Abstract
Artemisinin-based combination therapies (ACT) are currently used as a first-line malaria therapy in endemic countries worldwide. This systematic review aims at presenting the current scenario of drug resistance molecular markers, either selected or involved in treatment failures (TF) during in vivo ACT efficacy studies from sub-Saharan Africa (sSA) and India. Eight electronic databases were comprehensively used to search relevant articles and finally a total of 28 studies were included in the review, 21 from sSA and seven from India. On analysis, Artemether + lumefantrine (AL) and artesunate + sulfadoxine-pyrimethamine (AS + SP) are the main ACT in African and Indian regions with a 28-day efficacy range of 54.3–100% for AL and 63–100% for AS + SP respectively. It was observed that mutations in the Pfcrt (76T), Pfdhfr (51I, 59R, 108N), Pfdhps (437G) and Pfmdr1 (86Y, 184F, 1246Y) genes were involved in TF, which varied with respect to ACTs. Based on studies that have genotyped the Pfk13 gene, the reported TF cases, were mainly linked with mutations in genes associated with resistance to ACT partner drugs; indicating that the protection of the partner drug efficacy is crucial for maintaining the efficacy of ACT. This review reveals that ACT are largely efficacious in India and sSA despite the fact that some clinical efficacy and epidemiological studies have reported some validated mutations (i.e., 476I, 539T and 561H) in circulation in these two regions. Also, the role of PfATPase6 in ART resistance is controversial still, while P. falciparum plasmepsin 2 (Pfpm2) in piperaquine (PPQ) resistance and dihydroartemisinin (DHA) + PPQ failures is well documented in Southeast Asian countries but studied less in sSA. Hence, there is a need for continuous molecular surveillance of Pfk13 mutations for emergence of artemisinin (ART) resistance in these countries.
Keywords: Artemisinin-based combination therapy (ACT), Drug resistance molecular markers, Failure treatment, Sub-Saharan African countries, India
Graphical abstract
Highlights
-
•
Mutations in Pfcrt, Pfdhfr, Pfdhps and Pfmdr1 were involved in treatment failures.
-
•
The direction of selection patterns of these mutations depended on the ACT used in the therapy.
-
•
Pfk13 (476I, 539T, 561H) mutations were already reported from sSA and India.
-
•
More studies needed on PfATPase6 and Pfpm2, to explore their link with ART and PPQ resistance.
-
•
This review reveals that currently, ACT are still largely efficacious against malaria in India and sSA.
1. Introduction
Malaria is an infectious disease caused in humans by Plasmodium parasites transmitted through bites of infected female Anopheles mosquitoes during their blood meal (White et al., 2014). Plasmodium falciparum and Plasmodium vivax are the main human species involved in malaria cases and deaths, causing approximately 229 million cases and 409,000 deaths worldwide in 2019 (WHO, 2020). P. falciparum is the most dangerous between the two malarial species with an enormous threat to the public health in several endemic countries (WHO, 2020).
Early diagnosis and prompt treatment with effective antimalarial drugs are the two key components of malaria control and elimination (WHO, 2018a). Since the discovery of artemisinin (ART) by Professor Tu Youyou, these medicines have continuously gained importance as an alternate drug, for their high efficacy level against malaria parasites as there has been increased prevalence of resistance to older antimalarial drugs (chloroquine-CQ, sulfadoxine-pyrimethamine-SP) reported from different areas (Blasco et al., 2017; Tu, 2015). The malaria therapy currently includes ACT as first- and second-line treatment in most endemic countries which consists of ART combination (or one of its derivatives) with other drug (s) (or two other antimalarials) referred to as partner drugs (WHO, 2018a). The drug combinations in the ACT not only have different mechanism leading to their improved efficacy but also, the chances of emergence of drug resistance to each component drugs are lower (Ashley et al., 2018). Six ACT are currently recommended by the World Health Organization (WHO) for treating malaria cases worldwide: i) artesunate + amodiaquine (AS + AQ), ii) artemether + lumefantrine (AL), iii) artesunate + sulfadoxine-pyrimethamine (AS + SP), iv) artesunate + mefloquine (AS + MQ), v) artesunate + pyronaridine (AS + PY) and vi) dihydroartemisinin + piperaquine (DHA + PPQ) (WHO, 2019). These different ACT are an essential part of the treatment policies in several malaria endemic countries (Bennett et al., 2017; Recht et al., 2017).
P. falciparum has developed resistance mechanisms to most antimalarial drugs in vogue including the current ACT where the only known and validated ART resistance foci are the Greater Mekong subregion (GMS) in Southeast Asia (Antony and Parija, 2016; Menard and Dondorp, 2017). The emergence and spread of ACT resistant malaria parasites have strongly hindered the control efforts in this area and have led initially to increased morbidity and mortality (Chookajorn, 2018). The appearance and/or spread of malaria parasites resistant to ACT in highly malaria burdened parts of the world such as sSA countries and India are a major concern for the scientist community, along with all the stakeholders in the fight against malaria.
The WHO advocates total compliance of the malaria treatment policy by clinicians, program officers and patients in order to prevent the emergence and propagation of ART resistance (WHO, 2018a). With this view, as an essential part of the Global technical strategy (GTS) for malaria 2016–2030, WHO has outlined the need for a continuous monitoring of ACT efficacy through surveillance systems where one of the main targets in the GTS report is the elimination of malaria in 20–30 countries (which were endemic in 2015) by 2025–2030 (WHO, 2016). The evaluation of therapeutic efficacy is the reference method for monitoring the efficacy of antimalarial drugs and thereafter informing national malaria program for revisions in the treatment policy. The prevalence of polymorphisms in molecular markers associated with drug resistance can be analyzed in parallel to support findings from clinical studies (Maji, 2018).
Some reviews published earlier have focused on either the efficacy of ACT or the prevalence of polymorphisms in drug resistance molecular markers and only one reported study from Africa has both of these aspects but there are no reports yet from India (Adam et al., 2018; Conrad and Rosenthal, 2019; Gogtay et al., 2013; Menard and Dondorp, 2017; Price et al., 2014; Prosser et al., 2018; Whegang et al., 2019). In the present review, the current knowledge on antimalarial drug resistance and ART resistance is briefly summarized (WHO, 2018a). In addition, we conducted a systematic review (SR) to summarize findings from existing literature on the current scenario of molecular markers which were either selected or involved in treatment failures after ACT-based treatment. Finally, a comparative analysis of these findings between sSA countries and India is presented and discussed in this review.
2. Antimalarial drug resistance: definition, genetic basis and molecular markers
Antimalarial drug resistance is one of the greatest threat to the control and elimination of malaria globally (WHO, 2018a). The drug resistance is defined as the ability of a parasite strain to survive and/or multiply despite the administration and the absorption of a drug, given in doses equal to or higher than those usually recommended but within the tolerance of the subject (WHO, 1967). Resistance to antimalarial drugs has only been documented in P. falciparum and P. vivax and the major mechanism responsible for acquisition of this phenotype is due to the mutations in genes associated with drug resistance or increase in their gene copy number (Menard and Dondorp, 2017). P. falciparum resistance has been demonstrated to most classes of antimalarial drugs including quinolines, antifolate and ART derivatives while in P. vivax, resistance has been documented for CQ, primaquine and antifolate drugs in several endemic countries (Bansal et al., 2017; Haldar et al., 2018; Price et al., 2014). Regarding P. falciparum, the drug resistance-associated genes include chloroquine resistance transporter (Pfcrt), Pf multidrug resistance protein 1 (Pfmdr1), Pf dihydrofolate reductase (Pfdhfr), Pf dihydropteroate synthase (Pfdhps), Pf cytochrome B (Pfcytb) and Pf sodium–hydrogen exchanger gene (Pfnhe-1) (Table 1) (Antony and Parija, 2016; Menard and Dondorp, 2017). The well-known single nucleotide polymorphisms (SNPs) associated with different drug resistance-associated genes are 76T in Pfcrt, 51I, 59R and 108N in Pfdhfr, 437G and 540E in Pfdhps; 268N/S/C in Pfcytb, 86Y, 184Y and 1246Y in Pfmdr1 for P. falciparum (Table 1) (Adjekukor, 2018; Korsinczky et al., 2000; Peterson et al., 1988; Pickard et al., 2003; Singh Sidhu et al., 2002; Triglia et al., 1997). A recent review on the countries in the horn of Africa, revealed a high prevalence of Pfcrt mutations while mutations in Pfmdr1 and PfATPase6 genes were low in Ethiopia, Somalia, Djibouti and Eritrea (Jalei et al., 2018). Another report by Berzosa and colleagues showed a prevalence of 78% and 2% of Pfdhfr triple mutants (51I/59R/108N) and Pfmdr1 double mutants (86Y + 1246Y) respectively in Equatorial Guinea, a Central African country (Berzosa et al., 2017).
Table 1.
Summary of current molecular markers associated with antimalarial drug resistance.
| Genes (accession numbers) | Chromosome location | Common codon mutations linked to drug resistance in the various genes | Antimalarial drugs/Drug class |
|---|---|---|---|
| Pfcrt (PF3D7_0709000)§ | 7 | Mutations (76T) | Chloroquine and Amodiaquine |
| Pfmdr1 (PF3D7_0523000)# | 5 | Mutations (86Y, 124F, 1042D, 1246Y) | Chloroquine, Amodiaquine, Lumefantrine, Mefloquine |
| Mutations (184F) | Mefloquine, artesunate | ||
| Increased copy number | Mefloquine | ||
| Pfdhfr (PF3D7_0417200)α | 4 | Mutations (51I, 59R, S108N, 164L) | Pyrimethamine, Cycloguanil |
| Pfdhps (PF3D7_0810800)¥ | 8 | Mutations (436A, 437G, 540E, 581G, 613S/T) | Sulfonamide, Sulfadoxine, Sulfone, Dapsone |
| Pfmrp1 (PF3D7_0112200)∗ | 1 | Mutations (191H, 437S) | Chloroquine, quinine |
| Pfcytb (mal_mito_3)∗∗ | Mitochondrial genome | Mutations (268N/S/C) | Atovaquone |
| Pfnhe-1 (PF3D7_1303500)& | 13 | Repeat polymorphism (DNNND repeat motif in microsatellite ms470 region) | Quinine |
| PfATPase4 (PF3D7_1211900)¶ | 12 | Mutations (398F, 990R, 211T, 415D, 917L) | Spiroindolones, Pyrazoles, Dihydroisoquinolones |
| PfATPase6 (PF3D7_0106300)¶¶ | 1 | Mutations (263E, 431K, 623E, 769N) | Artemisinin and its derivatives |
| Pfpm 2 (PF3D7_1408000)‡ | 14 | Increased copy number | Piperaquine |
| Pfk13 (PF3D7_1343700)‡‡ | 13 | Mutations (446I, 458Y, 476I, 493H, 539T, 543T, 553L, 561H, 580Y) | ART and its derivatives |
ART: Artemisinin; Pf: Plasmodium falciparum; Pfatpase6: Pf Ca2+-ATPase; Pfcrt: Pf chloroquine resistance transporter; Pfdhfr: Pf dihydrofolate reductase; Pfcytb: Pf cytochrome B; Pfdhps: Pf dihydropteroate synthase; Pfk13: Pf Kelch gene; Pfmdr1: Pf multidrug resistance protein 1 (the protein is called Pgh1, P-glycoprotein homolog 1); Pfmrp1: Pf multidrug resistance protein 1; Pfnhe-1: Pf sodium–hydrogen exchanger; Pfpm: Pf Plasmepsin.
Information was based on Peterson et al., 1988α; Triglia et al. (1997)¥; Korsinczky et al., 2000∗∗; Singh Sidhu et al., 2002§; Mu et al., 2003∗; Jambou et al. (2005)¶¶; Sidhu et al., 2005#; Ferdig et al., 2004&; Henry et al., 2009&; Ariey et al., 2014‡‡; Chilongola et al. (2015)¶; Spillmarn and Kirk (2015)¶; Menard and Dondorp (2017); Bopp et al. (2018)‡.
Mutations proposed for PfATPase6 gene 6 are still not validated for drug resistance while the link between increased number in Pfpm2 gene and resistance to piperaquine is still controversial.
The Pfcrt K76T mutations have been reported from Eastern regions of India, especially from Arunachal Pradesh and Assam, along with mutations in the Pfmdr1 gene at the positions 86Y and 1246Y associated with reduced sensitivity to CQ (Das et al., 2014; Sharma et al., 2016). For Pfdhfr gene, mutations at codons 16V, 51I, 59R, 108N and 164L and in Pfdhps mutations at codons 436A, 437G, 540E, 581G and 613S/T are reported to be associated with resistance to SP in Arunachal Pradesh, Assam and several other areas too (Kumar et al., 2015; Patel et al., 2017).
3. Artemisinin resistance: mechanism, molecular markers and current scenario in sub-Saharan Africa and India
The first case of therapeutic failure with artesunate monotherapy was documented from Thailand-Cambodia border (Dondorp et al., 2009; Noedl et al., 2008) and there are also other studies which have observed delayed parasite clearance and increased recrudescence rates reported treatment with ACT (Witkowski et al, 2010, 2013). Two mechanisms viz parasitic quiescence and altered temporal response were shown to be responsible for ART resistance using in vitro studies initially and later confirmed by in vivo studies (Klonis et al., 2013; Paloque et al., 2016; Witkowski et al, 2010, 2013). The basic mechanism of ART resistance was explained by point mutations in the beta-propeller domain of the gene encoding the Kelch protein 13- Pfk13 probably responsible for reduced susceptibility (i.e., delayed parasite clearance time) to ART and its derivatives (Ariey et al., 2014; Menard and Dondorp, 2017; Ouji et al., 2018; Straimer et al., 2015).
The Pfk13 gene is located on the chromosome 13 with only one exon and encodes the kelch protein that has three domains namely Plasmodium-specific domain, BTB/POZ domain (Broad-Complex, Tramtrack and Bric à brac; Poxvirus and zinc finger respectively) and a C-terminal six-blade propeller domain (Pasupureddy et al., 2019). A summary of validated and putative mutations in the Pfk13 gene associated with ART resistance has been reported (Pasupureddy et al., 2019; WHO, 2018b). The resistance to ART and its derivatives is confirmed in the GMS, consisting of six countries - Cambodia, Thailand, Myanmar, Lao People's Democratic Republic, Vietnam and China where the prevalence of mutations in the reported gene is reported to be relatively high (WHO, 2018a). Ikeda and colleagues reported a novel mutation in the Pfk13 gene (675V) in Uganda which is currently categorized by the WHO as “associated/candidate” mutations (Ikeda et al., 2018). From sSA, validated mutations (i.e., 561H) in the Pfk13 gene responsible for ART resistance as well as some non-synonymous with few novel mutations have been reported (Ikeda et al., 2018; Lu et al., 2017; Ménard et al., 2016; Uwimana et al., 2020). In India, mutation at 561H of the Pfk13 gene currently validated by the WHO for ART resistance was reported from only one sample (Mishra et al., 2015) (Table 1).
