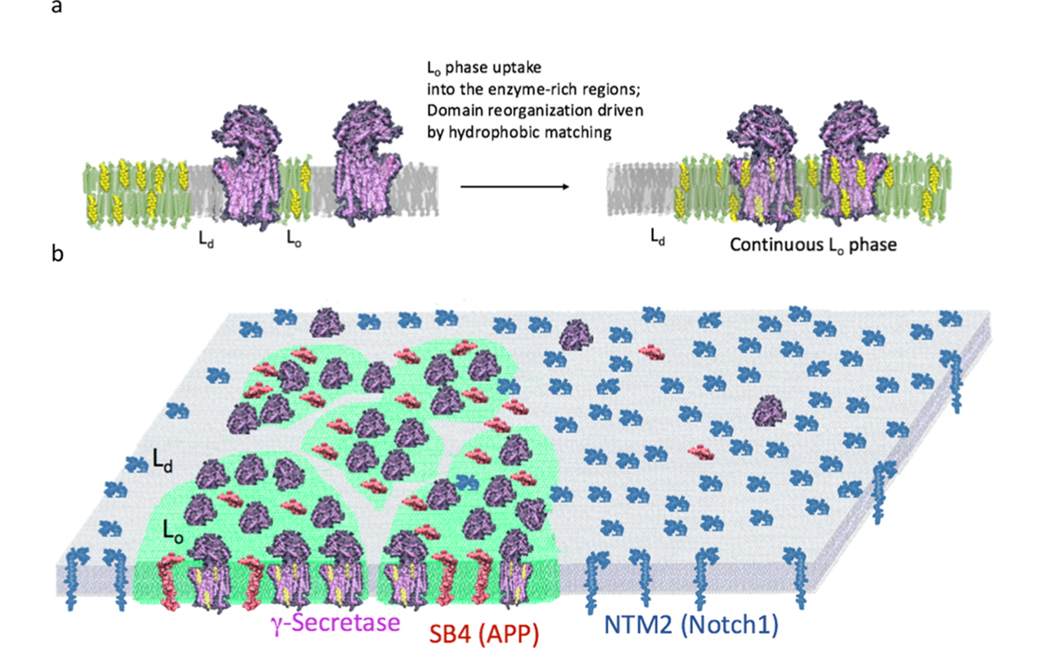
Abstract
γ-Secretase is a multisubunit complex that catalyzes intramembranous cleavage of transmembrane proteins. The lipid environment forms membrane microdomains that serve as spatio-temporal platforms for proteins to function properly. Despite substantial advances in the regulation of γ-secretase, the effect of the local membrane lipid microenvironment on the regulation of γ-secretase is poorly understood. Here, we characterized and quantified the partitioning of γ-secretase and its substrates, the amyloid precursor protein (APP) and Notch, into lipid bilayers using solid-supported model membranes. Notch substrate is preferentially localized in the liquid-disordered (Ld) lipid domains, whereas APP and γ-secretase partition as single or higher complex in both phases but highly favor the ordered phase, especially after recruiting lipids from the ordered phase, indicating that the activity and specificity of γ-secretase against these two substrates are modulated by membrane lateral organization. Moreover, time-elapse measurements reveal that γ-secretase can recruit specific membrane components from the cholesterol-rich Lo phase and thus creates a favorable lipid environment for substrate recognition and therefore activity. This work offers insight into how γ-secretase and lipid modulate each other and control its activity and specificity.
Graphical Abstract
INTRODUCTION
γ-Secretase is an aspartyl protease complex composed of four essential subunits: presenilin, nicastrin, Aph-1, and Pen-2.1 This enzyme catalyzes the intramembranous cleavage of over 90 substrates,2 including the amyloid precursor protein (APP) and Notch receptors. The sequential proteolysis of APP by BACE1 and γ-secretase generates amyloid β (Aβ) peptides, one of the main pathological hallmarks of Alzheimer’s disease. γ-Secretase also cleaves Notch that is involved in the development and human disorders.
Although significant advances in the understanding of γ-secretase−substrate interactions,3,4 enzymatic regulation, and catalysis have been made, the underlying molecular mechanisms of the cleavage and substrate recognition within the lipid membrane context still remain to be investigated. In particular, lipids are known to play an important role in defining membrane protein (MP) interactions. Those lipid−protein interactions are determined by the protein structure and physicochemical properties of the surrounding lipids that contribute to membrane lateral heterogeneity.5 Cholesterol tends to pack tightly, and therefore more ordered, to sphingomyelin and saturated lipids, which configurates a microscopic phase separation from the membrane fluid or disordered phase. Liquid-ordered and liquid-disordered regions can coexist as distinct phases in model membranes and vesicles extracted directly from the plasma membrane. The presence of raft microdomains6,7 and ordered phase enriched in sphingolipids and cholesterol, in the cellular membrane, further complicates the enzymology of the system.8
The effect of lipid composition and the role of lipid rafts in the cleavage of γ-secretase and distribution of APP has been extensively studied. Subtle changes in membrane lipid composition affect substrate processing.9−11 These studies were carried on in reconstituted γ-secretase proteoliposomes9 and also utilizing biochemical methods that demonstrated that cholesterol-enriched buoyant membranes are the major site of amyloid β production,11 it has been suggested that γ-secretase and APP are located in lipid rafts in cultured neuronal cells,12 mouse brain, and human brain membrane.13 Solution NMR studies in micelles and bicelles indicate that there is a cholesterol-binding motif built into the structure of the C-terminal fragment domain of APP, potentially promoting to its partitioning to raft domains.14−16 Analogous studies of the Notch transmembrane domain did not find any evidence for cholesterol-specific binding.17 Recent structural studies showed that both substrates have similar binding modes to γ-secretase.3,4 Nevertheless, all of these methodologies do not allow for the in situ visualization and quantification of enzyme and substrate partitioning in raft domains, which hinder our understanding of how these membrane proteins communicate with lipids and require novel means for such investigation.
In this study, to directly visualize the distribution of γ-secretase and its substrates in raft-containing membranes, we utilized atomic force microscopy (AFM) to study the enzyme and its substrates at the single-molecule level in planar membrane models. We quantified the phase partitioning of γ-secretase, APP, and Notch substrates and determined whether their distribution and activity are influenced by the membrane lateral organization. γ-Secretase is not only influenced by the membrane lateral organization but also is surprisingly able to recruit lipid from the adjacent ordered phase and remodel the Lo phase. This recruitment leads to encompassing and concentrating the complex into enzyme-rich regions to modulate its function. Moreover, we found that the APP and Notch substrates reside in different membrane domains, which affects γ-secretase activity and specificity. Our findings indicate that membrane proteins with a sufficiently large hydrophobic interaction area can alter local membrane environments for their function, in effect overriding and dominating local lipid thermodynamic driving forces, opening up new directions for the investigation of protein and membrane interactions and intramembrane proteolysis.
EXPERIMENTAL SECTION
Substrate Purification.
SB4 and NTM2 substrates were prepared as previously described.18,19 Briefly, biotinylated substrates NTM2 and SB4 were expressed in BL21(DE3) cells induced at 20 °C with 100 μM IPTG and in the presence of 50 μM biotin for 5 h. Bacteria were then harvested by centrifugation and lysed by French Press. The lysate was centrifuged at 12 000 rpm for 1 h at 4 °C, and the supernatant fraction was loaded into an amylose column and eluted with a gradient of maltose. Finally, the maltose-binding protein (MBP) tag was removed from the purified protein by overnight thrombin cleavage when required.
Purification of γ-Secretase.
Using a Pichia pastoris (yeast) protein expression system, the four subunits of the wild-type human γ-secretase complex were overexpressed using a protocol for protein expression following a similar strategy used in our previous study,20 and the tagged γ-secretase complex was isolated following centrifugation, solubilization, and affinity chromatography, as described previously.20
Lipids and Liposome Preparation.
1,2-Dioleoyl-sn-glycero-3-phosphocholine (DOPC), cholesterol (extracted from ovine wool), and sphingomyelin (extracted from porcine Brain) were purchased from Avanti Polar Lipids. Ternary lipid mixtures composed of DOPC/sphingomyelin/cholesterol at a 2:2:1 molar ratio were prepared from fresh lipid stocks in chloroform and dried for 2 h under vacuum. After drying, the lipid films were hydrated in a physiologically relevant buffer (150 mM NaCl, 20 mM Tris at pH 7.0, and 10 mM MgCl2) to a lipid concentration of ∼10 mg/mL and subsequently sonicated until clear lipid solution was obtained. The resulting liposome solution was utilized within the same day of preparation in the AFM experiments.
