Abstract
This study was conducted to investigate the effects of a high-fat diet (HFD) and high-fat and high-cholesterol diet (HFHCD) on glucose and lipid metabolism and on the intestinal microbiota of the host animal. A total of 30 four-week-old female C57BL/6 mice were randomly divided into three groups (n=10) and fed with a normal diet (ND), HFD, or HFHCD for 12 weeks, respectively. The HFD significantly increased body weight and visceral adipose accumulation and partly lowered oral glucose tolerance compared with the ND and HFHCD. The HFHCD increased liver weight, liver fat infiltration, liver triglycerides, and liver total cholesterol compared with the ND and HFD. Moreover, it increased serum high-density lipoprotein cholesterol, low-density lipoprotein cholesterol, and total cholesterol compared with the ND and HFD and upregulated alanine aminotransferase, aspartate aminotransferase, and alkaline phosphatase significantly. The HFHCD also significantly decreased the α-diversity of the fecal bacteria of the mice, to a greater extent than the HFD. The composition of fecal bacteria among the three groups was apparently different. Compared with the HFHCD-fed mice, the HFD-fed mice had more Oscillospira, Odoribacter, Bacteroides, and [Prevotella], but less [Ruminococcus] and Akkermansia. Cecal short-chain fatty acids were significantly decreased after the mice were fed the HFD or HFHCD for 12 weeks. Our findings indicate that an HFD and HFHCD can alter the glucose and lipid metabolism of the host animal differentially; modifications of intestinal microbiota and their metabolites may be an important underlying mechanism.
Keywords: glucose metabolism, high-fat and high-cholesterol diet, high-fat diet, intestinal microbiota, lipid metabolism
Introduction
In recent years, the incidence of various metabolic diseases, such as obesity and non-alcoholic fatty liver disease (NAFLD), has been increasing in various regions of the world [1, 2]. The occurrence of these chronic metabolic diseases is closely related to changes in dietary structure [3, 4], especially the intake ratio of the three major macronutrients (carbohydrates, fats, and proteins). Studies have demonstrated that a high-fat diet (HFD) could induce obesity. Intake of a high-fat and high-cholesterol diet (HFHCD) can cause steatohepatitis, inflammation, and fibrosis. This diet also causes severe weight loss, abnormal serum transaminase, and cholesterol as the main lipid in the liver [5].
Recently, many studies have demonstrated that the gut microbiota is closely related to obesity and NAFLD [6]. Bäckhed et al. elucidated the role of the gut microbiota in host energy metabolism and growth by showing that germ-free mice have lower body weights and levels of fat than do conventional mice [7]. The gut microbiota can regulate the host’s ability to harvest and store energy, which may lead to obesity [7, 8]. The occurrence of obesity is accompanied by changes in the structure and function of the gut microbiota. Animal and human studies both showed that the most common change in gut microbiota in obese individuals is an increased ratio of Firmicutes to Bacteroidetes (F/B). F/B is often used as an important indicator of malnutrition or metabolism-related diseases. The Western diet was demonstrated to cause changes in the composition of the gut microbiota, with decreased levels of Bacteroidetes and Bifidobacterium and increased levels of Firmicutes and Proteobacteria [9, 10]. Compared with conventionally bred mice, HFD-fed germ-free mice have lower lipid levels in the liver [11], suggesting that liver lipid accumulation is related to gut microbiota. Le Roy et al. transplanted the gut microbiota of fasting mice with hyperglycemia and insulinemia into germ-free mice, and this caused NAFLD in these animals [12]. Relative to healthy individuals, the gut microbiota of patients with NAFLD is significantly different, with higher levels of Proteobacteria, Enterobacteria, and Escherichia [13]. Moreover, in the progression of NAFLD, the content of Proteobacteria is increased, and the content of Firmicutes is decreased [14].
Gut microbiota metabolites significantly affect host metabolism and lead to the development of obesity and NAFLD [15]. Because of the existence of the intestinal-liver axis in the body, if the intestinal barrier is broken, the liver is the first organ in the body to encounter microorganisms, toxins, and microbial metabolites from the intestine [15]. Bile acids are synthesized from cholesterol in the liver and released into the intestines to aid in the digestion of dietary lipids. The gut microbiota can affect bile acid metabolism and reabsorption and play an important role in host health [16]. Bile acids and their metabolites help maintain the homeostasis of glycogen, cholesterol, and triglycerides. Clinical studies have shown that bile acids can promote the development of NAFLD by altering the signaling of the nuclear bile acid receptor farnesoid X receptor (FXR) [17, 18]. Gut microorganisms produce short-chain fatty acids (SCFAs) by digesting nondigestible carbohydrates. Clinical studies have demonstrated that SCFAs can affect the development of obesity and metabolic diseases by activating G-protein-coupled receptors (GPCRs) and can participate in the occurrence of NAFLD through a variety of mechanisms. However, the differential roles of gut microbiota and their metabolites in obesity and NAFLD have not been clarified fully.
The aim of this study was to investigate the differences in the effects of obesity-related diets and NAFLD-related diets on liver function, glucose and lipid metabolism, and gut microbiota in mice. The relationships among diet, gut microbiota, and metabolic function were explored. These results may provide a basis for interventions in diet-induced metabolic diseases by changing the structure of the gut microbiota.
Methods and Materials
Mice
Thirty 4-week-old female C57BL/6 mice were purchased from Chengdu Dashuo Experimental Animal Co., Ltd. All mice were housed in a specific pathogen-free facility at an ambient temperature of 23 ± 1°C and a humidity of 50–70% under a 12 h light/dark cycle with access to water and food ad libitum. After adaptive feeding for one week, the tested mice were randomly divided into three groups (n=10 per group) and fed either a normal diet (ND; 270 kcal/100 g; 10% of energy from fat, 20% from protein, and 70% from carbohydrates), HFD (521 kcal/100 g; 60% of energy from fat, 20% from protein, and 20% from carbohydrates; D12492; Research Diets, New Brunswick, NJ, USA), or HFHCD (453 kcal/100 g; 40% of energy from fat, 20% from protein, and 40% from carbohydrates; D12109C; Research Diets) for 12 weeks. The HFHCD also contained 1.25 g of cholesterol (1.25%) per 100 g. At the end of week 12, all of the tested mice were anesthetized by intraperitoneal injection of 1% pentobarbital sodium solution (50 mg/kg of body weight). Thereafter, blood samples were collected by eyeball extirpation and then the mice were sacrificed by cervical dislocation.
