Abstract
Cell walls are barriers found in almost all known bacterial cells. These structures establish a controlled interface between the external environment and vital cellular components. A primary component of cell wall is a highly crosslinked matrix called peptidoglycan (PG). PG crosslinking, carried out by transglycosylases and transpeptidases, is necessary for proper cell wall assembly. Transpeptidases, targets of β-lactam antibiotics, stitch together two neighboring PG stem peptides (acyl-donor and acyl-acceptor strands). We recently described a novel class of cellular PG probes that were processed exclusively as acyl-donor strands. Herein, we have accessed the other half of the transpeptidase reaction by developing probes that are processed exclusively as acyl-acceptor strands. The critical nature of the crossbridge on the PG peptide was demonstrated in live bacterial cells and surprising promiscuity in crossbridge primary sequence was found in various bacterial species. Additionally, acyl-acceptor probes provided insight into how chemical remodeling of the PG crossbridge (e.g., amidation) can modulate crosslinking levels, thus establishing a physiological role of PG structural variations. Together, the acyl-donor and -acceptor probes will provide a versatile platform to interrogate PG crosslinking in physiologically relevant settings.
Graphical Abstract
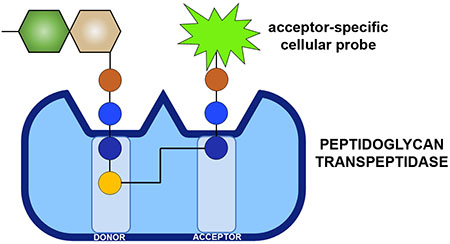
Gram-positive pathogens continue to impose a significant global public health burden.1 Among these organisms, Enterococci are one of the leading causes of nosocomial infections.2 With the rise in antibiotic drug resistance, it has become clear that more bacterial targets are needed to improve the therapeutic options to fight Enterococci infections. Peptidoglycan (PG), an essential component of bacterial cell walls, is one of the most important targets of antibiotics. Disaccharides of N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid (MurNAc) make up the basic PG unit, which is further decorated with a pentameric stem peptide (e.g., l-Ala-d-isoGlx-l-Lys-d-Ala-d-Ala).3–4 PG units are polymerized and crosslinked via transglycosylation and transpeptidation (TP) reactions that assemble the sugars and peptides, respectively. PG crosslinking is essential, and, therefore, molecules that block this reaction are traditionally valuable antibiotics (e.g., β-lactams and vancomycin) and continue to result in promising drug leads (e.g., teixobactin).5–7
Intense efforts have led to a greater understanding of the chemical makeup, mechanism of assembly, and essential proteins that construct the PG network. Moreover, there is some evidence that small structural modifications within the PG matrix can alter the overall plasticity and physical properties of the cell wall (e.g., resistance to lysozyme).8–10 Chemical composition of the stem peptide primary sequence can vary by the amidation/methylation of carboxylic groups, removal of amino acids, variation of amino acid character, and extensive alteration to the central lysine group. Yet, for the most part, their physiological impact remains elusive.11–14
The 3rd position with the PG stem peptide has defining structural characteristics. As an example, the difference of a carboxylic acid group on l-Lys verus meso-2,6-diaminopimelic acid (m-DAP) has important implications in the activation of the human innate immune system.15 In Enterococcus faecium
(E. faecium), the lysine sidechain is modified with d-iAsx while Enterococcus faecalis (E. faecalis) displays l-Ala-l-Ala. In addition, the nature of the crossbridge (the amino acids anchored onto the lysine residues) is critical based on its role in the nucleophilic step during crosslinking reactions. There are two main classes of PG crosslinking enzymes: d,d-TPs and l,d-TPs (Figure 1). Crosslinking by Penicillin Binding Protein (PBP) d,d-TPs generate 4-3 peptide crosslinks and l,d-TPs (Ldts) create 3-3 crosslinks within PG.
Figure 1.
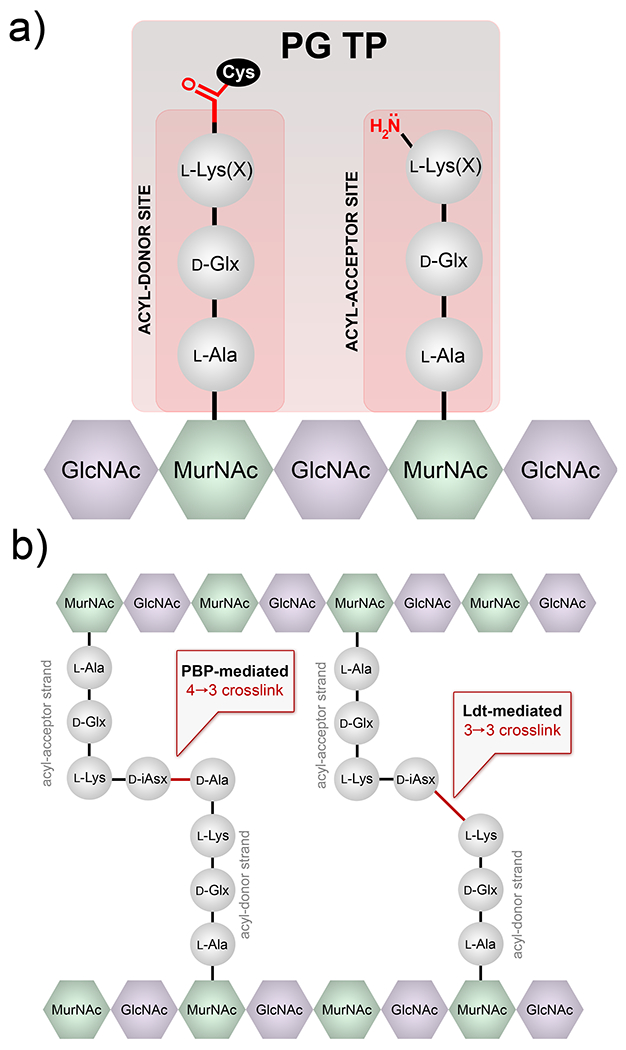
Crosslinking reaction by TPs. (a) Example of two binding sites (acyl-donor and -acceptor) at the intermediate step of the crosslinking reaction are depicted. The acyl-chain is covalently anchored on the TP, which is subsequently transferred to the amino group on the acyl-acceptor strand. X represents the crossbridging amino acids. (b) Schematic of PG crosslinking in E. faecium showing the 2 types of crosslinks observed.