4. Selection and implications of molecular markers in failure treatment after ACT treatment in India and sSA
4.1. Methods
4.1.1. Electronic databases and search strategy
The authors AA, LPKF and SC did comprehensive searching of eight electronic databases including PubMed, EMBASE, MEDLINE, Wiley, ResearchGate, African Journal Online, ScienceDirect and Scopus to identify all relevant papers for the present study. The search for potentially relevant articles was performed from May to July 2019. The main key terms were: “malaria”, “Plasmodium”, “Plasmodium falciparum”, “artemisinin-based combination therapies”, “artesunate”, “artemether”, “efficacy”, “in vivo”, “molecular markers”, “resistance”, “selection”, “selected”, “treatment”, “treatment failure”, “Africa”, “India”. The names of sSA countries or Indian states were also used in combination with other key terms. These search terms were used in English and French in combination with Boolean operator “AND”, “OR” and truncation element “∗“. The search strategy was adjusted according to the requirements of different databases (Supplemental material 1). A summary of the strategy of searching as outlined previously (Supplemental material 2 and 3) (Moher et al., 2009).
4.1.2. Screening strategy of papers retrieved from the electronic databases
Titles and abstracts of papers were independently reviewed by authors AA, LPKF and SC using the above mentioned search strategy. The full texts of potentially eligible publications were directly retrieved from databases if titles or abstracts were informative (i.e., giving all key terms useful to include or exclude a paper in the review). If not, the full text was retrieved and totally read in order to decide if the papers fulfilled the inclusion criteria. The publications not available free online were either purchased or requested from the corresponding author. The reference lists of relevant papers were also examined.
4.1.3. Eligibility criteria of studies included in the review
The studies included in the present study were considered only after they fulfilled the following four prerequisites: (i) Treatment efficacy studies (TES) having addressed either the effect of ACT on the selection of molecular markers associated with drug resistance or the involvement of these markers associated in treatment failure cases, (ii) studies conducted in India and sSA countries, (iii) articles written in English or French languages and (iv) articles peer-reviewed and published between January 2008 and December 2018 (a 11-year period was chosen in order to reduce the methodological variability between eligible studies. In addition, by the year 2008 ACT were already adopted and made available at affordable prices for treatment of all ages in public sector in most malaria endemic countries). All discrepancies between the authors AA, LPKF and SC were solved in discussions with the supervising author (VS).
4.1.4. Bias risk assessment
The methodological quality of studies was assessed using the Joanna Briggs Institute (JBI) critical appraisal tools for use in JBI systematic reviews checklist for randomized controlled trials available at: https://joannabriggs.org/critical-appraisal-tools (The Joanna Briggs Institute, 2020). This tools consists of 13 questions addressing different type of bias (selection, performance, attrition and reporting) related to different methodological aspect of the included studies (i.e., randomization process, blinding). The bias risk for each methodological element was classified as “high”, “low” and “unclear”. Quality of studies was independently evaluated and any disagreement was resolved through discussion.
4.1.5. Data extraction and verification for consistency
The collected information from the studies included the following: the first author name, the year of publication, the name of the African country or Indian state, the year of data collection, the evaluated ACT, the study population, the level of malarial endemicity, the PCR-corrected cure rate of ACT, the type of analysis used for computing cure rate (i.e., per protocol and intention-to-treat), the molecular markers evaluated, the nature of mutations and mutations involved in treatment failure cases and the prevalence of selected mutations. All the data were independently entered by three authors in the Excel spreadsheet (Microsoft Office, Excel, USA) (AA, LPKF and SC) and discussed extensively with the supervising author (VS).
4.2. Results and discussion
4.2.1. Selection process of the included studies
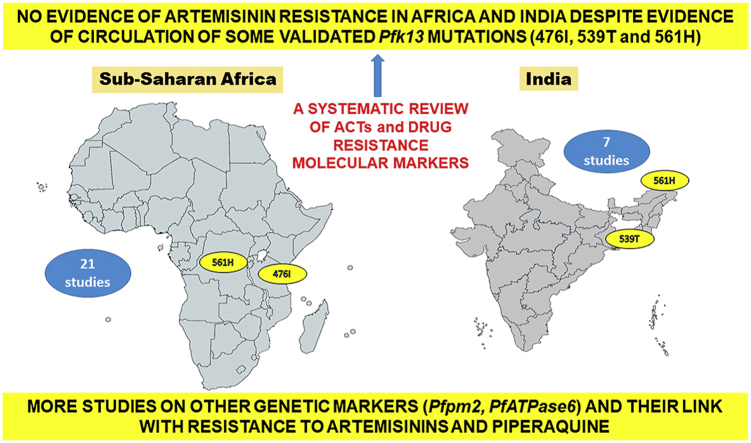
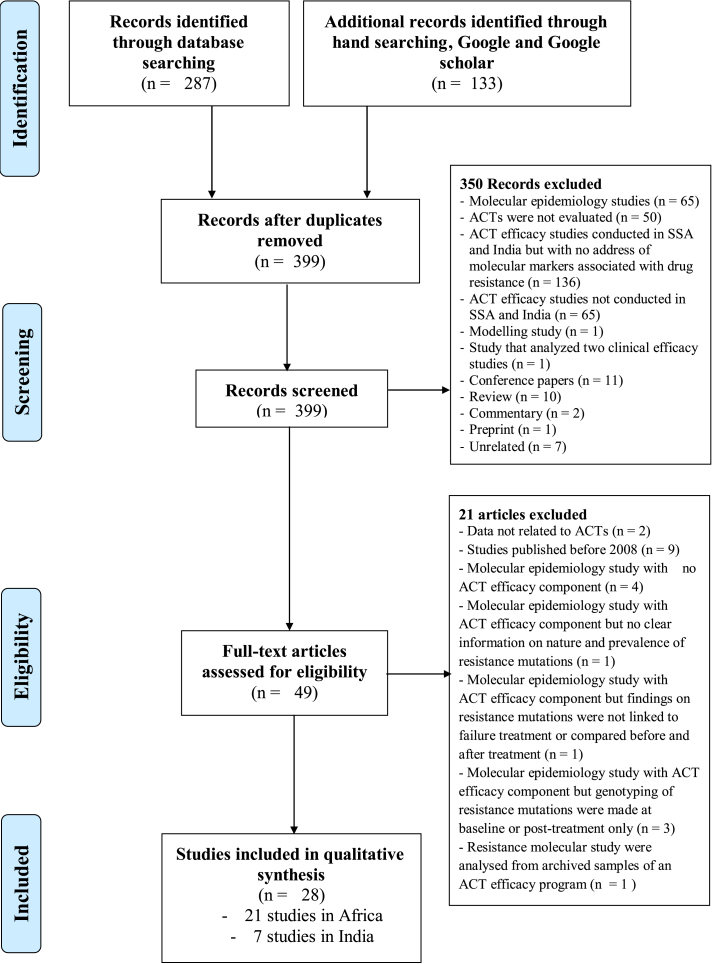
A total of 420 studies were retrieved from targeted electronic databases (n = 287) and by hand searching (n = 133) as presented in the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) flowchart (Fig. 1 and Fig. S3). Three hundred and ninety-nine studies were screened based on titles, abstracts and among them duplicates along with other studies from regions other than sSA and India were excluded from this systematic review. In addition, research papers such as reviews, conference papers, commentary and unrelated studies were also excluded. Finally, a total of 28 studies, 21 from sSA countries and seven from India, were shortlisted for this systematic review (Fig. 1). The list of excluded studies is presented in Supplemental material 4.
Fig. 1.
PRISMA chart of the selection steps of included studies.
4.2.2. Results of bias risk assessment
As shown in Fig. S5, bias risk assessment has varied from one study to another and was mainly found for “drug allocation concealment” and “blinding treatment to patients, caregivers and outcome assessors”. Few studies were designed as either single-blinded or open-label trial (Djimdé et al., 2008; Otienoburu et al., 2016; Saha et al., 2012; Somé et al., 2010; Sondo et al., 2016). It should be noted that the risk was « unclear » for several studies as the authors had not sufficiently given information on some methodological aspects including randomization, drug allocation concealment, comparability of treated groups for instance (Fig. S5). Some studies used only per protocol (PP) analysis to evaluate the ACT efficacy instead of intention-to-treat (ITT) analysis which is recommended by the JBI (Fig. S5) (Gadalla et al., 2011; Kamugisha et al., 2012; Kiaco et al., 2015).
4.2.3. Characteristics of the included studies
In Africa, the included studies were conducted in twelve countries namely Angola, Burkina Faso, Mali, Nigeria, Sierra Leone, Liberia, Sudan, Somalia, Tanzania, Rwanda, The Democratic Republic of Congo (DRC) and Uganda and were published in years during and after 2008 (Djimdé et al., 2008; Happi et al., 2009; Karema et al., 2010; Somé et al., 2010; Baliraine and Rosenthal, 2011; Gadalla et al., 2011, 2013; Ngasala et al., 2011a, 2011b; Kamugisha et al., 2012; Kiaco et al., 2015; Warsame et al., 2015, 2017; Otienoburu et al., 2016; Sondo et al., 2016; Yeka et al., 2016; Plucinski et al., 2017; Baraka et al., 2018; Davlantes et al., 2018; Kakolwa et al., 2018; Smith et al., 2018) (Table 2).
Table 2.
Characteristics of the studies included in the systematic review.
| Author_Year | Countries (Area) | Study period | Endemicity | Study population | Antimalarial drugs evaluated | Molecular markers investigated |
|---|---|---|---|---|---|---|
| AFRICAN COUNTRIES | ||||||
| Djimdé et al. (2008)a | Mali (Bougoula-Hameau) | 2002 to 2004 | Hyperendemic | ≥6 months | AS; AS + AQ; AS + SP | Pfcrt, Pfdhfr, Pfdhps, Pfmdr1 |
| Happi et al. (2009) | Nigeria (Ibadan) | 2006 to 2007 | Hyperendemic | ≤10 years | AL | Pfmdr1, PfATPase6 |
| Karema et al. (2010)a | Rwanda (Mashesha and Rukara) | 2006 | Not specified | 6–59 months | AS + CPG-DDS; AQ + SP | Pfdhps, Pfdhfr |
| Somé et al. (2010) | Burkina Faso (Bobo-Dioulasso) | Not specified | Holoendemic | ≥6 months | AL; AQ + SP; DHA + PPQ | Pfmdr1, Pfcrt, Pfdhps, Pfdhfr |
| Baliraine and Rosenthal (2011) | Uganda (Kampala) | 2004 to 2008 | Not specified | Children (1–10 years) | AL; AS + AQ; AQ + SP | Pfmdr1 |
| Gadalla et al. (2011)a | Sudan (Asar, Daraweesh, and Abu Adam) | 2012 | Not specified | All age | AL | Pfmdr1, PfATPase6 |
| Ngasala et al. (2011a)a | Tanzania (Fukayosi and Yombo) | 2007 to 2008 | Not specified | 6–59 months | AL | Pfmdr1, Pfcrt |
| Ngasala et al. (2011b)a | Tanzania (Ngeta and Mwanabwito) | 2007 | Not specified | 3–59 months | AL | Pfmdr1, Pfcrt |
| Kamugisha et al. (2012) | Tanzania (Igombe-Mwanza) | 2010 to 2011 | Not specified | 6–60 months | AL | Pfdhfr, Pfdhps, Pfmdr1, Pfcrt, PfATPase6 |
| Gadalla et al. (2013) | Sudan (Kassala) | 2012 | Moderate perennial | All age | AS + SP | Pfdhfr, Pfdhps, Pfmdr1 |
| Kiaco et al. (2015) | Angola (Luanda) | 2011 to 2013 | Mesoendemic | All age | AL | PfATPase6, Pfk13, Pfmdr1 |
| Warsame et al. (2015) | Somalia (Jamame, Janale and Jowhar) | 2011 | Not specified | ≥6 months | AS + SP | Pfdhfr, Pfdhps |
| Otienoburu et al. (2016) | Liberia (Nimba) | 2016 | Not specified | Not specified | AL; AS + AQ | Pfcrt, Pfmdr1 |
| Sondo et al. (2016) | Burkina Faso (Nanoro) | Not specified | Not specified | All age | AL; AS + AQ | Pfcrt, Pfmdr1 |
| Yeka et al. (2016) | Uganda (Apac, Mubende and Kanungu) | 2013 to 2014 | Mesoendemic | 6–59 months | AL; AS + AQ | Pfcrt, Pfmdr1 |
| Plucinski et al. (2017) | Angola (Benguela, Lunda Sul and Zaire) | 2015 | Mesoendemic, Hyperendemic | 6–59 months | AL; AS + AQ; DHA + PPQ | Pfcytb, Pfk13, Pfmdr1 |
| Warsame et al. (2017) | Somalia (Janale, Jowhar and Bosaso) | 2013 to 2015 | Moderate-to-high; Low | ≥6 months | AL; AS + SP | Pfdhfr, Pfdhps |
| Baraka et al. (2018) | The Democratic Republic of Congo (Lisungi), and Uganda (Kazo) | 2012–2014 | Not specified | 6–59 months | AL; AS + AQ | Pfmdr1 |
| Davlantes et al. (2018) | Angola (Benguela, Lunda Sul and Zaire) | 2017 | Mesoendemic, Hyperendemic | 6–59 months | AL; AS + AQ; DHA + PPQ | Pfcrt, Pfmdr1, Pfk13, Pfpm2 |
| Kakolwa et al. (2018) | Tanzania (Chamwino, Butimba, Kibaha, and Rufiji, Kyela, Kilombero, Muheza, Nagaga and Ujiji) | 2011, 2012, 2015 | Not specified | 6–59 months (2011), 6 months-10 yrs (2012), ≥6 months (2015) | AL | Pfk13, Pfpm2 |
| Smith et al. (2018) | Sierra Leone (Bo, Kenema and Makeni) | 2016 | Not specified | 6–59 months | AL; AS + AQ; DHA + PPQ | Pfdhfr, Pfdhps, Pfk13, Pfpm2 |
| INDIA | ||||||
| Saha et al. (2012)a | West Bengal, Jalpaiguri | 2009 to 2010 | Not specified | ≥6 months; > 5 yrs | AL; AS + SP; AS + MQ | Pfdhfr, Pfdhps, Pfcrt, PfATPase6 |
| Mishra et al. (2012) | 25 sentinel sites | 2009 to 2010 | Not specified | All age | AS + SP | Pfdhfr, Pfdhps |
| Ganguly et al. (2013)a | West Bengal (Purulia) | Not specified | Not specified | All age | AS + SP | Pfdhfr, Pfdhps |
| Srivastava et al. (2013) | Ranchi, Meghalaya and Keonjhar | 2007 to 2010 | Hyperendemic | All age | AS + SP | Pfdhfr, Pfdhps |
| Mishra et al. (2014) | Arunachal Pradesh (Changlang, Miao), Tripura (Gomati, Silachari), Mizoram (Lunglei, Tlabung) | 2012 | Not specified | All age | AS + SP | Pfdhfr, Pfdhps |
| Mishra et al. (2017) | Balaghat and Anuppur district, Madhya Pradesh | 2012 to 2014 | Not specified | ≥1 yrs | AS + SP | Pfdhfr, Pfdhps, Pfk13 |
| Das et al. (2018) | West Bengal | 2013 to 2014 | Not specified | Not specified | AS + SP | Pfk13 |
ACT: Artemisinin-based combination therapy; AL: Artemether + Lumefantrine; AS + AQ: artesunate + amodiaquine; AS + CPG-DDS: Artesunate + Chlorproguanil-Dapsone; AS + MQ: artesunate + mefloquine; AS + SP: artesunate + sulfadoxine-pyrimethamine; DHA + PPQ: dihydroartemisinin + piperaquine; GTS: Global technical strategy; Pf: Plasmodium falciparum; Pfatpase6: Pf Ca2+-ATPase; Pfcrt: Pf chloroquine resistance transporter; Pfdhfr: Pf dihydrofolate reductase; Pfcytb: Pf cytochrome B; Pfdhps: Pf dihydropteroate synthase; Pfk13: Pf Kelch gene; Pfmdr1: Pf multidrug resistance protein 1; Pfpm2: Pf plasmepsin 2.
