Abstract

Pyrazolo[1,5-a]pyrimidin-7(4H)-one was identified through high-throughput whole-cell screening as a potential antituberculosis lead. The core of this scaffold has been identified several times previously and has been associated with various modes of action against Mycobacterium tuberculosis (Mtb). We explored this scaffold through the synthesis of a focused library of analogues and identified key features of the pharmacophore while achieving substantial improvements in antitubercular activity. Our best hits had low cytotoxicity and showed promising activity against Mtb within macrophages. The mechanism of action of these compounds was not related to cell-wall biosynthesis, isoprene biosynthesis, or iron uptake as has been found for other compounds sharing this core structure. Resistance to these compounds was conferred by mutation of a flavin adenine dinucleotide (FAD)-dependent hydroxylase (Rv1751) that promoted compound catabolism by hydroxylation from molecular oxygen. Our results highlight the risks of chemical clustering without establishing mechanistic similarity of chemically related growth inhibitors.
Keywords: pyrazolo[1,5-a]pyrimidin-7(4H)-one; structure−activity relationship; tuberculosis; antitubercular activity
Tuberculosis (TB) is a bacterial infection caused by Mycobacterium tuberculosis (Mtb), which is spread through coughing.1,2 The World Health Organization (WHO) estimates that 1.7 billion people, almost one-quarter of the world’s population, are infected with Mtb, including both active and latent TB patients.3 The urgency of eradicating TB is therefore compelling not only because it is the single deadliest infectious disease but also due to its growing resistance toward available drugs. In 2018, there were about half a million new cases with resistance to rifampicin, of which 78% are multidrug-resistant TB (MDR-TB).3 Among cases of MDR-TB in 2018, 6.2% was estimated to have extensively drug-resistant TB (XDR-TB), requiring complex regimens and an especially long duration of treatment.3 However, treatment success remains low due to a limited selection of highly active drugs.4 There is, therefore, an urgent need for the development of new drug candidates exhibiting novel mechanisms of action to address drug resistance and shorten treatment duration.
To discover new antitubercular agents, high-throughput screening (HTS) against whole cells has produced most of the existing TB drugs and clinical candidates.5 An in-house HTS campaign of diverse small-molecule libraries yielded compound 1 (Figure 1) as a confirmed hit. This compound displays an embedded pyrazolo[1,5-a]pyrimidin-7(4H)-one scaffold. Several previous studies have reported pyrazolo[1,5-a]pyrimidin-7(4H)-one derivatives with moderate antitubercular activities with varying modes of action.6−8 From whole-cell screens against Mtb H37Rv, compounds 2 and 3, which have a similar pyrazolo[1,5-a]pyrimidin-7(4H)-one scaffold, were identified as hits.6 However, the mode of action profiling by selecting spontaneous resistant mutants revealed important differences. Resistant clones selected against 2 had mutations in eccB3 (Rv0283), which is a component of the ESX-3 type VII secretion system in the Mtb genome.6 Further study of this compound demonstrated that ESX-3 was likely not the real target but, rather, the molecule perturbs iron metabolism in which ESX-3 plays a crucial part.8 On the other hand, mutants selected for resistance to 3 exhibited insertions of a transposon (IS6110) in rv1685c, a transcription factor whose biological role and regulatory targets are not known.6 Furthermore, another analogue 4 (Figure 1) was reported to be an inhibitor of the Mtb 1-deoxy-d-xylulose 5-phosphate synthase (DXS) with a moderate MIC value.7,9 Strikingly, although all three compounds display a high degree of chemical similarity, each molecule has been shown to have a different mode of action.
Figure 1.
Chemical structure of the hit compound, 1, and analogues 2–4 that have been previously reported.6−8
In this work, we undertook a structure–activity relationship (SAR) study starting from pyrazolo[1,5-a]pyrimidin-7(4H)-one analogue 1 to shine light on the confusing landscape of potential targets and resistance mechanisms.
Results and Discussion
Synthesis of Analogues and Structural Elucidation by X-ray Crystallography
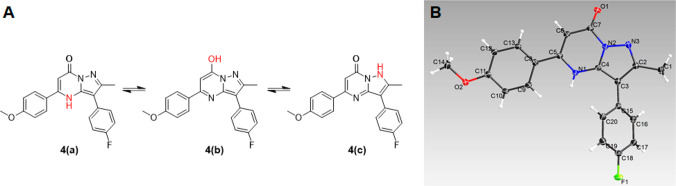
To identify the essential structural moieties for antitubercular activity, we designed and synthesized a set of modified analogues. The general synthesis of these fused heterocycle pyrazolo[1,5-a]pyrimidin-7(4H)-ones (1, 2, 4, P6, P8–P27) relies on the one-step cyclocondensation reaction of commercially available β-ketoesters 5 with aminopyrazoles 6 (Scheme 1). The core scaffold, pyrazolo[1,5-a]pyrimidin-7(4H)-one, has three plausible tautomeric structures (Figure 2A). Different tautomeric forms of small molecules are known to interact with different biological targets in unique ways; therefore, we wanted to establish which tautomer we were studying. The crystalline form of 4 generated by normal reaction conditions in this study was verified to have the structure shown as 4a in Figure 2A by single crystal X-ray diffraction (Cambridge Structural Database ID. CCDC 2034666). It was clear that the electron density around the two nitrogen atoms N1 and N3, along with the C=O bond length of 1.23 ± 0.01 Å, support the tautomer 4a as being dominant. This bond length is consistent with the expected sp2 C=O bond length observed in aldehydes and ketones and comfortably different from the ar C–O bond length of 1.36 Å expected for phenolic compounds.10 Therefore, we used this tautomer structure (4a) to depict the synthetic derivatives.
Scheme 1. General Synthetic Scheme of Pyrazolo[1,5-a]pyrimidin-7(4H)-one Derivatives 1, 2, 4, and P6, P8–P27.
Reagents and conditions: (a) toluene, reflux, overnight; (b) AcOH, reflux, overnight.
Figure 2.
(A) Three probable tautomeric structures (4a–c) of the pyrazolo[1,5-a]pyrimidin-7(4H)-one scaffold in 4; (B) X-ray crystal structure of 4.
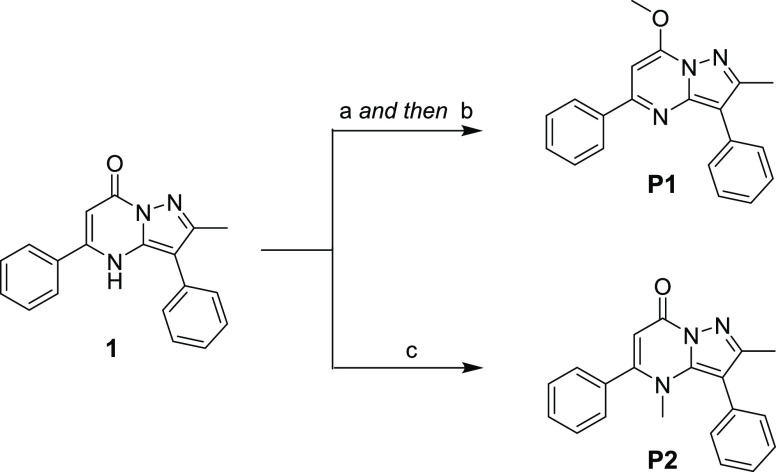
O-Methylation (P1) and N-methylation (P2) of hit 1 were performed to evaluate the importance of these different tautomeric configurations of the pyrazolopyrimidine core. Following conversion to the chlorinated intermediate by POCl3, quenching with sodium methoxide afforded P1.11 Direct methylation of 1 with methyl iodide in the presence of Cs2CO3 generated N-Me analogue P2 (Scheme 2).
Scheme 2. Synthesis of P1 and P2.
Reagents and conditions: (a) POCl3 (2.5 equiv), DIPEA (0.2 equiv), 90 °C, overnight; (b) NaOMe in MeOH (2.0 equiv), rt, overnight; (c) MeI (5.0 equiv), Cs2CO3 (3.0 equiv), rt, overnight.
To validate substituent effects in the C2 position, the replacement of methyl with a hydroxyl (P3) or an amino group (P4) was performed as described in Scheme 3. P3 was generated from methyl phenylcyanoacetate 9 through a key intermediate, amino pyrazolone 10, by the addition of hydrazine12 followed by intramolecular cyclization. Intermediate 9 in turn was prepared by coupling benzyl cyanide 7 with dimethyl carbonate 8. On the other hand, the coupling reaction between iodobenzene 11 and malononitrile 12 provided phenylmalononitrile 13,13 which generated the diamino pyrazole 14 after the reaction with hydrazine.12 The fused heterocyclic P4 was finally generated by an intramolecular condensation reaction after the reaction with β-ketoester 5.
Scheme 3. Synthesis of Analogues P3–P5.

Reagents and conditions: (a) 7 (1.0 equiv), 8 (10 equiv), NaOMe (2.0 equiv), 90 °C, overnight; (b) N2H4·H2O (2.0 equiv), EtOH, reflux, overnight; (c) 5 (1.0 equiv), EtOH, reflux, overnight; (d) 11 (1.0 equiv), 12 (2.0 equiv), K2CO3 (4.0 equiv), CuBr2 (0.1 equiv), DMSO, 120 °C, overnight; (e) methanolic HCl, rt, overnight; (f) ethyl chloroformate (1.2 equiv), K2CO3 (1.0 equiv), CH2Cl2, rt, 36 h; (g) EtOH, AcOH, reflux, 5 h.
The triazinone analogue P5 was synthesized for the evaluation of the main core scaffold, pyrazolo pyrimidinone. Benzonitrile 15 was reacted with methanol in acidic conditions to generate iminoester 16, and then, condensation with ethyl chloroformate resulted in the formation of imidate 17. After the reaction with aminopyrazole 6, the desired product, P5, was obtained.14
The trifluoromethylated analogue P7 was synthesized by a light-promoted reaction with trifluoromethyl iodide combined with Cs2CO3 (Scheme 4). Instead of blue LEDs as a light source,15 365 nm UV light was alternatively used for this photoactivation reaction, and the efficient and practical incorporation of a trifluoromethyl group into the α-carbonyl carbon was confirmed.
