Abstract

Schistosomiasis is a chronic and painful disease of poverty caused by the flatworm parasite Schistosoma. Drug discovery for antischistosomal compounds predominantly employs in vitro whole organism (phenotypic) screens against two developmental stages of Schistosoma mansoni, post-infective larvae (somules) and adults. We generated two rule books and associated scoring systems to normalize 3898 phenotypic data points to enable machine learning. The data were used to generate eight Bayesian machine learning models with the Assay Central software according to parasite’s developmental stage and experimental time point (≤24, 48, 72, and >72 h). The models helped predict 56 active and nonactive compounds from commercial compound libraries for testing. When these were screened against S. mansoni in vitro, the prediction accuracy for active and inactives was 61% and 56% for somules and adults, respectively; also, hit rates were 48% and 34%, respectively, far exceeding the typical 1–2% hit rate for traditional high throughput screens.
Keywords: Schistosoma, schistosomiasis, drug discovery, machine learning, Bayesian, phenotypic screen
Schistosomiasis is one of a number of parasitic infectious diseases associated with poverty that principally impact low- and middle-income countries.1,2 The disease is caused by various species of the flatworm parasite, Schistosoma, which lives in the blood vasculature and produces eggs that are responsible for a variety of pathologies. With more than 200 million people infected worldwide, the painful and often lifelong consequences of this disease can negatively impact the economic performance of the afflicted communities.3,4 Treatment relies solely on praziquantel (PZQ),5−9 which is safe, affordable, and reasonably effective in decreasing disease-associated morbidity. However, as the only drug available, there is concern regarding decreased efficacy or resistance, particularly as its use continues to expand.9−11 Moreover, there is a lack of pharmaceutical investment in new chemotherapies for schistosomiasis.
Academia remains key to the identification, characterization, and preclinical evaluation of antischistosomal small molecules.5,11 This process has involved small molecule screens of either validated targets or, more often, phenotypic (whole organism) screens of the schistosome parasite in culture.11,12 Although the amount of data accumulated is small relative to major areas of research such as cancer, it is still a valuable resource for the application of machine learning methods to the drug discovery process. Computational techniques are an attractive drug discovery and development modality, especially given the financially constrained environment for diseases like schistosomiasis.11,13 To date, however, there have been just a few efforts using these types of tools (e.g., docking and quantitative structure–activity relationship models) for schistosomiasis, in contrast to the more typical strategy of screening small molecule collections.11,14,15 Bayesian machine learning methods have convincingly demonstrated their applicability to predicting active compounds for other infectious diseases of poverty such as Chagas disease,16 Ebola,17,18 and tuberculosis.19−21
With respect to schistosomiasis, the phenotypic screening data in the literature have been generated using a plethora of quantitative or partially quantitative metrics for bioactivity and involved more than one developmental stage, most often post-infective larvae (schistosomula or somules) and adults of S. mansoni, the species best adapted to the laboratory environment.12,22−25 To render the disparate data potentially useful for machine learning methods, we developed two “rule books” with which the data identified in a literature search could be normalized. These data were used to generate Assay Central18,26−30 Bayesian machine learning models of antischistosomal activity for somules and adults over four different time points (eight models total). These Bayesian models were subsequently used to identify potential antischistosomal molecules in various chemical libraries. Using both manual and automated molecule selection techniques, a set of compounds was purchased and screened for bioactivity against somules and adults of S. mansoni. The eight Assay Central training data sets produced a high-quality, binary data set that can be utilized for additional machine learning methods. Also, each of the eight training data sets was applied to six other algorithms, and these model performances were compared to that of Assay Central.
Results
For the machine learning application of Assay Central, two rule books were developed. The first normalized phenotypic screening data for S. mansoni that reported single metric outputs (e.g., ED50 and % mortality; 19 articles between 1980 and 2019; Tables 1 and S1), and the second normalized data from screens mainly performed by the University of California, San Diego (UCSD) team (two published articles and 13 published and unpublished data sets) using an observational approach that describes and then enumerates the many phenotypic changes of which the schistosome is capable (Tables 2 and S2). In total, 3377 somule and 521 adult worm data points were curated for machine learning methods.
Table 1. Rule Book for Example Phenotypic Screen Data from the Literature That Report Single Metric Outputs.
| rule
book score (0–4)a associated with compound concentration (μM) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| reference | test parameter | developmental stageb | time (h) | conc. (μM) | 0 | 1 | 2 | 3 | 4 |
| (22) | EC50 (μM) | 49-Adult | 24 | NA | ≥50 | <50 | <25 | <10 | <5 |
| 48 | ≥25 | <25 | <12.5 | <7.5 | <5 | ||||
| 72 | ≥10 | <10 | <5 | <2.5 | <1 | ||||
| (31) | LD50 (μM) | 24-NTS | 24 | NA | ≥50 | <50 | <25 | <10 | <5 |
| (32) | percent killing | 49-Adult | 4 | 10 | <10 | <25 | <50 | <75 | <100 |
| 8 | <10 | <25 | <50 | <75 | <100 | ||||
| 16 | <10 | <25 | <50 | <75 | <100 | ||||
| 24 | <25 | <50 | <75 | <100 | |||||
| 48 | <50 | <75 | <100 | ||||||
| 72 | <75 | <100 | |||||||
| (33) | 100% mortality (μM) | 24-NTS | 48 | various fixed values | ≥25 | 25 | 12.5 | 6.25 | 3.125 |
| 49-Adult | 168 | >5 | ≤5 | ||||||
| (34) | percent motility reduction | 49-Adult | 120 | ∼30 | <100 | 100 | |||
| (35) | minimum active concentration (MAC; μM) | 46-Adult | 96 | NA | >10 | <5 | <2.5 | <1 | |
Compound activities have rule book scores that are scaled from 0 to 4 where 4 represents the most active compound.
Terms: NTS, newly transformed somules; 24-NTS, NTS allowed to acclimate to culture conditions overnight or for 24 h prior to screening; 46-Adult, 46-day-old adult; 49-Adult, 49-day-old adult. See Table S1 for full details.
Table 2. Rule Book for Example Screen Data Based on the Number and Severity of Phenotypic Changes Taking Place.
| rule
book score (0–4)a associated with the time (h) to appearance of phenotypic changes |
|||||||
|---|---|---|---|---|---|---|---|
| number of phenotypic changes recorded | developmental stage | concentration tested (μM) | 0 | 1 | 2 | 3 | 4 |
| 3 changes or D/deg/teg blebb | NTS and 42-Adults | 0.1 | >192 | <168 | <144 | <120 | <96 |
| 2 changes | >168 | <144 | <120 | <96 | <72 | ||
| 1 change | >144 | <120 | <96 | <72 | <24 | ||
| 3 changes or D/deg/teg bleb | 5 | >120 | <96 | <72 | <48 | <24 | |
| 2 changes | >96 | <72 | <48 | <24 | <6 | ||
| 1 change | >72 | <48 | <24 | <6 | <1 | ||
| 3 changes or D/deg/teg bleb | 10 | >120 | <96 | <72 | <48 | <24 | |
| 2 changes | >96 | <72 | <48 | <24 | <3 | ||
| 1 change | >72 | <48 | <24 | <3 | <1 | ||
Compound activities have rule book scores that are scaled from 0 to 4 where 4 represents the most active compound.
Terms: D, dead; deg, degenerating; teg bleb, damage (blebbing) to surface tegument of adult parasites; any one of these particular changes observed is awarded a rule book score of 4. See Table S2 for full details.
Data sets resulting from the two rule books were combined to develop eight Bayesian machine learning models with Assay Central over four time points (≤24, 48, 72, and >72 h) for both somules and adults (Figure 1). Active (hit) compounds were defined as those receiving rule book scores of 3 or 4. Five-fold cross-validation performance metrics for these machine learning models are presented in Table 3 and Figure S1. Of the approximately 3100 and 500 compounds screened in the literature against somules and adults, respectively, both sets had a similar 5–10% recovery of active molecules and covered a similar chemical space as measured by our domain score of 0.305–0.384 (which is calculated by reference to the ChEMBL database36). Predictive performance was assessed via receiver operating characteristic (ROC) scores, which fell within a tight range from 0.796 to 0.845 across all time points and developmental stages, thus suggesting that they are likely performing similarly. In general, all of the internal performance metrics were higher for adult models than somule models, particularly the F1-Score, Cohen’s kappa, and Matthews correlation coefficient.
Figure 1.

Workflow for the overall process of selecting compounds from machine learning models. Dotted lines represent processes, and solid lines represent outputs. See Methods section and Figures 2 and 3 for more information on the selection processes that resulted in the 56 compounds being tested for bioactivity against S. mansoni.
Table 3. Five-Fold Cross-Validation Results of Bayesian Machine Learning Models Used to Predict 56 Compounds for Screening against S. mansoni In Vitroa.
| time point (h) | life stage | no. active | no. total | ROC | F1-score | CK | MCC | domain |
|---|---|---|---|---|---|---|---|---|
| ≤24 | adults | 45 | 493 | 0.845 | 0.426 | 0.339 | 0.397 | 0.308 |
| ≤24 | somules | 227 | 3114 | 0.806 | 0.260 | 0.157 | 0.246 | 0.383 |
| 48 | adults | 35 | 450 | 0.832 | 0.310 | 0.210 | 0.307 | 0.307 |
| 48 | somules | 204 | 3101 | 0.818 | 0.276 | 0.187 | 0.276 | 0.382 |
| 72 | adults | 34 | 447 | 0.841 | 0.494 | 0.441 | 0.457 | 0.305 |
| 72 | somules | 175 | 2979 | 0.843 | 0.276 | 0.198 | 0.290 | 0.377 |
| >72 | adults | 85 | 509 | 0.815 | 0.533 | 0.410 | 0.432 | 0.308 |
| >72 | somules | 358 | 3151 | 0.796 | 0.366 | 0.232 | 0.302 | 0.384 |
Terms: ROC = receiver operator characteristic; CK = Cohen’s kappa; MCC = Matthews correlation coefficient.
In addition to the Bayesian algorithm of Assay Central, six other machine learning methods (random forest, k-Nearest Neighbors, support vector classification, naïve Bayesian, AdaBoosted decision trees, and deep learning) were applied to the eight training data sets arising from the implementation of the rule books. The same 5-fold cross-validation performance metrics output by Assay Central were generated to allow for an evaluation of machine learning algorithms on the same data sets. These metrics were compared as radar plots in Figure S2. Metrics were comparable between the algorithms, although recall and ROC (also referred to as area-under-the-curve) were generally greater for the Assay Central models (difference of ≥0.2), especially in the adult data sets. Independent and pairwise comparisons of these alternative algorithms are shown in Table S3, and Figure S3 depicts the rank normalized and “difference from the top” rank normalized score (ΔRNS) metrics. These comparisons suggest that there are no significant differences between most machine learning algorithms for these data sets even using more sophisticated and computationally intensive machine learning methods. The exception was the Adaboosted decision trees algorithm, which was significantly poorer in performance than the other algorithms.