Fundamental understanding of PG crosslinking is key to define the processes that control PG structure and may provide novel drug targets. Single d-amino acid PG probes have spurred valuable insight into PG crosslinking in live bacterial cells.16–21 Furthermore, elegant prior in vitro studies using synthetic PG analogs have provided insight into the substrate structural requirements of TPs.12, 22–25 However, fewer reports have described probes that decipher TP substrate requirements in live bacterial cells. A recent example of a synthetic l-Lys-d-Ala-d-Ala stem peptide mimic led to the demonstration of extraseptal TP activity in Staphylococcus aureus (S. aureus).26 Likewise, we recently reported the use of synthetic PG probes to establish crosslinking parameters based on the acyl-donor strand of Ldts.27 We have now built on those findings by developing complementary probes that selectively track and dissect acyl-acceptor strand processing by TPs in live bacterial cells.
We initially hypothesized that structural analogs of the two TP substrates could be developed to control active site loading.
In designing our previous acyl-donor probe 1, we mimicked the stem peptide fragment as a tetrapeptide and blocked the nucleophilic site on the acyl-acceptor strand (Figure 2a). In this work, the acyl-acceptor specific probe 2 was designed by installing the acyl-acceptor fragment (crossbridging d-iAsx in the case of E. faecium) and removing the terminal fragment recognized by the acyl-donor site (terminal d-Ala and/or D-Ala-D-Ala). Therefore, PG probes with the basic scaffold of tripeptide 2 should act solely as an acyl-acceptor strand. To track incorporation into the PG, a fluorescent handle was added to the N-terminus of the stem tripeptide analogs. Incubation of live bacterial cells with tripeptide probes is expected to result in their covalent incorporation into the PG scaffold by TPs (Figure 2b). Crosslinking can then be readily quantified and analyzed using standard fluorescence-based techniques, thus providing a mode to interrogate how primary sequences of the acyl-acceptor strand modulate PG crosslinking (Figure 2c).
Figure 2.
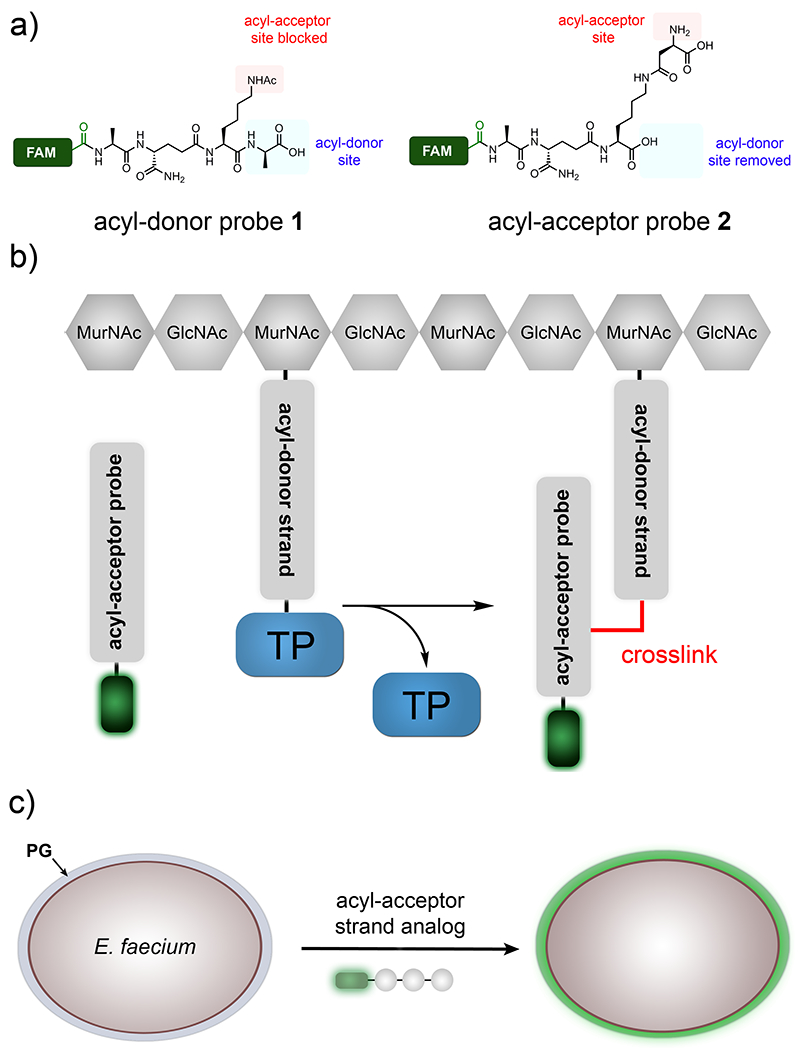
Design of tripeptide probes and the assay to assess acyl-acceptor strand in live cells. (a) Structure-based design of acyl-donor and -acceptor probes. (b) Schematic diagram showing how the fluorescently tagged tripeptide probe gets covalently crosslinked into the bacterial PG scaffold. (c) Schematic of the whole cell analysis that can be accessed by the tripeptide probes.