All experimental procedures were performed in accordance with the Guidelines for Animal Experiments at West China School of Public Health, Sichuan University. The animal experimental facility and animals used in this study were officially approved by the Experimental Animal Management Committee of the Sichuan Government (approval no. SYXK2018-011). The experimental protocols were approved by the Medical Ethics Committee of the West China School of Public Health, Sichuan University.
Histopathology
One square centimeter of the hepatic lobe was collected and fixed in 10% neutral phosphate-buffered saline formalin for 24 h. Subsequently, it was routinely stained with hematoxylin and eosin (H&E). One square centimeter of visceral adipose tissue were collected and fixed in fat fixative and stained with oil red O. Tissue sections were observed by a professional physician blinded to the experimental design.
Oral glucose tolerance test (OGTT)
An OGTT was performed on each mouse at the end of week 12 using the method described by Andrikopoulos et al. [19]. Mice were fasted for 8 h, and the fasting blood glucose levels were measured with a glucose meter using blood collected from the tip of the tail vein. Thereafter, each of the tested mice were fed with glucose (2 g/kg of body weight), and the blood glucose level was measured at 30, 60, 90, and 120 min after glucose gavage.
Serum and tissue analyses
The serum was collected by centrifugation at 2,000× g for 20 min. Serum high-density lipoprotein cholesterol (HDL-C), low-density lipoprotein cholesterol (LDL-C), total triglyceride (TG), total cholesterol (TC), alanine aminotransferase (ALT), aspartate aminotransferase (AST), and alkaline phosphatase (ALP) levels were assayed using corresponding commercial kits (Changchun Huili Biotech Co., Ltd., Changchun, China) by an automatic biochemical analyzer (Rayto Life and Analytical Sciences Co., Ltd., Shenzhen, China).
The liver tissue was homogenized and subjected to centrifugation, and the supernatant was then collected. Protein concentrations were determined by BCA assay and normalized to equal concentrations. TC and TG levels in the liver tissues were measured using commercial TC and TG assay kits (Changchun Huili) by an automatic biochemical analyzer (Rayto).
Bacterial DNA extraction and 16s rRNA sequencing
Fresh stool pellets from mice were collected at 12 weeks of feeding and frozen at −80°C. Total DNA was extracted using a TIANamp Stool DNA Kit (Tiangen Biotech Co., Ltd., Beijing, China) according to the manufacturer’s instructions.
Fecal microbiota communities were determined by 16S rRNA sequencing using universal primers (forward primer, 338F, 5’-ACTCCTACGGGAGGCAGCAG-3’; reverse primer, 806R, 5’-GGACTACHVGGGTWTCTAAT-3’) [20]. PCR amplification was performed in a 25 µl reaction mixture containing 50 ng of template DNA, 12.5 µl of Phusion Hot Start Flex 2× Master Mix (New England Biolabs Inc., Beverly, MA, USA), and 2.5 µl of each primer. ddH2O was used to adjust the final volume. The PCR cycling conditions were as follows: initial denaturation at 98°C for 30 s; followed by 35 cycles of denaturation at 98°C for 10 s, annealing at 54°C for 30 s, and extension at 72°C for 45 s; and a final extension at 72°C for 10 min. The amplicon pools were prepared for sequencing, and the size and quantity of the amplicon library were assessed. The libraries were sequenced on an Illumina MiSeq instrument (Illumina Inc., San Diego, CA, USA) using the 300 bp paired-end protocol and the standard Illumina sequencing primers.
Bioinformatics
The bioinformatics analysis was performed as described previously [20]. Briefly, the sequencing data were filtered, and the effective tags were clustered into operational taxonomic units (OTUs) with a similarity of 97% using the Uparse 7.0.1001 software (https://drive5.com/uparse/). The taxonomic assignment was performed with an SSU rRNA database, and the OTU abundance table was constructed using QIIME python scripts. Multiple sequence alignment was conducted using the MUSCLE3.8.31 software (https://www.drive5.com/muscle/). The α-diversity, β-diversity, and relative abundance of the microbes in each sample were calculated based on the normalized read count.
Cecal SCFA detection
Cecal contents (100 mg) were acidified with 15% phosphoric acid and fixed with an internal standard (isocaproic acid) solution and ether. The mixture was extracted, and the supernatant was collected. Detection was performed by Agilent 7890B gas chromatography (Agilent Technologies, Inc., Santa Clara, CA, USA).
Fecal bile acid detection
Feces collected at week 12 (10 mg) were added to methanol and then shaken, sonicated, and centrifuged. The supernatant (100 µl) was mixed with 900 µl of methanol and vortexed for 30 s. The filtrate was added to the test bottle. Detection was performed by an AB4000 triple quadrupole mass spectrometer (AB Sciex, Concord, Canada) coupled to a Waters ACQUITY UPLC liquid chromatography system (Waters Corporation, Milford, MA, USA).
Statistical analysis
GraphPad Prism 7.0 was used for statistical analyses (GraphPad Software, Inc., San Diego, CA, USA). Data are presented as the mean ± SD. One-way ANOVA or the Kruskal-Wallis nonparametric test was used to compare multiple groups of independent samples. Significance was set at P<0.05. All tests were two-tailed.