These studies were conducted in rural areas.
Two studies were from mesoendemic areas, three from hyperendemic areas, one each from holoendemic, mesoendemic and hyperendemic areas; though the level of endemicity was not specified in remaining studies (Davlantes et al., 2018; Djimdé et al., 2008; Happi et al., 2009; Kiaco et al., 2015; Plucinski et al., 2017; Somé et al., 2010) (Table 2).
The studies evaluated the efficacy of ACT between the years 2002-2017 in children, especially those aged <5 years, and/or both children and adults. Seven studies, six in sSA and one in India, collected data before 2008, the year of first documentation of ART-resistance (Baliraine and Rosenthal, 2011; Djimdé et al., 2008; Happi et al., 2009; Karema et al., 2010; Ngasala et al., 2011a, 2011b; Srivastava et al., 2013).
ACT evaluated in the studies included in AL, AS + AQ, AS + SP, AQ + SP and DHA + PPQ (Table 3). The included Indian studies evaluated efficacy of three ACT viz AS + SP, AS + MQ and AL between 2012 and 2014 in children, adolescents and adults, among which one study was conducted in a highly endemic area, while the level of endemicity was not specified in the remaining four studies (Das et al., 2018; Ganguly et al., 2013; Mishra et al, 2012, 2014, 2017; Saha et al., 2012; Srivastava et al., 2013).
Table 3.
PCR-corrected efficacy rates of ACT and non-ACT after 28 and 42 days of follow-up from studies included in the review.
| Authors_Year | ACT tested | Type of analysis | 28-day ACPR (%) | 42-day ACPR (%) |
|---|---|---|---|---|
| Djimdé et al. (2008) | AS + AQ | PP | 99.1 | – |
| AS + SP | PP | 100.0 | – | |
| Happi et al. (2009)a | AL | ITT | 95.6 | 82.2 |
| Karema et al. (2010)a | AS + CPG-DDS | ITT | 70.5–73.3 | – |
| AQ + SP | ITT | 38.1–87.8 | – | |
| Somé et al. (2010)a | AL | ITT | – | 68.8 |
| DHA + PPQ | ITT | – | 89.1–92.4 | |
| AQ + SP | ITT | – | 88.2 | |
| Baliraine and Rosenthal (2011)a | AL | ITT | 89.1 | – |
| AS + AQ | ITT | 79.3 | – | |
| AQ + SP | ITT | 71.1 | – | |
| Gadalla et al. (2011) | AL | PP | 95.0 | 95.0 |
| Ngasala et al. (2011a) | AL | ITT | 98.2–98.8 | 95.8–98.1 |
| Ngasala et al. (2011b) | AL | ITT | 95.1 | 93.0 |
| Kamugisha et al. (2012) | AL | PP | 96.0 | – |
| Gadalla et al. (2013) | AS + SP | PP | 90.5 | – |
| ITT | 85.3 | – | ||
| Kiaco et al. (2015) | AL | PP | 91.3 | – |
| Warsame et al. (2015) | AS + SP | PP | 77.8–99.0 | – |
| ITT | 79.3–99.0 | |||
| Otienoburu et al. (2016) | AL | – | – | – |
| AS + AQ | – | – | – | |
| Sondo et al. (2016)a | AL | PP | 77.8 | – |
| AS + AQ | PP | 84.1 | – | |
| Yeka et al. (2016) | AS + AQ | ITT | 70.7 | – |
| AL | ITT | 54.3 | – | |
| Plucinski et al. (2017) | AL | PP | 86.5–96.1 | – |
| AS + AQ | PP | 99.9–100.0 | – | |
| DHA + PPQ | PP | 98.8–100.0 | 98.8–100.0 | |
| AL | ITT | 88.1–96.3 | – | |
| AS + AQ | ITT | 99.9–100.0 | – | |
| DHA + PPQ | ITT | 98.8–100.0 | 98.8–100.0 | |
| Warsame et al. (2017) | AL | PP | 97.6–100.0 | – |
| ITT | 91.1–100b | |||
| AS + SP | PP | 87.7 | – | |
| ITT | 78.8b | |||
| Baraka et al. (2018)a | AL | PP | NE | NE |
| AS + AQ | PP | NE | NE | |
| Davlantes et al. (2018) | AL | PP | 95.5–96.4 | – |
| AS + AQ | PP | 93.0–100.0 | – | |
| DHA + PPQ | PP | – | 100.0 | |
| AL | ITT | 95.5–96.5 | – | |
| AS + AQ | ITT | 93.3–100.0 | – | |
| DHA + PPQ | ITT | 100.0 | 100.0 | |
| Kakolwa et al. (2018) | AS + AQ | PP | NE | – |
| AL | PP | NE | – | |
| DHA + PPQ | PP | – | NE | |
| Smith et al. (2018) | AS + AQ | PP | 100.0 | – |
| AL | PP | 100.0 | – | |
| DHA + PPQ | PP | – | 100.0 | |
| AS + AQ | ITT | 100.0 | – | |
| AL | ITT | 100.0 | – | |
| DHA + PPQ | ITT | – | 100.0 | |
| Saha et al. (2012) | AS + SP | PP | – | 90.6 |
| AL | PP | – | 95.6 | |
| AS + MQ | PP | – | 100 | |
| Mishra et al. (2012) | AS + SP | ITT | 98.8 | – |
| Ganguly et al. (2013) | AS + SP | PP | – | 97.0 |
| Srivastava et al. (2013) | AS + SP | ITT | 100.0 | – |
| Mishra et al. (2014) | AS + SP | ITT | – | 59.5–82.7 |
| Mishra et al. (2017) | AS + SP | PP | 98.7–100.0 | – |
| Das et al. (2018) | AS + SP | _ | _ | _ |
ACT: Artemisinin-based combination therapy; ACPR: adequate clinical and parasitological response; AL: artemether + lumefantrine; AS + AQ: artesunate + amodiaquine; AS + CPG-DDS: Artesunate + Chlorproguanil-Dapsone; AS + MQ: artesunate + mefloquine; AS + SP: artesunate + sulfadoxine-pyrimethamine; DHA + PPQ: dihydroartemisinin + piperaquine; PP: per protocol; ITT: Intention-to-treat; NE: Non extractible.
These studies were secondary analysis of previously published trials. Thus information was obtained from the primary studies.
Compute based on data presented in the study.
4.2.4. Efficacy of ACT
The PCR-corrected cure rates of different ACT showed large variations in studies from sSA. Based on ITT analysis, the cure rates of different ACTs were: 54.3–100% for AL, 70.7–100% for AS + AQ, 79.3–99.0% for AS + SP and 99.8–100.0% for DHA + PPQ after 28-day follow-up, 68.8–98.1% for AL, 89.1%–100% for DHA + PPQ after 42-day follow-up (Table 3). ACT were highly efficacious against malaria parasites (i.e., cure rate ≥ 95%) in most studies while reduced efficacy (i.e., cure rate < 90%) were reported in one study from Angola for AL in one of the study site (i.e. Zaire area) (Plucinski et al., 2017).
In India three ACT were evaluated in the studies included in the review namely AS + SP (six studies), AL (one study) and AS + MQ (one study). Based on ITT analysis, the efficacy of AS + SP ranged from 98.7 to 100% and from 90.6 to 97.0% after 28 days and 42 days of follow-up respectively (Table 3). ACT efficacy was >90% in all the Indian studies with only one exception for which AS + SP efficacy of 59.5–82.7% was found (Mishra et al., 2014).
ACTs were highly efficacious (i.e., cure rates > 90%) at the time of data gathering despite the fact that nine studies reported cure rates <90% (8 in sSA and 1 in India) (Baliraine and Rosenthal, 2011; Gadalla et al., 2013; Karema et al., 2010; Mishra et al., 2014; Plucinski et al., 2017; Sondo et al., 2016; Warsame et al, 2015, 2017; Yeka et al., 2016) (Supplementary file 6). At least three possible hypotheses could explain this low cure rates reported in these nine studies. Firstly, AS + SP was evaluated in one third of the studies (3/9) and the high TF rates following treatment with this ACT was statistically attributable to the presence of isolates with multiple mutations in Pfdhfr/Pfdhps genes (Mishra et al., 2014; Warsame et al, 2015, 2017)(Table 4). Secondly, distortions in the study design of the included studies as some of them were not designed: i) as randomized control trial (RCT), ii) open-label RCT and iii) had high bias risk for some aspects including “drug allocation concealment” and “blinding”. These factors can greatly influence the results of drug efficacy studies (Dettori, 2010). Thirdly, the possible presence of ART-resistant isolates among TF cases cannot be ruled out. We are unable to confirm or deny this statement as eight of out the nine studies were conducted before 2008 (ART resistance was discovered this year)(Dondorp et al., 2009; Noedl et al., 2008), conducted before 2014 (link between Pfk13 mutations in ART resistance was confirmed in this year) (Ariey et al., 2014), and none genotyped Pfk13 gene (Supplementary material 6).
Table 4.
Nature and direction of selected polymorphisms with regard to study and drugs (ACT and non-ACT) in SSA and India.
| Authors_Year | Drug tested | Molecular markers selected or involved in ACT treatment failure | Nature and direction of selected mutations (Main findings) |
|---|---|---|---|
| Djimdé et al. (2008) | AS + AQ | Pfcrt, Pfmdr1 | Pfcrt 76T (+) and Pfmdr1 86Y (+) |
| AS + SP | Pfdhfr, Pfdhps | Pfdhfr: Single mutants 59R (+) and 108N (+); Triple mutant (108N + 59R + 51I) (+), Quadruple mutant (Pfdhfr: 108N + 59R + 51I + Pfdhps: 437G) (+) | |
| Happi et al. (2009) | AL | Pfmdr1 | Wild type N86 (+), Y184 (−), Single mutant 184F (+), Haplotypes N86–184F-D1246 (+) and “No N86–184F-D1246”b (−) No evidence of selection for the rest of single mutations (86Y, D1246, 1246Y) |
| Karema et al. (2010) | AS + CPG-DDS | Pfdhps, Pfdhfr | The risk of TF significantly doubled for each additional resistance mutations in Pfdhfr (OR = 2.2; 95%CI: 1.34–3.7) |
| AQ + SP | Pfdhps, Pfdhfr | The risk of TF significantly doubled for each additional resistance mutations in Pfdhfr (OR = 2.4; 95%CI: 1.10–5.55) and Pfdhps (OR = 2.1; 95%CI: 1.34–3.54) | |
| Somé et al. (2010) | AL | Pfmdr1, Pfcrt, Pfdhps, Pfdhfr | Pfmdr1: Wild types N86 (−) and Y184 (−); Pfcrt: wild type K76 (−) |
| AQ + SP | Pfmdr1, Pfcrt, Pfdhps, Pfdhfr | Pfdhfr: Single mutants 51I (+), 59R (+) and 108N (+) | |
| DHA + PPQ | Pfmdr1, Pfcrt, Pfdhps, Pfdhfr | None | |
| Baliraine and Rosenthal. (2011) | AL | Pfmdr1 | Pfmdr1: Wild types N86 (+) and D1246 (+); Mutants Y184 (+), haplotypes N86–184F-D1246 (+) |
| AS + AQ | Pfmdr1 | No evidence of selection | |
| AQ + SP | Pfmdr1 | No evidence of selection | |
| Gadalla et al. (2011) | AL | Pfmdr1, PfATPase6 |
Pfmdr1: Haplotypes N86–184F-D1246 (+), Increased copy number PfATPase6: No evidence of selection |
| Ngasala et al. (2011a) | AL | Pfmdr1, Pfcrt | Pfmdr1: wild type N86 (+), Pfcrt: No selection of K76 wild type |
| Ngasala et al. (2011b) | AL | Pfmdr1, Pfcrt | Pfmdr1: wild type N86 (+), Pfcrt: wild type K76 (+) |
| Kamugisha et al. (2012) | AL | Pfmdr1, Pfcrt, PfATPase6, Pfdhfr, Pfdhps |
Pfmdr1: Single mutant 86Y (+); Pfcrt: No evidence of selection PfATPase6: Wild types (No selection); Pfdhfr: Single mutants 108N (−), 51I (−), 59R (−); Pfdhps: Mutants 437G (−), 540E (−) |
| Gadalla et al. (2013) | AS + SP | Pfmdr1, Pfcrt, Pfdhps, Pfdhfr |
Pfdhps haplotype SGEGA (436A, 437G, 540E, 581G, 613S) (associated to all TF) Pfmdr1: haplotype N86–184F-D1246 (+) Pfcrt: No evidence of selection |
| Kiaco et al. (2015)a | AL | Pfmdr1, PfATPase6, Pfk13 |
Pfmdr1: 86Y (found in 2 TF), increase in copy number (found in 1 TF); No selection of these mutations PfATPase6: No evidence of selection Pfk13: wild types found in TF (n = 9) |
| Warsame et al. (2015) | AS + SP | Pfdhfr, Pfdhps | TF significantly associated with the presence of double mutant Pfdhps 437G + 540E (OR = 22.4; 95%CI: 5.1–98.1), quadruple mutant Pfdhfr 51I/108N + Pfdhps 437G/540E (OR = 5.5, 95% CI: 2.3–13.6), quintuple mutant Pfdhfr 51I/59R/108N + Pfdhps 437G/540E (OR = 3.5, 95%CI: 1.4–8.8) |
| Otienoburu et al. (2016) | AL | Pfmdr1, Pfcrt |
Pfmdr1: single mutant 1246Y (+) and N86 (+); Pfcrt: single mutant 76T (+) |
| AS + AQ | Pfmdr1, Pfcrt |
Pfmdr1: single mutant 186Y (+); Haplotypes N86–Y184–1246Y (+), 86Y–Y184–1246Y (+), N86–Y184-D1246 (−), N86–184F-D1246 (+) and 86Y–184F-D1246 (−) Pfcrt: No evidence of selection |
|
| Sondo et al. (2016) | AS + AQ | Pfmdr1, Pfcrt | Pfmdr1: No selection of polymorphisms; Pfcrt: 76T (+) |
| AL | Pfmdr1, Pfcrt | Pfmdr1: No selection of polymorphisms; Pfcrt: 76T (−) | |
| Yeka et al. (2016) | AS + AQ | Pfmdr1, Pfcrt | Pfmdr1: single mutant 86Y (+) and 1246Y (+); Pfcrt: 76T (−) |
| AL | Pfmdr1, Pfcrt | Pfmdr1: D1246 (+); Pfcrt: No evidence of selection | |
| Plucinski et al. (2017) | AL | Pfk13, Pfcytb, Pfmdr1 | Pfk13 and Pfcytb: Wild types (+); Pfmdr1: Haplotypes N86–184F-D1246 and N86–Y184-D1246 found in most of TF |
| DHA + PPQ | Pfk13, Pfcytb, Pfmdr1 | Pfk13 and Pfcytb: Wild types (+);Pfmdr1: Haplotypes N86–184F-D1246 and N86–Y184-D1246 found in most of TF | |
| Warsame et al. (2017) | AL | Pfdhfr, Pfdhps | None |
| AS + SP | Pfdhfr, Pfdhps | Quintuple mutation (51I/108N + 437G/540E/581G) were found in all TF cases | |
| Baraka et al. (2018) | AL | Pfmdr1 |
Pfmdr1: F184 (Borderline significant directional selection in Uganda, P-value = 0.055); N86 and D1246 (No evidence of selection) 86Y was significantly associated with reduced risk of TF (RR = 0.34; 95%CI 0.11–1.05; p = 0.04) |
| AS + AQ | Pfmdr1 | Pfmdr1: N86 and D1246 (No evidence of selection) | |
| Davlantes et al. (2018) | AL | Pfcrt, Pfmdr1, Pfk13, Pfpm2 | All 33 TF in AL arm carried Pfmdr1 (N86–Y184-D1246, N86–184F-D1246 and 86Y–184F-D1246) or Pfcrt (72C–V73–74I-75E-76T) mutations associated with lumefantrine on day of failure Pfk13: Wild types found in all TF; 504V mutant found in one TF Pfpm2: No increase in copy number in all TF |
