Abstract
Background
The global rise of metabolic disorders, such as obesity, type 2 diabetes, and cardiovascular disease, demands a thorough molecular understanding of the cellular mechanisms that govern health or disease. The endoplasmic reticulum (ER) is a key organelle for cellular function and metabolic adaptation and, therefore disturbed ER function, known as “ER stress,” is a key feature of metabolic disorders.
Scope of review
As ER stress remains a poorly defined phenomenon, this review provides a general guide to understanding the nature, etiology, and consequences of ER stress in metabolic disorders. We define ER stress by its type of stressor, which is driven by proteotoxicity, lipotoxicity, and/or glucotoxicity. We discuss the implications of ER stress in metabolic disorders by reviewing evidence implicating ER phenotypes and organelle communication, protein quality control, calcium homeostasis, lipid and carbohydrate metabolism, and inflammation as key mechanisms in the development of ER stress and metabolic dysfunction.
Major conclusions
In mammalian biology, ER is a phenotypically and functionally diverse platform for nutrient sensing, which is critical for cell type-specific metabolic control by hepatocytes, adipocytes, muscle cells, and neurons. In these cells, ER stress is a distinct, transient state of functional imbalance, which is usually resolved by the activation of adaptive programs such as the unfolded protein response (UPR), ER-associated protein degradation (ERAD), or autophagy. However, challenges to proteostasis also impact lipid and glucose metabolism and vice versa. In the ER, sensing and adaptive measures are integrated and failure of the ER to adapt leads to aberrant metabolism, organelle dysfunction, insulin resistance, and inflammation. In conclusion, the ER is intricately linked to a wide spectrum of cellular functions and is a critical component in maintaining and restoring metabolic health.
Keywords: Obesity, Endoplasmic reticulum, Inflammation, Autophagy, UPR
Graphical abstract

Abbreviations
- 4-PBA
sodium phenylbutyrate
- ACAT
acyl-coenzyme Acholesterol transferase
- AGPAT
acylglycerin-3-phosphate-O-acyltransferases
- ATF
activating transcription factor
- ATG
autophagy-related proteins
- BAT
brown adipose tissue
- BiP
binding immunoglobulin protein
- C/EBP
CCAAT/enhancer-binding protein
- C53
CDK5RAP3-like protein
- CALCOCO1
calcium binding and coiled-coil domain 1
- CAMK2B
calcium/calmodulin-dependent protein kinase II beta
- CCL2
C–C motif chemokine ligand 2
- CDN
cyclic dinucleotides
- CE
cholesterol esters
- cGAMP
cyclic guanosine monophosphate-adenosine monophosphate
- cGAS
cyclic GMP-AMP synthase
- CHOP
C/EBP homologous protein
- ChREBP
carbohydrate-responsive element-binding protein
- CANX
calnexin
- COP2
coat protein complex 2
- CP
proteasome core particle
- CREBH
cyclic AMP-responsive element-binding protein 3-like protein 3
- CALR
calreticulin
- DDI2
DNA damage-inducible 1 homolog 2
- DDIT3
DNA damage-inducible transcript 3
- DGAT
diacylglycerol O-acyltransferase 1
- DRP1
dynamin-related protein-1
- DUB
deubiquitinating enzyme
- EIF2AK
eukaryotic translation initiation factor 2-alpha kinase
- EIF2B
eukaryotic translation initiation factor 2B
- EIF2α
eukaryotic translation initiation factor 2-alpha
- ER
endoplasmic reticulum
- ERAD
ER-associated degradation
- ERLEC1
endoplasmic reticulum lectin 1
- ER-phagy
ER-autophagy
- FBXO32
F-box only protein 32
- FFA
free fatty acid
- FITM2
fat storage-inducing transmembrane protein 2
- FOXO1
forkhead box protein O 1
- G6PC1
glucose-6-phosphatase catalytic subunit 1
- GCN2
general control non-derepressible 2
- GDP
guanosine diphosphate
- GEF
guanine nucleotide exchange factor
- GlcNAc
N-acetylglucosamine
- GP78
E3 ubiquitin-protein ligase GP78
- GRP-78
endoplasmic reticulum chaperone BiP
- HFD
high-fat diet
- HMGCR
3-hydroxy-3-methyl-glutaryl-coenzyme A reductase
- HRD1
hydroxymethylglutaryl reductase degradation protein 1
- HRI
heme-regulated eIF2α kinase
- IDOL
inducible degrader of LDL receptor protein
- IKK
inhibitor of nuclear factor kappa-B kinase
- IL
interleukin
- ITPR
inositol triphosphate receptor
- IRE1α
inositol-requiring enzyme 1-alpha
- IRF3
interferon regulatory factor 3
- IRS1
insulin receptor substrate 1
- ISR
integrated stress response
- JNK
c-Jun N-terminal kinase
- LDL
low-density lipoprotein
- MAM
mitochondria-associated membranes
- MBTPS1
membrane-bound transcription factor site-1 protease
- MBTPS2
membrane-bound transcription factor site-2 protease
- MFN
mitofusin
- MLXIPL
MLX interacting protein-like
- mtDNA
mitochondrial DNA
- mTORC1
mechanistic target of rapamycin complex 1
- NAFLD
non-alcoholic fatty liver disease
- NASH
non-alcoholic steatohepatitis
- NFE2L1
nuclear factor erythroid 2 related factor-1, also known as Nrf1
- NF-κB
nuclear factor-κB
- NGLY1
peptide-N(4)-(N-acetyl-beta-glucosaminyl) asparagine amidase
- NPC1
NPC intracellular cholesterol transporter 1
- OGT
O-linked N-acetylglucosamine transferase
- ORAI1
calcium release-activated calcium channel protein 1
- OS9
OS9 endoplasmic reticulum lectin
- p38-MAPK
p38 mitogen-activated protein kinase
- PSME
proteasome activator subunit
- PERK
PKR-like ER kinase
- Pik3r1
phosphoinositide-3-kinase regulatory subunit 1
- PKR
protein kinase RNA-activated
- RER
respiratory exchange ratio
- RETREG1
reticulophagy regulator 1
- RIDD
regulated IRE1-dependent decay
- ROS
reactive oxygen species
- RP
proteasome regulatory particle
- Rpn
26S proteasome non-ATPase regulatory subunit
- RYR
cardiac-type ryanodine receptor
- SCAP
SREBP-cleavage activating protein
- SDF2L1
stromal cell-derived factor 2-like 1
- SEL1L
SEL1L adaptor subunit of ERAD E3 ubiquitin ligase
- SERCA
sarco/endoplasmic-reticulum calcium ATPase
- SOAT1
sterol O-acyltransferase-1
- SOCE
store-operated calcium entry-associated regulatory factor
- SREBP1
sterol-regulatory element-binding protein-1
- STAT3
signal transducer and activator of transcription 3
- STIM
stromal interaction molecules
- STING1
stimulator of interferon response cGAMP interactor 1
- T2DM
type 2 diabetes mellitus
- TAG
triacylglycerol
- TBK1
TANK-binding kinase 1
- TNF-α
tumor necrosis factor alpha
- TRIM63
E3 ubiquitin-protein ligase TRIM63
- ULK1
Unc-51-like autophagy activating kinase 1
- UPR
unfolded protein response
- UPS
ubiquitin-proteasome system
- VCP
valosin-containing protein
- VDAC
voltage-dependent anion channels
- WAT
white adipose tissue
- WRS
Wolcott-Rallison syndrome
- XBP1
X-box-binding protein-1
1. Introduction
Metabolism is essential for converting nutrients into biochemical energy equivalents and building blocks for cellular growth, organismal development, activity, and health. The pressure to respond to fluctuations in nutrient availability has led to the development of an array of cellular and molecular mechanisms for metabolic adaptation. Multicellular organisms have an extra layer of complexity with the interplay of hormonal cues and metabolically active tissues. This allows the coordination of systemic metabolic adaptation in response to environmental challenges, such as changes in nutrient supply, temperature shifts, and escaping predators. While these evolutionary pressures have resulted in a remarkable metabolic adaptive capacity of the human body, modern society is a very different environment in which overnutrition and sedentary lifestyles are increasingly prevalent. Altogether, in this changed new metabolic world, the global increase in obesity and associated disorders such as type 2 diabetes mellitus (T2DM), non-alcoholic fatty liver disease (NAFLD), cardiovascular disease, and cancer represents one of the current major public health challenges. Obesity, the excessive accumulation of white adipose tissue (WAT), is a major risk factor for an array of pathologies. Obesity is a condition of chronic metabolic stress in which the conserved programs of metabolic adaptation fail or become maladaptive, ultimately resulting in a progressive loss of metabolic quality control mechanisms.
This review focuses on the endoplasmic reticulum (ER) as a central and universal platform of nutrient sensing, metabolic adaptation, and stress resistance. The ER is a key organelle with remarkable functional plasticity and structural diversity, regulating proteostasis, lipid metabolism, gluconeogenesis, and calcium signaling, all of which are critical components of obesity-linked metabolic disorders. Disturbances in ER homeostasis are usually referred to as “ER stress,” which is transient if mitigated by the activation of adaptive stress resistance mechanisms originating in the ER itself. Depending on the type of stressor, this is referred to as proteotoxicity, lipotoxicity, or glucotoxicity. However, it is important to realize that the type of stress and adaptive programs are intricately linked, dependent on each other, and occur in concert. Therefore, disturbances in proteostasis, for example, will impact lipid metabolism in the ER and vice versa. This is especially true in obesity, where continuous metabolic pressure creates strong challenges to most, if not all, pillars of ER function. The activation of adaptive stress-resistance mechanisms, also known as the integrated stress response (ISR), is evident in many tissues such as the brain, liver, and adipose tissue. In obesity, ER stress remains unresolved and is associated with ER dysfunction, which leads to compromised cellular health, tissue inflammation, and ultimately systemic metabolic collapse. As ER stress is a poorly defined and obscure term for a variety of diverse challenges to cell type-specific functions, we systematically review these in the context of the respective stressor and corresponding adaptive stress-resistance mechanisms in metabolic health and disease. In addition, we curated a comprehensive overview table (Supplementary Table 1) summarizing key regulators of ER stress with their function, gene/protein designation, and literature reference.
2. ER structure
The first key step to understanding the development of ER stress is acknowledging the highly diverse and complex nature of the ER. Depending on the organism, cell type, or physiological state, the ER is structurally and functionally highly dynamic and adaptive. The ER emerges from the nuclear envelope and consists of a variety of structural domains that are interconnected and contiguous, forming a vast network throughout cells. In principle, these structural domains are categorized as membrane cisternae (“sheets”) and tubules. ER sheets are mostly found in close proximity to the nucleus and are lined with ribosomes for protein biosynthesis (“rough ER”), whereas ER tubules are devoid of ribosomes, branched, and spread throughout the cytosol (“smooth ER”). Most of the basic principles of ER biology have been studied in model organisms such as budding yeast or immortalized cell lines [1], but in mammalian biology, several extreme and distinct ER phenotypes are found that deviate from most of the generalized concepts obtained in model systems. For example, the ER of exocrine acinar cells of the pancreas, which produce digestive enzymes, is almost exclusively composed of sheets and is the most prevalent constituent of these cells. Adipocytes, on the other hand, contain very little ER, which is mostly tubular. Hepatocytes in the liver have a mixed phenotype of both sheets and tubules. Muscle cells have a unique ER, also often known as sarcoplasmic ER, that is specialized to support contractility. An open gap in understanding ER function in the metabolism and ER stress in pathophysiology determines the definition of the structural-functional relationship of these specialized ER phenotypes. It is very well established that common chemical inducers of ER stress such as tunicamycin [2] or proteasome inhibitors [3] have a strong impact on the structural phenotype, although this remodeling is usually transient and once stress is resolved, the ER returns to its original state. These changes include dilation of the ER [2], resolution of sheet structures [3], formation of ER whorls [4], and diminished ER content. ER stress is also associated with lipid droplet biogenesis [3] and glycogen deposition [5]. In obesity, these ER phenotypes are linked to ER stress [2,3,6], but why these structural changes occur and if they contribute to insulin resistance, for example, remains largely unclear. The structure of the ER is maintained by scaffolding proteins and tethers that, in a regulated fashion, connect the ER to the cytoskeleton and other organelles. It is possible that a disturbed structural-functional relationship observed in obesity is caused by a discrete abnormality of the structural apparatus, and restoration of which might help reduce ER stress and improve metabolic health. Indeed, genetically restoring a “lean” ER phenotype in the livers of obese mice partially rescues their metabolic dysfunction [7]. Obviously, as the structure of ER is diverse in cell types involved in the onset of metabolic disease, it is also likely that therapeutic angles to manage ER deformation are diverse.
3. ER connections
An important factor of ER biology is that the peripheral tubular ER network forms abundant membrane contact sites with mitochondria, endosomes, and the plasma membrane, the latter called the cortical ER [8]. Likewise, the ER has contact sites with lipid droplets, peroxisomes, and the Golgi apparatus. These connections start at the point of organelle biogenesis, as organelles arise from ER membranes and therefore, at least at some point, share a membrane interface [8]. Membrane contact sites are defined as ribosome-free, 10 nm gaps between organelles with no direct membranous connection. They are important exchange platforms of lipids, small molecules, and second messengers between the ER and other organelles. This includes cholesterol transfer from endosomes/lysosomes to the ER via NPC intracellular cholesterol transporter 1 (NPC1) [9], calcium exchange via the voltage-dependent anion channels (VDACs)/inositol trisphosphate receptor (ITPRs) complex between the ER and mitochondria [2], transfer of triglycerides from the ER to lipid droplets via diacylglycerol O-acyltransferases (DGATs) [10], and exchange of various lipid species such as phospholipids, sphingolipids, and cardiolipins to the Golgi [11]. Thus, the ER is uniquely placed at the nexus of directing endogenous biosynthesis and sensing nutrients. As obesity is associated with broad changes in lipid metabolism, it is conceivable that alterations in the function of membrane contact sites contribute to the cellular dysfunction underlying metabolic disorders. Indeed, chronic enrichment of ER-mitochondrial contact sites in hepatocytes leads to mitochondrial calcium overload and insulin resistance in obesity [2]. In addition to this organelle communication, ER contact sites also provide a scaffold for organelle fusion and fission events. The ER wraps around mitochondria to facilitate mitochondrial dynamic remodeling and autophagy [12], key elements of mitochondrial quality control. Dynamin-related protein-1 (DRP1, encoded by DNM1L) and mitofusins (MFN1 and MFN2) are found in ER-mitochondria contact sites, and MFN2 has been shown to play a prominent role in obesity and metabolism in mice and humans [13]. MFN2 is downregulated in obesity, and using transgenic mouse models, we have learned that MFN2 in hepatocytes is necessary for maintaining glucose tolerance [14], in white adipocytes it controls adiposity [15], and in brown adipocytes it is required for thermogenesis [16,17]. Interestingly, one hallmark of MFN2 loss and disturbed mitochondrial quality control is that this defect does not occur in isolation but rather has a broad impact on organelle dynamics and function, as loss of MFN2 is associated with ER stress and abnormal lipid droplets [14,18]. In summary, the structural and connective aspects of ER biology are intricately linked to cellular metabolism. In the following section, we discuss the functional diversity of the ER and the fundamental mechanisms that either prevent or enhance ER stress in metabolic disorders.