Scheme 4. Synthesis of Analogue P7.
Reagents and conditions: (a) 1 (1.0 equiv), Cs2CO3 (3 equiv), CF3I in DMSO, 365 nm UV irradiation during overnight rt stirring.
Structure–Activity Relationship Study of Synthetic Analogues and Their Antitubercular Activities
Preliminary SAR studies to address important questions around the hit scaffold were focused on determining key moieties or functionalities that may play important roles in antitubercular activity. Minimum inhibitory concentrations were determined against Mtb strain H37Rv in the presence (ADC) or absence (GCas) of albumin. Lower MICs in GCas generally mean lower protein binding. A focused library containing 27 compounds (P1–P27) was constructed for the preliminary SAR (Tables 1 and 2). For comparative purposes, we also resynthesized the literature compounds 2 and 4. In order to explore the core pyrazolo[1,5-a]pyrimidin-7(4H)-one scaffold, the methylated analogues P1 with O-Me and P2 with N-Me were synthesized, resulting in loss of activity. Among the three different tautomeric configurations in Figure 2A, the structure of the O-methylated P1 derivative was constrained to the tautomer (4b) form, while the N-methylated P2 was confined to the tautomer (4a) and lost its hydrogen bond donor capability. Polar functional groups in the C2 position including hydroxyl (P3) or amino (P4) groups can cause significant changes in the original scaffold including conformational differences and hydrogen bonding characteristics. All of these changes resulted in the loss of antimicrobial activity. In addition, it was found that the triazinone derivative, P5, was almost inactive against whole cells (Table 1).
Table 1. Structure and Antitubercular Activities of Compounds 1, 2, 4, and P1–P5c.
MIC of compounds tested against Mtb H37Rv in Middlebrook 7H9/ADC/Tween.
MIC of compounds tested against Mtb H37Rv in 7H9/glucose/casitone/tyloxapol.
See the Methods section.
Table 2. Structural Aspects and Antitubercular Activities of Compounds P6–P27.

MIC of compound tested against Mtb H37Rv in Middlebrook 7H9/ADC/Tween.
MIC of compound tested against Mtb H37Rv in 7H9/glucose/casitone/tyloxapol. See the Methods section.
Cytotoxicity of compound tested against HepG2 cells in DMEM/10% FBS supplemented with glucose.
Cytotoxicity of compound tested against HepG2 cells in DMEM/10% FBS with galactose.
Not determined.
Compounds P6–P27 were used to evaluate four different positions around the core, pyrazolo[1,5-a]pyrimidin-7(4H)-one (Table 2). First, P6, which has a methyl group, and P7, which has a trifluoromethyl group at the R1 position, retained activities in ADC media but displayed slightly detrimental effects in albumin containing media, suggesting strong protein binding and no electronic effect in the R1 position compared to the initial hit. The next series, P8–P20, were derivatized specifically in the R2 position with diverse moieties and substituents. Benzyl (P8) instead of phenyl retained its activity in GCas media, but cyclohexylmethyl (P9), morpholinomethyl (P10), cyclohexyl (P11), and isobutyl (P12) were not tolerated at this position. However, P8 displayed a somewhat large discrepancy in the MIC with or without protein in the media as evidenced by the markedly increased MIC in 7H9/ADC media. Some heteroaryl groups including 3-picolyl (P13), 3-furyl (P14), and pyrazinyl (P15) also showed the same pattern of relatively big MIC differences in the two media. With the phenyl group at the R2 position, a fluoro substituent as a small electron-withdrawing group was confirmed to retain its original potency or generate slightly better activities regardless of the substitution pattern (P16–P18). Furthermore, the compounds embedding 3,4-difluoro (P19) or 3,4-dichloro (P20) were favorable for activity in 7H9/ADC due to less protein binding as seen by the 5-fold improvement in the MIC compared to the initial hit, 1.
To assess the importance of the phenyl group at the R3 position, P21 was synthesized, resulting in loss of antitubercular activity. Therefore, an aromatic ring in positions R2 and R3 was confirmed to be necessary for antitubercular activity in the main scaffold. To evaluate the effect of the methyl group at the R4 position, several analogues were designed (P22, P23, P26, and P27). The removal of the methyl group at position R4 (P22) was detrimental to the activity compared to 1. On the other hand, P26, the R4 des-methyl analogue of P25, showed 4-fold better potency. In addition, the isopropyl substituted P23 was inactive against Mtb. P19 showed improved potency (2.73 μM in 7H9/ADC and 1.56 μM in 7H9/GCas). Surprisingly, the R4 trifluoromethyl analogue of P19 and P27, completely lost activity, suggesting the possible effects of this electron withdrawing functionality on the properties of this heterocycle. After modifications of R2 and R3, P24 and P25 were found to retain their activities in GCas and were 8-fold more potent in the presence of protein than the initial hit without serious cytotoxicity. Most active derivatives were not cytotoxic against HepG2 cells and additionally did not exert overt mitochondrial toxicity as seen by the lack of cytotoxicity during the growth of these cells on galactose as the carbon source. Specifically, P25 showed a selectivity index of >60 in glucose media and >30 in galactose media, respectively (Table 2). In addition, our data indicated that the compounds were not highly vulnerable to host metabolism as seen by the moderate stability in the microsomal stability assays (Table S1). Therefore, active compounds without serious cytotoxicity against HepG2 cells including P19, P24, and P25 were selected and used for further biological studies.
In Vitro and Ex Vivo Antitubercular Activity of Pyrazolo[1,5-a]pyrimidin-7(4H)-one Derivatives
Targets in mycolyl-arabinogalactan biosynthesis as well as in the bc1 complex of the terminal respiratory oxidase have been reported to be promiscuous targets on the basis of high hit rates of compound libraries against these proteins.16 To generate an early understanding of the potential mechanism of action, compounds 1, 2, 4, and P19 were profiled using assays that report on the inhibition of these targets using a pini-LUX strain that informs on the inhibition of cell wall biosynthesis17 as well as a cydC::aph mutant that is hypersusceptible to QcrB inhibitors.18,19 These assays indicated that these compounds did not inhibit these apparently promiscuous targets (Table 3). In addition, the reporters for compounds that result in DNA damage indicated that the primary mechanism of action did not involve the modulation of DNA homeostasis.17 MIC determination under a variety of carbon sources with in vivo relevance including dipalmitoyl-phosphatidylcholine, cholesterol, and glucose showed that these compounds retained activity against Mtb under all these conditions. In addition, these compounds were active against Mtb, slowly replicating under the growth restriction imposed by low pH and nitrosative stress, suggesting that these compounds inhibit targets of functional importance under a wide diversity of growth conditions.
Table 3. Biological Evaluation of Pyrazolo[1,5-a]pyrimidin-7(4H)-one Derivatives.
| MIC (μM) |
reporter gene assay |
|||||||
|---|---|---|---|---|---|---|---|---|
| hit | 7H9a | GASTb | GBSA/DBSAc | nitrate/butyrated | Chole | CydKOf | cell wallg | DNA damageh |
| 1 | 12.5 | 2.3 | 12.5 | 12.5 | 9.40 | 12.5 | no | no |
| 2 | 19.0 | 19.0 | 4.70 | 6.25 | 12.5 | 25.0 | no | no |
| 4 | 12.5 | 3.13 | 9.40 | 6.25 | 12.5 | 9.40 | no | no |
| P19 | 3.13 | 1.20 | 3.13 | 1.56 | 3.13 | 3.13 | no | no |
| INH | 0.39 | 0.10 | 0.30 | N/Di | 0.20 | 0.30 | N/D | N/D |
| SQ109 | N/D | N/D | N/D | N/D | N/D | N/D | yes | N/D |
| MOX | N/D | N/D | N/D | N/D | N/D | N/D | N/D | yes |
MIC of compound against Mtb strain H37Rv in Middlebrook 7H9/BSA containing glucose/glycerol/Tween 80.
MIC of compound against Mtb strain H37Rv in GAST/Fe.
MIC of compound against Mtb strain H37Rv in Middlebrook 7H9/BSA/Tyloxapol with glucose (GBSA) or dipalmitoylphosphatidylcholine (DBSA) as the carbon source.
MIC of compound against Mtb strain H37Rv in Middlebrook 7H9/BSA/Tyloxapol/butyrate/0.1 mM nitrite/pH 6.0.
MIC of compound against Mtb strain H37Rv in Middlebrook 7H9/BSA/Tyloxapol/cholesterol.
MIC of compound against Mtb strain H37Rv in Middlebrook 7H9/ADC with cydC::aph.
Induction of the cell wall responsive iniBAC promoter as measured using the piniB-LUX reporter strain.
Induction of the DNA damage responsive recA and radA reporters using the precA-LUX and pradA-LUX reporter strains.
Not determined.
Prompted by the favorable selectivity indices obtained for several of the synthesized derivatives, the efficacy of the compounds against intracellular Mtb was determined. For that, the cytotoxicity of selected analogues was first evaluated against the murine macrophage cell line, J774. While largely tolerated by HepG2 cells, the tested compounds P19, P24, and P25 exhibited modest cytotoxicity against J774 cells (Figure 3A) with IC50 values of 29.6 ± 1.2, 22.5 ± 2.1, and 20.2 ± 0.8 μM respectively.
Figure 3.

(A) Murine macrophage cytotoxicity was measured using a standard luminescence-based assay. Curves represent the average of three trials done in duplicates. (B) J774 cells were infected with Mtb H37Rv (MOI 1:5) and treated with the corresponding compounds for 7 days. Survival of intracellular bacteria was measured by plating serial dilutions following J774 lysis. Bars: mean ± SEM obtained from two trials done in duplicates.