AFM.
Fluid imaging was acquired using an MFP-3D-BIO AFM (Asylum Research), with an Olympus BL-AC40TS AFM probe (Asylum Research) in tapping mode. Probes were tuned using an Auto Thermal method. Complete and defect-free solid-supported planar membranes were formed by incubating lipid vesicles in a freshly cleaved mica substrate at room temperature for 20 min followed by extensive buffer (150 mM NaCl, 20 mM Tris at pH 7.0, and 10 mM MgCl2) rinse to eliminate the excess of vesicles. Images from different locations of the lipid bilayer were also acquired to ensure that holes were not present before starting the experiment. After protein incubation, multiple square images were acquired (4 and 10 μm) at the same lipid bilayer location over a time course of approximately 2 h after protein loading at a scan rate of 0.6 Hz. A Petri dish heater for MFP-3D AFM was used on the AFM stage to maintain the temperature at 25 °C and to perform temperature-dependent experiments. In the temperature-dependent experiments, the lipid bilayer was heated in two steps, first to 31 °C and then to 37 °C until it reached thermal equilibrium. At 37 °C, the protein was added and the system was cooled down to 31 °C. γ-Secretase was incubated at a final detergent concentration 200-fold below the critical micelle concentrations (CMC) of n-dodecyl-β-D-maltoside (DDM). The protein final concentration was 10 nM for γ-secretase and 10 μM for SB4 and NTM2. Images of the neat lipid bilayer before and after protein was added were acquired at both 31 and 37 °C when the system reached steady thermal equilibrium, as well as at room temperature, as indicated (near 25 °C). Image analysis was performed using Igor Pro. To choose the image features consistent with single enzyme complexes, the circularity was chosen as a filter (circularity: the ratio of the square of the perimeter to (4 × π × area)). This value approaches 1 for a perfect circle. AFM feature volume analysis was carried out based on the polymer size versus chain length arguments,21,22 as outlined in the Supporting Information. To evaluate differences between the average feature volumes, the oneway analysis of variance test (ANOVA) was used, with a threshold of p < 0.05 to indicate statistically significant differences.
γ-Secretase Activity Assay.
The γ-secretase assay is adapted from methods previously described18,19 Briefly, the APP and Notch recombinant substrates were incubated with 0.004 μg/μL of purified γ-secretase and various lipid mixtures at 0.04 μg/μL in the presence of 0.25% CHAPSO in piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES) buffer (50 mM PIPES, pH 7.0, 150 mM KCl, 5 mM CaCl2, 5 mM MgCl2) at 37°C for 4 h. The reaction was also allowed to proceed in the presence or absence of 1 μM of the γ-secretase inhibitor. The cleavage products were quantified with cleavage site-specific antibodies, G2–10 for Aβ40 and SM320 for NICD, and the signal was quantified by the AlphaLISA technology (PerkinElmer Life Sciences).
RESULTS
Development of a Model System for γ-Secretase and Lipid Microdomain Interplay.
To investigate the interaction of γ-secretase and substrates with lipids, we developed a system that uses AFM to monitor the dynamics of lipid−protein interactions (Figure 1a). Spontaneous and direct insertion of purified γ-secretase and substrates into 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC)/sphingomyelin/cholesterol (2:2:1)-supported bilayers in planar format was monitored by AFM.23
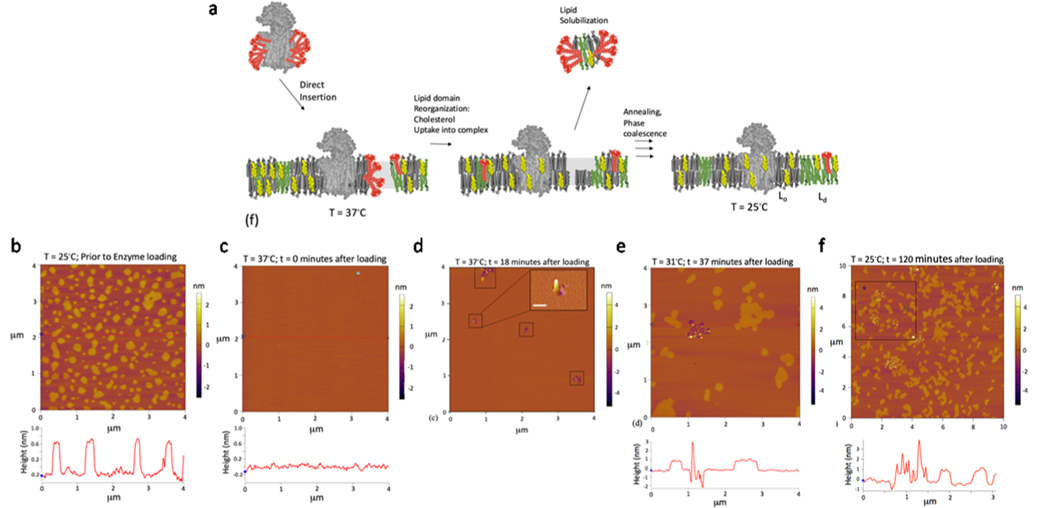
Figure 1.
Characterization of the direct insertion of γ-secretase into phase-separated planar-supported membrane systems with atomic force microscopy (AFM). (a) Schematic representation of the experimental procedure: direct insertion followed by lipid domain reorganization and then annealing and phase coalescence. (b−e) Time course of the direct insertion of γ-secretase into preformed DOPC/sphingomyelin/cholesterol (2:2:1)-supported bilayers on a glass-supported mica sheet. (b) Image at 25 °C prior to protein addition, and the image line profile below was obtained from the indicated line segment bisecting the image. (c) Image at 37 °C immediately after the addition of solubilized γ-secretase and the image line profile. (d) Image at 37 °C at t = 18 min after the addition of solubilized γ-secretase, and the boxed regions indicate where initial direct insertion has occurred. The three-dimensional (3D) image inset is zoomed in to show the 3D topography of an insertion event (scale bar 100 nm). (e) Image at 31 °C t = 37 min after the addition of solubilized γ-secretase and the image line profile. (f) Image (zoomed out to 10 × 10 μm2) at 25 °C after the addition of γ-secretase after 120 min of incubation time and cooling and the image line profile.
The starting supported lipid bilayer configuration was imaged at 25 °C prior to protein addition (Figure 1b). Phase-separated supported membrane was clearly observed as lighter Lo (liquid-ordered) cholesterol-rich domains against a darker Ld (liquid-disordered) phase. The height difference seen in the height profiles shown below the image between Lo and Ld phases is approximately 0.8 nm (Figure 1b), consistent with the previous reports.24,25 The temperature is then increased to 37 °C where there is no longer a clear distinction between Lo and Ld phases (Figure 1c). Once a constant temperature (37 °C) is reached, the purified γ-secretase complex is added. The AFM image displayed in Figure 1d was obtained at 18 min after loading. The boxed regions indicate where γ-secretase direct insertion initiated. We observed regions consistent with enzyme insertion, as well as adjacent regions where bilayer reorganization or lipid loss occurred. The inset of Figure 1d is zoomed into the indicated region of interest (ROI). This structural perturbation on the membrane is expected given the large hydrophobic surface area of the γ-secretase complex, which requires the rearrangement and packing of a large number of phospholipids to embed into the supported bilayer. As outlined in Figure S1, modeling analysis suggests that ∼68 phosphatidylcholine lipids form the lipid shell surrounding the hydrophobic region of the 3.4 Å cryo-EM structure of Bai et al. embedded into an explicit phosphatidylcholine (PC) bilayer using an automated protocol.26,27 In addition, γ-secretase inserted as a detergent-solubilized purified complex might have carried residual detergent during the direct insertion process, which has been shown to lead to the solubilization of small patches of bilayer leading to membrane defects with depth consistent with half leaflet of the bilayer. Furthermore, the height profile from this image is consistent with the removal of the top leaflet (<2.1 nm) (Figure S2), which has been observed in other studies of direct insertion of membrane proteins.23
The system is then cooled down in two steps. First to 31 °C (Figure 1e; 37 min after loading), where it is possible to distinguish lipid domains, and then down to 25 °C (Figure 1f; 120 min after loading). Figure 1a describes the sequence of events leading to fully intact bilayers with embedded MPs that is consistent with the AFM imagery. The cooling has resulted in the enzyme reorganizing into specific concentrated regions containing features consistent with further insertion and clustering of the enzyme. Also, the lipid domains are reorganizing and coalescing, and the defects arising from lipid solubilization and MP insertion are beginning to be filled in and annealed, ultimately leading to fully intact bilayers with embedded MPs. It is also important to note that during this time, the complex is undergoing continuous insertion and the lipid bilayer cholesterol-rich microdomains are continuously reorganizing.