A total of 56 compounds were selected and purchased for phenotypic screening of S. mansoni somules and adults (Figure S4). For each developmental stage, 10 predicted actives were chosen using each of the automated and manual methods (Figures 2 and 3), and eight predicted nonactives were chosen using the manual method. Although compounds were selected on a developmental stage-specific basis, all compounds were tested against both stages. The identities of the purchased compounds were blinded to the UCSD team performing the phenotypic screens until after the screen data were assembled. Results are summarized in Tables 4 and 5 for somules and adults, respectively, and the combined data for all compounds against both developmental stages are presented in Table S4. Table S5 presents the same 5-fold cross-validation metrics discussed above for the training data after integrating the phenotypic screening data for the 56 tested compounds. This inclusion of tested compounds had little, if any, consistent impact on the metrics compared to the original models.
Figure 2.

Workflow for the manual compound selection process that utilized only the raw predictions from Assay Central models. Dotted lines represent processes, and solid lines represent outputs. Assay Central honeycomb plots are described in the Methods section.
Figure 3.

Workflow for the automated compound selection process that utilized rank and consensus scripts with the diversity collection from Enamine. Dotted lines represent processes, and solid lines represent outputs.
Table 4. Screening of Automatically and Manually Predicted Actives, and Nonactives vs S. mansoni Somules In Vitro at 10 μM for the Time Points Indicated.
| somule
severity scorea (10 μM) |
||||||
|---|---|---|---|---|---|---|
| compound | SMILES | method | prediction | 24 h | 48 h | 72 h |
| Z304863612 | OC(CNC1=C2C=CC=CC2=NC(=N1)C3=CC=NC=C3)C4=CC=C(Cl)C=C4 | automated | active | 3 | 4 | 4 |
| Z56174662 | CC1=C(C(N2CCN(CC2)C3=CC=CC=C3)C4=CC=CS4)C5=C([NH]1)C=CC=C5 | automated | active | 2 | 4 | 4 |
| Z56175896 | CC1=C(C(N2CCN(CC2)C3=CC=C(F)C=C3)C4=NC=CC=C4)C5=C([NH]1)C=CC=C5 | automated | active | 0 | 0 | 3 |
| Z56978084 | ClC1=C(Cl)C=C(NC(=O)NC2=C(C(=O)N3CCOCC3)C4=C(CCCC4)S2)C=C1 | automated | active | 2 | 1 | 2 |
| Z133946058 | COC1=CC=C(C=C1)N2CCN(CCCN[S](=O)(=O)C3=C(C=C(Cl)C=C3)C(F)(F)F)CC2 | automated | active | 1 | 1 | 2 |
| Z204004384 | COC1=CC=C(CN2CCCN(CC2)C3=C4C5=C(CCC5)SC4=NC(=N3)C6=CN=CC=C6)C=C1 | automated | active | 0 | 0 | 2 |
| Z385159220 | COC1=C(OC)C=C(CCNC(=O)CCCNC2=C3C=CC(=CC3=NC=C2)Cl)C=C1 | automated | active | 0 | 1 | 2 |
| Z48867676 | CN(C)CCCNC1=NC(=CS1)C2=CC=C(C=C2)[S](=O)(=O)N3CCCCCC3 | automated | active | 1 | 0 | 1 |
| Z276431168 | FC1=C(C=CC=C1)C(=O)NC2=C3C=CC=CC3=NC4=C2CCCC4 | automated | active | 0 | 0 | 0 |
| Z89250915 | CC(NC1=CC=C(C=C1)N2CCN(CC2)CC3=CC=CC=C3)C(=O)NCC4=CC=CC=C4 | automated | active | 0 | 0 | 0 |
| (S)-duloxetine hydrochloride | CNCC[C@H](OC1=C2C=CC=CC2=CC=C1)C1=CC=CS1 | manual | active | 3 | 4 | 4 |
| revaprazan hydrochloride | c1cc(F)ccc1Nc2nc(C)c(C)c(n2)N3CCc4ccccc4C3C | manual | active | 2 | 4 | 4 |
| Z56872965 | CCCCC[N]1C(=C(C(=O)NCCN2CCOCC2)C3=C1N=C4C=CC=CC4=N3)N | manual | active | 1 | 4 | 4 |
| amsacrine hydrochloride | COC1=CC(=CC=C1NC2=C3C=CC=CC3=NC4=C2C=CC=C4)N[S](C)(=O)=O | manual | active | 1 | 2 | 4 |
| Z425126666 | OC(COC1=CC=CC=C1)C[N]2C(=NC3=C2C=CC=C3)CC4=NC5=C(S4)C=CC=C5 | manual | active | 0 | 0 | 2 |
| tyrphostin AG 1478 | COC1=C(OC)C=C2C(=NC=NC2=C1)NC3=CC(=CC=C3)Cl | manual | active | 1 | 1 | 1 |
| Org 27569 | CCC1=C([NH]C2=C1C=C(Cl)C=C2)C(=O)NCCC3=CC=C(C=C3)N4CCCCC4 | manual | active | 0 | 0 | 0 |
| Z367636216 | CC1=CC(=CC=C1)C2=NN=C(S)[N]2CC(=O)NCCCN3C4=C(SC5=C3C=CC=C5)C=CC=C4 | manual | active | 0 | 0 | 0 |
| caroverine hydrochloride monohydrate | CCN(CC)CCN1C(=O)C(=NC2=CC=CC=C12)CC3=CC=C(OC)C=C3 | manual | active | 0 | 0 | 0 |
| AGK2 | ClC1=CC=C(Cl)C(=C1)C2=CC=C(O2)\C=C(C#N)\C(=O)NC3=CC=CC4=C3C=CC=N4 | manual | active | 0 | 0 | 0 |
| U-73122 | COC1=CC2=C(C=C1)[C@H]1CC[C@]3(C)[C@H](CC[C@H]3[C@@H]1CC2)NCCCCCCN1C(=O)C=CC1=O | manual | nonactive | 4 | 4 | 4 |
| piperlongumine | COC1=CC(=CC(=C1OC)OC)/C=C/C(=O)N2CCC=CC2=O | manual | nonactive | 4 | 4 | 4 |
| tiamulin fumarate | CCN(CC)CCSCC(=O)O[C@@H]1C[C@@](C)(C=C)[C@@H](O)[C@H](C)[C@]23CCC(=O)[C@H]2[C@@]1(C)[C@H](C)CC3 | manual | nonactive | 2 | 1 | 3 |
| sivelestat sodium tetrahydrate | CC(C)(C)C(=O)OC1=CC=C(C=C1)[S](=O)(=O)NC2=CC=CC=C2C(=O)NCC(O)=O | manual | nonactive | 0 | 0 | 0 |
| PNU-282987 | O=C(N[C@H]1CN2CCC1CC2)c1ccc(Cl)cc1 | manual | nonactive | 0 | 0 | 0 |
| R(+)-IAA-94 | CC1(CC2=CC(=C(Cl)C(=C2C1=O)Cl)OCC(O)=O)C3CCCC3 | manual | nonactive | 0 | 0 | 0 |
| ecabet sodium | CC(C)C1=C(C=C2C(CC[C@H]3[C@@](C)(CCC[C@]23C)C(O)=O)=C1)S(O)(=O)=O | manual | nonactive | 0 | 0 | 0 |
| I-OMe-tyrphostin AG 538 | COC1=C(O)C(=CC(=C1)\C=C(C#N)\C(=O)C2=CC(=C(O)C=C2)O)I | manual | nonactive | 0 | 0 | 0 |
Compound activities have severity scores that are scaled from 0 to 4 where 4 represents the most active compound. Active compounds are those generating a severity score of ≥2. Compounds were tested in two experiments, each in duplicate, and representative data are shown. Structures and the descriptors associated with the severity scores are shown in Table S4 as are the phenotypic data arising from the use of 1 μM compound.
Table 5. Screening of Automatically and Manually Predicted Actives and Nonactives vs Adult S. mansoni In Vitro at 10 μM for the Time Points Indicated.
| adult
severity scorea (10 μM) |
|||||||
|---|---|---|---|---|---|---|---|
| compound | SMILES | method | prediction | 1 h | 5 h | 24 h | 48 h |
| Z288901226 | C[S](=O)(=O)NC1=C(F)C=CC(=C1)NC(=O)C2=C(NC3=CC(=CC=C3)C(F)(F)F)N=CC=C2 | automated | active | 1 | 1 | 3 | 4 |
| Z2241105867 | ClC1=CC(=CC=C1)C2=N[N](C=C2CNCC3=CC=CO3)C4=CC=CC=C4 | automated | active | 2 | 2 | 2 | 3 |
| Z827016000 | CC(NCC1=CC=C(O1)C2=CC=C(C=C2)C(F)(F)F)C3=CC=C(N[S](C)(=O)=O)C=C3 | automated | active | 2 | 2 | 2 | 2 |
| Z105384660 | FC1=CC=C(C=C1)C(N2CCN(CC2)CC3=CSC=N3)C4=CC=C(F)C=C4 | automated | active | 1 | 1 | 1 | 1 |
| Z44528364 | CC(N1CCN(CC1)C/C=C/C2=CC=CC=C2)C(=O)N(C)CC3=CC=CC=C3 | automated | active | 0 | 1 | 0 | 0 |
| Z827015296 | OCCCNCC1=CC=C(O1)C2=C(Cl)C=C(Cl)C=C2 | automated | active | 0 | 0 | 0 | 0 |
| Z225086696 | COC1=C(OC)C(=C(CN2CCN(CC2)C(=O)C3=C[NH]N=C3C4=CC=C(F)C=C4)C=C1)OC | automated | active | 0 | 0 | 0 | 0 |
| Z230347224 | CC(=O)NC1=C(Cl)C=C(NC(=O)C2=C[NH]N=C2C3=CC=C(F)C=C3)C=C1 | automated | active | 0 | 0 | 0 | 0 |
| Z56958732 | COC(=O)C1=C(C)NC(=C(C1C2=CC(=C(OC)C(=C2)Br)OC)C(=O)OC)C | automated | active | 0 | 0 | 0 | 0 |
| Z90192490 | O=C(NC1CC1)C(N2CCN(CC2)C/C=C/C3=CC=CC=C3)C4=CC=CC=C4 | automated | active | 0 | 0 | 0 | 0 |
| nemadipine-A | CCOC(=O)C1=C(C)NC(=C(C1C2=C(F)C(=C(F)C(=C2F)F)F)C(=O)OCC)C | manual | active | 4 | 4 | 4 | 4 |
| moxidectin | CO\N=C1\C[C@]2(C[C@@H]3C[C@@H](C\C=C(C)\C[C@@H](C)\C=C\C=C4/CO[C@@H]5[C@H](O)C(C)=C[C@@H](C(=O)O3)[C@]45O)O2)O[C@@H]([C@H]1C)C(\C)=C\C(C)C | manual | active | 2 | 2 | 3 | 4 |
| etravirine | CC1=C(OC2=NC(=NC(=C2Br)N)NC3=CC=C(C=C3)C#N)C(=CC(=C1)C#N)C | manual | active | 2 | 2 | 2 | 3 |
| Z53005631 | CN(CC(=O)NC1=C(SCC#N)C=CC=C1)CC2=CC=C(O2)C3=CC=C(Br)C=C3 | manual | active | 0 | 2 | 2 | 2 |
| SB202190 hydrochloride | OC1=CC=C(C=C1)C2=NC(=C([NH]2)C3=CC=NC=C3)C4=CC=C(F)C=C4 | manual | active | 0 | 0 | 2 | 2 |
| niflumic acid | OC(=O)C1=CC=CN=C1NC2=CC=CC(=C2)C(F)(F)F | manual | active | 0 | 0 | 1 | 1 |
| Z826994844 | FC1=C(CN(CC2=NC=CC=C2)[S](=O)(=O)C3=CC(=C(Cl)C=C3)C#N)C=CC(=C1)Br | manual | active | 0 | 0 | 0 | 0 |
| Z18885599 | FC(F)(F)C1=CC(=CC=C1)NC2=C(C=CC=N2)C(=O)OCC(=O)NC3=C(C=CC=C3)C#N | manual | active | 0 | 0 | 0 | 0 |
| trilostane | C[C@]12CC[C@H]3[C@@H](CC[C@@]45O[C@@H]4C(O)=C(C[C@]35C)C#N)[C@@H]1CC[C@@H]2O | manual | active | 0 | 0 | 0 | 0 |
| mycophenolic acid | COC1=C(C\C=C(C)\CCC(O)=O)C(=C2C(=O)OCC2=C1C)O | manual | active | 0 | 0 | 0 | 0 |
| itraconazole | CCC(C)N1N=CN(C1=O)C1=CC=C(C=C1)N1CCN(CC1)C1=CC=C(OC[C@H]2CO[C@@](CN3C=NC=N3)(O2)C2=C(Cl)C=C(Cl)C=C2)C=C1 | manual | nonactive | 0 | 0 | 3 | 3 |
| dabigatran etexilate | CCCCCCOC(=O)NC(=N)C1=CC=C(NCC2=NC3=CC(=CC=C3[N]2C)C(=O)N(CCC(=O)OCC)C4=NC=CC=C4)C=C1 | manual | nonactive | 1 | 1 | 1 | 1 |
| BIX 01294 trihydrochloride hydrate | COC1=CC2=NC(=NC(=C2C=C1OC)NC3CCN(CC3)CC4=CC=CC=C4)N5CCCN(C)CC5 | manual | nonactive | 1 | 0 | 0 | 0 |
| eletriptan | CN1CCC[C@@H]1CC1=CNC2=CC=C(CCS(=O)(=O)C3=CC=CC=C3)C=C12 | manual | nonactive | 1 | 0 | 0 | 0 |
| rutecarpine | O=C1N2CCC3=C([NH]C4=C3C=CC=C4)C2=NC5=C1C=CC=C5 | manual | nonactive | 0 | 0 | 0 | 0 |
| tetrabenazine | COC1=CC2=C(C=C1OC)C3CC(=O)C(CC(C)C)CN3CC2 | manual | nonactive | 0 | 0 | 0 | 0 |
| clindamycin 2-phosphate | CCC[C@@H]1C[C@H](N(C)C1)C(=O)N[C@H]([C@H](C)Cl)[C@H]1O[C@H](SC)[C@H](OP(O)(O)=O)[C@@H](O)[C@H]1O | manual | nonactive | 0 | 0 | 0 | 0 |
| ondansetron hydrochloride dihydrate | C[N]1C2=C(C(=O)C(CC2)C[N]3C=CN=C3C)C4=C1C=CC=C4 | manual | nonactive | 0 | 0 | 0 | 0 |
Compound activities have severity scores that are scaled from 0 to 4 where 4 represents the most active compound. Active compounds are those generating a severity score of ≥2. Compounds were tested in two experiments, each in duplicate, and representative data are shown. Structures and the descriptors associated with the severity scores are shown in Table S4.