4. Protein quality control
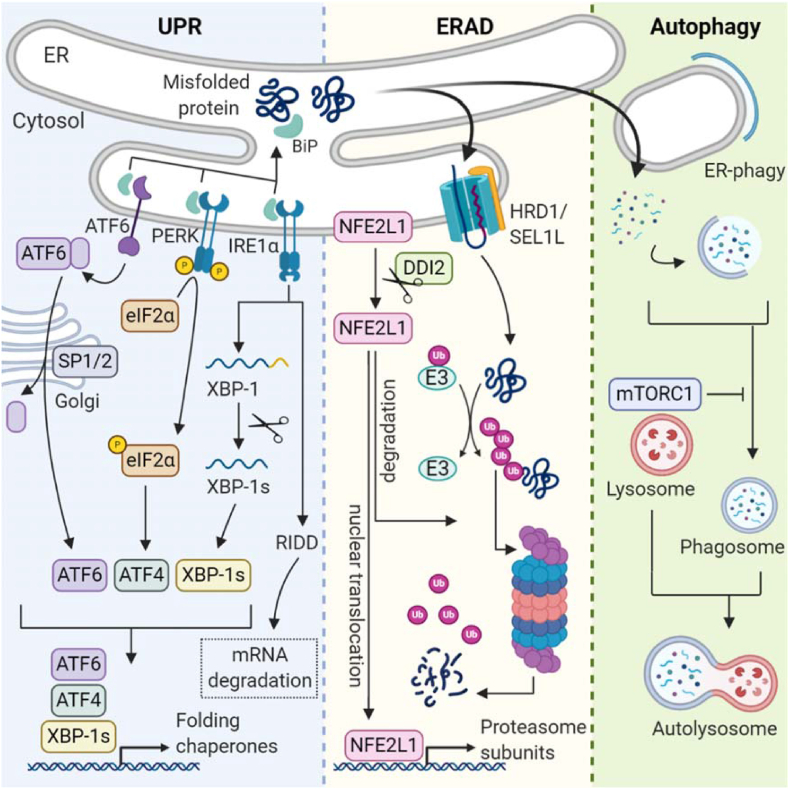
Amino acids are the fundamental building blocks of proteins, including metabolic enzymes and hormones, which play critical roles in growth, development, and metabolism throughout life. It is crucial that the turnover of amino acids and protein production is tightly regulated. Particularly, folding of the nascent polypeptide chain is a complex and relatively error-prone process [19]. Hence, this is supported by a large set of molecular chaperones and carefully safeguarded by cellular quality control mechanisms. Even under ideal conditions, the pool of proteins in an organism undergoes a constant turnover due to the natural half-life or if proteins become damaged or simply obsolete. In physiological conditions, the rates of protein synthesis and degradation fluctuate, for example, with fasting and feeding in the liver, which is controlled by the mechanistic target of the rapamycin complex 1 (mTORC1) pathway (reviewed in [20,21]). Disturbances in this delicate balance of protein life are managed by the activation of multiple adaptive mechanisms such as unfolded protein response (UPR), ER-associated degradation (ERAD), and autophagy, all serving the purpose of restoring cellular protein homeostasis or proteostasis (Figure 1) [22,23]. The major outcomes of UPR activation are inhibition of protein translation and increasing the folding capacity of the ER, both effective at eliminating proteotoxic stress and restoring proteostasis [24]. However, the last resort is suicide, UPR-induced cell death, if the resolution of ER stress and restoration of homeostasis are unsuccessful. The three classical main arms of the UPR are mediated through protein kinase R-like ER kinase (PERK, encoded by EIF2AK3), inositol-requiring enzyme 1-alpha (IRE1α, a serine/threonine protein kinase/endoribonuclease encoded by ERN1), and activating transcription factor-6 (ATF6), although this is a limited perspective, as there is evidence that more arms exist. PERK, IRE1α, and ATF6 are anchored in the ER membrane and activated by the presence of unfolded proteins. The current leading theories posit that unfolded proteins are sensed either through direct contact with the luminal domains of the receptors or indirectly through dissociation of chaperones such as binding immunoglobulin protein (BiP, also known as GRP-78, or HSPA5 encoded by HSPA5) from the receptors [25]. In any case, sensing luminal unfolded proteins leads to UPR activation and initiation of downstream events that facilitate the resolution of ER stress [26].
Figure 1.
Cellular protein quality control. Three major cellular programs are responsible for proteostasis. The UPR is activated by binding of unfolded proteins to chaperone proteins and the luminal parts of the UPR sensors. The ATF6, PERK, and IRE1α pathways attenuate proteostatic burden by promoting the transcription of folding chaperones and inhibiting protein translation via EIF2α. In ERAD, misfolded substrates are shuttled from the ER lumen into the cytosol, where they undergo ubiquitination degradation by the proteasome. During proteostatic stress, the transcription factor Nfe2l1 escapes proteasomal degradation and promotes transcription of proteasome subunits. In autophagy regulated by mTORC1, redundant proteins and organelles are degraded in autolysosomes.
4.1. Protein production and EIF2α kinases
The first line of ER stress response is halting protein translation through phosphorylation of eukaryotic translation initiation factor 2-alpha (EIF2α) by PERK. EIF2α is also phosphorylated by several other kinases and therefore is a key node on which multiple stress pathways converge to regulate proteostasis. The EIF2 is a trimer consisting of three subunits, alpha, beta, and gamma, and phosphorylation occurs on serine 51 residue of the alpha subunit. In turn, this phosphorylation inhibits guanine nucleotide exchange factor (GEF) of EIF2, namely eukaryotic translation initiation factor 2B (EIF2B), and thereby traps EIF in its inactive GDP-bound state, which halts protein translation [27,28]. Ingeniously, phosphorylated EIF2α tolerates alternative non-AUG translation of transcription factors for protein chaperones, for example, ATF4 (Figure 1) [29,30]. Disturbances in EIF2α phosphorylation have detrimental consequences for metabolic health. In beta cells, EIF2α is baseline highly phosphorylated. The deletion of PERK leads to loss of beta cells and T2DM [31,32]. Mice carrying a hepatocyte-specific deletion of PERK display defective gluconeogenesis among other metabolic abnormalities [33]. Furthermore, patients with Wolcott-Rallison syndrome (WRS), which is a rare autosomal recessive disorder characterized by a mutation in the gene coding for PERK, demonstrate symptoms such as T2DM, multiple epiphyseal dysplasia, osteoporosis, and growth retardation [34], emphasizing the importance of PERK and its complex roles in metabolic homeostasis.
Another EIF2α kinase with implications for metabolic regulation is protein kinase RNA-activated (PKR, encoded by EIF2AK2), which is named after its activation mechanism by double-stranded RNA introduced, for example, by viral infections [35]. However, PKR does not solely react to dsRNA derived from pathogens but also responds to organelle stress and lipotoxicity. In mouse models of obesity, PKR activity is increased, which is linked to insulin resistance [36]. Correspondingly, small molecule inhibitors of PKR improve both insulin sensitivity and glucose intolerance in obese mice [37]. However, whole-body deletion of PKR in mice does not necessarily result in a phenotype [38], indicating a context-dependent role of PKR. Nevertheless, PKR is a leading example of a kinase that operates at the nexus of metabolic disorders and pathogen-induced inflammation. The two other known EIF2α kinases are general control non-derepressible 2 (GCN2, encoded by EIF2AK4), which is induced upon amino acid deprivation, and heme-regulated EIF2α kinase (HRI, encoded by EIF2AK1), which is activated by heme deprivation [39,40]. While the significance of HRI for metabolic disorders remains largely unclear, GCN2 has multiple roles in nutrient sensing. Mice lacking GCN2 demonstrate a general sensitivity to diets lacking essential amino acids, for example, leucine [41]. Furthermore, in GCN2-deficient mice, muscle tissue is extensively degraded in an attempt to sustain amino acid levels. Also, while lipogenesis is repressed in livers of regular mice during leucine starvation, this adaptive regulation is absent in GCN2-deficient mice [42,43]. In summary, these examples highlight that translational control through eIF2α and its kinases is a key adaptive mechanism for nutrient sensing and stress resistance with an impact on metabolic diseases.
4.2. Protein folding via IRE1α-XBP1 and ATF6
The second major arm of the UPR is the activation of IRE1α. This bifunctional enzyme senses the accumulation of unfolded proteins by their binding to the luminal domain of IRE1α, which leads to its dimerization and autophosphorylation. This activates its cytosolic endoribonuclease activity, initiating two separate events to restore proteostasis. One is the degradation of mRNAs by regulated IRE1-dependent decay (RIDD) (Figure 1), by which IRE1α cleaves and destabilizes a set of ER-localized mRNAs, thus reducing the folding burden created by newly synthesized proteins entering the ER lumen. The other is the non-canonical-specific splicing of the mRNA of X-box-binding protein-1 (XBP1) [44]. This mRNA encodes the transcription factor XBP1s, which controls chaperone gene expression, but also has non-coding functions [45]. XBP1s is also regulated on the protein level through phosphorylation by mitogen-activated protein kinase 14 (MAPK14, also known as p38-MAPK) signaling [46] and interaction with several nuclear proteins with established roles in metabolism such as forkhead box protein O 1 (FOXO1) [47] and the regulatory subunits of phosphoinositide-3-kinase regulatory subunit 1 (Pik3r1, also known as PI3K), p85 alpha, and p85 beta [48]. Mice deficient in the IRE1α-XBP1 mechanism show multiple metabolic abnormalities depending on the cell type involved. Mice with hepatocyte-specific deletion of XBP1 develop insulin resistance [49]. In adipose tissue, XBP1 is involved in tissue remodeling during lactation [50], but is dispensable for non-shivering thermogenesis in BAT [3]. Deletion of the IRE1α-XBP1 pathway in beta cells results in severe cellular dysfunction, including decreased insulin secretion, diminished insulin and proinsulin contents in cells, and less oxidative folding of proinsulin [51,52]. In the brain, XBP1s in pro-opiomelanocortin neurons mitigates obesity-induced ER stress with beneficial effects on energy balance and glucose homeostasis [53]. Clearly, the IRE1α-XBP1 arm of the UPR is a key element of metabolic homeostasis, but in obesity, this mechanism is compromised in several ways. The inflammatory environment associated with obesity directly impairs the endoribonuclease activity of IRE1α. This results in lower levels of processed XBP1 and a reduced activation of UPR target genes in the liver [54]. In addition, reduced overall p38-MAPK signaling hinders the nuclear translocation and full activation of XBP1, which prevents the resolution of ER stress and restoration of glucose homeostasis [46]. Indeed, forced activation of XBP1 is associated with multiple metabolic benefits [53,55,56], and pharmacologically targeting IRE1α may help reduce ER stress and inflammation therapeutically.
The third arm of the UPR is mediated through ATF6. The accumulation of unfolded proteins leads to ATF6 interaction with coat protein complex 2 (COP2) vesicles, which then transport ATF6 from the ER to the Golgi [57,58]. There, ATF6 is cleaved by membrane-bound transcription factor site-1 protease (MBTPS1, also known as S1P) and membrane-bound transcription factor site-1 protease (MBTPS2, also known as S2P), creating the active transcription factor (Figure 1) [59]. This ATF6 fragment initiates the transcription of chaperones involved in protein folding to alleviate ER stress [60]. The ATF6 pathway is activated in most tissues of obesity, which is most evident in beta cells of pancreatic islets [61]. However, in mice, genetic deletion of ATF6 shows no apparent major phenotype, unless the mice are bred on a susceptible background or treated with ER stress-inducing compounds. The role of ATF6 seems to be complex, as treating ATF6-deficient mice with tunicamycin leads to liver dysfunction and steatosis [62], but in beta cells, ATF6 seems to promote the development of diabetes [62]. This demonstrates that UPR activation is context- and cell type-dependent and potentially other UPR arms compensate or interact with the ATF6 pathway to mitigate ER stress or accelerate maladaptive responses. In summary, UPR is a versatile mechanism for the resolution of ER stress and tightly linked to metabolic status. While one major outcome of successful UPR activation is the restoration of proteostasis by halting synthesis and “fixing” troubled proteins, it may also lead to elimination of cells once UPR is set in overdrive by triggering apoptosis via DNA damage-inducible transcript 3 (DDIT3, also known as CHOP) [63,64]. However, in maintaining proteostasis, UPR is intricately linked to alternative proteostatic mechanisms that operate in concert and may compensate for each other during the restoration of ER homeostasis, thus preventing fatal outcomes for cells.
4.3. ER-associated protein degradation
Instead of “repairing” proteins by assisting their folding via the UPR, there is also the alternate possibility of directly degrading troubled proteins to avoid their accumulation and ER stress. ERAD is a protective mechanism facilitating the elimination of these proteins through the UPS, which is highly evolutionarily conserved from yeast to humans. In yeast, it is well established that if UPR fails, ERAD is activated and will compensate to restore proteostasis [65]. In mammalian systems, failure of ERAD has detrimental effects on metabolism. Human genetic studies have found associations between ERAD gene variations and metabolic disease, such as familial hypercholesterolemia and diabetes as well as neurological and autoimmune disorders [66]. In particular, beta cells can be severely affected by changes in ERAD, as the precursor of insulin, and proinsulin, which is prone to misfolding with up to 20% of proteins failing to reach conformation, is concomitantly eliminated by ERAD [67]. As germline deletion of any of the key ERAD genes in mice is lethal [68], most studies used cell models or at best tissue-specific and/or inducible knock-out models to gain in vivo insight. ERAD follows a complex series of events that extends from the ER into the cytoplasm. First, unfolded or misfolded proteins in the ER lumen or membrane are recognized by lectins, such as OS9 endoplasmic reticulum lectin (OS9), endoplasmic reticulum lectin 1 (ERLEC1, also known as XTP3-B), and chaperones such as BiP [69]. Second, ERAD substrates undergo active retro-translocation across the lipid bilayer through adaptor–dislocon complexes, for example, through the SEL1L adaptor subunit of ERAD E3 ubiquitin ligase (SEL1L) and hydroxymethylglutaryl reductase degradation protein 1 (HRD1, encoded by SYVN1, also known as synovolin) complex [68,70]. Third, ATPase valosin-containing protein (VCP, also known as p97) translocates substrates out of the ER membrane, breaking up their tertiary structure [71].
4.4. Ubiquitin-proteasome system
After transferal to the cytosol, ERAD substrates concomitantly enter the ubiquitin-proteasome system (UPS) machinery, where they undergo polyubiquitination by E3 ubiquitin ligases and subsequent degradation by the proteasome [72]. Of note, most of the knowledge about the mechanisms of ERAD and UPS is built on experiments using cellular models and compared to UPR, but very little is known about the actual mammalian biology of ERAD. This highlights the unmet need to explore the molecular basis of ERAD and UPS in a tissue-specific and physiology-focused manner. In that vein, while it was long assumed that the structure, regulation, and function of the proteasome is relatively static and generalizable to all mammalian cells, we know that proteasomes form microdomains within cells, for example, in the nucleus [73] or in close proximity to rough ER [74]. The relevance of these microdomains for ERAD and proteostasis, especially in tissues linked to metabolic disorders, is unknown. In many cell lines, the proteasome is a macromolecular complex that consists of a 20S core particle (CP) and one or two 19S regulatory particles (RPs) that close off access to the CP [75]. The nature of how proteasomes exist in vivo remains largely unclear. However, there is recent evidence that both the structure and activity of the proteasome might be acutely regulated by hormonal and metabolic status, for example, by the cAMP level and glucagon stimulation in brown adipocytes, hepatocytes, and muscle cells [3,76]. During this process, ubiquitinated proteins are recognized by ubiquitin receptors on the RP, namely Rpn1, Rpn10, and Rpn13 [77]. Deubiquitinating enzymes (DUBs) remove ubiquitin and may also undo the recognition of substrates by the proteasome [72], providing an extra layer of regulation with potential roles in metabolic disorders. Once the substrate is irreversibly bound to the proteasome, it is translocated into the proteolytic chamber of the CP, where it then degrades. While we have outlined several distinct layers of potential regulation herein, the prevailing view is that the UPS is mainly driven by the activity of ubiquitin E3 ligases, enzymes that catalyze the final step of ubiquitination. There are few examples of global E3 ubiquitin-protein ligase TRIM63 (TRIM63, also known as MURF1), F-box only protein 32 (FBXO32, also known as MAFbx) in muscle atrophy [78], and targeted HMGCR by HRD1 [79] and GP78 [80,81] or the LDL receptor by IDOL [82] protein degradation in metabolism. However, how cells respond to global changes in ubiquitination upon fluctuations in metabolism is less understood.