J774 cells were then infected with Mtb H37Rv at a multiplicity of infection of 5 (MOI 1:5) and treated with P19 or P25 for 7 days. Taking into consideration both their in vitro MICs and corresponding IC50 values against J774 cells, we settled on a low dose of 7.5 μM and a high dose of 15 μM for both compounds, concentrations above the MIC but below the cytotoxic threshold for J774 cells. It was found that both P19 and P25 possess cidal activity against intramacrophage Mtb H37Rv (Figure 3B) at the tested concentrations. High doses of P19 reduced the bacterial burden by ∼0.5 logs while P25 reduced bacterial counts by 1.5 and 2 orders of magnitude at 7.5 and 15 μM, respectively.
Biological Evaluation of Pyrazolo[1,5-a]pyrimidin-7(4H)-ones
Compound 2, which was previously shown to interfere with iron homeostasis in mycobacteria by directly binding to intracellular Fe2+, shares a similar scaffold with 1, 4, P19, and P24.8 To deplete the iron levels in the medium, we added 100 μM desferrioxamine (DFO; MIC = 250 μM), a siderophore produced by Streptomyces that cannot be utilized by mycobacteria.20 While siderophores selectively bind to Fe3+, the loss of extracellular Fe3+ due to chelation would lead to the depletion of intracellular Fe2+ during growth. We also supplemented the iron levels by the addition of 250 μM FeCl3, half the concentration of the inorganic salt’s MIC (500 μM). We utilized in-house controls, the antitubercular benzoxazoles (593 and 596), compounds that interfere with iron metabolism as evidenced by transcriptional upregulation of mycobactin biosynthetic genes and the esx3 operon (unpublished results), as positive controls to qualify the assay. The benzoxazoles 593 and 596 were 8- to 16-fold more potent under iron-depleted conditions (Figure 4A). Similarly, it was confirmed that 2 had greater efficacy under iron-limited conditions in accordance to previously published results.8 In contrast, neither 4 nor the newly described pyrazolo[1,5-a]pyrimidin-7(4H)-ones (P19 and P24) were affected by iron depletion, producing at best, a negligible 2-fold decrease in MIC. Excess iron in the medium led to neither activation nor attenuation of any of the drugs tested, indicating that none of these compounds act as ionophores that shuttle iron into the cell.
Figure 4.

(A) Effect of iron levels on the potency of test compounds. MICs were measured in the presence of 100 μM desferrioxamine (DFO) or 250 μM FeCl3. The increase in potency of the compounds is expressed as MIC ratios equal to MIC in pure 7H9 media divided by MIC in 7H9 supplemented with either DFO or FeCl3. Bars represent mean ± SEM (n = 2). (B) Effect of 1 in the iron uptake of Mtb in vitro. Cells treated with the specified compound were fed 55Fe, and the radioactivity inside H37Rv was measured over a 72 h time course. Data represent mean ± SD (n = 3). CPM, counts per minute.
We further determined the effect of the hit compound 1 directly on iron uptake by Mtb, on the basis of the reported accumulation of intracellular iron in M. smegmatis following treatment with 2.8 We treated Mtb cells with 1 in a growth medium labeled with 55Fe and tracked the influx of radioactivity into the cells. As shown in Figure 4B, compound 1 did not inhibit iron uptake unlike DFO, which completely inhibited uptake. Taken together with the result of our iron limitation experiments, the antitubercular activities of these pyrazolo[1,5-a]pyrimidin-7(4H)-one analogues based on hit 1 were not related to the perturbations in iron homeostasis.
To determine the mechanism of action of the most active analogues, we independently raised mutants against P19 and P24 at 5× and 10× of their in vitro MIC. Despite having minimal structural differences, we were only able to raise resistant mutants against P19. These clones arose at a frequency of 3 × 10–8 at 5× MIC and 4 × 10–9 at 10× MIC.
We measured resistance of the mutants (P19R) against P19, P24, 1, 2, and 4. We found that mutants raised against 5× MIC of P19 were 4- to 7-fold more resistant than the WT, and those that were raised against 10× MIC were highly resistant (≥8-fold) to P19. Unsurprisingly, the resistant mutants exhibited similar resistance profiles against P24. P19R strains were also highly resistant to the original hit compound, 1. On the other hand, the resistant mutants exhibited relatively small increased MICs against 2 or 4, values that are within the margin of error of typical MIC measurements, indicating a lack of cross-resistance of P19R strains against 2 and 4. This data suggested that 1, P19, and P24 had distinct targets or resistance mechanisms from those of 2 and 4 (Table 4).
Table 4. Spontaneous Resistant Mutants Raised against P19 and Their Corresponding MICs.
| SNPs identified
in P19 resistant mutants
in rv1751 (# of independent mutants isolated) |
||||||||
|---|---|---|---|---|---|---|---|---|
| Cpd | MIC (μM) | WT | L399W (7) | V96L (6) | H97Y/N (2) | L250Q (2) | P152S (1) | A434T (1) |
| P19 | MIC | 0.78 | 12.5 | 2.3 | 1.56 | 2.3 | 1.56 | 1.56 |
| fold change | 16 | 3 | 2 | 3 | 2 | 2 | ||
| P24 | MIC | 1.56 | 18.8 | 7.82 | 6.25 | 6.26 | 3.90 | 3.12 |
| fold change | 12 | 5 | 4 | 4 | 2.5 | 2 | ||
| 1 | MIC | 1.2 | 12.5 | 2.3 | 1.56 | 3.13 | 2.3 | 1.56 |
| fold change | 10 | 2 | 1.3 | 2.6 | 2 | 1.3 | ||
| 2 | MIC | 0.39 | 0.39 | 1.2 | 0.39 | 1.2 | 1.2 | 1.2 |
| fold change | 1 | 3 | 1 | 3 | 3 | 3 | ||
| 4 | MIC | 1.56 | 3.13 | 2.3 | 1.56 | 4.7 | 2.3 | 2.3 |
| fold change | 2 | 1.5 | 1 | 3 | 1.5 | 1.5 | ||
| INH | MIC | 0.04 | 0.04 | 0.04 | 0.04 | 0.04 | 0.04 | 0.04 |
The genomic DNA of the isolated P19R strains was extracted and sequenced, and it was found that all single-nucleotide polymorphisms (SNPs) mapped onto rv1751, a nonessential gene annotated as a probable oxidoreductase but whose function has not been definitively demonstrated. A closer examination of the SNPs and their corresponding MICs showed that the level of resistance can be classified into three categories: (1) mutations in P152, L250, and A434 resulting in low-level resistance, (2) mutations in V96 and H97 imparting intermediate resistance, and (3) mutations in L399 conferring high-level resistance to P19 and P24. Sequence similarity searching showed primarily flavin adenine dinucleotide (FAD)-dependent hydroxylases and monooxygenases from different organisms (a logical result considering Rv1751 is also predicted to have a FAD-binding domain) whose known substrates are aromatic compounds (e.g., phenols and hydroxybenzoates).21−23 One of the more similar proteins was Rox, a rifampicin monooxygenase from Streptomyces venezuelae (PDB ID: 5VQB).24 We modeled Rv1751 using the crystal structure of Rox as a template (Figure 5A). In this modeled structure, the SNPs that conferred intermediate- and high-level resistance were located proximal to the RIF binding site, while those that lead to low-level resistance were distal to this site (Figure 5B). H97 and L399 both point directly toward the RIF binding site, suggesting that mutations in these positions will likely lead to a change in the substrate binding affinity, in this case, a change in the binding affinity of P19 to Rv1751 (Figure 5B). Taken together, these results suggested that the resistant clones have mutations that affected the catabolism of P19, resulting in decreased susceptibility.
Figure 5.
(A) Calculated structure of Rv1751 (red ribbons) superimposed on crystal structure of rifampicin monooxygenase (Rox, green ribbons) from S. venezuelae. Rifampicin (RIF) is shown in magenta, and FAD is shown in cyan. (B) Co-crystal structures of RIF and FAD are superimposed onto the calculated structure of Rv1751 (red ribbons). Residues mutated in P19R are highlighted in yellow (conferring high-level resistance) and cyan (conferring low-level resistance).
To determine the potential role of Rv1751 in the resistance toward P19, the degree of P19 degradation when exposed to Mtb was measured via a LC/MS-based assay. Under conditions that saturated the compound with Mtb cells (P19 at 10× MIC added to 109 CFUs of H37Rv), it was found that there was about 10% degradation of P19 by Mtb after 24 h of exposure. When we performed a similar experiment using the P19R–intermediate Rv1751 V96L mutant, almost 30% of the compound was degraded, and when the L399W mutant was used, nearly 50% of P19 was metabolized (Figures 6A and S1). Interestingly, the degree to which P19 was catabolized by the various forms of Rv1751 matches the level of resistance that these polymorphisms impart to cells. That is, the higher level of resistance (higher MICs) of specific strains are associated with their greater catabolic capability (Figure 6B). Finally, in accordance with the slight cross-resistance of the P19R strains to 4, there were only small differences in the degree of degradation of 4 when it was exposed to either the WT or the P19R strains. To identify the major metabolites of P19 generated in mutant cells, a heavy oxygen (18O) stable isotope labeling experiment coupled with MS-based analysis was performed. On the basis of the preliminary LC/MS analysis of the metabolites (Figure S1), we speculated that one or two oxygen atoms were incorporated into some of the major metabolites. Due to the fact that the incorporation test with 18O-enriched water (H218O) did not provide any meaningful information, P19 was incubated with the L399W mutant cells under an 18O2 environment for 2 days, and then, the media was analyzed by a triple quadrupole tandem MS fragmentation technique to characterize the degradation pattern. As shown in Figures S2–S4, L399W mutant cells accelerated the catabolic degradation of P19 by the hydroxylation of R1, hydroxylation of the phenyl ring in the R3 position, and/or N-methylation of the pyrimidinone ring.
Figure 6.
(A) Degree of compound degradation upon exposure to WT H37Rv and the indicated P19R strains was measured via LC/MS. Bars represent mean ± SEM (n = 3). (B) The correlation between compound degradation by Mtb and in vitro MIC shows the dependence of antimycobacterial activity to compound metabolism.
