Abstract
Recent studies suggest that rare variants exhibit stronger effect sizes and might play a crucial role in the etiology of lung cancers (LC). Whole exome plus targeted sequencing of germline DNA was performed on 1045 LC cases and 885 controls in the discovery set. To unveil the inherited causal variants, we focused on rare and predicted deleterious variants and small indels enriched in cases or controls. Promising candidates were further validated in a series of 26,803 LCs and 555,107 controls. During discovery, we identified 25 rare deleterious variants associated with LC susceptibility, including 13 reported in ClinVar. Of the five validated candidates, we discovered two pathogenic variants in known LC susceptibility loci, ATM p.V2716A (Odds Ratio [OR] 19.55, 95%CI 5.04–75.6) and MPZL2 p.I24M frameshift deletion (OR 3.88, 95%CI 1.71–8.8); and three in novel LC susceptibility genes, POMC c.*28delT at 3′ UTR (OR 4.33, 95%CI 2.03–9.24), STAU2 p.N364M frameshift deletion (OR 4.48, 95%CI 1.73–11.55), and MLNR p.Q334V frameshift deletion (OR 2.69, 95%CI 1.33–5.43). The potential cancer-promoting role of selected candidate genes and variants was further supported by endogenous DNA damage assays. Our analyses led to the identification of new rare deleterious variants with LC susceptibility. However, in-depth mechanistic studies are still needed to evaluate the pathogenic effects of these specific alleles.
Subject terms: Non-small-cell lung cancer, Cancer epidemiology
Introduction
Lung cancer (LC), the leading cause of cancer mortality in the US, has recently shown substantial drops in mortality, largely attributed to reduced smoking rates and improvement in new treatments such as immunotherapy1. Prior genome-wide association studies (GWAS) identified novel genetic factors influencing LC risk, which are sometimes modulated by smoking behavior2. Notably, in the 15q25.1 region that shows the most significant and consistent genetic signal, a missense p.D398N and a 22-bp deletion (del) in the core promoter region of CHRNA5 have been identified that affect the function and expression3,4. Carriers of these variants find quitting smoking more difficult than noncarriers5 and may benefit from a targeted smoking cessation intervention6.
Previous studies have estimated heritability of LC to be 18%7. Recent genetic studies suggest that rare variants (minor allele frequency [MAF] < 1%) that are functionally deleterious, exhibit far larger effect sizes than common variants8–10 as they display signs of stronger selective pressure11,12, and could account for missing heritability unexplained by common variants11. Fewer than 3% of protein-coding single nucleotide variants (SNVs) corresponding to approximately 300 genes per genome are predicted to result in loss of protein function (LoF) through the introduction of stop-gain, frameshift, or the disruption of an essential splice site13. Insertions (ins) or deletions (indels) have been understudied, though they are the second most abundant source of human genetic variation. Selected indels have been identified as playing a key role in causing LC, such as p.E746_A750del in EGFR14–16.
Supporting the hypothesis that deleterious mutations will show lower MAF are recent identifications of several rare missense variants that have a moderate-to-large effect on LC risk, for example, PARK2 p.R275W (OR 5.24)17, BRCA2 p.K3326X (OR 2.47), CHEK2 p.I157T (OR 0.38)18, LTB p.L87F (OR 7.52), P3H2 p.Q185H (OR 5.39)19, DBH p.V26M20, and ATM p.L2307F (OR 8.82)21. Because of the stronger evolutionary pressure and weak linkage disequilibrium (LD) with common SNPs used in GWAS, finding these rare variants through population-based studies can be challenging22. To maximize the potential for the detection of large-effect, rare deleterious variants (SNVs and small indels ≤21 bp), we employed whole exome sequencing (WES) plus targeted sequencing on healthy controls and selected high-risk LC cases enriched with the highest genetic risk of LC, for example, early-onset or family history of LC (FHLC)7,23,24.
Results
Demographics of study subjects
As shown in Table 1, the vast majority of subjects in the discovery study ─ Transdisciplinary Research in Cancer of the Lung (TRICL; 1,045 LCs vs. 885 controls) ─ and the validation sets (26,803 LCs and 555,107 controls) were primarily of European-descent (Supplementary Fig. 1). LC cases were significantly more likely to be smokers and with higher pack-years than controls (P-value < 0.0001). The TRICL and Genetic Epidemiology of LC Consortium (GELCC) cases were enriched for having FHLC.
Table 1.
Basic characteristics of LC cases and controls in the discovery and validations sets.
| Discovery | Validation# | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Characteristics | TRICL | GELCC | COPDGene | TCGA | gnomAD | OncoArray | Affymetrix | UKB | ||||
| Platform | WES | WES | WES | WES | WES + WGS | Genotyping | Exome array | Genotyping | ||||
| N (%)& | LC Case n = 1045 | Control n = 885 | LC case n = 380 | Controls n = 318 | LC cases n = 1015 | Controls n = 134,187 | LC cases n = 17,878 | Controls n = 13,425 | LC case n = 5364 | Controls n = 5724 | LC Case n = 2166 | Controls n = 401,453 |
| Ethnicity | P < 0.0001 | P < 0.0001 | ||||||||||
| White | 909 (87%) | 830 (94%) | 372 (98%) | 318 (100%) | 742 (73%) | 94,134 (70%) | 13,876 (78%) | 11,011 (82%) | 3086 (58%) | 3550 (62%) | 2094 (97%) | 375,894 (94%) |
| Other† | 136 (13%) | 55 (6%) | 6 (2%) | 0 | 273 (27%) | 40, 053 (30%) | 210 (1%) | 128 (1%) | 625 (12%) | 652 (11%) | 65 (3%) | 24,055 (6%) |
| Age, yr. | P = 0.006 | P < 0.0001 | ||||||||||
| Mean (range) | 63 (24–91) | 61 (20–90) | 64 (30–87) | 63 (55–80) | 65 (30–90) | 54 (18–90) | 64 (19–95) | 62 (18–97) | 61 (30–95) | 59 (31–91) | 62 (40–70) | 56 (37–73) |
| <60 yr. | 418 (40%) | 356 (40%) | 102 (27%) | 88 (28%) | 214 (21%) | – | 6036 (43%) | 5303 (40%) | 2335 (43%) | 3063 (53%) | 624 (29%) | 242,687 (60%) |
| Sex | P < 0.0001 | |||||||||||
| Male | 614 (59%) | 515 (58%) | 232 (61%) | 172 (54%) | 563 (59%) | 73,370 (55%) | 11,147 (62%) | 8274 (62%) | 2930 (55%) | 3125 (55%) | 1182 (55%) | 186,083 (46%) |
| Female | 431 (41%) | 370 (42%) | 171 (45%) | 146 (46%) | 452 (41%) | 60,817 (45%) | 6731 (38%) | 5151 (38%) | 2434 (45%) | 2599 (45%) | 984 (45%) | 215,370 (54%) |
| Smoking | P < 0.0001 | P < 0.0001 | P < 0.0001 | P < 0.0001 | ||||||||
| Never | 125 (12%) | 308 (35%) | 31 (8%) | 0 | 173 (17%) | – | 1720 (10%) | 4152 (31%) | 572 (11%) | 1726 (30%) | 203 (10%) | 236,246 (59%) |
| Ever | 918 (88%) | 576 (65%) | 346 (91%) | 318 (100%) | 742 (73%) | – | 15,889 (89%) | 8998 (67%) | 4675 (87%) | 3972 (69%) | 1945 (90%) | 163,226 (41%) |
| Mean PY (range) | 42 (0–196) | 23 (0–133) | 46 (0–165) | 54 (10–97) | 42 (0–154) | – | 46(0–315) | 33 (0–260) | 45 (0–231) | 34 (0–218) | 40 (0–220) | 23 (0–301) |
| FHLC | P < 0.0001 | P < 0.0001 | ||||||||||
| Yes | 506 (48%) | 72 (8%) | 122 (33%) | – | – | – | – | – | – | 457 (21%) | 49,104 (12%) | |
| No | 359 (34%) | 306 (35%) | 258 (67%) | – | – | – | – | – | – | 1709 (79%) | 352,349 (88%) | |
| Histology | ||||||||||||
| AD | 459 (44%) | – | 182 (48%) | – | 577 (57%) | – | 6568 (37%) | – | 2106 (39%) | – | 781 (36%) | – |
| SCC | 342 (33%) | – | 118 (31%) | – | 438 (43%) | – | 4284 (24%) | – | 1131 (21%) | – | 461 (21%) | – |
| Other | 244 (23%) | – | 80 (21%) | – | 0 | – | 7026 (39%) | – | 2127 (40%) | – | 924 (43%) | – |
TRICL Transdisciplinary Research in Cancer of the Lung, WES whole-exome sequencing, WGS whole-genome sequencing, LC lung cancer, PY pack-year, FHLC family history of LC (first degree), AD adenocarcinoma, SCC squamous cell carcinoma.
&Numbers do not add up due to missing data.
†Other ethnicities in TRICL (one African control subject and 190 unknown), TCGA (8% African American 2% East Asian, and 17% unknown), gnomAD (8.8% African, 7.2% East Asian, 11.4% South Asian, and 2.5% other). Genetic ancestry analysis of TRICL subjects shows most of the subjects of the “unknown” race were located between the European- and Asian-ancestry clusters (Supplemental Fig. 1). Genetic ancestry analysis of TCGA patients shows the vast majority of subjects with “unknown” race were primarily genetic European ancestry (i.e., 90% TCGA-LCs were genetically Europeans)82.
#The validation sets include 26,803 LCs and 555,107 controls: (1) Genetic Epidemiology of LC (GELCC) WES data for 380 LCs (258 sporadic and 122 FLC were selected from high-risk LC families with at least two first-degree relatives affected with LC); (2) COPDGene WES data for 318 controls with normal lung function; (3) TCGA (The Cancer Genome Atlas) germline WES data for 1015 LCs; (4) GnomAD (genome aggregation database, v2.1) WES and WGS data for 134,187 non-cancer controls (excluded individuals from cancer cohort studies, such as the TCGA cohort). (5) OncoArray genotyping data for 17,878 LCs vs. 13,425 controls; (6) Affymetrix exome array data for 5364 LCs vs. 5724 controls; (7) UK Biobank (UKB) genotyping data for 2166 LCs vs. 401,453 controls.