4.5. NFE2L1 links UPR, ERAD, and UPS
The first clues to understanding how global ubiquitination is sensed and controlled came from studying proteasome inhibitors, which are in clinical use for the treatment of multiple myeloma. Cells possess an ingenious mechanism by which global levels of ubiquitinated proteins in an elegant bounce-back mechanism are transcriptionally coupled to proteasomal activity [83]. When levels of ubiquitinated proteins exceed the capacity of proteasomes, cells increase the synthesis of proteasomal subunits to overcome this relative deficiency, for example, upon treatment with proteasome inhibitors [83]. This adaptive proteostatic mechanism is mediated by nuclear factor erythroid 2 related factor-1 (NFE2L1) transcription factor. NFE2L1 is anchored in the ER membrane and heavily post-translationally modified before being transcriptionally active. In a complex process, NFE2L1 undergoes deglycosylation by peptide-N(4)-N-acetyl-beta-glucosaminyl) asparagine amidase NGLY1 [84], translocates across the ER membrane by VCP [85], ubiquitination by HRD1 [86], and proteolytic cleavage by protein DNA damage-inducible 1 homolog 2 (DDI2) [87], which generates a b-ZIP-DNA-binding domain-containing fragment of NFE2L1 (Figure 1). Under normal cellular circumstances, when proteasomal activity is sufficient, this fragment is virtually absent as it is immediately degraded by the proteasome as it forms. However, when ubiquitin levels rise above the proteasomal capacity, NFE2L1 escapes degradation and initiates transcription of proteasome subunits and other ERAD components in the nucleus [3,88]. Interestingly, there is recent evidence that this adaptive component of proteasomal activity is an important regulator of proteostasis and ER stress in physiology and obesity. In brown adipocytes, NFE2L1-mediated proteasomal activity is induced during cold acclimatization but dispensable at thermoneutrality [3], matching the cold-induced increase in global brown fat ubiquitination that could partly be mediated by HRD1 [89]. Disruption of NFE2L1-mediated proteasomal activity either by loss of NFE2L1 or proteasome inhibitors causes cellular hyperubiquitination, ER stress, tissue inflammation, diminished non-shivering thermogenesis, and insulin resistance [3]. Similarly, adipose tissue browning [3] and white adipocyte function [3,90] are also affected by the loss of NFE2L1-mediated proteasomal activity, but the relevance in obesity, especially in humans, remains unclear. Next to adipose tissue, the liver ERAD/UPS play a prominent role in metabolic regulation. Proteasome dysfunction through deleting the proteasome activator subunit 3 (PSME3) results in hyperubiquitination, ER stress, steatosis, and hepatic insulin resistance [91]. A similar NASH phenotype has been described in hepatic NFE2L1 deficiency [92,93]. Particularly, NFE2L1 links proteasomal activity to fasting and feeding cycles [94] as well as the response to certain nutrients in the liver [93]. Another noteworthy player linking ERAD, ER stress, and metabolism is stromal cell-derived factor 2-like 1 (SDF2L1), an ER-localized protein with a potential function in ERAD substrate recognition [95]. Suppression of SDF2L1 in the liver results in ER stress, steatosis, and insulin resistance [96]. In summary, deficiency of ERAD-related genes compromises proteostasis and disrupts systemic metabolic health. Interestingly, several studies have suggested that inhibition of specific ERAD-related genes could be beneficial for treating metabolic disorders. Inhibition of hepatic HRD1 [97] or full-body deletion [98] protects against obesity in mice. However, it is important to distinguish between the ERAD process, which is adaptive in nature, and the effects of removing an essential component of the machinery. While many projects claim to have studied ERAD itself, they only analyzed the secondary effects of ERAD loss, which may or may not have overlapping relevance. In this light, it is not surprising that liver-specific disruption of ERAD compromises liver function and even a “beneficial” effect on body weight regulation and protection from obesity in these models [[97], [98], [99], [100], [101]], reflecting a general sickness of the organism. Any future approaches aimed at exploiting ERAD as a therapeutic should therefore focus on the adaptive component of ERAD, which is mediated by NFE2L1 [3,83,93,94], as it has a unique and diverse biology with multiple angles for molecular targeting [102].
4.6. Autophagy
Macroautophagy (hereafter autophagy) is a highly conserved cellular adaptation process discovered in yeast as a survival mechanism in response to starvation [103,104]. The recycling of endogenous cellular parts during nutrient deprivation is an ancient and powerful ability for the maintenance and preservation of organismal life. Autophagy involves several different autophagy-related (ATG) proteins and is initiated by activation of the unc-51-like autophagy activating kinase 1 (ULK1/ATG1 in yeast), which forms the phagophore [105,106]. This has a double membrane structure that further expands to engulf cytoplasmic material and finally evolves into a closed structure, the autophagosome. Subsequently, autophagosomes fuse with lysosomes to form autolysosomes and enzymatically “digest” their cargo into their building blocks, such as amino acids, lipids, and carbohydrates (Figure 1). Aside from regulating energy levels and cellular proteostasis, autophagy also plays a critical role in cellular stress responses [107,108], which are often induced by the accumulation of unwanted materials. Failure to eliminate this “protein waste” is associated with several storage diseases with relevance for human disease, including metabolic derangements. A critical key node on which autophagy and most proteostatic pathways converge is mTORC1, which is a multicomponent kinase complex that is regulated by insulin, growth factors, amino acid levels, oxygen, and energy levels. The biology of mTORC1 and its implications for metabolic disease are vast and were reviewed in detail elsewhere [109]. In healthy cells in a nutrient-rich environment and unstressed conditions, mTORC1 represses autophagy and stimulates biosynthesis, cell cycle progression, and growth (Figure 1). However, upon nutrient stress, mTORC1 is inactivated and autophagic gene transcription increases [105,110]. Interestingly, mTORC1 is also connected to transcriptional activation of NFE2L1 and proteasomal protein degradation via sterol-regulatory element binding protein-1 (SREBP1) [94,111]. This SREBP1-related arm of mTORC1 activity also regulates lipogenesis [112], linking nutrient sensing by mTORC1 to multiple relevant sources of ER stress. This crosstalk of autophagic flux and ER stress has been demonstrated in many cell types critical for metabolic disorders, for example, in pancreatic beta cells, adipocytes, and cardiomyocytes [113]. In principle, in most tissues, obesity is a state of relative autophagic deficiency, as autophagy is downregulated in genetic and dietary models of obesity, which is particularly caused by autophagy-related-7 protein (ATG7) levels, a critical component in the initiation of the autophagosome formation. Suppression of ATG7 in the liver results in defective insulin signaling and elevated ER stress [114]. However, when ATG7 expression is restored, ER stress is alleviated, enhancing hepatic insulin action and systemic glucose tolerance in obese mice [114]. Furthermore, deletion of ATG7 disturbs lipid metabolism by decreasing triglyceride breakdown, which results in aberrant lipid accumulation and compromised metabolic function [115]. In contrast to most other tissues, in pancreatic beta cells, autophagy is upregulated in insulin resistance, and deletion of ATG7 in beta cells impairs insulin secretion and inhibits the compensatory increase in beta cell mass in obesity [116]. Another important link between the autophagic machinery and ER function is the process of so-called ER-phagy, by which parts of the ER network are engulfed by autophagosomes through specific ER-phagy receptors, for example, reticulophagy regulator 1 (RETREG1) [117] and then removed by lysosomal degradation. ER-phagy contributes to the adaptive remodeling of the ER and has been connected to the ER stress response, specifically to the UPR [118,119], and ribosome stalling via the newly discovered CDK5RAP3-like protein (C53) [120]. The soluble ER-phagy receptor calcium binding and coiled-coil domain 1 (CALCOCO1) has recently been identified to be induced by IRE1 and helps degrade tubular ER during ER stress [121]. Mutant prohormones, such as the insulin-deficient Akita proinsulin mutant, forms aggregates that are degraded by ER-phagy [122,123]. Aside from this anecdotal observation of ER-phagy, its role in physiology and metabolic disorders is relatively unexplored. In summary, autophagy is a major pillar of proteostasis and intricately linked to ER function and stress through multiple signaling key nodes and physical interactions.
5. Calcium homeostasis
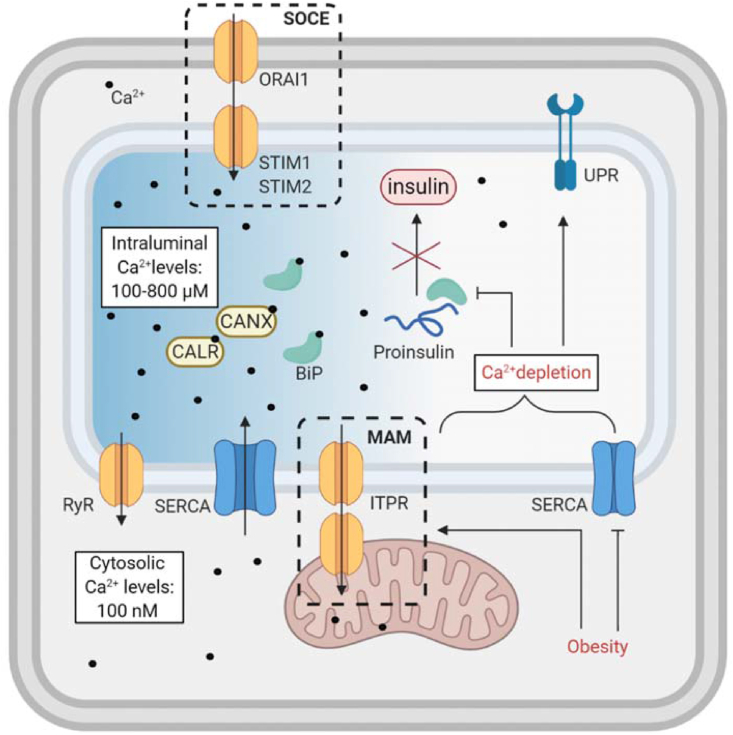
Obesity and metabolic disease are associated with disturbances in ER-calcium homeostasis [124,125], which have severe consequences for ER function and proteostasis. Calcium is an intracellular signaling molecule that is implicated in a wide spectrum of processes, including energy metabolism and protein synthesis. The ER is the largest intracellular calcium store; it has Ca2+ luminal concentrations of 100–800 μM compared to 100 nM Ca2+ in the cytosol (Figure 2) [126]. This steep gradient is maintained by several mechanisms (see [126,127]). Intraluminal chaperones, such as calnexin (CANX), calreticulin (CALR), and BiP, act as buffers by directly binding to calcium [128]. Luminal calcium is essential for protein folding and maturation [126,129], as calcium binds to ER folding chaperones and is required for their activity [126]. Aberrant calcium levels prevent proper folding of proteins and consequently activate the UPR and ERAD pathways [130]. In particular, folding of the already-vulnerable proinsulin is disturbed by calcium depletion, which can lead to insulin secretion deficiencies [131,132]. Calcium is continuously imported into the ER by sarco/endoplasmic-reticulum Ca2+ ATPase (SERCA) proteins. Obesity and high levels of free fatty acids (FFA) reduce SERCA activity, which results in ER-calcium depletion (Figure 2) [125,133,134]. Store-operated calcium entry-associated regulatory factor (SOCE) regulates ER-calcium uptake in response to decreased luminal calcium levels by allowing the direct flux of calcium ions from the extracellular space into the ER lumen through direct membrane connection between the ER and plasma membrane. Stromal-interaction molecules (STIMs) sense luminal calcium depletion and facilitate SOCE by forming plasma membrane-ER bridges with calcium release-activated calcium channel proteins (ORAI1). SERCA inhibition by thapsigargin leads to a decrease in luminal calcium, immediate experimental ER stress, and SOCE. Obesity is associated with alterations in the SOCE machinery in hepatocytes, which contributes to ER stress and causes glucose intolerance in mouse models [135]. Pharmacological blocking of SOCE also impairs pancreatic insulin secretion [136]. Conversely, in response to high luminal calcium levels, calcium is released from the ER by ryanodine receptors (RYRs) and ITPR channels. ITPR in the liver is found in mitochondrial-associated membranes (MAMs) and facilitates ER-mitochondrial communication and lipid droplet formation [137]. ITPR has different isoforms and is generally overactivated in obesity, which results in higher cytosolic Ca2+, inflammation and insulin resistance [7]. In general,@@ ER-calcium mediates inter-organelle communication, as BiP and other chaperones reside close to ER membrane contact sites and facilitate calcium transfer between organelles [8,138]. However, excessive and uncontrolled flux of calcium into mitochondria stimulates ATP production, leading to increased reactive oxygen species (ROS) levels, and pro-apoptotic pathways. Obesity, T2DM, and high-fat diet (HFD) all modulate MAM sites, which disturbs calcium metabolism and can aggravate insulin resistance [2,139,140]. Through these mechanisms, the ER also controls cytosolic calcium levels and high levels of calcium in the cytosol induce stress response pathways, which may result in apoptosis. Sorcin is a calcium sensor protein involved in maintaining ER-calcium by inhibiting RYR activity, thus terminating calcium-induced calcium release into the cytosol [141]. In the liver, obesity-induced lipotoxicity promotes calcium efflux [142] and by stimulating calcium/calmodulin-dependent protein kinase II beta (CAMK2B) [143], chronic cytosolic calcium overload is linked to inflammation and insulin resistance [143,144]. Lipotoxicity, on which we elaborate in the following chapter, also disturbs ER-calcium homeostasis [133]. In summary, disturbances in ER-calcium levels compromise metabolic function, and, conversely, metabolic disorders enhance cellular stress through disturbing calcium homeostasis through multiple mechanisms.
Figure 2.
Calcium homeostasis in the ER. The high calcium-ion gradient across the ER membrane between lumen and the cytosol is maintained through several mechanisms. SOCE allows calcium transfer from the extracellular environment to the ER. SERCA continuously “pumps” calcium into the ER lumen. RyR reduces high levels of ER-calcium. ITPR shuttles calcium to the mitochondria through MAM. Obesity leads to ER-calcium depletion, which disrupts ER homeostasis and hinders proper folding of proteins. These disturbances promote ER stress and cellular dysfunction.