Conclusions
Although the pyrazolo[1,5-a]pyrimidin-7(4H)-one scaffold has been found in several antitubercular agents with different mode of actions, there are no reports regarding the exploration of the SAR based on the evaluation of activity against Mtb cells. We confirmed the exact tautomeric structure by small-molecule X-ray crystallography and explored the SAR from 1 by the design and synthesis of a focused library containing 26 analogues in which we modified each moiety in the original pyrazolo[1,5-a]pyrimidin-7(4H)-one scaffold to understand its contribution to whole-cell activity. The compounds were found to be active against Mtb under a variety of in vitro growth conditions that reflect the carbon sources thought to be important for in vivo pathogenesis of the organism. The compounds were found to have favorable selectivity indices against Mtb compared to eukaryotic cells with greater than 10-fold selectivity for the pathogen than HepG2 cells. The most promising analogues including P19, P24, and P25 were found to exert growth inhibition of Mtb during parasitism of host macrophages. These compounds were selected and evaluated for further biological studies, which indicated that the compounds did not inhibit the targets in cell wall biosynthesis of respiration that have previously been reported to be apparently promiscuous targets. Despite being nearly identical in structure to a known iron chelator, these compounds did not exert their antitubercular effect by perturbation of iron homeostasis. We were, however, unable to decipher the intracellular target of this scaffold since resistant mutants mapped to rv1751 encoding a putative FAD-dependent hydroxylase, which we could confirm, led to increased catabolism of the compound. These studies suggest that structural similarity does not necessarily imply mechanistic similarity. Highly similar scaffolds with completely different targets have been reported previously; for example, very closely related indazole sulfonamides have been reported to be potent inhibitors of inosine monophosphate dehydrogenase25 and the β-ketoacyl-ACP synthase involved in mycolic acid biosynthesis.26 Such profound changes in the target with closely related structures suggests that the chemical clustering of the hit series, as is commonly done in Mtb whole-cell-based screening programs, must be done with additional care when selecting a hit series and interpreting the SAR in the absence of mechanistic information.
Methods
Chemistry
1H and 13C NMR spectra were recorded on a Varian Mercury-300 NMR spectrometer or a Bruker AVANCE III HD NanoBay 400 MHz spectrometer, and chemical shifts were measured in ppm relative to the specific solvent signal. Routine mass and purity analyses (LRMS) were performed on an HP Agilent LC/MS series 1100 system equipped with a reverse phase column (Agilent Poroshell 120 EC-C18, 2.7 μm, 50 × 2.1 mm) and photodiode array detector coupled to an Agilent 1946 DSL quadrupole mass selective detector using electrospray ionization (ESI). The gradient mobile phase consisting of acetonitrile/water with 0.1% formic acid and UV detection at 254 and 210 nm were used to confirm all final products to be ≥95%. The melting point was measured on an Electrothermal 9100 apparatus. Most reagents used in the synthetic procedure were purchased from Sigma-Aldrich, Alfa Aesar, and TCI. The progress of the reaction was monitored using thin-layer chromatography (TLC) (silica gel 60 F254 0.25 mm), and the products were visualized by UV light (254 and 365 nm). SiliaFlash P60 (40–60 μm) used in flash column chromatography was purchased from Silicycle Inc. Other solvents were purchased from commercial vendors and used without further purification unless otherwise mentioned.
Synthesis for the Compounds and Their Characterization Data
General syntheses of 1, 2, 4, and P6–P27 and the final cyclocondensation reactions for P3–P5 were performed using the previously reported procedure with some modifications.27−29 The synthetic procedures of P1(11,29) and P2(30) were followed by the previously reported procedure with some modifications. The key intermediate 10 for the synthesis of P3,12,31 intermediate 14 for the precursor of P4,12,13 and intermediate 17 for the P5(14) were prepared using the previously reported procedure with some modifications.
2-Methyl-3,5-diphenylpyrazolo[1,5-a]pyrimidin-7(4H)-one (1)
Off-white solid: mp = 293 °C; 1H NMR (300 MHz, DMSO-d6) δ 12.16 (s, 1H), 7.73 (d, J = 8.4 Hz, 2H), 7.51–7.42 (m, 7H), 7.35–7.30 (m, 1H), 5.92 (s, 1H), 2.32 (s, 3H); 13C NMR (75 MHz, DMSO-d6) δ 155.9, 151.0, 150.4, 138.8, 132.8, 130.8, 130.4, 129.5, 128.35, 128.31, 127.9, 126.5, 104.0, 94.9, 13.1; LRMS (ESI) m/z 302 [M + H]+.
3-(4-Chlorophenyl)-5-(cyclohexylmethyl)pyrazolo[1,5-a]pyrimidin-7(4H)-one (2)
Off-white solid: 1H NMR (300 MHz, DMSO-d6) δ 11.54 (bs, 1H), 7.82 (s, 1H), 7.40–7.31 (m, 4H), 5.53 (s, 1H), 2.43 (d, J = 6.6 Hz, 2H), 1.68–1.58 (m, 6H), 1.19–1.06 (m, 3H), 0.99–0.92 (m, 2H); 13C NMR (75 MHz, DMSO-d6) δ 156.4, 153.2, 141.5, 137.4, 131.6, 129.1, 128.9, 128.3, 103.5, 95.9, 40.0, 37.0, 32.2, 25.6, 25.3; LRMS (ESI) m/z 342 [M + H]+.
3-(4-Fluorophenyl)-5-(4-methoxyphenyl)-2-methylpyrazolo[1,5-a]pyrimidin-7(4H)-one (4)
White solid: mp = 271 °C; 1H NMR (300 MHz, DMSO-d6) δ 11.97 (s, 1H), 7.73 (d, J = 8.7 Hz, 2H), 7.54–7.50 (m, 2H), 7.28 (t, J = 9.0 Hz, 2H), 7.06 (d, J = 9.0 Hz, 2H), 5.91 (s, 1H), 3.82 (s, 3H), 2.27 (s, 3H); 13C NMR (75 MHz, DMSO-d6) δ 162.8, 161.3, 159.6, 155.9, 150.6, 150.2, 139.0, 131.7, 131.6, 129.6, 127.31, 127.27, 124.8, 115.4, 115.1, 114.0, 103.0, 94.0, 55.4, 13.0; LRMS (ESI) m/z 350 [M + H]+.
7-Methoxy-2-methyl-3,5-diphenylpyrazolo[1,5-a]pyrimidine (P1)
Pale-yellow solid: 1H NMR (300 MHz, DMSO-d6) δ 8.28–8.25 (m, 2H), 7.86–7.83 (m, 2H), 7.59–7.47 (m, 5H), 7.32–7.27 (m, 1H), 7.15 (s, 1H), 4.31 (s, 3H), 2.57 (s, 3H); 13C NMR (75 MHz, DMSO-d6) δ 157.8, 156.3, 152.6, 147.7, 138.0, 133.2, 131.0, 129.5, 129.1, 128.9, 128.0, 126.5, 108.2, 85.2, 58.4, 58.3, 15.2; LRMS (ESI) m/z 316 [M + H]+.
2,4-Dimethyl-3,5-diphenylpyrazolo[1,5-a]pyrimidin-7(4H)-one (P2)
Pale-yellow solid: 1H NMR (300 MHz, DMSO-d6) δ 8.06–8.02 (m, 2H), 7.71 (d, J = 7.8 Hz, 2H), 7.53 (t, J = 7.8 Hz, 2H), 7.48–7.37 (m, 4H), 6.50 (s, 1H), 4.27 (s, 3H), 2.52 (s, 3H); 13C NMR (75 MHz, DMSO-d6) δ 160.3, 158.0, 150.9, 147.8, 138.1, 130.8, 130.5, 130.2, 129.2, 129.1, 127.8, 127.5, 110.0, 96.9, 36.7, 11.3; LRMS (ESI) m/z 316 [M + H]+.
3,5-Diphenylpyrazolo[1,5-a]pyrimidine-2,7(1H,4H)-dione (P3)
Pale-yellow solid: 1H NMR (300 MHz, DMSO-d6) δ 12.0 (s, 1H), 7.77 (s, 2H), 7.64 (d, J = 7.2 Hz, 2H), 7.55 (m, 3H), 7.43 (t, J = 7.4 Hz, 2H), 7.26 (t, J = 6.9 Hz, 1H), 5.95 (s, 1H); 13C NMR (75 MHz, DMSO-d6) δ 161.9, 155.2, 150.4, 138.7, 132.9, 130.7, 130.2, 128.6, 128.5, 128.3, 128.0, 125.9, 95.8, 90.7, 56.1, 18.6; LRMS (ESI) m/z 304 [M + H]+.
2-Amino-3,5-diphenylpyrazolo[1,5-a]pyrimidin-7(4H)-one (P4)
Off-white solid: 1H NMR (300 MHz, DMSO-d6) δ 11.79 (s, 1H), 7.74 (s, 2H), 7.52–7.43 (m, 7H), 7.30 (t, J = 7.0 Hz, 1H), 5.85 (s, 1H), 5.29 (bs, 2H); 13C NMR (75 MHz, DMSO-d6) δ 156.0, 155.3, 149.5, 138.8, 133.0, 130.7, 130.4, 128.7, 128.5, 127.8, 126.2; LRMS (ESI) m/z 303 [M + H]+.
7-Methyl-2,8-diphenylpyrazolo[1,5-a][1,3,5]triazin-4(1H)-one (P5)
Pale-yellow solid: 1H NMR (300 MHz, DMSO-d6) δ 8.11 (d, J = 8.1 Hz, 2H), 7.70 (d, J = 7.8 Hz, 2H), 7.60–7.43 (m, 5H), 7.33–7.28 (m, 1H), 2.45 (s, 3H); 13C NMR (75 MHz, DMSO-d6) δ 153.0, 151.8, 145.4, 144.3, 132.0, 131.34, 131.27, 128.7, 128.5, 127.9, 126.6, 111.4, 14.4; LRMS (ESI) m/z 303 [M + H]+.