Identification of rare and deleterious variants in the TRICL discovery set
In the discovery set, a total of 2,182,753 variants were detected. Applying a three-step filtering method based on allele frequency (MAF < 1% in non-Finnish European [NFE] population from the Genome Aggregation Database [gnomAD]), variant class (missense, protein-truncating and regulatory), and functional effects (predicted deleterious and or with clinical significance from ClinVar), we identified 67,470 rare and putatively deleterious variants: 63% missense, 16% frameshift (fs), 12% in-frame indels, 6% regulatory (untranslated region [UTR] and splice acceptor/donor), and 3% stop-gain. Single variant association analysis identified 75 potential candidates.
Given the known challenge of excessive false-positive indel detection rates caused by the high frequency of homopolymer-associated sequencing errors25–28, we subjected these 75 potential candidates to additional filtering and manual inspection using Genome Browser (Supplementary Table 1). Twenty-five of the 75 were high-confidence putative candidates (two SNVs, four ins, and 19 del). Supplementary Fig. 2 shows the variant visualization map for the candidates and variant carriers (read alignment and depth). Thirteen out of the 25 candidates (in 24 genes) reported clinical significance in ClinVar, and eight were classified as pathogenic. Also, 5/24 genes were mapped to known LC-GWAS loci, such as 3q28 TP6329, 5q31.1 TXNDC1530, 11q22.3 ATM21, 11q23.3 MPZL231, and 22q12.1 CHEK218. Three mapped in known GWAS loci for COPD/ PF (pulmonary function): 1p34.3 BMP8A32,33, 1p36.31 PHF1332, and 14q23.1 TALPID3/KIAA058634.
We next assessed the dose-effect of the 25 candidates: 16 were enriched in LCs (risk-conferring alleles) and 9 were enriched in controls (protective alleles). Compared with subjects with zero risk- and protective-alleles, the groups carrying one, and two risk-alleles (5 LCs) showed a progressively increased risk, whereas groups carry one, and two protective-alleles (6 controls) demonstrated a gradually reduced risk (Supplementary Table 2). All 6 controls harbored MOB3A p.F69_I75del, whereas 4/5 LCs harbored STAU2 p.N364M fs*67del.
Studying the demographics of the mutation carriers, there was no significant difference in smoking (status and pack-years) or FHLC between carriers and non-carriers. Notably, 5/6 two-protective-alleles carriers were male, whereas 4/5 two-risk-alleles carriers were female and had adenocarcinoma (AD). Overall, age did not differ significantly between carriers and non-carriers (Supplementary Fig. 3). However, in LC cases, onset-age in risk-allele carriers (54 yrs for two-risk-alleles carriers, 62 yrs for one-risk-allele carriers) were significantly younger than the protective-allele carriers (69 yrs; Supplementary Table 2).
Further gene-environment (G×E) interaction analysis showed that two variants interacted with smoking behavior (Supplementary Table 1). Specifically, the risk MLNR p.Q334V fs*3del interacted with pack-years (P-value 0.0035); the protective-effect associated with the MOB3A p.F69_I75del is substantial and significant among males (10/11 control carriers were male, whereas 0/2 LCs carriers were male; P-value 0.042), smokers (6/11 control carriers were smokers, whereas 0/2 LCs carriers were smokers; P-value 0.016), and pack-years (P-value 0.0036). We also identified that the protective variant TXNDC15 p.E9G fs*68del interacted with FHLC, as 5/7 of LC carriers with FHLC, compared to 0/21 controls (P-value 0.035).
We subsequently conducted gene-based rare variant burden tests for the 24 genes harboring potential candidates, five genes, namely, MLNR, CCDC105, BMP8A, MME, and NPHP3, showed suggestive associations (Table 2). We also performed exome wide gene-based tests, however, none showed strong association after multiple testing corrections (Supplementary Fig. 4).
Table 2.
Gene-based association tests in the TRICL study, ranked by P-value from the combined multivariate and collapsing test.
| Genes | N. rare deleterious variants* | N. multi-marker genotypes | N. carriers LC /Control | KBAC test P-value | CMC test P-value | CMC test OR (95% CI) | Gene constraint LoF o/e (90% CI)& | Gene PhoRank to phenotype# |
|---|---|---|---|---|---|---|---|---|
| Risk genes | ||||||||
| CCDC105 | 20 | 11 | 28/5 | 0.012 | 0.013 | 5.63 (0.87–31.4) | 0.71 (0.46–1.12) | 0.12 _ PF |
| BMP8A | 3 | 4 | 11/2 | 0.014 | 0.014 | 4.22 (1.14–36.3) | 0.8 (0.49–1.35) | 0.32 _ PF |
| MME/CD10 | 5 | 6 | 7/0 | 0.014 | 0.015 | 1.85 (0.65–11.16) | 0.7 (0.54–0.92) | 0.83 _ LC |
| NPHP3 | 6 | 7 | 7/0 | 0.015 | 0.015 | 1.68 (0.76–15.4) | 0.5 (0.38–0.65) | 0.68 _ PF |
| MLNR | 5 | 6 | 11/2 | 0.005 | 0.022 | 4.19 (1.12–38.9) | 0.51 (0.5–1.16) | 0.09 _ PF |
| NKX6-1 | 9 | 11 | 47/31 | 0.064 | 0.048 | 1.30 (0.82–2.06) | 0.39 (0.18–1.0) | 0.28 _ LC |
| ENAM | 7 | 8 | 9/2 | 0.043 | 0.065 | 2.94 (0.79–9.07) | 0.6 (0.44–0.84)` | 0.32 _ PF |
| ATM | 15 | 15 | 16/11 | 0.591 | 0.098 | 1.58 (0.69–7.04) | 0.60 (0.51–0.71) | 0.95 _ LC |
| RHBDD3 | 9 | 10 | 11/4 | 0.101 | 0.102 | 1.64 (0.85–23.6) | 0.71 (0.41–1.27) | 0.16 _ PF |
| STAU2 | 27 | 31 | 107/76 | 0.141 | 0.213 | 1.21 (0.89–1.65) | 0.14 (0.07–0.32) | 0.23 _ LC |
| TALPID3 | 11 | 12 | 17/8 | 0.139 | 0.403 | 1.64 (0.93–2.24) | 0.54 (0.42–0.72) | 0.61 _ PF |
| MPZL2 | 6 | 7 | 7/3 | 0.153 | 0.403 | 1.34 (0.77–2.13) | 1.34 (0.9–1.86) | 0.12 _ LC |
| TP63 | 6 | 7 | 9/3 | 0.396 | 0.539 | 1.20 (0.59–12.3) | 0.13 (0.07–0.27) | 0.87 _ LC |
| POMC | 6 | 7 | 7/5 | 0.744 | 0.790 | 1.45 (0.57–3.71) | 0.74 (0.42–1.38) | 0.30 _ PF |
| F13B | 5 | 6 | 7/5 | 0.965 | 0.905 | 1.02 (0.81–1.27) | 0.59 (0.41–0.85) | 0.34 _ PF |
| Protective genes | ||||||||
| TXNDC15 | 11 | 12 | 10/27 | 0.746 | 0.001 | 0.31 (0.15–0.64) | 0.38 (0.21–0.76) | 0.60 _ PF |
| GJB6 | 2 | 3 | 0/6 | 0.877 | 0.008 | 0.12 (0.02–0.66) | 1.07 (0.66–1.74) | 0.31 _ LC |
| MOB3A | 3 | 4 | 3/12 | 0.587 | 0.008 | 0.21 (0.05–0.67) | 0.96 (0.54–1.7) | 0.10 _ LC |
| CASQ2 | 2 | 4 | 15/26 | 0.955 | 0.013 | 0.73 (0.44–1.23) | 0.94 (0.65–1.38) | 0.36 _ PF |
| OR51J1 | 2 | 3 | 1/6 | 0.351 | 0.037 | 0.14 (0.03–0.84) | 0.19 (0.07–0.88) | 0.10 _ PF |
| FAM111A | 5 | 6 | 11/21 | 0.406 | 0.076 | 0.42 (0.20–0.92) | 2.09 (0.66–1.95) | 0.29 _ LC |
| PHF13 | 9 | 10 | 16/12 | 0.097 | 0.742 | 1.23 (0.53–5.17) | 0.01 (0–0.25) | 0.28 _ LC + PF |
| MLKL | 17 | 19 | 30/28 | 0.055 | 0.689 | 0.95 (0.54–3.52) | 0.87 (0.63–1.24) | 0.20 _ PF |
| CHEK2 | 8 | 9 | 7/5 | 0.484 | 0.811 | 1.11 (0.64–2.02) | 1.15 (0.87–1.53) | 0.97 _ LC |
TRICL Transdisciplinary Research in Cancer of the Lung, CMC Combined Multivariate and Collapsing, KBAC Kernel-Based Adaptive Cluster, LoF loss of function, LC lung cancer, PF pulmonary function, OR odds ratio, CI confidence interval, o/e observed/expected.
*Number of rare deleterious variants within the genes. False discovery rate (FDR) adjusted P-value was reported.
&Gene constraint LoF o/e values developed with gnomAD: observed counts are based on sequencing data from gnomAD, expected counts are based on a mutational model that takes sequence context and coverage into account. Lower o/e, in particular, the upper bound of the CI < 0.35 are indicative of strong intolerance (disease-causing). The top three genes with the lowest o/e were bolded: PHF13, TP63, and STAU2.
#Genes Phevo PhoRank is based on gene functions relevant to the disease phenotype (LC, COPD/PF) from diverse biomedical ontologies. Disease-associated genes have a higher Phevor score. The top four genes with the highest scores were bolded: CHEK2, ATM, TP63, and MME/CD10.
Meta-analyses of the discovery and validation sets
In the seven validation datasets, of the 25 candidates, 100% were covered by the gnomAD, 22 (88%) in TCGA, 16 (64%) in COPDGene, nine (36%) in GELCC, and nine (36%) were covered in one of the three case–control studies (OncoArray, Affymetrix, and UKB) with genotyping data. Table 3 summarizes the top five candidates with consistent associations from the meta-analysis.
Table 3.