Insertion and Clustering of the γ-Secretase Complex into Lipid Bilayer Lead to the Redistribution of Membrane Microdomains.
Using this system, we examined γ-secretase dynamics of insertion and lipid microdomain reorganization at 25 °C by imaging the same area over a time lapse of ∼3 h (Figure 2a –d). The images are consistent with the direct insertion of γ-secretase as protrusions with the heights consistent with the size of nicastrin, the γ-secretase ectodomain, are evidenced (vide infra) (Figure 2a). When the same area is imaged 17 min later (Figure 2b; 164 min after loading), considerable growth of Lo regions is observed mainly in regions adjacent to γ-secretase and in a lesser extent in enzyme-free areas. Furthermore, Lo domains have begun to migrate toward the inserted enzyme regions. Also, the lipid loss defect regions have appeared to coalesce due to lipid rearrangements and the formation of larger protein clusters. Over time, it is clear that the Lo domains closest to the areas of enzyme insertion have moved to the immediate vicinity or are in direct contact with the enzyme-enriched regions. The features above the enzyme-dense region (rectangle box) in Figure 2c,d were further analyzed and resulted in Figure 2e,f. This region lost the Lo area and transitioned from one continuous domain into three separate adjacent domains by redistribution of lipids and cholesterol. Considering molecular geometry arguments and an average Lo single lipid area of ∼0.41 nm2,28−30 the change in the area of the indicated region amounted to ∼180 000 molecules per leaflet, a number in great excess of the number needed to form an annular boundary lipid shell around the γ-secretase complexes present in the enzyme-rich region containing ∼80 enzymes (∼2700 annular shell lipids per leaflet).
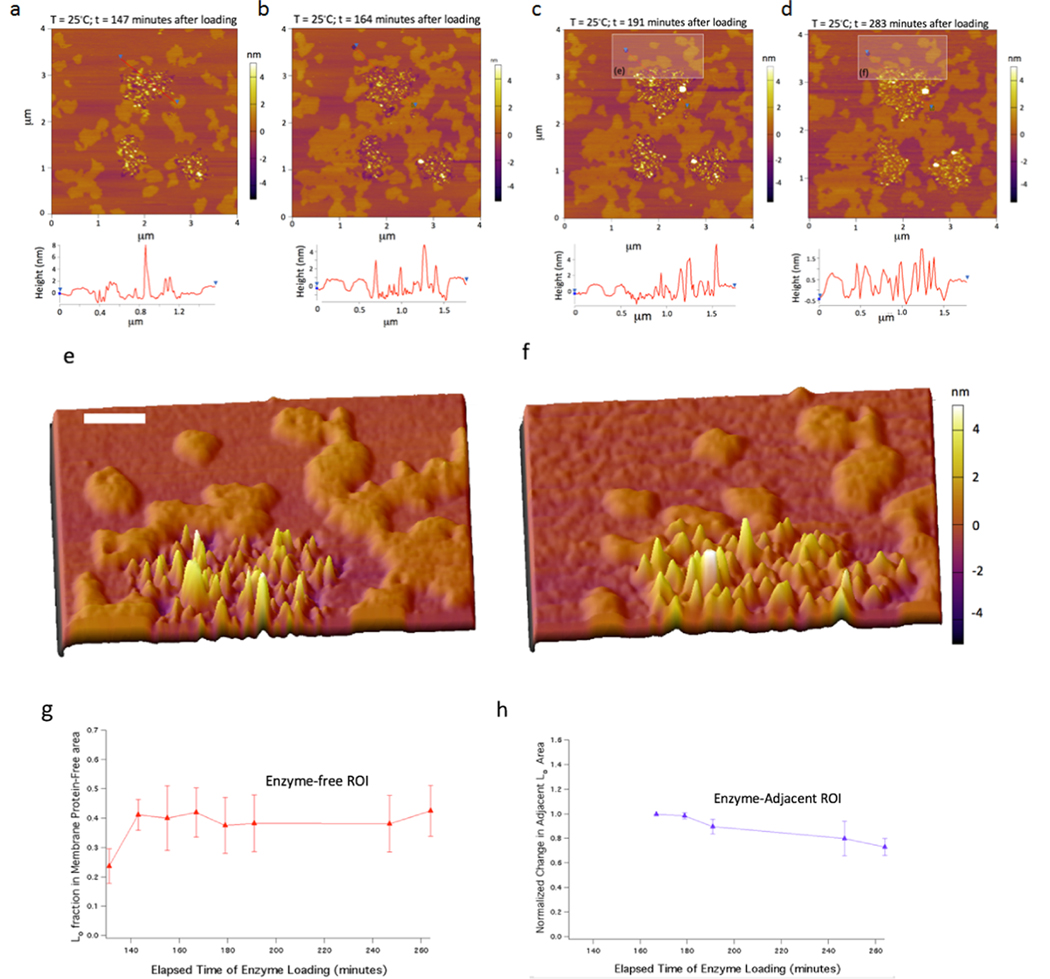
Figure 2.
Insertion of γ-secretase into model membranes induces the redistribution of microdomains. (a−d) Time course (t = 147, 164, 191, and 283 min at 25 °C after loading) of reconstitution of γ-secretase into preformed DOPC/sphingomyelin/cholesterol (2:2:1)-supported bilayers on a glass-supported mica sheet. (e, f) 3D images zoomed into the rectangular ROIs in the previous two images (scale bar 100 nm). (g, h) Normalized local Lo domain changes over the course of the incubation, comparing enzyme-free areas (red; n = 3 ROIs) versus areas adjacent to high-density enzyme areas (blue; n = 4 ROIs).
A few other differences are apparent when comparing the AFM images over time (Figure 2b,d). The first is that the lipid loss defect dark regions in Figure 2b have mostly disappeared in Figure 2d due to bilayer reorganization after ∼1.5 h at 25 °C. Second, the interstitial regions between the enzymes appear to be completely filled by lipids of height consistent with Lo microdomains. To further quantify this Lo domain reorganization phenomena, we examined both (1) enzyme-free area Lo domain areas and (2) Lo domain areas positioned adjacent to enzyme-rich areas over the time points for images 2a−d, displayed in Figure 2g,h. The area occupancy of the Lo phase calculated at different time points suggests that Lo phases are unchanged after 3 h in the enzyme-free areas (red trace points in Figure 2g), implying that Lo/Ld phases reached an equilibrium over this period. However, in contrast, in the enzyme-adjacent Lo, blue trace points in Figure 2h show a marked decrease of ∼20% indicative of shrinkage of the adjacent Lo domains where the lipids and cholesterol are transported into the enzyme-rich regions (Figure S3). The lipid domain rearrangement suggests that the enzyme−lipid bilayer system elicits molecular reorganization to minimize the local free energy in the enzyme-rich regions. Furthermore, we compare two successive time points after the addition of blank γ-secretase buffer (the same detergent-containing buffer but without γ-secretase) to a preformed Lo/Ld-supported lipid bilayer (at 18 and 143 min of incubation) (Figure S4). Inspection of the overlays shows that only minimal differences in domain structure occurred at these successive time points and no defect structures resulted from blank γ-secretase buffer incubation (Figure S4). These control experiments using the same buffer without γ-secretase have no effect on domain reorganization, supporting our conclusion that the direct insertion of γ-secretase into lipid bilayer induces the phase-separated lipid reorganization.
In addition, we measured the hydrodynamic radius of γ-secretase by dynamic light scattering (Figure S5). We found that the majority of the complexes have a diameter of 10 nm consistent with the monomeric size of the γ-secretase complex.31
γ-Secretase Complex Is Spatially Distributed in Both Lo and Ld Phases as a Single Enzyme Complex or High-Order Assemblies After Direct Insertion.
