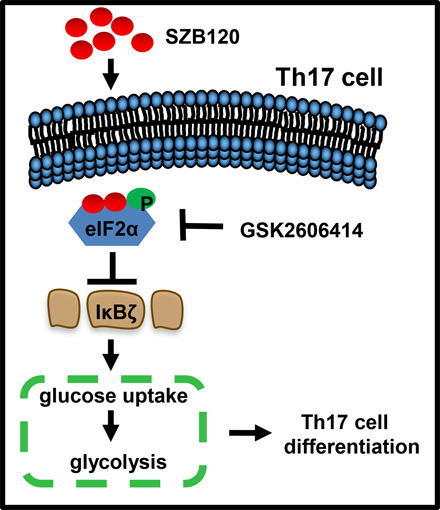
Key Points
SZB120 inhibits Th17 cell differentiation by enhancing eIF2α phosphorylation.
SZB120 efficiently alleviates autoimmune diseases involving the CNS and skin.
Visual Abstract
Abstract
IL-17–secreting Th17 cells play an important role in the pathogenesis of various inflammatory and autoimmune diseases. IL-17–targeted biologics and small molecules are becoming promising treatments for these diseases. In this study, we report that SZB120, a derivative of the natural compound 3-acetyl-β-boswellic acid, inhibits murine Th17 cell differentiation by interacting with the α-subunit of eukaryotic initiation factor 2 (eIF2α). We showed that SZB120 directly interacts with eIF2α and contributes to serine 51 phosphorylation of eIF2α. The suppressive effect of SZB120 on Th17 cell differentiation was reversed by GSK2606414, an inhibitor of eIF2α phosphokinase. Phosphorylation of eIF2α induced by SZB120 decreased the protein expression of IκBζ, which is important for Th17 cell differentiation. Notably, interaction with eIF2α by SZB120 also impaired glucose uptake and glycolysis in T cells. In vivo, SZB120 treatment of C57BL/6 mice significantly attenuated IL-17/Th17–mediated autoimmune disease. Our study indicates that SZB120 is a promising drug candidate for IL-17/Th17–mediated inflammatory diseases.
Introduction
T helper 17 cells were identified in 2005 and were distinguished by their ability to produce multiple proinflammatory cytokines, including IL-17 (IL-17A), IL-17F, and IL-22 (1). Thus, Th17 cells play critical roles in mucosal defense and the development of inflammatory diseases such as psoriasis, multiple sclerosis, and Crohn disease (2, 3). The complete differentiation of Th17 cells requires networks of multiple transcription factors (TFs), including RORγt, STAT3, IFN regulatory factor 4, BATF, IκBζ, c-Maf, Runx1, and Ahr (4–10). In addition, naive cells that develop into effector or regulatory T cell lineages require distinct metabolic pathways. Notably, metabolic characteristics, such as enhanced glycolysis, de novo fatty acid synthesis, and glutaminolysis, favor Th17 cell differentiation (11–13). Similarly, activation of mammalian target of rapamycin (mTOR) or mTOR-dependent hypoxia-inducible factor 1α (HIF-1α), which results in increased aerobic glycolysis, leads to specific induction of Th17 cells (14). Treatment with an activator of AMP-activated kinase (AMPK), another metabolic sensor with functions opposing those of mTOR, strongly impaired Th17 cell generation (15). The specific molecular signaling network and cellular metabolism signatures provide ways to manipulate Th17 cell differentiation.
Eukaryotic initiation factor 2 (eIF2) α is one subunit of eIF2. Phosphorylation of the eIF2α subunit (p-eIF2α), which represses the initiation phase of protein synthesis, is a central mechanism for translational control for all eukaryotic cells (16). Beyond its well-known effects, the most important function of p-eIF2α is the downregulation of protein synthesis and translation. This subunit is also related to cell cycle progression and memory consolidation (17). p-eIF2α is implicated in a variety of major diseases, such as cancers, diabetes, and neurodegenerative diseases (18–20). However, the relationship between eIF2α and immune cells is largely unknown.
Boswellic acids (BAs), the main bioactive elements of frankincense, have been used as traditional medicine to treat rheumatic disorders, asthma, Crohn disease, and collagenous colitis for centuries (21). The six major BAs are αBAs and βBAs, acetylated αBAs and acetylated βΒΑs, 11-keto-β-BA (KBA), and 3-acetyl-KBA (AKBA), which have therapeutic potential because of their anti-inflammatory and anticancer activities (22). To date, most studies on their mechanisms of action have focused on AKBA and KBA. For example, AKBA inhibits several enzymes, including MAT2A, 5-lipoxygenase, and cyclooxygenase-1 (23–25), and interferes with signaling pathways such as NF-κB and Ca2+ signaling (26). However, acetylated αBA and acetylated βBA (AβBA) shows strong clinical potential with a higher peak concentration value than KBA and AKBA after oral administration (27). Therefore, a series of ABA derivatives were designed and synthesized to improve the biological activity and bioavailability. In this study, the molecular target and mechanism of action of an AβBA derivative, which we termed SZB120, were characterized, and the anti-inflammatory activities were evaluated in vitro and in vivo. Our data showed that SZB120 inhibits Th17 cell differentiation by directly binding to eIF2α and elevating p-eIF2α. We also found SZB120 significantly restrains the glucose uptake capacity and glycolytic metabolism of Th17 cells in a dose-dependent manner. SZB120 effectively diminished IL-17 production in Th17 cells in two mouse models, the experimental autoimmune encephalomyelitis (EAE) model, an animal model of multiple sclerosis, and an imiquimod (IMQ)-induced mouse model of psoriasis. These findings indicate the clinical potential of SZB120 in IL-17/Th17–relevant autoimmune and autoinflammatory diseases and identify eIF2α as a potential target for regulating Th17 cell differentiation.
Materials and Methods
SZB120
A mixture of AKBA (0.5 g, 1 mmol) and 3 ml of thionyl chloride was placed in a round-bottom flask. The reaction mixture was heated at 90°C for 1 h and dried under a high vacuum. Dichloromethane (10 ml) and triethylamine (0.3 g, 3 mmol) were then added to the reaction mixture, and 2-amino-1,3-propanediol (0.27 g, 3 mmol) was slowly added to the reaction mixture at 0°C. Next, the mixture was stirred until the reaction was completed at 25°C. After concentration in vacuo, the residue was subjected to silica gel column chromatography to obtain SZB120 as a white solid (0.35 g, 61.4%). We characterized SZB120 as follows: 1H-nuclear magnetic resonance (400 MHz, CDCl3); δ: 6.37–6.35 (doublet, 1 proton), 5.36 (triplet, 1 proton), 5.15 (triplet, 1 proton), 3.95–3.93 (multiplet, 1 proton), 3.87–3.82 (multiplet, 2 protons), 3.1 (broad, 2 protons), 3.76–3.70 (multiplet, 2 protons), 2.23 (multiplet, 1 proton), 2.10 (singlet, 3 protons), 2.05–1.97 (multiplet, 2 protons), 1.97–1.75 (multiplet, 6 protons), 1.65–1.58 (multiplet, 3 protons), 1.52–1.38 (multiplet, 6 protons), 1.33–1.26 (multiplet, 5 protons), 1.24–1.15 (multiplet, 5 protons), 1.12 (singlet, 3 protons), 1.05–0.99 (multiplet, 4 protons), 0.97–0.81 (multiplet, 9 protons), 0.80 (singlet, 3 protons), 0.79 (singlet, 3 protons); 13C-nuclear magnetic resonance (400 MHz, CDCl3); δ: 176.89, 170.40, 139.42, 124.63, 73.86, 63.34, 63.05, 59.18, 52.22, 50.37, 46.91, 46.82, 42.30, 41.51, 40.12, 39.77, 39.60, 37.51, 34.90, 33.82, 33.41, 31.26, 28.78, 28.12, 26.91, 26.52, 24.70, 23.87, 23.37, 23.24, 21.38, 20.04, 17.46, 16.85, and 13.33; liquid chromatography–mass spectrometry (electrospray ionization source) calculated (mass + proton), 572.5 m/z.
Mice
C57BL/6 (wild-type) mice (7–12 wk) were purchased from Shanghai Laboratory Animal Center. All mice were kept under specific pathogen–free conditions. IL-17A–GFP mice were purchased from the Jackson Laboratory (018472). All mice were randomly used for all experiments in compliance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals with the approval (SYXK-2003-0026) of the Scientific Investigation Board of Shanghai Jiao Tong University School of Medicine, Shanghai, China. To ameliorate any suffering of mice observed throughout these experimental studies, we euthanized the mice by CO2 inhalation.
IMQ-induced psoriatic mouse model
Male C57BL/6 mice received a daily topical dose of 55 mg if IMQ cream (5%) (Med-Shine Pharmaceutical, Chengdu, China) on the shaved back (2 × 2.5 cm2) or 25 mg per ear for four to six consecutive days to induce psoriasiform lesions. For SZB120 treatment, the mice were treated daily with 10 mg/kg/d SZB120 dissolved in 10% DMSO i.p. for four to six consecutive days. The control mice received the same volume of DMSO. On the fifth day, single-cell suspensions generated from the ear skin of the IMQ-treated mice were collected for the following analysis. On the seventh day, the mice were sacrificed, and splenocytes and skin samples on the back were collected.