Results
Body weight and blood glucose
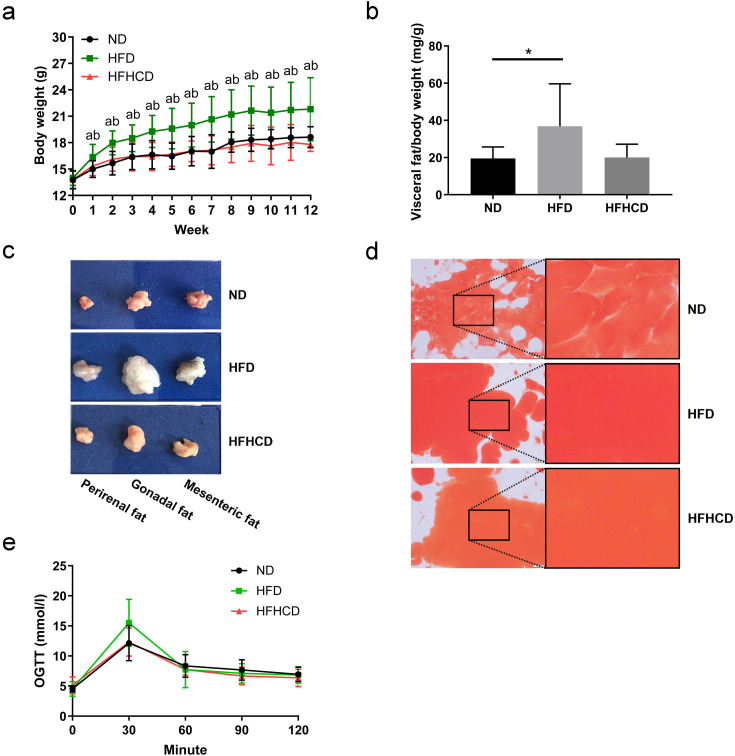
During the whole experiment, the body weights of mice in the HFD group were significantly higher than those of mice in the ND and HFHCD groups (P<0.05). There was no significant difference in body weight between the ND and HFHCD groups (P>0.05; Fig. 1a). The visceral adipose tissue accumulation in the HFD group was greater than that observed in the ND (P=0.0360) and HFHCD (P=0.0540) groups (Fig. 1b), especially in the gonadal adipose tissue (Fig. 1c). Oil red O staining revealed greater adipose accumulation in the HFD and HFHCD groups (Fig. 1d). However, no significant difference was found in visceral adipose tissue content between the ND and HFHCD groups. As shown in Fig. 1e, there was no significant difference in blood glucose during the whole OGTT test in the tested mice among three groups, although an increased tendency was found in the HFD group at 30 min compared with the ND group (P=0.0813).
Fig. 1.
Body weight, adipose tissue weight, and the glucose response of mice (n=6–10 per group). (a) The body weights of mice during feeding with different diets. aP<0.05 compared with the ND group. bP<0.05 compared with the HFHCD group. (b) Visceral fat/body weight ratios of mice. *P<0.05. (c) Representative images for visceral adipose tissue of mice in the three groups. (d) Oil red O staining of the visceral adipose tissue. Scale bars: 500 µm. (e) An oral glucose tolerance test (OGTT) was performed on mice at week 12. ND, normal diet; HFD, high-fat diet; HFHCD, high-fat and high-cholesterol diet.
Liver tissue and lipid metabolism
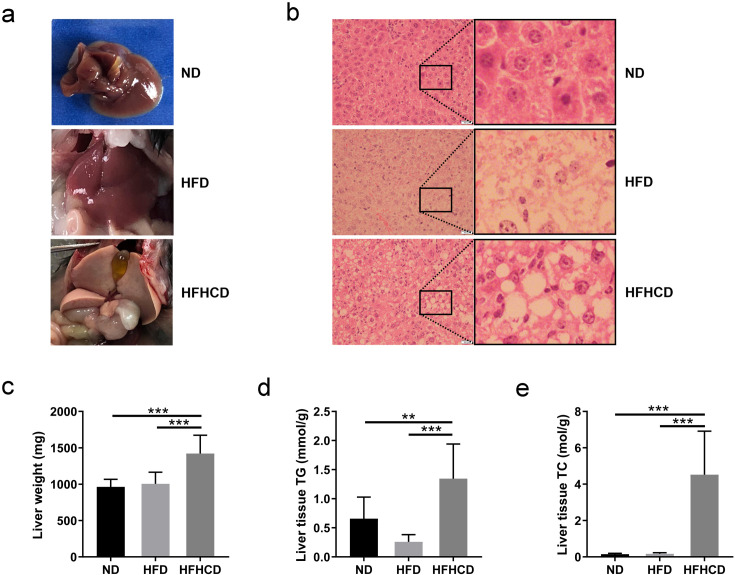
As shown in Fig. 2a, the livers of ND-fed mice and HFD-fed mice were similar and relatively normal. However, the livers of the HFHCD-fed mice were obviously whiter and larger. The profiles of H&E staining (Fig. 2b) showed that fat infiltration occurred in the livers of the HFD- and HFHCD-fed mice and that obvious fat microvesicles and macrovesicles were present in the livers of the HFHCD-fed mice. As shown in Figs. 2c–e, HFHCD-fed mice exhibited significantly higher liver weights and liver tissue TG and TC levels than the ND- and HFD-fed mice (P<0.01).
Fig. 2.
Liver parameters of mice at week 12 of feeding (n=6–10 per group). (a) Representative images of livers of mice in the three groups. (b) The H&E staining profiles of liver tissues. Scale bars: 500 µm. (c) Liver weight, (d) liver tissue triglyceride (TG) levels and (e) liver tissue total cholesterol (TC) levels of each group. *P<0.05; **P<0.01; ***P<0.001. ND, normal diet; HFD, high-fat diet; HFHCD, high-fat and high-cholesterol diet.
Blood biochemical indicators
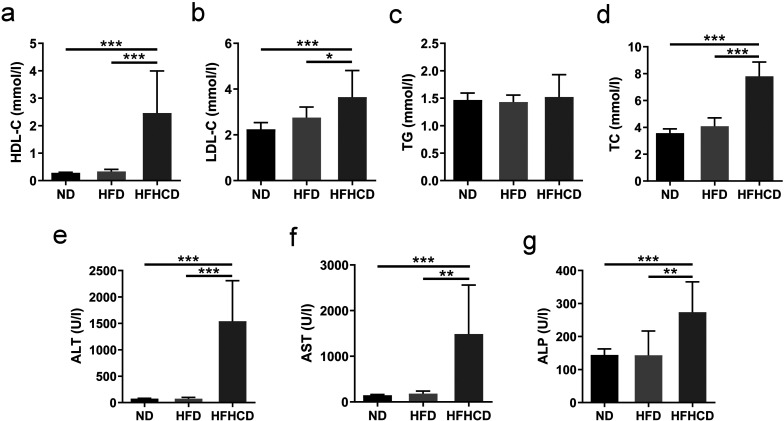
After 12 weeks of feeding, HFHCD-fed mice had significantly higher serum HDL-C, LDL-C, TC, ALT, AST, and ALP levels than the ND- and HFD-fed mice (P<0.05; Figs. 3a–g). There was no significant difference in any of the tested serum biochemical indicators between the ND and HFD groups (P>0.05).
Fig. 3.
