Measuring protein stability in the GroEL chaperonin cage reveals massive destabilization. eLife 9, e56511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Vaitheeswaran S, and Thirumalai D (2008) Interactions between amino acid side chains in cylindrical hydrophobic nanopores with applications to peptide stability. Proc. Natl. Acad. Sci. U. S. A 105, 17636–17641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Fedorov AN, and Baldwin TO (1997) GroE modulates kinetic partitioning of folding intermediates between alternative states to maximize the yield of biologically active protein. J. Mol. Biol 268, 712–723. [DOI] [PubMed] [Google Scholar]
- (43).Koculi E, Horst R, Horwich AL, and Wuthrich K (2011) Nuclear magnetic resonance spectroscopy with the stringent substrate rhodanese bound to the single-ring variant SR1 of the E. coli chaperonin GroEL. Protein Sci. 20, 1380–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Weissman JS, Hohl CM, Kovalenko O, Kashi Y, Chen S, Braig K, Saibil HR, Fenton WA, and Norwich AL (1995) Mechanism of GroEL action: productive release of polypeptide from a sequestered position under GroES. Cell 83, 577–587. [DOI] [PubMed] [Google Scholar]
- (45).Rye HS, Burston SG, Fenton WA, Beechem JM, Xu Z, Sigler PB, and Horwich AL (1997) Distinct actions of cis and trans ATP within the double ring of the chaperonin GroEL. Nature 388, 792–798. [DOI] [PubMed] [Google Scholar]