6. Lipid metabolism
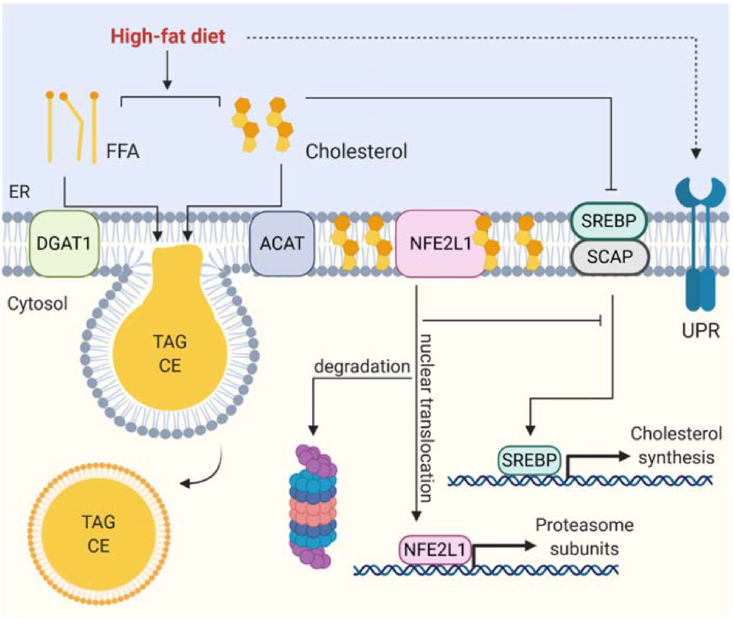
The ER is the primary site of cellular lipid synthesis, coordinates transfer of lipids to the cytoplasm and other organelles, and is the origin of lipid droplet biogenesis (Figure 3) [10,[145], [146], [147]]. It is important to note that total plasma or tissue lipidome analysis might be misleading as there are compartmentalized changes in lipid composition in physiology and obesity that take place at the cellular or organelle level [[148], [149], [150]]. It is well established that ER dysfunction is a pivotal driver on the road to aberrant lipid metabolism associated with obesity and metabolic disorders. Excess lipid exposure leads to ER stress and, conversely, ER stressors disrupt lipid metabolism, eventually overpowering the protective mechanisms of the ER [49,151]. A major link between metabolic disorders, lipid metabolism, and ER stress is the response to modern diets that are rich in processed nutrients and unhealthy fats. Chronic overnutrition and excess lipid exposure impose significant challenges on the adaptive mechanism of the ER. First, this is manifested in whole-body nutrient utilization, as diurnal shifts in the respiratory exchange ratio (RER) are lost and obesity is associated with low RER, indicating chronic lipid over carbohydrate utilization. Second, nutrient homeostasis during fasting and feeding cycles are lost. Surprisingly, obesity is a state of relative fasting and incomplete nutrient utilization. Third, the body weight gain associated with a chronic positive energy balance eventually overwhelms the storage capacity of adipose tissue, resulting in adipose tissue dysfunction and ectopic lipid deposition, which are strongly linked to insulin resistance and diabetes. Chronic consumption of HFDs is associated with lipotoxicity, which is the exposure to high levels of cholesterol and FFAs [93,132,[152], [153], [154], [155], [156], [157]]. While these potentially harmful lipids are usually safely stored in adipocytes or lipid droplets in other cells, obesity is a condition in which lipids escape and overflow in other organelles, cells, and tissues. Particularly, saturated FFAs such as palmitate are lipotoxic in high amounts [154,155,157] because biophysically they have a high packing density and usually cells very tightly control the amount of palmitate and other saturated fatty acids in all lipid classes. Similar biochemical and biophysical considerations apply to cellular cholesterol, the concentration of which is also tightly regulated. One regulatory mechanism is the esterification of cholesterol by sterol O acyltransferase-1 (SOAT1, also known as ACAT), enabling safe storage of cholesterol fatty acid esters in lipid droplets [158]. In response to low cholesterol levels, sterol regulatory element-binding protein (SREBP) and SREBP cleavage-activating protein (SCAP) are liberated from the ER, processed in the Golgi, and initiate transcription of cholesterol synthesis genes in the nucleus [159]. Studies found that ER stress increases the expression of SREBP-dependent lipogenesis markers through the UPR [160,161] and reduces cholesterol efflux by lowering the expression of lipoprotein receptors in the liver [162]. ER stress initiated by these lipid imbalances is a fundamental driver of lipotoxicity and related pathologies [[160], [161], [162], [163], [164]]. Prolonged lipotoxicity in hepatocytes contributes to non-alcoholic fatty liver disease (NAFLD) [165] and in the pancreas drives beta cell dysfunction and insulin insufficiency [132,155]. These pathologies are intricately linked to the UPR and in most cases activation of the UPR is protective against these HFD-induced disorders [[166], [167], [168]]. In addition to direct diet-induced lipotoxicity arising from nutrients, obesity is also associated with aberrant de novo lipogenesis, which is reduced in adipose tissue but increased in the liver, where lipogenesis results in lipotoxicity [148,[169], [170], [171]]. Somewhat paradoxically, lipogenesis in the liver is linked to a phenomenon referred to as selective insulin resistance, as particularly SREBP and MLX interacting protein-like (MLXIPL, also known as ChREBP)-mediated lipogenesis pathways are upregulated in obesity, while other insulin-responsive mechanisms are not [171]. However, lipogenesis is functionally diverse, as the ER both senses and synthesizes structural lipids, including phospholipids, sphingolipids, and cholesterol. Obesity is associated with a disrupted phospholipid ratio of the ER membrane lipids, which interferes with SERCA function, induces ER stress, and exacerbates liver dysfunction [133]. However, most lipids are stored in lipid droplets, including those for the storage of chemical energy and triacylglycerols (TAGs) [145]. DGAT1 and DGAT2 play essential roles in TAG biogenesis and transfer to lipid droplets [172]. In times of energetic needs, ER is involved in lipolysis, buffering liberated fatty acids from TAGs, which naturally causes a transient ER stress response via IRE1-XBP1. Failure in re-esterification by DGAT1 or inactivation of the UPR to respond under these conditions causes chronic ER stress and cellular dysfunction [173]. Another emerging player is fat storage-inducing transmembrane protein 2 (FITM2), an ER membrane fatty acid-CoA diphosphatase that is involved in lipid droplet biogenesis and ER structure [174]. In parallel, the ER has other UPR-independent mechanisms in place to attenuate lipotoxicity in cells. These are related to the UPS and cholesterol regulation. First, key enzymes of lipogenesis such as HMGCR or AGPATs underlie regulated turnover by the UPS [3]. In addition, there is an extra layer of regulation through NFE2L1. Above a certain cholesterol concentration in the ER membrane, NFE2L1 proteolytic cleavage is inhibited and less NFE2L1 is found in the nucleus. This leads to de-repression of important genetic cholesterol detoxification programs and hepatic cholesterol excretion, thus serving as a complementary player in the SREBP system [93]. In the liver and BAT, NFE2L1 promotes proteostasis and prevents lipotoxicity [3,93]. In conclusion, lipotoxicity and aberrant lipid metabolism disrupt cellular and tissue health. When discussing ER stress and metabolic disease, lipotoxicity and proteostasis should be acknowledged as interacting factors.
Figure 3.
ER lipid metabolism in obesity. As a result of high-fat diet consumption, cells are exposed to high levels of (saturated) fatty acids and cholesterol. High levels of membrane fatty acids and cholesterol change the membrane composition and lead to ER stress. Cells have several mechanisms to attenuate lipotoxicity: DGAT1 re-esterifies fatty acids and ACAT esterifies cholesterol for safe storage in lipid droplets as triglycerides or cholesterol esters, respectively. While the SREBP/SCAP system responds to low levels of ER cholesterol, high levels of ER cholesterol block NFE2L1 proteolytic cleavage and lead to fewer proteasome genes and de-repression of inflammation genes.
7. Carbohydrate metabolism and glycosylation
Carbohydrates are important nutrients and a vital energy source with critical roles for metabolic disorders. Balanced glucose levels are crucial for proper metabolic function. Uptake and utilization of this metabolite depend on multiple factors such as dietary intake, glycogenolysis, and gluconeogenesis in the liver, kidney, and intestine. The latter is designed to generate glucose from non-carbohydrate precursors such as amino acids, lipids, pyruvate, and lactate during fasting. The upregulation of gluconeogenesis in the liver of patients with T2DM is considered a major contributor to hyperglycemia and subsequent diabetic organ damage, which is also caused by glycation. Next to glucagon and incretins, insulin is a key hormone in glucose metabolism, so insulin resistance results in uncontrolled gluconeogenesis and diminished glucose clearance [175]. Gluconeogenesis is linked to ER function on several levels. For example, hepatocytes harbor an ER-located transcription factor, the cyclic AMP-responsive element-binding protein 3-like protein 3 (CREBH, encoded by CREB3L3), which has structural and mechanistic similarities to ATF6 [176]. This protein is activated through intramembrane proteolysis under a variety of cellular stress signals, most notably nutrient deprivation. Upon activation, CREBH transits from the ER to the Golgi apparatus, where it is processed to release transcription factor CREB that drives expression of key genes encoding metabolic regulators or enzymes involved in glucose metabolism [60]. Furthermore, the final step of gluconeogenesis, the formation of glucose from glucose-6-phosphate by the glucose-6-phosphatase catalytic subunit 1 (G6PC1), is carried out in the ER lumen. G6PC is a multimeric protein complex embedded in the ER membrane with its catalytic site facing the ER lumen. It makes sense that glucose is produced in the ER lumen, as complex carbohydrate structures are not just nutrients but also critical post- or co-translational modifications of proteins and lipids. More than half of the eukaryotic proteome carries carbohydrate residues in various structures and quantities. This post-translational modification is important for protein folding, binding affinity, and signal transduction pathways. Folding of these glycoproteins in the ER is closely regulated by chaperones and enzymes to maintain cellular homeostasis [177]. Tunicamycin is a well-established chemical inducer of ER stress by inhibiting N-linked glycosylation. N-linked glycosylation starts with the formation of dolichol-linked N-acetylglucosamine (GlcNAc), which then is transferred to folded proteins on the luminal side of the ER. Upon transferring the completed glycan onto the nascent polypeptide, two glucose residues are removed from the structure by exoglycosidases. This glycosylation serves as a quality control step in the ER, and once the protein is properly folded, the removal of the final third glucose residue signals that the cargo is ready for transit to the Golgi for further processing and cellular distribution. Thus, glucose metabolism is linked to protein folding, translocation, and overall proteostasis. Therefore, an imbalance in glucose levels is linked to ER stress and UPR activation. All three main UPR branches have been linked to glucose metabolism. One of the first findings in genetic models of UPR deficiency was defective gluconeogenesis in the livers of PERK-deficient mice or animals harboring a homozygous mutation that eliminates EIF2α phosphorylation on serine 51 [29,33]. Moreover, the UPR and glucose homeostasis in pancreatic beta cells are closely connected to insulin regulation [178]. For example, IRE1α phosphorylation not only activates the UPR but also promotes insulin biosynthesis in beta cells [179]. These are only highlights of the complex interaction of glucose, glycosylation, and UPR activation. Post-translational glycosylation is also directly involved in the regulation of proteostasis through NFE2L1, which undergoes glycosylation while anchored in the ER membrane. Deglycosylation by NGLY1 and cleavage by protease DDI2 are necessary steps in the maturation of active NFE2L1, which controls the adaptive component of proteasomal activity [87]. Mutants mimicking the deglycosylated state of NFE2L1 result in enhanced transcriptional activity of the protein [64] and increased proteostasis. Another direct regulation of glycosylation is the hexosamine signaling pathway, in which the nutrient-dependent synthesis of UDP-GlcNAc is coupled to O-GlcNAc modification of Ser/Thr residues of key nuclear and cytoplasmic targets. O-GlcNAc modification has emerged as a versatile cellular mechanism modulating signaling during growth, metabolism, stress, circadian rhythm, and host–pathogen interactions. This is especially evident in neurons and adipocytes, as O-linked N-acetylglucosamine transferase (OGT) has been shown to modulate energy balance and obesity through multiple cellular effector mechanisms [[180], [181], [182]]. In summary, healthy ER function depends on homeostasis of all three major classes of nutrients, proteins, lipids, and glucose. In metabolic disorders, a critical hallmark of ER stress-associated cellular dysfunction is the activation of inflammatory signaling pathways and chronic tissue inflammation.
8. Inflammation
Obesity is a state of chronic low-grade inflammation that is characterized by aberrant production of cytokines and infiltration of immune cells in metabolic organs [49,175,183]. What is the cause of inflammation in this metabolic context? This is a major topic of “immunometabolism” or “metaflammation” research [184], which unlike proposals by some in the field does not directly relate to the metabolism of immune cells during inflammation (which is simply metabolism, not immunometabolism) but rather studies the intersection where metabolic and inflammatory pathways converge to regulate cellular function. Inflammation is normally a complex reaction of the body to cope with pathogens, dying cells, and other harmful irregularities, but in metaflammation, this process is sterile and driven by aberrant metabolism. One of the first conceptual formulations that metabolic disorders are in fact inflammatory diseases was the notion that atherosclerosis is an inflammatory response to the lipid-driven injury of the vessel wall. It is well established that obesity and associated disorders including type 1 diabetes have a strong inflammatory component. A classic example is the impact of cytokines such as tumor necrosis factor alpha (TNF-α) on metabolic function. TNF-alpha leads to activation of c-Jun N-terminal kinase (JNK), which inhibits insulin receptor signaling through serine phosphorylation of insulin receptor substrate 1 (IRS1). This modification inactivates IRS1, which ultimately contributes to insulin resistance [185]. In this immunometabolic context, ER stress has various effects on cellular function in isolated cells and an impact on systemic metabolism [49]. This directly involves altered utilization of nutrients by parenchymal cells and endocrine regulation of metabolism, most notably insulin resistance. In metabolic disorders, ER stress is unresolved despite the activation of proteostatic countermeasures. This chronic stress response is closely linked to inflammation and the immune response, and in turn, chronic inflammation hampers the resolution of ER stress [54]. ER stress in parenchymal and immune cells leads to the production of cytokines and an anti-inflammatory response, as the UPR is linked to the activation of cytosolic stress kinases such as JNK [186], PKR [187], and IκBα kinase (IKK) [188]. Each of these key signaling nodes has been implicated in obesity and associated disorders as explained as follows. While whole-body JNK deficiency protects from weight gain [189], there are isoform and cell type-specific differences in the impact of JNK signaling on metabolic disorders [[190], [191], [192], [193], [194], [195]], although JNK activation principally aggravates tissue inflammation. PKR is activated by double-stranded RNA and modulates metabolic homeostasis, although the outcome of genetic PKR deletion depends on the mouse model and background [[36], [37], [38],196,197]. In addition, the IKK family is implicated in inflammatory signaling and IKK-e and TBK1 have been shown to impact metabolic homeostasis [198,199]. Downstream of these immunometabolic key nodes is the inflammatory transcriptional response. For instance, the UPR is extensively involved in the signal transduction of inflammatory responses. Signal transducer and activator of transcription 3 (STAT3) is a transcription factor activated via phosphorylation upon cytokine signaling and has been linked to the ER stress response. In mouse hepatocytes, STAT3 is activated after it is bound by IRE1 and subsequent phosphorylation to promote tissue repair in response to liver damage (Figure 4) [200]. PERK-mediated eIF2α phosphorylation halts protein synthesis and favors nuclear factor-κB (NFκB) activation to induce pro-inflammatory genes [60,201]. A key event on the road to tissue inflammation in metabolic disorders is the infiltration of the tissue with immune cells, especially macrophages. This is evident almost everywhere in the body, most prominently in adipose tissue, the liver, and vessel walls. It is important to note that the processes described here in the context of atherosclerosis principally also take place in adipose tissues and the liver, as these are fundamental principles of immunometabolic stress. Atherogenesis is driven by the formation of foam cells from infiltrating macrophages that engulf modified LDL particles in the subintimal space. Foam cells are macrophages loaded with cholesterol esters (CE) that are formed by cholesterol esterification through ACAT, an ER membrane enzyme (Figure 4) [[202], [203], [204]]. While the storage of CE in lipid droplets is safe, free cholesterol accumulation in the ER membrane leads to calcium-related ER stress and engages PERK-mediated apoptosis. By treating mice with phenylbutyrate (4-PBA), a chemical chaperone, PERK activation in macrophages is inhibited, apoptosis prevented, and their function preserved, ultimately resulting in reduced atherosclerosis [205]. IRE1α is then activated by pattern-recognition receptors of macrophages, leading to a pro-inflammatory response [206]. Depletion of IRE1α from the macrophages in obesity leads to a shift from pro-inflammatory toward anti-inflammatory tissue phenotypes [183]. Moreover, the IRE1α pathway strongly influences the production of several proatherogenic cytokines, chemokines, and their receptors, including interleukin (IL)-1β or C–C motif chemokine ligand 2 (CCL2). By inhibition of IRE1 in vivo, hyperlipidemia-induced IL-1β and IL-18 production decreases, which leads to a reduction in atherosclerotic plaque size. IRE1 inhibition also alters the cellular composition of plaque by reducing the numbers of macrophages in atherosclerotic lesions. Most likely this effect is due to the reduction of the strong macrophage chemoattractant CCL2 [207]. Interestingly, inflammation-induced protein nitrosylation also impairs IRE1 function directly, which has consequences for UPR activation and the resolution of ER stress in the context of NAFLD [54]. This can be interpreted as a vicious circle of inflammation and ER stress. Another important link between inflammation and ER stress is via the NFE2L1 pathway. In brown fat, NFE2L1-mediated proteasomal protein quality control is required to mitigate cold-induced ER stress [3]. Small molecule proteasome inhibitors and lack of NFE2L1 induce ER stress, chemokine and cytokine production, and ultimately result in severe brown fat inflammation, so much so that even the plasma inflammatory mediators such as TNFɑ are elevated [3]. However, these outcomes strongly depend on environmental challenges, as they are not observed at thermoneutrality and only lead to insulin resistance when mice are fed a HFD. This highlights the adaptive and dynamic nature of this NFE2L1-dependent immunometabolic mechanism [3]. Another important link of ER function and inflammation is the cGAS-cGAMP-STING pathway, which is also upregulated in adipose tissue of rodents on a HFD. In obesity, mitochondrial DNA (mtDNA) is sensed by cyclic GMP-AMP synthase (cGAS) and triggers its oligomerization and activation. Activated cGAS catalyzes the formation of cyclic dinucleotides, most notably cyclic guanosine monophosphate-adenosine monophosphate (2′3′-cGAMP), which in turn bind to the ER-localized stimulator of interferon response cGAMP interactor 1 (STING1). STING1 dimerizes and activates and recruits interferon regulatory factor 3 (IRF3) and NF-kB, thus initiating the production of type 1 interferons and other inflammatory cytokines, which results in insulin resistance [208]. Interestingly, STING1 activity is modulated by cellular ROS levels [209], which could have important implications as ROS is a feature of aberrant metabolism. In summary, these examples demonstrate that one major outcome of ER stress is cellular and tissue inflammation. In the absence of pathogens, pathways and cells of the immune system are activated by aberrant metabolism in obesity. This intimate relationship of ER stress and inflammation is involved in the restoration of cellular homeostasis, but also leads to metabolic diseases if sustained unresolved and chronic.