At first, we synthesized a panel of tripeptide probes that were designed to investigate acyl-acceptor strand recognition in live E. faecium (Figure 3). d-iAsx is the canonical crossbridge amino acid in E. faecium, where a nucleophilic attack by the sidechain N-terminus nitrogen leads to formation of PG crosslinks. All probes were synthesized using standard solid phase peptide chemistry and included a carboxy-fluorescein tag (FAM). E. faecium cells were treated with individual probes and cellular fluorescence levels were measured after overnight incubation. The first two stem peptide mimics, TriQK and TriQD, were designed to test the role of crossbridging amino acids is the acyl-capture step of TP. Our results showed that when cells were treated with TriQK, which lacks the crossbridging amino acid, fluorescence levels were near background (Figure 3a). Introduction of the crossbridge d-iAsp (TriQD) led to ~7- fold fluorescence increase over background. These results provide in vivo evidence for the importance of the crossbridge for robust crosslinking of the PG scaffold and suggest that the enzyme responsible for d-iAsp installation, Aslfm,28 is a potential narrow-spectrum drug target. Given that only approximately 60% of the lysines are modified with d-iAsx in E. faecium29, and that we have evidence that unmodified lysine residues do not participate in TP reactions as acyl acceptors, these results also suggest a mode of controlling PG crosslinking based on Aslfm processing of PG precursors in the cytosol of cells prior to exposure to TPs.
Figure 3.
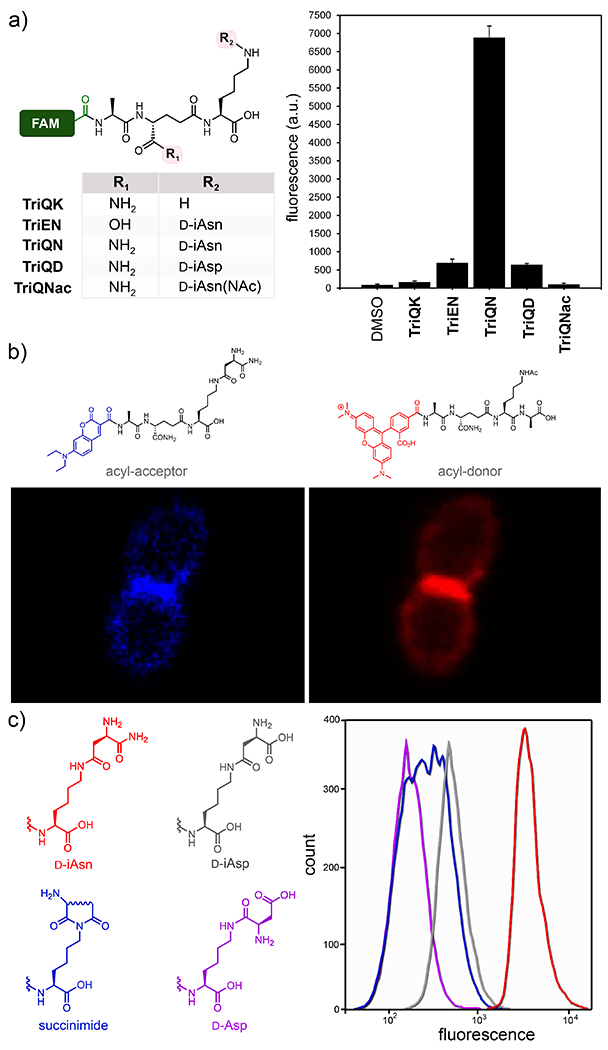
Crosslinking of tripeptide probes in live E. faecium cells. (a) Chemical series tripeptide probes based on the modifications at the d-iGlx and lysine sidechain; FAM = carboxyfluorescein. Flow cytometry analysis of E. faecium (D344s) treated overnight with 100 ¼M of tri-peptide probes. Data are represented as mean + SD (n = 3). (b) Confocal microscopy of E. faecium cells labeled with the designated probes (100 μM) for 5 min. Scale bar = 2 μm. (c) Labeling of cells similar to (a) with a focused library of re-arranged crossbriges.