Serum biochemical parameters of mice at week 12 of feeding (n=6–10 per group). (a) High-density lipoprotein cholesterol (HDL-C) levels. (b) Low-density lipoprotein cholesterol (LDL-C) levels. (c) Triglyceride (TG) levels. (d) Total cholesterol (TC) levels. (e) Alanine aminotransferase (ALT) levels. (f) Aspartate aminotransferase (AST) levels. (g) Alkaline phosphatase (ALP) levels. *P<0.05; **P<0.01; ***P<0.001. ND, normal diet; HFD, high-fat diet; HFHCD, high-fat and high-cholesterol diet.
Fecal bile acid and cecal SCFAs
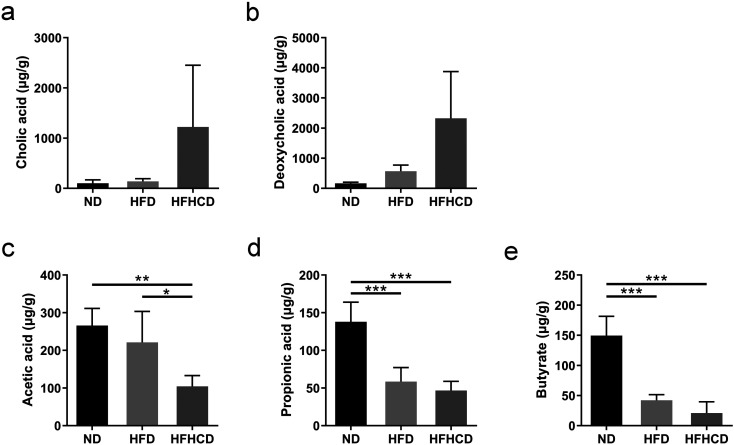
As shown in Figs. 4a and b, there was no significant difference in fecal cholic acid (CA) and deoxycholic acid (DCA) levels among the three groups, whereas the HFHCD group exhibited an increasing trend. As for cecal SCFAs levels, HFHCD-fed mice had a significantly lower acetic acid levels than ND- and HFD-fed mice (P<0.05; Fig. 4c). The levels of propionic acid and butyric acid were decreased significantly after the mice were fed the HFD or HFHCD for 12 weeks (P<0.001; Figs. 4d and e).
Fig. 4.
Fecal bile acid levels and cecal SCFAs levels of mice at week 12 of feeding (µg/g wet weight of feces; n=6–10 per group) (a) Cholic acid levels in fecal samples. (b) Deoxycholic acid levels in fecal samples. (c) Cecal acetic acid levels. (d) Cecal propionic acid levels. (e) Cecal butyric acid levels. *P<0.05; **P<0.01; ***P<0.001. ND, normal diet; HFD, high-fat diet; HFHCD, high-fat and high-cholesterol diet.
Microbiota analysis
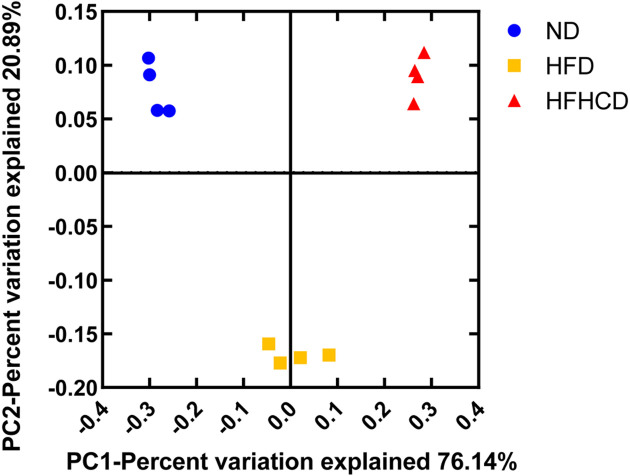
At week 12 of feeding, the analysis of microbial α-diversity using the Observed_species, Chao 1, Shannon, and PD_whole_tree indices revealed a significantly lower species richness and diversity in the HFHCD-fed mice compared with the ND- and HFD-fed mice (P<0.05; Table 1). The Observed_species, Shannon, and PD_whole_tree indices were lower in the HFD-fed mice compared with the ND-fed mice (P<0.05; Table 1). According to a principal coordinates analysis (PCoA), the compositions of the fecal microbiotas of the three groups were clearly different (Fig. 5).
Table 1. The α-diversity of fecal microbiota in mice at week 12 (n=4 per group).
| Group | ND | HFD | HFHCD |
|---|---|---|---|
| Observed_species | 692.43 ± 31.43 | 570.65 ± 27.94a | 430.50 ± 11.17ab |
| Chao1 | 994.80 ± 59.34 | 938.60 ± 49.68 | 715.00 ± 42.82ab |
| Shannon | 6.91 ± 0.15 | 6.47 ± 0.17a | 4.87 ± 0.27ab |
| PD_whole_tree | 30.24 ± 1.10 | 24.72 ± 0.82a | 21.24 ± 0.34ab |
aP<0.05 compared with the ND group. bP<0.05 compared with the HFD group. ND, normal diet; HFD, high-fat diet; HFHCD, high-fat and high-cholesterol diet.
Fig. 5.
Principal coordinates analysis (PCoA) of fecal microbiota of mice at week 12 of feeding (n=4 per group). ND, normal diet; HFD, high-fat diet; HFHCD, high-fat and high-cholesterol diet.
The modifications of the intestinal microbiota composition at two different taxonomic levels are summarized in Tables 2 and 3. At the phylum level (Table 2), the HFD significantly reduced the relative abundance of Bacteroidetes, whereas it increased the relative abundances of Firmicutes and Proteobacteria and increased the F/B value in the fecal microbiota of mice at week 12 (P<0.05; compared with the ND group). The HFHCD significantly reduced the relative abundances of Bacteroidetes and Firmicutes, whereas it increased the relative abundance of Proteobacteria and increased the F/B value compared with both the ND and HFD groups (P<0.05).