Figure 4.
Immunometabolic interplay in obesity. In obesity, immune cells and parenchymal cells are in crosstalk as they infiltrate metabolic tissues. Excess fatty acids and glucose are sensed by TLRs on macrophages and trigger an IREα response, which releases pro-inflammatory cytokines. Metabolic disorders are also characterized by the formation of foam cells, which carry a surplus of cholesterol esters and other lipids. Cytokines produced by the immune cells are sensed by parenchymal cells and trigger the activation of stress kinases such as JNK, which are directly linked to insulin resistance. IREα in parenchymal cells is also activated by cytokines and leads to transcription of pro-inflammatory mediators. The cGAS-cGAMP-STING pathway is triggered by HFD and enhances inflammatory responses via NF-κB.
9. Summary and translational perspective
The ER is a dynamic and diverse organelle that is in close physical and functional connection with all critical parts of cells. Hence, the ER is the central platform for coordinating nutrient homeostasis and orchestrating stress responses arising from within and outside the ER. This ER stress is a very complex phenomenon triggered by environmental challenges that is usually resolved by the adaptive programs of the integrated stress response, including proteostatic, lipidostatic, and glycostatic countermeasures. Neither these ER stress phenotypes nor the adaptive programs occur in isolation but rather are intricately connected and depend on each other. In the context of obesity and associated metabolic disorders, excessive exposure to high-calorie nutrients such as lipids and glucose is associated with the chronic activation of adaptive stress resistance mechanisms, but these fail to restore ER homeostasis, resulting in tissue inflammation and endocrine dysregulation. In light of the global epidemic of overweight and obesity, understanding the fundamental biology of metabolic adaptation in the ER may hold novel therapeutic approaches for metabolic disorders. While natural compounds and toxins have helped discover mechanisms of ER function, applying aspects of this knowledge pharmacologically to treat human disease is hampered by several roadblocks. These challenges include the pleiotropic nature of ER stress manipulation, as pharmacological targeting with compounds may come with undesired off-target side effects. In addition, ER homeostasis is maintained by delicately balanced programs as outlined in this review and targeting one arm might lead to maladaptive activation of ER stress pathways. However, we have made important steps toward exploiting key regulators of ER function. There is progress in preclinical animal models, for example, small targeting of ER stress in the liver or beta cells alleviates metabolic dysfunction. We are optimistic that the next decade will bring more discoveries and therapeutic breakthroughs for resolving ER stress and curing metabolic disorders.
Author contributions
A.B., I.L.L., and N.W. conceptualized and wrote the manuscript with input from N.H.
Competing financial interests
The authors declare no competing financial interests.
Acknowledgments
We acknowledge the scientific environment in the Bartelt lab and thank all lab members for their critical input and discussions. A.B. was supported by the Deutsche Forschungsgemeinschaft (SFB1123-B10), the German Center for Cardiovascular Research (DZHK) Junior Research Group Excellence Grant, and the European Research Council Starting Grant 852742 PROTEOFIT. The figures were created with BioRender. We apologize to colleagues whose work we could not cite due to space limitations.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2021.101169.
Conflict of interest
None declared.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Westrate L.M., Lee J.E., Prinz W.A., Voeltz G.K. Form follows function: the importance of endoplasmic reticulum shape. Annual Review of Biochemistry. 2015;84(1):791–811. doi: 10.1146/annurev-biochem-072711-163501. [DOI] [PubMed] [Google Scholar]
- 2.Arruda A.P., Pers B.M., Parlakgül G., Güney E., Inouye K., Hotamisligil G.S. Chronic enrichment of hepatic endoplasmic reticulum-mitochondria contact leads to mitochondrial dysfunction in obesity. Nature Medicine. 2014;20(12):1427–1435. doi: 10.1038/nm.3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartelt A., Widenmaier S.B., Schlein C., Johann K., Goncalves R.L.S.S., Eguchi K. Brown adipose tissue thermogenic adaptation requires Nrf1-mediated proteasomal activity. Nature Medicine. 2018;24(3):292–303. doi: 10.1038/nm.4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu F., Du W., Zou Q., Wang Y., Zhang X., Xing X. COPII mitigates ER stress by promoting formation of ER whorls. Cell Research. 2020 doi: 10.1038/s41422-020-00416-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lytridou A.A., Demetriadou A., Christou M., Potamiti L., Mastroyiannopoulos N.P., Kyriacou K. Stbd1 promotes glycogen clustering during endoplasmic reticulum stress and supports survival of mouse myoblasts. Journal of Cell Science. 2020;133(20) doi: 10.1242/jcs.244855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tersey S.A., Nishiki Y., Templin A.T., Cabrera S.M., Stull N.D., Colvin S.C. Islet β-cell endoplasmic reticulum stress precedes the onset of type 1 diabetes in the nonobese diabetic mouse model. Diabetes. 2012;61(4):818–827. doi: 10.2337/db11-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parlakgül G., Arruda A.P., Cagampan E., Pang S., Güney E., Lee Y. 2020. High resolution 3D imaging of liver reveals a central role for subcellular architectural organization in metabolism.Https://Www.Biorxiv.Org/Content/10.1101/2020.11.18.387803v3 [DOI] [Google Scholar]
- 8.Phillips M.J., Voeltz G.K. Structure and function of ER membrane contact sites with other organelles. Nature Reviews Molecular Cell Biology. 2016;17(2):69–82. doi: 10.1038/nrm.2015.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Höglinger D., Burgoyne T., Sanchez-Heras E., Hartwig P., Colaco A., Newton J. NPC1 regulates ER contacts with endocytic organelles to mediate cholesterol egress. Nature Communications. 2019;10(1):4276. doi: 10.1038/s41467-019-12152-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilfling F., Wang H., Haas J.T., Krahmer N., Gould T.J., Uchida A. Triacylglycerol synthesis enzymes mediate lipid droplet growth by relocalizing from the ER to lipid droplets. Developmental Cell. 2013;24(4):384–399. doi: 10.1016/j.devcel.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mesmin B., Kovacs D., D'Angelo G. Lipid exchange and signaling at ER-Golgi contact sites. Current Opinion in Cell Biology. 2019;57:8–15. doi: 10.1016/j.ceb.2018.10.002. [DOI] [PubMed] [Google Scholar]
- 12.Friedman J.R., Lackner L.L., West M., DiBenedetto J.R., Nunnari J., Voeltz G.K. ER tubules mark sites of mitochondrial division. Science (New York, N.Y.) 2011;334(6054):358–362. doi: 10.1126/science.1207385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bach D., Pich S., Soriano F.X., Vega N., Baumgartner B., Oriola J. Mitofusin-2 determines mitochondrial network architecture and mitochondrial metabolism. A novel regulatory mechanism altered in obesity. Journal of Biological Chemistry. 2003;278(19):17190–17197. doi: 10.1074/jbc.M212754200. [DOI] [PubMed] [Google Scholar]
- 14.Sebastián D., Hernández-Alvarez M.I., Segalés J., Sorianello E., Muñoz J.P., Sala D. Mitofusin 2 (Mfn2) links mitochondrial and endoplasmic reticulum function with insulin signaling and is essential for normal glucose homeostasis. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(14):5523–5528. doi: 10.1073/pnas.1108220109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mancini G., Pirruccio K., Yang X., Blüher M., Rodeheffer M., Horvath T.L. Mitofusin 2 in mature adipocytes controls adiposity and body weight. Cell Reports. 2019;26(11):2849–2858. doi: 10.1016/j.celrep.2019.02.039. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mahdaviani K., Benador I.Y., Su S., Gharakhanian R.A., Stiles L., Trudeau K.M. Mfn2 deletion in brown adipose tissue protects from insulin resistance and impairs thermogenesis. EMBO Reports. 2017;18(7):1123–1138. doi: 10.15252/embr.201643827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boutant M., Kulkarni S.S., Joffraud M., Ratajczak J., Valera-Alberni M., Combe R. Mfn2 is critical for brown adipose tissue thermogenic function. The EMBO Journal. 2017;36(11):1543–1558. doi: 10.15252/embj.201694914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schneeberger M., Dietrich M.O., Sebastián D., Imbernón M., Castaño C., Garcia A. Mitofusin 2 in POMC neurons connects ER stress with leptin resistance and energy imbalance. Cell. 2013;155(1):172–187. doi: 10.1016/j.cell.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hartl F.U., Hayer-Hartl M. Converging concepts of protein folding in vitro and in vivo. Nature Structural & Molecular Biology. 2009;16(6):574–581. doi: 10.1038/nsmb.1591. [DOI] [PubMed] [Google Scholar]
- 20.Saxton R.A., Sabatini D.M. mTOR signaling in growth, metabolism, and disease. Cell. 2017;168(6):960–976. doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu G.Y., Sabatini D.M. mTOR at the nexus of nutrition, growth, ageing and disease. Nature Reviews Molecular Cell Biology. 2020;21(4):183–203. doi: 10.1038/s41580-019-0199-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bartelt A., Widenmaier S.B. Proteostasis in thermogenesis and obesity. Biological Chemistry. 2020;401(9):1019–1030. doi: 10.1515/hsz-2019-0427. [DOI] [PubMed] [Google Scholar]
- 23.Metcalf M.G., Higuchi-Sanabria R., Garcia G., Tsui C.K., Dillin A. Beyond the cell factory: homeostatic regulation of and by the UPR ER. Science Advances. 2020;6(29) doi: 10.1126/sciadv.abb9614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sage A.T., Holtby-Ottenhof S., Shi Y., Damjanovic S., Sharma A.M., Werstuck G.H. Metabolic syndrome and acute hyperglycemia are associated with endoplasmic reticulum stress in human mononuclear cells. Obesity. 2012;20(4):748–755. doi: 10.1038/oby.2011.144. [DOI] [PubMed] [Google Scholar]
- 25.Adams C.J., Kopp M.C., Larburu N., Nowak P.R., Ali M.M.U. Structure and molecular mechanism of ER stress signaling by the unfolded protein response signal activator IRE1. Frontiers in Molecular Biosciences. 2019;6:11. doi: 10.3389/fmolb.2019.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nature Reviews Molecular Cell Biology. 2012;13(2):89–102. doi: 10.1038/nrm3270. [DOI] [PubMed] [Google Scholar]
- 27.Nika J., Rippel S., Hannig E.M. Biochemical analysis of the eIF2beta gamma complex reveals a structural function for eIF2alpha in catalyzed nucleotide exchange. Journal of Biological Chemistry. 2001;276(2):1051–1056. doi: 10.1074/jbc.M007398200. [DOI] [PubMed] [Google Scholar]
- 28.Gordiyenko Y., Llácer J.L., Ramakrishnan V. Structural basis for the inhibition of translation through eIF2α phosphorylation. Nature Communications. 2019;10(1):2640. doi: 10.1038/s41467-019-10606-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harding H.P., Novoa I., Zhang Y., Zeng H., Wek R., Schapira M. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Molecular Cell. 2000;6(5):1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- 30.Harding H.P., Zhang Y., Bertolotti A., Zeng H., Ron D. Perk is essential for translational regulation and cell survival during the unfolded protein response. Molecular Cell. 2000;5(5):897–904. doi: 10.1016/s1097-2765(00)80330-5. [DOI] [PubMed] [Google Scholar]
- 31.Zhang P., McGrath B., Li S., Frank A., Zambito F., Reinert J. The PERK eukaryotic initiation factor 2 alpha kinase is required for the development of the skeletal system, postnatal growth, and the function and viability of the pancreas. Molecular and Cellular Biology. 2002;22(11):3864–3874. doi: 10.1128/mcb.22.11.3864-3874.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harding H.P., Zeng H., Zhang Y., Jungries R., Chung P., Plesken H. Diabetes mellitus and exocrine pancreatic dysfunction in perk-/- mice reveals a role for translational control in secretory cell survival. Molecular Cell. 2001;7(6):1153–1163. doi: 10.1016/S1097-2765(01)00264-7. [DOI] [PubMed] [Google Scholar]
- 33.Hotamisligil G.S. Endoplasmic reticulum stress and the inflammatory basis of metabolic disease. Cell. 2010;140(6):900–917. doi: 10.1016/j.cell.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wolcott C.D., Rallison M.L. Infancy-onset diabetes mellitus and multiple epiphyseal dysplasia. The Journal of Pediatrics. 1972;80(2):292–297. doi: 10.1016/S0022-3476(72)80596-1. [DOI] [PubMed] [Google Scholar]
- 35.Holcik M., Sonenberg N. Translational control in stress and apoptosis. Nature Reviews Molecular Cell Biology. 2005;6(4):318–327. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- 36.Nakamura T., Furuhashi M., Li P., Cao H., Tuncman G., Sonenberg N. Double-stranded RNA-dependent protein kinase links pathogen sensing with stress and metabolic homeostasis. Cell. 2010;140(3):338–348. doi: 10.1016/j.cell.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakamura T., Arduini A., Baccaro B., Furuhashi M., Hotamisligil G.S. Small-molecule inhibitors of PKR improve glucose homeostasis in obese diabetic mice. Diabetes. 2014;63(2):526–534. doi: 10.2337/db13-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lancaster G.I., Kammoun H.L., Kraakman M.J., Kowalski G.M., Bruce C.R., Febbraio M.A. PKR is not obligatory for high-fat diet-induced obesity and its associated metabolic and inflammatory complications. Nature Communications. 2016;7(1):10626. doi: 10.1038/ncomms10626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Han A.-P.P., Yu C., Lu L., Fujiwara Y., Browne C., Chin G. Heme-regulated eIF2alpha kinase (HRI) is required for translational regulation and survival of erythroid precursors in iron deficiency. The EMBO Journal. 2001;20(23):6909–6918. doi: 10.1093/emboj/20.23.6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abdel-Nour M., Carneiro L.A.M., Downey J., Tsalikis J., Outlioua A., Prescott D. The heme-regulated inhibitor is a cytosolic sensor of protein misfolding that controls innate immune signaling. Science (New York, N.Y.) 2019;365(6448):eaaw4144. doi: 10.1126/science.aaw4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wek R.C.C., Jiang H.-Y.H.-Y., Anthony T.G.G. Coping with stress: eIF2 kinases and translational control. Biochemical Society Transactions. 2006;34(1):7. doi: 10.1042/BST20060007. [DOI] [PubMed] [Google Scholar]