We then analyzed the role of amidation in the 2nd position d-iGlu in the crosslinking step. Recent genetic analyses revealed the essential nature of the genes encoding the enzymes that carry out the amidation of d-iGlu to d-iGln in S. aureus and Streptococcus pneumoniae.30–31 Subsequent in vitro characterization suggested that lack of d-iGlu amidation impairs PG crosslinking.11, 23 Yet, it remained unresolved whether d-iGlu amidation impacts processing at the acyl-donor, acyl-acceptor, or both sites. We recently demonstrated that amidation of d-iGlu is critical for the acyl-donor strand for both classes of TPs.27 To test these concepts in the acyl-acceptor substrate, E. faecium cells were treated with TriEN. In sharp contrast to the d-iGln variant (TriQN), cellular fluorescence levels were significantly lower in cells treated with the d-iGlu variant (TriEN).
We next evaluated how chemical remodeling of the acyl-acceptor strand may influence TP crosslinking in E. faecium. While the sidechain of the 3rd position lysine is initially loaded with a d-iAsp crossbridge, its carboxylic acid can be biochemically amidated to generate d-iAsn crossbridges to varying levels in E. faecium.29 In fact, for similar organisms like Lactococcus lactis (L. lactis) it has been estimated that ~75% of crossbridges d-iAsp are amidated to d-iAsn.32 Yet, the physiological and TP-processing consequences of d-iAsp amidation remain unresolved. We hypothesized that we could establish the role of amidation using synthetic analogs in live bacteria. Quite strikingly, treatment of E. faecium with TriQN resulted in considerably higher crosslinking levels relative to TriQD (Figure 3a). These results suggest that amidation of d-iAsp within the acyl-acceptor strand play a critical role in controlling PG crosslinking levels. Moreover, we demonstrated that the crossbridge of TriQN is, in fact, the acyl-acceptor site by building a control probe (TriQNAc) in which the nucleophilic amino group is acetylated to block acyl-capture. Fluorescence levels for cells treated with TriQNAc were near background, a result that is consistent with the site of acyl-capture.
A different strain of E. faecium showed a similar labeling profile as the parental drug-sensitive strain (Figure S1), which suggests that a different strain within type of bacteria has similar acyl-acceptor substrate preferences. Moreover, cell labeling was not entirely disrupted in an isogenic strain lacking Ldt TP gene (ldtfm), which suggests that the tripeptide probe is likely incorporated into the PG scaffold via l,d-TPs and d,d-TPs (Figure S1). Confocal microscopy was then used to delineate the incorporation of the probes within the PG scaffold of live cells. For these experiments, a short pulse labeling step was performed with the acyl-acceptor probe modified with a blue fluorophore and the acyl-donor probe modified with a red fluorophore (Figure 3b. From our results, it is clear that there is some overlap between PG incorporation of these probes and we established that the septal region is the primary site of PG incorporation.
Having shown that amidation of the crossbridge (d-iAsp to d-iAsn) had significant consequences in crosslinking levels in E. faecium, we tested the commonality these observations across other species with similarly structured crossbridges.32 Consistently, labeling levels in L. lactis and Lactococcus casei were considerably lower with TriQD relative to TriQN (Figure S2). Interestingly, a recent transposon mutagenesis screen revealed a single gene responsible for the immune-augmenting activity of L. casei: asnH whose protein product amidates d-iAsp to D-iAsn.33 It was observed that the mutant strain not expressing AsnH lacked thick peptidoglycan in electron microscopy analysis, a finding that is consistent with d-iAsp amidation having an significant effect in PG crosslinking. Together, our data point to a three-level biochemical control (addition of crossbridge, d-iGlu to d-iGln, and d-iAsp to D-iAsn) of the acyl-acceptor substrate structure that likely combine to tune crosslinking level in E. faecium.
A possible consequence of d-iAsp amidation is spontaneous re-arrangement of isoasparagine. These types of re-arrangements are well documented in mammalian proteins and also play important roles in intein splicing.34–37 Isoasparagine re-arrangement is initiated by the internal attack of the amide nitrogen onto the carboxamide of isoasparagine yielding a cyclic succinimide intermediate, which can be hydrolyzed into either aspartic acid or isoaspartic acid (Figure S3). A solid-state NMR analysis of E. faecium PG revealed fingerprints consistent with all such re-arrangement products.29 We wondered if spontaneous d-iAsn re-arrangement products could still serve as competent crossbridges for PG crosslinking, and, therefore, built a secondary library that mapped out three of the possible products (Figure 3c and Figure S4. E. faecium labeling results showed that all re-arranged products were considerablypoorer TP substrates. It is intriguing to speculate about the biological impact in the nonenzymatic deamidation of d-iAsn considering its potential deleterious effect in PG crosslinking. Perhaps, it could serve as a molecular timer in a similar context as what has been described to deamidation of asparagine in human proteins. A possibility may be that d-iAsn re-arrangement could provide a competitive advantage by reducing susceptibility against bacteriophage endolysins that specifically cleave d-iAsn but not d-Asp crosslinks.38
To extend our results to another type of Enterococci, acyl-acceptors probes were adopted for E. faecalis. In the case of E. faecalis, ligases BppA1 and BppA2 are responsible for transferring l-Ala from Ala-tRNA to the first and 2nd positions of the ε-amino group of lysine of PG precursors, respectively.39 We synthesized TriQAA, which mimics the crossbridge in E. faecalis, to initially test acyl-acceptor processing (Figure 4a). Incubation of E. faecalis cells with TriQAA led to a ~10.5 fold increase over the unmodified lysine side chain TriQK, thus again demonstrating the necessity for a crossbridging unit for proper acyl capture strand in Enterococci PG crosslinking. Inverting the stereochemistry of the crossbridge in TriQAA to TriQaa also led to baseline fluorescence, indicating that TPs are stereospecific at the acyl-acceptor position. Acetylation of the E. faecalis probe, TriQAAac, which blocks acyl-acceptor nucleophilic site, led to baseline fluorescence levels. This finding is consistent with the amino group of the terminal l-alanine as the nucleophile for covalent crosslinking into the PG scaffold. Surprisingly, lack of amidation at the second position (TriEAA) only resulted in a ~1.3-fold decrease in fluorescence, showing that amidation may not be an absolute requirement for the acyl-acceptor recognition in E. faecalis.