Table 2. Relative abundance of the top six OTUs of species at the phylum level in fecal samples at week 12 (n=4 per group).
| Group | ND | HFD | HFHCD |
|---|---|---|---|
| Bacteroidetes (%) | 56.4 ± 4.84 | 27.90 ± 6.41a | 6.10 ± 0.73ab |
| Firmicutes (%) | 36.29 ± 5.68 | 52.35 ± 2.55a | 25.99 ± 4.38ab |
| Proteobacteria (%) | 5.95 ± 1.70 | 14.59 ± 2.86a | 53.35 ± 4.17ab |
| TM7 (%) | 0.63 ± 0.15 | 0.21 ± 0.07a | 0ab |
| Actinobacteria (%) | 0. 35 ± 0.13 | 0.24 ± 0.14 | 0.06 ± 0.01a |
| Deferribacteres (%) | 0.14 ± 0.03 | 3.96 ± 1.32a | 0.56 ± 0.27b |
| F/B | 0.65 ± 0.16 | 1.98 ± 0.61a | 4.25 ± 0.25ab |
aP<0.05 compared with the ND group. bP<0.05 compared with the HFD group. ND, normal diet; HFD, high-fat diet; HFHCD, high-fat and high-cholesterol diet; F/B, the ratio of Firmicutes to Bacteroidetes.
Table 3. Relative abundance of the top ten OTUs of species at the genus level in fecal samples at week 12 (n=4 per group).
| Group (%) | ND | HFD | HFHCD |
|---|---|---|---|
| Lactobacillus | 5.13 ± 1.45 | 0.97 ± 0.17a | 1.25 ± 0.29a |
| Oscillospira | 3.02 ± 0.55 | 9.05 ± 1.79a | 3.48 ± 1.00b |
| Odoribacter | 0.20 ± 0.07 | 2.38 ± 0.65a | 0.10 ± 0.02b |
| [Prevotella] | 5.08 ± 0.80 | 2.64 ± 1.10a | 0.22 ± 0.04ab |
| Bacteroides | 1.87 ± 0.58 | 5.41 ± 1.89a | 1.15 ± 0.06b |
| Dorea | 0.13 ± 0.03 | 0.84 ± 0.33a | 0.33 ± 0.10b |
| [Ruminococcus] | 0.18 ± 0.08 | 1.43 ± 0.23a | 2.74 ± 0.67ab |
| Sutterella | 3.56 ± 1.54 | 0.01 ± 0.01a | 0.93 ± 0.20a |
| Desulfovibrio | 0.40 ± 0.12 | 0.88 ± 0.14a | 0.70 ± 0.05a |
| Akkermansia | 0.02 ± 0.01 | 0.01 ± 0.01 | 13.47 ± 1.60ab |
aP<0.05 compared with the ND group. bP<0.05 compared with the HFD group. ND, normal diet; HFD, high-fat diet; HFHCD, high-fat and high-cholesterol diet. Brackets indicate that the sequence of a bacterium is recorded in the Greengenes database but is not recorded by NCBI.
At the genus level (Table 3), the HFD-fed mice had higher relative abundances of Oscillospira, Odoribacter, Bacteroides, and [Ruminococcus] and lower relative abundances of Lactobacillus and [Prevotella] (P<0.05; compared with the ND group), whereas the HFHCD-fed mice had higher relative abundances of [Ruminococcus] and Akkermansia and lower relative abundances Lactobacillus and [Prevotella] (P<0.05; compared with the ND group). Compared with the HFHCD group, the HFD-fed mice had higher relative abundances of Oscillospira, Odoribacter, Bacteroides, and [Prevotella] and lower relative abundances of [Ruminococcus] and Akkermansia (P<0.05). Brackets indicate that the nucleotide sequence of the bacterium is recorded in the Greengenes database but is not recorded by NCBI.
Discussion
Accumulating scientific evidence from many well-designed clinical and animal studies have demonstrated the strong relationship between obesity and intestinal microbiota; moreover, these studies have indicated that several specific gut microbes can regulate metabolic syndrome (MS), especially obesity, one the most important aspects of MS [21, 22]. Furthermore, the possible relationship between NAFLD and intestinal microbiota has also been the focus of several recent studies, which found that intestinal bacteria produce a G-protein-coupled receptor ligand protein [23], release FXR signals to regulate gut-liver axis activity [24], and change intestinal permeability [25] to regulate the progress of NAFLD. Therefore, it would be interesting to profile the microbiota related to obesity and NAFLD and characterize or differentiate the function of intestinal microbes related to obesity and NAFLD, respectively. So two types of test diet, HFD and HFHCD, were used to cause obesity and NAFLD, and comparative studies were conducted to characterize their differential effects on glucose and lipid metabolism via gut microbiota.
In the present study, the HFD significantly increased the body weights of the tested mice and caused more visceral adipose tissue than the HFHCD. The tested HFD also tended to damage, at least partly, the glucose metabolism of mice based on the results of the OGTT at 30 min. However, it did not significantly change the blood and liver biochemical parameters of the tested mice that are indicative of serum lipid metabolism and liver function. These results indicate that the mice that were fed the HFD were overweight or obese without a significant metabolism disorder in the serum or liver. Similar results were found in studies with Wistar rats fed an HFD (40–75% of the diet energy) [26, 27], while conflicting results were also found in mice fed an HFD (60% of the diet energy), with both overweight and dyslipidemia observed in the mice [28]. These conflicting findings indicate that excessive dietary fat and energy intake cannot fully explain the abnormal changes in serum lipids in obese people. Kübeck et al. [29] highlighted that the functional signals generated from interactions between intestinal microbiota and dietary components could play an important role in host energy homeostasis and the development of obesity. The intestinal microbiota and its metabolites might be important targets in the management and prevention of diet-induced obesity and related metabolism disorders.
Conversely, in this study, the HFHCD significantly increased serum TC, HDL-C, and LDL-C levels, as well as liver weight and liver TG and TC levels. Furthermore, it also apparently increased liver ALT, AST, and ALP levels. All of these parameters are deeply associated with the health of the host animal. For example, a higher content of LDL-C is related to the incidence and severity of cardiovascular disease [30], and elevated liver ALT, AST, and ALP levels are considered hallmarks of liver damage [31]. Our results showed that the HFHCD negatively altered serum lipid metabolism and liver function in the tested mice but did not increase body weight and body fat, which was consistent with a similar study [26]. These results demonstrated well that an HFHCD may induce nonalcoholic steatohepatitis, as pointed out in previous studies [1]. An HFHCD might negatively affect the health of the host animal to a greater extent and faster than an HFD.