Automated and Manual Predictions for Somules
For somules, regardless of whether manual or automated predictions were made, the predicted actives possessed structural moieties in common with the active compounds in the training set. This is to be expected. These features include fused aromatic ring systems such as phenothiazines, indoles, and piperazines, nitrogen heterocycles such as 4-anilinoquinazoline, and peripheral substituents such as chlorine and fluorine (examples in Figure 4). Overall, 61% of those compounds predicted to be either active (yielding a severity score of ≥2) or inactive against somules was indeed confirmed as such in the phenotypic screening assay (Table 4). Further, 27 of the 56 (48%) compounds tested vs somules were active.
Figure 4.

Compounds selected for phenotypic screening against S. mansoni somules and that exemplify the trends observed in the raw predictions from multiple vendor libraries for this developmental stage. Salts were removed for clarity of the parent compound.
Seven of the ten automatically predicted active compounds against somules were experimentally confirmed, i.e., severity scores of ≥2 at 10 μM after 72 h (Table 4). Three were strong hits with severity scores of 3 or 4, namely, Z304863612, Z56174662, and Z56175896, whereas the other four, Z56978084, Z133946058, Z204004384, and Z385159220, yielded scores of 2. Notably, the top hit, Z304862612, was also a strong hit at 1 μM with a severity score of 4 after 72 h (Table S4). Furthermore, two compounds, Z56174662 and Z56978084, were active against the adults with scores of 4 and 2, respectively, after 48 h (Table S4).
Five of the ten predicted actives chosen manually for somules were confirmed experimentally (Table 4). Four of these were strongly active with severity scores of 4 after 72 h (i.e., the antidepressant (S)-duloxetine hydrochloride; the proton pump inhibitor revaprazan hydrochloride; the antineoplastic amsacrine hydrochloride; and Z56872965). In contrast, Z425126666 yielded a score of 2. The two top hits, (S)-duloxetine and revaprazan hydrochloride, were also active at 1 μM, generating severity scores of 4 and 3, respectively, after 72 h (Table S4). Furthermore, five compounds, namely, revaprazan hydrochloride, Z56872965, Z425126666, Org 27569, and Z367636216, were active against adults with severity scores between 2 and 4 after 48 h (Table S4).
Three of the eight compounds manually selected as nonactive compounds against somules were, in fact, strongly active at 10 μM: the phosphoinositide-specific phospholipase C inhibitor U-73122, the natural product piperlongumine, and the antibiotic tiamulin fumarate. The other five predicted nonactives, sivelestat sodium, PNU-282987, R(+)-IAA-94, ecabet sodium, and I-OMe-Tyrphostin AG 538, were confirmed as inactive (Table 4). Two of the active compounds, U-73122 and piperlongumine, were also active against adults with severity scores of 4 after 48 h (Table S4).
Automated and Manual Predictions for Adults
Similar to somules and regardless of whether manual or automated predictions were employed, the predicted adult active compounds possessed many structural moieties observed in the active training data compounds. Also, the prediction of hits for adults included those substituents seen in somules such piperazine rings and halogens (namely, trifluorine and bromine) as well as nitrile and carbonyl moieties (examples in Figure 5). Other chemistries not seen in the somule outputs included dihydropyridine analogs and steroids as well as compounds with multiple methoxy substituents. Overall, 56% of those compounds predicted to be either active (yielding a severity score of ≥2) or inactive against adults was indeed confirmed as such in the phenotypic screening assay (Table 5). Further, 19 of the 56 (34%) compounds tested vs adults were active.
Figure 5.

Compounds selected for in vitro testing against adult S. mansoni and that exemplify the trends observed in the raw predictions from multiple vendor libraries for this developmental stage. Salts were removed for clarity of the parent compound.
Of the ten automated active predictions for adult worms, three were confirmed as active, i.e., severity scores of ≥2 at 10 μM after 48 h with Z827016000, Z2241105867, and Z288901226 generating severity scores of 2, 3, and 4, respectively (Table 5). Two of these, Z827016000 and Z2241105867, and a third adult-inactive compound, Z827015296, were active against somules at 10 μM after 72 h with scores of 3 or 4 (Table S4).
Five of the ten manually predicted adult actives were confirmed as active (Table 5). Specifically, nemadipine-A, an L-type calcium channel blocker, generated the maximum severity score of 4 at all time points measured. Moxidectin, an antinematode macrocyclic lactone, was also strongly active with a score of 4 after 48 h. Both bioactivities are consistent with previously published data for these compounds.37,38 The non-nucleoside reverse transcriptase inhibitor etravirine39,40 was active with a score of 3, whereas two other active compounds, Z53005631 and the p38 MAPK inhibitor SB202190 hydrochloride, each yielded scores of 2. The same five compounds were also active against somules at 10 μM after 72 h with scores between 2 and 4 (Table S4), as was one additional nonadult active compound, Z826994844, with a score of 2 after 72 h.
Finally, of the eight manually predicted nonactive compounds, only the antifungal itraconazole was active against adult worms with a score of 3 after 48 h (Table 5). The same compound plus two others, dabigatran etexilate and BIX 01294 trihydrochloride hydrate, were also active against somules at 10 μM after 72 h with scores of 2 and 4, respectively (Table S4).
Compound Prioritization Process for Future Antischistosomal Studies
Because adult worms are ultimately responsible for disease in humans via the eggs they produce,4 nine bioactive compounds were initially prioritized for further investigation based on the generation of severity scores of 3 or 4 against adults after 24 h. These were itraconazole, moxidectin, piperlongumine, nemadipine-A, the benzimidazole Z425126666, revaprazan hydrochloride, the indole Z56174662, the pyridine-containing Z288901226, and U-73122. Prioritization was also influenced by activity against somules and, with the exception of Z288901226, the nine chosen compounds generated severity scores of ≥2 after 72 h (Tables 4 and S4).
During the screening assays, precipitation was noted for itraconazole, and the compound was not considered further. Three other compounds are known to have antischistosomal effects, including in some cases in vivo activity, namely, moxidectin22,37 piperlongumine,41−43 and nemadipine-A.38 Due to our desire to identify novel starting points for treatments, these were also removed from further consideration.
The remaining five compounds (Figure 6) were evaluated with other Assay Central models for stability,44 permeability,45 and cytotoxicity46 (Table S6 and Figure S5). From these predictions, Z425126666 scored the best, i.e., was active for stability and permeability but inactive for cytotoxicity. The other four compounds were scored as inactive for stability and active for permeability and cytotoxicity.
Figure 6.

Five antischistosomal compounds prioritized for future studies. Salts were removed for clarity of the parent compound.
Discussion
For machine learning, we developed two rule books to normalize the disparate literature data arising from small molecule, in vitro phenotypic screens of S. mansoni (Tables 1 and 2; Tables S1 and S2). The parsing and normalization of the data for the rule books were manually intensive and time-consuming yet necessary to develop the machine learning models. The rule books are also a first step toward developing a unified database of antischistosomal compounds.
The scores derived from both rule books were pooled and applied to eight Bayesian machine learning methods with Assay Central. These models produced favorable 5-fold cross-validation metrics with ROC scores exceeding 0.8 (Table 3, Figure S1). Although less literature data were available for adult worms (the largest model totaled 509 versus 3151 compounds for somules), there were generally higher 5-fold cross-validation performance metrics for these models over the somule counterparts. The more diverse and larger somule sets have a lower ratio of actives to total compounds compared with the adult data sets (approximately 1–5% less), which likely impacted the internal performance of the models. Machine learning models are only as good as the data that comprises them, so with less active compounds to learn bioactivity features from, the less likely predictions will be accurate.