Figure 4.
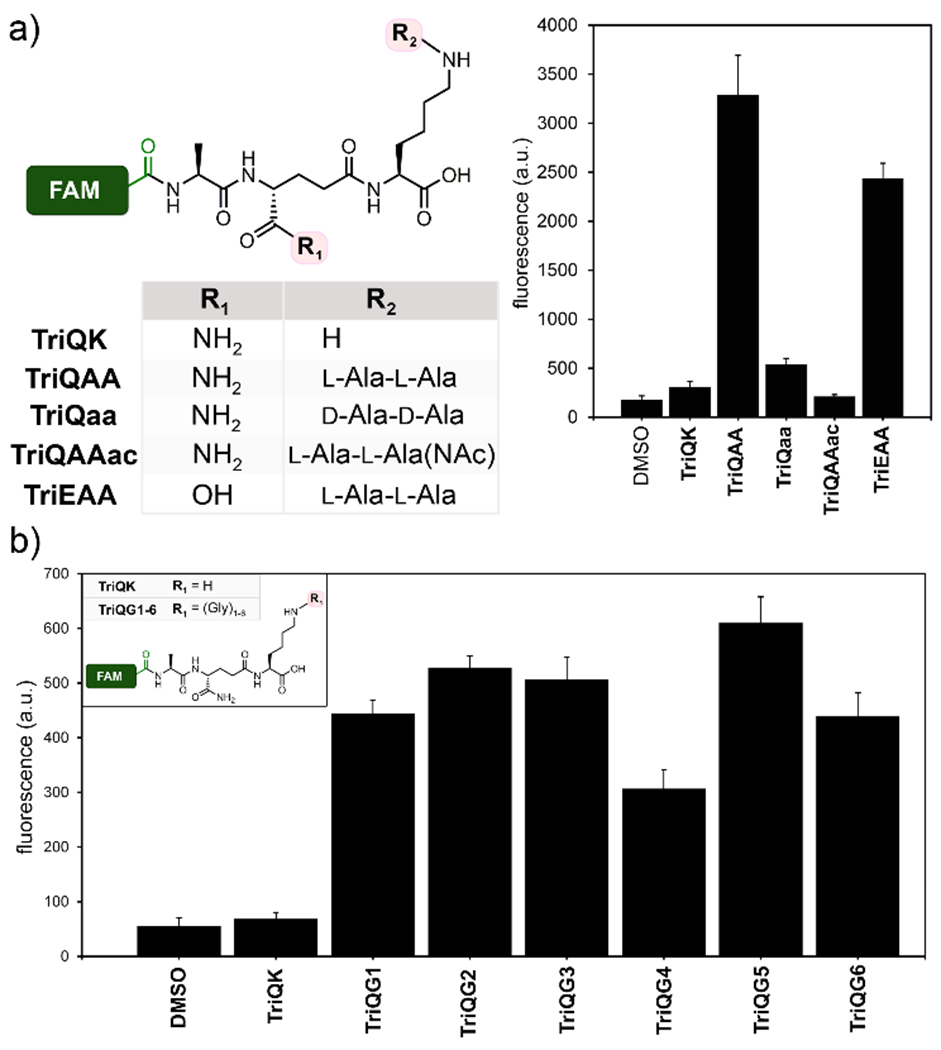
Crosslinking of tripeptide probes in live E. faecalis and S. aureus cells. Chemical series tripeptide probes based corresponding to the stem peptide of (a) E. faecalis and (b) S. aureus. Flow cytometry analysis of cells treated overnight with 100 ¼M of tri-peptide probes. Data are represented as mean + SD (n = 3).
We next turned our attention to the problematic human pathogen S. aureus. In S. aureus, peptidyl transferases FemX, FemA, and FemB catalyze the stepwise addition of five glycines to the lysine residue on the 3rd position of the PG precursor.40–42 Recently, Walker and co-workers isolated PG precursors corresponding to one, three, and five glycine units and showed in vitro that PBP transpeptidases have varying levels of preference for the length of the glycine chain.12 We built a library of tripeptide S. aureus probes that complement their approach by assessing the acyl-acceptor preference in live bacterial cells. We synthesized TriQG1-6 (Figure 4b), corresponding to glycine chains from 1 to 6 units, and measured their incorporation into the PG scaffold of S. aureus cells.
Consistent with our previous results, the absence of glycine modification (TriQK) resulted in background levels of PG incorporation. Interestingly, addition of a single glycine (TriQG1) led to ~7-fold increase in cellular fluorescence levels.