- 42.Guo F., Cavener D.R. The GCN2 eIF2alpha kinase regulates fatty-acid homeostasis in the liver during deprivation of an essential amino acid. Cell Metabolism. 2007;5(2):103–114. doi: 10.1016/j.cmet.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 43.Anthony T.G., McDaniel B.J., Byerley R.L., McGrath B.C., Cavener D.R., McNurlan M.A. Preservation of liver protein synthesis during dietary leucine deprivation occurs at the expense of skeletal muscle mass in mice deleted for eIF2 kinase GCN2. Journal of Biological Chemistry. 2004;279(35):36553–36561. doi: 10.1074/jbc.M404559200. [DOI] [PubMed] [Google Scholar]
- 44.Ron D., Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nature Reviews Molecular Cell Biology. 2007;8(7):519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 45.Liu X., Beaudoin J.-D., Davison C.A., Kosmaczewski S.G., Meyer B.I., Giraldez A.J. A functional non-coding RNA is produced from xbp-1 mRNA. Neuron. 2020;107(5):854–863. doi: 10.1016/j.neuron.2020.06.015. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee J.J., Sun C., Zhou Y., Lee J.J., Gokalp D., Herrema H. p38 MAPK-mediated regulation of Xbp1s is crucial for glucose homeostasis. Nature Medicine. 2011;17(10):1251–1260. doi: 10.1038/nm.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou Y., Lee J., Reno C.M., Sun C., Park S.W., Chung J. Regulation of glucose homeostasis through a XBP-1-FoxO1 interaction. Nature Medicine. 2011;17(3):356–365. doi: 10.1038/nm.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Park S.W., Zhou Y., Lee J., Lu A., Sun C., Chung J. The regulatory subunits of PI3K, p85alpha and p85beta, interact with XBP-1 and increase its nuclear translocation. Nature Medicine. 2010;16(4):429–437. doi: 10.1038/nm.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ozcan U., Cao Q., Yilmaz E., Lee A.-H., Iwakoshi N.N., Ozdelen E. Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science. 2004;306(5695):457–461. doi: 10.1126/science.1103160. [DOI] [PubMed] [Google Scholar]
- 50.Gregor M.F., Misch E.S., Yang L., Hummasti S., Inouye K.E., Lee A.-H. The role of adipocyte XBP1 in metabolic regulation during lactation. Cell Reports. 2013;3(5):1430–1439. doi: 10.1016/j.celrep.2013.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tsuchiya Y., Saito M., Kadokura H., Miyazaki J.-I., Tashiro F., Imagawa Y. IRE1-XBP1 pathway regulates oxidative proinsulin folding in pancreatic β cells. The Journal of Cell Biology. 2018;217(4):1287–1301. doi: 10.1083/jcb.201707143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hassler J.R., Scheuner D.L., Wang S., Han J., Kodali V.K., Li P. The IRE1α/XBP1s pathway is essential for the glucose response and protection of β cells. PLoS Biology. 2015;13(10) doi: 10.1371/journal.pbio.1002277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Williams K.W., Liu T.T.T., Kong X., Fukuda M., Deng Y., Berglund E.D. Xbp1s in Pomc neurons connects ER stress with energy balance and glucose homeostasis. Cell Metabolism. 2014;20(3):471–482. doi: 10.1016/j.cmet.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang L., Calay E.S., Fan J., Arduini A., Kunz R.C., Gygi S.P. METABOLISM. S-Nitrosylation links obesity-associated inflammation to endoplasmic reticulum dysfunction. Science (New York, N.Y.) 2015;349(6247):500–506. doi: 10.1126/science.aaa0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Deng Y., Wang Z.V., Tao C., Gao N., Holland W.L., Ferdous A. The Xbp1s/GalE axis links ER stress to postprandial hepatic metabolism. Journal of Clinical Investigation. 2013;123(1):455–468. doi: 10.1172/JCI62819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Deng Y., Wang Z.V., Gordillo R., Zhu Y., Ali A., Zhang C. Adipocyte Xbp1s overexpression drives uridine production and reduces obesity. Molecular Metabolism. 2018;11:1–17. doi: 10.1016/j.molmet.2018.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Westrate L.M., Hoyer M.J., Nash M.J., Voeltz G.K. Vesicular and uncoated Rab1-dependent cargo carriers facilitate ER to Golgi transport. Journal of Cell Science. 2020;133(14) doi: 10.1242/jcs.239814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schindler A.J., Schekman R. In vitro reconstitution of ER-stress induced ATF6 transport in COPII vesicles. Proceedings of the National Academy of Sciences. 2009;106(42):17775–17780. doi: 10.1073/pnas.0910342106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ye J., Rawson R.B., Komuro R., Chen X., Davé U.P., Prywes R. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Molecular Cell. 2000;6(6):1355–1364. doi: 10.1016/S1097-2765(00)00133-7. [DOI] [PubMed] [Google Scholar]
- 60.Hetz C., Zhang K., Kaufman R.J., Zhang K., Kaufman R.J. Mechanisms, regulation and functions of the unfolded protein response. Nature Reviews Molecular Cell Biology. 2020;21(8):421–438. doi: 10.1038/s41580-020-0250-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Engin F., Nguyen T., Yermalovich A., Hotamisligil G.S. Aberrant islet unfolded protein response in type 2 diabetes. Scientific Reports. 2014;4:4054. doi: 10.1038/srep04054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Usui M., Yamaguchi S., Tanji Y., Tominaga R., Ishigaki Y., Fukumoto M. Atf6α-null mice are glucose intolerant due to pancreatic β-cell failure on a high-fat diet but partially resistant to diet-induced insulin resistance. Metabolism: Clinical and Experimental. 2012;61(8):1118–1128. doi: 10.1016/j.metabol.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 63.González-Quiroz M., Blondel A., Sagredo A., Hetz C., Chevet E., Pedeux R. When endoplasmic reticulum proteostasis meets the DNA damage response. Trends in Cell Biology. 2020;30(11):881–891. doi: 10.1016/j.tcb.2020.09.002. [DOI] [PubMed] [Google Scholar]
- 64.Koizumi S., Hamazaki J., Murata S. Transcriptional regulation of the 26S proteasome by Nrf1. Proceedings of the Japan Academy. Series B, Physical and Biological Sciences. 2018;94(8):325–336. doi: 10.2183/pjab.94.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Travers K.J., Patil C.K., Wodicka L., Lockhart D.J., Weissman J.S., Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101(3):249–258. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- 66.Guerriero C.J., Brodsky J.L. The delicate balance between secreted protein folding and endoplasmic reticulum-associated degradation in human physiology. Physiological Reviews. 2012;92(2):537–576. doi: 10.1152/physrev.00027.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guo H., Xiong Y., Witkowski P., Cui J., Wang L., Sun J. Inefficient translocation of preproinsulin contributes to pancreatic β cell failure and late-onset diabetes. Journal of Biological Chemistry. 2014;289(23):16290–16302. doi: 10.1074/jbc.M114.562355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bhattacharya A., Qi L. ER-associated degradation in health and disease - from substrate to organism. Journal of Cell Science. 2019;132(23) doi: 10.1242/jcs.232850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Olzmann J.A., Kopito R.R., Christianson J.C. The mammalian endoplasmic reticulum-associated degradation system. Cold Spring Harbor Perspectives in Biology. 2013;5(9) doi: 10.1101/cshperspect.a013185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wu X., Siggel M., Ovchinnikov S., Mi W., Svetlov V., Nudler E. Structural basis of ER-associated protein degradation mediated by the Hrd1 ubiquitin ligase complex. Science (New York, N.Y.) 2020;368(6489) doi: 10.1126/science.aaz2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.van den Boom J., Meyer H. VCP/p97-Mediated unfolding as a principle in protein homeostasis and signaling. Molecular Cell. 2018;69(2):182–194. doi: 10.1016/j.molcel.2017.10.028. [DOI] [PubMed] [Google Scholar]
- 72.Collins G.A., Goldberg A.L. The logic of the 26S proteasome. Cell. 2017;169(5):792–806. doi: 10.1016/j.cell.2017.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Albert S., Schaffer M., Beck F., Mosalaganti S., Asano S., Thomas H.F. Proteasomes tether to two distinct sites at the nuclear pore complex. Proceedings of the National Academy of Sciences of the United States of America. 2017;114(52):13726–13731. doi: 10.1073/pnas.1716305114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Albert S., Wietrzynski W., Lee C.-W., Schaffer M., Beck F., Schuller J.M. Direct visualization of degradation microcompartments at the ER membrane. Proceedings of the National Academy of Sciences of the United States of America. 2020;117(2):1069–1080. doi: 10.1073/pnas.1905641117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rousseau A., Bertolotti A. Regulation of proteasome assembly and activity in health and disease. Nature Reviews Molecular Cell Biology. 2018;19(11):697–712. doi: 10.1038/s41580-018-0040-z. [DOI] [PubMed] [Google Scholar]
- 76.VerPlank J.J.S., Lokireddy S., Zhao J., Goldberg A.L. 26S Proteasomes are rapidly activated by diverse hormones and physiological states that raise cAMP and cause Rpn6 phosphorylation. Proceedings of the National Academy of Sciences. 2019;116(10):4228–4237. doi: 10.1073/pnas.1809254116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Peth A., Uchiki T., Goldberg A.L. ATP-dependent steps in the binding of ubiquitin conjugates to the 26S proteasome that commit to degradation. Molecular Cell. 2010;40(4):671–681. doi: 10.1016/j.molcel.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bodine S.C. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science. 2001;294(5547):1704–1708. doi: 10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- 79.Hampton R.Y., Gardner R.G., Rine J. Role of 26S proteasome and HRD genes in the degradation of 3-hydroxy-3-methylglutaryl-CoA reductase, an integral endoplasmic reticulum membrane protein. Molecular Biology of the Cell. 1996;7(12):2029–2044. doi: 10.1091/mbc.7.12.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Song B.-L., Sever N., DeBose-Boyd R.A. Gp78, a membrane-anchored ubiquitin ligase, associates with Insig-1 and couples sterol-regulated ubiquitination to degradation of HMG CoA reductase. Molecular Cell. 2005;19(6):829–840. doi: 10.1016/j.molcel.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 81.Zhang T., Xu Y., Liu Y., Ye Y. gp78 functions downstream of Hrd1 to promote degradation of misfolded proteins of the endoplasmic reticulum. Molecular Biology of the Cell. 2015;26(24):4438–4450. doi: 10.1091/mbc.E15-06-0354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zelcer N., Hong C., Boyadjian R., Tontonoz P. LXR regulates cholesterol uptake through idol-dependent ubiquitination of the LDL receptor. Science. 2009;325(5936):100–104. doi: 10.1126/science.1168974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Steffen J., Seeger M., Koch A., Krüger E. Proteasomal degradation is transcriptionally controlled by TCF11 via an ERAD-dependent feedback loop. Molecular Cell. 2010;40(1):147–158. doi: 10.1016/j.molcel.2010.09.012. [DOI] [PubMed] [Google Scholar]
- 84.Tomlin F.M., Gerling-Driessen U.I.M., Liu Y.-C., Flynn R.A., Vangala J.R., Lentz C.S. Inhibition of NGLY1 inactivates the transcription factor Nrf1 and potentiates proteasome inhibitor cytotoxicity. ACS Central Science. 2017;3(11):1143–1155. doi: 10.1021/acscentsci.7b00224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Radhakrishnan S.K., den Besten W., Deshaies R.J. p97-dependent retrotranslocation and proteolytic processing govern formation of active Nrf1 upon proteasome inhibition. ELife. 2014;3 doi: 10.7554/eLife.01856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tsuchiya Y., Morita T., Kim M., Iemura S., Natsume T., Yamamoto M. Dual regulation of the transcriptional activity of Nrf1 by β-TrCP- and Hrd1-dependent degradation mechanisms. Molecular and Cellular Biology. 2011;31(22):4500–4512. doi: 10.1128/MCB.05663-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Koizumi S., Irie T., Hirayama S., Sakurai Y., Yashiroda H., Naguro I. The aspartyl protease DDI2 activates Nrf1 to compensate for proteasome dysfunction. ELife. 2016;5 doi: 10.7554/eLife.18357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Radhakrishnan S.K., Lee C.S., Young P., Beskow A., Chan J.Y., Deshaies R.J. Transcription factor Nrf1 mediates the proteasome recovery pathway after proteasome inhibition in mammalian cells. Molecular Cell. 2010;38(1):17–28. doi: 10.1016/j.molcel.2010.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhou Z., Torres M., Sha H., Halbrook C.J., Van den Bergh F., Reinert R.B. Endoplasmic reticulum-associated degradation regulates mitochondrial dynamics in brown adipocytes. Science (New York, N.Y.) 2020;368(6486):54–60. doi: 10.1126/science.aay2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hou Y., Liu Z., Zuo Z., Gao T., Fu J., Wang H. Adipocyte-specific deficiency of Nfe2l1 disrupts plasticity of white adipose tissues and metabolic homeostasis in mice. Biochemical and Biophysical Research Communications. 2018;503(1):264–270. doi: 10.1016/j.bbrc.2018.06.013. [DOI] [PubMed] [Google Scholar]
- 91.Otoda T., Takamura T., Misu H., Ota T., Murata S., Hayashi H. Proteasome dysfunction mediates obesity-induced endoplasmic reticulum stress and insulin resistance in the liver. Diabetes. 2013;62(3):811–824. doi: 10.2337/db11-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Xu Z., Chen L., Leung L., Yen T.S.B., Lee C., Chan J.Y. Liver-specific inactivation of the Nrf1 gene in adult mouse leads to nonalcoholic steatohepatitis and hepatic neoplasia. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(11):4120–4125. doi: 10.1073/pnas.0500660102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Widenmaier S.B., Snyder N.A., Nguyen T.B., Arduini A., Lee G.Y., Arruda A.P. NRF1 is an ER membrane sensor that is central to cholesterol homeostasis. Cell. 2017;171(5):1094–1109. doi: 10.1016/j.cell.2017.10.003. e15. [DOI] [PubMed] [Google Scholar]
- 94.Zhang Y., Nicholatos J., Dreier J.R., Ricoult S.J.H., Widenmaier S.B., Hotamisligil G.S. Coordinated regulation of protein synthesis and degradation by mTORC1. Nature. 2014;513(7518):440–443. doi: 10.1038/nature13492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tiwari A., Schuiki I., Zhang L., Allister E.M., Wheeler M.B., Volchuk A. SDF2L1 interacts with the ER-associated degradation machinery and retards the degradation of mutant proinsulin in pancreatic β-cells. Journal of Cell Science. 2013;126(Pt 9):1962–1968. doi: 10.1242/jcs.117374. [DOI] [PubMed] [Google Scholar]