Comparisons between the machine learning methods (Figures S2 and S3, Table S3) suggest that the more advanced methods like deep learning and support vector classification do not significantly improve the internal predictive performance of the resulting models. This is a similar outcome to previous comparisons of the same algorithms using data sets for tuberculosis and HIV infection.28,30 This could be related to the data set size, balance of the data set, or other factors such as model hyperparameter optimization. Lacking an algorithm with a clear and significant performance increase, the Bayesian method utilized by Assay Central is faster in generating models compared to the other algorithms like deep learning and can be implemented quickly on an average desktop computer, a major advantage in the constrained drug discovery research environment for diseases of poverty.11
Several libraries of compounds from commercial vendors were virtually screened with the Assay Central Bayesian machine learning models to select both predicted-active and -inactive compounds for in vitro phenotypic assays of S. mansoni somules and adults. These predictions were performed either manually or in an automated manner (Figures 1–3), and 56 compounds were selected and purchased. Bioactivity against the parasite as a function of time and/or concentration was presented as severity scores, in accordance with previous studies (Tables 4 and 5).47−49 Nine active compounds were initially prioritized on the basis of severity scores of 3 or 4 against adult worms after 24 h; of these, eight were also active against somules.
After triaging for in-assay precipitation problems and prior evidence of antischistosomal activity, we settled on five compounds for future follow up studies: revaprazan hydrochloride, U-73122, Z425126666, Z56174662, and Z288901226 (Figure 6). All possess common antischistosomal chemical moieties seen in adult and somule predictions, including indole (Z56174662) and pyrimidine rings (revaprazan hydrochloride) as well as fluorine substituents (Z288901226 and revaprazan hydrochloride). U-73122 does not possess these somule-specific moieties but instead has a steroid core that is common in adult active predictions. This may explain why it was selected manually as a potential developmental stage-specific compound. Both revaprazan hydrochloride and U-73122 have the advantage of known mechanisms of action (i.e., acid pump antagonist and phospholipase C inhibitor, respectively).50−53 Only one compound, Z425126666, was predicted favorably by the stability, permeability, and cytotoxicity models (Table S6). This may indicate the need for further optimization of the molecular properties in future studies.
Both the automated and manual compound prediction methods demonstrated advantages and disadvantages in this study. The automated approach was efficient in selecting compounds with established antischistosomal chemical features such as halogen substituents, piperazine, and quinazolines, which is valuable for finding hit compounds based on known chemistries. However, this method did not produce novel chemistries for testing. In contrast, the manual compound selection method for active compounds, although more time-consuming, allowed us to pick “underdog” compounds that diverge from the more established chemistries. For somules, both the automated and manual prediction methods were reasonably accurate in selecting active and inactive compounds as evidenced by the 70%, 50%, and 63% correct prediction return for the automated and manual actives, and manual inactives, respectively. For adults, prediction accuracy was somewhat less in relation to actives predicted automatically (30%) or manually (50%), whereas the prediction of manual inactives was 87.5% accurate. Together, the methods employed are less time-consuming and more likely to yield active compounds than the screening of large libraries, as indicated by our 48% and 34% hit rates for somules and adults, respectively, from just 56 molecules. Future developments of an automated compound selection method may include molecular property and toxicity predictions as well as expanding the number of libraries utilized. The manual selection process could be improved with a more defined selection of chemical diversity rather than tediously judging structures.
Repurposing approved drugs is a means to fast-tracking a drug to the clinic54 and has been applied in the context of infectious diseases of poverty, including schistosomiasis.22,47 One compound to emerge as strongly bioactive against both somules and adults was revaprazan hydrochloride. Revaprazan is a reversible proton pump inhibitor that reduces gastric acid secretions50 but is also known to activate the serotonin receptor 4b.51 The compound is approved in South Korea and India (under the trade name Revanex) to treat excess gastric acid secretion and gastritis and is used at a daily dose of 200 mg/day. Although the drug has poor water solubility and a relatively low oral bioavailability,53 it is well tolerated in rats after oral administration (50–100 mg/kg).55In vitro studies in Caco-2 cells suggest that the uptake is mediated by a nucleobase transport system, which may contribute to the dose-dependent bioavailability when saturated.56 This compound is a good example of repurposing an already-approved drug.
A number of the compounds that were identified as bioactive vs adults and/or somules are already known for their antischistosomal activity, e.g., moxidectin22,37,57 and piperlongumine.41−43 These studies were not found in our initial literature search and, thus, were not included in our training data but offer an opportunity for further validation of the prediction and experimental approaches herein. In a previous in vitro study, moxidectin was considered to be active (at 10 μM for 72 h) against somules and moderately active against adults (at 33.3 μM for 24 h).22 The drug has also shown some efficacy in patients infected with S. mansoni, particular in decreasing egg burdens.37,57 In our own screens, 10 μM moxidectin produced degenerative changes in both developmental stages by 48 h.
We also tested piperlongumine as part of the predicted nonactive compounds for somules. Contrary to the prediction, piperlongumine was in fact strongly active against somules and adults (dead or dying parasites by 48 h; Tables 4, 5, and S4). Our experimental data are consistent with other in vitro studies whereby adult worms were dead by 24 h at 15 μM and 7-day old somules were killed within 48 h at the same concentration.41 The rediscovery (confirmation) of active compounds is a familiar issue in machine learning, as predictions are limited by the training data available. Refinement of the rule books and the development of a comprehensive database will limit the future rediscovery of active compounds.
Lago et al. screened 73 nonsteroidal anti-inflammatory drugs, including a compound screened by us here, niflumic acid, against adult S. mansoni in vitro for 72 h at 50 μM.58 Niflumic acid was not active in the initial in vitro screen, but other analogs had modest activity (LC50 values ranged from 20.6 to 37.4 μM), the best of which, mefenamic acid, generated an LC50 of 11.1 μM. An inspection of the rule book based on these published single metric data (Table 1) shows that these compounds would be considered inactive (rule book scores of zero). We had selected niflumic acid manually as an adult active because it possesses the attractive feature of a known mechanism of action (a cyclooxygenase-2 inhibitor). Experimentally, however, it was essentially inactive (a severity score of 1 after 48 h at 10 μM), i.e., consistent with the data from Lago et al. Interestingly, however, the analog Z288901226 (Figure S4B), which was predicted to be active using the automated selection method, was lethal to adults by 48 h at 10 μM. Thus, even though our prediction of activity for niflumic acid was incorrect, our identification of the active analog Z288901226 provides a novel starting point for further exploration of this 3-(trifluoromethyl)anilino-3-pyridine chemotype.
Conclusion
We have described a process to curate and normalize the disparate phenotypic screening data for S. mansoni using two rule books. Once standardized, these data sets were interrogated by the proprietary software Assay Central to generate a total of eight Bayesian machine learning models. From these models, 56 predicted active and nonactive small molecules were selected for in vitro phenotypic screening against S. mansoni somules and adults; we identified five actives for future optimization studies. The prediction accuracy was 61% and 56% for somules and adults, respectively, with hit rates of 48% and 34%, respectively. Thus, the return on the time and effort invested exceeds the typical 1–2% hit rate from high throughput screens,59,60 which is especially attractive when working with schistosomes given the need for small animal hosts to propagate the parasite and the finite numbers of parasites that can be recovered per host. Finally, the rule books represent a first step toward a unified database of antischistosomal activity. We will continue the iterative feedback process of generating and assembling new data to improve our machine learning models.
Methods
Literature Search for S. mansoni Phenotypic Screening Data
We performed a literature search for reports of in vitro phenotypic screens of S. mansoni. Using PubMed, we employed a combination of search terms that included “schistosome screen”, “Schistosoma screen”, “schistosome phenotypic screen”, and/or “antischistosomal in vitro” and identified 19 publications that were published between 1980 and 201922,23,31−35,47,61−74,24,33,47,75 (Tables S1 and S2). Articles were excluded when (i) screens did not include the flatworm itself, (ii) information to unequivocally identify the tested compound was lacking, or (iii) the compounds tested were not compatible with our machine learning methods, e.g., metal-coordinating complexes.
Development of Two Rule Books to Normalize Phenotypic Screening Data in the Literature
Reports of in vitro antischistosomal activity in the literature have typically employed two phenotypic screen approaches: (i) those that reported single metric outputs (ED50, LD50, % mortality, % survival, etc.) either derived from an observationally based adjudication system or the measurement of a biochemical marker (e.g., ATP or NADPH) at fixed time points and (ii) those that involve the observational assessment and enumeration of the phenotypic changes that the schistosome parasite is capable of (changes relating to motility, size, and density) as a function of time and/or concentration. For each approach, we developed a “rule book” that employs a sliding scale of scores 0 (no activity) to 4 (most activity) whereby potent compounds that act quickly and at low concentrations receive higher scores than those that take more time to act and/or act at higher concentrations.
In the first rule book (Table 1; full details in Table S1) for example, ED50 values measured at 24 h in the range of 10–25 and <5 μM would generate rule book scores of 2 and 4, respectively. However, to achieve the same scores at the longer time point of 72 h, the ED50 values would be necessarily more stringent, i.e., 2.5–5 and <1 μM, respectively.
For the second rule book, phenotypic changes (principally shape, motility, and density) were counted up to a maximum of three to provide a partially quantitative assessment of overall severity (Table 2; full details in Table S2). Severe changes that involved degenerating parasites, damage to the outer tegument (specific to adult worms), and worm death were given the same weighting as three phenotypic changes. In addition to the number of phenotypic changes, the time to appearance of these changes was considered such that those that occurred in shorter time frames received a higher rule book score. Finally, the concentration at which the changes were observed (between 0.1 and 10 μM) also contributed to the final rule book score. For example, degenerate or dead parasites observed at <72 h in the presence of 0.5 μM compound would result in a rule book score of 4, whereas in the presence of 10 μM compound, the score would be 2. Data incorporated as part of the second phenotypic screening approach are derived from peer-reviewed resources,24,47 the CHEMBL database, and unpublished screens performed by the UCSD authors.
Data Set Organization for Machine Learning
Upon application of the two rule book scoring systems, the resulting data sets were pooled for generating machine learning models with Assay Central (Figure 1). Models were generated according to the screened development stage (somule or adult) and experimental time point for modeling (≤24, 48, 72, and >72 h). For building somule models, data for both newly transformed somules (NTS) and somules that had been allowed to acclimate overnight prior to screening were consolidated. Likewise, for building adult models, data for adults that had been harvested at 37 days post-infection or at later time points were consolidated.
The same activity thresholds were applied to all individual models: compounds generating a rule book score of 3 or 4 were considered active whereas those that yielded a score of 0–2 were considered inactive. This threshold was chosen with a view to finding strongly active compounds. For any given time-point model, inactive compounds at longer time points were included to maximize chemical diversity; e.g., inactive compounds at 72 h were included in the 48 h model. When duplicate compounds between articles were observed, the binary activities reflecting the rule book score, i.e., a value of 1 for rule book scores 3–4 and a value of 0 for rule book scores 0–2, were averaged and rounded to classify the compound as active or inactive. A > 72 h model was also generated to consider a compound’s activity over all recorded time points by applying a binary classification. Thus, if a compound was inactive between 24 and 72 h but active at 168 h, the compound was considered active in the >72 h model but inactive in the other models. Four models were built from standardized time points (≤24, 48, 72, and >72 h postexposure) for each developmental stage (eight total).