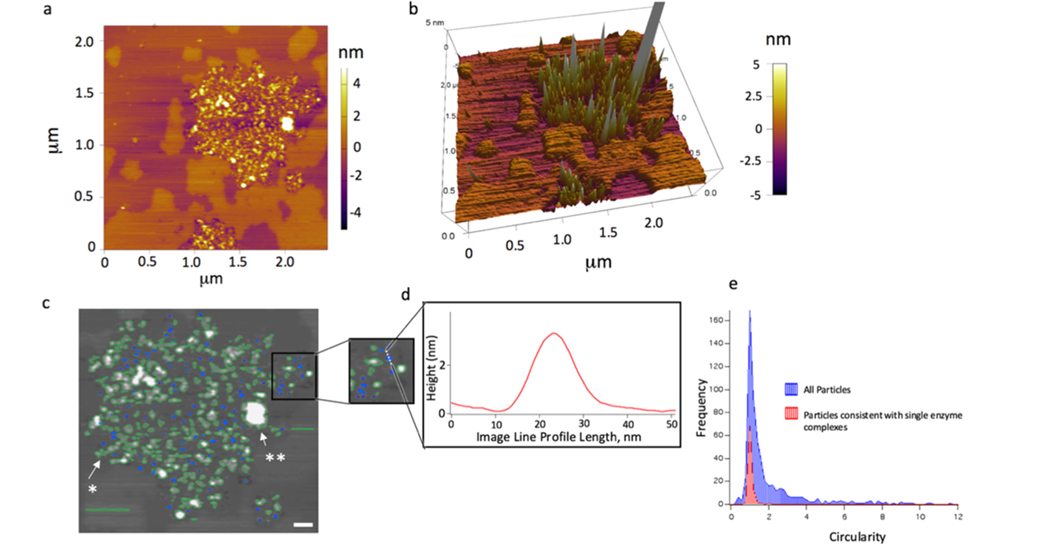
Furthermore, analysis of the distribution of γ-secretase single enzyme complexes shows the presence in both the Lo and Ld phases of γ-secretase (Figure 3a,b). In particular, the molecular volume of protein particles protruding from the supported membrane surface was determined from tapping mode AFM images. A molecular volume analysis framework was assessed based on the heights and widths of the protruding protein features that are compared to predicted volumes from the molecular mass of the extramembrane protein portion.21,22 Under conditions of direct membrane protein insertion, two subsets of features are observed: (1) single membrane protein complexes and (2) high-order assemblies (Figure 3c). A region of direct insertion of γ-secretase into preformed DOPC/SM/ chol (2:2:1)-supported bilayers on a glass-supported mica sheet is overlaid with the results of a particle localization analysis tool in Igor Pro (Figure 3c; blue overlay). The subsets of blue labeled features are small, circular features consistent with single γ-secretase complexes. The feature shown in the inset and height profile has an approximate volume of 149.5 nm3 (Figure 3d). To choose the features consistent with single enzyme complexes, the circularity was chosen as a filter. A histogram of the distribution of enzyme circularity shows that only a small fraction is consistent with monomeric γ-secretase (Figure 3e).
Figure 3.
Feature volume analysis of AFM height images of the γ-secretase complex. (a) Dispersion of enzyme complexes within a larger area at longer incubation time (t = 283 min of incubation time at 25 °C). (b) 3D rendering graph of dispersed single enzyme complexes and their positioning in both the Lo and Ld phases. (c) Overlaid images with the results of a particle localization analysis tool in Igor Pro (blue and green overlays). (d) Inset is zoomed in to a smaller ROI and an image line profile of the dotted line in the inset. The feature shown has an approximate volume of 149.5 nm3. The blue labeled features are small, circular, and consistent with single γ-secretase complexes (scale bar 100 nm). To choose the features consistent with the single enzyme complex, the circularity was chosen as a filter (circularity: the ratio of the square of the perimeter to (4 × π × area)). This value approaches 1 for a perfect circle. (e) Histogram of the single enzyme feature circularity (red) with the histogram of all of the features present in a 4 × 4 μm2 image (blue).
In contrast, some representative features consistent with multimembrane protein oligomers and chains are indicated by a solid arrow and a single asterisk, and large-scale aggregates are indicated by an arrow and double asterisks (Figure 3c). Oligomers, in this case, pertain to a chain of approximately 5−10 enzymes with similar heights, whereas the large-scale aggregate feature has an extended height and area that most likely results from >50 enzymes that form a structure that is adsorbing more complexes over time and growing in size. We note that the chain features are also consistent with individual enzymes at a near-direct contact at this lateral resolution.
APP and Notch Substrates Partition into Different Domains of Lipid Bilayer.
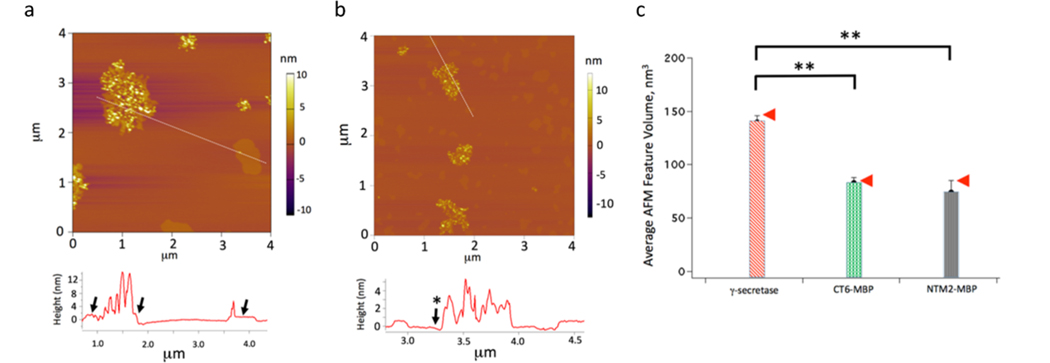
Direct insertion of the substrates SB4 (APP substrate) and NTM2 (Notch1 substrate) fused with maltose-binding protein (MBP)19 was imaged at the same experimental conditions as for γ-secretase−substrate buffer. The MBP-SB4 and MBP-NTM2 substrates show remarkably different distributions (Figure 4a,b). It is clear that SB4 is surrounded by the Lo phase, while NTM2 favors the Ld phase, which is highlighted when generating the height profiles (Figure 4a). The region with a high concentration of MBP-NTM2 exists standalone (Figure 4b) and does not contain the perceptible Lo phase at the edges as it was seen for MBP-SB4 (Figure 4a and accompanying image height profile). Furthermore, essentially none of the ∼50 Lo phase regions in Figure 4b contain MBP-NTM2, whereas all but one in Figure 4a contain MBP-SB4 with the flat Lo features adjacent to the substrate. This is supported by the line profile in Figure 4a, where the arrows indicate the Lo regions adjacent to the regions where the MBP-SB4 is localized. In contrast, these Lo regions are absent in the NTM2-MBP case, as indicated by the line profile in Figure 4b by the arrow with the asterisk.
Figure 4.
APP and Notch substrates locate in different domains of the lipid bilayer. (a) Insertion of MBP-SB4 into the model membrane; the image was taken at 31 °C after the addition of the substrate for 150 min. The image line profile is obtained from the indicated line segment. (b) Insertion of MBP-NTM2 into the model membrane; the image was taken at 31 °C after the addition of the substrate for 150 min. (c) Bar chart obtained from computing the averae AFM feature volume for γ-secretase (n = 86), MBP-SB4 (n = 50), and MBP-NTM2 (n = 50) for the subset of features consistent with arising from single molecules protruding from the supported membrane. The arrows indicate, for reference, the predicted volume of protruding γ-secretase subunit nicastrin (NCT; 147 nm3/molecule) and MBP (85 nm3/molecule) based on polymer size arguments (Figure S6, the Supporting Information section). The double asterisks in Figure 4c indicate statistically significant differences between the average feature volumes (p < 0.05).