Top five hits from discovery and validation association analysis.
| Candidates | n. (freq.%) carriers | Meta-analysis& | |||||
|---|---|---|---|---|---|---|---|
| (ClinVar) | TRICL: 1045 LC, 885 Control | GELCC: 380 LC, COPDGene: 318 Control | TCGA: 1015 LC, GnomAD: 134,187 Control | GWAS studies @ OncoArray/Affymetrix | Total freq.% carrier LC/Control | OR (95% CI) | P-value |
| ATM missense SNV p.V2716A, rs587782652 (Pathogenic) | 2 (0.19%) / 0 | 0 / 0 | 2 (0.20%)/ 5 (0.004%) | 5 (0.03%) / 0 @ OncoArray | 0.05% / 0.003% | 19.55 (5.04–75.6) | 1.7e-05 |
| POMC 3′ UTR deletion c.*28delT, rs756770132 (VUS) | 6 (0.57%) / 0 | 4 (1.05%) 1 FLC / 0 | 6 (0.59%) / 207 (0.17%) | - | 0.66% / 0.15% | 4.33 (2.03–9.24) | 0.00015 |
| STAU2 LoF deletion p.N364M fs*67, rs746501298 | 21 (2.01%) / 4 (0.45%) | 4 (1.05%) 3 FLC / 0 | 0 / 25 (0.02%) | - | 1.02% / 0.02% | 4.48 (1.73–11.55) | 0.0019 |
| MPZL2 LoF deletion p.I24M fs*22, rs752672077 (Pathogenic) | 3 (0.29%) / 0 | 4 (1.05%) 2 FLC / 0 | 5 (0.49%) / 189 (0.15%) | - | 0.49% / 0.14% | 3.88 (1.71–8.8) | 0.0012 |
| MLNR LoF deletion, p.Q334V fs*3, rs563947699 | 9 (0.86%) 3 FHLC/ 0 | 6 (1.58%) 2 FLC / 0 | 7 (0.69%) / 431 (0.35%) | 29 (0.54%) / 49 (0.86%) @ Affymetrix | 0.65% / 0.34% | 2.69 (1.33–5.43) | 0.0060 |
TRICL Transdisciplinary Research in Cancer of the Lung, SNV single nucleotide variants, Indels insertion (ins)/deletion (del), LoF loss of function, fs frameshift, VUS variant of uncertain significance from ClinVar, NA not available, LC lung cancer, AD adenocarcinoma, FLC familiar lung cancer, OR odds ratio, CI confidence interval.
#The validation sets include 26,803 LCs and 555,107 controls: 1) Genetic Epidemiology of LC (GELCC) WES data for 380 LCs (122 FLC and 258 sporadic); 2) COPDGene WES data for 318 controls; 3) TCGA (The Cancer Genome Atlas) WES data for 1015 LCs; 4) GnomAD (genome aggregation database) WES and WGS data for 134,187 non-cancer controls; 5) OncoArray genotyping data for 17,878 LCs vs. 13,425 controls; 6) Affymetrix exome array data for 5364 LCs vs. 5724 controls; 7) UK Biobank (UKB) genotyping data for 2166 LCs vs. 401,453 controls.
@The ATM p.V2716A genotype was from the OncoArray study; the MLNR p.Q334V deletion genotype was from the Affymetrix study.
&The freq.% of carriers were based on the available cases and controls. False discovery rate (FDR) adjusted P-values indicate significant associations based on a fixed-effect meta-analysis.
The topmost risk-conferring variant is a missense SNV, p.V2716A, in the phosphatidylinositol 3-kinase (PI3K) catalytic domain of ATM (Ataxia telangiectasia mutated; OMIM 607585, UniProt Q13315). This pathogenic variant (rs587782652) is exceedingly rare in the gnomAD, with MAF 0.0021% and 0.0054% in non-cancer controls and NFE population, respectively. In our combined datasets, this variant presented in 0.05% of LCs and 0.003% controls, with remarkably high effect sizes (OR 19.55, 95%CI 5.04–75.6; P-value 1.7e-05). LC carriers of this variant were predominately enriched in smokers (8/9 carriers), AD (7/9 carriers), and early-onset (6/9 carriers; mean 55 yrs). Further, four additional rare deleterious variants were observed in ATM (Fig. 1 and Supplementary Table 3). No LD is present among these variants and the candidate p.V2716A (Supplementary Table 4).
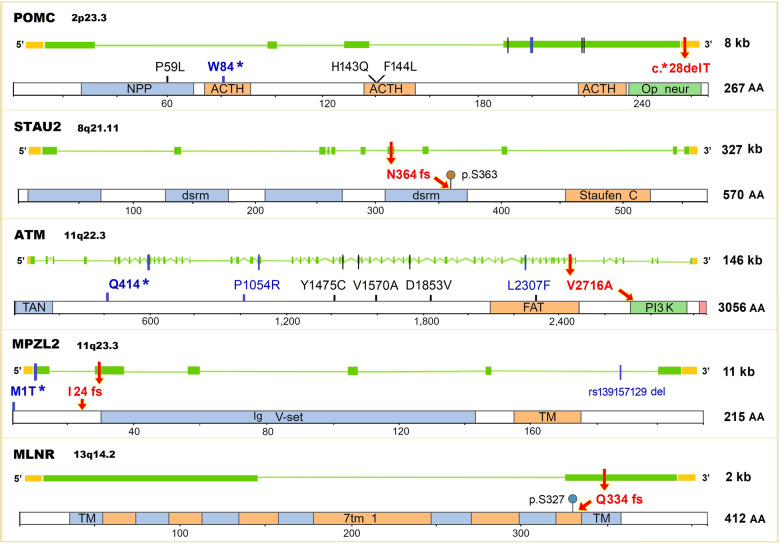
Fig. 1. Gene exons, protein domains, and rare deleterious variants of the candidate genes.
The top five candidate variants (red arrows): 1) POMC c.*28 deletion (del) located at target sites of several miRNAs in 3′ UTR; 2) STAU2 p.N364M fs*67del located in the double-stranded RNA-binding motif (dsrm), and next to a phosphorylation site p.S363; 3) ATM V2716A located in the PI3-kinase (PI3K) catalytic domain; 4) MPZL2 p.I24M fs*22del was close to the antibody variable domain of immunoglobulins (Ig-V); 5) MLNR p.Q334V fs*3del located in the transmembrane receptor domain (TM), and close to a phosphorylation site p.S327. The color vertical bars represent different types of variants: ClinVar pathogenic variants (bold blue: POMC W84* stop-gain, ATM Q414* stop-gain, and MPZL2 M1T* start-loss), previous reported LC-associated variants (blue: ATM P1054R and L2307F, and MPZL2 deletion rs13915729), and ClinVar variants of uncertain significance (black). Gene exons (green blocks), introns (horizontal green lines), untranslated regions (UTRs, orange blocks), and protein domain/motif (framed rectangles) are shown. The length of the gene (kb) and protein (number of amino acids, AA) are shown to the right.
The second risk variant is c.*28delT in the 3′ UTR of POMC (Pro-opiomelanocortin; OMIM 176830, UniProt # P01189). The MAF of this 2 bp del (rs756770132) were 0.086%/0.17% in gnomAD non-cancer/NFE controls; while in our dataset presented in 0.66% of LCs and 0.15% of controls, conferring a 4-fold risk for carriers (OR 4.33, 95%CI 2.03–9.24; P-value 0.00015). Although reported as VUS in ClinVar, this 3′ UTR del is located in a critical site computationally predicted to be targets of several miRNAs by the TargetScan35, including hsa-miR-149-3p and hsa-mir-625-5p. We also observed four additional rare deleterious variants in the TRICL set (Fig. 1 and Supplementary Table 3).
The third novel risk variant is p.N364M fs*67del in STAU2 (Staufen homolog 2; OMIM 605920, UniProt Q9NUL3). This del (rs746501298) is very rare in gnomAD (MAF 0.011%/0.0027% in non-cancer/NFE population controls), but presented in 1.02% of LCs and 0.02% of non-cancer controls (OR 4.48, 95%CI 1.73–11.55; P-value 0.0019). It was predicted to disrupt the double-stranded RNA-binding motif (DSRM; Fig. 1) which plays a critical role in RNA editing. This del is also reported in the Catalogue of Somatic Mutations In Cancer (COSMIC, # COSM253104).
The fourth and fifth variants are two pathogenic, truncating deletions ─ p.I24M fs*22del (rs752672077) in MPZL2 (Myelin protein zero-like protein 2, or Epithelial v-like antigen 1 [EVA1]; OMIM 604873, UniProt O60487), and p.Q334V fs*3del (rs563947699) in MLNR (Motilin receptor; OMIM 602885, UniProt O43193) ─ with effects sizes of 3.88 (95% CI 1.71–8.8) and 2.69 (95% CI 1.33–5.43), respectively. The MPZL2 deletion was close to the Immunoglobulin-like antibody Variable domain (Ig-V; Fig. 1) which is involved in thymocyte development36. In gnomAD, MAF was the highest in the Ashkenazi Jewish (AJ, 0.38%) than other populations, including NFE (0.123%), Latino (0.028%), and African (0.012%). Additionally, a start-loss p.M1T of MPZL2 was present in two LCs (Fig. 1 and Supplementary Table 3).
Other interesting candidates from the discovery (Supplementary Table 1), include 1) two VUS ins, TP63 c.*2550insT (rs772929136) and CHEK2 c.*2insC (rs749257861), both were located in the 3′ UTR; however, no genotype data/coverage were available in validation sets; 2) a protective effect pathogenic variant, CHEK2 p.S428F (rs137853011), that was non-significant in the meta-analysis (OR 0.41, 95% CI 0.13–1.31, P-value 0.13).
Candidate gene prioritization
As shown in Table 2, of the 24 candidate genes, the most evolutionarily constrained (intolerance) genes with the lowest LoF observed/expected (o/e) values were PHF13, TP63, and STAU2; whereas the genes with the highest LC-correlated PhoRank scores were CHEK2, ATM, TP63, and MME. The most interesting protein interaction network consists of eight genes and is centered on three known DNA damage response genes, CHEK2-ATM-TP63, linking five other genes (Supplementary Fig. 5). GO enrichment analysis highlighted genes involved in replicative senescence (which triggers a DNA damage response); whereas KEGG pathway analysis revealed that genes were involved in small cell LC (Supplementary Table 5).
Endogenous DNA damage assay
Large conserved networks of E. coli and human proteins were recently discovered to promote endogenous DNA damage when overproduced37. These networks are known as DNA damageome proteins (DDPs)37. The DNA damageome also includes LoF variants that show DNA damage-up phenotypes38, most of which are not directly related to DNA repair but rather participate in the DNA damage production. We selected six prioritized genes for the assay: CHEK2, ATM, MPZL2, MLNR, POMC, and MME. We discovered the knockdown of five genes, overproduction of the mutant MLNR p.Q334V fs*3del and wildtype POMC promote DNA damage. Specifically, we first used pooled small interfering RNAs (siRNAs) that minimize off-target effects, and observed significantly increased DNA damage levels (γH2AX) for 5/6 genes (Fig. 2a–c), including two well-known DNA repair genes (CHEK2 and ATM) and three newly discovered DDPs (POMC, MLNR, and MME). By contrast, the knockdown of MPZL2 did not affect DNA damage. For the three newly discovered DDPs, we further validated their DNA damage phenotypes using different individual siRNAs (Fig. 2d–f). Moreover, overproducing the mutant MLNR p.Q334V fs*3del and the wildtype POMC open reading frame (ORF) from the plasmid promote DNA damage in the lung fibroblast-derived cell line (Fig. 2g–i).