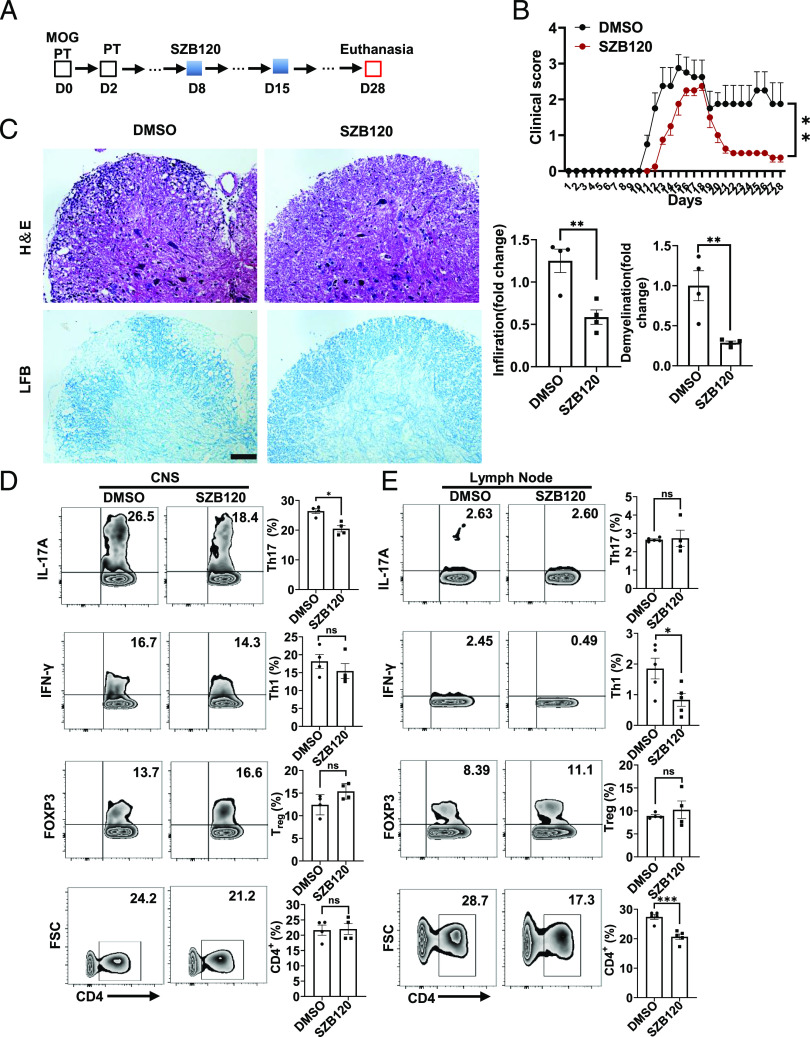
EAE induction and treatment
Female C57BL/6 mice (8–10 wk) were immunized s.c. with CFA (Sigma-Aldrich)–myelin oligodendrocyte glycoprotein (MOG) 35–55 peptide (ChinaPeptides). A total of 200 ng of pertussis toxin (Calbiochem) was administered i.p. on days 0 and 2 after immunization. Mice were scored daily according to the following scale: 0, normal; 1, paralyzed tail; 2, mildly paralyzed hind legs; 3, totally paralyzed hind legs; 3.5, paraplegia with mildly paralyzed forelimbs; 4, paraplegia with paralyzed forelimbs; 4.5, moribund; and 5, death. SZB120 (20 mg/kg/d) and DMSO treatments were carried out every day from the eighth day of immunization.
Histological analysis and immunohistochemistry
For histologic examination, back skin samples of the IMQ-treated mice were fixed with 4% neutral paraformaldehyde and then embedded in paraffin. Sections along the median plane of the back with a thickness of 6 μm were prepared and subjected to staining with H&E. On the seventh day, skin thickness was measured using a micrometer (Mitutoyo). Epidermal hyperplasia (acanthosis) and the number of dermal-infiltrating cells were assessed as previously reported (28). For immunohistochemistry, Ki-67 (proliferation) and CD4 (T cells) expression was evaluated in skin sections using anti-mouse Ki-67 mAb (Thermo Fisher Scientific) and anti-CD4 mAb (Abcam), respectively, following the manufacturer’s instructions. For the EAE model, the lumbar spinal cord of the mice with EAE was carefully removed for H&E and Luxol fast blue staining. The number of infiltrating cells and the percentage of myelin loss in inflammatory foci per section were determined in ImageJ software.
T cell differentiation
Spleens from C57BL/6 mice or IL-17A–GFP mice were directly crushed on cell strainers to obtain single-cell suspensions, and RBCs were lysed. Naive CD4+ T cells were sorted by using a Mouse Naive CD4+ T Cell Isolation Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions and determined with flow cytometry based on cell surface markers (CD4+ CD25− CD62Lhi). Cell purity was >94%. For the cultures of all T cells, RPMI 1640 (catalog no. 11875-093, supplemented with 10% FBS [catalog no. 10099-141], 100× penicillin, streptomycin, fungizone [catalog no. 15240062] and 2 mM l-glutamine [catalog no. 25030-149] was used (all from Life Technologies); 10 mM HEPES buffer (catalog no. 25-060-CI; Corning, Manassas, VA), 55 μM 2-ME (catalog no. 21985-023; Life Technologies, Grand Island, NY), 1 mM Sodium pyruvate (catalog no. 25-000-CIR; Corning) and MEM Non-Essential Amino Acids (catalog no. 11140-050; Life Technologies) were also used. Approximately 0.1 million cells were plated in 96-well plates coated with anti-mouse CD3 (2.5 μg/ml, 145-2C11; BD Biosciences) and soluble anti-mouse CD28 (2 μg/ml, clone 37.51; BD Biosciences). The Th1 cell differentiation condition included 10 ng/ml recombinant murine (rm)IL-12 (R&D Systems) and 10 μg/ml anti–IL-4 (11B11; BioLegend). The Th2 cell differentiation condition included 5 ng/ml rmIL-2 (R&D Systems), 50 ng/ml rmIL-4 (R&D Systems), and 10 μg/ml anti–IFN-γ (XMG1.2; eBioscience). The Th17 cell differentiation condition included 10 ng/ml rmIL-6 (R&D Systems), 5 ng/ml rmTGF-β1 (R&D Systems), 10 ng/ml rmIL-23 (R&D Systems), 5 μg/ml anti–IL-4 (11B11; BioLegend) and 10 μg/ml anti–IFN-γ (XMG1.2; eBioscience). The inducible T regulatory cell (iTreg) differentiation condition included 5 ng/ml rmTGF-β (R&D Systems) and 10 ng/ml rmIL-2 (R&D Systems). Different concentrations of SZB120 and AβBA (MedChemExpress) were added to the T cell culture from the beginning unless specified.
Flow cytometry and intracellular cytokine staining
The purity of isolated cells was investigated by FACS using anti-mouse CD4-allophycocyanin (catalog no. 17-0041-83), anti-mouse CD25-PE (catalog no. E011551630), and PE-CF594 rat anti-mouse CD62L (catalog no. 562404; BD Biosciences, Franklin Lakes, NJ). After 72 h, the cells were analyzed for IL-17A–GFP, or the cells were collected and restimulated for 4–6 h with Cell Stimulation Cocktail (00-4975-93; Thermo Fisher Scientific) for intracellular staining in the absence of SZB120. Cells were fixed and permeabilized with the Foxp3 Staining Buffer Set (eBioscience) or BD Cytofix/Cytoperm (BD Biosciences) and stained with fluorescent Abs. After washing, stained cells were assayed with a BD LSRFortessa X-20 flow cytometer, and the data were analyzed with FlowJo software. Splenocytes, single-cell suspensions from the skin of the IMQ-treated mice, lymph nodes, and CNS infiltrates of the mice with EAE were analyzed by following similar steps. For flow cytometry, CD4 (clone GK1.5), IL-17A (clone eBio17B7), IFN-γ (clone XMG1.2), IL-4 (clone eBio 11B11), Ly-6G (product no. 45-5931-80), CD11b (product no. 25-0112-82), CD3 (product no MA1-80640), IκBζ (product no. 46-6801-80) and Foxp3 (clone FJK-16s) were purchased from eBioscience.
mRNA expression analysis by quantitative PCR
T cells were activated as described above and were collected on the fourth day. Total RNA of the cells was extracted using TRIzol Reagent (Invitrogen). RNA was quantified spectrophotometrically, and 1 μg of total RNA was reverse transcribed into cDNA with HiScript II Q RT SuperMix for qPCR (+gDNA Wiper) (catalog no. R223-01; Vazyme Biotech, Nanjing, China). The cDNA samples were distributed on plates at 200 ng/well and run in triplicate. qPCR was carried out with FastStart Universal SYBR Green Master Mix (Roche) in an Applied Biosystems 7500 Fast Real-Time PCR System or a ViiA 7 Real-Time PCR System (Applied Biosystems). All gene expression results were normalized to the expression of the housekeeping gene GAPDH. Primer sequences are listed as follows: IL-23R forward, 5′-GACTCAGCCAACTCCTCCAGCCAG-3′ and reverse, 5′-TTGGCACTAAGGGCTCAGTCAGA-3′; IL-17A forward, 5′-TCCAGAAGGCCCTCAGACTA-3′ and reverse, 5′-TCGACCCTGAAAGTGAAGG-3′; IL-17F forward, 5′-TGCTACTGTTGATGTTGGGAC-3′ and reverse, 5′-ATGCCCTGGTTTTGGTTGAA-3′; IL-22 forward: 5′-CGTCAACCGCACCTTTAT-3′ and reverse, 5′-AGGGCTGGAACCTGTCTG-3′; CCL20 forward, 5′-AGCAGCAAGCAACTACGACT-3′ and reverse, 5′-GGATCAGCGCACACAGATT-3′; CCR6 forward, 5′-CAACTTCACAGTTCTAATAGTCCTC-3′ and reverse: 5′-GAGGTGAAGCCCAGAAAATGC-3′; GAPDH forward, 5′-AGGTCGGTGTGAACGGATTTG-3′ and reverse, 5′-GGGGTCGTTGATGGCAAC-3′.