2,6-Dimethyl-3,5-diphenylpyrazolo[1,5-a]pyrimidin-7(4H)-one (P6)
Off-white solid: mp = 270–280 °C (decomposition); 1H NMR (300 MHz, DMSO-d6) δ 12.03 (s, 1H), 7.49 (s, 5H), 7.40–7.38 (m, 3H), 7.31–7.24 (m, 1H), 2.29 (s, 3H), 1.86 (s, 3H); 13C NMR (75 MHz, DMSO-d6) δ 157.0, 150.3, 147.8, 138.2, 133.7, 130.9, 129.6, 129.5, 129.1, 128.4, 128.2, 126.5, 102.6, 101.8, 13.2, 11.9; LRMS (ESI) m/z 316 [M + H]+.
2-Methyl-3,5-diphenyl-6-(trifluoromethyl)pyrazolo[1,5-a]pyrimidin-7(4H)-one (P7)
Off-white solid: 1H NMR (400 MHz, DMSO-d6) δ 12.91 (bs, 1H), 7.55–7.47 (m, 5H), 7.44–7.41 (m, 4H), 7.37–7.32 (m, 1H), 2.31 (s, 3H); 13C NMR (100 MHz, DMSO-d6) δ 153.5, 152.8, 151.6, 137.3, 132.9, 129.94, 129.85, 128.4, 128.2, 127.9, 127.1, 125.2, 122.5, 105, 5, 96.8, 96.5, 13.0; LRMS (ESI) m/z 370 [M + H]+.
5-Benzyl-2-methyl-3-phenylpyrazolo[1,5-a]pyrimidin-7(4H)-one (P8)
Pale-yellow solid: 1H NMR (300 MHz, DMSO-d6) δ 12.00 (s, 1H), 7.51–7.34 (m, 9H), 7.30–7.24 (m, 1H), 5.47 (s, 1H), 3.96 (s, 3H), 2.26 (s, 3H); 13C NMR (75 MHz, DMSO-d6) δ 155.9, 153.2, 150.0, 138.5, 137.1, 130.7, 129.4, 128.8, 128.7, 128.6, 126.9, 126.8, 103.1, 95.8, 37.5, 13.0; LRMS (ESI) m/z 316 [M + H]+.
5-(Cyclohexylmethyl)-2-methyl-3-phenylpyrazolo[1,5-a]pyrimidin-7(4H)-one (P9)
White solid: 1H NMR (300 MHz, DMSO-d6/CDCl3) δ 11.64 (s, 1H), 7.48–7.33 (m, 5H), 5.49 (s, 1H), 2.43 (d, J = 4.8 Hz, 2H), 2.27 (s, 3H), 1.68–1.65 (m, 6H), 1.22–1.15 (m, 3H), 1.00–0.94 (m, 2H); 13C NMR (75 MHz, DMSO-d6/CDCl3) δ 155.8, 152.8, 149.8, 138.4, 130.7, 129.3, 128.4, 126.5, 102.9, 95.6, 95.5, 36.9, 32.2, 25.7, 25.4, 12.9; LRMS (ESI) m/z 322 [M + H]+.
2-Methyl-5-(morpholinomethyl)-3-phenylpyrazolo[1,5-a]pyrimidin-7(4H)-one (P10)
Pale-green solid: 1H NMR (300 MHz, CDCl3) δ 7.41–7.31 (m, 6H), 5.70 (bs, 1H), 3.68 (bs, 4H), 3.52 (bs, 2H), 2.55 (bs, 4H), 2.40 (bs, 3H); 13C NMR (75 MHz, CDCl3) δ 157.0, 151.2, 148.6, 138.3, 131.2, 129.2, 128.2, 127.1, 103.8, 95.5, 67.0, 58.4, 53.5, 29.8, 13.4; LRMS (ESI) m/z 325 [M + H]+.
5-Cyclohexyl-2-methyl-3-phenylpyrazolo[1,5-a]pyrimidin-7(4H)-one (P11)
White solid: 1H NMR (300 MHz, DMSO-d6/CDCl3) δ 11.60 (s, 1H), 7.47–7.30 (m, 5H), 5.52 (s, 1H), 2.65–2.58 (m, 1H), 2.27 (s, 3H), 1.91–1.70 (m, 5H), 1.44–1.23 (m, 5H); 13C NMR (75 MHz, DMSO-d6/CDCl3) δ 158.8, 156.4, 149.9, 138.4, 130.8, 129.4, 128.4, 126.5, 102.9, 92.1, 40.4, 31.5, 25.7, 25.1, 12.9; LRMS (ESI) m/z 308 [M + H]+.
5-Isobutyl-2-methyl-3-phenylpyrazolo[1,5-a]pyrimidin-7(4H)-one (P12)
Off-white solid: 1H NMR (300 MHz, DMSO-d6) δ 11.69 (s, 1H), 7.51–7.33 (m, 5H), 5.56 (s, 1H), 2.44 (d, J = 7.5 Hz, 2H), 2.26 (s, 3H), 2.04–1.91 (m, 1H), 0.91 (d, J = 6.6 Hz, 6H); 13C NMR (75 MHz, DMSO-d6) δ 155.8, 153.3, 149.8, 138.6, 130.8, 129.4, 128.7, 126.8, 102.9, 95.7, 95.6, 40.8, 27.8, 21.9, 13.0; LRMS (ESI) m/z 282 [M + H]+.
2-Methyl-3-phenyl-5-(pyridin-3-yl)pyrazolo[1,5-a]pyrimidin-7(4H)-one (P13)
Off-white solid: 1H NMR (300 MHz, DMSO-d6) δ 12.26 (bs, 1H), 8.95 (s, 1H), 8.72 (d, J = 4.5 Hz, 1H), 8.18 (d, J = 7.5 Hz, 1H), 7.57–7.46 (m, 5H), 7.38–7.30 (m, 1H); 13C NMR (75 MHz, DMSO-d6) δ 156.3, 151.9, 151.2, 149.3, 139.7, 136.6, 131.4, 130.2, 129.3, 127.5, 124.1, 104.9, 96.4, 13.8; LRMS (ESI) m/z 303 [M + H]+.
5-(Furan-3-yl)-2-methyl-3-phenylpyrazolo[1,5-a]pyrimidin-7(4H)-one (P14)
Pale-yellow solid: 1H NMR (300 MHz, DMSO-d6) δ 11.78 (s, 1H), 8.47 (s, 1H), 7.81 (t, J = 1.8 Hz, 1H), 7.46–7.45 (m, 4H), 7.37–7.32 (m, 1H), 7.10 (s, 1H), 6.08 (s, 1H), 2.26 (s, 3H); 13C NMR (75 MHz, DMSO-d6) δ 156.6, 151.1, 145.2, 144.5, 143.9, 139.2, 131.6, 130.3, 129.3, 127.5, 120.0, 110.0, 104.5, 94.3, 13.9; LRMS (ESI) m/z 292 [M + H]+.
2-Methyl-3-phenyl-5-(pyrazin-2-yl)pyrazolo[1,5-a]pyrimidin-7(4H)-one (P15)
Pale-yellow solid: 1H NMR (300 MHz, CDCl3) δ 9.24 (s, 1H), 8.76 (d, J = 2.4 Hz, 1H), 8.66 (t, J = 1.8 Hz, 1H), 7.57–7.38 (m, 6H), 6.58 (s, 1H), 2.51 (s, 3H); 13C NMR (75 MHz, CDCl3) δ 156.7, 152.6, 146.8, 143.7, 143.1, 142.8, 142.0, 137.8, 130.9, 129.8, 128.6, 127.7, 105.2, 95.1, 13.6; LRMS (ESI) m/z 304 [M + H]+.
5-(2-Fluorophenyl)-2-methyl-3-phenylpyrazolo[1,5-a]pyrimidin-7(4H)-one (P16)
White solid: 1H NMR (300 MHz, DMSO-d6) δ 12.35 (s, 1H), 7.69–7.56 (m, 2H), 7.47–7.45 (m, 4H), 7.40–7.32 (m, 3H), 5.84 (s, 1H), 2.32 (s, 3H); 13C NMR (75 MHz, DMSO-d6) δ 160.8, 157.5, 155.6, 150.4, 145.9, 138.7, 132.6, 132.5, 131.0, 130.6, 129.5, 128.6, 126.8, 124.64, 124.60, 121.4, 116.1, 115.8, 130.8, 97.2, 13.2; LRMS (ESI) m/z 320 [M + H]+.
5-(3-Fluorophenyl)-2-methyl-3-phenylpyrazolo[1,5-a]pyrimidin-7(4H)-one (P17)
Pale-yellow solid: 1H NMR (300 MHz, DMSO-d6) δ 12.17 (bs, 1H), 7.69–7.33 (m, 9H), 6.02 (s, 1H), 2.32 (s, 3H); 13C NMR (75 MHz, DMSO-d6) δ 163.4, 160.2, 155.7, 150.5, 149.7, 138.8, 135.0, 130.8, 130.7, 130.6, 129.5, 128.5, 126.7, 124.3, 117.5, 117.2, 115.3, 115.0, 104.3, 95.4, 13.1; LRMS (ESI) m/z 320 [M + H]+.
5-(4-Fluorophenyl)-2-methyl-3-phenylpyrazolo[1,5-a]pyrimidin-7(4H)-one (P18)
White solid: 1H NMR (300 MHz, DMSO-d6) δ 12.13 (s, 1H), 7.81 (bs, 2H), 7.47–7.42 (m, 4H), 7.37–7.32 (m, 3H), 5.92 (s, 1H), 2.29 (s, 3H); 13C NMR (75 MHz, DMSO-d6) δ 165.2, 161.9, 155.8, 150.3, 150.1, 138.9, 130.9, 130.6, 130.5, 129.5, 128.5, 126.7, 115.6, 115.3, 104.1, 95.0, 13.1; LRMS (ESI) m/z 320 [M + H]+.