Fig. 2. Discovery of DNA damageome genes/proteins and variants.
a siRNA knockdown endogenous DNA damage assay scheme. b Increased DNA damage (γH2AX) levels in five out of the six genes knockdowns (mean ± SEM, n = 2~4), MLNR, CHEK2, POMC, ATM, and MME, compared with non-targeting (NT) siRNA control. There is no increasing DNA damage in MPZL2 knockdown cells. c Representative flow histograms showing higher γH2AX levels in gene knockdowns. d–f MLNR, MME, and POMC knockdown by two individual siRNAs confirmed the DNA damage-up phenotypes by pooled siRNAs in b. DNA damage quantified by d median fluorescence intensity or e DNA-damage positive subpopulation. f Examples of flow cytometry dot plots showing DNA-damage positive subpopulation. g Overproduction endogenous DNA damage assay scheme. h Wildtype POMC and mutant MLNR p.Q334V fs*3del overproduction promote DNA damage. GFP-Tubulin as a control. i Representative histograms of (g). *P-value < 0.05, **P-value < 0.01, n.s not significant (P-value > 0.05).
Discussion
Our analyses led to the identification of 25 rare deleterious candidates (in 24 genes) that may be associated with LC susceptibility. Of the five validated variants, we rediscovered two pathogenic variants mapped to known LC susceptibility loci, ATM p.V2716A and MPZL2 p.I24M fs*22del; and identified three deletions in novel LC susceptibility genes, POMC 3′ UTR c.*28delT, STAU2 p.N364M fs*67del, and MLNR p.Q334V fs*3del. Our GxE analysis also suggests some of these associations may be further modified by smoking (MLNR p.Q334V fs*3del and MOB3A p.F69_I75del) and FHLC (TXNDC15 p.E9G fs*68del). Additionally, our assays of cellular DNA damage identified POMC and MLNR as part of the DNA damageome, and confirmed a double-strand break repair role of ATM.
This study confirms a robust association between LC susceptibility and ATM and discovered a new pathogenic p.V2716A, that reside in the PI3K catalytic domain. We also found this association is more evident in AD, which is consistent with several previous studies21,39,40. ATM is a critical first responder to DNA damage in the cell and essential for genome stability. Several association studies have indicated that common variants of ATM are linked to cancer susceptibility, including LC41–43. Expression of the PI3K domain in ataxia-telangiectasia cells resulted in complemented radiosensitivity and reduced chromosomal breakage after irradiation44–46, suggesting the PI3K domain contains many of the significant activity of ATM47. Our DNA damage assay also shows elevated DNA damage in lung fibroblasts confirming the previous finding that ATM defective cells accumulate more double-strand breaks48. Further, the presence of additional rare deleterious variants, together with previously identified p.P1054R31 and p.L2307F21, strongly suggests that the ATM gene plays a role in LC susceptibility.
Another known LC locus we rediscovered is MPZL2 (also called Epithelial v-like antigen 1, EVA), and the pathogenic frameshift p.I24M fs*22del. MPZL2 is located at 11q23.3, a known GWAS locus for LC31,49 and hearing loss50,51. MPZL2 is one of the top candidate target genes at this locus based on the expression quantitative trait loci (eQTLs) mapping31. MPZL2 is a member of the immunoglobulin superfamily, preferentially expressed in lung and thymus epithelium with a potential role as a favorable prognostic marker in thyroid cancer52. Interestingly, the MAF of p.I24M fs*22del in the AJ population was 5-fold higher than the general population in gnomAD. There are several examples where rare causal variants (e.g., variants in the P53, CFTR, and BRCA1/2) have higher frequencies within the AJ population53–56. In our DNA damage assay, MPZL2 expression levels do not affect endogenous DNA damage in lung fibroblasts, implying the need to investigate alternative mechanisms in future functional studies.
The most consistent and interesting findings are two new deletions: POMC 3′ UTR c.*28delT and MLNR p.Q334V fs*3del. POMC encodes a polypeptide hormone precursor that regulating energy metabolism, nicotinic-induced weight loss, and immune reactions57–59. In particular, POMC plays a role in UV-induced DNA damage through interactions with TP53 and is associated with skin cancer susceptibility60–64. Abnormal expression of POMC was a poor prognostic marker for LC65–68. Using in vitro models, Derghal et al. evaluated putative miRNA (i.e., miR-383, miR-384-3p, and miR-488) and found them physically bind to the 3′ UTR mRNA and regulate POMC expression in several neuronal subtypes69. Our DNA damage assay showed both downregulation and overproduction of wildtype POMC promotes endogenous DNA damage. Whether and how the c.*28delT affects POMC expression and their putative role to LC risk merit further mechanistic investigation. MLNR is a member of the G-protein coupled receptor 1 family, and known for regulating gastrointestinal activity70. MLNR variants and dysregulation have been implicated in lung occult small cell carcinoma, bile duct cancer71, and head and neck cancer72. Our overproduction results of the MLNR p.Q334V fs*3del suggest a dominant-negative role in terms of DNA damage promotion. Collectively, these findings suggesting POMC and MLNR, while both functions in multiple cellular processes, might also share their various effects on DNA damage.
Although the pathogenic variant, CHEK2 p.S428F with lower LC risk is not statistically significant in the meta-analysis, its protective effect is consistent with another known pathogenic low-frequency variant, CHEK2 p.I157T, associated with reduced risk of smoking-related cancers (lung, laryngeal, urinary, and upper aerodigestive tract)18,73–75. In contrast, both p.I157T and p.S428F showed an increased risk of breast cancer75–79. The mechanism underlying this effect is an ongoing question with unknown impact, perhaps related to smoking exposure and cell cycle checkpoint signaling/apoptosis75. STAU2 is a double-stranded RNA-binding protein and a major regulator of mRNA transport, decay, and translation80. It was reported that STAU2 downregulation enhances levels of DNA damage (γH2AX) and promotes apoptosis (PARP1 cleavage) in camptothecin-treated cells81,82. The role of STAU2 in LC requires future investigations.
A main strength of the study is the focus on LC patients with extreme phenotypes of known risk factors (i.e., early-onset, FHLC, or familial cases in high-risk families), which provide >5 times statistical power10. Another strength was the relatively large sample size, which is by far the largest collection of LC rare variant analysis to our knowledge. It should be noted however that our study still has limited power to detect association for ultra-rare variants and those candidates (16/25) that could not be assessed in the validation. Third, our exome plus customized captures (50 Mb + 250 kb) in the discovery offers an efficient method for analyzing known susceptibility regions at greater depth and better coverage, particularly for indels that are often poorly captured in GWAS. Last, we have focused on the investigation of predicted LoF variants which provide directionality of effect. Notably, 14/25 candidates we identified were frameshift deletions that result in either truncated proteins or nonsense-mediated mRNA decay. In the discovery, we observed non-coding variants reside in regulatory regions that may influence target gene expression; however, the lack of population frequency information and insufficient coverage in the validation, limits our ability to explore this aspect for some non-coding variants.
There exist various challenges using the gnomAD as controls, including lack of individual-level data, inability to perform GxE interaction, gene-burden tests, and differences in platforms/coverage. Additionally, there were some racial differences in non-white between TCGA cases (27%) and gnomAD controls (30%), that could cause biased effect sizes in the meta-analysis. Genetic ancestry analysis shows 90% TCGA-LCs were inferred as genetic European ancestry83. However, it is possible that a small portion of European ancestry TCGA-patients has AJ origin, given that 7% of ovarian cancer84 and 24% of endometrial cancer85 are of AJ heritage. It is of note that in our dataset, none of the variant allele carriers of the 25 candidates were found to have African-ancestry. Therefore, we expect this potential population stratification effect to be relatively small on rare variant associations, particularly in non-Africans that have not experienced severe population bottlenecks86–88.
Although we demonstrated strong joint-effect of the 25 potential candidates (Supplementary Table 2), it is challenging to detect tissue-specific eQTL effects, identify mutational signatures, or construct polygenic risk score (PRS) based on these rare or ultra-rare candidates, due to their low frequencies and weak LD among rare or with common variants. We found some lung-tissue specific eQTL variants from The Genotype-Tissue Expression project (GTEx): three SNPs for ATM, 61 SNPs for POMC, 75 SNPs for MPZL2, and 141 SNPs for STAU2; but none of them overlap or are in LD with the 25 candidates we are reporting. Future studies could integrate single-cell transcriptomic sequencing and epigenomic maps in cells and tissues relevant to LC, to establish mutation signatures (i.e., DNA mismatch repair) and explore the application of PRS to clinical care.
In conclusion, our results provide evidence that rare deleterious variants with moderate to large effect sizes, in particular ATM p.V2716A, MPZL2 p.I24M fs*22del, STAU2 p.N364M fs*67del, POMC 3′ UTR c.*28delT, and MLNR p.Q334V fs*3del, contribute to LC susceptibility. Additional targeted studies using CRISPR/Cas9 mutagenesis could be performed for each variant, to evaluate more comprehensively what its effects are on gene functions and the underlying molecular mechanisms. Future extremely large-scale multi-ancestry studies may also provide additional opportunities to assess ancestry-specific predisposing variants, and discover new genetic alterations with relatively large attributable risk for LC.
Methods
Study population in the discovery set
The discovery set included 1094 LC cases and 933 controls from the TRICL study89. All study subjects and biospecimens were collected with informed consent under institutional review board (IRB) approved protocols. Subjects were selected from four sites: Harvard School of Public Health (HSPH), International Agency for Research on Cancer (IARC), University of Liverpool, and Mount Sinai Hospital and Princess Margaret Hospital (MSH-PMH) in Toronto89. Cases were selected because they reported FHLC (first-degree) or were early-onset (<60 yrs) or had specimens available (Table 1). Never smokers were defined as persons who had smoked fewer than 100 cigarettes in their lifetimes. The ethnicities were inferred using FastPop90.