These results are consistent with a previously reported viable S. aureus femAB-null strain in which one glycine crossbridges were observed, albeit with severe crosslinking defects and impaired growth.42–43 Our results clearly demonstrate the physiological consequence of a lack of a single glycine unit and point to FemX is an particularly attractive target for therapeutic development, something that had been suggested but had not been observed in live bacterial cells due to the lethality of femX-deletion.44 Crossbridges of 2 to 5 glycine resulted in comparable labeling levels. Extending beyond the natural pentaglycine chain (TriQG6) did not significantly impact crosslinking, which suggests that there is a high level of flexibility by the acyl-acceptor strand in S. aureus.
Having individually established the structural preference of the acyl-acceptor strand for various bacteria, we next set out to examine how crossbridges are tolerated by TPs across different species. Because TPs are frequent targets of β-lactams and other potent antibiotics, these enzymes play central roles in the drug resistance among several classes of medically relevant bacteria.45 Moreover, mobile elements carrying TP genetic information can be a powerful modality for the acquisition of drug resistant phenotypes. The mecA gene underpinning the drug resistant phenotype of Methicillin-resistant Staphylococcus aureus (MRSA) has been proposed to originate from Staphylococcus sciuri (S. sciuri).46 Therefore, the ability of TPs to process diverse PG architectures may have implications on the potential of TP to cross polinate drug resistance phenotypes. As an example, it was recently shown that commensal streptococci can serve as a reservoir for PBPs that can cross pollinate drug resistant phenotypes.47
To investigate the tolerance of non-native crossbridges in PG crosslinking reactions, the three canonical crossbridges (TriQN, TriQAA, and TriQG5) were incubated with E. faecium, E. faecalis, and S. aureus. S. aureus showed some promiscuity in accepting both TriQAA and TriQN (Figure 5 and Figure S5 for raw values) while still demonstrating some preference for pentaglycine crossbridge. Conversely, E. faecium showed strong preference for its natural acyl-acceptor strand TriQN. Treatment of E. faecium cells with TriQAA or TriQG5 led to background fluorescence levels. Strikingly, E. faecalis showed little preference in the acyl-acceptor side chain and labeled well with all three tripeptide probes. . These findings are consistent with previous in vitro studies that illustrated the promiscuity of E. faecalis TPs in accepting non-native crossbridges.48–49 Our results provide in vivo evidence that some bacterial strains possess a high degree of tolerance for exogenous crossbridges, which may ultimately provide a potentially viable drug resistance modality.
Figure 5.
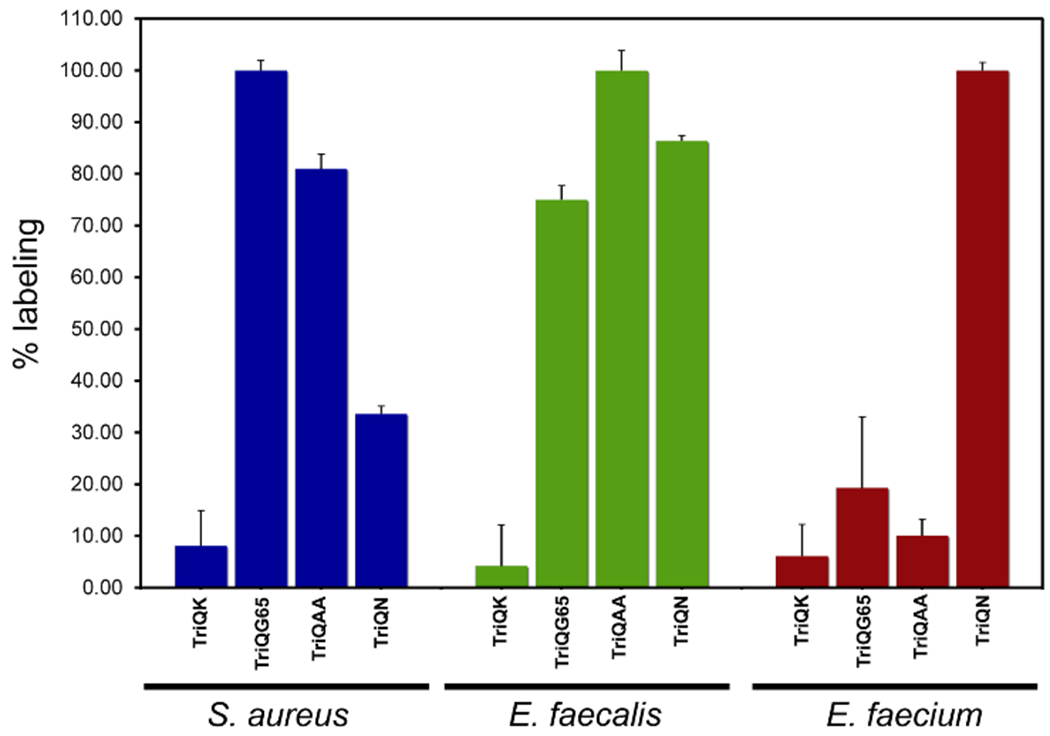
Crossbridge preferences across bacterial species. Flow cytometry analysis of cells treated overnight with 100 ¼M of tri-peptide probes. Data are represented as mean + SD (n = 3).