5-(3,4-Difluorophenyl)-2-methyl-3-phenylpyrazolo[1,5-a]pyrimidin-7(4H)-one (P19)
Off-white solid: mp = 255 °C; 1H NMR (300 MHz, DMSO-d6) δ 12.13 (bs, 1H), 7.94 (t, J = 9.3 Hz, 1H), 7.73–7.45 (m, 6H), 7.37–7.32 (m, 1H), 6.00 (bs, 1H), 2.32 (s, 3H); 13C NMR (75 MHz, DMSO-d6) δ 155.6, 152.5, 152.4, 150.8, 150.6, 149.2, 149.1, 147.5, 147.4, 138.7, 130.8, 129.5, 128.5, 126.8, 125.5, 117.8, 117.6, 104.2, 95.4, 13.1; LRMS (ESI) m/z 338 [M + H]+.
5-(3,4-Dichlorophenyl)-2-methyl-3-phenylpyrazolo[1,5-a]pyrimidin-7(4H)-one (P20)
Pale-yellow solid: mp = 280 °C; 1H NMR (300 MHz, DMSO-d6) δ 12.17 (bs, 1H), 8.08 (bs, 1H), 7.78 (bs, 2H), 7.52–7.46 (m, 4H), 7.38–7.33 (m, 1H), 6.05 (bs, 1H), 2.32 (s, 3H); 13C NMR (75 MHz, DMSO-d6) δ 155.6, 150.6, 148.6, 138.8, 133.3, 131.2, 130.7, 130.6, 130.1, 129.5, 128.6, 128.4, 126.8, 104.2, 95.7, 13.1; LRMS (ESI) m/z 370 [M + H]+.
2,3-Dimethyl-5-phenylpyrazolo[1,5-a]pyrimidin-7(4H)-one (P21)
Off-white solid: 1H NMR (300 MHz, DMSO-d6) δ 11.90 (s, 1H), 7.76–7.74 (m, 2H), 7.56–7.54 (m, 3H), 5.81 (s, 1H), 2.22 (s, 3H), 2.10 (s, 3H); 13C NMR (75 MHz, DMSO-d6) δ 156.7, 152.3, 150.7, 139.9, 133.6, 131.4, 129.5, 128.5, 97.2, 94.52, 94.47, 13.0, 7.4; LRMS (ESI) m/z 240 [M + H]+.
3,5-Diphenylpyrazolo[1,5-a]pyrimidin-7(4H)-one (P22)
Off-white solid: 1H NMR (300 MHz, DMSO-d6) δ 12.26 (bs, 1H), 8.22 (s, 1H), 7.84 (d, J = 6.3 Hz, 2H), 7.69 (d, J = 6.3 Hz, 2H), 7.58–7.53 (m, 3H), 7.49–7.43 (m, 2H), 7.33–7.29 (m, 1H), 6.05 (s, 1H); 13C NMR (75 MHz, DMSO-d6) δ 156.9, 152.4, 143.1, 138.7, 133.6, 131.5, 129.3, 128.8, 128.3, 127.2, 106.5, 95.8; LRMS (ESI) m/z 288 [M + H]+.
2-Isopropyl-3,5-diphenylpyrazolo[1,5-a]pyrimidin-7(4H)-one (P23)
Off-white solid: 1H NMR (300 MHz, DMSO-d6) δ 12.15 (s, 1H), 7.73 (d, J = 7.8 Hz, 2H), 7.56–7.44 (m, 7H), 7.38–7.33 (m, 1H), 5.92 (s, 1H), 3.10–3.01 (m, 1H), 1.18 (d, J = 6.9 Hz, 6H); 13C NMR (75 MHz, DMSO-d6) δ 159.2, 156.1, 151.1, 139.0, 132.9, 130.9, 130.6, 130.2, 128.5, 128.1, 127.0, 103.2, 94.9, 26.0, 22.1; LRMS (ESI) m/z 330 [M + H]+.
5-(3,4-Dichlorophenyl)-3-(4-fluorophenyl)-2-methylpyrazolo[1,5-a]pyrimidin-7(4H)-one (P24)
Off-white solid: 1H NMR (300 MHz, DMSO-d6) δ 12.14 (bs, 1H), 8.07 (s, 1H), 7.77 (s, 2H), 7.56 (bs, 2H), 7.30 (t, J = 8.7 Hz, 2H), 6.06 (s, 1H), 2.29 (s, 3H); 13C NMR (75 MHz, DMSO-d6) δ 162.9, 159.7, 155.5, 150.3, 148.7, 139.1, 133.3, 131.7, 131.6, 131.3, 130.7, 130.0, 128.9, 128.3, 128.2, 127.1, 127.0, 125.3, 115.5, 115.2, 103.5, 95.6, 12.9; LRMS (ESI) m/z 388 [M + H]+.
3-(4-Chlorophenyl)-5-(3,4-difluorophenyl)-2-methylpyrazolo[1,5-a]pyrimidin-7(4H)-one (P25)
Pale-yellow solid: 1H NMR (300 MHz, DMSO-d6) δ 12.11 (bs, 1H), 7.95 (t, J = 9.6 Hz, 1H), 7.67–7.50 (m, 6H), 6.06 (s, 1H), 2.32 (s, 3H); 13C NMR (75 MHz, DMSO-d6) δ 155.6, 152.6, 152.4, 150.8, 150.7, 149.2, 149.1, 147.6, 147.4, 139.2, 131.5, 131.3, 129.7, 128.5, 125.5, 117.9, 117.8, 117.6, 117.5, 103.2, 95.4, 13.0; LRMS (ESI) m/z 372 [M + H]+.
3-(4-Chlorophenyl)-5-(3,4-difluorophenyl)pyrazolo[1,5-a]pyrimidin-7(4H)-one (P26)
Off-white solid: 1H NMR (300 MHz, DMSO-d6) δ 12.18 (bs, 1H), 8.25 (s, 1H), 8.08–8.01 (m, 1H), 7.75 (bs, 2H), 7.69–7.60 (m, 2H), 7.51 (d, J = 6.9 Hz, 2H), 6.15 (s, 1H); 13C NMR (75 MHz, DMSO-d6) δ 155.9, 152.7, 152.5, 150.9, 150.8, 149.4, 149.2, 147.7, 147.5, 142.1, 138.6, 131.1, 129.6, 129.3, 128.6, 125.6, 117.9, 117.7, 105.0, 95.6; LRMS (ESI) m/z 358 [M + H]+.
5-(3,4-Difluorophenyl)-3-phenyl-2-(trifluoromethyl)pyrazolo[1,5-a]pyrimidin-7(4H)-one (P27)
Pale-yellow solid: 1H NMR (300 MHz, DMSO-d6) δ 7.94–7.87 (m, 1H), 7.63–7.60 (m, 2H), 7.51–7.44 (m, 4H), 6.21 (s, 1H); 13C NMR (75 MHz, DMSO-d6) δ 155.4, 152.8, 150.8, 150.6, 149.4, 149.3, 147.5, 147.3, 141.6, 141.1, 140.6, 130.4, 129.9, 128.5, 128.2, 128.0, 126.0, 119.6, 118.2, 117.9, 117.7, 104.1, 96.3; LRMS (ESI) m/z 392 [M + H]+.
X-ray Crystallography
A crystal of compound 4 of approximate dimensions 0.100 × 0.200 × 0.800 mm was used for X-ray crystallographic analysis at T = 100 K. The X-ray data were collected on a Bruker D8 QUEST system equipped with a multilayer mirror monochromator using a Cu Kα microfocus sealed tube. A total of 3013 frames were collected. The total exposure time was 3.20 h. The frames were integrated with the Bruker SAINT software package using a narrow-frame algorithm. The integration of the data using a monoclinic unit cell yielded a total of 55 109 reflections to a maximum θ angle of 61.24° (0.88 Å resolution), of which 2473 were independent (average redundancy of 22.284, completeness = 99.4%, Rint = 5.22%, Rsig = 1.59%) and 2299 (92.96%) were greater than 2σ(F2). The final cell constants of a = 12.9908(5) Å, b = 10.2770(4) Å, c = 12.2626(5) Å, β = 98.196(2)°, and volume = 1620.41(11) Å3 are based upon the refinement of the XYZ-centroids of 165 reflections above 20σ(I) with 11.33° < 2θ < 118.2°. Data were corrected for absorption effects using the Multi-Scan method (SADABS). The ratio of minimum to maximum apparent transmission was 0.904. The calculated minimum and maximum transmission coefficients (based on crystal size) are 0.5500 and 0.9200.
The structure was solved and refined using the Bruker SHELXTL Software Package, using the space group P121/c1, with Z = 4 for the formula unit, C20H16FN3O2. The final anisotropic full-matrix least-squares refinement on F2 with 238 variables converged at R1 = 3.76% for the observed data and wR2 = 10.14% for all data. The goodness-of-fit was 1.167. The largest peak in the final difference electron density synthesis was 0.545 e–/Å3, and the largest hole was −0.395 e–/Å3 with an RMS deviation of 0.133 e–/Å3. On the basis of the final model, the calculated density was 1.432 g/cm3 and F(000), 728 e–.
Crystallographic data has been deposited in the Cambridge Structural Database under CCDC 2034666. These data can be obtained free of charge from FIZ Karlsruhe via www.ccdc.cam.ac.uk/structures.
Biology
Efficacy and Validating Inhibition of Pyrazolo[1,5-a]pyrimidin-7(4H)-one Series against Mtb In Vitro and Ex Vivo
The antimicrobial susceptibility testing against Mtb H37Rv and the P19R strains was performed in either 7H9/ADC (4.7 g/L 7H9 base, 5 g/L albumin fraction V, 2 g/L glucose, 0.81 g/L NaCl, 0.02% glycerol, and 0.05% Tween 80) or 7H9/GCas (4.7 g/L 7H9 base, 4 g/L glucose, 0.81 g/L NaCl, 0.3 g/L Casitone, and 0.05% Tyloxapol). Bacteria were grown in the corresponding media up to an OD650nm of 0.2–0.4. A 2-fold serial dilution series of the test compounds was placed in each well of a sterile 96-well round-bottom plate, and then, 50 μL of H37Rv diluted to an OD650nm of 0.0002 was added. Plates were incubated at 37 °C for 2 weeks prior to determining the MIC. The MIC is defined here as the drug concentration that completely inhibits the growth of the cells. The MICs under iron-replete or -deplete conditions were done by supplementing the growth medium with either 100 μM of DFO (deplete) or 250 μM FeCl3 (replete).