| AS + AQ | Pfcrt, Ppfmdr1, Pfk13, Pfpm2 | All 33 TF in AS + AQ arm carried Pfmdr1 (N86–Y184-D1246, N86–184F-D1246 and 86Y–184F-D1246) or Pfcrt (72C–V73–74I-75E-76T) mutations associated with AQ resistance on day of failure Pfk13: Wild types found in all TF Pfpm2: No increase in copy number in all TF |
|
| DHA + PPQ | Pfcrt, Ppfmdr1, Pfk13, Pfpm2 | None | |
| Kakolwa et al. (2018) | AS + AQ | Pfk13, Pfpm2 | Pfk13: Not applicable as the PCR-corrected cure rate was 100% after treatment |
| AL | Pfk13, Pfpm2 | Pfk13: Not applicable as the PCR-corrected cure rate was 100% after treatment | |
| DHA + PPQ | Pfk13, Pfpm2 | Pfpm2: 3 Mutants with multiple copies (Not associated with TF) | |
| Smith et al. (2018) | AS + AQ | Pfk13, Pfpm2, Pfdhfr, Pfdhps | Not applicable as the PCR-corrected cure rate was 100% after treatment |
| AL | Pfk13, Pfpm2, Pfdhfr, Pfdhps | ||
| DHA + PPQ | Pfk13, Pfpm2, Pfdhfr, Pfdhps | ||
| Saha et al. (2012)a | AS + SP | PfATPase6, Pfcrt, Pfdhfr, Pfdhps | Quadruple mutant (Pfdhfr- 51I + 59R + 108N, Pfdhps-437G) (found in 2 TF); Quintuple mutant (Pfdhfr- 51I + 59R + 108N, Pfdhps-436A + 540E) (found in 2 TF) and (Pfdhfr- 59R + 108N + 164L, Pfdhps-436A + 540E) (found in 1 TF) |
| AL | Pfdhfr, Pfdhps | Quintuple mutant (Pfdhfr-59R + 108N + 164L, Pfdhps-436A + 540E) (found in 2 TF) | |
| AS + MQ | Pfdhfr, Pfdhps | Not applicable (Failure to genotype strains) | |
| Mishra et al. (2012)a | AS + SP | Pfdhfr, Pfdhps |
Pfdhfr: double mutant (59R + 108N) (found in 12 TF) Pfdhfr: single mutant 108N (found in 4 TF) No association was detected between infection with parasites harbouring three or more mutations in the genes investigated (i.e. dhfr and dhps) and failure of AS+SP treatment |
| Ganguly et al. (2013)a | AS + SP | Pfdhfr, Pfdhps | Quintuple mutant (Pfdhfr: 51I + 59R + 180N, Pfdhps: 437G + 540E) (found in 2 TF) |
| Srivastava et al. (2013)a | AS + SP | Pfdhfr, Pfdhps | At 0 day Pfdhfr-Double mutant (59R + 108N) and at day of failure addition mutation 59R + 108N + 450E (found in one TF) and 59R + 108N + 581G (found in one TF) |
| Mishra et al. (2014)a | AS + SP | Pfdhfr, Pfdhps | Presence of Pfdhfr plus Pfdhps quintuple mutation was observed predominantly in TF samples |
| Mishra et al. (2017) | AS + SP | Pfk13, Pfdhfr, Pfdhps |
Pfdhps- and Pfdhfr wild type, Pfdhfr-double mutant (59R + 108N), Pfdhfr-single mutant 108N (found in 32 TF) Identification of two non-synonymous mutations in Pfk13 gene (579T, 657H) |
| Das et al. (2018) | AS + SP | Pfk13 | Identification of 539T mutation (found in 1 TF) along with a novel 625R mutation (found in 4 other TF cases) |
(+): Positively selected (i.e., the prevalence of the molecular marker significantly increased post-treatment); (−): Negatively selected (i.e., the prevalence of the molecular marker significantly lowered post-treatment); ACT: Artemisinin-based combination therapy; AL: artemether + lumefantrine; AS + AQ: artesunate + amodiaquine; AS + CPG-DDS: Artesunate + Chlorproguanil-Dapsone; AS + MQ: artesunate + mefloquine; AS + SP: artesunate + sulfadoxine-pyrimethamine; DHA + PPQ: dihydroartemisinin + piperaquine; GTS: Global technical strategy; Pf: Plasmodium falciparum; Pfatpase6: Pf Ca2+-ATPase; Pfcrt: Pf chloroquine resistance transporter; Pfdhfr: Pf dihydrofolate reductase; Pfcytb: Pf cytochrome B; Pfdhps: Pf dihydropteroate synthase; Pfk13: Pf Kelch gene; Pfmdr1: Pf multidrug resistance protein 1; Pfpm2: Pf plasmepsin 2; TF: Treatment failure.
Mutations in the genes were found in treatment failure case.
“No N86–184F-D1246” refers to haplotypes different from N86–184F-D1246.
Our finding on an overall high efficacy of ACTs in sSA and India is supported by several recently published RCTs (Beavogui et al., 2020; Diallo et al., 2020; Lingani et al., 2020), thereby outlining that ACT are efficacious in these two areas till date. In parallel, it should be noted that a recent epidemiological study, conducted in Rwanda, reported the circulation of the 561H mutation (Pfk13), now validated for ART resistance (Uwimana et al., 2020). This underlines the necessity for continuous ACTs efficacy studies in order to make decisions in a timely way for necessary to changes in ACTs-based treatment policies in sSA and India.
4.2.5. Molecular markers with implications in treatment failure
The patterns of molecular markers related to partner drugs for resistance in the ACT were different depending on the investigated marker.
4.2.5.1. Pfk13
Five studies from Africa evaluated the selective pressure of ACT on the Pfk13 propeller gene and three found wild or mutant types of this gene with absence of any selective pressure (Davlantes et al., 2018; Kakolwa et al., 2018; Kiaco et al., 2015; Plucinski et al., 2017; Smith et al., 2018). Some mutations, located in the propeller domain of the Pfk13 gene, were reported to be non-synonymous (463S, 476I, 496S, 510M, 540A, 556K, 562T, 578S, 602D and 646T) in three of these studies (Davlantes et al., 2018; Kakolwa et al., 2018; Smith et al., 2018); where the 476I mutation is validated with resistance or reduced susceptibility to ART (Table 4) (WHO, 2018). This is consistent with molecular epidemiology studies on the Pfk13 that reported the presence of other validated mutations (i.e., 561H in DRC and India) (Ménard et al., 2016; Mishra et al., 2015). The mutation 561H was found in one of the >9,000 samples from several sSA countries analyzed previously (Ménard et al., 2016).
The ART mode of action still remains elusive and controversial, though it has been shown that they cause oxidative stress by the production of reactive oxygen species (ROS) with likely impact on the multiple cellular targets (Kavishe et al., 2017). A molecule called ferriprotoporphyrin (FP) is a product of hemoglobin digestion, which thereafter polymerizes to hemozoin (malarial pigment). When ARTs are administered, these react with the FP iron (FP–Fe2+) to form activated drugs and alongside form ROS causing attacks on several cellular targets including mitochondrial membrane and transport chain, hemoglobin digesting proteases and many cytosolic targets including the cysteine-containing redox system of the parasite (Kavishe et al., 2017). In addition, it is likely that the seemingly promiscuous nature of ART and its derivatives may have slowed the emergence of resistance (Wang et al., 2015). The authors Mbengue and colleagues established the crucial role of P. falciparum phosphatidylinositol-3-phosphate (PfPI3K) in acquisition of ART resistance (Mbengue et al., 2015) and found a strong association between increased level in PfPI3K and C580Y mutation in the Pfk13 gene, which revealed that K13 protein act as adapter of PfPI3K to control its expression levels (Kavishe et al., 2017). In addition, PfPI3K may impact functions of the apicoplast and food vacuole, host remodeling and cell survival via DNA repair and transcriptional pathways which have also been implicated in ART resistance (Bhattacharjee et al., 2012; Cheeseman et al., 2012; Tawk et al., 2010). In addition, other mechanisms of actions for ART have been proposed recently (Birnbaum et al., 2020; Hou et al., 2019; Yang et al., 2019).
Among the included studies in this review, 15 studies (10 from sSA and five from India) were published before the involvement of the Pfk13 gene in ART resistance was established (Ariey et al., 2014). Thus, none of these studies performed genotyping of this gene among TF cases and as a consequence it is impossible to confirm the causative role of Pfk13 mutations in these studies. Though, one Indian study genotyped the Pfk13 gene and the authors found a novel mutation 625R in four TF and 539T mutation (validated for ART resistance) in one TF (Das et al., 2018). Another study by the same research group, reported the presence of the same mutation (539T) in patients treated with AS + SP (Das et al., 2019). The 539T mutation is recently validated by the WHO for ART resistance (WHO, 2019). Also, the WHO and experts outlined that results of these two studies should be interpreted with caution and concluded that “it is premature to claim that ART resistance has emerged in India” (Rasmussen et al., 2019; WHO, 2019).
4.2.5.2. PfATPase6
Five studies, three from Africa and one from India, included in the review addressed the P. falciparum sarcoplasmic/endoplasmic reticulum Ca2+-ATPase gene (PfATPase6) (Table 2, Table 4) (Gadalla et al., 2011; Happi et al., 2009; Kamugisha et al., 2012; Kiaco et al., 2015; Saha et al., 2012). The PfATPase6 gene codes a protein enzyme, sarcoplasmic reticulum calcium ATPase (SERCA), which plays a crucial role in a large number of vital functions through regulation of cytoplasmic calcium in Plasmodium parasites (Berridge et al., 1998; Kimura et al., 1993). Few other studies found some polymorphisms in this gene, although the association of SNPs in PfATPase6 with ART resistance and the underlying mechanism remains to be confirmed (Table 1) (Chilongola et al., 2015; Li et al., 2016). Jambou and coworkers showed mutation at the position 769N of the PfATPase6 gene was associated with increased 50% inhibitory concentration (>30 nmol/L) following treatment with artemether (Jambou et al., 2005). In contrast, a study found a lack of association between this mutation in the PfATPase6 gene and ART resistance (Cui et al., 2012). These contradictory results imply a need for further studies addressing PfATPase6 and ART resistance. However, keeping in mind that the ART could act at different locations inside the parasite, it is likely that polymorphisms in the PfATPase6 gene or other genes modulate the susceptibility to ART and its derivatives (Cui and Su, 2009; Kavishe et al., 2017).
4.2.5.3. Pfmdr1 and Pfcrt
It was showed that orthologue genes of Pfmdr1encode a putative ATP-binding cassette transporter mediating multidrug resistance in Candida albicans and mammalian cell lines (Duraisingh and Cowman, 2005). The single mutations (86Y, 184Y, 1246Y) and haplotypes (N86–184F-D1246; haplotypes different from N86–184F-D1246 termed as “No N86–184F-D1246”) in Pfmdr1 were found to be selected significantly by the ACT evaluated in the included studies (Table 4). The mutation at 86Y in Pfmdr1 have been shown to be associated with increased sensitivity to structurally unrelated antimalarial drugs (MQ and ART) (Duraisingh et al., 2000). It was also reported that the 86Y mutation in this gene was positively selected by either AS + AQ or AL in three included studies (Table 4) where one study has recently shown to reduce the risk of AL failure with carriage of mutant alleles 86Y compared to wild type N86 in the Pfmdr1 gene (Baraka et al., 2018; Djimdé et al., 2008; Kamugisha et al., 2012; Yeka et al., 2016). This same pattern was found for Pfmdr1 1246Y in two studies included in the review (Otienoburu et al., 2016; Yeka et al., 2016). It was shown that the mutant alleles 86Y, 184Y and 1246Y in the Pfmdr1 gene mediate the reduced susceptibility to CQ and amodiaquine (AQ), but increased in vitro sensitivity to MQ, lumefantrine and ART (Table 1) (Wurtz et al., 2014). It was reported that the haplotype N86–184F-D1246 was positively selected by AL in Nigerian children where the parasite strains carrying this haplotype were known to tolerate high concentrations of lumefantrine (Malmberg et al., 2013). Similarly, in this reported review in four included studies; three of them revealed that this haplotype was positively selected by AL, and in the remaining one study it was shown to be positively selected by AS + SP (Baliraine and Rosenthal, 2011; Gadalla et al., 2013; Happi et al., 2009; Otienoburu et al., 2016).
PfCRT is a phosphoprotein located in the membrane of P. falciparum digestive vacuole and plays a critical role in CQ resistance by directly mediating the efflux of CQ from the parasite digestive vacuole (Bray et al., 2005). Positive selection of wild types in the Pfcrt (K76) and Pfmdr1 (N86 and Y184) genes after AL treatment (Table 4) was found in several studies (Baliraine and Rosenthal, 2011; Happi et al., 2009; Ngasala et al., 2011a, 2011b; Otienoburu et al., 2016). The reappearance of wild type alleles in parasite population is considered as a good indicator of their return to CQ sensitivity as shown by Laufler and coworkers in Malawi (Laufer et al., 2006), though with reduced sensitivity to lumefantrine and two other structurally similar drugs viz MQ and halofantrine (McCarthy and Price, 2015).