The computed average AFM structure volume for γ-secretase, MBP-SB4, and MBP-NTM2 for the subset of features arising from single MBP structures protruding from the supported membrane is shown in Figure 4c. The red arrows indicate, for reference, the predicted volume of protruding γ-secretase subunit nicastrin (147 nm3/molecule) and MBP (85 nm3/molecule) based on polymer size versus chain length arguments.21,22 In each instance, we see that the average volume of the features is consistent with the predicted volume of nicastrin and MBP in the γ-secretase, MBP-SB4, and MBP-NTM2 cases, 139.6 ± 6.1, 82.8 ± 5.2, and 73.9 ± 11.2 nm3, respectively. Thus, we have achieved the direct measurement of individual γ-secretase and substrate molecules embedded in supported bilayers for the first time, to the best of our knowledge, and determined their distribution within local membrane environments.
γ-Secretase Has Distinct Activity for APP and Notch Substrates in Various Lipid Membrane Environments.
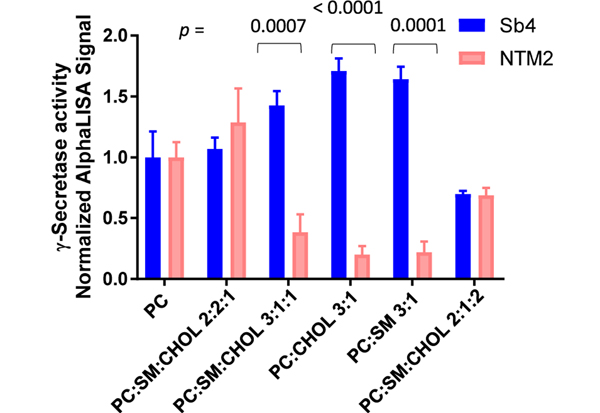
To directly examine the effect of lipids on γ-secretase activity toward APP and Notch, NTM2 and SB4 are added to the same reaction. The purified γ-secretase activity was measured for both substrates and normalized by the total activity in the DOPC-only mixture. Lipid environment has a major influence on γ-secretase activity, an effect previously observed.9,10 Interestingly, we were able to identify the preference of γ-secretase to cleave one of the substrates contingent on which lipid mixture the system was embedded in (Figure 5). In lipid mixtures containing cholesterol, sphingomyelin, or both at concentrations lower than a molar ratio of 1:4, γ-secretase preferentially cleaves APP. That result is consistent with our observation that APP substrates partition preferentially in cholesterol-rich membrane phases or more ordered systems. When the lipid mixture mimics a more fluid system, the cleavage of the Notch substrate becomes more favorable.
Figure 5.
Effect of different lipid mixtures on γ-secretase for APP and Notch1 substrates. The assay was assayed as previously described. (Experiments were performed as triplicates, mean ± SD; p values are displayed on top of the graph.).
DISCUSSION
Multiple lines of evidence, from the organism scale to the molecular level, have implicated changes in the lipid microenvironment and cholesterol-rich membrane microdomains, termed lipid rafts, in the regulation of substrate cleavage by γ-secretase.32−34 Enzyme activity and cleavage specificity have been shown to be cholesterol- and lipid-dependent,9,35 but it is unclear whether the interactions are specific, involving tight binding to well-defined regions of the enzyme complex or if the effect is through physical changes of the surrounding membrane microenvironment. We note that previous methods do not allow for the direct in situ quantification of enzyme and substrate partitioning in the context of intact membranes with cholesterol-containing phase-separated lipid domains. Therefore, in this work, we merged cholesterol-rich domain formation with our methods to investigate γ-secretase and its substrates in supported membranes.36,37 This has enabled the visualization of the phase partitioning of active γ-secretase and its substrates in micron-scale domains in model-supported membranes using AFM. Using this system, we have found that (1) the APP and Notch substrates preferably partition into Lo and Ld domains of the lipid bilayer, respectively; (2) γ-secretase exists as a single enzyme complex or high-order assemblies that localizes in both Lo and Ld domains; however, γ-secretase favors the Lo phase; and (3) regions of high γ-secretase concentration elicit lipid reorganization and cause the depletion of adjacent Lo domains. Finally, we have shown that γ-secretase specificity against APP and Notch substrates can be modulated by lipids.
Direct insertion combined with AFM provides a novel window into examining how γ-secretase sequesters lipids and partitions into a phase-separated model Lo/Ld system. It is strikingly apparent from these experiments that the phase partitioning of these MPs is distinctly different and there is a microenvironmental selection preference built into the sequence and structure of the substrates and also the enzyme itself.38 It makes intuitive sense that nature would encode and employ such a regulation mechanism, especially given that γ-secretase is known to cleave a variety of transmembrane proteins with no apparent transmembrane sequence dependence. Considering the extremely large hydrophobic interaction area of the enzyme, γ-secretase itself alters its local membrane microenvironment as part of its catalytic function as we observed.
Since γ-secretase−substrate cleavage is not dictated by protein sequence, regulation via raft domain sequestration is also likely to play an important role in function. Our finding reveals that γ-secretase strongly influences local membrane microdomain structure and it follows that the phase heterogeneity and enzyme local concentration cannot be ascribed to the influence of lipids alone in phase-separated biomembranes. There has been much debate about this interplay of lipid thermodynamics versus membrane protein influence in dominating and dictating phase separation and protein sequestration.39−42 It has been established that phase sequestration of lipid-anchored raft proteins such as RAS is indeed likely dominated by lipid thermodynamics, as evidenced in AFM studies in model systems where these proteins sequester at Lo/Ld interfaces as “lineactants” in some cases.43,44 However, due to the massive hydrophobic interaction area of the γ-secretase complex, we see direct evidence in our studies that the presence of this membrane protein is dominating the thermodynamics of the local biomembrane microenvironment, leading to domain reorganization and concentration in the Lo phase.
Furthermore, in parallel, since there are different thicknesses between the Lo and Ld domains and the supported bilayers contain regions consistent with lipid loss from the direct insertion process, these discontinuities of the bilayer at Lo/Ld domain edges and loss defects result in regions of high interfacial line tension. Line tension at phase boundaries and defects has been recognized as an essential parameter that controls the sizes of lipid domains and the kinetics of phase separation and thus influences membrane protein sequestration.5,45 It has been postulated that biological systems make use of modulations in line tension to co-ordinate cell processes by regulation of compositional heterogeneity. For the γ-secretase system, this line tension along with protein−lipid and protein−protein interactions modulates the thermodynamics of lipid mixing and phase separation leading to moderately packed enzyme complexes within cholesterol-rich Lo domains. Our data is consistent with the idea that hydrophobic matching could be involved in driving enzyme concentration in the Lo phase, and thus the process of equilibration from a temperature above Lo/Ld phase coexistence causes domain reorganization that leads to the uptake and incorporation of local Lo domains adjacent to the enzyme-rich areas. In contrast, we note that in the NTM2 substrate-partitioning case, even though the substrates are densely packed, the MPs are located in the Ld domain, as indicated by the interstitial thicknesses.
The interactions of γ-secretase with lipids and cholesterol have been probed beyond biochemical detergent solubilization assays through other methods, including cleavage assays, cryo-EM structure studies, and molecular dynamics simulations. This data, taken in the context of the lipid bilayer elasticity theory,46 indicates that there are processes that work in parallel involving these interactions that lead to adjacent Lo domain uptake and incorporation to form enzyme complex-rich areas. The first process involves protein−lipid interactions that occur when the replacement and configuration of the lipid chains in the first or annular boundary shell at the protein perimeter are achieved after the direct insertion of the enzyme complexes. In this scenario, the native, cell-derived lipids have been, for the most part, depleted by detergent solubilization and purification prior to direct insertion. The annular boundary lipid shell number or protein-to-lipid stoichiometry, NS, is ∼68 when estimated by counting the number in a model formed from the structure of Bai et al.26 This is a large number and greater than 1.4 times the number estimated by the relation introduced by Marsh47 that is based on the number of membrane-spanning helices (nα = 18; NS = 48) and greater than that of even large MPs such as cytochrome c oxidase48 and the nicotinic acetylcholine receptor,49 since presumably essentially all of the helices of the complex interact with lipid. However, under these conditions, given the sheer number of molecules involved in the adjacent Lo domain reorganization, this mechanism is contributing but not likely driving the process.