Western blotting
Cells were lysed in RIPA lysis buffer (Beyotime, Jiangsu, China) supplemented with Protease and Phosphatase Inhibitor Cocktails (Thermo Fisher Scientific). Abs used in Western blotting included the following: rabbit anti-eIF2α Ab (1:1000; Cell Signaling Technology), rabbit anti-IκBζ Ab (1:1000; Cell Signaling Technology), rabbit anti-eIF2α (phospho-S51) Ab (1:1000; Abcam, Cambridge, U.K.), mouse anti–β-actin Ab (1:2000; Proteintech, Wuhan, China), HRP-labeled anti-rabbit IgG (H+L), or anti-mouse IgG (H+L) (1:5000; Jackson Immunoresearch Laboratories). Images have been cropped for presentation.
T cell proliferation and apoptosis
The cell cycle was measured using a PE-conjugated Ab against Ki-67 (Thermo Fisher Scientific) after 3 d under Th17 cell differentiation. Cell apoptosis was measured by staining cells with 7-aminoactinomycin D (7AAD) and annexin V (MultiSciences, Zhejiang, China) according to the manufacturer’s instructions. Cell proliferation was measured by loading cells with 5 μM CellTrace Violet (Invitrogen) followed by stimulation with Th17 culture conditions for 3 d.
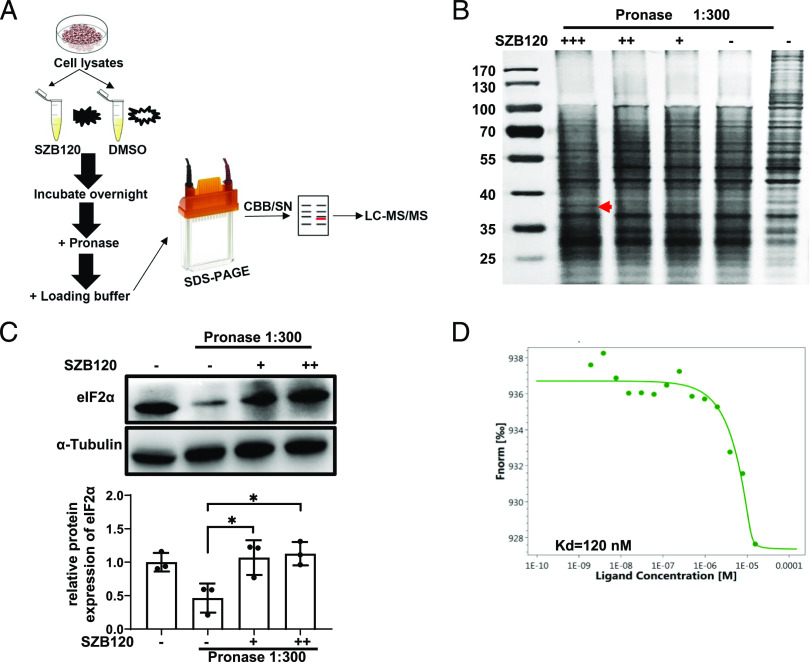
Drug affinity responsive target stability assay
A drug affinity responsive target stability (DARTS) assay for identifying the targets of SZB120 were performed according to the protocol by Lomenick et al. (29, 30). In brief, Jurkat cells or T cells in Th17 cell differentiation conditions were lysed, and protein concentrations were determined by the BCA Protein Assay Kit (Thermo Fisher Scientific) to ensure an equal amount of protein lysate per sample. Cell lysates were treated either with various concentrations of SZB120 (1–100 μM) or with DMSO alone. After incubation overnight at 4°C, digestion was performed using Pronase (Roche). Each cellular lysate sample was proteolyzed at room temperature for 10 min with 0.05 mg/ml Pronase and stopped using excess protease inhibitors. The samples were then loaded on SDS-PAGE gels and stained with Silver Stain (Thermo Fisher Scientific). Finally, the bands with increased staining from the SZB120 lane and the matching area of the control lane were cut out, digested with in-gel trypsin, and subjected to liquid chromatography-tandem mass spectrometry analysis.
eIF2α protein expression and purification
The mouse eIF2α expression plasmid in the pET-28a (+) vector was transformed into Escherichia coli BL21 (DE3)-competent cells and expressed as a fusion protein containing an N-terminal tobacco etch virus protease cleavable 6His-tag. Cells were grown at 37°C in Luria–Bertani medium until OD600 = 4.0–6.0, and expression was induced by the addition of 0.5 mM isopropyl β-D-1-thiogalactopyranoside overnight at 20°C. Cells were pelleted and resuspended in cold lysis buffer (25 mM HEPES [pH 7.5], 0.5 M NaCl, 5% glycerol, 5 mM imidazole, and 5 mM 2-ME). The cell suspension was lysed with an ultra-high pressure cell disruptor, and insoluble debris was removed by centrifugation at 14,000 rpm at 4°C for 1 h. Proteins were eluted with 250 mM imidazole in lysis buffer. The 6His-tag was removed by tobacco etch virus protease cleavage overnight by dialysis against 25 mM HEPES (pH 7.5), 200 mM NaCl, 5% glycerol, and 5 mM 2-ME, which was followed by nickel reverse chromatography and dialysis to desalt. Untagged proteins were loaded onto a 5 ml HiTrap Q HP column (GE Healthcare). Proteins were eluted with a linear NaCl gradient (25–500 mM). The eluted proteins were further purified by size-exclusion chromatography using a HiPrep 26/60 Superdex 200 (GE Healthcare). Proteins for biochemical assays were stored at −80°C in 25 mM HEPES (pH 7.5), 150 mM NaCl, 1 mM MgCl2, 5 mM KCl, 5% glycerol, and 2 mM DTT. Site-directed mutagenesis was performed with a KOD-Plus-Mutagenesis Kit (Toyobo) using pET28a-eIF2α as a template.
Microscale thermophoresis assay
For determination of the binding affinity of SZB120 and the eIF2α protein, a microscale thermophoresis (MST) assay was conducted by using Monolith NT.115 (NanoTemper Technologies). Proteins were labeled with the Monolith NT Protein Labeling Kit RED (NanoTemper Technologies) according to the supplied labeling protocol. Labeled proteins were used at a concentration of 400 nM. SZB120 was titrated in a 1:1 dilution beginning at 2.5 mM, which contained 5% (v/v) DMSO. Samples were diluted in MST assay buffer that contained 100 mM Tris-HCl (pH 8.0), 20 mM MgCl2, and 200 mM KCl supplemented with DMSO at a final concentration of 5% to make sure that all samples contained the same DMSO concentration. For measurement, the samples were added into Premium Coated Capillaries.
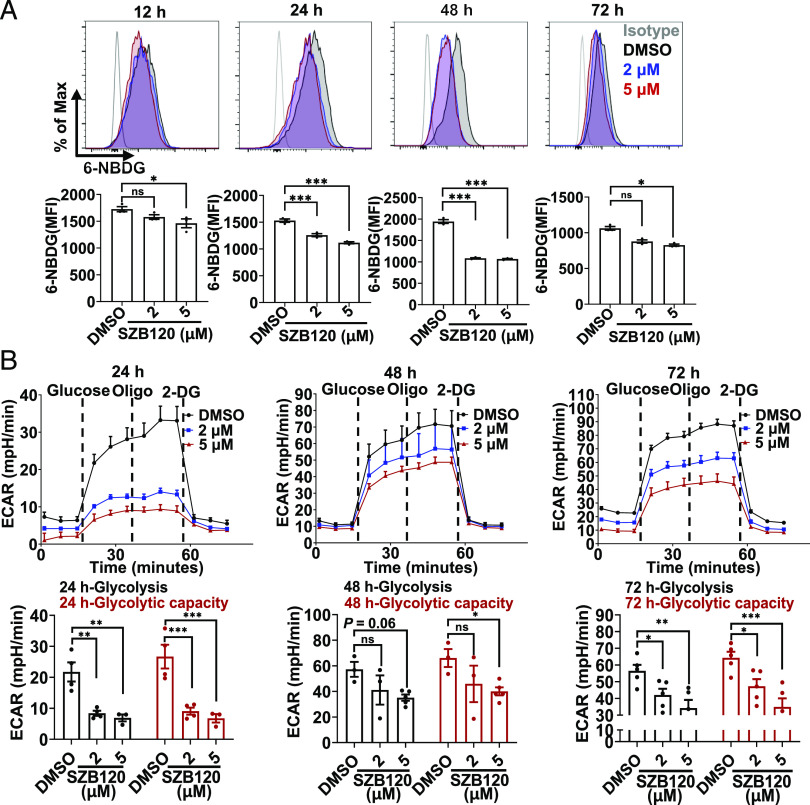
Glucose uptake
For glucose uptake experiments, the internal solution containing 6-NBDG (100 μg/ml) (Thermo Fisher Scientific) was used according to the manuscript. Briefly, naive CD4+ T cells were cultured under Th17 cell differentiation conditions and harvested at the indicated times. Cells were washed once using PBS. The cells were then incubated with a 6-NBDG solution that was dissolved in glucose-free DMEM in a 5% CO2 incubator at 37°C for 10 min. This procedure needed to be performed in a dark place. After washing, stained cells were assayed with BD LSRFortessa X-20 as soon as possible.