WES and variant calling in the discovery set
WES was performed using captures with Agilent SureSelect v5 (50 Mb, Agilent Technologies) and custom capture targeted known LC-GWAS region91,92 (250 kb). Germline DNA was sequenced at the Center for Inherited Disease Research. The mean on-target coverage was 52x for each sequencing experiment and greater than 97% of on-target bases had a depth greater than 10x. Sequence reads were mapped to the human reference GRCh37/hg19 using the Burrows-Wheeler Aligner. SNVs and indels were called based on the union of raw GATK v3.3-0 and Atlas2. QC process involved the following user-definable criteria: i) low-complexity repeats and segmental duplications were filtered out; ii) quality score ≥20, depth ≥10, and AB ≥ 0.2 for heterozygous calls; iii) call rate ≥0.85; and iv) samples with abnormal heterozygosity rate, sex discordance, <95% completion rates, and unexpected relatedness (identity-by-state >10%) were filtered out.
Rare variant filtering and functional annotation in the discovery set
Following variant calling, rare variants were further enriched by the application of three-steps: i) Variant with MAF < 1% in the gnomAD (NFE ancestry, v2.1); ii) Variants class, including missense, protein-truncating, and regulatory; and iii) Mutation effects, i.e., variant results in protein truncation and predicted to be deleterious from 4/6 prediction tools (SIFT, Polyphen-2, MutationTaster, MutationAssessor, FATHMM, and FATHMM-MKL). The miRNAs putatively bound to the sequence containing UTR variants were identified by the TargetScan35. We additionally incorporated rare variants classified as pathogenic, likely pathogenic, or VUS from the ClinVar database, which compiles clinically observed human variants.
Single variant association test in the discovery set
For variants derived from the above automated filtering schema, we conducted the association test using Fisher’s exact test. We used the Genome Browser (Golden Helix) visualization tool to verify the presence of the potential candidates in each carrier. By manual review of the variants’ coverage plot (read depth) and pile-up plot (read alignment), we rule out low-confidence variants resulting from mapping error, strand bias, and weak exon conservation.
Gene–environment interaction and gene-based burden analysis in the discovery set
For the candidates identified from the association test, we performed G×E interaction (i.e., age-onset, sex, smoking status, pack-years, and FHLC), using the mixed linear regression model. To measure the cumulative effect of the rare deleterious variants within the gene, we performed collapsing tests using the CMC and the KBAC tests93,94.
Study populations in the validation sets and meta-analysis
The candidate variants were further examined in seven validation datasets, aggregated from different centers and across several platforms (four WES data and three genome-wide genotyping datasets as shown in Table 1). We tabulated the variant carrier counts per candidate and performed meta-analyses using the inverse-variance-weighted fixed-effects (assume the true effect size is the same in all studies).
GELCC study (Genetic Epidemiology of LC Consortium, 380 LCs): This included 122 familial and 258 sporadic LC cases. i) Familial LC Study Subjects (dbGaP phs000629.v1.p1). The familial cases were selected from high-risk LC families with at least two first-degree relatives affected with LC95. The GELCC study population and recruitment scheme have been described in detail previously96. Samples and data were collected by the familial LC recruitment sites of the GELCC, that included the University of Cincinnati, University of Colorado Health Science Center, Karmanos Cancer Institute at Wayne State University, Louisiana State University Health Sciences Center-New Orleans, Mayo Clinic, University of Toledo, Johns Hopkins University, and Saccomanno Research Institute. ii) Sporadic LC Study Subjects. The sporadic LC patients were selected from our previous WES study19,20, including samples from the HSPH, Baylor College of Medicine (BCM), and MD Anderson Cancer Center (MDACC). Germline DNA was sequenced utilizing NimbleGen VCRome 2.1 (Roche)19,20, and HumanOmniExpressExome (Illumina)95.
TCGA (The Cancer Genome Atlas cohort, 1015 LCs): this public germline WES dataset includes non-tumor DNA from 577 AD and 438 SCC (dbGaP Phs000178.v9.p8), using Agilent SureSelect (Agilent Technologies) and NimbleGen SeqCap (Roche).
COPDGene (Genetic Epidemiology of COPD Study97, 318 controls): controls were selected to be white, smokers with normal lung function data (defined as post-bronchodilator Forced Expiratory Volume in 1 s [FEV1] ≥ 0 80% predicted, FEV1/FVC ≥ 0.7), and with smoking histories ≥10 pack-years; WES utilized NimbleGen VCRome 2.1 (Roche)19,20.
GnomAD (the Genome Aggregation Database, 134,187 controls): we restricted our analyses to non-cancer individuals (excluded individuals from cancer cohort studies, such as the TCGA cohort), resulting in a data subset of 118,479 exomes and 15,708 whole genomes; multiple exome captures were utilized including Nimblegen SeqCap (Roche), Agilent SureSelect (Agilent Technologies), and Illumina Exome BeadChip (Illumina).
Oncoarray case–control study (17,878 LCs vs. 13,425 controls; dbGaP phs001273): The OncoArray consortium is a network created to increase understanding of the genetic architecture of common cancers. We restricted our analyses to European descent subjects (Supplementary Fig. 1)98–100; participants were obtained from 29 LC studies across North America and Europe, and genotyped on OncoArray-500K BeadChip (Illumina). There were 1162 participants in the OncoArray consortium who were also exome-sequenced in the TRICL discovery, and therefore these samples were excluded from the analysis in the validation phase.
Affymetrix case–control studies (5364 LCs vs. 5724 controls; dbGaP phs001681.v1.p1). This is a large pooled sample was assembled consisting of 10 independent case–control studies which previously described elsewhere99,101. Study participants were genotyped on an Axiom Exome Plus Array (Affymetrix)99,101, which contains a custom panel of key LC GWAS markers, and rare coding SNVs and indels102. There were 992 participants in the Affymetrix that were also exome-sequenced in the TRICL discovery, and therefore these samples were excluded from the analysis in the validation phase.
UKB (UK Biobank cohort103; 2166 LCs vs. 401,453 controls): we restricted our analyses to non-cancer controls and LC cases; individuals were genotyped on UK BiLEVE Axiom Array and UK Biobank Axiom Array (Affymetrix)103,104.
Gene prioritization based on functional annotations and protein interactions network
To better reprioritize genes and candidates, we used three prioritization tools: 1) Gene evolutionary constraint to LoF variation, which using the o/e ratio from the gnomAD. 2) Phevor PhoRank algorithm105, which ranks the genes based on their phenotypic relevance as defined by diverse biomedical ontologies. 3) Protein–Protein interactions (PPI) network using the STRING database106, with an interaction score cut-off ≥0.15 (low confidence).
Functional evaluation of candidate genes using endogenous DNA damage assay
Endogenous DNA damage is proposed to drive cancers by genome instability — a hallmark of cancer37,38. To test whether knockdown or overexpression of the candidate genes or variants induces endogenous DNA damage, we performed flow cytometric assays to measure γH2AX levels, a DNA double-strand-break marker107, following siRNA knockdown and overproduction of GFP fusions of proteins of interest.
Human cell lines and reagents. MRC5-SV40, a human lung fibroblasts derived cell line was maintained in standard Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum, 2 mM L-glutamine, 100 μg/mL penicillin, and 100 μg/mL streptomycin37,38. The cell line was authenticated by ATCC STR analysis and routinely check to be mycoplasma-free. MLNR p.Q334V fs*3del, MME p.P156L fs, MPZL2 p.I24M fs*22del, and full-length wildtype POMC entry clones for gateway cloning was synthesized, sequence-verified, and cloned into pDONR223 (Invitrogen) by Genscipt. All the above clones were further subcloned into an N-terminal GFP tagged vector (pcDNA6.2/N-EmGFP-DEST, Invitrogen), using Gateway LR Clonase II Enzyme Mix (Invitrogen). Overexpression plasmids transfections were performed using GenJet In Vitro DNA Transfection Reagent Ver. II (# SL100489, SignaGen). Non-targeting pool siRNA (D-001810-10), SMARTpool siRNAs each containing four targeting sequences of MME, MLNR, POMC, ATM, CHEK2, and MPZL2, sets of 4 siRNAs targeting MME, MLNR, and POMC were purchased from Dharmacon. The target sequences for MME, MLNR, and POMC are as follows: #1 MME (GGAGGCUGGUUGAAACGUA), #2 MME (GAACCUAUAGGCCAGAGUA), #3 MME (AAAGAUGAGUGGAUAAGUG), #4 MME (GACAGCACCUUAAUGGAAU); #1 MLNR (GCGCUAACGUGAAGACGAU), #2 MLNR (GCGCAUCUAUCAACCCAAU), #3 MLNR (CAUCGUCGCUCUGCAACUU), #4 MLNR (GAAGAUUCGCGGAUGAUGU); #1 POMC (GACAAGCGCUACGGCGGUU), #2 POMC (CAGUGAAGGUGUACCCUAA), #3 POMC (GGCCGAGACUCCCAUGUUC), #4 POMC (CUACAAGAAGGGCGAGUGA). siRNA transfections were carried out with lipofectamine RNAiMax Transfection Reagent (#13778075, Invitrogen), following the manufacturer’s recommendations. SMARTpool ON-TARGETplus siRNA was designed and modified for greater specificity and reduce off-targets up to 90% utilizing a dual-strand modification.
Real-time quantitative reverse transcription PCR (RT-qPCR). Knockdown efficiency was quantified by RT-qPCR and shown in Supplementary Fig. 6. RNeasy mini kit (Qiagen #74106) was used to extract total RNA from cells 72 h post siRNA transfection or protein overproduction. 300 ng of total RNA from each sample was used to synthesize cDNA by the Superscript III first-strand synthesis system (Invitrogen, #18080051). The qPCR reactions were performed using iTaq Universal SYBR Green Supermix (BioRad #172-5121) on a QuantStudio 3 Real-Time PCR System (Applied Biosystems). For each gene, three replicates were analyzed and the average threshold cycle (Ct) was calculated. The relative expression levels were calculated with the 2–ΔΔCt method108. Primers used included GAPDH (housekeeping gene) forward: CAA TGA CCC CTT CAT TGA CC; GAPDH reverse: GAT CTC GCT CCT GGA AGA TG; POMC forward: GCC AGT GTC AGG ACC TCA C; POMC reverse: GGG AAC ATG GGA GTC TCG G; CHEK2 forward: TCT CGG GAG TCG GAT GTT GAG; CHEK2 reverse: CCT GAG TGG ACA CTG TCT CTA A; ATM forward: GGC TAT TCA GTG TGC GAG ACA; ATM reverse: TGG CTC CTT TCG GAT GAT GGA; MPZL2 forward: TTA ATG GGA CAG ATG CTC GGT; MPZL2 reverse: AAG ACA CCC GGT CCT TAA ACC; MME forward: AGA AGA AAC AGC GAT GGA CTC C; MME reverse: CAT AGA GTG CGA TCA TTG TCA CA; MLNR forward (siRNA): CTG AGC GCA TCT ATC AAC CCA; MLNR reverse (siRNA): TCC CAT CGT CTT CAC GTT AGC; MLNR forward (overexpression): GTG GTG ACC GTG ATG CTG AT; MLNR reverse (overexpression): AGC AGG ATG AGT AGG TCG GA.