In the present study, the HFD and HFHCD were compared with regard to their abilities to affect the intestinal microbiota of host animals. The HFHCD significantly decreased the α-diversity index of the fecal bacteria of mice, to a greater extent than the HFD. A decline in the diversity of intestinal microbiota is considered to be characteristic of obese people [32]. An observational study of adolescents showed that lower α-diversity was associated with higher liver fat accumulation and that there was no significant association between the tested dietary components and hepatic steatosis [33]. The sharp decrease in microbial richness and diversity caused by the HFHCD may be one of the important causes of NAFLD. Additionally, the decrease of α-diversity in the HFD-fed mice in this study was smaller than in similar studies [28], which partly explains why the HFD did not cause severe dyslipidemia in mice. Furthermore, the β-diversity parameter clearly distinguished the fecal bacteria of mice fed with the ND, HFD, and HFHCD. These results indicated that the HFHCD and HFD dynamically damaged the richness, diversity, and composition of the intestinal microbiota in different manners.
Regarding the composition of the fecal microbiota among the tested mice at the taxonomic phylum level, both the HFD and HFHCD significantly increased the F/B value and relative abundance of Proteobacteria in the intestinal microbiota. It is well known that a higher abundance of Firmicutes than Bacteroidetes is associated with obesity and MS. Individuals with high F/B values have a high ability to obtain energy from food [34]. Additionally, the excessive intake of fat can lead to elevated levels of Proteobacteria, as well as obesity [9, 10]. In the present study, the tested mice fed with the HFHCD showed higher F/B ratios, with much more Proteobacteria, but they did not become obese like the HFD-fed mice. Additional dietary fat from the HFHCD accumulates in the liver of mice, rather than in visceral adipose tissues. Hildebrandt et al. [10] demonstrated the importance of diet as a determinant of intestinal microbiome composition independent of obesity using RELMβ knockout mice. We believe that dietary components, such as cholesterol in the HFHCD and saturated fatty acid in the HFD, may interact with intestinal microbiota in a variety of manners, leading to different physiological outcomes. The characteristic alternation in the diversity and composition of the intestinal microbiota may be one of the underlying mechanisms of obesity and NAFLD induced by the HFD and HFHCD respectively.
Regarding the composition of the fecal bacteria in the tested mice at the taxonomic genus level, both the HFD and HFHCD significantly decreased the abundance of Lactobacillus, an active uncoupling bacterium in bile acids. Lactobacilli play an important role in the hepatobiliary circulation of the host animal, in particular in the excretion and absorption of bile acids in the small intestine by maintaining the homeostasis of the intestinal epithelium [16]. Fecal lactobacilli also possess bile salt hydrolase, which is associated with increased resistance to bile toxicity in host animals [35, 36]. The depletion in intestinal lactobacilli might be related to the abilities of the HFD and HFHCD to affect lipid metabolism and liver function. In addition to lactobacilli, the HFD and HFHCD also caused a significant decrease in fecal [Prevotella] and a significant increase in fecal [Ruminococcus]. Previous studies have demonstrated that [Prevotella] is related to the ability to decompose carbohydrates [37, 38] and that [Ruminococcus] is related to the occurrence of atherosclerosis [39]. Moreover, the HFD and HFHCD have different effects on the amount of Oscillospira, Odoribacter, Bacteroides, and Akkermansia. This may explain why the two diets caused different metabolic reactions. Further studies should be conducted to analyze how an HFD or HFHCD affects the health of the animal host through each of these intestinal microbes.
Dietary fiber can be converted into SCFAs via fermentation by intestinal microorganisms, which could be generally used as an energy source and may be involved in a variety of metabolic pathways in the host animal, including gluconeogenesis [40, 41] and adipogenesis [5]. Rodent studies have shown that SCFAs can increase AMPK activity in the liver and skeletal muscle [42, 43], increase the amount of PGC1-α and UCP1 in brown fat [42], and promote thermogenesis and fatty acid oxidation to prevent diet-induced obesity [44]. The reduction of SCFAs may cause weakened glucose and lipid metabolism, decreased immunity, and decreased changes in the structure of the gut microbiota. In the present study, the HFHCD significantly decreased acetic acid, propionic acid, and butyric acid, whereas the HFD also decreased propionic acid and butyric acid. These results indicate that both the tested HFD and HFHCD could influence intestinal SCFAs differentially, which might be the mechanism underlying the triggering of obesity, abnormal serum and lipid metabolism, and abnormal liver function by the HFD and HFHCD. The difference in damage to SCFA metabolism caused by the HFD or HFHCD could result from the characteristic effects of the HFD or HFHCD on intestinal microbiota diversity and composition. On the other hand, as other important metabolites of intestinal microbiota, bile acids and their metabolites help maintain the homeostasis of glycogen, cholesterol, and triglycerides. Increased levels of intestinal bile acids were found to increase the F/B ratio and alter the gut microbiota composition, partly due to their antimicrobial activity. The imbalanced regulation between bile acids and intestinal microorganisms is involved in the pathogenesis of obesity and NAFLD [45]. The HFHCD used in this study tended to increase intestinal primary and secondary bile acids, and this might be the mechanism underlying the damaging of microbiota and the triggering of NAFLD.
In conclusion, the intake of an HFD can cause overweight or obesity without a significant metabolism disorder in the serum and liver, whereas an HFHCD can negatively alter serum lipid metabolism and liver function without increasing body weight and body fat, which may affect the health of the host animal to a greater extent. The HFHCD in this study damaged the richness, diversity, and composition of the intestinal microbiota much more severer than the HFD and affected the metabolism of SCFAs and bile acids more significantly. Each of the characteristic diet-microbiota interactions may play an important and different role in metabolic diseases, such as obesity and NAFLD. Further studies focusing upon a specific bacterium and the associated signaling pathways involved in different metabolic diseases might be of critical significance.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant number 81973042). We are grateful to Dr. Gaku Harata (Technical Research Laboratory, Takanashi Milk Products Co., Ltd., Yokohama, Japan) for help in the 16S rRNA sequencing and bioinformatics analysis. We also appreciate the support of the Public health and Preventive Medicine Provincial Experiment Teaching Center at Sichuan University and Food Safety Monitoring and Risk Assessment Key Laboratory of Sichuan Province. The authors would like to thank Enago (http://www.enago.jp) for the English language review.