4.2.5.4. Pfdhfr and Pfdhps
Resistance to SP is conferred by polymorphisms in Pfdhfr and Pfdhps genes encoding enzymes involved in the folate-pathway essential for P. falciparum survival (Gregson and Plowe, 2005). The Pfdhfr (51I, 59R and 108N) and Pfdhps (437G) genes with single and multiple mutations were positively selected after treatment with AQ + SP and AS + SP, as revealed in two studies reported from Mali and Burkina Faso (Djimdé et al., 2008; Somé et al., 2010) (Table 4) which were also reported in Sudanese and Somalian TF patients treated with AS + SP (Gadalla et al., 2013; Warsame et al., 2017). These reported mutations are known to elicit resistance to SP in malaria parasites (Naidoo and Roper, 2013; Okell et al., 2017a). This could be due to the high selective pressure exerted by the exposition to the therapeutic combination of SP which is largely used as intermittent preventive treatment in pregnancy (IPTp), seasonal malaria chemotherapy in children and ITP in infants in most African countries where malaria is endemic (Group et al., 2017; WHO, 2018a). However, reports from Indian subcontinent stated that all the Pfdhfr and Pfdhps mutations were of multiple kind (double, triple, quadruple and quintuple) in TF with AS + SP as compared to the included African studies where mainly single mutations were found (Baliraine and Rosenthal, 2011; Kamugisha et al., 2012; Otienoburu et al., 2016; Somé et al., 2010; Yeka et al., 2016). In addition, all cases of Pfdhfr/Pfdhps multiple mutations from the included African studies were reported only from the West part of the continent (Djimdé et al., 2008; Otienoburu et al., 2016; Warsame et al., 2015). These two findings from our study are not in line with previous report showing a high circulation of Pfdhfr/Pfdhps multiple mutations, quintuple and even sextuple mutations, in all regions of Africa (Okell et al., 2017b; Ruizendaal et al., 2017; Sridaran et al., 2010). This discrepancy can be due to the fact that we focused on studies that linked results of ACT efficacy and molecular markers associated with drug resistance. Thus, studies that only addressed the epidemiology of molecular markers associated with drug resistance got excluded from our analysis. The molecular epidemiological studies observed that Pfdhfr/Pfdhps multiple mutations, especially double mutations 437G + 540E are more commonly found in East African countries than in West African countries (Sridaran et al., 2010). Higher drug pressure due to the higher utilization of AS + SP in East African countries could be responsible for this increase in the polymorphisms (WHO, 2019). Also, parasites with multiple Pfdhfr and Pfdhps mutations have a higher level of in vitro resistance and have been shown to be a strong predictor for TF as compared to their single counterparts (Mombo-ngoma et al., 2011; Ndiaye et al., 2013). The presence of high prevalence of parasites with multiple Pfdhfr and Pfdhps mutations found in TF cases with AS + SP from Indian studies indicates the need for an eventual switch to other ACT in the future.
4.2.4.5. Pfpm2 and 3
P. falciparum plasmepsin (Pfpm) 2 and 3 are located in the food vacuole and involved in the degradation of hemoglobin into yielding amino acids for protein synthesis with toxic byproduct heme (Moura et al., 2009). Treatment with PPQ leads to a depletion of ribosomes and swelling of the food vacuole with undigested hemoglobin vesicles and an accumulation of free heme in the parasite (Moura et al., 2009). Although the free heme is toxic for the parasite, inhibition of hemoglobin degradation will starve the parasite and increased Pfpm2 and 3 might help the parasite to maintain amino-acid production when hemoglobin degradation is otherwise inhibited by PPQ (Amato et al., 2017).
In our review, three studies investigated the selective pressure of DHA + PPQ on the Pfpm2 gene but found an absence of such selection and one of these studies found multiple copies of Pfpm2 but no association with DHA + PPQ failures was reported (Table 4) (Davlantes et al., 2018; Kakolwa et al., 2018; Smith et al., 2018). Witkowski and colleagues demonstrated a strong link between an increase in the proportion of multicopy Pfpm2 parasites and DHA + PPQ related TF rates in Cambodia (Witkowski et al., 2017). It should be noted that some strains were already ART resistant and this could explain this strong link found by these authors. Two other studies showed an increased sensitivity to PPQ in malaria strains in which Pfpm2 and 3 were inactivated (Amato et al., 2017; Mukherjee et al., 2018). In contrast, a more recent study showed conflicting results using genetically engineered laboratory malarial strains. The authors found no major changes in susceptibility to PPQ due to increased number of copies the Pfpm2 and Pfpm3 genes (Loesbanluechai et al., 2019). Two other studies concluded that an increase in copy number of Pfpm2 is not required to render P. falciparum laboratory strains (Dd2) and clinical isolates resistant to PPQ (Boonyalai et al., 2020; Ross et al., 2018).
All these above discussed data suggest that the Pfpm2 would not be the only gene involved in resistance to PPQ and DHA + PPQ failures. This is supported by findings from genetic engineering-based studies that have suggested that mutations in the P. falciparum exonuclease (Pfexo) and Pfcrt genes are associated with PPQ resistance and DHA + PPQ failures (Amato et al., 2017; Boonyalai et al., 2020; Ross et al., 2018).
To date, increased copy number of Pfpm2 is considered and commonly used as a marker of PPQ resistance and DHA + PPQ failures, particularly in Southeast Asian countries as above presented (Boonyalai et al., 2020; Huang et al., 2020; van der Pluijm et al., 2019). In contrast, studies on mutations in Pfpm2 and its link with PPQ resistance is increasingly available in Africa where mutations in this gene have been documented in high transmission areas of the continent, while in India, there is not much available information about the polymorphisms of this gene (Inoue et al., 2018). A study conducted in several sSA countries reported a low prevalence of P. falciparum isolates with reduced susceptibility to PPQ, and none of these isolates had multiple copies of Pfpm2 (Robert et al., 2018). Thus, it is necessary to study the role of Pfpm2 on PPQ resistance and DHA + PPQ failures in sSA as this ACT is being included in national treatment policies of several African countries (such as Cameroon, Senegal) (Ministere de la Sante Publique, 2019).
4.2.5. ACT and risk of drug resistance
The evidence of selection of mutant variants associated with drug resistance in the ACT suggests the importance of follow up in order to know the prevalence of sensitive strains circulating in malarious patients. In the present review it was observed that all selected and/or involved polymorphisms in TF were related to partner drug of the ART derivative; thereby stressing the importance to preserve the efficacy of partner drugs (Nsanzabana, 2019). Also, using a meta-analysis, Zhou and colleagues found a higher risk of drug resistance emergence using AS + SP compared to AL while the risk was lower in DHA + PPQ compared to AL (Zhou et al., 2016) with no difference among the following different ACT viz AL with AS + MQ or AS + AQ, and ii) AS + AQ and AS + SP (Zhou et al., 2016). Thus, a possibility to prevent the appearance of ACT resistance could be the utilization of DHA + PPQ, which has the lowest risk of ART resistance emergence, in countries where this ACT is not used. Another approach to the protection of ACT efficacy would be the cyclic utilization of other non- ACT antimalarial drugs such as CQ where decreased efficacy of ACT is reported. Some authors reported return to plasmodial sensitivity to CQ in Malawi a few years after withdrawal of this antimalarial drug thereby suggesting its reutilization, in combination with other drug as seen with ACT, as first-line treatment could be interesting approach in these areas (Laufer et al., 2006). Besides, the possibility to use ACT containing more than one partner drug (termed as “triple ACTs”), although currently controversial, could be an alternative approach as recently modelled and debated (Dini et al., 2018; Krishna, 2019; White, 2019). All these suggested steps taken together could help in achieving malaria elimination objectives of the WHO in sSA and India.
5. Limitations of the review
One of the limitations of this review could be a selection bias as only published articles written in English or French were considered for this review analysis. There is a possibility that some studies were missed even though a rigorous strategy was used to search for relevant papers. The other important limitation was lack of coverage of all African countries where malaria is highly endemic but no studies are reported from these countries.
6. Conclusions
In conclusion, ART resistance is the main threat to malaria control in endemic countries worldwide. Firstly, in this review high efficacy of ACT in Africa and India was observed thereby, supporting the need for protection of this efficacy over space and time. Secondly, the study noted the non-synonymous mutations in the Pfk13 gene and ART resistance has not emerged in sSA and India yet, despite the fact that some clinical efficacy and epidemiological studies have reported the circulation of some validated mutations (i.e., 476I, 539T and 561H) in these two highly malaria endemic areas. This review also highlights a selective action of ACT on some molecular markers associated with resistance to partner drugs and thereby supporting the fact that TF does not necessarily mean ART resistance (Krishna and Kremsner, 2013). Finally, we outline the need to pursue researches on other possible molecular markers of ART resistance such as PfATPase6 for which the role in ART resistance is still controversial. Also, the link between multiple copies of Pfpm2, PPQ resistance and DHA + PPQ failures in malaria endemic areas other than Southeast Asia area needs more research. Given the fact that DHA + PPQ pressure is increasing in sSA as this ACT is being adopted by several sSA countries, it is important to have studies on African P. falciparum isolates for PPQ resistance. It is hence crucial to implement, maintain and scale up the therapeutic efficacy surveillance systems in endemic countries in order to induce changes in treatment policies to protect the efficacy of ACT with special emphasis on partner drugs. All these strategies coupled together will thereby limit the chance of emergence and spread of ART resistance and likely achieve the elimination targets by 2030 in India and most African countries.
Funding
The present study has received no financial support.
Author's contributions
Aditi Arya: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Data curation, Writing-Original draft, Writing- Review and Editing.
Loick Pradel Kojom Foko: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Data curation, Writing-Original draft, Writing- Review and Editing.
Shewta Chaudhry: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Data curation, Writing-Original draft, Writing- Review and Editing.
Amit Sharma: Critical review and final editing.
Vineeta Singh: Conceptualization, Methodology, Validation, Investigation, Writing- Review and Editing, Supervision, Project administration.
Data availability statement
The authors confirm that the data supporting the findings of this study are available within the article and in the attached supplementary files.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijpddr.2020.11.006.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Adam I., Ibrahim Y., Gasim G.I. Efficacy and safety of artemisinin-based combination therapy for uncomplicated Plasmodium falciparum malaria in Sudan: a systematic review and meta - analysis. Malar. J. 2018;17:110. doi: 10.1186/s12936-018-2265-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adjekukor C.U. Past and current findings in antimalarial drug resistance molecular markers in endemic areas of Africa. Int J Trop Dis Health. 2018;30:1–11. doi: 10.9734/IJTDH/2018/41204. [DOI] [Google Scholar]
- Amato R., Lim P., Miotto O., Neal A.T., Sreng S., Suon S., Drury E. Genetic markers associated with dihydroartemisinin–piperaquine failure in Plasmodium falciparum malaria in Cambodia: a genotype-phenotype association study. Lancet Infect. Dis. 2017;17:164–173. doi: 10.1016/S1473-3099(16)30409-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antony H.A., Parija S.C. Antimalarial drug resistance: an overview. Tropenmed. Parasitol. 2016;6:30–41. doi: 10.4103/2229-5070.175081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariey F., Witkowski B., Amaratunga C., Beghain J., Langlois A.-C., Khim N., Duru V., Bouchier C., Ma L., Lim P. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature. 2014;505:50–55. doi: 10.1007/s40778-014-0003-z.Genome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashley E.A., Phyo A.P., Woodrow C.J. Malaria. Lancet. 2018;391:1608–1621. doi: 10.1016/S0140-6736(18)30324-6. [DOI] [PubMed] [Google Scholar]
- Baliraine F.N., Rosenthal P.J. Prolonged selection of pfmdr1 polymorphisms after treatment of falciparum malaria with Artemether-Lumefantrine in Uganda. J. Infect. Dis. 2011;204:1120–1124. doi: 10.1093/infdis/jir486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal D., Acharya A., Bharti P.K., Abdelraheem M.H., Elmalik A., Abosalah S., Khan F.Y., ElKhalifa M., Kaur H., Mohapatra P.K., Sehgal R., Idris M.A., Mahanta J., Singh N., Babiker H.A., Sultan A.A. Distribution of mutations associated with antifolate and chloroquine resistance among imported Plasmodium vivax in the State of Qatar. Am. J. Trop. Med. Hyg. 2017;97:1797–1803. doi: 10.4269/ajtmh.17-0436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baraka V., Mavoko H.M., Nabasumba C., Francis F., Lutumba P., Alifrangis M., Van Geertruyden J.-P. Impact of treatment and re-treatment with artemether-lumefantrine and artesunate- amodiaquine on selection of Plasmodium falciparum multidrug resistance gene-1 polymorphisms in the Democratic Republic of Congo and Uganda. PloS One. 2018;13 doi: 10.5061/dryad.61m9p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beavogui A.H., Camara A., Delamou A., Diallo M.S., Doumbouya A., Kourouma K., Bouedouno P., Guilavogui T., Dos Santos Souza S., Kelley J., Talundzic E., Fofana A., Plucinski M.M. Efficacy and safety of artesunate-amodiaquine and artemether-lumefantrine and prevalence of molecular markers associated with resistance, Guinea: an open-label two-arm randomised controlled trial. Malar. J. 2020;19:223. doi: 10.1186/s12936-020-03290-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett A., Bisanzio D., Yukich J.O., Mappin B., Fergus C.A., Lynch M., Cibulskis R.E., Bhatt S. Population coverage of artemisinin-based combination treatment in children younger than 5 years with fever and Plasmodium falciparum infection in Africa, 2003–2015: a modelling study using data from national surveys. Lancet Glob Health. 2017;5:e418–e427. doi: 10.1016/S2214-109X(17)30076-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berridge M.J., Bootman M.D., Lipp P. Molecular biology: calcium - a life and death signal. Nature. 1998;395:645–648. doi: 10.1038/27094. [DOI] [PubMed] [Google Scholar]