Flow-cytometric DNA damage assays. Sensitive DNA damage assays by flow cytometry were performed as previously described37,38. γH2AX primary antibody (Sigma, Catalog #05-636) and goat anti-mouse secondary antibody, Alexa Fluor 647 (Thermo Fisher, Catalog #A21236) were used to stain cells. Stained cells were then analyzed by a BD LSRFortessa flow cytometer. FCS files were analyzed by FlowJo 10.5 software. For siRNA experiments, cells were collected 72 h post transfection and median fluorescence intensity was quantified. Also, to quantify the DNA-damage positive subpopulations, 0.5% of the mock cells were gated as the γH2AX threshold as previously demonstrated. The percentage of γH2AX positive cells in each sample was calculated and compared to its corresponding non-targeting siRNA control. For overproduction experiments, mock-transfected cells were used to set the gates to determine the GFP and γH2AX positive cells. 0.5% of the mock cells were gated as the γH2AX threshold. The DNA-damage ratios by protein overproduction for 72 h are calculated as described. Briefly, the damage ratio is defined as (Q2/Q3)/(Q1/Q4), where Q2 is the portion of transfected γH2AX-positive cells; Q3 is the portion of transfected, γH2AX -negative cells; Q1 is the portion of untransfected, γH2AX-positive cells; and Q4 is the portion of untransfected, γH2AX-negative cells. The DNA damage ratios by candidate protein overproduction were compared with GFP-Tubulin as previously described.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
We would like to thank all individuals who participated in this study. This work was supported by grants from the National Institutes of Health (R01CA127219, R01CA141769, R01CA060691, R01CA87895, R01CA80127, R01CA84354, R01CA134682, R01CA134433, R01CA074386, R01CA092824, R01CA250905, R01HL113264, R01HL082487, R01HL110883, R03CA77118, P20GM103534, P30CA125123, P30CA023108, P30CA022453, P30ES006096, P50CA090578, U01CA243483, U01HL089856, U01HL089897, U01CA76293, U19CA148127, U01CA209414, K07CA181480, N01-HG-65404, HHSN268200782096C, HHSN261201300011I, HHSN268201100011, HHSN268201 200007 C, DP1-CA174424, DP1-AG072751, CA125123, RR024574, Intramural Research Program of the National Human Genome Research Institute (JEB-W), and Herrick Foundation. Dr. Amos is an Established Research Scholar of the Cancer Prevention Research Institute of Texas (RR170048). We also want to acknowledge the Cytometry and Cell Sorting Core support by the Cancer Prevention and Research Institute of Texas Core Facility (RP180672). At Toronto, the study is supported by The Canadian Cancer Society Research Institute (# 020214) to R. H., Ontario Institute for Cancer Research to R. H, and the Alan Brown Chair to G. L. and Lusi Wong Programs at the Princess Margaret Hospital Foundation. The Liverpool Lung Project is supported by Roy Castle Lung Cancer Foundation.
Author contributions
Drafted the Paper: Y.L., J.X., and C.I.A. Project Coordination: C.I.A., R.J.H., D.C.C., and P.B. Statistical Analysis: Y.L., S.T., X.X., D.Z., C.W.P., C.M.L., and C.I.A. Genetic validation analysis: Y.L., S.T., X.X., C.C., Y.Li., J.B., D.Z., W.H., C.W.P., C.M.L., and C.I.A. Functional DNA damage assay analysis: J.X., Z.S., and S.M.R. Sample collection, exome sequencing, and development of the epidemiological studies: J.M., M.R.S., M.E.S., F.K., C.M.L., A.G.S., I.I.W., M.H.C., E.K.S., J.B.W., S.M.P., M.A., E.K., C.G., D.M., M.Y., M.dA., P.Y., T, M.P.A.D., J.L., B.S., D.Z., A.M., V.J., I.H., D.M., J.S., G.S., P.B., G.L., J.K.F., R.J.H., D.C.C., and C.I.A.
Data availability
The data generated and/or analyzed during the related study are described in the figshare metadata record: 10.6084/m9.figshare.13280387109. The data that support the findings of this study are available via the dbGaP (database of genotypes and phenotypes) repository. The data are controlled-access, so interested parties will need to request access — information on how to do so can be found on pages linked to below. The access numbers are https://identifiers.org/dbgap:phs000878.v2.p1110 for Transdisciplinary Research in Cancer of the Lung (TRICL) study, https://identifiers.org/dbgap:phs001273.v1.p1111 for the OncoArray study, https://identifiers.org/dbgap:phs001681.v1.p1112 for the Affymetrix study, https://identifiers.org/dbgap:phs000629.v1.p1113 for part of the Genetic Epidemiology of Lung Cancer Consortium (GELCC) study, and https://identifiers.org/dbgap:phs000178.v9.p8114 for The Cancer Genome Atlas (TCGA) study. Two files are not publicly available in order to protect patient privacy. These are: ‘TRICL WES.xlsx’ (underlying Supplementary Table 2 and Supplementary Fig. 3) and ‘TRICL WES.bam’ (underlying Supplementary Fig. 2). These data are only available to authorized researchers who have submitted an IRB application. Please email the corresponding author for access. Data underlying Supplementary Table 5 and Supplementary Fig. 5 are a publicly available resource available from the STRING (Search Tool for the Retrieval of Interacting Genes) website: http://string-db.org/. The file used in this study was ‘Protein-Protein Interaction Networks Functional Enrichment Analysis-STRING.txt’.
Sources of other datasets used in this study are: the UKB dataset is accessible to approved researchers and applications through ukbgene at www.ukbiobank.ac.uk. The GnomAD dataset can be downloaded from the Genome Aggregation Database at https://gnomad.broadinstitute.org/.
Competing interests
E.K.S. reports institutional grant funding from Bayer and GlaxoSmithKline. Other authors declare no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Yanhong Liu, Jun Xia.
Supplementary information
The online version contains supplementary material available at 10.1038/s41698-021-00146-7.
References
- 1.Rizvi NA, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bosse Y, Amos CI. A decade of GWAS results in lung cancer. Cancer Epidemiol. Biomark. Prev. 2018;27:363–379. doi: 10.1158/1055-9965.EPI-16-0794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wei C, et al. A case-control study of a sex-specific association between a 15q25 variant and lung cancer risk. Cancer Epidemiol. Biomark. Prev. 2011;20:2603–2609. doi: 10.1158/1055-9965.EPI-11-0749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bierut LJ, et al. Variants in nicotinic receptors and risk for nicotine dependence. Am. J. Psychiatry. 2008;165:1163–1171. doi: 10.1176/appi.ajp.2008.07111711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, L. S., et al. CHRNA5 risk variant predicts delayed smoking cessation and earlier lung cancer diagnosis–a meta-analysis. J. Natl Cancer Inst.107, djv100 (2015). [DOI] [PMC free article] [PubMed]
- 6.Chen LS, et al. Interplay of genetic risk factors (CHRNA5-CHRNA3-CHRNB4) and cessation treatments in smoking cessation success. Am. J. Psychiatry. 2012;169:735–742. doi: 10.1176/appi.ajp.2012.11101545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mucci LA, et al. Familial risk and heritability of cancer among twins in Nordic countries. JAMA. 2016;315:68–76. doi: 10.1001/jama.2015.17703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kang G, Lin D, Hakonarson H, Chen J. Two-stage extreme phenotype sequencing design for discovering and testing common and rare genetic variants: efficiency and power. Hum. Hered. 2012;73:139–147. doi: 10.1159/000337300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lamina C. Digging into the extremes: a useful approach for the analysis of rare variants with continuous traits? BMC Proc. 2011;5(Suppl. 9):S105. doi: 10.1186/1753-6561-5-S9-S105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li D, Lewinger JP, Gauderman WJ, Murcray CE, Conti D. Using extreme phenotype sampling to identify the rare causal variants of quantitative traits in association studies. Genet. Epidemiol. 2011;35:790–799. doi: 10.1002/gepi.20628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gorlov IP, Gorlova OY, Sunyaev SR, Spitz MR, Amos CI. Shifting paradigm of association studies: value of rare single-nucleotide polymorphisms. Am. J. Hum. Genet. 2008;82:100–112. doi: 10.1016/j.ajhg.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gorlov IP, Gorlova OY, Frazier ML, Spitz MR, Amos CI. Evolutionary evidence of the effect of rare variants on disease etiology. Clin. Genet. 2011;79:199–206. doi: 10.1111/j.1399-0004.2010.01535.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tennessen JA, et al. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337:64–69. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi YW, et al. EGFR exon 19 deletion is associated with favorable overall survival after first-line gefitinib therapy in advanced non-small cell lung cancer patients. Am. J. Clin. Oncol. 2018;41:385–390. doi: 10.1097/COC.0000000000000282. [DOI] [PubMed] [Google Scholar]
- 15.Sequist LV, et al. First-line gefitinib in patients with advanced non-small-cell lung cancer harboring somatic EGFR mutations. J. Clin. Oncol. 2008;26:2442–2449. doi: 10.1200/JCO.2007.14.8494. [DOI] [PubMed] [Google Scholar]
- 16.Tian Y, et al. Different subtypes of EGFR exon19 mutation can affect prognosis of patients with non-small cell lung adenocarcinoma. PLoS ONE. 2018;13:e0201682. doi: 10.1371/journal.pone.0201682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiong D, et al. A recurrent mutation in PARK2 is associated with familial lung cancer. Am. J. Hum. Genet. 2015;96:301–308. doi: 10.1016/j.ajhg.2014.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y, et al. Rare variants of large effect in BRCA2 and CHEK2 affect risk of lung cancer. Nat. Genet. 2014;46:736–741. doi: 10.1038/ng.3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Y, et al. Rare variants in known susceptibility loci and their contribution to risk of lung cancer. J. Thorac. Oncol. 2018;13:1483–1495. doi: 10.1016/j.jtho.2018.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y, et al. Focused analysis of exome sequencing data for rare germline mutations in familial and sporadic lung cancer. J. Thorac. Oncol. 2016;11:52–61. doi: 10.1016/j.jtho.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ji X, et al. Protein-altering germline mutations implicate novel genes related to lung cancer development. Nat. Commun. 2020;11:2220. doi: 10.1038/s41467-020-15905-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peng B, Li B, Han Y, Amos CI. Power analysis for case-control association studies of samples with known family histories. Hum. Genet. 2010;127:699–704. doi: 10.1007/s00439-010-0824-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Osann KE. Lung cancer in women: the importance of smoking, family history of cancer, and medical history of respiratory disease. Cancer Res. 1991;51:4893–4897. [PubMed] [Google Scholar]