- 96.Sasako T., Ohsugi M., Kubota N., Itoh S., Okazaki Y., Terai A. Hepatic Sdf2l1 controls feeding-induced ER stress and regulates metabolism. Nature Communications. 2019;10(1):947. doi: 10.1038/s41467-019-08591-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bhattacharya A., Sun S., Wang H., Liu M., Long Q., Yin L. Hepatic Sel1L-Hrd1 ER-associated degradation (ERAD) manages FGF21 levels and systemic metabolism via CREBH. The EMBO Journal. 2018;37(22) doi: 10.15252/embj.201899277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fujita H., Yagishita N., Aratani S., Saito-Fujita T., Morota S., Yamano Y. The E3 ligase synoviolin controls body weight and mitochondrial biogenesis through negative regulation of PGC-1β. The EMBO Journal. 2015;34(8):1042–1055. doi: 10.15252/embj.201489897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ghosh S., Taylor J.L., Mendoza T.M., Dang T., Burk D.H., Yu Y. Siah2 modulates sex-dependent metabolic and inflammatory responses in adipose tissue to a high-fat diet challenge. Biology of Sex Differences. 2019;10(1):19. doi: 10.1186/s13293-019-0233-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sun F., Liao Y., Qu X., Xiao X., Hou S., Chen Z. Hepatic DNAJB9 drives anabolic biasing to reduce steatosis and obesity. Cell Reports. 2020;30(6):1835–1847. doi: 10.1016/j.celrep.2020.01.043. e9. [DOI] [PubMed] [Google Scholar]
- 101.Sun X., Zhao D., Lu F., Peng S., Yu M., Liu N. Hydrogen sulfide regulates muscle RING finger-1 protein S-sulfhydration at Cys44 to prevent cardiac structural damage in diabetic cardiomyopathy. British Journal of Pharmacology. 2020;177(4):836–856. doi: 10.1111/bph.14601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kim H.M., Han J.W., Chan J.Y. Nuclear factor erythroid-2 like 1 (NFE2L1): structure, function and regulation. Gene. 2016;584(1):17–25. doi: 10.1016/j.gene.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ohsumi Y. Historical landmarks of autophagy research. Cell Research. 2014;24(1):9–23. doi: 10.1038/cr.2013.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hansen M., Rubinsztein D.C., Walker D.W. Autophagy as a promoter of longevity: insights from model organisms. Nature Reviews Molecular Cell Biology. 2018;19(9):579–593. doi: 10.1038/s41580-018-0033-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pohl C., Dikic I. Cellular quality control by the ubiquitin-proteasome system and autophagy. Science. 2019;366(6467):818–822. doi: 10.1126/science.aax3769. [DOI] [PubMed] [Google Scholar]
- 106.Stolz A., Ernst A., Dikic I. Cargo recognition and trafficking in selective autophagy. Nature Cell Biology. 2014;16(6):495–501. doi: 10.1038/ncb2979. [DOI] [PubMed] [Google Scholar]
- 107.Velázquez A.P., Tatsuta T., Ghillebert R., Drescher I., Graef M. Lipid droplet–mediated ER homeostasis regulates autophagy and cell survival during starvation. Journal of Cell Biology. 2016;212(6):621–631. doi: 10.1083/jcb.201508102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Madrigal-Matute J., Cuervo A.M. Regulation of liver metabolism by autophagy. Gastroenterology. 2016;150(2):328–339. doi: 10.1053/j.gastro.2015.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Laplante M., Sabatini D.M. mTOR signaling in growth control and disease. Cell. 2012;149(2):274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Dikic I. Proteasomal and autophagic degradation systems. Annual Review of Biochemistry. 2017;86(1):193–224. doi: 10.1146/annurev-biochem-061516-044908. [DOI] [PubMed] [Google Scholar]
- 111.Zhao J., Garcia G.A., Goldberg A.L. Control of proteasomal proteolysis by mTOR. Nature. 2016;529(7586):E1–E2. doi: 10.1038/nature16472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Li S., Brown M.S., Goldstein J.L. Bifurcation of insulin signaling pathway in rat liver: mTORC1 required for stimulation of lipogenesis, but not inhibition of gluconeogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(8):3441–3446. doi: 10.1073/pnas.0914798107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhu Q. The ER stress-autophagy axis: implications for cognitive dysfunction in diabetes mellitus. Clinical Science (London, England: 1979) 2020;134(11):1255–1258. doi: 10.1042/CS20200235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yang L., Li P., Fu S., Calay E.S., Hotamisligil G.S. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metabolism. 2010;11(6):467–478. doi: 10.1016/j.cmet.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Singh R., Kaushik S., Wang Y., Xiang Y., Novak I., Komatsu M. Autophagy regulates lipid metabolism. Nature. 2009;458(7242):1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ebato C., Uchida T., Arakawa M., Komatsu M., Ueno T., Komiya K. Autophagy is important in islet homeostasis and compensatory increase of beta cell mass in response to high-fat diet. Cell Metabolism. 2008;8(4):325–332. doi: 10.1016/j.cmet.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 117.Khaminets A., Heinrich T., Mari M., Grumati P., Huebner A.K., Akutsu M. Regulation of endoplasmic reticulum turnover by selective autophagy. Nature. 2015;522(7556):354–358. doi: 10.1038/nature14498. [DOI] [PubMed] [Google Scholar]
- 118.Wilkinson S. ER-phagy: shaping up and destressing the endoplasmic reticulum. FEBS Journal. 2019;286(14):14932. doi: 10.1111/febs.14932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Strzyz P. Foundations of ER-phagy regulation. Nature Reviews Molecular Cell Biology. 2020;21(5) doi: 10.1038/s41580-020-0238-8. 251–251. [DOI] [PubMed] [Google Scholar]
- 120.Stephani M., Picchianti L., Gajic A., Beveridge R., Skarwan E., Sanchez de Medina Hernandez V. A cross-kingdom conserved ER-phagy receptor maintains endoplasmic reticulum homeostasis during stress. ELife. 2020;9 doi: 10.7554/eLife.58396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Nthiga T.M., Kumar Shrestha B., Sjøttem E., Bruun J.-A., Bowitz Larsen K., Bhujabal Z. CALCOCO1 acts with VAMP-associated proteins to mediate ER-phagy. The EMBO Journal. 2020;39(15) doi: 10.15252/embj.2019103649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hübner C.A., Dikic I. ER-phagy and human diseases. Cell Death & Differentiation. 2020;27(3):833–842. doi: 10.1038/s41418-019-0444-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Izumi T., Yokota-Hashimoto H., Zhao S., Wang J., Halban P.A., Takeuchi T. Dominant negative pathogenesis by mutant proinsulin in the Akita diabetic mouse. Diabetes. 2003;52(2):409–416. doi: 10.2337/diabetes.52.2.409. [DOI] [PubMed] [Google Scholar]
- 124.Arruda A.P., Hotamisligil G.S. Calcium homeostasis and organelle function in the pathogenesis of obesity and diabetes. Cell Metabolism. 2015;22(3):381–397. doi: 10.1016/j.cmet.2015.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Guerrero-Hernandez A., Verkhratsky A. Calcium signalling in diabetes. Cell Calcium. 2014;56(5):297–301. doi: 10.1016/j.ceca.2014.08.009. [DOI] [PubMed] [Google Scholar]
- 126.Görlach A., Klappa P., Kietzmann T. The endoplasmic reticulum: folding, calcium homeostasis, signaling, and redox control. Antioxidants and Redox Signaling. 2006;8(9–10):1391–1418. doi: 10.1089/ars.2006.8.1391. [DOI] [PubMed] [Google Scholar]
- 127.Clapham D.E. Calcium signaling. Cell. 2007;131(6):1047–1058. doi: 10.1016/j.cell.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 128.Lièvremont J.P., Rizzuto R., Hendershot L., Meldolesi J. BiP, a major chaperone protein of the endoplasmic reticulum lumen, plays a direct and important role in the storage of the rapidly exchanging pool of Ca2+ Journal of Biological Chemistry. 1997;272(49):30873–30879. doi: 10.1074/jbc.272.49.30873. [DOI] [PubMed] [Google Scholar]
- 129.Carreras-Sureda A., Pihán P., Hetz C. Calcium signaling at the endoplasmic reticulum: fine-tuning stress responses. Cell Calcium. 2018;70:24–31. doi: 10.1016/j.ceca.2017.08.004. [DOI] [PubMed] [Google Scholar]
- 130.Coe H., Michalak M. Calcium binding chaperones of the endoplasmic reticulum. General Physiology and Biophysics. 2009;28 Spec No: F96–103. [PubMed] [Google Scholar]
- 131.Guest P.C., Bailyes E.M., Hutton J.C. Endoplasmic reticulum Ca2+ is important for the proteolytic processing and intracellular transport of proinsulin in the pancreatic β-cell. Biochemical Journal. 1997;323(2):445–450. doi: 10.1042/bj3230445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Karunakaran U., Kim H.-J., Kim J.-Y., Lee I.-K. Guards and culprits in the endoplasmic reticulum: glucolipotoxicity and β-cell failure in type II diabetes. Experimental Diabetes Research. 2012;2012:639762. doi: 10.1155/2012/639762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Fu S., Yang L., Li P., Hofmann O., Dicker L., Hide W. Aberrant lipid metabolism disrupts calcium homeostasis causing liver endoplasmic reticulum stress in obesity. Nature. 2011;473(7348):528–531. doi: 10.1038/nature09968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Gwiazda K.S., Yang T.-L.B., Lin Y., Johnson J.D. Effects of palmitate on ER and cytosolic Ca2+ homeostasis in beta-cells. American Journal of Physiology. Endocrinology and Metabolism. 2009;296(4):E690–E701. doi: 10.1152/ajpendo.90525.2008. [DOI] [PubMed] [Google Scholar]
- 135.Arruda A.P., Pers B.M., Parlakgul G., Güney E., Goh T., Cagampan E. Defective STIM-mediated store operated Ca2+ entry in hepatocytes leads to metabolic dysfunction in obesity. ELife. 2017;6 doi: 10.7554/eLife.29968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kono T., Tong X., Taleb S., Bone R.N., Iida H., Lee C.-C. Impaired store-operated calcium entry and STIM1 loss lead to reduced insulin secretion and increased endoplasmic reticulum stress in the diabetic β-cell. Diabetes. 2018;67(11):2293–2304. doi: 10.2337/db17-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Feriod C.N., Gustavo Oliveira A., Guerra M.T., Nguyen L., Mitchell Richards K., Jurczak M.J. Hepatic inositol 1,4,5 trisphosphate receptor type 1 mediates fatty liver. Hepatology Communications. 2017;1(1):23–35. doi: 10.1002/hep4.1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hayashi T., Su T.-P. Sigma-1 receptor chaperones at the ER-mitochondrion interface regulate Ca(2+) signaling and cell survival. Cell. 2007;131(3):596–610. doi: 10.1016/j.cell.2007.08.036. [DOI] [PubMed] [Google Scholar]
- 139.Shinjo S., Jiang S., Nameta M., Suzuki T., Kanai M., Nomura Y. Disruption of the mitochondria-associated ER membrane (MAM) plays a central role in palmitic acid-induced insulin resistance. Experimental Cell Research. 2017;359(1):86–93. doi: 10.1016/j.yexcr.2017.08.006. [DOI] [PubMed] [Google Scholar]
- 140.Ma J.H., Shen S., Wang J.J., He Z., Poon A., Li J. Comparative proteomic analysis of the mitochondria-associated ER membrane (MAM) in a long-term type 2 diabetic rodent model. Scientific Reports. 2017;7(1):2062. doi: 10.1038/s41598-017-02213-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Marmugi A., Parnis J., Chen X., Carmichael L., Hardy J., Mannan N. Sorcin links pancreatic β-cell lipotoxicity to ER Ca2+ stores. Diabetes. 2016;65(4):1009–1021. doi: 10.2337/db15-1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wires E.S., Trychta K.A., Bäck S., Sulima A., Rice K.C., Harvey B.K. High fat diet disrupts endoplasmic reticulum calcium homeostasis in the rat liver. Journal of Hepatology. 2017;67(5):1009–1017. doi: 10.1016/j.jhep.2017.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ozcan L., Wong C.C.L., Li G., Xu T., Pajvani U., Park S.K.R. Calcium signaling through CaMKII regulates hepatic glucose production in fasting and obesity. Cell Metabolism. 2012;15(5):739–751. doi: 10.1016/j.cmet.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ersoy B.A., Maner-Smith K.M., Li Y., Alpertunga I., Cohen D.E. Thioesterase-mediated control of cellular calcium homeostasis enables hepatic ER stress. Journal of Clinical Investigation. 2018;128(1):141–156. doi: 10.1172/JCI93123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Jacquemyn J., Cascalho A., Goodchild R.E. The ins and outs of endoplasmic reticulum-controlled lipid biosynthesis. EMBO Reports. 2017;18(11):1905–1921. doi: 10.15252/embr.201643426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kuerschner L., Moessinger C., Thiele C. Imaging of lipid biosynthesis: how a neutral lipid enters lipid droplets. Traffic (Copenhagen, Denmark) 2008;9(3):338–352. doi: 10.1111/j.1600-0854.2007.00689.x. [DOI] [PubMed] [Google Scholar]
- 147.Jacquier N., Choudhary V., Mari M., Toulmay A., Reggiori F., Schneiter R. Lipid droplets are functionally connected to the endoplasmic reticulum in Saccharomyces cerevisiae. Journal of Cell Science. 2011;124(Pt 14):2424–2437. doi: 10.1242/jcs.076836. [DOI] [PubMed] [Google Scholar]
- 148.Eissing L., Scherer T., Tödter K., Knippschild U., Greve J.W., Buurman W.A. De novo lipogenesis in human fat and liver is linked to ChREBP-β and metabolic health. Nature Communications. 2013;4:1528. doi: 10.1038/ncomms2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Bartelt A., Weigelt C., Cherradi M.L., Niemeier A., Tödter K., Heeren J. Effects of adipocyte lipoprotein lipase on de novo lipogenesis and white adipose tissue browning. Biochimica et Biophysica Acta. 2013;1831(5):934–942. doi: 10.1016/j.bbalip.2012.11.011. [DOI] [PubMed] [Google Scholar]
- 150.Bartelt A., John C., Schaltenberg N., Berbée J.F.P., Worthmann A., Cherradi M.L. Thermogenic adipocytes promote HDL turnover and reverse cholesterol transport. Nature Communications. 2017;8:15010. doi: 10.1038/ncomms15010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Song M.J., Malhi H. The unfolded protein response and hepatic lipid metabolism in non alcoholic fatty liver disease. Pharmacology & Therapeutics. 2019;203:107401. doi: 10.1016/j.pharmthera.2019.107401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kong F.-J., Wu J.-H., Sun S.-Y., Zhou J.-Q. The endoplasmic reticulum stress/autophagy pathway is involved in cholesterol-induced pancreatic β-cell injury. Scientific Reports. 2017;7:44746. doi: 10.1038/srep44746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Shimabukuro M., Zhou Y.T., Levi M., Unger R.H. Fatty acid-induced beta cell apoptosis: a link between obesity and diabetes. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(5):2498–2502. doi: 10.1073/pnas.95.5.2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Plötz T., von Hanstein A.S., Krümmel B., Laporte A., Mehmeti I., Lenzen S. Structure-toxicity relationships of saturated and unsaturated free fatty acids for elucidating the lipotoxic effects in human EndoC-βH1 beta-cells. Biochimica et Biophysica Acta. Molecular Basis of Disease. 2019;1865(11):165525. doi: 10.1016/j.bbadis.2019.08.001. [DOI] [PubMed] [Google Scholar]
- 155.Laybutt D.R., Preston A.M., Akerfeldt M.C., Kench J.G., Busch A.K., Biankin A.V. Endoplasmic reticulum stress contributes to beta cell apoptosis in type 2 diabetes. Diabetologia. 2007;50(4):752–763. doi: 10.1007/s00125-006-0590-z. [DOI] [PubMed] [Google Scholar]