The second process is the selective sorting of Lo phase lipids and cholesterol to fill in the interstitial regions between enzyme complexes driven by hydrophobic forces and thickness matching. It has been shown by both theory and experiment that deformations in lipid bilayer thickness induced by proteins can yield energetically favorable bilayer-mediated interactions between integral membrane proteins.50 Furthermore, this process has been shown to lead to the large-scale organization of integral membrane proteins into protein-rich regions like the ones evidenced here. Precedents of clustering or concentration of membrane-associated proteins into phase-separated domains have been seen in various systems studied with AFM.44,51,52 We note that the γ-secretase AFM results obtained by direct insertion reported here bear a striking resemblance to the negatively stained EM of the enzyme reconstituted in exogeneous Escherichia coli polar lipids by Ayciriex et al.53 In both cases, the enzyme is reconstituted into the membranes into moderately packed and individually identifiable complexes in regions with area fractions greater than ∼50%. However, in our measurements, we are able to visualize single enzyme complexes in both the Lo and Ld phases with interstitial thicknesses experimentally observed for the first time (see Figure 3). The other mechanism that could be driving the direct adjacent Lo domain uptake and reorganization is the direct binding of cholesterol or lipids to the complex.54,55 Of the six structures obtained so far by cryo-EM at high resolution, even though the structures were not obtained in intact lipid bilayers, a number of them contain bound lipids and cholesterol in the structure.56,57 Most notably, the Notch-bound γ-secretase structure contains three cholesterols and two PC molecules.3 These components that have remained bound to the detergent-solubilized protein during purification are therefore likely candidates for structurally or functionally significant interactions and would be resequestered by the enzyme complex after insertion if they were lost during purification. Molecular dynamics has been used to expand on the high-resolution structural data and use these structures as starting conformations to observe the preferential membrane thicknesses and lipid- and cholesterol-binding propensities.58,59 It should be noted that these MD studies have so far been limited to configurations of individual γ-secretase enzymes with substrates at tens of picosecond time durations in atomistic and coarse-grained simulations with set compositions in small regions. Nonetheless, in recent MD studies by Aguayo-Ortiz et al., up to seven potential cholesterol-binding sites were identified by per amino acid occupancy contacts of cholesterol over the duration of the simulations, some of which overlapped with the four phosphatidylcholine lipid-binding sites identified using the same methodology.59 These findings, along with the cryo-EM-binding site evidence, potentially provide an additional underlying cholesterol-binding mechanistic reason for the adjacent Lo domain uptake and incorporation to form protein-rich areas evidenced in our AFM time-lapse data. Our studies underscore how AFM is highly complementary to cryo-EM and MD, as this method enables studies of large numbers of enzymes over variable temperatures in large (10 × 10 μm2) regions at a single-molecule resolution over hours in intact lipid bilayers, allowing for capturing the time evolution of the phase sequestration and domain reorganization process, as well as the lipid bilayer thickness adjacent to the enzymes and substrates.
With regard to the substrates, solution NMR studies of unaltered MPs in micelles and bicelles indicate that there is a cholesterol-binding motif built into the structure of the β-C-terminal fragment (β-CTF) of APP, presumably leading to cholesterol phase partitioning of the substrate.16 In contrast, Notch-TMD shows stark and γ-secretase (detected in the form of PS1-NTF (presenilin-1)) resides in the raft domains.32,60 It is important to point out that in this work we are using distinctly different species in terms of both substrate primary sequence and the aforementioned presence of detection moieties based on MBP. SB4 is based on C100 and contains an N-terminal AviTag sequence for biotin ligation and a FLAG epitope tag at the C-terminus, truncating the native sequence close to the proposed cholesterol-binding region of C99.61 In addition, our Notch NTM2 substrate contains the Notch intracellular domain (NICD) and an AviTag at the C-terminus for biotin ligation.62 Nevertheless, we do detect major differences in the SB4 and NTM2 phase partitionings in the AFM studies. The APP-derived MBP-SB4 distinctly partitions within the Lo phase and accumulates Lo phase lipid domains on the perimeter of the MP-rich region in the AFM imagery. Conversely, the MBP-NTM2 forms concentrated regions isolated as islands within Ld phase domains. More subtle factors related to the extramembrane substrate sequence play a role in phase partitioning as native sequences of both Notch1 and APP. For example, the cytosolic C-terminal domain of Notch1 contains a tentative membrane re-entrant segment (LWF)17 and APP (C99) contains a short surface-associated amphipathic helix that is connected to the TMD with a water-exposed loop in addition to the cholesterol-binding region.62
SUMMARY AND CONCLUSIONS
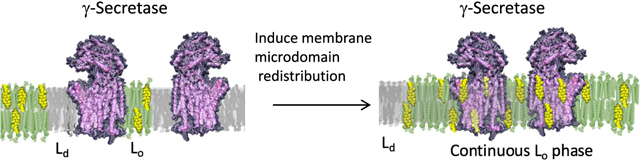
This work that directly visualizes γ-secretase and its substrates by AFM leads to important findings with major functional implications for intramembrane proteolysis. The direct insertion and equilibration of γ-secretase in Lo/Ld model systems lead to Lo phase uptake and concentrated regions of the enzyme in the ordered phase (Figure 6a,b). In essence, the enzyme complex’s hydrophobic interactions with cholesterol and lipids dominate the thermodynamics of the local biomembrane microenvironment, leading to domain reorganization and partitioning into the Lo phase. Furthermore, given that SB4(APP) and NTM2(Notch1) have stark differences in their association with the Lo phase suggests a thermodynamic regulation mechanism that would manifest in the sorting of SB4(APP) to regions proximal to the enzyme complex in the Lo phase and the exclusion of NTM2 (Notch2) from the Lo phase (Figure 6b). This is the first direct evidence of a lipid microdomain sorting mechanism that serves to modulate intramembrane proteolysis of diverse substrates in intact biomembranes. Overall, this work adds substantial direct visual evidence for a regulatory role of the lipid environment on γ-secretase activity from the standpoint of differential partitioning of substrates that conceivably leads to the control of activity and specificity.
Figure 6.
Model for γ-secretase sorting of substrates mediated by lipid microdomain. (a) Direct enzyme insertion followed by lipid reorganization and packing of enzyme complexes primarily in the Lo phase. (b) Subsequent incorporation and concentration of enzyme complex into the Lo phase (green) guide proteolysis by recruiting the cholesterol-rich ordered phase. Overall, the direct visualization of the phase localization of enzyme and substrates suggests a thermodynamic regulatory mechanism that manifests in the Lo/Ld sorting of enzyme and SB4(APP) shown in red and the exclusion of NTM2 (Notch2) shown in blue from the Lo phase.
Supplementary Material
ACKNOWLEDGMENTS
The authors acknowledge the National Institutes of Health and The National Science Foundation for the support of this research (NSF 1207480 and NIH S06GM008168–28 (M.L.G.), R01NS096275 (Y.-M.L.), RF1AG057593 (Y.-M.L.), U54CA137788/U54CA132378 (M.L.G. and Y.-M.L.), the JPB Foundation (Y.-M.L.), and The Cure Alzheimer’s Fund (S.L.W. and Y.-M.L.)). The authors also acknowledge William H. Goodwin and Alice Goodwin and the Commonwealth Foundation for Cancer Research, the Experimental Therapeutics Center of MSKCC, and the William Randolph Hearst Fund in Experimental Therapeutics.
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.langmuir.0c01178.
Estimation of the lipid shell surrounding γ-secretase (Figure S1); closer inspection of direct insertion events for the enzyme at 37 °C (Figure S2); direct observation of enzyme-adjacent Lo domain reorganization by AFM (Figure S3); AFM imagery of the impact of blank γ-secretase buffer on an existing Lo/Ld SLB (Figure S4); dynamic light scattering of solubilized γ-secretase in solution (Figure S5); and feature volume analysis of AFM height images of γ-secretase substrates (Figure S6) (PDF)
Complete contact information is available at: https://pubs.acs.org/10.1021/acs.langmuir.0c01178
The authors declare the following competing financial interest(s): LYM is co-inventor of intellectual property (assay for gamma secretase activity and screening method for gamma secretase inhibitors) licensed to Jiangsu Continental Medical Development.
Contributor Information
Marilia Barros, Chemical Biology Program, Memorial Sloan-Kettering Cancer Center, New York, New York 10065, United States.
William J. Houlihan, Department of Chemical Engineering and the Department of Biomedical Engineering, The City College of the City University of New York, New York, New York 10031, United States
Chelsea J. Paresi, Chemical Biology Program, Memorial Sloan-Kettering Cancer Center, New York, New York 10065, United States; Pharmacology Graduate Program, Weill Graduate School of Medical Sciences of Cornell University, New York, New York 10021, United States
Matthew Brendel, Molecular Cytology Core, Memorial Sloan-Kettering Cancer Center, New York, New York 10065, United States.