- 24.Cote ML, et al. Increased risk of lung cancer in individuals with a family history of the disease: a pooled analysis from the International Lung Cancer Consortium. Eur. J. Cancer. 2012;48:1957–1968. doi: 10.1016/j.ejca.2012.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loman NJ, et al. Performance comparison of benchtop high-throughput sequencing platforms. Nat. Biotechnol. 2012;30:434–439. doi: 10.1038/nbt.2198. [DOI] [PubMed] [Google Scholar]
- 26.Albers CA, et al. Dindel: accurate indel calls from short-read data. Genome Res. 2011;21:961–973. doi: 10.1101/gr.112326.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Minoche AE, Dohm JC, Himmelbauer H. Evaluation of genomic high-throughput sequencing data generated on Illumina HiSeq and genome analyzer systems. Genome Biol. 2011;12:R112. doi: 10.1186/gb-2011-12-11-r112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Balzer S, Malde K, Jonassen I. Systematic exploration of error sources in pyrosequencing flowgram data. Bioinformatics. 2011;27:i304–i309. doi: 10.1093/bioinformatics/btr251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y, et al. Deciphering associations for lung cancer risk through imputation and analysis of 12,316 cases and 16,831 controls. Eur. J. Hum. Genet. 2015;23:1723–1728. doi: 10.1038/ejhg.2015.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dong J, et al. Association analyses identify multiple new lung cancer susceptibility loci and their interactions with smoking in the Chinese population. Nat. Genet. 2012;44:895–899. doi: 10.1038/ng.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McKay JD, et al. Large-scale association analysis identifies new lung cancer susceptibility loci and heterogeneity in genetic susceptibility across histological subtypes. Nat. Genet. 2017;49:1126–1132. doi: 10.1038/ng.3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shrine N, et al. New genetic signals for lung function highlight pathways and chronic obstructive pulmonary disease associations across multiple ancestries. Nat. Genet. 2019;51:481–493. doi: 10.1038/s41588-018-0321-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu Z, et al. Genetic overlap of chronic obstructive pulmonary disease and cardiovascular disease-related traits: a large-scale genome-wide cross-trait analysis. Respir. Res. 2019;20:64. doi: 10.1186/s12931-019-1036-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kichaev G, et al. Leveraging polygenic functional enrichment to improve GWAS power. Am. J. Hum. Genet. 2019;104:65–75. doi: 10.1016/j.ajhg.2018.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 36.Fagerberg L, et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell Proteom. 2014;13:397–406. doi: 10.1074/mcp.M113.035600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xia J, et al. Bacteria-to-human protein networks reveal origins of endogenous DNA damage. Cell. 2019;176:127–143 e124. doi: 10.1016/j.cell.2018.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bosse Y, et al. Transcriptome-wide association study reveals candidate causal genes for lung cancer. Int. J. Cancer. 2020;146:1862–1878. doi: 10.1002/ijc.32771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Selvan ME, et al. Inherited rare, deleterious variants in ATM increase lung adenocarcinoma risk. J. Thorac. Oncol. 2020;15:1871–1879. doi: 10.1016/j.jtho.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parry EM, et al. Germline mutations in DNA repair genes in lung adenocarcinoma. J. Thorac. Oncol. 2017;12:1673–1678. doi: 10.1016/j.jtho.2017.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang H, et al. ATM sequence variants associate with susceptibility to non-small cell lung cancer. Int. J. Cancer. 2007;121:2254–2259. doi: 10.1002/ijc.22918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lo YL, et al. ATM polymorphisms and risk of lung cancer among never smokers. Lung Cancer. 2010;69:148–154. doi: 10.1016/j.lungcan.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 43.Hsia TC, et al. Effects of ataxia telangiectasia mutated (ATM) genotypes and smoking habits on lung cancer risk in Taiwan. Anticancer Res. 2013;33:4067–4071. [PubMed] [Google Scholar]
- 44.Chenevix-Trench G, et al. Dominant negative ATM mutations in breast cancer families. J. Natl Cancer Inst. 2002;94:205–215. doi: 10.1093/jnci/94.3.205. [DOI] [PubMed] [Google Scholar]
- 45.Morgan SE, Lovly C, Pandita TK, Shiloh Y, Kastan MB. Fragments of ATM which have dominant-negative or complementing activity. Mol. Cell Biol. 1997;17:2020–2029. doi: 10.1128/MCB.17.4.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 47.Scott SP, et al. Missense mutations but not allelic variants alter the function of ATM by dominant interference in patients with breast cancer. Proc. Natl Acad. Sci. USA. 2002;99:925–930. doi: 10.1073/pnas.012329699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kuhne M, et al. A double-strand break repair defect in ATM-deficient cells contributes to radiosensitivity. Cancer Res. 2004;64:500–508. doi: 10.1158/0008-5472.CAN-03-2384. [DOI] [PubMed] [Google Scholar]
- 49.Dai J, et al. Genome-wide association study of INDELs identified four novel susceptibility loci associated with lung cancer risk. Int. J. Cancer. 2020;146:2855–2864. doi: 10.1002/ijc.32698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bademci G, et al. MPZL2 is a novel gene associated with autosomal recessive nonsyndromic moderate hearing loss. Hum. Genet. 2018;137:479–486. doi: 10.1007/s00439-018-1901-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wesdorp M, et al. MPZL2, encoding the epithelial junctional protein myelin protein zero-like 2, is essential for hearing in man and mouse. Am. J. Hum. Genet. 2018;103:74–88. doi: 10.1016/j.ajhg.2018.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guttinger M, et al. Epithelial V-like antigen (EVA), a novel member of the immunoglobulin superfamily, expressed in embryonic epithelia with a potential role as homotypic adhesion molecule in thymus histogenesis. J. Cell Biol. 1998;141:1061–1071. doi: 10.1083/jcb.141.4.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Einhorn Y, et al. Differential analysis of mutations in the Jewish population and their implications for diseases. Genet. Res. 2017;99:e3. doi: 10.1017/S0016672317000015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shi L, et al. Comprehensive population screening in the Ashkenazi Jewish population for recurrent disease-causing variants. Clin. Genet. 2017;91:599–604. doi: 10.1111/cge.12834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kerem B, Chiba-Falek O, Kerem E. Cystic fibrosis in Jews: frequency and mutation distribution. Genet. Test. 1997;1:35–39. doi: 10.1089/gte.1997.1.35. [DOI] [PubMed] [Google Scholar]
- 56.Powers J, et al. A rare TP53 mutation predominant in Ashkenazi Jews confers risk of multiple cancers. Cancer Res. 2020;80:3732–3744. doi: 10.1158/0008-5472.CAN-20-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Picciotto MR, Mineur YS. Molecules and circuits involved in nicotine addiction: the many faces of smoking. Neuropharmacology. 2014;76 Pt B:545–553. doi: 10.1016/j.neuropharm.2013.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang H, Xu Y, van den Pol AN. Nicotine excites hypothalamic arcuate anorexigenic proopiomelanocortin neurons and orexigenic neuropeptide Y neurons: similarities and differences. J. Neurophysiol. 2011;106:1191–1202. doi: 10.1152/jn.00740.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mineur YS, et al. Nicotine decreases food intake through activation of POMC neurons. Science. 2011;332:1330–1332. doi: 10.1126/science.1201889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wenczl E, et al. (Pheo)melanin photosensitizes UVA-induced DNA damage in cultured human melanocytes. J. Invest. Dermatol. 1998;111:678–682. doi: 10.1046/j.1523-1747.1998.00357.x. [DOI] [PubMed] [Google Scholar]
- 61.Cui R, et al. Central role of p53 in the suntan response and pathologic hyperpigmentation. Cell. 2007;128:853–864. doi: 10.1016/j.cell.2006.12.045. [DOI] [PubMed] [Google Scholar]
- 62.Suzuki I, et al. Increase of pro-opiomelanocortin mRNA prior to tyrosinase, tyrosinase-related protein 1, dopachrome tautomerase, Pmel-17/gp100, and P-protein mRNA in human skin after ultraviolet B irradiation. J. Invest. Dermatol. 2002;118:73–78. doi: 10.1046/j.1523-1747.2002.01647.x. [DOI] [PubMed] [Google Scholar]
- 63.Slominski A, Tobin DJ, Paus R. Does p53 regulate skin pigmentation by controlling proopiomelanocortin gene transcription? Pigment Cell Res. 2007;20:307–308. doi: 10.1111/j.1600-0749.2007.00390.x. [DOI] [PubMed] [Google Scholar]
- 64.Krude H, et al. Severe early-onset obesity, adrenal insufficiency and red hair pigmentation caused by POMC mutations in humans. Nat. Genet. 1998;19:155–157. doi: 10.1038/509. [DOI] [PubMed] [Google Scholar]
- 65.Tsai HE, et al. Downregulation of hepatoma-derived growth factor contributes to retarded lung metastasis via inhibition of epithelial-mesenchymal transition by systemic POMC gene delivery in melanoma. Mol. Cancer Ther. 2013;12:1016–1025. doi: 10.1158/1535-7163.MCT-12-0832. [DOI] [PubMed] [Google Scholar]
- 66.Stovold R, et al. Neuroendocrine and epithelial phenotypes in small-cell lung cancer: implications for metastasis and survival in patients. Br. J. Cancer. 2013;108:1704–1711. doi: 10.1038/bjc.2013.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Meredith SL, et al. Irradiation decreases the neuroendocrine biomarker pro-opiomelanocortin in small cell lung cancer cells in vitro and in vivo. PLoS ONE. 2016;11:e0148404. doi: 10.1371/journal.pone.0148404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hao L, Zhao X, Zhang B, Li C, Wang C. Positive expression of pro-opiomelanocortin (POMC) is a novel independent poor prognostic marker in surgically resected non-small cell lung cancer. Tumour Biol. 2015;36:1811–1817. doi: 10.1007/s13277-014-2784-1. [DOI] [PubMed] [Google Scholar]
- 69.Derghal A, et al. Leptin modulates the expression of miRNAs-targeting POMC mRNA by the JAK2-STAT3 and PI3K-Akt pathways. J. Clin. Med. 2019;8:2213–2224. doi: 10.3390/jcm8122213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Feighner SD, et al. Receptor for motilin identified in the human gastrointestinal system. Science. 1999;284:2184–2188. doi: 10.1126/science.284.5423.2184. [DOI] [PubMed] [Google Scholar]