For the ex vivo efficacy test, J774 cells (2 × 104 cells/well) were seeded in flat-bottom 24 well plates (Corning Inc.) in DMEM GlutaMAX (Gibco/ThermoFisher Scientific) supplemented with 10% fetal bovine serum, 20 mM HEPES + 0.5 mM sodium pyruvate (hereafter abbreviated DMEM/FBS). Cells were infected with Mtb (1 × 105 cells/well, MOI 1:5) for 24 h, followed by medium removal and washing (2×) with an equal volume of Dulbecco’s PBS. Infected cells were fed DMEM/FBS and exposed to test compounds at the specified concentrations in fresh growth medium (1000 μL/well). Cells were incubated at 37 °C and 95% humidity in 5% CO2 incubator for 7 days. The medium was changed every 2 days, and the infected cells were treated with drugs each time. After 7 days of incubation, 0.1% SDS was added in each well to lyse the J774 cells. After 5 min, the lysate was thoroughly mixed and diluted in 7H9/ADC, and the appropriate dilutions were plated in duplicate on 7H11/OADC plates to calculate most probable number of bacteria. Colonies were counted manually in each plate following 6–8 weeks of incubation at 37 °C.
Cytotoxicity Testing against Mammalian Cells
The in vitro cytotoxicity of the pyrazolopyrimidinones were measured in DMEM supplemented with 10% FBS using either glucose (5 mM) or galactose (10 mM) as the carbon source. Briefly, 1 × 104 cells (HepG2 or J774) were seeded onto each well of a sterile 96-well tissue culture treated flask and left to attach overnight. The next day, media was aspirated and replaced with a 2-fold serial dilution of compounds in the same media. Cells were treated for 24 h, prior to viability determination using CellTiter-Glo (Promega) according to the manufacturer’s instructions.
Iron Uptake Studies
To determine the role of the test compounds in the iron uptake of Mtb, logarithmically growing cells were labeled with 55Fe metabolically by supplementing the growth medium with 0.75 μM 55FeCl3. The cells were then treated with either DFO (39 μM) or 1 (10 μM) for 24 h prior to collecting the cells and washing exhaustively in PBS. The cells were pelleted and resuspended in 200 μL of scintillation fluid, and counts per minute (CPM) was counted by a scintillation counter (Beckman Coulter LS6500).
Generation and Characterization of Pyrazolo[1,5-a]pyrimidin-7(4H)-one Resistant Mutants
To generate mutants against the pyrazolo[1,5-a]pyrimidin-7(4H)-one series, 107, 108, and 109 cells of Mtb H37Rv were plated on 7H11/OADC plates containing with 5× and 10× MIC of the drugs, with drug-free plates used to enumerate bacterial load. The plates were incubated at 37 °C for 4–6 weeks. After 4 weeks, colonies on both the 5× and 10× MIC of P19 were picked and inoculated in 7H9/ADC/Tween medium and the MIC was tested to confirm their resistance against P19. Genomic DNA of mutants was isolated by the cetyl trimethyl ammonium bromide method. Whole genome sequencing was performed and analyzed as described.25,32
Determination of Drug Metabolism in Live Bacteria
To determine the degree of drug metabolism in live Mtb and P19R strains, the LC/MS-based protocol following compound exposure was employed. Cells were grown to an OD of 1.0, washed twice with PBS, and resuspended using PBS at 1/3 the original culture volume (final OD ∼ 3.0). Then, 1 mL of the cell suspension was added to enough compound in DMSO to create a final compound concentration of 10× MIC. A 200 μL aliquot was removed for the 0 h time point and mixed with an equal volume of acetonitrile and kept at 4 °C. After 24 and 48 h, another 200 μL aliquot was removed and mixed with acetonitrile. The cells in these aliquots were pelleted, and the supernatant was filtered through a 0.2 μm centrifuge filter prior to injecting directly into the LC/MS. The LC gradient used to separate the metabolites was 5–95% acetonitrile in H2O over 20 min. Extracted ion chromatograms were used to determine the % degradation, which was calculated by comparing 0, 24, and 48 h time points. The structural evaluation of the P19 catabolism into the metabolites was performed on an Agilent 1290 Infinity HPLC with a 6460C triple quadrupole mass selective detector in product ion mode. Separation was achieved with an Agilent EclipsePlus C18 column (2.1 × 50 mm, 1.8 μm) with a linear gradient of 95% aq. 0.1% formic acid to 95% acetonitrile (with 0.1% formic acid) over 5 min with a 0.8 mL/min flow rate. Energy resolved mass spectrometry profiles were created in product ion mode with collision energy ranging from 0 to 80 V (x-axis is the center-of-mass calculation with nitrogen gas) with electro-spray ionization in positive mode. For high resolution mass spectrometry (HRMS) by Waters Xevo-G XS QTof, ions were produced using positive ion electrospray at a capillary voltage of 2.8 kV. The ESI source temperature was 280 °C. The LC system was a Waters Acquity Class I UPLC. Samples were stream injected using the loop method into the mobile phase consisting of MeOH/acetonitrile = 1/1 with 0.2% formic acid and 0.1% trifluoroacetic acid added. The flow rate was 0.2 mL/min. The MS was operated at a resolution of at least 25 000, and accurate mass assignments were determined using the internal standard method.
Acknowledgments
This work was funded in part by the Intramural Research Program of NIAID (AI000693-25) and by grants from the Foundation for the National Institutes of Health (BARRY11HTB0) with support from the Bill & Melinda Gates Foundation (OPP1024021).
Glossary
Abbreviations
- MIC
minimum inhibitory concentration
- AcOH
acetic acid
- DIPEA
N,N-diisopropylethylamine
- DMSO
dimethyl sulfoxide
- Me
methyl
- Ph
phenyl
- Bn
benzyl
- isoPr
isopropyl
- INH
isoniazid
- MOX
moxifloxacin
- ETH
ethionamide
- NMR
nuclear magnetic resonance
- LRMS
low resolution mass spectrometry
- HRMS
high resolution mass spectrometry
- LC/MS
liquid chromatography/mass spectrometry
- ESI
electrospray ionization
- BSA
bovine serum albumin
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsinfecdis.0c00851.
Author Present Address
○ M.D.J.L.: Infectious Disease & Vaccines, Merck Research Laboratories, Merck & Co., West Point, Pennsylvania 19486, United States.
The authors declare no competing financial interest.
Supplementary Material
References
- Barry C. E. 3rd; Boshoff H. I. M.; Dartois V.; Dick T.; Ehrt S.; Flynn J.; Schnappinger D.; Wilkinson R. J.; Young D. (2009) The spectrum of latent tuberculosis: rethinking the biology and intervention strategies. Nat. Rev. Microbiol. 7, 845–855. 10.1038/nrmicro2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dheda K.; Barry C. E. 3rd; Maartens G. (2016) Tuberculosis. Lancet 387, 1211–26. 10.1016/S0140-6736(15)00151-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (2019) Global tuberculosis report 2019, WHO, Geneva. [Google Scholar]
- Dheda K.; Gumbo T.; Maartens G.; Dooley K. E.; Murray M.; Furin J.; Nardell E. A.; Warren R. M.; (2019) The Lancet Respiratory Medicine Commission: 2019 update: epidemiology, pathogenesis, transmission, diagnosis, and management of multidrug-resistant and incurable tuberculosis. Lancet Respir. Med. 7, 820–826. 10.1016/S2213-2600(19)30263-2. [DOI] [PubMed] [Google Scholar]
- Lechartier B.; Rybniker J.; Zumla A.; Cole S. T. (2014) Tuberculosis drug discovery in the post-post-genomic era. EMBO Mol. Med. 6, 158–168. 10.1002/emmm.201201772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioerger T. R.; O’Malley T.; Liao R.; Guinn K. M.; Hickey M. J.; Mohaideen N.; Murphy K. C.; Boshoff H. I.; Mizrahi V.; Rubin E. J.; et al. (2013) Identification of new drug targets and resistance mechanisms in Mycobacterium tuberculosis. PLoS One 8, e75245 10.1371/journal.pone.0075245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao J.; Eoh H.; He R.; Wang Y.; Wan B.; Franzblau S. G.; Crick D. C.; Kozikowski A. P. (2008) Structure-activity relationships of compounds targeting Mycobacterium tuberculosis 1-deoxy-D-xylulose 5-phosphate synthase. Bioorg. Med. Chem. Lett. 18, 5320–5323. 10.1016/j.bmcl.2008.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragset M. S.; Poce G.; Alfonso S.; Padilla-Benavides T.; Ioerger T. R.; Kaneko T.; Sacchettini J. C.; Biava M.; Parish T.; Argüello J. M.; Steigedal M.; Rubin E. J. (2015) A novel antimycobacterial compound acts as an intracellular iron chelator. Antimicrob. Agents Chemother. 59, 2256–2264. 10.1128/AAC.05114-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masini T.; Lacy B.; Monjas L.; Hawksley D.; de Voogd A.; Illarionov B.; Iqbal A.; Leeper F.; Fischer M.; Kontoyianni M.; Hirsch A. K. H. (2015) Validation of a homology model of Mycobacterium tuberculosis DXS: rationalization of observed activities of thiamine derivatives as potent inhibitors of two orthologues of DXS. Org. Biomol. Chem. 13, 11263–11277. 10.1039/C5OB01666E. [DOI] [PubMed] [Google Scholar]