Metabolic assay
The metabolic function of naive CD4+ T cells cultured for the indicated times in vitro with a Th17 cell differentiation system was analyzed by measuring the extracellular acidification rate (ECAR) using an XF96 Extracellular Flux Analyzer (Seahorse Bioscience). The cells were kept in XF media (Seahorse Bioscience) supplemented with 2 mM glutamine (Life Technologies) and subjected to the glycolysis stress test protocol using sequential addition of 10 mM glucose, 1 μM oligomycin, and 50 mM 2-deoxy-glucose. Glycolysis and glycolytic capacity were calculated as follows: glycolysis equals maximal ECAR after glucose treatment minus ECAR before glucose (nonglycolytic acidification); glycolytic capacity equals maximal ECAR after-oligomycin treatment minus ECAR before glucose (nonglycolytic acidification). Analysis of data was performed using the Seahorse Wave 2.4.0 software package (Seahorse Bioscience).
Statistics
The data were analyzed with GraphPad Prism 8 and are presented as the mean ± SEM. Unpaired t tests (two-tailed) were used when two groups were compared, and ANOVA with Bonferroni correction was used for multiple comparisons (one-way or two-way). The p values <0.05 were considered significant (*p < 0.05, **p < 0.01, ***p < 0.001). Error bars depict the SEM.
Results
SZB120 selectively inhibits Th17 cell differentiation
We first screened modified small molecules by Th17 cell differentiation assays in vitro. For this, naive CD4+ T cells were isolated from IL-17A–GFP mouse spleens and induced under Th17 cell differentiation conditions in the presence of modified molecules. SZB120 showed strong efficacy and was chosen for further research (Fig. 1A). Compared with AβBA, SZB120 could modulate Th17 cell differentiation at a relatively lower concentration (Fig. 1B). The inhibition of Th17 cell differentiation was most pronounced when SZB120 was added within the first 24 h, and this inhibitory effect was sustained for a significant period of time (Supplemental Fig. 1A, 1B). SZB120-treated cells decreased the mRNA expression of IL-17A, IL-17F, IL-22, IL-23R, CCL20, and CCR6 in a dose-dependent manner (Supplemental Fig. 1C). Moreover, the regulation of Th17 cell differentiation by SZB120 was independent of cell apoptosis and proliferation (Fig. 1C, 1D, Supplemental Fig. 1D). We then explored whether SZB120 treatment would dampen the other subsets of T cells. Naive CD4+ T cells were isolated from wild-type mouse spleens and cultured under Th1, Th2, and iTreg conditions in the presence of SZB120 and DMSO. Low concentrations of SZB120, which can impair Th17 cell differentiation did not influence Th1, Th2, or iTreg differentiation (Fig. 1E, Supplemental Fig. 1E). As demonstrated above, treatment with SZB120 can selectively inhibit Th17 cell differentiation in vitro.
FIGURE 1.
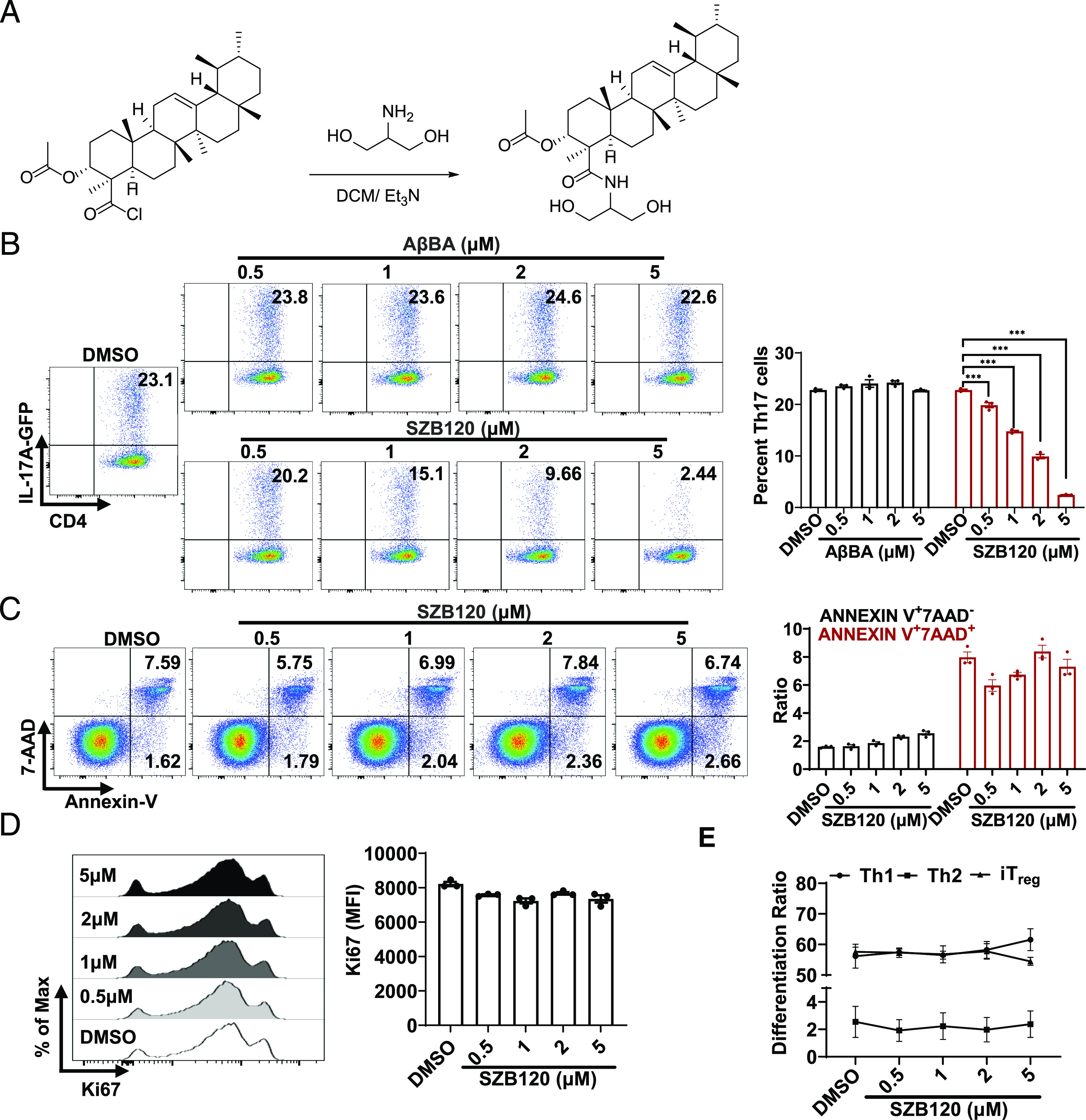
SZB120 selectively inhibits Th17 cell differentiation. (A) The synthesis of SZB120. (B) Mouse primary naive CD4+ T cells from IL-17A–GFP mice were differentiated under Th17 cell polarization conditions for 72 h with DMSO and different concentrations of AβBA (0.5, 1, 2, 5 μM) or SZB120 (0.5, 1, 2, 5 μM) (n = 3). (C) Naive CD4+ T cells were differentiated under Th17 polarization conditions for 72 h with DMSO and SZB120 (0.5, 1, 2, 5 μM). CD4+ annexin V+ 7AAD+ cells were analyzed. The black bar represents the percentage of annexin V+ 7AAD− cells (early apoptosis). The red bar indicates the percentage of annexin V+ 7AAD+ cells (late apoptosis) (n = 3). (D) The proliferation marker Ki67 was tested under the Th17 cell differentiation conditions mentioned above with DMSO and SZB120 (0.5, 1, 2, 5 μM). Cells were gated for CD4+ and stained for Ki67. Black bar indicates the mean fluorescence intensity (MFI) of Ki67+ cells (n = 3). (E) T cells were activated under Th1, Th2, and iTreg cell polarization conditions in the presence of DMSO and 0.5, 1, 2, and 5 μM SZB120 (n = 3–6). Data are the mean ± SEM by two-way ANOVA (B and C) or one-way ANOVA (D and E) with Bonferroni correction for multiple comparisons. *p < 0.05, ***p < 0.001, compared with DMSO. ns, no significance
Identification of eIF2α as the direct interacting protein target of SZB120
To investigate how SZB120 regulates Th17 cell function, we aimed to elucidate the precise molecular target of SZB120 responsible for its effects on Th17 cells. For unbiased target identification, we used Jurkat cells for DARTS assays (Fig. 2A). The DARTS technique is a relatively quick strategy to determine the direct molecular target of small molecules. The greatest advantage is the ability to use the native small molecule without having to immobilize or modify it (29). The DARTS results revealed a strongly protected band of ∼38 kDa in the SZB120-treated sample, and mass spectrometry identified eIF2α as one of the most abundant and enriched proteins (Fig. 2B, Supplemental Table I). The interaction between SZB120 and eIF2α was verified by Western blotting in Jurkat cells and in vitro–differentiated Th17 cells (Fig. 2C, Supplemental Fig. 2A). An MST assay was performed to further confirm the interaction between SZB120 and eIF2α, and the equilibrium Kd of the SZB120-eIF2α interaction was 120 nM (Fig. 2D). These results showed that SZB120 could directly target eIF2α with high affinity.