Assay Central
The associated rule book scores were used with the Assay Central technology to predict compounds for in vitro screening against S. mansoni. The Assay Central software has been described in detail elsewhere.18,26−30 Briefly, all screening data were collated within Molecular Notebook (Molecular Materials Informatics, Inc. in Montreal, Canada). The underlying framework applies a series of molecular standardization scripts for thorough curation, including removing salts and flagging abnormal valences and mixtures, to generate high-quality (i.e., machine learning-ready) data sets and Bayesian models that are capable of bioactivity predictions.45,76 These models employ extended-connectivity fingerprints of a maximum diameter of 6 (ECFP6) that are generated from the Chemistry Development Kit library77 by applying the Morgan algorithm. The ECFP6 descriptors are well-known for their ability to map structure–activity relationships.45 All Assay Central models include several metrics45 to evaluate and compare predictive performance, including ROC, recall, precision, F1-Score, Cohen’s kappa,78,79 and Matthews correlation coefficient80 scores. A Domain metric was also generated for each model to provide a measure of chemical coverage of the training data in relation to the chemical space of the entire ChEMBL database (comprising nearly two million compounds) ranging from 0 (no overlap) to 1 (total overlap).30
The generation of probability-like prediction scores from Bayesian models within the Assay Central software has also been previously described.45,76 Briefly, this score sums the “contributions” of molecular fingerprints to an active classification, determined by the ratio of its presence in active and inactive training data. Bayesian predictions were evaluated using the standard probability cutoff45 so that a chemical receiving a score of ≥0.5 is classified as active, i.e., a hit compound, at the modeled target. Predictions also included an applicability score whereby a higher score indicates that more of the predicted molecule’s fingerprints are present in the training data. There is no standard cutoff for the applicability score, rather this serves to increase confidence in the prediction score.
Three Assay Central prediction methods were conducted to either manually select active and nonactive compounds or, using an automated workflow (discussed in more depth in the following section), select active compounds (Figure 1). A “raw” prediction outputs a user-defined number of top-scoring molecules with no consideration of the applicability score or diversity. Compounds identified as present in the training data are also excluded from raw prediction outputs to avoid testing compounds with known (according to our literature search) bioactivity. In contrast, a “ranked” prediction is initially identical with the raw prediction but outputs a user-defined subset of diverse compounds (according to Tanimoto similarity and molecular fingerprints) from a user-defined number of top-scoring compounds, for example, the most diverse 10 compounds from the top-scoring 100. Finally, a “consensus” prediction considers multiple models to output a consolidated score that is calculated from the average of the component prediction scores (constrained between 0 and 1) multiplied by the component applicability scores for a given molecule. A raw prediction was applied for the manual selection of active and nonactive compounds (Figure 2), whereas both ranked and consensus prediction methods were applied in the automated workflow to select active compounds (Figure 3).
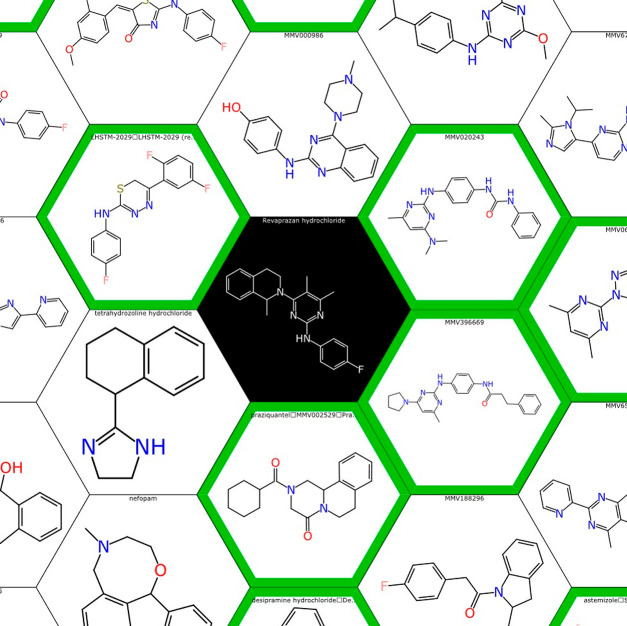
The honeycomb visualization feature of Assay Central (Figure 7)81 allows users to investigate, in a visually intuitive manner, the similarity of predicted compounds to those found in the training data sets. This feature depicts an external compound as the central point of the plot and builds the training compounds around it, so that the increasing distance is proportional to the decreasing similarity with the central compound.
Figure 7.
An example image of the Assay Central honeycomb visualization feature.81 This was employed in the manual selection of compounds for in vitro testing. Training molecules (white background) are organized in relation to a central predicted molecule (black background) such that increasing distance is proportional to decreasing structural similarity (evaluated by the Tanimoto coefficient and ECFP6) with the central molecule (revaprazan hydrochloride).
Comparison of Assay Central with Other Machine Learning Algorithms
The eight Bayesian models generated within Assay Central were compared to six other machine learning algorithms, namely, random forest, k-Nearest Neighbors, support vector classification, naïve Bayesian, AdaBoosted decision trees, and deep learning.28,30,82 Briefly, deep learning was implemented using Keras (https://keras.io/) and Tensorflow (www.tensorflow.org) backend, and hyperparameter optimization was performed with three layers and the Scikit-learn grid search method. Other algorithms were built using the open source Scikit-learn (http://scikit-learn.org/stable/) python library. All alternative algorithms employed the ECFP6 molecular descriptor as used in Assay Central for a straightforward comparison of algorithms using the same data sets and descriptors. The 5-fold cross-validation performance metrics were compared using a rank normalized score as performed previously.30,83,84 Rank normalized scores were evaluated using a pairwise comparison to compare per training set, and an independent comparison was used to give a more general comparison. A “difference from the top” (ΔRNS) metric30,83,84 gave a rank normalized score for each algorithm subtracted from the highest rank normalized score for a specific training set. The ΔRNS metric retains the pairwise results from each training set cross-validation score by algorithm, allowing a direct performance comparison of two algorithms (using all of the available model quality metrics) without losing information from the other algorithms.
Compound Selection for Phenotypic Screens of S. mansoni
The described machine learning models were applied to vendor libraries (see below) to select compounds for in vitro phenotypic screening against S. mansoni somules and adults. For each developmental stage, predicted active compounds were chosen manually and automatically (Figure 1). Predicted nonactives for each developmental stage were chosen using the manual method only; the automated method was considered a proof-of-concept, so only active predictions were deemed of consequence.
The manual approach to predicting active and inactive compounds for each developmental stage was as follows (Figure 2). First, a raw prediction was generated against all models within several small molecule collections: (i) an internally curated collection of 1355 FDA-approved drugs from 2016 to 2018, (ii) a lead-like and chemically diverse collection from Enamine containing over 50 000 compounds,85 (iii) the Library of Pharmacologically Active Compounds or LOPAC1280 from Sigma-Aldrich,86 and (iv) a screening library from Selleck Chemicals of over 1600 natural products.87 When prioritizing compounds, more consideration was given to predictions from the ≤24 h and >72 h models so as to capture fast-action and chemical diversity, respectively. Compounds were filtered on the basis of multiple criteria including known compound targets, cost, and liabilities; for example, compounds known to elicit serious side effects were deprioritized. As the goal of this study was to discover novel antischistosomal chemistries, the predictions were further filtered so that compounds that were dissimilar to the training data were considered more desirable candidates for testing, as determined by Assay Central applicability scores and honeycomb plots (Figure 7).81 Ten active and eight inactive compounds were manually predicted for in vitro phenotypic screens of adults and somules (36 compounds total).
For the automated approach to predicting active compounds (Figure 3), only the diversity collection from Enamine85 was interrogated for both simplicity of purchase and the drug-like nature of this collection. First, a ranked prediction was applied to each of the four time point models in a given developmental stage to output the ten most diverse compounds from the top-scoring 50 (totaling 40 compounds per life stage). Then, a consensus prediction was applied to each deduplicated subset across all time point models to output the ten predicted active compounds for each developmental stage. These were then purchased.
Sources of Compounds
A total of 56 compounds (Figure S4, Table S4) was selected by both the manual and automated methods for screening of S. mansoni adults and somules. The following compounds were purchased from Cayman Chemical: U-73122, BIX01294 hydrochloride hydrate, tyrphostin AG-1478, sivelestat sodium tetrahydrate, AGK2, trilostane, itraconazole, dabigatran etexilate, SB202190 hydrochloride, niflumic acid, amsacrine hydrochloride, R(+)-IAA-94, piperlongumine, tiamulin fumarate, PNU-282987, tetrabenazine, nemadipine-A, mycophenolic acid, etravirine, (S)-duloxetine hydrochloride, moxidectin, and rutecarpine. The following compounds were purchased from Sigma-Aldrich: clindamycin 2-phosphate, tyrphostin I-OMe-AG-538, and escabet sodium. The following compounds were purchased from Selleck Chemicals: revaprazan hydrochloride and Org 27569. The following compounds were purchased from Enamine: Z385159220, Z18885599, Z276431168, Z2241105867, Z827016000, Z48867676, Z53005631, Z90192490, Z56174662, Z105384660, Z44528364, Z827015296, Z304863612, and Z56958732. Finally, the following compounds were purchased from Toronto Research Chemical: eletriptan and ondansetron hydrochloride dihydrate. Powders were stored according to vendor specifications. Compounds were then dissolved at 10 mM in fresh DMSO and shipped to UCSD on dry ice for storage at −80 °C until use.
Life Cycle of S. mansoni and Screening of Compounds Predicted by Assay Central
S. mansoni (NMRI isolate) was maintained by passage through Biomphalaria glabrata snails (NMRI line) and 3–5 week-old, male Golden Syrian hamsters as intermediate and definitive hosts, respectively. Somules were generated from infectious larvae (cercariae) that were harvested from infected snails, and adult parasites were harvested from hamsters, as described.47,88 Somules were used for screening within 2 h of their preparation from cercariae (otherwise known as NTS).
For phenotypic screens of somules,47 parasites (40 animals/well in clear, u-bottomed 96-well plates) were incubated in 100 μL of Basch medium89 supplemented with 4% heat-inactivated FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin. Compounds predicted by Assay Central were then added at 2× of the final concentrations of 1 and 10 μM. The same medium (100 μL) was immediately added to mix the compound with a final concentration of 0.5% DMSO. Compounds were tested in two experiments each in duplicate. Incubations were maintained at 37 °C in a 5% CO2 environment, and phenotypic changes were noted at 24, 48, and 72 h. A compound was considered active when it generated a severity score of ≥2 after 72 h (see below).
Adult parasites (five males and approximately two pairs per well in 24-well plates) were maintained in 2 mL of the same medium under the same conditions in the presence of 10 μM compound and a final concentration of 0.1% DMSO.47 Phenotypic changes were noted at 1, 5, 24, and 48 h. Compounds were tested in two experiments, each in duplicate. A compound was considered active when it generated a severity score of ≥2 after 48 h (see below).
Phenotypic changes in both developmental stages were observed using a Zeiss Axiovert A1 inverted microscope. The parasite’s phenotypic changes in shape, density, and motility were recorded using a constrained nomenclature of simple and, where possible, self-explanatory descriptors.47−49 To allow for the partially quantitative comparisons of compound effects, each descriptor was typically given a value of 1 and these were summed to generate a “severity score” with a maximum value of 4. Descriptors recording severe phenotypes, i.e., death, degeneracy or, for adult parasites specifically, damage to the surface tegument, were given the maximum value of 4.