References
- 1.Afshin A, Reitsma MB, Murray CJL. Health Effects of Overweight and Obesity in 195 Countries. N Engl J Med. 2017; 377: 1496–1497. [DOI] [PubMed] [Google Scholar]
- 2.Price AJ, Crampin AC, Amberbir A, Kayuni-Chihana N, Musicha C, Tafatatha T, et al. Prevalence of obesity, hypertension, and diabetes, and cascade of care in sub-Saharan Africa: a cross-sectional, population-based study in rural and urban Malawi. Lancet Diabetes Endocrinol. 2018; 6: 208–222. doi: 10.1016/S2213-8587(17)30432-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kostic AD, Gevers D, Siljander H, Vatanen T, Hyötyläinen T, Hämäläinen AM, et al. DIABIMMUNE Study Group. The dynamics of the human infant gut microbiome in development and in progression toward type 1 diabetes. Cell Host Microbe. 2015; 17: 260–273. doi: 10.1016/j.chom.2015.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Okada H, Kuhn C, Feillet H, Bach JF. The ‘hygiene hypothesis’ for autoimmune and allergic diseases: an update. Clin Exp Immunol. 2010; 160: 1–9. doi: 10.1111/j.1365-2249.2010.04139.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singh V, Chassaing B, Zhang L, San Yeoh B, Xiao X, Kumar M, et al. Microbiota-Dependent Hepatic Lipogenesis Mediated by Stearoyl CoA Desaturase 1 (SCD1) Promotes Metabolic Syndrome in TLR5-Deficient Mice. Cell Metab. 2015; 22: 983–996. doi: 10.1016/j.cmet.2015.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fändriks L. Roles of the gut in the metabolic syndrome: an overview. J Intern Med. 2017; 281: 319–336. doi: 10.1111/joim.12584 [DOI] [PubMed] [Google Scholar]
- 7.Bäckhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, et al. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci USA. 2004; 101: 15718–15723. doi: 10.1073/pnas.0407076101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cox AJ, West NP, Cripps AW. Obesity, inflammation, and the gut microbiota. Lancet Diabetes Endocrinol. 2015; 3: 207–215. doi: 10.1016/S2213-8587(14)70134-2 [DOI] [PubMed] [Google Scholar]
- 9.Chang CJ, Lin CS, Lu CC, Martel J, Ko YF, Ojcius DM, et al. Ganoderma lucidum reduces obesity in mice by modulating the composition of the gut microbiota. Nat Commun. 2015; 6: 7489. doi: 10.1038/ncomms8489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hildebrandt MA, Hoffmann C, Sherrill-Mix SA, Keilbaugh SA, Hamady M, Chen YY, et al. High-fat diet determines the composition of the murine gut microbiome independently of obesity. Gastroenterology. 2009; 137: 1716–1724.e2. doi: 10.1053/j.gastro.2009.08.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rabot S, Membrez M, Bruneau A, Gérard P, Harach T, Moser M, et al. Germ-free C57BL/6J mice are resistant to high-fat-diet-induced insulin resistance and have altered cholesterol metabolism. FASEB J. 2010; 24: 4948–4959. [DOI] [PubMed] [Google Scholar]
- 12.Le Roy T, Llopis M, Lepage P, Bruneau A, Rabot S, Bevilacqua C, et al. Intestinal microbiota determines development of non-alcoholic fatty liver disease in mice. Gut. 2013; 62: 1787–1794. doi: 10.1136/gutjnl-2012-303816 [DOI] [PubMed] [Google Scholar]
- 13.Boursier J, Mueller O, Barret M, Machado M, Fizanne L, Araujo-Perez F, et al. The severity of nonalcoholic fatty liver disease is associated with gut dysbiosis and shift in the metabolic function of the gut microbiota. Hepatology. 2016; 63: 764–775. doi: 10.1002/hep.28356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loomba R, Seguritan V, Li W, Long T, Klitgord N, Bhatt A, et al. Gut Microbiome-Based Metagenomic Signature for Non-invasive Detection of Advanced Fibrosis in Human Nonalcoholic Fatty Liver Disease. Cell Metab. 2017; 25: 1054–1062.e5. doi: 10.1016/j.cmet.2017.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chu H, Duan Y, Yang L, Schnabl B. Small metabolites, possible big changes: a microbiota-centered view of non-alcoholic fatty liver disease. Gut. 2019; 68: 359–370. doi: 10.1136/gutjnl-2018-316307 [DOI] [PubMed] [Google Scholar]
- 16.Long SL, Gahan CGM, Joyce SA. Interactions between gut bacteria and bile in health and disease. Mol Aspects Med. 2017; 56: 54–65. doi: 10.1016/j.mam.2017.06.002 [DOI] [PubMed] [Google Scholar]
- 17.Fuchs C, Claudel T, Trauner M. Bile acid-mediated control of liver triglycerides. Semin Liver Dis. 2013; 33: 330–342. doi: 10.1055/s-0033-1358520 [DOI] [PubMed] [Google Scholar]
- 18.Jiao N, Baker SS, Chapa-Rodriguez A, Liu W, Nugent CA, Tsompana M, et al. Suppressed hepatic bile acid signalling despite elevated production of primary and secondary bile acids in NAFLD. Gut. 2018; 67: 1881–1891. doi: 10.1136/gutjnl-2017-314307 [DOI] [PubMed] [Google Scholar]
- 19.Andrikopoulos S, Blair AR, Deluca N, Fam BC, Proietto J. Evaluating the glucose tolerance test in mice. Am J Physiol Endocrinol Metab. 2008; 295: E1323–E1332. doi: 10.1152/ajpendo.90617.2008 [DOI] [PubMed] [Google Scholar]
- 20.Cheng RY, Yao JR, Wan Q, Guo JW, Pu FF, Shi L, et al. Oral administration of Bifidobacterium bifidum TMC3115 to neonatal mice may alleviate IgE-mediated allergic risk in adulthood. Benef Microbes. 2018; 9: 815–828. doi: 10.3920/BM2018.0005 [DOI] [PubMed] [Google Scholar]
- 21.Higashikawa F, Noda M, Awaya T, Danshiitsoodol N, Matoba Y, Kumagai T, et al. Antiobesity effect of Pediococcus pentosaceus LP28 on overweight subjects: a randomized, double-blind, placebo-controlled clinical trial. Eur J Clin Nutr. 2016; 70: 582–587. doi: 10.1038/ejcn.2016.17 [DOI] [PubMed] [Google Scholar]
- 22.Shi L, Li M, Miyazawa K, Li Y, Hiramatsu M, Xu J, et al. Effects of heat-inactivated Lactobacillus gasseri TMC0356 on metabolic characteristics and immunity of rats with the metabolic syndrome. Br J Nutr. 2013; 109: 263–272. doi: 10.1017/S000711451200116X [DOI] [PubMed] [Google Scholar]