FIGURE 2.
Identification of eIF2α as a direct target protein of SZB120 by DARTS. (A) The schematic illustration of the DARTS technique. (B) The cell lysates were treated with DMSO or SZB120 (1, 10, 100 μM) overnight at 4°C, followed by SDS-PAGE analysis and Silver Staining. The red arrow indicates a protective band by SZB120 of ∼38 kDa. (C) DARTS and Western blotting to confirm the potential binding target eIF2α of SZB120 (10 and 100 μM). The lower panel is the quantitative results. Data are shown as a fold of α-tubulin (n = 3). (D) MST analysis of the binding affinity between SZB120 and eIF2α. The fitted Kd value is shown. *p < 0.05.
SZB120 inhibits Th17 cell differentiation by enhancing eIF2α phosphorylation
It is well-acknowledged that eIF2α regulates translation, and its phosphorylation level varies in response to different environmental challenges (17). However, the role of eIF2α in Th17 cells is less commonly reported. We first tested the impact of SZB120 on p-eIF2α. As shown in Fig. 3A, SZB120 promoted p-eIF2α after 1 h, and the regulatory effect was dose-dependent (Supplemental Fig. 2B). IκBζ is one member of the nuclear IκB family. This molecule was reported to be a TF required for Th17 cell development in mice (31). Bambouskova et al. noted that dimethyl itaconate markedly increased p-eIF2α and caused specific suppression of IκBζ (32). Consistent with their findings, we also observed an obvious decrease in IκBζ expression in Th17 cells in response to SZB120 treatment by using flow cytometry (Fig. 3B). Moreover, we obtained a similar result by using Western blotting (Supplemental Fig. 2C). To further confirm that the inhibitory effect of SZB120 on Th17 cell differentiation was p-eIF2α dependent, we used the protein kinase R (PKR)–like endoplasmic reticulum kinase inhibitor GSK2606414, which could downregulate p-eIF2α. The effect of SZB120 on Th17 cell differentiation was impaired in the presence of GSK2606414 (Fig. 3C). We also found that GSK2606414 reduced p-eIF2α and enhanced the expression of IκBζ triggered by SZB120 under Th17 cell differentiation conditions (Fig. 3D, 3E). These results suggested that the suppression of Th17 cell differentiation by SZB120 is highly related to eIF2α/IκBζ, and GSK2606414 could weaken the inhibitory effect of SZB120.
FIGURE 3.
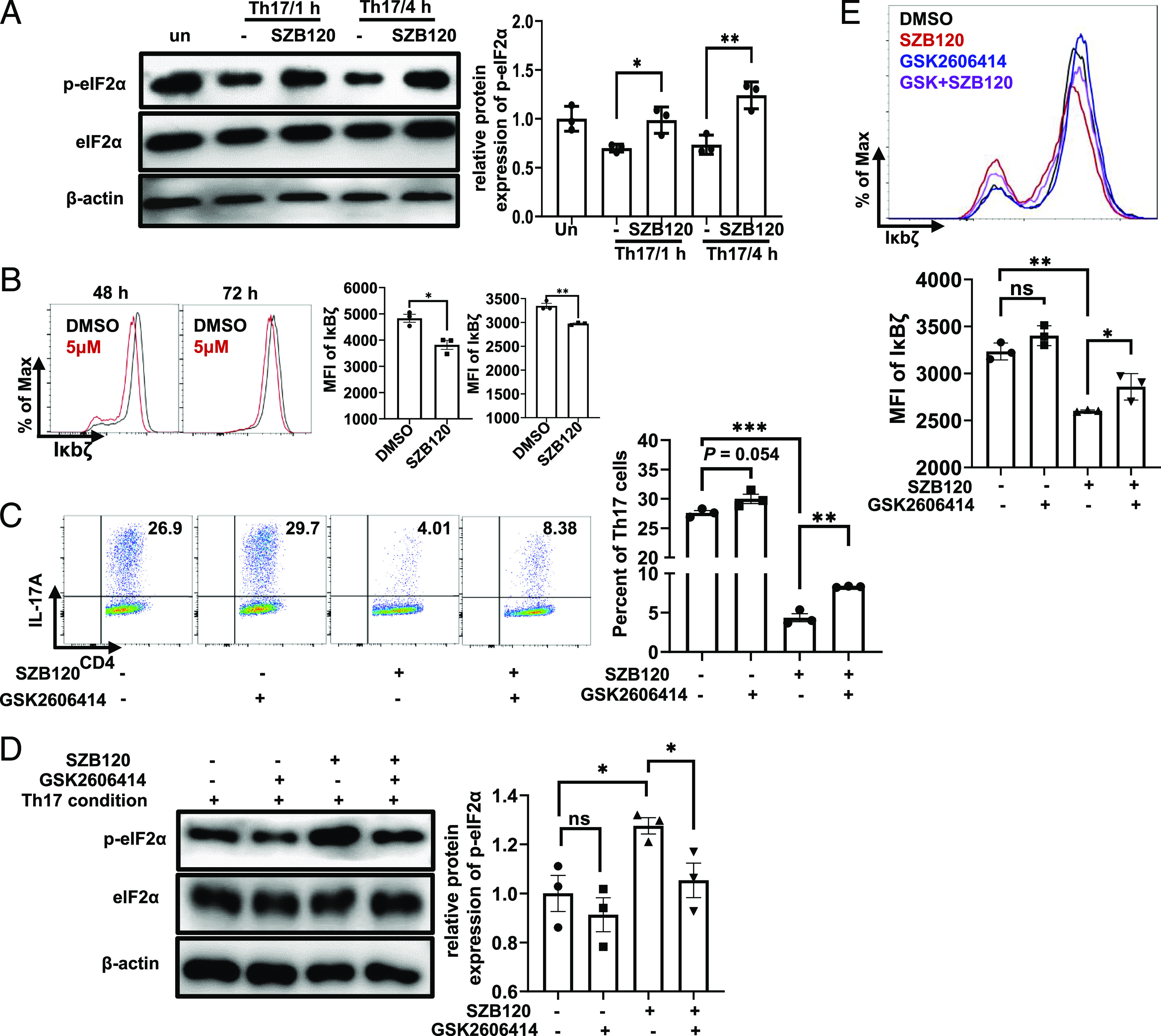
SZB120 inhibits IL-17A secretion in an eIF2α phosphorylation-dependent manner. (A) Unstimulated or TCR-activated Th17 cells were treated with DMSO and 5 μM SZB120 and lysed for Western blotting of p-eIF2α and eIF2α after 1 and 4 h. The right panel is the quantitative results. The data are shown as a fold of eIF2α (n = 3). (B) Naive CD4+ T cells were activated under Th17 cell polarization conditions and treated with DMSO and 5 μM SZB120. Cells were measured by flow cytometry to determine the IκBζ mean fluorescence intensity (MFI) of CD4+ T cells at the indicated times (n = 3). Data are the mean ± SEM by unpaired t tests. (C) T cells activated under Th17 polarizing conditions were treated with DMSO, SZB120 (5 μM), GSK2606414 (4 nM), or SZB120 plus GSK2606414 and stained for IL-17A intracellular cytokines (n = 3). (D) Unstimulated or TCR-activated Th17 cells were treated with the four groups as described in (C), and lysates were analyzed after 4 h by Western blotting of p-eIF2α and eIF2α. The quantitative results are shown in the right panel. Data are shown as a fold of eIF2α (n = 3). (E) Naive CD4+ T cells treated as described in (C) were differentiated under Th17 polarization conditions for 72 h. Cells were measured by flow cytometry to determine the IκBζ MFI of CD4+ T cells. The quantitative results are shown in the lower panel. All data represent at least three similar experiments with three samples per group in each. Data are the mean ± SEM by one-way ANOVA (A and C–E) with Bonferroni correction for multiple comparisons or the mean ± SEM by unpaired t tests (two-tailed) (B). *p < 0.05, **p < 0.01, ***p < 0.001.
SZB120 affects the glucose metabolism of Th17 cells
Because IκBζ could modulate glucose uptake via IL-17 signaling (33), we wondered whether SZB120 could affect the glucose uptake of Th17 cells. For this, 6-NBDG, a fluorescent glucose analogue, was used to measure glucose uptake by CD4+ T cells under Th17-polarized conditions. As expected, glucose uptake was downregulated from 12 h after activation (Fig. 4A), and this inhibitory effect was most obvious at 48 h postactivation. Interestingly, we found that SZB120 had no obvious effect on genes related to glucose transport (data not shown). Notably, increased glucose uptake can facilitate a higher rate of glycolysis (34). We then measured the ECAR of Th17 cells treated with different concentrations of SZB120 and DMSO at different time points. We found obvious changes in glycolysis and the glycolytic capacity of Th17 cells treated with 2 and 5 μM of SZB120 from the early phase (Fig. 4B). Together, these data indicated that SZB120 could dramatically impair glucose uptake and glycolysis in the progression of Th17 cell differentiation.
FIGURE 4.