Ethics Statement
The use of hamsters in support of the S. mansoni life cycle was in accordance with a protocol approved by UC San Diego’s Institutional Animal Care and Use Committee. The committee derives its authority for its activities from the United States Public Health Service (PHS) Policy on Humane Care and Use of Laboratory Animals and the Animal Welfare Act and Regulations (AWAR).
Acknowledgments
K.M.Z. and S.E. acknowledge the National Institutes of Health (NIH) funding to develop the software from 1R43GM122196-01 and R44GM122196-02A1 “Centralized assay datasets for modelling support of small drug discovery organizations” from the National Institute of General Medical Sciences. C.R.C. acknowledges R21AI126296 from the NIH and OPP1171488 from the Bill and Melinda Gates Foundation. S.S., C.L.M, and E.K.C. were supported by Skaggs Scholarships from the UC San Diego Skaggs School of Pharmacy and Pharmaceutical Sciences. K.M. was supported in part by the Eunice Kennedy Shriver National Institute of Child Health and Human Development of the National Institutes of Health under the award number T35HD064385. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. S. mansoni-infected hamsters were provided in part by the National Institute of Allergy and Infectious Diseases (NIAID) Schistosomiasis Resource Center of the Biomedical Research Institute (Rockville, MD, USA) through the National Institutes of Health (NIH)-NIAID Contract HHSN272201700014I for distribution through BEI Resources. We thank Dr. Alex M. Clark (Molecular Materials Informatics, Inc.) for Assay Central support.
Glossary
Abbreviations
- PZQ
praziquantel
- ECFP6
extended-connectivity fingerprints of maximum diameter 6
- ROC
receiver operating characteristic
- ΔRNS
“difference from the top” rank normalized score
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsinfecdis.0c00754.
Further details on the models, structures of public molecules, and computational models (PDF)
Table S1: Rule book and associated single metric screening data from the literature (XLSX)
Table S2: Rule book and associated data for multivariate phenotypic screens performed at UCSD (XLSX)
Table S4: Multivariate phenotypic screens performed at UCSD (XLSX)
Author Contributions
∇ K.M.Z., S.S., and C.L.M. are joint first authors.
The authors declare the following competing financial interest(s): S.E. is the owner and K.M.Z., D.H.F., and T.R.L. are employees of Collaborations Pharmaceuticals, Inc. All other authors are associates of the University of California, San Diego.
Supplementary Material
References
- Hotez P. J. (2012) Preface. In Parasitic Helminths Targets, Screens, Drugs and Vaccines (Caffrey C. R., Ed.) p XI, Wiley-Blackwell, Weinheim. [Google Scholar]
- WHO Neglected tropical diseases, https://www.who.int/neglected_diseases/diseases/en/.
- McManus D. P.; Dunne D. W.; Sacko M.; Utzinger J.; Vennervald B. J.; Zhou X. N. (2018) Schistosomiasis. Nat. Rev. Dis Primers 4 (1), 13. 10.1038/s41572-018-0013-8. [DOI] [PubMed] [Google Scholar]
- Colley D. G.; Bustinduy A. L.; Secor W. E.; King C. H. (2014) Human schistosomiasis. Lancet 383 (9936), 2253–64. 10.1016/S0140-6736(13)61949-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiser J.; Utzinger J. (2010) The drugs we have and the drugs we need against major helminth infections. Adv. Parasitol. 73, 197–230. 10.1016/S0065-308X(10)73008-6. [DOI] [PubMed] [Google Scholar]
- Gabrielli A. F.; Montresor A.; Chitsulo L.; Engels D.; Savioli L. (2011) Preventive chemotherapy in human helminthiasis: theoretical and operational aspects. Trans. R. Soc. Trop. Med. Hyg. 105 (12), 683–93. 10.1016/j.trstmh.2011.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Enden E. (2009) Pharmacotherapy of helminth infection. Expert Opin. Pharmacother. 10 (3), 435–51. 10.1517/14656560902722463. [DOI] [PubMed] [Google Scholar]
- Caffrey C. R. (2012) Parasitic Helminths: Targets, Screens, Drugs and Vaccines, Vol. 3, Wiley-Blackwell, Weinheim. [Google Scholar]
- Caffrey C. R. (2015) Schistosomiasis and its treatment. Future Med. Chem. 7 (6), 675–6. 10.4155/fmc.15.27. [DOI] [PubMed] [Google Scholar]
- Bustinduy A. L.; Friedman J. F.; Kjetland E. F.; Ezeamama A. E.; Kabatereine N. B.; Stothard J. R.; King C. H. (2016) Expanding Praziquantel (PZQ) Access beyond Mass Drug Administration Programs: Paving a Way Forward for a Pediatric PZQ Formulation for Schistosomiasis. PLoS Neglected Trop. Dis. 10 (9), e0004946 10.1371/journal.pntd.0004946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffrey C. R., El-Sakkary N., Mader P., Krieg R., Becker K., Schlitzer M., Drewry D. H., Vennerstrom J. L., and Grevelding C. G. (2019) Drug Discovery and Development for Schistosomiasis. In Neglected tropical diseases: drug disscovery and development (Swinnery D., Pollastri M., Mannhold R., Buschmann H., and Holenz J., Eds.) pp 187–226, Wiley-VCH Verlag GmbH & Co. KGaA, 10.1002/9783527808656.ch8. [DOI] [Google Scholar]
- Panic G.; Flores D.; Ingram-Sieber K.; Keiser J. (2015) Fluorescence/luminescence-based markers for the assessment of Schistosoma mansoni schistosomula drug assays. Parasites Vectors 8, 624. 10.1186/s13071-015-1233-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponder E. L.; Freundlich J. S.; Sarker M.; Ekins S. (2014) Computational models for neglected diseases: gaps and opportunities. Pharm. Res. 31 (2), 271–7. 10.1007/s11095-013-1170-9. [DOI] [PubMed] [Google Scholar]
- Gaba S.; Jamal S.; Scaria V. (2014) Cheminformatics models for inhibitors of Schistosoma mansoni thioredoxin glutathione reductase. Sci. World J. 2014, 957107. 10.1155/2014/957107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez H. W.; Soeung M.; Zorn K. M.; Ashoura N.; Mottin M.; Andrade C. H.; Caffrey C. R.; de Siqueira-Neto J. L.; Ekins S. (2019) High Throughput and Computational Repurposing for Neglected Diseases. Pharm. Res. 36 (2), 27. 10.1007/s11095-018-2558-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekins S.; de Siqueira-Neto J. L.; McCall L. I.; Sarker M.; Yadav M.; Ponder E. L.; Kallel E. A.; Kellar D.; Chen S.; Arkin M.; Bunin B. A.; McKerrow J. H.; Talcott C. (2015) Machine Learning Models and Pathway Genome Data Base for Trypanosoma cruzi Drug Discovery. PLoS Neglected Trop. Dis. 9 (6), e0003878 10.1371/journal.pntd.0003878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekins S.; Freundlich J. S.; Clark A. M.; Anantpadma M.; Davey R. A.; Madrid P. (2015) Machine learning models identify molecules active against the Ebola virus in vitro. F1000Research 4, 1091. 10.12688/f1000research.7217.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anantpadma M.; Lane T.; Zorn K. M.; Lingerfelt M. A.; Clark A. M.; Freundlich J. S.; Davey R. A.; Madrid P. B.; Ekins S. (2019) Ebola Virus Bayesian Machine Learning Models Enable New in Vitro Leads. ACS Omega 4 (1), 2353–2361. 10.1021/acsomega.8b02948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekins S.; Reynolds R. C.; Kim H.; Koo M. S.; Ekonomidis M.; Talaue M.; Paget S. D.; Woolhiser L. K.; Lenaerts A. J.; Bunin B. A.; Connell N.; Freundlich J. S. (2013) Bayesian models leveraging bioactivity and cytotoxicity information for drug discovery. Chem. Biol. 20 (3), 370–8. 10.1016/j.chembiol.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekins S.; Reynolds R. C.; Franzblau S. G.; Wan B.; Freundlich J. S.; Bunin B. A. (2013) Enhancing hit identification in Mycobacterium tuberculosis drug discovery using validated dual-event Bayesian models. PLoS One 8 (5), e63240 10.1371/journal.pone.0063240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekins S.; Casey A. C.; Roberts D.; Parish T.; Bunin B. A. (2014) Bayesian models for screening and TB Mobile for target inference with Mycobacterium tuberculosis. Tuberculosis (Oxford, U. K.) 94 (2), 162–9. 10.1016/j.tube.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panic G.; Vargas M.; Scandale I.; Keiser J. (2015) Activity Profile of an FDA-Approved Compound Library against Schistosoma mansoni. PLoS Neglected Trop. Dis. 9 (7), e0003962 10.1371/journal.pntd.0003962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks J. C.; Roberts W. M.; Leasure C.; Suzuki B. M.; Robinson K. J.; Currey H.; Wangchuk P.; Eichenberger R. M.; Saxton A. D.; Bird T. D.; Kraemer B. C.; Loukas A.; Hawdon J. M.; Caffrey C. R.; Liachko N. F. (2018) Sertraline, Paroxetine, and Chlorpromazine Are Rapidly Acting Anthelmintic Drugs Capable of Clinical Repurposing. Sci. Rep. 8 (1), 975. 10.1038/s41598-017-18457-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maccesi M.; Aguiar P. H. N.; Pasche V.; Padilla M.; Suzuki B. M.; Montefusco S.; Abagyan R.; Keiser J.; Mourao M. M.; Caffrey C. R. (2019) Multi-center screening of the Pathogen Box collection for schistosomiasis drug discovery. Parasites Vectors 12 (1), 493. 10.1186/s13071-019-3747-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R.; Beasley R.; Long T.; Caffrey C. R. (2018) Algorithmic Mapping and Characterization of the Drug-Induced Phenotypic-Response Space of Parasites Causing Schistosomiasis. IEEE/ACM Trans. Comput. Biol. Bioinf. 15, 469–481. 10.1109/TCBB.2016.2550444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalecki A. G.; Zorn K. M.; Clark A. M.; Ekins S.; Narmore W. T.; Tower N.; Rasmussen L.; Bostwick R.; Kutsch O.; Wolschendorf F. (2019) High-throughput screening and Bayesian machine learning for copper-dependent inhibitors of Staphylococcus aureus. Metallomics 11 (3), 696–706. 10.1039/C8MT00342D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekins S.; Gerlach J.; Zorn K. M.; Antonio B. M.; Lin Z.; Gerlach A. (2019) Repurposing Approved Drugs as Inhibitors of Kv7.1 and Nav1.8 to Treat Pitt Hopkins Syndrome. Pharm. Res. 36 (9), 137. 10.1007/s11095-019-2671-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane T.; Russo D. P.; Zorn K. M.; Clark A. M.; Korotcov A.; Tkachenko V.; Reynolds R. C.; Perryman A. L.; Freundlich J. S.; Ekins S. (2018) Comparing and Validating Machine Learning Models for Mycobacterium tuberculosis Drug Discovery. Mol. Pharmaceutics 15 (10), 4346–4360. 10.1021/acs.molpharmaceut.8b00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval P. J.; Zorn K. M.; Clark A. M.; Ekins S.; Wright S. H. (2018) Assessment of Substrate-Dependent Ligand Interactions at the Organic Cation Transporter OCT2 Using Six Model Substrates. Mol. Pharmacol. 94 (3), 1057–1068. 10.1124/mol.117.111443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorn K. M.; Lane T. R.; Russo D. P.; Clark A. M.; Makarov V.; Ekins S. (2019) Multiple Machine Learning Comparisons of HIV Cell-based and Reverse Transcriptase Data Sets. Mol. Pharmaceutics 16 (4), 1620–1632. 10.1021/acs.molpharmaceut.8b01297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidi A.; Lalli C.; Gimmelli R.; Nizi E.; Andreini M.; Gennari N.; Saccoccia F.; Harper S.; Bresciani A.; Ruberti G. (2017) Discovery by organism based high-throughput screening of new multi-stage compounds affecting Schistosoma mansoni viability, egg formation and production. PLoS Neglected Trop. Dis. 11 (10), e0005994 10.1371/journal.pntd.0005994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai G.; Sayed A. A.; Lea W. A.; Luecke H. F.; Chakrapani H.; Prast-Nielsen S.; Jadhav A.; Leister W.; Shen M.; Inglese J.; Austin C. P.; Keefer L.; Arner E. S.; Simeonov A.; Maloney D. J.; Williams D. L.; Thomas C. J. (2009) Structure mechanism insights and the role of nitric oxide donation guide the development of oxadiazole-2-oxides as therapeutic agents against schistosomiasis. J. Med. Chem. 52 (20), 6474–83. 10.1021/jm901021k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T.; Ziniel P. D.; He P. Q.; Kommer V. P.; Crowther G. J.; He M.; Liu Q.; Van Voorhis W. C.; Williams D. L.; Wang M. W. (2015) High-throughput screening against thioredoxin glutathione reductase identifies novel inhibitors with potential therapeutic value for schistosomiasis. Infect Dis Poverty 4, 40. 10.1186/s40249-015-0071-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y.; Chollet J.; Vargas M.; Mansour N. R.; Bickle Q.; Alnouti Y.; Huang J.; Keiser J.; Vennerstrom J. L. (2010) Praziquantel analogs with activity against juvenile Schistosoma mansoni. Bioorg. Med. Chem. Lett. 20 (8), 2481–4. 10.1016/j.bmcl.2010.03.001. [DOI] [PubMed] [Google Scholar]
- Senga K.; Novinson T.; Wilson H. R.; Robins R. K. (1981) Synthesis and antischistosomal activity of certain pyrazolo[1,5-a]pyrimidines. J. Med. Chem. 24 (5), 610–3. 10.1021/jm00137a023. [DOI] [PubMed] [Google Scholar]
- Anon ChEMBL , https://chembl.gitbook.io/chembl-interface-documentation/downloads.