Kevin D. Rynearson, Department of Neurosciences, University of California, San Diego, California 92093, United States
Chang-wook Lee, Skaggs School of Pharmacy and Pharmaceutical Sciences and, University of California, San Diego, California 92093, United States.
Olga Prikhodko, Department of Neurosciences, University of California, San Diego, California 92093, United States.
Cristina Cregger, Department of Neurosciences, University of California, San Diego, California 92093, United States.
Geoffrey Chang, Skaggs School of Pharmacy and Pharmaceutical Sciences and and Department of Pharmacology, University of California, San Diego, California 92093, United States.
Steven L. Wagner, Department of Neurosciences, University of California, San Diego, California 92093, United States; Research Biologist, VA San Diego Healthcare System, La Jolla, California 92161, United States
M. Lane Gilchrist, Department of Chemical Engineering and the Department of Biomedical Engineering, The City College of the City University of New York, New York, New York 10031, United States.
Yue-Ming Li, Chemical Biology Program, Memorial Sloan-Kettering Cancer Center, New York, New York 10065, United States; Pharmacology Graduate Program, Weill Graduate School of Medical Sciences of Cornell University, New York, New York 10021, United States.
REFERENCES
- (1).De Strooper B. Aph-1, Pen-2, and Nicastrin with Presenilin generate an active gamma-Secretase complex. Neuron 2003, 38, 9–12. [DOI] [PubMed] [Google Scholar]
- (2).Haapasalo A; Kovacs DM The many substrates of presenilin/ gamma-secretase. J. Alzheimer’s Dis. 2011, 25, 3–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang G; Zhou R; Zhou Q; Guo X; Yan C; Ke M; Lei J; Shi Y. Structural basis of Notch recognition by human gamma-secretase. Nature 2019, 565, 192–197. [DOI] [PubMed] [Google Scholar]
- 4.Zhou R; Yang G; Guo X; Zhou Q; Lei J; Shi Y. Recognition of the amyloid precursor protein by human gamma-secretase. Science 2019, 363, No. eaaw0930. [DOI] [PubMed] [Google Scholar]
- 5.Lorent JH; Levental I. Structural determinants of protein partitioning into ordered membrane domains and lipid rafts. Chem. Phys. Lipids 2015, 192, 23–32. [DOI] [PubMed] [Google Scholar]
- 6.Pike LJ Lipid rafts: bringing order to chaos. J. Lipid Res. 2003, 44, 655–667. [DOI] [PubMed] [Google Scholar]
- 7.Levental KR; Lorent JH; Lin X; Skinkle AD; Surma MA; Stockenbojer EA; Gorfe AA; Levental I. Polyunsaturated Lipids Regulate Membrane Domain Stability by Tuning Membrane Order. Biophys. J. 2016, 110, 1800–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vetrivel KS; Thinakaran G. Membrane rafts in Alzheimer’s disease beta-amyloid production. Biochim. Biophys. Acta, Mol. Cell Biol. Lipids 2010, 1801, 860–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holmes O; Paturi S; Ye W; Wolfe MS; Selkoe DJ Effects of membrane lipids on the activity and processivity of purified gamma-secretase. Biochemistry 2012, 51, 3565–3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Osenkowski P; Ye W; Wang R; Wolfe MS; Selkoe DJ Direct and potent regulation of gamma-secretase by its lipid microenvironment. J. Biol. Chem. 2008, 283, 22529–22540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wahrle S; Das P; Nyborg AC; McLendon C; Shoji M; Kawarabayashi T; Younkin LH; Younkin SG; Golde TE Cholesterol-dependent gamma-secretase activity in buoyant cholesterol-rich membrane microdomains. Neurobiol. Dis. 2002, 9, 11–23. [DOI] [PubMed] [Google Scholar]
- 12.Ledesma MD; Abad-Rodriguez J; Galvan C; Biondi E; Navarro P; Delacourte A; Dingwall C; Dotti CG Raft disorganization leads to reduced plasmin activity in Alzheimer’s disease brains. EMBO Rep. 2003, 4, 1190–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hur JY; Teranishi Y; Kihara T; Yamamoto NG; Inoue M; Hosia W; Hashimoto M; Winblad B; Frykman S; Tjernberg LO Identification of novel gamma-secretase-associated proteins in detergent-resistant membranes from brain. J. Biol. Chem. 2012, 287, 11991–12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song Y; Mittendorf KF; Lu Z; Sanders CR Impact of bilayer lipid composition on the structure and topology of the transmembrane amyloid precursor C99 protein. J. Am. Chem. Soc. 2014, 136, 4093–4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song Y; Hustedt EJ; Brandon S; Sanders CR Competition between homodimerization and cholesterol binding to the C99 domain of the amyloid precursor protein. Biochemistry 2013, 52, 5051–5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barrett PJ; Song Y; Van Horn WD; Hustedt EJ; Schafer JM; Hadziselimovic A; Beel AJ; Sanders CR The amyloid precursor protein has a flexible transmembrane domain and binds cholesterol. Science 2012, 336, 1168–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deatherage CL; Lu Z; Kroncke BM; Ma S; Smith JA; Voehler MW; McFeeters RL; Sanders CR Structural and biochemical differences between the Notch and the amyloid precursor protein transmembrane domains. Sci. Adv. 2017, 3, No. e1602794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tian Y; Bassit B; Chau D; Li YM An APP inhibitory domain containing the Flemish mutation residue modulates gamma-secretase activity for Abeta production. Nat. Struct. Mol. Biol. 2010, 17, 151–158. [DOI] [PubMed] [Google Scholar]
- 19.Chau DM; Crump CJ; Villa JC; Scheinberg DA; Li YM Familial Alzheimer Disease Presenilin-1 Mutations Alter the Active Site Conformation of gamma-secretase. J. Biol. Chem. 2012, 287, 17288–17296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aller SG; Yu J; Ward A; Weng Y; Chittaboina S; Zhuo R; Harrell PM; Trinh YT; Zhang Q; Urbatsch IL; Chang G. Structure of P-glycoprotein reveals a molecular basis for poly-specific drug binding. Science 2009, 323, 1718–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schneider S; Larmer J; Henderson R; Oberleithner H. Molecular weights of individual proteins correlate with molecular volumes measured by atomic force microscopy. Pfluegers Arch. 1998, 435, 362–367. [DOI] [PubMed] [Google Scholar]
- 22.Saslowsky DE; Lawrence J; Ren X; Brown DA; Henderson RM; Edwardson JM Placental alkaline phosphatase is efficiently targeted to rafts in supported lipid bilayers. J. Biol. Chem. 2002, 277, 26966–26970. [DOI] [PubMed] [Google Scholar]
- 23.Milhiet PE; Gubellini F; Berquand A; Dosset P; Rigaud JL; Le Grimellec C; Levy D. High-resolution AFM of membrane proteins directly incorporated at high density in planar lipid bilayer. Biophys. J. 2006, 91, 3268–3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khadka NK; Ho CS; Pan J. Macroscopic and Nanoscopic Heterogeneous Structures in a Three-Component Lipid Bilayer Mixtures Determined by Atomic Force Microscopy. Langmuir 2015, 31, 12417–12425. [DOI] [PubMed] [Google Scholar]
- 25.Heberle FA; Petruzielo RS; Pan J; Drazba P; Kucerka N; Standaert RF; Feigenson GW; Katsaras J. Bilayer thickness mismatch controls domain size in model membranes. J. Am. Chem. Soc. 2013, 135, 6853–6859. [DOI] [PubMed] [Google Scholar]