Targeting the enzymatic processes that control PG assembly is a powerful method of disabling bacterial pathogens. Overall, our results demonstrate the requirement of the crossbridge in acyl-acceptors for proper crosslinking by TPs in three different types of bacteria, thus reinforcing the concept that lysine modification is pivotal in tuning the PG crosslinking machinery. Future investigations will center on finding out how flexibility in PG crosslinking may promote the evolution of drug resistance. In developing a comprehensive structural analysis of both the acyl-donor and -acceptor strands, we anticipate that these results may guide drug design and open new avenues of therapeutic modalities.
Supplementary Material
ACKNOWLEDGMENT
This study was supported by GM124893-01 (M.P).
Footnotes
Supporting Information
Additional experimental details (methods, characterization and synthesis of peptide probes) and figures. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interests.
REFERENCES
- 1.Andersson DI; Hughes D, Antibiotic resistance and its cost: is it possible to reverse resistance? Nat Rev Microbiol 2010, 8 (4), 260–71. [DOI] [PubMed] [Google Scholar]
- 2.Arias CA; Murray BE, The rise of the Enterococcus: beyond vancomycin resistance. Nat Rev Microbiol 2012, 10 (4), 266–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vollmer W; Blanot D; de Pedro MA, Peptidoglycan structure and architecture. FEMS Microbiol Rev 2008, 32 (2), 149–67. [DOI] [PubMed] [Google Scholar]
- 4.Silhavy TJ; Kahne D; Walker S, The bacterial cell envelope. Cold Spring Harb Perspect Biol 2010, 2 (5), a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ling LL; Schneider T; Peoples AJ; Spoering AL; Engels I; Conlon BP; Mueller A; Schaberle TF; Hughes DE; Epstein S; Jones M; Lazarides L; Steadman VA; Cohen DR; Felix CR; Fetterman KA; Millett WP; Nitti AG; Zullo AM; Chen C; Lewis K, A new antibiotic kills pathogens without detectable resistance. Nature 2015, 517 (7535), 455–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Breukink E; de Kruijff B, Lipid II as a target for antibiotics. Nat Rev Drug Discov 2006, 5 (4), 321–32. [DOI] [PubMed] [Google Scholar]
- 7.Bugg TD; Braddick D; Dowson CG; Roper DI, Bacterial cell wall assembly: still an attractive antibacterial target. Trends Biotechnol 2011, 29 (4), 167–73. [DOI] [PubMed] [Google Scholar]
- 8.Wang G; Lo LF; Forsberg LS; Maier RJ, Helicobacter pylori peptidoglycan modifications confer lysozyme resistance and contribute to survival in the host. MBio 2012, 3 (6), e00409–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Juan C; Torrens G; Barcelo IM; Oliver A, Interplay between Peptidoglycan Biology and Virulence in Gram-Negative Pathogens. Microbiol Mol Biol Rev 2018, 82 (4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vijayrajratnam S; Pushkaran AC; Balakrishnan A; Vasudevan AK; Biswas R; Mohan CG, Bacterial peptidoglycan with amidated meso-diaminopimelic acid evades NOD1 recognition: an insight into NOD1 structure-recognition. Biochem J 2016, 473 (24), 4573–4592. [DOI] [PubMed] [Google Scholar]
- 11.Zapun A; Philippe J; Abrahams KA; Signor L; Roper DI; Breukink E; Vernet T, In vitro reconstitution of peptidoglycan assembly from the Gram-positive pathogen Streptococcus pneumoniae. ACS Chem Biol 2013, 8 (12), 2688–96. [DOI] [PubMed] [Google Scholar]
- 12.Srisuknimit V; Qiao Y; Schaefer K; Kahne D; Walker S, Peptidoglycan Cross-Linking Preferences of Staphylococcus aureus Penicillin-Binding Proteins Have Implications for Treating MRSA Infections. J Am Chem Soc 2017, 139 (29), 9791–9794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morlot C; Straume D; Peters K; Hegnar OA; Simon N; Villard AM; Contreras-Martel C; Leisico F; Breukink E; Gravier-Pelletier C; Le Corre L; Vollmer W; Pietrancosta N; Havarstein LS; Zapun A, Structure of the essential peptidoglycan amidotransferase MurT/GatD complex from Streptococcus pneumoniae. Nat Commun 2018, 9 (1), 3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumar JK, Lysostaphin: an antistaphylococcal agent. Appl Microbiol Biotechnol 2008, 80 (4), 555–61. [DOI] [PubMed] [Google Scholar]