- Berzosa P., Cantos A.E., García L., González V., Navarro M., Fernández T., Barja M.R., Herrador Z., Rubio J.M., Ncogo P. Profile of molecular mutations in pfdhfr, pfdhps, pfmdr1, and pfcrt genes of Plasmodium falciparum related to resistance to different anti-malarial drugs in the Bata District (Equatorial Guinea) Malar. J. 2017;16:28. doi: 10.1186/s12936-016-1672-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee S., Stahelin R.V., Speicher K.D., Speicher D.W., Haldar K. Endoplasmic reticulum PI(3)P lipid binding targets malaria proteins to the host cell. Cell. 2012;148:201–212. doi: 10.1016/j.cell.2011.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum J., Scharf S., Schmidt S., Jonscher E., Maria Hoeijmakers W.A., Flemming S., Toenhake C.G., Schmitt M., Sabitzki R., Bergmann B., Fröhlke U., Mesén-Ramírez P., Soares A.B., Herrmann H., Bártfai R., Spielmann T. A Kelch13-defined endocytosis pathway mediates artemisinin resistance in malaria parasites. Science. 2020;367:51–59. doi: 10.1126/science.aax4735. [DOI] [PubMed] [Google Scholar]
- Blasco B., Leroy D., Fidock D.A., Diseases I. Antimalarial drug resistance: linking Plasmodium falciparum parasite biology to the clinic. Nat. Med. 2017;23:917–928. doi: 10.1038/nm.4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonyalai N., Vesely B.A., Thamnurak C., Praditpol C., Fagnark W., Kirativanich K., Saingam P., Chaisatit C., Lertsethtakarn P., Gosi P., Kuntawunginn W., Vanachayangkul P., Spring M.D., Fukuda M.M., Lon C., Smith P.L., Waters N.C., Saunders D.L., Wojnarski M. Piperaquine resistant Cambodian Plasmodium falciparum clinical isolates: in vitro genotypic and phenotypic characterization. Malar. J. 2020;19:269. doi: 10.1186/s12936-020-03339-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bopp S., Magistrado P., Wong W., Schaffner S.F., Mukherjee A., Lim P., Dhorda M., Amaratunga C., Woodrow C.J., Ashley E.A., White N.J., Dondorp A.M., Fairhurst R.M., Ariey F., Menard D., Wirth D.F., Volkman S.K. Plasmepsin II – III copy number accounts for bimodal. Nat. Commun. 2018;9:1769. doi: 10.1038/s41467-018-04104-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray P.G., Martin R.E., Tilley L., Ward S.A., Kirk K., Fidock D.A. Defining the role of PfCRT in Plasmodium falciparum chloroquine resistance. Mol. Microbiol. 2005;56:323–333. doi: 10.1111/j.1365-2958.2005.04556.x. [DOI] [PubMed] [Google Scholar]
- Cheeseman I.H., Miller B.A., Nair S., Nkhoma S., Tan A., Tan J.C., Al Saai S., Phyo A.P., Ler Moo C., Lwin K.M., McGready R., Ashley E., Imwong M., Stepniewska K., Yi P., Dondorp A.M., Mayxay M., Newton P.N., White N.J., Nosten F., Ferdig M.T., Anderson T.J.C. A major genome region underlying artemisinin resistance in malaria. Science. 2012;335:79–82. doi: 10.1126/science.1215966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chilongola J., Ndaro A., Tarimo H., Shedrack T., Barthazary S., Kaaya R., Masokoto A., Kajeguka D., Kavishe R.A., Lusingu J. Occurrence of pfatpase6 single nucleotide polymorphisms associated with Artemisinin resistance among field isolates of Plasmodium falciparum in North-Eastern Tanzania. Malar. Res. Treat. 2015;2015:1–7. doi: 10.1155/2015/279028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chookajorn T. How to combat emerging artemisinin resistance : lessons from “ the three little pigs. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1006923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad M.D., Rosenthal P.J. Antimalarial drug resistance in Africa: the calm before the storm? Lancet Infect. Dis. 2019;19:e338–e351. doi: 10.1016/S1473-3099(19)30261-0. [DOI] [PubMed] [Google Scholar]
- Cui L., Su X. Discovery, mechanisms of action and combination therapy of artemisinin. Expert Rev. Anti Infect. Ther. 2009;7:999–1013. doi: 10.1038/jid.2014.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Long, Wang Z., Jiang H., Parker D., Wang H., Su X.Z., Cui Liwang. Lack of association of the S769N mutation in Plasmodium falciparum SERCA (PfATP6) with resistance to artemisinins. Antimicrob. Agents Chemother. 2012;56:2546–2552. doi: 10.1128/AAC.05943-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S., Mahapatra S.K., Tripathy S., Chattopadhyay S., Dash S.K., Mandal D., Das B., Hati A.K., Roy S. Double mutation in the pfmdr1 gene is associated with emergence of chloroquine-resistant Plasmodium falciparum malaria in Eastern India. Antimicrob. Agents Chemother. 2014;58:5909–5915. doi: 10.1128/AAC.02762-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S., Manna S., Saha B., Hati A.K., Roy S. Novel pfkelch13 Gene polymorphism associates with Artemisinin resistance in Eastern India. Clin. Infect. Dis. 2019;69:1144–1152. doi: 10.1093/cid/ciy1038. [DOI] [PubMed] [Google Scholar]
- Das S., Saha B., Hati A.K., Roy S. Evidence of artemisinin-resistant plasmodium falciparum malaria in eastern India. N. Engl. J. Med. 2018;379:1962–1964. doi: 10.1056/NEJMc1713777. [DOI] [PubMed] [Google Scholar]
- Davlantes E., Dimbu P.R., Ferreira C.M., Florinda Joao M., Pode D., Félix J., Sanhangala E., Andrade B.N., Dos Santos Souza S., Talundzic E., Udhayakumar V., Owens C., Mbounga E., Wiesner L., Halsey E.S., Martins J.F., Fortes F., Plucinski M.M. Efficacy and safety of artemether-lumefantrine, artesunate-amodiaquine, and dihydroartemisinin-piperaquine for the treatment of uncomplicated Plasmodium falciparum malaria in three provinces in Angola, 2017. Malar. J. 2018;17:144. doi: 10.1186/s12936-018-2290-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettori J. The random allocation process: two things you need to know. Evid. Base Spine Care J. 2010;1:7–9. doi: 10.1055/s-0030-1267062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diallo M.A., Yade M.S., Ndiaye Y.D., Diallo I., Diongue K., Sy S.A., Sy M., Seck M.C., Ndiaye M., Dieye B., Gomis J.F., Sow D., Dème A.B., Badiane A.S., Ndiaye D. Efficacy and safety of artemisinin-based combination therapy and the implications of Pfkelch13 and Pfcoronin molecular markers in treatment failure in Senegal. Sci. Rep. 2020;10:8907. doi: 10.1038/s41598-020-65553-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dini S., Zaloumis S., Cao P., Price R.N., Fowkes F.J.I., Pluijm R.W. Van Der. Investigating the efficacy of triple artemisinin-based combination therapies for treating Plasmodium falciparum malaria patients using mathematical modeling. Antimicrob. Agents Chemother. 2018;62 doi: 10.1128/AAC.01068-18. e01068-01018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djimdé A.A., Fofana B., Sagara I., Sidibe B., Toure S., Dembele D. Efficacy, safety, and selection of molecular markers of drug resistance by two ACTs in Mali. Am. J. Trop. Med. Hyg. 2008;78:455–461. [PubMed] [Google Scholar]
- Dondorp A.M., Nosten F., Yi P., Das D., Phyo A.P., Tarning J., Ph D., Lwin K.M., Ariey F., Hanpithakpong W., Lee S.J., Ringwald P., Silamut K., Herdman T., An S.S., Yeung S., Socheat D., White N.J. Artemisinin resistance in plasmodium falciparum malaria. N. Engl. J. Med. 2009;361:455–467. doi: 10.1086/657120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duraisingh M.T., Cowman A.F. Contribution of the pfmdr1 gene to antimalarial drug-resistance. Acta Trop. 2005;94:181–190. doi: 10.1016/j.actatropica.2005.04.008. [DOI] [PubMed] [Google Scholar]
- Duraisingh M.T., Jones P., Sambou I., Von Seidlein L., Pinder M., Warhurst D.C. The tyrosine-86 allele of the pfmdr1 gene of Plasmodium falciparum is associated with increased sensitivity to the anti-malarials mefloquine and artemisinin. Mol. Microbiol. 2000;108:13–23. doi: 10.1016/S0166-6851(00)00201-2. [DOI] [PubMed] [Google Scholar]
- Ferdig M.T., Cooper R.A., Mu J., Deng B., Joy D.A., Su X.Z., Wellems T.E. Dissecting the loci of low-level quinine resistance in malaria parasites. Mol. Microbiol. 2004;52:985–997. doi: 10.1111/j.1365-2958.2004.04035.x. [DOI] [PubMed] [Google Scholar]
- Gadalla N.B., Abdallah T.M., Atwal S., Sutherland C.J., Adam I. Selection of pfdhfr/pfdhps alleles and declining artesunate/sulphadoxine-pyrimethamine efficacy against Plasmodium falciparum eight years after deployment in eastern Sudan. Malar. J. 2013;12:255. doi: 10.1186/1475-2875-12-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadalla N.B., Adam I., Elzaki S., Bashir S., Mukhtar I., Oguike M., Gadalla A., Mansour F., Warhurst D., El-sayed B.B., Sutherland C.J. Increased pfmdr1 copy number and sequence polymorphisms in Plasmodium falciparum isolates from Sudanese malaria patients treated with Artemether-Lumefantrine. Antimicrob. Agents Chemother. 2011;55:5408–5411. doi: 10.1128/AAC.05102-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly S., Saha P., Guha S.K., Biswas A., Das S., Kundu P.K., Maji A.K. High prevalence of asymptomatic Malaria in a tribal population in Eastern India. J. Clin. Microbiol. 2013;51:1439–1444. doi: 10.1128/JCM.03437-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogtay N., Kannan S., Um T., Pl O., Sinclair D. Artemisinin-based combination therapy for treating uncomplicated Plasmodium vivax malaria (Review) 2013. Cochrane Database Syst Rev CD008492. [DOI] [PMC free article] [PubMed]
- Gregson A., Plowe C.V. Mechanisms of resistance of malaria parasites to antifolates. Pharmacol. Rev. 2005;57:117–145. doi: 10.1124/pr.57.1.4. [DOI] [PubMed] [Google Scholar]
- Group Act, Tougher S., Hanson K., Goodman C. What happened to anti-malarial markets after the Affordable Medicines Facility-malaria pilot? Trends in ACT availability, price and market share from five African countries under continuation of the private sector co-payment mechanism. Malar. J. 2017;16:173. doi: 10.1186/s12936-017-1814-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldar K., Bhattacharjee S., Safeukui I. Drug resistance in plasmodium. Nat. Rev. Microbiol. 2018;16:156–170. doi: 10.1038/nrmicro.2017.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Happi C.T., Gbotosho G.O., Folarin O.A., Sowunmi A., Hudson T., Neil M.O., Milhous W., Wirth D.F., Oduola A.M.J. Selection of Plasmodium falciparum multidrug resistance gene 1 alleles in asexual stages and gametocytes by Artemether-Lumefantrine in Nigerian children with uncomplicated falciparum malaria. Antimicrob. Agents Chemother. 2009;53:888–895. doi: 10.1128/AAC.00968-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry M., Briolant S., Zettor A., Pelleau S., Baragatti M., Baret E., Mosnier J., Amalvict R., Fusai T., Rogier C., Pradines B. Plasmodium falciparum Na+/H+ exchanger 1 transporter is involved in reduced susceptibility to quinine. Antimicrob Agents Chemother. 2009;53:1926–1930. doi: 10.1128/AAC.01243-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou H., Zhang G., MA L., Su P., Zhang Z., Dai B., Ye Z. Effects and mechanism of action of Artemisinin on mitochondria of Plasmodium berghei. Chin. J. Integr. Med. 2019;26:277–282. doi: 10.1007/s11655-019-3164-x. [DOI] [PubMed] [Google Scholar]
- Huang F., Shrestha B., Liu H., Tang L.H., Zhou S. Sen, Zhou X.N., Takala-Harrison S., Ringwald P., Nyunt M.M., Plowe C.V. No evidence of amplified Plasmodium falciparum plasmepsin II gene copy number in an area with artemisinin-resistant malaria along the China-Myanmar border. Malar. J. 2020;19:334. doi: 10.1186/s12936-020-03410-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M., Kaneko M., Tachibana S.-I., Balikagala B., Sakurai-Yatsushiro M., Yatsushiro S., Takahashi N., Yamauchi M., Sekihara M., Hashimoto M., Katuro O.T., Olia A., Obwoya P.S., Auma M.A., Anywar D.A., Odongo-aginya E.I., Okello-onen J., Hirai M., Ohashi J., Palacpac N.M.Q., Kataoka M., Tsuboi T., Kimura E., Horii T., Mita T. Artemisinin-resistant Plasmodium falciparum with high survival rates, Uganda, 2014-2016. Emerg. Infect. Dis. 2018;24:718–726. doi: 10.3201/eid2404.170141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue J., Silva M., Fofana B., Sanogo K., Martensson A., Sagara I., Bjorkman A., Veiga M.I., Ferreira P.E., Djimdé A.A., Gil J.P. Plasmodium falciparum plasmepsin 2 duplications, West Africa. Emerg. Infect. Dis. 2018;24:1591–1593. doi: 10.3201/eid2408.180370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalei A.A., Chaijaroenkul W., Na-bangchang K. Plasmodium falciparum drug resistance gene status in the Horn of Africa : a systematic review. Afr. J. Pharm. Pharmacol. 2018;12:361–373. doi: 10.5897/AJPP2018.4942. [DOI] [Google Scholar]
- Jambou R., Legrand E., Niang M., Khim N., Lim P., Volney B., Ekala M.T., Bouchier C., Esterre P., Fandeur T., Mercereau-Puijalon O. Resistance of Plasmodium falciparum field isolates to in-vitro artemether and point mutations of the SERCA-type PfATPase6. Lancet. 2005;366:1960–1963. doi: 10.1016/S0140-6736(05)67787-2. [DOI] [PubMed] [Google Scholar]
- Kakolwa M.A., Mahende M.K., Ishengoma D.S., Mandara C.I., Ngasala B., Kamugisha E., Kataraihya J.B., Mandike R., Mkude S., Chacky F., Njau R., Premji Z., Lemnge M.M., Warsame M., Menard D., Kabanywanyi A.M. Efficacy and safety of artemisinin-based combination therapy, and molecular markers for artemisinin and piperaquine resistance in Mainland Tanzania. Malar. J. 2018;17:369. doi: 10.1186/s12936-018-2524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamugisha E., Jing S., Minde M., Kataraihya J., Kongola G., Kironde F., Swedberg G. Efficacy of artemether-lumefantrine in treatment of malaria among under-fives and prevalence of drug resistance markers in Igombe-Mwanza, north-western Tanzania. Malar. J. 2012;11:58. doi: 10.1186/PREACCEPT-6142726864727876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karema C., Imwong M., Fanello C.I., Stepniewska K., Uwimana A., Nakeesathit S., Dondorp A., Day N.P., White N.J. Molecular correlates of high-level antifolate resistance in Rwandan children with Plasmodium falciparum malari. Antimicrob. Agents Chemother. 2010;54:477–483. doi: 10.1128/AAC.00498-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavishe R.A., Koenderink J.B., Alifrangis M. Oxidative stress in malaria and artemisinin combination therapy : pros and Cons. FEBS J. 2017;284:2579–2591. doi: 10.1111/febs.14097. [DOI] [PubMed] [Google Scholar]