- Barda B.; Coulibaly J. T.; Puchkov M.; Huwyler J.; Hattendorf J.; Keiser J. (2016) Efficacy and Safety of Moxidectin, Synriam, Synriam-Praziquantel versus Praziquantel against Schistosoma haematobium and S. mansoni Infections: A Randomized, Exploratory Phase 2 Trial. PLoS Neglected Trop. Dis. 10 (9), e0005008 10.1371/journal.pntd.0005008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva-Moraes V.; Couto F. F.; Vasconcelos M. M.; Araujo N.; Coelho P. M.; Katz N.; Grenfell R. F. (2013) Antischistosomal activity of a calcium channel antagonist on schistosomula and adult Schistosoma mansoni worms. Mem Inst Oswaldo Cruz 108 (5), 600–4. 10.1590/0074-0276108052013011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das K.; Clark A. D. Jr.; Lewi P. J.; Heeres J.; De Jonge M. R.; Koymans L. M.; Vinkers H. M.; Daeyaert F.; Ludovici D. W.; Kukla M. J.; De Corte B.; Kavash R. W.; Ho C. Y.; Ye H.; Lichtenstein M. A.; Andries K.; Pauwels R.; De Bethune M. P.; Boyer P. L.; Clark P.; Hughes S. H.; Janssen P. A.; Arnold E. (2004) Roles of conformational and positional adaptability in structure-based design of TMC125-R165335 (etravirine) and related non-nucleoside reverse transcriptase inhibitors that are highly potent and effective against wild-type and drug-resistant HIV-1 variants. J. Med. Chem. 47 (10), 2550–60. 10.1021/jm030558s. [DOI] [PubMed] [Google Scholar]
- Andries K.; Azijn H.; Thielemans T.; Ludovici D.; Kukla M.; Heeres J.; Janssen P.; De Corte B.; Vingerhoets J.; Pauwels R.; de Bethune M. P. (2004) TMC125, a novel next-generation nonnucleoside reverse transcriptase inhibitor active against nonnucleoside reverse transcriptase inhibitor-resistant human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 48 (12), 4680–6. 10.1128/AAC.48.12.4680-4686.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraes J.; Nascimento C.; Lopes P. O.; Nakano E.; Yamaguchi L. F.; Kato M. J.; Kawano T. (2011) Schistosoma mansoni: In vitro schistosomicidal activity of piplartine. Exp. Parasitol. 127 (2), 357–64. 10.1016/j.exppara.2010.08.021. [DOI] [PubMed] [Google Scholar]
- de Moraes J.; Nascimento C.; Yamaguchi L. F.; Kato M. J.; Nakano E. (2012) Schistosoma mansoni: in vitro schistosomicidal activity and tegumental alterations induced by piplartine on schistosomula. Exp. Parasitol. 132 (2), 222–7. 10.1016/j.exppara.2012.07.004. [DOI] [PubMed] [Google Scholar]
- Mengarda A. C.; Mendonca P. S.; Morais C. S.; Cogo R. M.; Mazloum S. F.; Salvadori M. C.; Teixeira F. S.; Morais T. R.; Antar G. M.; Lago J. H. G.; Moraes J. (2020) Antiparasitic activity of piplartine (piperlongumine) in a mouse model of schistosomiasis. Acta Trop. 205, 105350. 10.1016/j.actatropica.2020.105350. [DOI] [PubMed] [Google Scholar]
- Perryman A. L.; Stratton T. P.; Ekins S.; Freundlich J. S. (2016) Predicting Mouse Liver Microsomal Stability with ″Pruned″ Machine Learning Models and Public Data. Pharm. Res. 33 (2), 433–49. 10.1007/s11095-015-1800-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark A. M.; Dole K.; Coulon-Spektor A.; McNutt A.; Grass G.; Freundlich J. S.; Reynolds R. C.; Ekins S. (2015) Open Source Bayesian Models. 1. Application to ADME/Tox and Drug Discovery Datasets. J. Chem. Inf. Model. 55 (6), 1231–45. 10.1021/acs.jcim.5b00143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perryman A. L.; Patel J. S.; Russo R.; Singleton E.; Connell N.; Ekins S.; Freundlich J. S. (2018) Naive Bayesian Models for Vero Cell Cytotoxicity. Pharm. Res. 35 (9), 170. 10.1007/s11095-018-2439-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdulla M. H.; Ruelas D. S.; Wolff B.; Snedecor J.; Lim K. C.; Xu F.; Renslo A. R.; Williams J.; McKerrow J. H.; Caffrey C. R. (2009) Drug discovery for schistosomiasis: hit and lead compounds identified in a library of known drugs by medium-throughput phenotypic screening. PLoS Neglected Trop. Dis. 3 (7), e478 10.1371/journal.pntd.0000478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long T.; Neitz R. J.; Beasley R.; Kalyanaraman C.; Suzuki B. M.; Jacobson M. P.; Dissous C.; McKerrow J. H.; Drewry D. H.; Zuercher W. J.; Singh R.; Caffrey C. R. (2016) Structure-Bioactivity Relationship for Benzimidazole Thiophene Inhibitors of Polo-Like Kinase 1 (PLK1), a Potential Drug Target in Schistosoma mansoni. PLoS Neglected Trop. Dis. 10 (1), e0004356 10.1371/journal.pntd.0004356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long T.; Rojo-Arreola L.; Shi D.; El-Sakkary N.; Jarnagin K.; Rock F.; Meewan M.; Rascon A. A. Jr.; Lin L.; Cunningham K. A.; Lemieux G. A.; Podust L.; Abagyan R.; Ashrafi K.; McKerrow J. H.; Caffrey C. R. (2017) Phenotypic, chemical and functional characterization of cyclic nucleotide phosphodiesterase 4 (PDE4) as a potential anthelmintic drug target. PLoS Neglected Trop. Dis. 11 (7), e0005680 10.1371/journal.pntd.0005680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. S.; Cho J. Y.; Song H.; Kim E. H.; Hahm K. B. (2012) Revaprazan, a novel acid pump antagonist, exerts anti-inflammatory action against Helicobacter pylori-induced COX-2 expression by inactivating Akt signaling. J. Clin. Biochem. Nutr. 51 (2), 77–83. 10.3164/jcbn.11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasi E. A.; Allen A. A.; Sugianto W.; Peralta-Yahya P. (2019) Identification of Three Antimicrobials Activating Serotonin Receptor 4 in Colon Cells. ACS Synth. Biol. 8 (12), 2710–2717. 10.1021/acssynbio.9b00310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleasdale J. E.; Thakur N. R.; Gremban R. S.; Bundy G. L.; Fitzpatrick F. A.; Smith R. J.; Bunting S. (1990) Selective inhibition of receptor-coupled phospholipase C-dependent processes in human platelets and polymorphonuclear neutrophils. J. Pharmacol Exp Ther 255 (2), 756–768. [PubMed] [Google Scholar]
- Li W.; Yang Y.; Tian Y.; Xu X.; Chen Y.; Mu L.; Zhang Y.; Fang L. (2011) Preparation and in vitro/in vivo evaluation of revaprazan hydrochloride nanosuspension. Int. J. Pharm. 408 (1–2), 157–62. 10.1016/j.ijpharm.2011.01.059. [DOI] [PubMed] [Google Scholar]
- Baker N. C.; Ekins S.; Williams A. J.; Tropsha A. (2018) A bibliometric review of drug repurposing. Drug Discovery Today 23 (3), 661–672. 10.1016/j.drudis.2018.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K. S.; Kim Y. G.; Yoo J. K.; Lee J. W.; Lee M. G. (1998) Pharmacokinetics of a new reversible proton pump inhibitor, YH1885, after intravenous and oral administrations to rats and dogs: hepatic first-pass effect in rats. Biopharm. Drug Dispos. 19 (8), 493–500. . [DOI] [PubMed] [Google Scholar]
- Li H.; Chung S. J.; Kim D. C.; Kim H. S.; Lee J. W.; Shim C. K. (2001) The transport of a reversible proton pump antagonist, 5, 6-dimethyl-2-(4-Fluorophenylamino)-4-(1-methyl-1,2,3, 4-tetrahydroisoquinoline-2-yl) pyrimidine hydrochloride (YH1885), across caco-2 cell monolayers. Drug Metab. Dispos. 29 (1), 54–59. [PubMed] [Google Scholar]
- Attah S. K., Kataliko K., Mutro M. N., Kpawor M., Opuku N. O., and Kanza E. (2014) Effect of a single dose of 8 mg moxi- dectin or 150 μg/kg ivermectin on intestinal helminths in participants of a clinical trial conducted in Northeast DRC, Liberia and Ghana. In 63rd Annual Meeting of the ASTMH, New Orleans, LA, USA.