- 23.Cohen LJ, Esterhazy D, Kim SH, Lemetre C, Aguilar RR, Gordon EA, et al. Commensal bacteria make GPCR ligands that mimic human signalling molecules. Nature. 2017; 549: 48–53. doi: 10.1038/nature23874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schubert K, Olde Damink SWM, von Bergen M, Schaap FG. Interactions between bile salts, gut microbiota, and hepatic innate immunity. Immunol Rev. 2017; 279: 23–35. doi: 10.1111/imr.12579 [DOI] [PubMed] [Google Scholar]
- 25.Leung C, Rivera L, Furness JB, Angus PW. The role of the gut microbiota in NAFLD. Nat Rev Gastroenterol Hepatol. 2016; 13: 412–425. doi: 10.1038/nrgastro.2016.85 [DOI] [PubMed] [Google Scholar]
- 26.Muniz LB, Alves-Santos AM, Camargo F, Martins DB, Celes MRN, Naves MMV. High-Lard and High-Cholesterol Diet, but not High-Lard Diet, Leads to Metabolic Disorders in a Modified Dyslipidemia Model. Arq Bras Cardiol. 2019; 113: 896–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Picchi MG, Mattos AM, Barbosa MR, Duarte CP, Gandini MA, Portari GV, et al. A high-fat diet as a model of fatty liver disease in rats. Acta Cir Bras. 2011; 26:(Suppl 2): 25–30. doi: 10.1590/S0102-86502011000800006 [DOI] [PubMed] [Google Scholar]
- 28.Campbell CL, Yu R, Li F, Zhou Q, Chen D, Qi C, et al. Modulation of fat metabolism and gut microbiota by resveratrol on high-fat diet-induced obese mice. Diabetes Metab Syndr Obes. 2019; 12: 97–107. doi: 10.2147/DMSO.S192228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kübeck R, Bonet-Ripoll C, Hoffmann C, Walker A, Müller VM, Schüppel VL, et al. Dietary fat and gut microbiota interactions determine diet-induced obesity in mice. Mol Metab. 2016; 5: 1162–1174. doi: 10.1016/j.molmet.2016.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dimova LG, Zlatkov N, Verkade HJ, Uhlin BE, Tietge UJF. High-cholesterol diet does not alter gut microbiota composition in mice. Nutr Metab (Lond). 2017; 14: 15. doi: 10.1186/s12986-017-0170-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Refaie AA, Ramadan A, Sabry NM, Khalil WKB, Mossa AH. Over-gene expression in the apoptotic, oxidative damage and liver injure in female rats exposed to butralin. Environ Sci Pollut Res Int. 2020; 27: 31383–31393. doi: 10.1007/s11356-020-09416-6 [DOI] [PubMed] [Google Scholar]
- 32.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, et al. A core gut microbiome in obese and lean twins. Nature. 2009; 457: 480–484. doi: 10.1038/nature07540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stanislawski MA, Lozupone CA, Wagner BD, Eggesbø M, Sontag MK, Nusbacher NM, et al. Gut microbiota in adolescents and the association with fatty liver: the EPOCH study. Pediatr Res. 2018; 84: 219–227. doi: 10.1038/pr.2018.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009; 1: 6ra14. doi: 10.1126/scitranslmed.3000322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gilliland SE, Speck ML. Deconjugation of bile acids by intestinal lactobacilli. Appl Environ Microbiol. 1977; 33: 15–18. doi: 10.1128/AEM.33.1.15-18.1977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones BV, Begley M, Hill C, Gahan CG, Marchesi JR. Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc Natl Acad Sci USA. 2008; 105: 13580–13585. doi: 10.1073/pnas.0804437105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chumpitazi BP, Cope JL, Hollister EB, Tsai CM, McMeans AR, Luna RA, et al. Randomised clinical trial: gut microbiome biomarkers are associated with clinical response to a low FODMAP diet in children with the irritable bowel syndrome. Aliment Pharmacol Ther. 2015; 42: 418–427. doi: 10.1111/apt.13286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kovatcheva-Datchary P, Nilsson A, Akrami R, Lee YS, De Vadder F, Arora T, et al. Dietary Fiber-Induced Improvement in Glucose Metabolism Is Associated with Increased Abundance of Prevotella. Cell Metab. 2015; 22: 971–982. doi: 10.1016/j.cmet.2015.10.001 [DOI] [PubMed] [Google Scholar]
- 39.Karlsson FH, Fåk F, Nookaew I, Tremaroli V, Fagerberg B, Petranovic D, et al. Symptomatic atherosclerosis is associated with an altered gut metagenome. Nat Commun. 2012; 3: 1245. doi: 10.1038/ncomms2266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.De Vadder F, Kovatcheva-Datchary P, Goncalves D, Vinera J, Zitoun C, Duchampt A, et al. Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell. 2014; 156: 84–96. doi: 10.1016/j.cell.2013.12.016 [DOI] [PubMed] [Google Scholar]
- 41.Rémésy C, Demigné C, Chartier F. Origin and utilization of volatile fatty acids in the rat. Reprod Nutr Dev. 1980; 20:(4B): 1339–1349. doi: 10.1051/rnd:19800725 [DOI] [PubMed] [Google Scholar]
- 42.Gao Z, Yin J, Zhang J, Ward RE, Martin RJ, Lefevre M, et al. Butyrate improves insulin sensitivity and increases energy expenditure in mice. Diabetes. 2009; 58: 1509–1517. doi: 10.2337/db08-1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zydowo MM, Smoleński RT, Swierczyński J. Acetate-induced changes of adenine nucleotide levels in rat liver. Metabolism. 1993; 42: 644–648. doi: 10.1016/0026-0495(93)90225-D [DOI] [PubMed] [Google Scholar]
- 44.Broussard JL, Devkota S. The changing microbial landscape of Western society: Diet, dwellings and discordance. Mol Metab. 2016; 5: 737–742. doi: 10.1016/j.molmet.2016.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Watanabe M, Fukiya S, Yokota A. Comprehensive evaluation of the bactericidal activities of free bile acids in the large intestine of humans and rodents. J Lipid Res. 2017; 58: 1143–1152. doi: 10.1194/jlr.M075143 [DOI] [PMC free article] [PubMed] [Google Scholar]