- Kiaco K., Teixeira J., Machado M., Rosário V., Lopes D. Evaluation of artemether-lumefantrine efficacy in the treatment of uncomplicated malaria and its association with pfmdr1, pfatpase6 and K13-propeller polymorphisms in Luanda, Angola. Malar. J. 2015;14:504. doi: 10.1186/s12936-015-1018-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M., Yamaguchi Y., Takada S., Tanabe K. Cloning of a Ca2+-ATPase gene of Plasmodium falciparum and comparison with vertebrate Ca2+-ATPases. J. Cell Sci. 1993;104:1129–1136. doi: 10.1242/jcs.104.4.1129. [DOI] [PubMed] [Google Scholar]
- Klonis N., Xie S.C., McCaw J.M., Crespo-Ortiz M.P., Zaloumis S.G., Simpson J.A., Tilley L. Altered temporal response of malaria parasites determines differential sensitivity to artemisinin. Proc. Natl. Acad. Sci. U.S.A. 2013;110:5157–5162. doi: 10.1073/pnas.1217452110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korsinczky M., Chen N., Kotecka B., Saul A., Rieckmann K., Cheng Q. Mutations in Plasmodium falciparum cytochrome b that are associated with atovaquone resistance are located at a putative drug-binding site. Antimicrob. Agents Chemother. 2000;44:2100–2108. doi: 10.1128/AAC.44.8.2100-2108.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna S. Triple artemisinin-containing combination anti-malarial treatments should be implemented now to delay the emergence of resistance: the case against. Malar. J. 2019;18:339. doi: 10.1186/s12936-019-2976-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna S., Kremsner P.G. Antidogmatic approaches to artemisinin resistance: reappraisal as treatment failure with artemisinin combination therapy. Trends Parasitol. 2013;29:313–317. doi: 10.1016/j.pt.2013.04.001. [DOI] [PubMed] [Google Scholar]
- Kumar A., Moirangthem R., Gahlawat S.K., Chandra J., Gupta P., Valecha N., Anvikar A., Singh V. Emergence of sulfadoxine-pyrimethamine resistance in Indian isolates of Plasmodium falciparum in the last two decades. Infect. Genet. Evol. 2015;36:190–198. doi: 10.1016/j.meegid.2015.08.027. [DOI] [PubMed] [Google Scholar]
- Laufer M.K., Thesing P.C., Eddington N.D., Masonga R., Dzinjalamala F.K., Takala S.L., Taylor T.E., Plowe C.V. Return of chloroquine antimalarial efficacy in Malawi. N. Engl. J. Med. 2006;355:1959–1966. doi: 10.1056/NEJMoa062032. [DOI] [PubMed] [Google Scholar]
- Li J., Chen J., Xie D., Monsuy U., Apicante R., Miko M., Obono O., Sala C., Yang L., Yang H., Lin M. Limited artemisinin resistance-associated polymorphisms in Plasmodium falciparum K13-propeller and PfATPase6 gene isolated from Bioko Island, Equatorial Guinea. Int. J. Parasitol.: Drugs Drug Resist. 2016;6:54–59. doi: 10.1016/j.ijpddr.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingani M., Bonkian L.N., Yerbanga I., Kazienga A., Valéa I., Sorgho H., Ouédraogo J.B., Mens P.F., Schallig H.D.F.H., Ravinetto R., D'AlesSAndro U., Tinto H. In vivo/ex vivo efficacy of artemether-lumefantrine and artesunate-amodiaquine as first-line treatment for uncomplicated falciparum malaria in children: an open label randomized controlled trial in Burkina Faso. Malar. J. 2020;19:8. doi: 10.1186/s12936-019-3089-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loesbanluechai D., Kotanan N., Cozar C. De, Kochakarn T., Ansbro M.R., Chotivanich K., White N.J., Wilairat P., Lee M.C.S., Javier F., Maria L., Chookajorn T., Kümpornsin K. Overexpression of plasmepsin II and plasmepsin III does not directly cause reduction in Plasmodium falciparum sensitivity to artesunate, chloroquine and piperaquine. Int. J. Parasitol.: Drugs Drug Resist. 2019;9:16–22. doi: 10.1016/j.ijpddr.2018.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu F., Culleton R., Zhang M., Ramaprasad A., Seidlein von L., Zhou H., Zhu G., Tang J., Liu Y., Wang W., Cao Y., Xu S., Gu Y., Li J., Zhang C., Gao Q., Menard D., Pain A., Yang H., Zhang Q., Cao J. Emergence of indigenous artemisinin-resistant plasmodium falciparum in Africa. N. Engl. J. Med. 2017;376:991–993. doi: 10.1056/NEJMc1612765. [DOI] [PubMed] [Google Scholar]
- Maji A.K. Drug susceptibility testing methods of antimalarial agents. Tropenmed. Parasitol. 2018;8:70–76. doi: 10.4103/2229-5070.248695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmberg M., Ferreira P.E., Tarning J., Ursing J., Ngasala B., Björkman A., Mårtensson A., Gil J.P. Plasmodium falciparum drug resistance phenotype as assessed by patient antimalarial drug levels and its association with pfmdr1 polymorphisms. J. Infect. Dis. 2013;207:842–847. doi: 10.1093/infdis/jis747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbengue A., Bhattacharjee S., Pandharkar T., Liu H., Estiu G., Stahelin R.V., Rizk S.S., Njimoh D.L., Ryan Y., Chotivanich K., Nguon C., Ghorbal M., Lopez-Rubio J.J., Pfrender M., Emrich S., Mohandas N., Dondorp A.M., Wiest O., Haldar K. A molecular mechanism of artemisinin resistance in Plasmodium falciparum malaria. Nature. 2015;520:683–687. doi: 10.1038/nature14412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy J., Price R. Antimalarial drugs. In: Bennett John E., D R., B M.J., editors. Mandell, Douglas, and Bennett's Principles and Practice of Infectious Diseases. eighth ed. 2015. p. 4320. [Google Scholar]
- Menard D., Dondorp A. Antimalarial drug resistance: a threat to malaria elimination. Cold Spring Harb Perspect Med. 2017;7:a025619. doi: 10.1101/cshperspect.a025619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ménard D., Khim N., Beghain J., Adegnika A.A., Shafiul-Alam M., Amodu O., Rahim-Awab G., Barnadas C., Berry A., Boum Y., Bustos M.D., Cao J., Chen J.-H., Collet L., Cui L., Thakur G.-D., Dieye A., Djallé D., Dorkenoo M.A., Eboumbou-Moukoko C.E., Espino F.-E.-C.J., Fandeur T., Ferreira-da-Cruz M.-F., Fola A.A., Fuehrer H.-P., HasSAn A.M., Herrera S., Hongvanthong B., Houzé S., Ibrahim M.L., Jahirul-Karim M., Jiang L., Kano S., Ali-Khan W., Khanthavong M., Kremsner P.G., Lacerda M., Leang R., Leelawong M., Li M., Lin K., Mazarati J.-B., Ménard S., Morlais I., Muhindo-Mavoko H., Musset L., Na-Bangchang K., Nambozi M., Niaré K., Noedl H., Ouédraogo J.-B., Pillai D.R., Pradines B., Quang-Phuc B., Ramharter M., Randrianarivelojosia M., Sattabongkot J., Sheikh-Omar A., Silué K.D., Sirima S.B., Sutherland C., Syafruddin D., Tahar R., Tang L.-H., Touré O.A., Tshibangu-wa-Tshibangu P., Vigan-Womas I., Warsame M., Wini L., Zakeri S., Kim S., Eam R., Berne L., Khean C., Chy S., Ken M., Loch K., Canier L., Duru V., Legrand E., Barale J.-C., Stokes B., Straimer J., Witkowski B., Fidock D.A., Rogier C., Ringwald P., Ariey F., Mercereau-Puijalon O. A worldwide map of Plasmodium falciparum K13-Propeller polymorphisms. N. Engl. J. Med. 2016;374:2453–2464. doi: 10.1056/nejmoa1513137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ministere de la Sante Publique . 2019. Guide de prise en charge du paludisme au Cameroun. A l’usage du personnel de santé. Yaoundé. [Google Scholar]
- Mishra N., Kaitholia K., Srivastava B., Shah N.K., Narayan J.P., Dev V., Phookan S., Anvikar A.R., Rana R., Bharti R.S., Sonal G.S. Declining efficacy of artesunate plus sulphadoxine-pyrimethamine in northeastern India. Malar. J. 2014;13:284. doi: 10.1186/1475-2875-13-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra N., Narayan P., Srivastava B., Arora U., Shah N.K., Ghosh S.K. Monitoring antimalarial drug resistance in India via sentinel sites: outcomes and risk factors for treatment failure, 2009 – 2010. Bull. World Health Organ. 2012;90:895–904. doi: 10.2471/BLT.12.109124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra N., Prajapati K., Kaitholia K., Bharti S., Srivastava B., Phookan S. Surveillance of artemisinin resistance in Plasmodium falciparum in India using the kelch13 molecular marker. Antimicrob. Agents Chemother. 2015;59:2548–2553. doi: 10.1128/AAC.04632-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra S., Bharti P.K., Shukla M.M., Ali N.A., Kashyotia S.S., Kumar A., Dhariwal A.C., Singh N. Clinical and molecular monitoring of Plasmodium falciparum resistance to antimalarial drug (artesunate+sulphadoxine-pyrimethamine) in two highly malarious district of Madhya Pradesh, Central India from 2012–2014. Pathog. Glob. Health. 2017;111:186–194. doi: 10.1080/20477724.2017.1331875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moher D., Liberati A., Tetzlaff J., Altman D.G., Altman D., Antes G., Atkins D., Barbour V., Barrowman N., Berlin J.A., Clark J., Clarke M., Cook D., D'Amico R., Deeks J.J., Devereaux P.J., Dickersin K., Egger M., Ernst E., Gøtzsche P.C., Grimshaw J., Guyatt G., Higgins J., Ioannidis J.P.A., Kleijnen J., Lang T., Magrini N., McNamee D., Moja L., Mulrow C., Napoli M., Oxman A., Pham B., Rennie D., Sampson M., Schulz K.F., Shekelle P.G., Tovey D., Tugwell P. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6 doi: 10.3736/jcim20090918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mombo-ngoma G., Oyakhirome S., Ord R., Gabor J.J., Greutélaers K.C., Profanter K., Greutélaers B., Kurth F., Lell B., Kun J.F.J., Issifou S., Roper C., Kremsner P.G., Grobusch M.P. High prevalence of dhfr triple mutant and correlation with high rates of sulphadoxine- pyrimethamine treatment failures in vivo in Gabonese children. Malar. J. 2011;10:123. doi: 10.1186/1475-2875-10-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moura P.A., Dame J.B., Fidock D.A. Role of Plasmodium falciparum digestive vacuole plasmepsins in the specificity and antimalarial mode of action of cysteine and aspartic protease inhibitors. Antimicrob. Agents Chemother. 2009;53:4968–4978. doi: 10.1128/AAC.00882-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A., Gagnon D., Wirth D.F. Inactivation of plasmepsins 2 and 3 sensitizes Plasmodium falciparum to the antimalarial drug piperaquine. Antimicrob. Agents Chemother. 2018;62:e02309–e02317. doi: 10.1128/AAC.02309-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naidoo I., Roper C. Mapping ‘ partially resistant ’, ‘ fully resistant ’, and ‘ super resistant ’ malaria. Trends Parasitol. 2013;29:505–515. doi: 10.1016/j.pt.2013.08.002. [DOI] [PubMed] [Google Scholar]
- Ndiaye D., Dieye B., Ndiaye Y.D., Tyne D. Van, Daniels R., Bei A.K., Mbaye A., Valim C., Lukens A., Mboup S., Ndir O., Wirth D.F., Volkman S. Polymorphism in dhfr/dhps genes, parasite density and ex vivo response to pyrimethamine in Plasmodium falciparum malaria parasites in Thies, Senegal. Int. J. Parasitol.: Drugs Drug Resist. 2013;3:135–142. doi: 10.1016/j.ijpddr.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngasala B., Malmberg M., Carlsson A.M., Ferreira P.E., Petzold M.G., Blessborn D., Bergqvist Y., Gil J.P., Premji Z., Björkman A., Mårtensson A. Efficacy and effectiveness of artemether-lumefantrine after initial and repeated treatment in children < 5 years of age with acute uncomplicated plasmodium falciparum malaria in rural Tanzania: a randomized trial. Clin. Infect. Dis. 2011;52:873–882. doi: 10.1093/cid/cir066. [DOI] [PubMed] [Google Scholar]
- Ngasala B., Malmberg M., Carlsson A.M., Ferreira P.E., Petzold M.G., Blessborn D., Bergqvist Y., Gil J.P., Premji Z., Mårtensson A. Effectiveness of artemether-lumefantrine provided by community health workers in under-five children with uncomplicated malaria in rural Tanzania : an open label prospective study. Malar. J. 2011;10:64. doi: 10.1186/1475-2875-10-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noedl H., Se Y., Schaecher K., Smith B.L., Socheat D., Fukuda M.M. Evidence of artemisinin-resistant malaria in Western Cambodia. N. Engl. J. Med. 2008;359:2619–2620. doi: 10.1056/NEJMc0805011. [DOI] [PubMed] [Google Scholar]
- Nsanzabana C. Resistance to artemisinin combination therapies (ACTs): do not forget the partner drug ! Trav. Med. Infect. Dis. 2019;4:26. doi: 10.3390/tropicalmed4010026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okell L.C., Griffin J.T., Roper C. Mapping sulphadoxine- Plasmodium falciparum malaria in infected humans and in parasite populations in Africa. Sci. Rep. 2017;7:7389. doi: 10.1038/s41598-017-06708-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okell L.C., Griffin J.T., Roper C. Mapping sulphadoxine-pyrimethamine-resistant Plasmodium falciparum malaria in infected humans and in parasite populations in Africa. Sci. Rep. 2017;7:7389. doi: 10.1038/s41598-017-06708-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otienoburu S.D., Ascofaré O.M., Schramm B., Jullien V., Jones J.J., Zolia Y.M., Houzé P., Ashley E.A., Kiechel J.R., Guérin P.J. Selection of Plasmodium falciparum pfcrt and pfmdr1 polymorphisms after treatment with artesunate – amodiaquine fixed dose combination or artemether – lumefantrine in Liberia. Malar. J. 2016;15:452. doi: 10.1186/s12936-016-1503-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouji M., Augereau J., Paloque L., Benoit-vical F. Plasmodium falciparum resistance to artemisinin-based combination therapies : a sword of Damocles in the path toward malaria elimination. Parasite. 2018;24:24. doi: 10.1051/parasite/2018021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paloque L., Ramadani A.P., Puijalon O.M., Augereau J.M., Vical F.B. Plasmodium falciparum : multifaceted resistance to artemisinins. Malar. J. 2016;15:149. doi: 10.1186/s12936-016-1206-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasupureddy R., Yadav A., Seshadri S., Pande V., Dixit R., Pandey K.C. Current scenario and future strategies to fight artemisinin resistance. Parasitol. Res. 2019;118:29–42. doi: 10.1007/s00436-018-6126-x. [DOI] [PubMed] [Google Scholar]