- 156.Naito H., Yoshikawa-Bando Y., Yuan Y., Hashimoto S., Kitamori K., Yatsuya H. High-fat and high-cholesterol diet decreases phosphorylated inositol-requiring kinase-1 and inhibits autophagy process in rat liver. Scientific Reports. 2019;9(1):12514. doi: 10.1038/s41598-019-48973-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Cnop M., Hannaert J.C., Hoorens A., Eizirik D.L., Pipeleers D.G. Inverse relationship between cytotoxicity of free fatty acids in pancreatic islet cells and cellular triglyceride accumulation. Diabetes. 2001;50(8):1771–1777. doi: 10.2337/diabetes.50.8.1771. [DOI] [PubMed] [Google Scholar]
- 158.Feng B., Yao P.M., Li Y., Devlin C.M., Zhang D., Harding H.P. The endoplasmic reticulum is the site of cholesterol-induced cytotoxicity in macrophages. Nature Cell Biology. 2003;5(9):781–792. doi: 10.1038/ncb1035. [DOI] [PubMed] [Google Scholar]
- 159.Maxfield F.R., Tabas I. Role of cholesterol and lipid organization in disease. Nature. 2005;438(7068):612–621. doi: 10.1038/nature04399. [DOI] [PubMed] [Google Scholar]
- 160.Lee J.N., Ye J. Proteolytic activation of sterol regulatory element-binding protein induced by cellular stress through depletion of Insig-1. Journal of Biological Chemistry. 2004;279(43):45257–45265. doi: 10.1074/jbc.M408235200. [DOI] [PubMed] [Google Scholar]
- 161.Bobrovnikova-Marjon E., Hatzivassiliou G., Grigoriadou C., Romero M., Cavener D.R., Thompson C.B. PERK-dependent regulation of lipogenesis during mouse mammary gland development and adipocyte differentiation. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(42):16314–16319. doi: 10.1073/pnas.0808517105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Wang T., Zhao Y., You Z., Li X., Xiong M., Li H. Endoplasmic reticulum stress affects cholesterol homeostasis by inhibiting LXRα expression in hepatocytes and macrophages. Nutrients. 2020;12(10) doi: 10.3390/nu12103088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Jo H., Choe S.S., Shin K.C., Jang H., Lee J.H., Seong J.K. Endoplasmic reticulum stress induces hepatic steatosis via increased expression of the hepatic very low-density lipoprotein receptor. Hepatology (Baltimore, Md.) 2013;57(4):1366–1377. doi: 10.1002/hep.26126. [DOI] [PubMed] [Google Scholar]
- 164.Sun Y., Zhang D., Liu X., Li X., Liu F., Yu Y. Endoplasmic reticulum stress affects lipid metabolism in atherosclerosis via CHOP activation and over-expression of miR-33. Cellular Physiology and Biochemistry: International Journal of Experimental Cellular Physiology, Biochemistry, and Pharmacology. 2018;48(5):1995–2010. doi: 10.1159/000492522. [DOI] [PubMed] [Google Scholar]
- 165.Brookheart R.T., Michel C.I., Schaffer J.E. As a matter of fat. Cell Metabolism. 2009;10(1):9–12. doi: 10.1016/j.cmet.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Rokbi B., Mazarin V., Maitre-Wilmotte G., Quentin-Millet M.J. Identification of two major families of transferrin receptors among Neisseria meningitidis strains based on antigenic and genomic features. FEMS Microbiology Letters. 1993;110(1):51–57. doi: 10.1111/j.1574-6968.1993.tb06294.x. [DOI] [PubMed] [Google Scholar]
- 167.Ye R., Jung D.Y., Jun J.Y., Li J., Luo S., Ko H.J. Grp78 heterozygosity promotes adaptive unfolded protein response and attenuates diet-induced obesity and insulin resistance. Diabetes. 2010;59(1):6–16. doi: 10.2337/db09-0755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Kammoun H.L., Chabanon H., Hainault I., Luquet S., Magnan C., Koike T. GRP78 expression inhibits insulin and ER stress-induced SREBP-1c activation and reduces hepatic steatosis in mice. Journal of Clinical Investigation. 2009;119(5):1201–1215. doi: 10.1172/JCI37007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Stringer D.M., Zahradka P., Declercq V.C., Ryz N.R., Diakiw R., Burr L.L. Modulation of lipid droplet size and lipid droplet proteins by trans-10,cis-12 conjugated linoleic acid parallels improvements in hepatic steatosis in obese, insulin-resistant rats. Biochimica et Biophysica Acta. 2010;1801(12):1375–1385. doi: 10.1016/j.bbalip.2010.08.011. [DOI] [PubMed] [Google Scholar]
- 170.Papazyan R., Sun Z., Kim Y.H., Titchenell P.M., Hill D.A., Lu W. Physiological suppression of lipotoxic liver damage by complementary actions of HDAC3 and SCAP/SREBP. Cell Metabolism. 2016;24(6):863–874. doi: 10.1016/j.cmet.2016.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Smith G.I., Shankaran M., Yoshino M., Schweitzer G.G., Chondronikola M., Beals J.W. Insulin resistance drives hepatic de novo lipogenesis in nonalcoholic fatty liver disease. Journal of Clinical Investigation. 2020;130(3):1453–1460. doi: 10.1172/JCI134165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Liu Q., Siloto R.M.P., Lehner R., Stone S.J., Weselake R.J. Acyl-CoA:diacylglycerol acyltransferase: molecular biology, biochemistry and biotechnology. Progress in Lipid Research. 2012;51(4):350–377. doi: 10.1016/j.plipres.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 173.Chitraju C., Mejhert N., Haas J.T., Diaz-Ramirez L.G., Grueter C.A., Imbriglio J.E. Triglyceride synthesis by DGAT1 protects adipocytes from lipid-induced ER stress during lipolysis. Cell Metabolism. 2017;26(2):407–418. doi: 10.1016/j.cmet.2017.07.012. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Becuwe M., Bond L.M., Pinto A.F.M., Boland S., Mejhert N., Elliott S.D. FIT2 is an acyl-coenzyme A diphosphatase crucial for endoplasmic reticulum homeostasis. The Journal of Cell Biology. 2020;219(10) doi: 10.1083/jcb.202006111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Hatting M., Tavares C.D.J.J., Sharabi K., Rines A.K., Puigserver P. Insulin regulation of gluconeogenesis. Annals of the New York Academy of Sciences. 2018;1411(1):21–35. doi: 10.1111/nyas.13435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Zhang K., Shen X., Wu J., Sakaki K., Saunders T., Rutkowski D.T. Endoplasmic reticulum stress activates cleavage of CREBH to induce a systemic inflammatory response. Cell. 2006;124(3):587–599. doi: 10.1016/j.cell.2005.11.040. [DOI] [PubMed] [Google Scholar]
- 177.Aebi M., Bernasconi R., Clerc S., Molinari M. N-glycan structures: recognition and processing in the ER. Trends in Biochemical Sciences. 2010;35(2):74–82. doi: 10.1016/j.tibs.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 178.Wagner M., Moore D.D. Endoplasmic reticulum stress and glucose homeostasis. Current Opinion in Clinical Nutrition and Metabolic Care. 2011;14(4):367–373. doi: 10.1097/MCO.0b013e32834778d4. [DOI] [PubMed] [Google Scholar]
- 179.Lipson K.L., Fonseca S.G., Ishigaki S., Nguyen L.X., Foss E., Bortell R. Regulation of insulin biosynthesis in pancreatic beta cells by an endoplasmic reticulum-resident protein kinase IRE1. Cell Metabolism. 2006;4(3):245–254. doi: 10.1016/j.cmet.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 180.Ruan H.-B., Dietrich M.O., Liu Z.-W., Zimmer M.R., Li M.-D., Singh J.P. O-GlcNAc transferase enables AgRP neurons to suppress browning of white fat. Cell. 2014;159(2):306–317. doi: 10.1016/j.cell.2014.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Li M.-D., Vera N.B., Yang Y., Zhang B., Ni W., Ziso-Qejvanaj E. Adipocyte OGT governs diet-induced hyperphagia and obesity. Nature Communications. 2018;9(1):5103. doi: 10.1038/s41467-018-07461-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Yang Y., Fu M., Li M.-D., Zhang K., Zhang B., Wang S. O-GlcNAc transferase inhibits visceral fat lipolysis and promotes diet-induced obesity. Nature Communications. 2020;11(1):181. doi: 10.1038/s41467-019-13914-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Shan B., Wang X., Wu Y., Xu C., Xia Z., Dai J. The metabolic ER stress sensor IRE1α suppresses alternative activation of macrophages and impairs energy expenditure in obesity. Nature Immunology. 2017;18(5):519–529. doi: 10.1038/ni.3709. [DOI] [PubMed] [Google Scholar]
- 184.Hotamisligil G.S. Inflammation, metaflammation and immunometabolic disorders. Nature. 2017;542(7640):177–185. doi: 10.1038/nature21363. [DOI] [PubMed] [Google Scholar]
- 185.Rui L., Aguirre V., Kim J.K., Shulman G.I., Lee A., Corbould A. Insulin/IGF-1 and TNF-alpha stimulate phosphorylation of IRS-1 at inhibitory Ser307 via distinct pathways. Journal of Clinical Investigation. 2001;107(2):181–189. doi: 10.1172/JCI10934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Kouroku Y., Fujita E., Jimbo A., Kikuchi T., Yamagata T., Momoi M.Y. Polyglutamine aggregates stimulate ER stress signals and caspase-12 activation. Human Molecular Genetics. 2002;11(13):1505–1515. doi: 10.1093/hmg/11.13.1505. [DOI] [PubMed] [Google Scholar]
- 187.Hamanaka R.B., Bennett B.S., Cullinan S.B., Diehl J.A. PERK and GCN2 contribute to eIF2α phosphorylation and cell cycle arrest after activation of the unfolded protein response pathway. Molecular Biology of the Cell. 2005;16(12):5493–5501. doi: 10.1091/mbc.e05-03-0268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Tam A.B., Mercado E.L., Hoffmann A., Niwa M. ER stress activates NF-κB by integrating functions of basal IKK activity, IRE1 and PERK. PloS One. 2012;7(10) doi: 10.1371/journal.pone.0045078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Hirosumi J., Tuncman G., Chang L., Görgün C.Z., Uysal K.T., Maeda K. A central role for JNK in obesity and insulin resistance. Nature. 2002;420(6913):333–336. doi: 10.1038/nature01137. [DOI] [PubMed] [Google Scholar]
- 190.Sabio G., Das M., Mora A., Zhang Z., Jun J.Y., Ko H.J. A stress signaling pathway in adipose tissue regulates hepatic insulin resistance. Science (New York, N.Y.) 2008;322(5907):1539–1543. doi: 10.1126/science.1160794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Das M., Sabio G., Jiang F., Rincón M., Flavell R.A., Davis R.J. Induction of hepatitis by JNK-mediated expression of TNF-alpha. Cell. 2009;136(2):249–260. doi: 10.1016/j.cell.2008.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Sabio G., Kennedy N.J., Cavanagh-Kyros J., Jung D.Y., Ko H.J., Ong H. Role of muscle c-Jun NH2-terminal kinase 1 in obesity-induced insulin resistance. Molecular and Cellular Biology. 2010;30(1):106–115. doi: 10.1128/MCB.01162-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Sabio G., Cavanagh-Kyros J., Ko H.J., Jung D.Y., Gray S., Jun J.Y. Prevention of steatosis by hepatic JNK1. Cell Metabolism. 2009;10(6):491–498. doi: 10.1016/j.cmet.2009.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Sabio G., Cavanagh-Kyros J., Barrett T., Jung D.Y., Ko H.J., Ong H. Role of the hypothalamic-pituitary-thyroid axis in metabolic regulation by JNK1. Genes & Development. 2010;24(3):256–264. doi: 10.1101/gad.1878510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Han M.S., Jung D.Y., Morel C., Lakhani S.A., Kim J.K., Flavell R.A. JNK expression by macrophages promotes obesity-induced insulin resistance and inflammation. Science (New York, N.Y.) 2013;339(6116):218–222. doi: 10.1126/science.1227568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Carvalho-Filho M.A., Carvalho B.M., Oliveira A.G., Guadagnini D., Ueno M., Dias M.M. Double-stranded RNA-activated protein kinase is a key modulator of insulin sensitivity in physiological conditions and in obesity in mice. Endocrinology. 2012;153(11):5261–5274. doi: 10.1210/en.2012-1400. [DOI] [PubMed] [Google Scholar]
- 197.Nakamura T., Kunz R.C., Zhang C., Kimura T., Yuan C.L., Baccaro B. A critical role for PKR complexes with TRBP in Immunometabolic regulation and eIF2α phosphorylation in obesity. Cell Reports. 2015;11(2):295–307. doi: 10.1016/j.celrep.2015.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Reilly S.M., Ahmadian M., Zamarron B.F., Chang L., Uhm M., Poirier B. A subcutaneous adipose tissue-liver signalling axis controls hepatic gluconeogenesis. Nature Communications. 2015;6:6047. doi: 10.1038/ncomms7047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Reilly S.M., Chiang S.-H., Decker S.J., Chang L., Uhm M., Larsen M.J. An inhibitor of the protein kinases TBK1 and IKK-ɛ improves obesity-related metabolic dysfunctions in mice. Nature Medicine. 2013;19(3):313–321. doi: 10.1038/nm.3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Liu Y.Y.Y., Shao M., Wu Y., Yan C., Jiang S., Liu J. Role for the endoplasmic reticulum stress sensor IRE1α in liver regenerative responses. Journal of Hepatology. 2015;62(3):590–598. doi: 10.1016/j.jhep.2014.10.022. [DOI] [PubMed] [Google Scholar]
- 201.Di Conza G., Ho P.-C. ER stress responses: an emerging modulator for innate immunity. Cells. 2020;9(3):695. doi: 10.3390/cells9030695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Chang C., Dong R., Miyazaki A., Sakashita N., Zhang Y., Liu J. Human acyl-CoA:cholesterol acyltransferase (ACAT) and its potential as a target for pharmaceutical intervention against atherosclerosis. Acta Biochimica et Biophysica Sinica. 2006;38(3):151–156. doi: 10.1111/j.1745-7270.2006.00154.x. [DOI] [PubMed] [Google Scholar]
- 203.Leon C., Hill J.S., Wasan K.M. Potential role of acyl-coenzyme A:cholesterol transferase (ACAT) Inhibitors as hypolipidemic and antiatherosclerosis drugs. Pharmaceutical Research. 2005;22(10):1578–1588. doi: 10.1007/s11095-005-6306-0. [DOI] [PubMed] [Google Scholar]
- 204.Sekiya M., Osuga J.-I., Igarashi M., Okazaki H., Ishibashi S. The role of neutral cholesterol ester hydrolysis in macrophage foam cells. Journal of Atherosclerosis and Thrombosis. 2011;18(5):359–364. doi: 10.5551/jat.7013. [DOI] [PubMed] [Google Scholar]
- 205.Erbay E., Babaev V.R., Mayers J.R., Makowski L., Charles K.N., Snitow M.E. Reducing endoplasmic reticulum stress through a macrophage lipid chaperone alleviates atherosclerosis. Nature Medicine. 2009;15(12):1383–1391. doi: 10.1038/nm.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Martinon F., Chen X., Lee A.-H., Glimcher L.H. TLR activation of the transcription factor XBP1 regulates innate immune responses in macrophages. Nature Immunology. 2010;11(5):411–418. doi: 10.1038/ni.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Tufanli O., Telkoparan Akillilar P., Acosta-Alvear D., Kocaturk B., Onat U.I., Hamid S.M. Targeting IRE1 with small molecules counteracts progression of atherosclerosis. Proceedings of the National Academy of Sciences of the United States of America. 2017;114(8):E1395–E1404. doi: 10.1073/pnas.1621188114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Bai J., Liu F. The cGAS-cGAMP-STING pathway: a molecular link between immunity and metabolism. Diabetes. 2019;68(6):1099–1108. doi: 10.2337/dbi18-0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Tao L., Lemoff A., Wang G., Zarek C., Lowe A., Yan N. Reactive oxygen species oxidize STING and suppress interferon production. ELife. 2020;9 doi: 10.7554/eLife.57837. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.