SZB120 affects glucose uptake and glycolysis in Th17 cells. (A) Representative flow cytometric analysis of Th17 cells treated with DMSO or SZB120 (2 or 5 μM). Cells were collected for 100 μM 6-NBDG dilution at the indicated times (n = 3). (B) The effect of SZB120 on glycolysis in Th17 cells was evaluated by monitoring the ECAR using an XF96 Extracellular Flux Analyzer. Glucose (10 mM), the oxidative phosphorylation inhibitor (oligomycin 1 μM), and the glycolytic inhibitor 2-deoxy-glucose (2-DG, 50 mM) were sequentially injected into each well at the indicated time points (n = 3–5). Data are the mean ± SEM by one-way (A) or two-way ANOVA (B) with Bonferroni correction for multiple comparisons. *p < 0.05, **p < 0.01, ***p < 0.001 compared with DMSO.
SZB120 ameliorates clinical symptoms of EAE
To evaluate the effect of SZB120 on Th17 cell–mediated autoimmune disease in vivo, we used an induced EAE model. SZB120 and DMSO were administered starting at day 8 postimmunization (Fig. 5A). Compared with the control treatment, SZB120 treatment resulted in a significant reduction in EAE severity (Fig. 5B). Lumbar spinal cord samples in each group were collected on day 15 and day 28. Histological analyses showed mitigated inflammation and demyelination in the spinal cords of the SZB120-treated mice (Fig. 5C, Supplemental Fig. 3A). In EAE, CD4+ T cells and their subsets play key roles during the course of the disease (5, 35). Mononuclear cells from the CNS and inguinal lymph nodes were collected on days 15 and 28. Decreasing Th17 cells were observed in the CNS of the SZB120-treated group despite no change in CD4+ T cells, Th1 cells, or iTreg during the acute phase (Fig. 5D). For lymph node cells, the percentages of CD4+ T cells and Th1 cells but not Th17 cells or iTreg decreased significantly in the same period (Fig. 5E). Similar results were obtained in the chronic phase (day 28); both the percentages of Th17 cells and CD4+ T cells were reduced in the CNS, whereas Th17 cells and Th1 cells were reduced in the lymph nodes (Supplemental Fig. 3B, 3C). Taken together, these results suggest that SZB120 plays a suppressive role in Th17 cells in the EAE model.
FIGURE 5.
SZB120 inhibits Th17 development in the mouse model of EAE. (A) Schematic diagram of the EAE model. (B) Clinical score of the mice with EAE treated with 20 mg/kg/d SZB120 and an equivalent volume of DMSO (n = 4). (C) Representative images of H&E staining and Luxol fast blue staining of paraffin-embedded sections of spinal cords isolated from the DMSO- or SZB120-treated EAE model on day 15 postimmunization. Scale bar, 30 μm. Quantification of CNS infiltrates and the amount of demyelination are shown in the right panel. (D and E) Flow cytometric analysis of Th17 cells, Th1 cells, and regulatory cells percentages gated on CD4+ T cells, which were taken from the CNS or lymph node samples of the DMSO- or SZB120-treated mouse model of EAE on day 15 after immunization (n = 4, 5). Data are the mean ± SEM. *p < 0.05, ***p < 0.001.
SZB120 inhibits IL-17 in IMQ-induced psoriasiform skin inflammation
To further evaluate the inhibitory effect of SZB120 on Th17 development in vivo, we used an IMQ-induced mouse model of psoriasis that is IL-17/Th17 related. We measured the frequency of IL-17+ cells in CD4+ T cell populations in the ear dermis and spleens of mice via flow cytometric analysis. We found that the SZB120-treated mice exhibited a lower frequency of CD4+ IL-17+–expressing cells than the control mice (Fig. 6A, 6B). SZB120 also decreased the infiltration of neutrophils in the dermis of IMQ-induced skin inflammation (Fig. 6C). Immunohistochemistry revealed an obvious decrease in CD4+ T cell infiltration as well as Ki67+ cells in the skin of the SZB120-treated mice compared with the DMSO-treated mice (Supplemental Fig. 3D). Because γδT17 cells are the main producer of IL-17 in psoriasiform plaques induced by IMQ (36), we then tested its proportion in the mouse ear dermis. We found that SZB120 substantially impaired not only γδT cells but also γδT17 cells (Fig. 6D). Consequently, SZB120 treatment prevented thickening of the skin and alleviated inflammatory phenotypes (erythema, thickness, and scaling) compared with the DMSO control (Fig. 6E, 6F). Histological analysis showed that SZB120 suppressed epidermal hyperplasia and dermal cellular infiltration (Fig. 6G, 6H). These data indicated that SZB120 treatment reduced the percentages of IL-17+–secreting CD4+ T cells in the IMQ-induced mouse model of psoriasis. Although other mechanisms were involved in the anti-inflammatory effect of SZB120 in the IMQ model, these data together indicated the therapeutic potential of SZB120 in the treatment of psoriasis.
FIGURE 6.
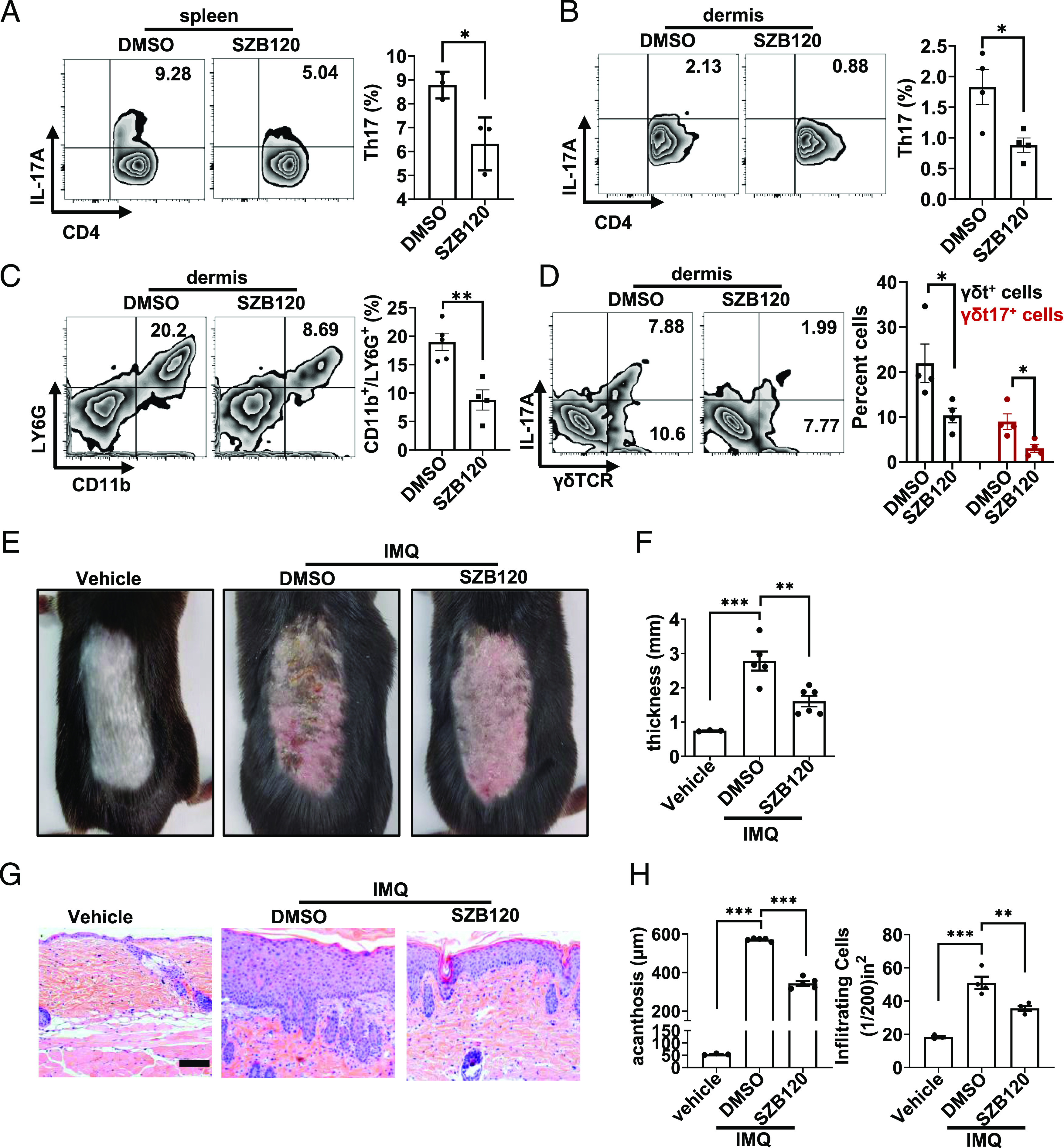
SZB120 represses IMQ-induced skin inflammation in mice. (A and B) The percentage of CD4+ IL-17+ splenocytes and ear dermis cells was measured (n = 3). (C) The percentage of CD11b+Ly-6G+ cells in each group was measured (n = 3). (D) The percentage of γδT+IL-17+ cells in ear dermis cells was measured (n = 3), and CD3+ cells were gated. (E) Representative phenotypes of the vehicle-, solvent control-, and SZB120-treated mice. The experiment was repeated three times independently. (F) Skin thickness was measured on the seventh day and compared with DMSO (n = 3–6). (G) Representative H&E staining of every group. Scale bar, 50 μm. (H) Acanthosis and dermal cellular infiltrate of skin samples of every group (n = 3–5). Data are the mean ± SEM by unpaired t test (A–C) and one-way ANOVA (D, F, and H). *p < 0.05, **p < 0.01, ***p < 0.001.