- 71.Xu HL, et al. Variants in motilin, somatostatin and their receptor genes and risk of biliary tract cancers and stones in Shanghai, China. Meta Gene. 2014;2:418–426. doi: 10.1016/j.mgene.2014.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Misawa K, et al. Neuropeptide receptor genes GHSR and NMUR1 are candidate epigenetic biomarkers and predictors for surgically treated patients with oropharyngeal cancer. Sci. Rep. 2020;10:1007. doi: 10.1038/s41598-020-57920-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Delahaye-Sourdeix M, et al. A rare truncating BRCA2 variant and genetic susceptibility to upper aerodigestive tract cancer. J. Natl Cancer Inst. 2015;107:djv037. doi: 10.1093/jnci/djv037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cybulski C, et al. Constitutional CHEK2 mutations are associated with a decreased risk of lung and laryngeal cancers. Carcinogenesis. 2008;29:762–765. doi: 10.1093/carcin/bgn044. [DOI] [PubMed] [Google Scholar]
- 75.Brennan P, et al. Uncommon CHEK2 mis-sense variant and reduced risk of tobacco-related cancers: case control study. Hum. Mol. Genet. 2007;16:1794–1801. doi: 10.1093/hmg/ddm127. [DOI] [PubMed] [Google Scholar]
- 76.Shaag A, et al. Functional and genomic approaches reveal an ancient CHEK2 allele associated with breast cancer in the Ashkenazi Jewish population. Hum. Mol. Genet. 2005;14:555–563. doi: 10.1093/hmg/ddi052. [DOI] [PubMed] [Google Scholar]
- 77.Roeb W, Higgins J, King MC. Response to DNA damage of CHEK2 missense mutations in familial breast cancer. Hum. Mol. Genet. 2012;21:2738–2744. doi: 10.1093/hmg/dds101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kilpivaara O, et al. CHEK2 variant I157T may be associated with increased breast cancer risk. Int. J. Cancer. 2004;111:543–547. doi: 10.1002/ijc.20299. [DOI] [PubMed] [Google Scholar]
- 79.Apostolou P, Papasotiriou I. Current perspectives on CHEK2 mutations in breast cancer. Breast Cancer. 2017;9:331–335. doi: 10.2147/BCTT.S111394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Furic L, Maher-Laporte M, DesGroseillers L. A genome-wide approach identifies distinct but overlapping subsets of cellular mRNAs associated with Staufen1- and Staufen2-containing ribonucleoprotein complexes. RNA. 2008;14:324–335. doi: 10.1261/rna.720308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang X, et al. The downregulation of the RNA-binding protein Staufen2 in response to DNA damage promotes apoptosis. Nucleic Acids Res. 2016;44:3695–3712. doi: 10.1093/nar/gkw057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Conde, L., Beaujois, R. & DesGroseillers, L. STAU2 protein level is controlled by caspases and the CHK1 pathway and regulates cell cycle progression in the non-transformed hTERT-RPE1 cells. Preprint from Research Square, 10.21203/rs.21203.rs-60003/v21201 PPR: PPR206819 (2020). [DOI] [PMC free article] [PubMed]
- 83.Yuan J, et al. Integrated analysis of genetic ancestry and genomic alterations across cancers. Cancer Cell. 2018;34:549–560.e549. doi: 10.1016/j.ccell.2018.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yang D, et al. Association of BRCA1 and BRCA2 mutations with survival, chemotherapy sensitivity, and gene mutator phenotype in patients with ovarian cancer. JAMA. 2011;306:1557–1565. doi: 10.1001/jama.2011.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cadoo KA. Understanding inherited risk in unselected newly diagnosed patients with endometrial cancer. JCO Precis. Oncol. 2019;3:473–474. doi: 10.1200/PO.18.00338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.O’Connor TD, et al. Fine-scale patterns of population stratification confound rare variant association tests. PLoS ONE. 2013;8:e65834. doi: 10.1371/journal.pone.0065834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lek M, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Genomes Project C, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang, Z., et al. Multi-omics analysis reveals a HIF network and Hub gene EPAS1 associated with lung adenocarcinoma. EBioMedicine, 93–101 (2018). [DOI] [PMC free article] [PubMed]
- 90.Li Y, et al. FastPop: a rapid principal component derived method to infer intercontinental ancestry using genetic data. BMC Bioinform. 2016;17:122. doi: 10.1186/s12859-016-0965-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bainbridge MN, et al. Targeted enrichment beyond the consensus coding DNA sequence exome reveals exons with higher variant densities. Genome Biol. 2011;12:R68. doi: 10.1186/gb-2011-12-7-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lupski JR, et al. Exome sequencing resolves apparent incidental findings and reveals further complexity of SH3TC2 variant alleles causing Charcot-Marie-Tooth neuropathy. Genome Med. 2013;5:57. doi: 10.1186/gm461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li B, Leal SM. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am. J. Hum. Genet. 2008;83:311–321. doi: 10.1016/j.ajhg.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Liu DJ, Leal SM. A novel adaptive method for the analysis of next-generation sequencing data to detect complex trait associations with rare variants due to gene main effects and interactions. PLoS Genet. 2010;6:e1001156. doi: 10.1371/journal.pgen.1001156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Musolf AM, et al. Whole exome sequencing of highly aggregated lung cancer families reveals linked loci for increased cancer risk on chromosomes 12q, 7p, and 4q. Cancer Epidemiol. Biomark. Prev. 2020;29:434–442. doi: 10.1158/1055-9965.EPI-19-0887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Liu P, et al. Familial aggregation of common sequence variants on 15q24-25.1 in lung cancer. J. Natl Cancer Inst. 2008;100:1326–1330. doi: 10.1093/jnci/djn268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Regan EA, et al. Genetic epidemiology of COPD (COPDGene) study design. COPD. 2010;7:32–43. doi: 10.3109/15412550903499522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ji X, et al. Identification of susceptibility pathways for the role of chromosome 15q25.1 in modifying lung cancer risk. Nat. Commun. 2018;9:3221. doi: 10.1038/s41467-018-05074-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Li Y, et al. Genetic interaction analysis among oncogenesis-related genes revealed novel genes and networks in lung cancer development. Oncotarget. 2019;10:1760–1774. doi: 10.18632/oncotarget.26678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Byun J, et al. Genome-wide association study of familial lung cancer. Carcinogenesis. 2018;39:1135–1140. doi: 10.1093/carcin/bgy080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kachuri L, et al. Fine mapping of chromosome 5p15.33 based on a targeted deep sequencing and high density genotyping identifies novel lung cancer susceptibility loci. Carcinogenesis. 2016;37:96–105. doi: 10.1093/carcin/bgv165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zuzarte PC, et al. A two-dimensional pooling strategy for rare variant detection on next-generation sequencing platforms. PLoS ONE. 2014;9:e93455. doi: 10.1371/journal.pone.0093455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Matthews PM, Sudlow C. The UK Biobank. Brain. 2015;138:3463–3465. doi: 10.1093/brain/awv335. [DOI] [PubMed] [Google Scholar]
- 104.Bycroft C, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Singleton MV, et al. Phevor combines multiple biomedical ontologies for accurate identification of disease-causing alleles in single individuals and small nuclear families. Am. J. Hum. Genet. 2014;94:599–610. doi: 10.1016/j.ajhg.2014.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Szklarczyk D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kinner A, Wu W, Staudt C, Iliakis G. Gamma-H2AX in recognition and signaling of DNA double-strand breaks in the context of chromatin. Nucleic Acids Res. 2008;36:5678–5694. doi: 10.1093/nar/gkn550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 109.Liu, Y. Metadata record for the manuscript: rare deleterious germline variants and risk of lung cancer. figshare10.6084/m9.figshare.13280387 (2020).
- 110.Transdisciplinary Research Into Cancer of the Lung (TRICL) - Exome Plus Targeted Sequencing. dbGaPhttps://identifiers.org/dbgap:phs000878.v2.p1.
- 111.Oncoarray Consortium - Lung Cancer Studies. dbGaPhttps://identifiers.org/dbgap:phs001273.v1.p1.
- 112.Transdisciplinary Research Into Cancer of the Lung (TRICL) – Affymetrix. dbGaPhttps://identifiers.org/dbgap:phs001681.v1.p1.
- 113.Genetic Epidemiology of Lung Cancer Consortium GWAS of Familial Lung Cancer. dbGaPhttps://identifiers.org/dbgap:phs000629.v1.p1.
- 114.National Institutes of Health The Cancer Genome Atlas (TCGA). dbGaPhttps://identifiers.org/dbgap:phs000178.v9.p8.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated and/or analyzed during the related study are described in the figshare metadata record: 10.6084/m9.figshare.13280387109. The data that support the findings of this study are available via the dbGaP (database of genotypes and phenotypes) repository. The data are controlled-access, so interested parties will need to request access — information on how to do so can be found on pages linked to below. The access numbers are https://identifiers.org/dbgap:phs000878.v2.p1110 for Transdisciplinary Research in Cancer of the Lung (TRICL) study, https://identifiers.org/dbgap:phs001273.v1.p1111 for the OncoArray study, https://identifiers.org/dbgap:phs001681.v1.p1112 for the Affymetrix study, https://identifiers.org/dbgap:phs000629.v1.p1113 for part of the Genetic Epidemiology of Lung Cancer Consortium (GELCC) study, and https://identifiers.org/dbgap:phs000178.v9.p8114 for The Cancer Genome Atlas (TCGA) study. Two files are not publicly available in order to protect patient privacy. These are: ‘TRICL WES.xlsx’ (underlying Supplementary Table 2 and Supplementary Fig. 3) and ‘TRICL WES.bam’ (underlying Supplementary Fig. 2). These data are only available to authorized researchers who have submitted an IRB application. Please email the corresponding author for access. Data underlying Supplementary Table 5 and Supplementary Fig. 5 are a publicly available resource available from the STRING (Search Tool for the Retrieval of Interacting Genes) website: http://string-db.org/. The file used in this study was ‘Protein-Protein Interaction Networks Functional Enrichment Analysis-STRING.txt’.
Sources of other datasets used in this study are: the UKB dataset is accessible to approved researchers and applications through ukbgene at www.ukbiobank.ac.uk. The GnomAD dataset can be downloaded from the Genome Aggregation Database at https://gnomad.broadinstitute.org/.