- Allen F. H.; Kennard O.; Watson D. G.; Brammer L.; Orpen A. G.; Taylor R. (1987) Tables of Bond Lengths Determined by X-Ray and Neutron-Diffraction. Part 1. Bond Lengths in Organic-Compounds. J. Chem. Soc., Perkin Trans. 2 S1–S19. 10.1039/p298700000s1. [DOI] [Google Scholar]
- Robins R. K.; Revankar G. R.; Obrien D. E.; Springer R. H.; Novinson T.; Albert A.; Senga K.; Miller J. P.; Streeter D. G. (1985) Purine Analog Inhibitors of Xanthine-Oxidase - Structure Activity Relationships and Proposed Binding of the Molybdenum Cofactor. J. Heterocycl. Chem. 22, 601–634. 10.1002/jhet.5570220303. [DOI] [Google Scholar]
- Wilson N. S.; Osuma A. T.; Van Camp J. A.; Xu X. D. (2012) A scalable approach to diaminopyrazoles using flow chemistry. Tetrahedron Lett. 53, 4498–4501. 10.1016/j.tetlet.2012.05.152. [DOI] [Google Scholar]
- Puente Á.; Ofial A. R.; Mayr H. (2017) Nucleophilic Reactivities of Bis-Acceptor-Substituted Benzyl Anions. Eur. J. Org. Chem. 2017, 1196–1202. 10.1002/ejoc.201601513. [DOI] [Google Scholar]
- Hamdi N.; Fischmeister C.; Dixneuf P. H.; Nievas A. R. (2006) Regioselective synthesis of a new [1,2,3]-triazoles directly from imidates. J. Heterocyclic Chem. 43, 499–501. 10.1002/jhet.5570430238. [DOI] [Google Scholar]
- Huang Y.; Lei Y. Y.; Zhao L.; Gu J. W.; Yao Q. L.; Wang Z.; Li X. F.; Zhang X. G.; He C. Y. (2018) Catalyst-free and visible light promoted trifluoromethylation and perfluoroalkylation of uracils and cytosines. Chem. Commun. 54, 13662–13665. 10.1039/C8CC07759B. [DOI] [PubMed] [Google Scholar]
- Cole S. T. (2016) Inhibiting Mycobacterium tuberculosis within and without. Philos. Trans. R. Soc., B 371, 20150506. 10.1098/rstb.2015.0506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naran K.; Moosa A.; Barry C. E. 3rd; Boshoff H. I.; Mizrahi V.; Warner D. F. (2016) Bioluminescent Reporters for Rapid Mechanism of Action Assessment in Tuberculosis Drug Discovery. Antimicrob. Agents Chemother. 60, 6748–6757. 10.1128/AAC.01178-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moosa A.; Lamprecht D. A.; Arora K.; Barry C. E. 3rd; Boshoff H. I. M.; Ioerger T. R.; Steyn A. J. C.; Mizrahi V.; Warner D. F. (2017) Susceptibility of Mycobacterium tuberculosis Cytochrome bd Oxidase Mutants to Compounds Targeting the Terminal Respiratory Oxidase, Cytochrome c. Antimicrob. Agents Chemother. 61, e01338-17 10.1128/AAC.01338-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora K.; Ochoa-Montaño B.; Tsang P. S.; Blundell T. L.; Dawes S. S.; Mizrahi V.; Bayliss T.; Mackenzie C. J.; Cleghorn L. A.; Ray P. C.; et al. (2014) Respiratory flexibility in response to inhibition of cytochrome C oxidase in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 58, 6962–6965. 10.1128/AAC.03486-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codd R.; Richardson-Sanchez T.; Telfer T. J.; Gotsbacher M. P. (2018) Advances in the Chemical Biology of Desferrioxamine B. ACS Chem. Biol. 13, 11–25. 10.1021/acschembio.7b00851. [DOI] [PubMed] [Google Scholar]
- Lah M. S.; Palfey B. A.; Schreuder H. A.; Ludwig M. L. (1994) Crystal structures of mutant Pseudomonas aeruginosa p-hydroxybenzoate hydroxylases: the Tyr201Phe, Tyr385Phe, and Asn300Asp variants. Biochemistry 33, 1555–64. 10.1021/bi00172a036. [DOI] [PubMed] [Google Scholar]
- Eppink M. H.; Schreuder H. A.; van Berkel W. J. (1998) Lys42 and Ser42 variants of p-hydroxybenzoate hydroxylase from Pseudomonas fluorescens reveal that Arg42 is essential for NADPH binding. Eur. J. Biochem. 253, 194–201. 10.1046/j.1432-1327.1998.2530194.x. [DOI] [PubMed] [Google Scholar]
- Beam M. P.; Bosserman M. A.; Noinaj N.; Wehenkel M.; Rohr J. (2009) Crystal structure of Baeyer-Villiger monooxygenase MtmOIV, the key enzyme of the mithramycin biosynthetic pathway. Biochemistry 48, 4476–87. 10.1021/bi8023509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koteva K.; Cox G.; Kelso J. K.; Surette M. D.; Zubyk H. L.; Ejim L.; Stogios P.; Savchenko A.; Sorensen D.; Wright G. D. (2018) Rox, a Rifamycin Resistance Enzyme with an Unprecedented Mechanism of Action. Cell Chem. Biol. 25, 403–412. 10.1016/j.chembiol.2018.01.009. [DOI] [PubMed] [Google Scholar]
- Park Y.; Pacitto A.; Bayliss T.; Cleghorn L. A.; Wang Z.; Hartman T.; Arora K.; Ioerger T. R.; Sacchettini J.; Rizzi M.; Donini S.; Blundell T. L.; Ascher D. B.; Rhee K.; Breda A.; Zhou N.; Dartois V.; Jonnala S. R.; Via L. E.; Mizrahi V.; Epemolu O.; Stojanovski L.; Simeons F.; Osuna-Cabello M.; Ellis L.; MacKenzie C. J.; Smith A. R.; Davis S. H.; Murugesan D.; Buchanan K. I.; Turner P. A.; Huggett M.; Zuccotto F.; Rebollo-Lopez M. J.; Lafuente-Monasterio M. J.; Sanz O.; Diaz G. S.; Lelievre J.; Ballell L.; Selenski C.; Axtman M.; Ghidelli-Disse S.; Pflaumer H.; Bosche M.; Drewes G.; Freiberg G. M.; Kurnick M. D.; Srikumaran M.; Kempf D. J.; Green S. R.; Ray P. C.; Read K.; Wyatt P.; Barry C. E. 3rd; Boshoff H. I. (2017) Essential but Not Vulnerable: Indazole Sulfonamides Targeting Inosine Monophosphate Dehydrogenase as Potential Leads against Mycobacterium tuberculosis. ACS Infect. Dis. 3, 18–33. 10.1021/acsinfecdis.6b00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P.; Capodagli G. C.; Awasthi D.; Shrestha R.; Maharaja K.; Sukheja P.; Li S. G.; Inoyama D.; Zimmerman M.; Ho Liang H. P.; Sarathy J.; Mina M.; Rasic G.; Russo R.; Perryman A. L.; Richmann T.; Gupta A.; Singleton E.; Verma S.; Husain S.; Soteropoulos P.; Wang Z.; Morris R.; Porter G.; Agnihotri G.; Salgame P.; Ekins S.; Rhee K. Y.; Connell N.; Dartois V.; Neiditch M. B.; Freundlich J. S.; Alland D. (2018) Synergistic Lethality of a Binary Inhibitor of Mycobacterium tuberculosis KasA. mBio 9, e02101-17 10.1128/mBio.02101-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senga K.; Novinson T.; Wilson H. R.; Robins R. K. (1981) Synthesis and antischistosomal activity of certain pyrazolo[1,5-a]pyrimidines. J. Med. Chem. 24, 610–613. 10.1021/jm00137a023. [DOI] [PubMed] [Google Scholar]
- Qi J.; Zhang F.; Mi Y.; Fu Y.; Xu W.; Zhang D.; Wu Y.; Du X.; Jia Q.; Wang K.; et al. (2011) Design, synthesis and biological activity of pyrazolo [1,5-a] pyrimidin-7(4H)-ones as novel Kv7/KCNQ potassium channel activators. Eur. J. Med. Chem. 46, 934–943. 10.1016/j.ejmech.2011.01.010. [DOI] [PubMed] [Google Scholar]
- Hwang J. Y.; Windisch M. P.; Jo S.; Kim K.; Kong S.; Kim H. C.; Kim S.; Kim H.; Lee M. E.; Kim Y.; Choi J.; Park D. S.; Park E.; Kwon J.; Nam J.; Ahn S.; Cechetto J.; Kim J.; Liuzzi M.; No Z.; Lee J. (2012) Discovery and characterization of a novel 7-aminopyrazolo[1,5-a]pyrimidine analog as a potent hepatitis C virus inhibitor. Bioorg. Med. Chem. Lett. 22, 7297–301. 10.1016/j.bmcl.2012.10.123. [DOI] [PubMed] [Google Scholar]
- Konteatis Z. D., Sui Z., Travins J. M., and Ye Z. (2019) Inhibitors of cellular metabolic processes. US 20180079753 A1.
- Taylor M. S.; Jacobsen E. N. (2003) Enantioselective Michael additions to alpha, beta-unsaturated imides catalyzed by a Salen-Al complex. J. Am. Chem. Soc. 125, 11204–11205. 10.1021/ja037177o. [DOI] [PubMed] [Google Scholar]
- Oh S.; Park Y.; Engelhart C. A.; Wallach J. B.; Schnappinger D.; Arora K.; Manikkam M.; Gac B.; Wang H.; Murgolo N.; Olsen D. B.; Goodwin M.; Sutphin M.; Weiner D. M.; Via L. E.; Boshoff H. I. M.; Barry C. E. 3rd (2018) Discovery and Structure-Activity-Relationship Study of N-Alkyl-5-hydroxypyrimidinone Carboxamides as Novel Antitubercular Agents Targeting Decaprenylphosphoryl-β-D-ribose 2′-Oxidase. J. Med. Chem. 61, 9952–9965. 10.1021/acs.jmedchem.8b00883. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.