- Lago E. M.; Silva M. P.; Queiroz T. G.; Mazloum S. F.; Rodrigues V. C.; Carnauba P. U.; Pinto P. L.; Rocha J. A.; Ferreira L. L. G.; Andricopulo A. D.; de Moraes J. (2019) Phenotypic screening of nonsteroidal anti-inflammatory drugs identified mefenamic acid as a drug for the treatment of schistosomiasis. EBioMedicine 43, 370–379. 10.1016/j.ebiom.2019.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Y.; Chen K.; Zhong Y.; Zhou B.; Ainscow E.; Wu Y. T.; Zhou Y. (2016) An Automatic Quality Control Pipeline for High-Throughput Screening Hit Identification. J. Biomol. Screening 21 (8), 832–41. 10.1177/1087057116654274. [DOI] [PubMed] [Google Scholar]
- Clare R. H.; Bardelle C.; Harper P.; Hong W. D.; Borjesson U.; Johnston K. L.; Collier M.; Myhill L.; Cassidy A.; Plant D.; Plant H.; Clark R.; Cook D. A. N.; Steven A.; Archer J.; McGillan P.; Charoensutthivarakul S.; Bibby J.; Sharma R.; Nixon G. L.; Slatko B. E.; Cantin L.; Wu B.; Turner J.; Ford L.; Rich K.; Wigglesworth M.; Berry N. G.; O’Neill P. M.; Taylor M. J.; Ward S. A. (2019) Industrial scale high-throughput screening delivers multiple fast acting macrofilaricides. Nat. Commun. 10 (1), 11. 10.1038/s41467-018-07826-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour N. R.; Paveley R.; Gardner J. M.; Bell A. S.; Parkinson T.; Bickle Q. (2016) High Throughput Screening Identifies Novel Lead Compounds with Activity against Larval, Juvenile and Adult Schistosoma mansoni. PLoS Neglected Trop. Dis. 10 (4), e0004659 10.1371/journal.pntd.0004659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram-Sieber K.; Cowan N.; Panic G.; Vargas M.; Mansour N. R.; Bickle Q. D.; Wells T. N.; Spangenberg T.; Keiser J. (2014) Orally active antischistosomal early leads identified from the open access malaria box. PLoS Neglected Trop. Dis. 8 (1), e2610 10.1371/journal.pntd.0002610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guglielmo S.; Cortese D.; Vottero F.; Rolando B.; Kommer V. P.; Williams D. L.; Fruttero R.; Gasco A. (2014) New praziquantel derivatives containing NO-donor furoxans and related furazans as active agents against Schistosoma mansoni. Eur. J. Med. Chem. 84, 135–45. 10.1016/j.ejmech.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziniel P. D.; Karumudi B.; Barnard A. H.; Fisher E. M.; Thatcher G. R.; Podust L. M.; Williams D. L. (2015) The Schistosoma mansoni Cytochrome P450 (CYP3050A1) Is Essential for Worm Survival and Egg Development. PLoS Neglected Trop. Dis. 9 (12), e0004279 10.1371/journal.pntd.0004279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nwaka S.; Besson D.; Ramirez B.; Maes L.; Matheeussen A.; Bickle Q.; Mansour N. R.; Yousif F.; Townson S.; Gokool S.; Cho-Ngwa F.; Samje M.; Misra-Bhattacharya S.; Murthy P. K.; Fakorede F.; Paris J. M.; Yeates C.; Ridley R.; Van Voorhis W. C.; Geary T. (2011) Integrated dataset of screening hits against multiple neglected disease pathogens. PLoS Neglected Trop. Dis. 5 (12), e1412 10.1371/journal.pntd.0001412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heimburg T.; Chakrabarti A.; Lancelot J.; Marek M.; Melesina J.; Hauser A. T.; Shaik T. B.; Duclaud S.; Robaa D.; Erdmann F.; Schmidt M.; Romier C.; Pierce R. J.; Jung M.; Sippl W. (2016) Structure-Based Design and Synthesis of Novel Inhibitors Targeting HDAC8 from Schistosoma mansoni for the Treatment of Schistosomiasis. J. Med. Chem. 59 (6), 2423–35. 10.1021/acs.jmedchem.5b01478. [DOI] [PubMed] [Google Scholar]
- Cowan N.; Yaremenko I. A.; Krylov I. B.; Terent’ev A. O.; Keiser J. (2015) Elucidation of the in vitro and in vivo activities of bridged 1,2,4-trioxolanes, bridged 1,2,4,5-tetraoxanes, tricyclic monoperoxides, silyl peroxides, and hydroxylamine derivatives against Schistosoma mansoni. Bioorg. Med. Chem. 23 (16), 5175–81. 10.1016/j.bmc.2015.02.010. [DOI] [PubMed] [Google Scholar]
- Sadhu P. S.; Kumar S. N.; Chandrasekharam M.; Pica-Mattoccia L.; Cioli D.; Rao V. J. (2012) Synthesis of new praziquantel analogues: potential candidates for the treatment of schistosomiasis. Bioorg. Med. Chem. Lett. 22 (2), 1103–6. 10.1016/j.bmcl.2011.11.108. [DOI] [PubMed] [Google Scholar]
- Caffrey C. R.; Steverding D.; Swenerton R. K.; Kelly B.; Walshe D.; Debnath A.; Zhou Y. M.; Doyle P. S.; Fafarman A. T.; Zorn J. A.; Land K. M.; Beauchene J.; Schreiber K.; Moll H.; Ponte-Sucre A.; Schirmeister T.; Saravanamuthu A.; Fairlamb A. H.; Cohen F. E.; McKerrow J. H.; Weisman J. L.; May B. C. (2007) Bis-acridines as lead antiparasitic agents: structure-activity analysis of a discrete compound library in vitro. Antimicrob. Agents Chemother. 51 (6), 2164–72. 10.1128/AAC.01418-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Sayed I.; Ramzy F.; William S.; El Bahanasawy M.; Abd El Satter M. M. (2012) Neocryptolepine analogues containing N-substituted side-chains at C-11: synthesis and antischistosomicidal activity. Med. Chem. Res. 21, 4219–4229. 10.1007/s00044-011-9934-4. [DOI] [Google Scholar]
- Ingram K.; Yaremenko I. A.; Krylov I. B.; Hofer L.; Terent’ev A. O.; Keiser J. (2012) Identification of antischistosomal leads by evaluating bridged 1,2,4,5-tetraoxanes, alphaperoxides, and tricyclic monoperoxides. J. Med. Chem. 55 (20), 8700–11. 10.1021/jm3009184. [DOI] [PubMed] [Google Scholar]
- Mahajan A.; Kumar V.; Mansour N. R.; Bickle Q.; Chibale K. (2008) Meclonazepam analogues as potential new antihelmintic agents. Bioorg. Med. Chem. Lett. 18 (7), 2333–6. 10.1016/j.bmcl.2008.02.077. [DOI] [PubMed] [Google Scholar]
- McAllister P. R.; Dotson M. J.; Grim S. O.; Hillman G. R. (1980) Effects of phosphonium compounds on Schistosoma mansoni. J. Med. Chem. 23 (8), 862–5. 10.1021/jm00182a010. [DOI] [PubMed] [Google Scholar]
- Easmon J.; Purstinger G.; Thies K. S.; Heinisch G.; Hofmann J. (2006) Synthesis, structure-activity relationships, and antitumor studies of 2-benzoxazolyl hydrazones derived from alpha-(N)-acyl heteroaromatics. J. Med. Chem. 49 (21), 6343–50. 10.1021/jm060232u. [DOI] [PubMed] [Google Scholar]
- Archer S.; Pica-Mattoccia L.; Cioli D.; Seyed-Mozaffari A.; Zayed A. H. (1988) Preparation and antischistosomal and antitumor activity of hycanthone and some of its congeners. Evidence for the mode of action of hycanthone. J. Med. Chem. 31 (1), 254–60. 10.1021/jm00396a040. [DOI] [PubMed] [Google Scholar]
- Clark A. M.; Ekins S. (2015) Open Source Bayesian Models. 2. Mining a ″Big Dataset″ To Create and Validate Models with ChEMBL. J. Chem. Inf. Model. 55 (6), 1246–60. 10.1021/acs.jcim.5b00144. [DOI] [PubMed] [Google Scholar]
- Willighagen E. L.; Mayfield J. W.; Alvarsson J.; Berg A.; Carlsson L.; Jeliazkova N.; Kuhn S.; Pluskal T.; Rojas-Cherto M.; Spjuth O.; Torrance G.; Evelo C. T.; Guha R.; Steinbeck C. (2017) The Chemistry Development Kit (CDK) v2.0: atom typing, depiction, molecular formulas, and substructure searching. J. Cheminf. 9 (1), 33. 10.1186/s13321-017-0220-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carletta J. (1996) Assessing agreement on classification tasks: The kappa statistic. Computational linguistics 22, 249–254. [Google Scholar]
- Cohen J. (1960) A coefficient of agreement for nominal scales. Educ. Psychol. Meas. 20, 37–46. 10.1177/001316446002000104. [DOI] [Google Scholar]
- Matthews B. W. (1975) Comparison of the predicted and observed secondary structure of T4 phage lysozyme. Biochim. Biophys. Acta, Protein Struct. 405 (2), 442–51. 10.1016/0005-2795(75)90109-9. [DOI] [PubMed] [Google Scholar]
- Ekins S.; Perryman A. L.; Clark A. M.; Reynolds R. C.; Freundlich J. S. (2016) Machine Learning Model Analysis and Data Visualization with Small Molecules Tested in a Mouse Model of Mycobacterium tuberculosis Infection (2014–2015). J. Chem. Inf. Model. 56 (7), 1332–43. 10.1021/acs.jcim.6b00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo D. P.; Zorn K. M.; Clark A. M.; Zhu H.; Ekins S. (2018) Comparing Multiple Machine Learning Algorithms and Metrics for Estrogen Receptor Binding Prediction. Mol. Pharmaceutics 15 (10), 4361–4370. 10.1021/acs.molpharmaceut.8b00546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korotcov A.; Tkachenko V.; Russo D. P.; Ekins S. (2017) Comparison of Deep Learning With Multiple Machine Learning Methods and Metrics Using Diverse Drug Discovery Data Sets. Mol. Pharmaceutics 14 (12), 4462–4475. 10.1021/acs.molpharmaceut.7b00578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruana R., and Niculescu-Mizil A. (2006) An empirical comparison of supervised learning algorithms. In 23rd International Conference on Machine Learning, Pittsburgh, PA.
- Enamine Discovery Diversity Set, https://enamine.net/hit-finding/diversity-libraries/dds-50240.
- LOPAC 1280 – The Library of Pharmacologically Active Compounds, https://www.sigmaaldrich.com/life-science/cell-biology/bioactive-small-molecules/lopac1280-navigator.html.
- Selleckchem. Natural Product Library, https://www.selleckchem.com/screening/natural-product-library.html.
- Abdulla M. H.; Lim K. C.; Sajid M.; McKerrow J. H.; Caffrey C. R. (2007) Schistosomiasis mansoni: novel chemotherapy using a cysteine protease inhibitor. PLoS Med. 4 (1), e14 10.1371/journal.pmed.0040014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basch P. F. (1981) Cultivation of Schistosoma mansoni in vitro. I. Establishment of cultures from cercariae and development until pairing. J. Parasitol. 67 (2), 179–85. 10.2307/3280632. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.