Discussion
BAs, mainly KBA and AKBA, have prominent antiproliferative and anti-inflammatory activity (24, 37). However, there are few reports on AβBA and its derivatives in vivo or in vitro. In this study, we report SZB120, an AβBA derivative, selectively inhibits the differentiation of Th17 cells from naive CD4+ T cells rather than other T cell subsets and shows therapeutic activity in mouse models of IL-17/Th17–mediated multiple sclerosis and psoriasis.
We identified eIF2α as a direct target of SZB120 by DARTS analysis and further verified this direct binding by Western blotting and MST assays. Our results showed that SZB120 treatment rapidly phosphorylated eIF2α (∼1 h postactivation) in a dose-dependent manner, and the inhibitory effect on Th17 cell differentiation was attenuated when the PERK inhibitor GSK2606414 was added. Sundrud et al. (38) reported that halofuginone-treated Th17 cells rapidly exhibit p-eIF2α and show a low differentiation ratio. Mice conditionally lacking Ser51 eIF2α phosphorylation in epithelial cells exhibited enhanced weight loss and elevated Th17 cell responses relative to littermate controls (39). Moreover, recent studies have indicated that p-eIF2α selectively inhibits translation (40–42). RORγt and IκBζ, which are induced by TGF-β and IL-6, are both essential TFs for Th17 cells, and either molecule alone was insufficient to induce Th17 cell differentiation ex vivo (31). Our data showed that p-eIF2α induced by SZB120 resulted in a substantial decrease in the IκBζ protein. IκBζ is required for Th17 cell development and IL-17 production in mice (31, 43). We speculate that the cooperation between RORγt and IκBζ was impaired by reduced IκBζ expression, thus inhibiting Th17 cell differentiation. IκBζ has also been reported to modulate glucose uptake (33), which prompted us to determine whether SZB120 affects glucose uptake of Th17 cells. Consistent with our assumptions, SZB120 significantly downregulated the glucose uptake and glycolysis in Th17 cells. Accumulating evidence indicates that metabolic processes are crucial players in T cell activation and differentiation (44). The differentiation of naive CD4+ T cells into a spectrum of subsets relies on distinct metabolic pathways (45), and Th17 cells have been found to primarily rely on glycolysis for their development and maintenance (46). Our results indicated that SZB120 inhibits Th17 cell differentiation by interfering with the eIF2α–IκBζ axis and reducing glycolysis, which supports Th17 cell differentiation.
To determine whether the efficacy observed in vitro will be translated into in vivo models, we tested SZB120 in two inflammatory autoimmune mouse models. In a Th17 cell–mediated mouse model of MOG–EAE (47), SZB120 significantly relieved inflammation and demyelination as well as the clinical score of MOG-induced EAE. More impressively, this treatment strongly reduced the percentage of Th17 cells in the CNS but did not have much effect on other types of CD4+ T cells at the peak or late stages of the disease. Furthermore, the SZB120-treated group showed some changes in the percentages of Th17 cells, Th1 cells, and CD4+ T cells in the lymph node cells.
Although IMQ-induced psoriasis-like inflammation is reported to be more a RORγt (+) innate lymphocyte- and γδT cell–dependent skin inflammation model than Th17 cells (48), we still use it to evaluate the anti–IL-17/Th-17 effect of SZB120. As expected, SZB120 treatment significantly suppressed CD4+ IL-17+ T cells in both the dermis and spleen of the IMQ-induced group compared with the DMSO group. Additionally, SZB120 reduced the percentages of IL-17–producing γδT cells in the dermis of IMQ-induced skin. This finding is probably because IκBζ is also a critical TF for the full function of γδT cells, whose expression is downregulated by SZB120. Moreover, the analysis of single-cell suspensions isolated from the mouse psoriatic dermis revealed that neutrophilic infiltration was strongly inhibited by SZB120. Collectively, SZB120 treatment had promising efficacy in ameliorating epidermal hyperplasia and dermal inflammation in IMQ-induced psoriasis-like skin inflammation. Considering that an excessive Th17 cell response plays a central role in autoimmune diseases, such as psoriasis (49), SZB120 shows great promise for clinical application in Th17-relevant autoimmune disease therapy.
In summary, we present SZB120, a derivative of AβBA, with a significant inhibitory effect on Th17 cell differentiation. SZB120 directly binds to and phosphorylates eIF2α, leading to decreased downstream IκBζ expression and impaired glucose metabolism, thereby affecting the development and maintenance of Th17 cells. SZB120 efficiently alleviates autoimmune diseases involving the CNS and skin in two mouse models, which makes it a potential drug candidate for IL-17–mediated autoimmune diseases.
Supplementary Material
Acknowledgments
We thank the members of the National Facility for Protein Science (Shanghai, China) for use of the large-scale protein preparation system and the members of the Zhangjiang Laboratory (Shanghai, China) for providing technical support and assistance in data collection and analysis.
This work was supported by National Natural Science Foundation of China Grants 81725018, 81930088, 82073428, 81703118, and 81803123.
The online version of this article contains supplemental material.
- 7AAD
- 7-aminoactinomycin D
- AβBA
- acetylated βBA
- AKBA
- 3-acetyl-11-keto-β-BA
- BA
- boswellic acid
- DARTS
- drug affinity responsive target stability
- EAE
- experimental autoimmune encephalomyelitis
- ECAR
- extracellular acidification rate
- IMQ
- imiquimod
- iTreg
- inducible T regulatory cell
- KBA
- 11-keto-β-BA
- MOG
- myelin oligodendrocyte glycoprotein
- MST
- microscale thermophoresis
- p-eIF2α
- phosphorylation of the eIF2a subunit
- rm
- recombinant murine
- TF
- transcription factor.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.El-Behi, M., Ciric B., Dai H., Yan Y., Cullimore M., Safavi F., Zhang G. X., Dittel B. N., Rostami A. 2011. The encephalitogenicity of T(H)17 cells is dependent on IL-1- and IL-23-induced production of the cytokine GM-CSF. Nat. Immunol. 12: 568–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harrington, L. E., Hatton R. D., Mangan P. R., Turner H., Murphy T. L., Murphy K. M., Weaver C. T. 2005. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat. Immunol. 6: 1123–1132. [DOI] [PubMed] [Google Scholar]
- 3.Miossec, P., Kolls J. K. 2012. Targeting IL-17 and TH17 cells in chronic inflammation. Nat. Rev. Drug Discov. 11: 763–776. [DOI] [PubMed] [Google Scholar]
- 4.Brüstle, A., Heink S., Huber M., Rosenplänter C., Stadelmann C., Yu P., Arpaia E., Mak T. W., Kamradt T., Lohoff M. 2007. The development of inflammatory T(H)-17 cells requires interferon-regulatory factor 4. Nat. Immunol. 8: 958–966. [DOI] [PubMed] [Google Scholar]
- 5.Aranami, T., Yamamura T. 2008. Th17 cells and autoimmune encephalomyelitis (EAE/MS). Allergol. Int. 57: 115–120. [DOI] [PubMed] [Google Scholar]
- 6.Schraml, B. U., Hildner K., Ise W., Lee W. L., Smith W. A., Solomon B., Sahota G., Sim J., Mukasa R., Cemerski S., et al. 2009. The AP-1 transcription factor Batf controls T(H)17 differentiation. Nature 460: 405–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Quigley, M., Huang X., Yang Y. 2007. Extent of stimulation controls the formation of memory CD8 T cells. J. Immunol. 179: 5768–5777. [DOI] [PubMed] [Google Scholar]
- 8.Bauquet, A. T., Jin H., Paterson A. M., Mitsdoerffer M., Ho I. C., Sharpe A. H., Kuchroo V. K. 2009. The costimulatory molecule ICOS regulates the expression of c-Maf and IL-21 in the development of follicular T helper cells and TH-17 cells. Nat. Immunol. 10: 167–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Veldhoen, M., Hirota K., Westendorf A. M., Buer J., Dumoutier L., Renauld J. C., Stockinger B. 2008. The aryl hydrocarbon receptor links TH17-cell-mediated autoimmunity to environmental toxins. Nature 453: 106–109. [DOI] [PubMed] [Google Scholar]
- 10.Zhang, F., Meng G., Strober W. 2008. Interactions among the transcription factors Runx1, RORγt and Foxp3 regulate the differentiation of interleukin 17–producing T cells. [Published erratum appears in 2009 Nat. Immunol. 10: 223.] Nat. Immunol. 9: 1297–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barbi, J., Pardoll D., Pan F. 2013. Metabolic control of the Treg/Th17 axis. Immunol. Rev. 252: 52–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berod, L., Friedrich C., Nandan A., Freitag J., Hagemann S., Harmrolfs K., Sandouk A., Hesse C., Castro C. N., Bähre H., et al. 2014. De novo fatty acid synthesis controls the fate between regulatory T and T helper 17 cells. [Published erratum appears in 2015 Nat. Med. 21: 414.] Nat. Med. 20: 1327–1333. [DOI] [PubMed] [Google Scholar]