- 26.Bai XC; Yan C; Yang G; Lu P; Ma D; Sun L; Zhou R; Scheres SH; Shi Y. An atomic structure of human gamma-secretase. Nature 2015, 525, 212–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stansfeld PJ; Goose JE; Caffrey M; Carpenter EP; Parker JL; Newstead S; Sansom MS MemProtMD: Automated Insertion of Membrane Protein Structures into Explicit Lipid Membranes. Structure 2015, 23, 1350–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Edholm O; Nagle JF Areas of molecules in membranes consisting of mixtures. Biophys. J. 2005, 89, 1827–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hofsäß C; Lindahl E; Edholm O. Molecular dynamics simulations of phospholipid bilayers with cholesterol. Biophys. J. 2003, 84, 2192–2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pandit SA; Vasudevan S; Chiu SW; Mashl RJ; Jakobsson E; Scott HL Sphingomyelin-cholesterol domains in phospholipid membranes: atomistic simulation. Biophys. J. 2004, 87, 1092–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun L; Zhao L; Yang G; Yan C; Zhou R; Zhou X; Xie T; Zhao Y; Wu S; Li X; Shi Y. Structural basis of human gamma-secretase assembly. Proc. Natl. Acad. Sci. U.S.A. 2015, 112, 6003–6008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vetrivel KS; Cheng H; Kim SH; Chen Y; Barnes NY; Parent AT; Sisodia SS; Thinakaran G. Spatial segregation of gamma-secretase and substrates in distinct membrane domains. J. Biol. Chem. 2005, 280, 25892–25900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hur JY; Welander H; Behbahani H; Aoki M; Franberg J; Winblad B; Frykman S; Tjernberg LO Active gamma-secretase is localized to detergent-resistant membranes in human brain. FEBS J. 2008, 275, 1174–1187. [DOI] [PubMed] [Google Scholar]
- 34.Urano Y; Hayashi I; Isoo N; Reid PC; Shibasaki Y; Noguchi N; Tomita T; Iwatsubo T; Hamakubo T; Kodama T. Association of active gamma-secretase complex with lipid rafts. J. Lipid Res. 2005, 46, 904–912. [DOI] [PubMed] [Google Scholar]
- 35.Winkler E; Kamp F; Scheuring J; Ebke A; Fukumori A; Steiner H. Generation of Alzheimer disease-associated amyloid β42/ 43 peptide by γ-secretase can be inhibited directly by modulation of membrane thickness. J. Biol. Chem. 2012, 287, 21326–21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ahn K; Shelton CC; Tian Y; Zhang X; Gilchrist ML; Sisodia SS; Li YM Activation and intrinsic gamma-secretase activity of presenilin 1. Proc. Natl. Acad. Sci. U.S.A. 2010, 107, 21435–21440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gilchrist ML; Ahn K; Li YM Imaging and Functional Analysis of γ-Secretase and Substrate in a Microsphere-Supported Biomembrane System. Anal. Chem. 2016, 88, 1303–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lorent JH; Diaz-Rohrer B; Lin X; Spring K; Gorfe AA; Levental KR; Levental I. Structural determinants and functional consequences of protein affinity for membrane rafts. Nat. Commun. 2017, 8, No. 1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leslie M. Do Lipid Rafts Exist?; American Association for the Advancement of Science, 2011. [Google Scholar]
- 40.Gurry T; Kahramanoğulları O; Endres RG Biophysical mechanism for ras-nanocluster formation and signaling in plasma membrane. PLoS One 2009, 4, No. e6148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Destainville N. An alternative scenario for the formation of specialized protein nano-domains (cluster phases) in biomembranes. Europhys. Lett. 2010, 91, No. 58001. [Google Scholar]
- 42.Schmid F. Physical mechanisms of micro-and nanodomain formation in multicomponent lipid membranes. Biochim. Biophys. Acta, Biomembr. 2017, 1859, 509–528. [DOI] [PubMed] [Google Scholar]
- 43.Weise K; Kapoor S; Denter C; Nikolaus Jr.; Opitz N; Koch S; Triola G; Herrmann A; Waldmann H; Winter R. Membrane-mediated induction and sorting of K-Ras microdomain signaling platforms. J. Am. Chem. Soc. 2011, 133, 880–887. [DOI] [PubMed] [Google Scholar]
- 44.Erwin N; Sperlich B; Garivet G; Waldmann H; Weise K; Winter R. Lipoprotein insertion into membranes of various complexity: lipid sorting, interfacial adsorption and protein clustering. Phys. Chem. Chem. Phys. 2016, 18, 8954–8962. [DOI] [PubMed] [Google Scholar]
- 45.García-Sáez AJ; Chiantia S; Schwille P. Effect of line tension on the lateral organization of lipid membranes. J. Biol. Chem. 2007, 282, 33537–33544. [DOI] [PubMed] [Google Scholar]
- 46.Seifert U. Configurations of fluid membranes and vesicles. Adv. Phys. 1997, 46, 13–137. [Google Scholar]
- 47.Marsh D. Stoichiometry of lipid-protein interaction and integral membrane protein structure. Eur. Biophys. J. 1997, 26, 203–208. [Google Scholar]
- 48.Knowles PF; Watts A; Marsh D. Spin-label studies of lipid immobilization in dimyristoylphosphatidylcholine-substituted cytochrome oxidase. Biochemistry 1979, 18, 4480–4487. [DOI] [PubMed] [Google Scholar]
- 49.Ellena JF; Blazing MA; McNamee MG Lipid-protein interactions in reconstituted membranes containing acetylcholine receptor. Biochemistry 1983, 22, 5523–5535. [DOI] [PubMed] [Google Scholar]
- 50.Kahraman O; Koch PD; Klug WS; Haselwandter CA Bilayer-thickness-mediated interactions between integral membrane proteins. Phys. Rev. E 2016, 93, No. 042410. [DOI] [PubMed] [Google Scholar]
- 51.Milhiet PE; Giocondi MC; Baghdadi O; Ronzon F; Roux B; Le Grimellec C. Spontaneous insertion and partitioning of alkaline phosphatase into model lipid rafts. EMBO Rep. 2002, 3, 485–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Seeger HM; Bortolotti CA; Alessandrini A; Facci P. Phase-transition-induced protein redistribution in lipid bilayers. J. Phys. Chem. B 2009, 113, 16654–16659. [DOI] [PubMed] [Google Scholar]
- 53.Ayciriex S; Gerber H; Osuna GM; Chami M; Stahlberg H; Shevchenko A; Fraering PC The lipidome associated with the γ-secretase complex is required for its integrity and activity. Biochem. J. 2016, 473, 321–334. [DOI] [PubMed] [Google Scholar]
- 54.Smith AW Lipid−protein interactions in biological membranes: a dynamic perspective. Biochim. Biophys. Acta, Biomembr. 2012, 1818, 172–177. [DOI] [PubMed] [Google Scholar]
- 55.Jung JI; Price AR; Ladd TB; Ran Y; Park H-J; Ceballos-Diaz C; Smithson LA; Hochhaus G; Tang Y; Akula R; et al. Cholestenoic acid, an endogenous cholesterol metabolite, is a potent γ-secretase modulator. Mol. Neurodegener. 2015, 10, No. 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lu P; Bai X.-c.; Ma D; Xie T; Yan C; Sun L; Yang G; Zhao Y; Zhou R; Scheres SH Three-dimensional structure of human γ-secretase. Nature 2014, 512, 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sun L; Zhao L; Yang G; Yan C; Zhou R; Zhou X; Xie T; Zhao Y; Wu S; Li X; Shi Y. Structural basis of human γ-secretase assembly. Proc. Natl. Acad. Sci. U.S.A. 2015, 112, 6003–6008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Grouleff J; Irudayam SJ; Skeby KK; Schiøtt B. The influence of cholesterol on membrane protein structure, function, and dynamics studied by molecular dynamics simulations. Biochim. Biophys. Acta, Biomembr. 2015, 1848, 1783–1795. [DOI] [PubMed] [Google Scholar]
- 59.Aguayo-Ortiz R; Straub JE; Dominguez L. Influence of membrane lipid composition on the structure and activity of γ-secretase. Phys. Chem. Chem. Phys. 2018, 20, 27294–27304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vetrivel KS; Cheng H; Lin W; Sakurai T; Li T; Nukina N; Wong PC; Xu H; Thinakaran G. Association of gamma-secretase with lipid rafts in post-Golgi and endosome membranes. J. Biol. Chem. 2004, 279, 44945–44954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shelton CC; Tian Y; Frattini MG; Li YM An exo-cell assay for examining real-time gamma-secretase activity and inhibition. Mol. Neurodegener. 2009, 4, No. 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chau D; Shum D; Radu C; Bhinder B; Gin D; Gilchrist ML; Djaballah H; Li Y-M A Novel High-Throughput 1536-well Notch1 γ-Secretase AlphaLISA Assay. Comb. Chem. High Throughput Screening 2013, 16, 415–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.