- Patel P., Bharti P.K., Bansal D., Ali N.A., Raman R.K., Mohapatra P.K., Sehgal R., Mahanta J., Sultan A.A., Singh N. Prevalence of mutations linked to antimalarial resistance in Plasmodium falciparum from Chhattisgarh, Central India: a malaria elimination point of view. Sci. Rep. 2017;7:16690. doi: 10.1038/s41598-017-16866-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson D.S., Walliker D., Wellems T.E. Evidence that a point mutation in dihydrofolate reductase-thymidylate synthase confers resistance to pyrimethamine in falciparum malaria. Proc. Natl. Acad. Sci. U.S.A. 1988;85:9114–9118. doi: 10.1073/pnas.85.23.9114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickard A.L., Wongsrichanalai C., Purfield A., Kamwendo D., Emery K., Zalewski C., Kawamoto F., Miller R.S., Meshnick S.R. Resistance to antimalarials in Southeast Asia and genetic polymorphisms in pfmdr1. Antimicrob. Agents Chemother. 2003;47:2418–2423. doi: 10.1128/AAC.47.8.2418-2423.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plucinski M.M., Dimbu P.R., Macaia A.P., Ferreira C.M., Samutondo C., Quivinja J., Afonso M., Kiniffo R., Mbounga E., Kelley J.S., Patel D.S., He Y., Talundzic E., Garrett D.O., Halsey E.S., Udhayakumar V., Ringwald P., Fortes F. Efficacy of artemether –lumefantrine, artesunate-amodiaquine, and dihydroartemisinin-piperaquine for treatment of uncomplicated Plasmodium falciparum malaria in Angola. Malar. J. 2017;16:62. doi: 10.1186/s12936-017-1712-4. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price R.N., von Seidlein L., Valecha N., Nosten F., Baird J.K., White N.J. Global extent of chloroquine-resistant Plasmodium vivax: a systematic review and meta-analysis. Lancet Infect. Dis. 2014;14:982–991. doi: 10.1016/S1473-3099(14)70855-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser C., Meyer W., Ellis J., Lee R. Evolutionary ARMS race: antimalarial resistance molecular surveillance. Trends Parasitol. 2018;34:322–334. doi: 10.1016/j.pt.2018.01.001. [DOI] [PubMed] [Google Scholar]
- Rasmussen C., Valecha N., Rin Lack of convincing evidence of artemisinin resistance in India. Clin. Infect. Dis. 2019;69:1461–1462. doi: 10.1093/cid/ciz166. [DOI] [PubMed] [Google Scholar]
- Recht J., Siqueira A.M., Monteiro W.M., Herrera S.M., Herrera S., Lacerda M.V.G. Malaria in Brazil, Colombia, Peru and Venezuela: current challenges in malaria control and elimination. Malar. J. 2017;16:273. doi: 10.1186/s12936-017-1925-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert M.G., Tsombeng F.F., Gendrot M., Mosnier J., Amalvict R., Benoit N., Torrentino-Madamet M., Pradines B. Absence of a high level of duplication of the plasmepsin II gene in Africa. Antimicrob. Agents Chemother. 2018;62:1–8. doi: 10.1128/AAC.00374-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross L.S., Dhingra S.K., Mok S., Yeo T., Wicht K.J., Kümpornsin K., Takala-Harrison S., Witkowski B., Fairhurst R.M., Ariey F., Menard D., Fidock D.A. Emerging Southeast Asian PfCRT mutations confer Plasmodium falciparum resistance to the first-line antimalarial piperaquine. Nat. Commun. 2018;9:25–28. doi: 10.1038/s41467-018-05652-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruizendaal E., Tahita M.C., Traoré-Coulibaly M., Tinto H., Schallig H.D.F.H., Mens P.F. Presence of quintuple dhfr N51, C59, S108 – dhps A437, K540 mutations in Plasmodium falciparum isolates from pregnant women and the general population in Nanoro, Burkina Faso. Mol. Biochem. Parasitol. 2017;217:13–15. doi: 10.1016/j.molbiopara.2017.08.003. [DOI] [PubMed] [Google Scholar]
- Saha P., Guha S.K., Das S., Mullick S., Ganguly S., Biswas A., Bera D.K., Chattopadhyay G., Das M., Kundu P.K., Ray K., Maji A.K. Comparative efficacies of Artemisinin combination therapies in Plasmodium falciparum malaria and polymorphism of pfATPase6, pfcrt , pfdhfr , and pfdhps genes in Tea Gardens of Jalpaiguri District, India. Antimicrob. Agents Chemother. 2012;56:2511–2517. doi: 10.1128/AAC.05388-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidhu A.B.S., Valderramos S.G., Fidock D.A. pfmdr1 mutations contribute to quinine resistance and enhance mefloquine and artemisinin sensitivity in Plasmodium falciparum. Mol. Biol. 2005;57:913–926. doi: 10.1111/j.1365-2958.2005.04729.x. [DOI] [PubMed] [Google Scholar]
- Sharma J., Soni M., Dutta P., Khan S., Mahanta J. Mutational prevalence of chloroquine resistance transporter gene among Plasmodium falciparum field isolates in Assam and Arunachal Pradesh, India. Indian J. Med. Microbiol. 2016;34:193–197. doi: 10.4103/0255-0857.180298. [DOI] [PubMed] [Google Scholar]
- Singh Sidhu A.B., Verdier-Pinard D., Fidock D.A. Chloroquine resistance in Plasmodium falciparum malaria parasites conferred by pfcrt mutations. Science. 2002;298:210–213. doi: 10.1126/science.1074045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S.J., Kamara A.R.Y., Sahr F., Samai M., Swaray A.S., Menard D., Warsame M. Efficacy of artemisinin-based combination therapies and prevalence of molecular markers associated with artemisinin, piperaquine and sulfadoxine-pyrimethamine resistance in Sierra Leone. Acta Trop. 2018;185:363–370. doi: 10.1016/j.actatropica.2018.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somé A.F., Séré Y.Y., Dokomajilar C., Zongo I. Selection of known Plasmodium falciparum resistance-mediating polymorphisms by artemether-lumefantrine and amodiaquine/sulfadoxine-pyrimethamine , but not by dihydroartemisinin-piperaquine in Burkina Faso. Antimicrob. Agents Chemother. 2010;54:1949–1954. doi: 10.1128/AAC.01413-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sondo P., Derra K., Nakanabo S.D., Tarnagda Z., Kazienga A., Zampa O., Valéa I., Sorgho H., Owusu-Dabo E., Ouédraogo J.B., Guiguemdé T.R., Tinto H. Artesunate-amodiaquine and artemether-lumefantrine therapies and selection of pfcrt and Pfmdr1 alleles in Nanoro, Burkina Faso. PloS One. 2016;11 doi: 10.1371/journal.pone.0151565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spillman N.J., Kirk K. The malaria parasite cation ATPase PfATP4 and its role in the mechanism of action of a new arsenal of antimalarial drugs. Int J Parasitol Drugs Drug Resist. 2015;27:149–162. doi: 10.1016/j.ijpddr.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridaran S., Mcclintock S.K., Syphard L.M., Herman K.M., Barnwell J.W. Anti-folate drug resistance in Africa : meta-analysis of reported dihydrofolate reductase (dhfr) and dihydropteroate synthase (dhps) mutant genotype frequencies in African Plasmodium falciparum parasite populations. Malar. J. 2010;9:247. doi: 10.1186/1475-2875-9-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava P., Ratha J., Shah N.K., Mishra N., Anvikar A.R., Sharma S.K., Das M.K., Srivastava B., Valecha N. A clinical and molecular study of artesunate + sulphadoxine-pyrimethamine in three districts of central and eastern India. Malar. J. 2013;12:247. doi: 10.1186/1475-2875-12-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straimer J., Gnadig N.F., Witkowski B., Amaratunga C., Duru V., Ramadani A.P., Dacheux M., Khim N., Zhang L., Lam S., Gregory P.D., Urnov F.D., Mercereau-Puijalon O., Benoit-Vical F., Fairhurst R.M., Ménard D., Fidock D.A. K13-propeller mutations confer artemisinin resistance in Plasmodium falciparum clinical isolates. Science. 2015;347:428–431. doi: 10.1126/science.1260867.K13-propeller. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tawk L., Chicanne G., Dubremetz J.F., Richard V., Payrastre B., Vial H.J., Roy C., Wengelnik K. Phosphatidylinositol 3-phosphate, an essential lipid in Plasmodium, localizes to the food vacuole membrane and the apicoplast. Eukaryot. Cell. 2010;9:1519–1530. doi: 10.1128/EC.00124-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Joanna Briggs Institute CheCklist for randomized controlled trials: critical appraisal tools for use in JBI systematic reviews. JBI Syst. Rev. 2020 Adelaide. [Google Scholar]
- Triglia T., Menting J.G.T., Wilson C., Cowman A.F. Mutations in dihydropteroate synthase are responsible for sulfone and sulfonamide resistance in Plasmodium falciparum. Proc. Natl. Acad. Sci. U.S.A. 1997;94:13944–13949. doi: 10.1073/pnas.94.25.13944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Y. 2015. Artemisinin — A Gift from Traditional Chinese Medicine to the World. [DOI] [PubMed] [Google Scholar]
- Uwimana A., Legrand E., Stokes B.H., Ndikumana J.L.M., Warsame M., Umulisa N., Ngamije D., Munyaneza T., Mazarati J.B., Munguti K., Campagne P., Criscuolo A., Ariey F., Murindahabi M., Ringwald P., Fidock D.A., Mbituyumuremyi A., Menard D. Emergence and clonal expansion of in vitro artemisinin-resistant Plasmodium falciparum kelch13 R561H mutant parasites in Rwanda. Nat. Med. 2020 doi: 10.1038/s41591-020-1005-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Pluijm R.W., Imwong M., Chau N.H., Hoa N.T., Thuy-Nhien N.T., Thanh N.V., Jittamala P., Hanboonkunupakarn B., Chutasmit K., Saelow C., Runjarern R., Kaewmok W., Tripura R., Peto T.J., Yok S., Suon S., Sreng S., Mao S., Oun S., Yen S., Amaratunga C., Lek D., Huy R., Dhorda M., Chotivanich K., Ashley E.A., Mukaka M., Waithira N., Cheah P.Y., Maude R.J., Amato R., Pearson R.D., Gonçalves S., Jacob C.G., Hamilton W.L., Fairhurst R.M., Tarning J., Winterberg M., Kwiatkowski D.P., Pukrittayakamee S., Hien T.T., Day N.P., Miotto O., White N.J., Dondorp A.M. Determinants of dihydroartemisinin-piperaquine treatment failure in Plasmodium falciparum malaria in Cambodia, Thailand, and Vietnam: a prospective clinical, pharmacological, and genetic study. Lancet Infect. Dis. 2019;19:952–961. doi: 10.1016/S1473-3099(19)30391-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Zhang C.J., Chia W.N., Loh C.C.Y., Li Z., Lee Y.M., He Y., Yuan L.X., Lim T.K., Liu M., Liew C.X., Lee Y.Q., Zhang J., Lu N., Lim C.T., Hua Z.C., Liu B., Shen H.M., Tan K.S.W., Lin Q. Haem-activated promiscuous targeting of artemisinin in Plasmodium falciparum. Nat. Commun. 2015;6:10111. doi: 10.1038/ncomms10111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warsame M., HasSAn A.M., Barrette A., Jibril A.M., Elmi H.H., Arale A.M., Mohammady H. El, Nada R.A., Ghilan J., Amran H., Muse A., Yusuf F.E., Omar A.S. Treatment of uncomplicated malaria with artesunate plus sulfadoxine – pyrimethamine is failing in Somalia : evidence from therapeutic efficacy studies and Pfdhfr and Pfdhps mutant alleles. Trop. Med. Int. Health. 2015;20:510–517. doi: 10.1111/tmi.12458. [DOI] [PubMed] [Google Scholar]
- Warsame M., HasSAn H.A., HasSAn A.M., Mohamed Arale A., Jibril A.M., Mohamud S.A., Barrette A., Yusuf Muse A., Yusuf F.E., Nada R.A., Amran J.G.H. Efficacy of artesunate + sulphadoxine/pyrimethamine and artemether + lumefantrine and dhfr and dhps mutations in Somalia: evidence for updating the malaria treatment policy. Trop. Med. Int. 2017;22:415–422. doi: 10.1111/ijlh.12426. [DOI] [PubMed] [Google Scholar]
- Whegang S., Andreas Y., Basco L.K. Monitoring the efficacy and safety of artemisinin-based combination therapies: a review and network meta-analysis of antimalarial therapeutic efficacy trials in Cameroon. Drugs R. 2019;19:1–14. doi: 10.1007/s40268-018-0259-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White N.J. Triple artemisinin-containing combination anti-malarial treatments should be implemented now to delay the emergence of resistance. Malar. J. 2019;18:338. doi: 10.1186/s12936-019-2955-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White N.J., Pukrittayakamee S., Tinh Hien T., Abul Faiz M., Mokuolu O.A., Dondorp A.M. Malaria. Lancet. 2014;391:1608–1621. doi: 10.1016/S0140-6736(18)30324-6. [DOI] [PubMed] [Google Scholar]
- WHO . 2020. World Malaria Report 2020; p. 166. Geneva, Switzerland. [Google Scholar]
- WHO . 2019. World Malaria Report 2019; p. 232. Geneva, Switzerland. [Google Scholar]
- WHO . 2018. World Malaria Report 2018; p. 210. Geneva, Switzerland. [Google Scholar]
- WHO . 2018. Artemisinin Resistance and Artemisinin-Based Combination Therapy Efficacy. [Google Scholar]
- WHO . 2016. Global Technical Strategy for Malaria 2016–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . 1967. Technical Report Series. Chemotherapy of Malaria. [Google Scholar]
- Witkowski B., Duru V., Khim N., Ross L.S., Saintpierre B., Chy S. A surrogate marker of piperaquine-resistant Plasmodium falciparum malaria: a phenotype–genotype association study. Lancet Infect. Dis. 2017;17:174–183. doi: 10.1016/S1473-3099(16)30415-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkowski B., Khim N., Chim P., Kim S., Ke S., Kloeung N., Chy S., Duong S. Reduced artemisinin susceptibility of Plasmodium falciparum ring stages in Western Cambodia. Antimicrob. Agents Chemother. 2013;57:914–923. doi: 10.1128/AAC.01868-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkowski B., Lelie J., Jose M., Laurent V., Su X., Berry A. Increased tolerance to Artemisinin in Plasmodium falciparum is mediated by a quiescence mechanism. Antimicrob. Agents Chemother. 2010;54:1872–1877. doi: 10.1128/AAC.01636-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . 2018. Artemisinin Resistance and Artemisinin-Based Combination Therapy Efficacy. [Google Scholar]
- Wurtz N., Fall M., Baret E., Camara C., Nakoulima A., Diatta B., Fall K.B., Mbaye P.S., Bercion R., Wade B., Pradines B. Role of Pfmdr1 in in vitro Plasmodium falciparum susceptibility to chloroquine, quinine, monodesethylamodiaquine, mefloquine, lumefantrine, and dihydroartemisinin. Antimicrob. Agents Chemother. 2014;58:7032–7040. doi: 10.1128/AAC.03494-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T., Yeoh L.M., Tutor M.V., Dixon M.W., McMillan P.J., Xie S.C., Bridgford J.L., Gillett D.L., Duffy M.F., Ralph S.A., McConville M.J., Tilley L., Cobbold S.A. Decreased K13 abundance reduces hemoglobin catabolism and proteotoxic stress, underpinning artemisinin resistance. Cell Rep. 2019;29:2917–2928. doi: 10.1016/j.celrep.2019.10.095. e5. [DOI] [PubMed] [Google Scholar]
- Yeka A., Kigozi R., Conrad M.D., Lugemwa M., Okui P., Katureebe C., Belay K., Kapella B.K., Chang M.A., Kamya M.R., Staedke S.G., Dorsey G., Rosenthal P.J. Artesunate/Amodiaquine versus Artemether/Lumefantrine for the treatment of uncomplicated malaria in Uganda : a randomized trial. J. Infect. Dis. 2016;213:1134–1142. doi: 10.1093/infdis/jiv551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Xia J., Wei H., Liu X., Peng H. Risk of drug resistance in Plasmodium falciparum malaria therapy — a systematic review and meta-analysis. Parasitol. Res. 2016;116:781–788. doi: 10.1007/s00436-016-5353-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article and in the attached supplementary files.