- 13.Xu, T., Stewart K. M., Wang X., Liu K., Xie M., Ryu J. K., Li K., Ma T., Wang H., Ni L., et al. 2017. Metabolic control of TH17 and induced Treg cell balance by an epigenetic mechanism. Nature 548: 228–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi, L. Z., Wang R., Huang G., Vogel P., Neale G., Green D. R., Chi H. 2011. HIF1alpha-dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells. J. Exp. Med. 208: 1367–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gualdoni, G. A., Mayer K. A., Göschl L., Boucheron N., Ellmeier W., Zlabinger G. J. 2016. The AMP analog AICAR modulates the Treg/Th17 axis through enhancement of fatty acid oxidation. FASEB J. 30: 3800–3809. [DOI] [PubMed] [Google Scholar]
- 16.Jackson, R. J., Hellen C. U., Pestova T. V. 2010. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 11: 113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boye, E., Grallert B. 2020. eIF2α phosphorylation and the regulation of translation. Curr. Genet. 66: 293–297. [DOI] [PubMed] [Google Scholar]
- 18.Hart, L. S., Cunningham J. T., Datta T., Dey S., Tameire F., Lehman S. L., Qiu B., Zhang H., Cerniglia G., Bi M., et al. 2012. ER stress-mediated autophagy promotes Myc-dependent transformation and tumor growth. J. Clin. Invest. 122: 4621–4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moon, S. L., Sonenberg N., Parker R. 2018. Neuronal regulation of eIF2α function in health and neurological disorders. Trends Mol. Med. 24: 575–589. [DOI] [PubMed] [Google Scholar]
- 20.Ozcan, L., Cristina de Souza J., Harari A. A., Backs J., Olson E. N., Tabas I. 2013. Activation of calcium/calmodulin-dependent protein kinase II in obesity mediates suppression of hepatic insulin signaling. Cell Metab. 18: 803–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ernst, E. 2008. Frankincense: systematic review. BMJ 337(dec17 2): a2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharma, T., Jana S. 2020. Investigation of molecular properties that influence the permeability and oral bioavailability of major β-Boswellic acids. Eur. J. Drug Metab. Pharmacokinet. 45: 243–255. [DOI] [PubMed] [Google Scholar]
- 23.Bai, J., Gao Y., Chen L., Yin Q., Lou F., Wang Z., Xu Z., Zhou H., Li Q., Cai W., et al. 2019. Identification of a natural inhibitor of methionine adenosyltransferase 2A regulating one-carbon metabolism in keratinocytes. EBioMedicine 39: 575–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Safayhi, H., Mack T., Sabieraj J., Anazodo M. I., Subramanian L. R., Ammon H. P. 1992. Boswellic acids: novel, specific, nonredox inhibitors of 5-lipoxygenase. J. Pharmacol. Exp. Ther. 261: 1143–1146. [PubMed] [Google Scholar]
- 25.Siemoneit, U., Hofmann B., Kather N., Lamkemeyer T., Madlung J., Franke L., Schneider G., Jauch J., Poeckel D., Werz O. 2008. Identification and functional analysis of cyclooxygenase-1 as a molecular target of boswellic acids. Biochem. Pharmacol. 75: 503–513. [DOI] [PubMed] [Google Scholar]
- 26.Roy, N. K., Deka A., Bordoloi D., Mishra S., Kumar A. P., Sethi G., Kunnumakkara A. B. 2016. The potential role of boswellic acids in cancer prevention and treatment. Cancer Lett. 377: 74–86. [DOI] [PubMed] [Google Scholar]
- 27.Reising, K., Meins J., Bastian B., Eckert G., Mueller W. E., Schubert-Zsilavecz M., Abdel-Tawab M. 2005. Determination of boswellic acids in brain and plasma by high-performance liquid chromatography/tandem mass spectrometry. Anal. Chem. 77: 6640–6645. [DOI] [PubMed] [Google Scholar]
- 28.Yan, S., Xu Z., Lou F., Zhang L., Ke F., Bai J., Liu Z., Liu J., Wang H., Zhu H., et al. 2015. NF-κB-induced microRNA-31 promotes epidermal hyperplasia by repressing protein phosphatase 6 in psoriasis. Nat. Commun. 6: 7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lomenick, B., Hao R., Jonai N., Chin R. M., Aghajan M., Warburton S., Wang J., Wu R. P., Gomez F., Loo J. A., et al. 2009. Target identification using drug affinity responsive target stability (DARTS). Proc. Natl. Acad. Sci. USA 106: 21984–21989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lomenick, B., Olsen R. W., Huang J. 2011. Identification of direct protein targets of small molecules. ACS Chem. Biol. 6: 34–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Okamoto, K., Iwai Y., Oh-Hora M., Yamamoto M., Morio T., Aoki K., Ohya K., Jetten A. M., Akira S., Muta T., Takayanagi H. 2010. IkappaBzeta regulates T(H)17 development by cooperating with ROR nuclear receptors. Nature 464: 1381–1385. [DOI] [PubMed] [Google Scholar]
- 32.Bambouskova, M., Gorvel L., Lampropoulou V., Sergushichev A., Loginicheva E., Johnson K., Korenfeld D., Mathyer M. E., Kim H., Huang L. H., et al. 2018. Electrophilic properties of itaconate and derivatives regulate the IκBζ-ATF3 inflammatory axis. Nature 556: 501–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Majumder, S., Amatya N., Revu S., Jawale C. V., Wu D., Rittenhouse N., Menk A., Kupul S., Du F., Raphael I., et al. 2019. IL-17 metabolically reprograms activated fibroblastic reticular cells for proliferation and survival. Nat. Immunol. 20: 534–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ganapathy-Kanniappan, S., Geschwind J.-F. H. 2013. Tumor glycolysis as a target for cancer therapy: progress and prospects. Mol. Cancer 12: 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goverman, J. 2009. Autoimmune T cell responses in the central nervous system. Nat. Rev. Immunol. 9: 393–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mabuchi, T., Takekoshi T., Hwang S. T. 2011. Epidermal CCR6+ γδ T cells are major producers of IL-22 and IL-17 in a murine model of psoriasiform dermatitis. J. Immunol. 187: 5026–5031. [DOI] [PubMed] [Google Scholar]
- 37.Gerbeth, K., Hüsch J., Fricker G., Werz O., Schubert-Zsilavecz M., Abdel-Tawab M. 2013. In vitro metabolism, permeation, and brain availability of six major boswellic acids from Boswellia serrata gum resins. Fitoterapia 84: 99–106. [DOI] [PubMed] [Google Scholar]
- 38.Sundrud, M. S., Koralov S. B., Feuerer M., Calado D. P., Kozhaya A. E., Rhule-Smith A., Lefebvre R. E., Unutmaz D., Mazitschek R., Waldner H., et al. 2009. Halofuginone inhibits TH17 cell differentiation by activating the amino acid starvation response. Science 324: 1334–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ravindran, R., Loebbermann J., Nakaya H. I., Khan N., Ma H., Gama L., Machiah D. K., Lawson B., Hakimpour P., Wang Y. C., et al. 2016. The amino acid sensor GCN2 controls gut inflammation by inhibiting inflammasome activation. Nature 531: 523–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stumpf, C. R., Moreno M. V., Olshen A. B., Taylor B. S., Ruggero D. 2013. The translational landscape of the mammalian cell cycle. Mol. Cell 52: 574–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shuda, M., Chang Y., Moore P. S. 2015. Mitotic 4E-BP1 hyperphosphorylation and cap-dependent translation. Cell Cycle 14: 3005–3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tanenbaum, M. E., Stern-Ginossar N., Weissman J. S., Vale R. D. 2015. Regulation of mRNA translation during mitosis. Elife 4: e07957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jeltsch, K. M., Hu D., Brenner S., Zöller J., Heinz G. A., Nagel D., Vogel K. U., Rehage N., Warth S. C., Edelmann S. L., et al. 2014. Cleavage of roquin and regnase-1 by the paracaspase MALT1 releases their cooperatively repressed targets to promote T(H)17 differentiation. Nat. Immunol. 15: 1079–1089. [DOI] [PubMed] [Google Scholar]
- 44.Almeida, L., Lochner M., Berod L., Sparwasser T. 2016. Metabolic pathways in T cell activation and lineage differentiation. Semin. Immunol. 28: 514–524. [DOI] [PubMed] [Google Scholar]
- 45.Geltink, R. I. K., Kyle R. L., Pearce E. L. 2018. Unraveling the complex interplay between T cell metabolism and function. Annu. Rev. Immunol. 36: 461–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dang, E. V., Barbi J., Yang H. Y., Jinasena D., Yu H., Zheng Y., Bordman Z., Fu J., Kim Y., Yen H. R., et al. 2011. Control of T(H)17/T(reg) balance by hypoxia-inducible factor 1. Cell 146: 772–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jäger, A., Dardalhon V., Sobel R. A., Bettelli E., Kuchroo V. K. 2009. Th1, Th17, and Th9 effector cells induce experimental autoimmune encephalomyelitis with different pathological phenotypes. J. Immunol. 183: 7169–7177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pantelyushin, S., Haak S., Ingold B., Kulig P., Heppner F. L., Navarini A. A., Becher B. 2012. Rorγt+ innate lymphocytes and γδ T cells initiate psoriasiform plaque formation in mice. J. Clin. Invest. 122: 2252–2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cai, Y., Fleming C., Yan J. 2012. New insights of T cells in the pathogenesis of psoriasis. Cell. Mol. Immunol. 9: 302–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.