- 15.Franchi L; Warner N; Viani K; Nunez G, Function of Nod-like receptors in microbial recognition and host defense. Immunol Rev 2009, 227 (1), 106–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuru E; Hughes HV; Brown PJ; Hall E; Tekkam S; Cava F; de Pedro MA; Brun YV; VanNieuwenhze MS, In Situ probing of newly synthesized peptidoglycan in live bacteria with fluorescent D-amino acids. Angew Chem Int Ed Engl 2012, 51 (50), 12519–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuru E; Tekkam S; Hall E; Brun YV; Van Nieuwenhze MS, Synthesis of fluorescent D-amino acids and their use for probing peptidoglycan synthesis and bacterial growth in situ. Nat Protoc 2015, 10 (1), 33–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Siegrist MS; Whiteside S; Jewett JC; Aditham A; Cava F; Bertozzi CR, (D)-Amino acid chemical reporters reveal peptidoglycan dynamics of an intracellular pathogen. ACS Chem Biol 2013, 8 (3), 500–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Siegrist MS; Swarts BM; Fox DM; Lim SA; Bertozzi CR, Illumination of growth, division and secretion by metabolic labeling of the bacterial cell surface. FEMS Microbiol Rev 2015, 39 (2), 184–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lebar MD; May JM; Meeske AJ; Leiman SA; Lupoli TJ; Tsukamoto H; Losick R; Rudner DZ; Walker S; Kahne D, Reconstitution of peptidoglycan cross-linking leads to improved fluorescent probes of cell wall synthesis. J Am Chem Soc 2014, 136 (31), 10874–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lupoli TJ; Tsukamoto H; Doud EH; Wang TS; Walker S; Kahne D, Transpeptidase-mediated incorporation of D-amino acids into bacterial peptidoglycan. J Am Chem Soc 2011, 133 (28), 10748–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lebar MD; Lupoli TJ; Tsukamoto H; May JM; Walker S; Kahne D, Forming cross-linked peptidoglycan from synthetic gram-negative Lipid II. J Am Chem Soc 2013, 135 (12), 4632–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ngadjeua F; Braud E; Saidjalolov S; Iannazzo L; Schnappinger D; Ehrt S; Hugonnet JE; Mengin-Lecreulx D; Patin D; Etheve-Quelquejeu M; Fonvielle M; Arthur M, Critical Impact of Peptidoglycan Precursor Amidation on the Activity of l,d-Transpeptidases from Enterococcus faecium and Mycobacterium tuberculosis. Chemistry 2018, 24 (22), 5743–5747. [DOI] [PubMed] [Google Scholar]
- 24.Welsh MA; Taguchi A; Schaefer K; Van Tyne D; Lebreton F; Gilmore MS; Kahne D; Walker S, Identification of a Functionally Unique Family of Penicillin-Binding Proteins. J Am Chem Soc 2017, 139 (49), 17727–17730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Triboulet S; Bougault CM; Laguri C; Hugonnet JE; Arthur M; Simorre JP, Acyl acceptor recognition by Enterococcus faecium L,D-transpeptidase Ldtfm. Mol Microbiol 2015, 98 (1), 90–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gautam S; Kim T; Shoda T; Sen S; Deep D; Luthra R; Ferreira MT; Pinho MG; Spiegel DA, An Activity-Based Probe for Studying Crosslinking in Live Bacteria. Angew Chem Int Ed Engl 2015, 54 (36), 10492–6. [DOI] [PubMed] [Google Scholar]
- 27.Pidgeon SE; Apostolos AJ; Nelson JM; Shaku M; Rimal B; Islam MN; Crick DC; Kim SJ; Pavelka MS; Kana BD; Pires MM, L,D-transpeptidase Specific Probe Reveals Spatial Activity of Peptidoglycan Crosslinking. ACS Chem Biol 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bellais S; Arthur M; Dubost L; Hugonnet JE; Gutmann L; van Heijenoort J; Legrand R; Brouard JP; Rice L; Mainardi JL, Aslfm, the D-aspartate ligase responsible for the addition of D-aspartic acid onto the peptidoglycan precursor of Enterococcus faecium. J Biol Chem 2006, 281 (17), 11586–94. [DOI] [PubMed] [Google Scholar]
- 29.Patti GJ; Kim SJ; Schaefer J, Characterization of the peptidoglycan of vancomycin-susceptible Enterococcus faecium. Biochemistry 2008, 47 (32), 8378–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Figueiredo TA; Sobral RG; Ludovice AM; Almeida JM; Bui NK; Vollmer W; de Lencastre H; Tomasz A, Identification of genetic determinants and enzymes involved with the amidation of glutamic acid residues in the peptidoglycan of Staphylococcus aureus. PLoS Pathog 2012, 8 (1), e1002508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu X; Gallay C; Kjos M; Domenech A; Slager J; van Kessel SP; Knoops K; Sorg RA; Zhang JR; Veening JW, High-throughput CRISPRi phenotyping identifies new essential genes in Streptococcus pneumoniae. Mol Syst Biol 2017, 13 (5), 931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Courtin P; Miranda G; Guillot A; Wessner F; Mezange C; Domakova E; Kulakauskas S; Chapot-Chartier MP, Peptidoglycan structure analysis of Lactococcus lactis reveals the presence of an L,D-carboxypeptidase involved in peptidoglycan maturation. J Bacteriol 2006, 188 (14), 5293–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ito M; Kim YG; Tsuji H; Takahashi T; Kiwaki M; Nomoto K; Danbara H; Okada N, Transposon mutagenesis of probiotic Lactobacillus casei identifies asnH, an asparagine synthetase gene involved in its immune-activating capacity. PLoS One 2014, 9 (1), e83876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Corti A; Curnis F, Isoaspartate-dependent molecular switches for integrin-ligand recognition. J Cell Sci 2011, 124 (Pt 4), 515–22. [DOI] [PubMed] [Google Scholar]
- 35.Weintraub SJ; Manson SR, Asparagine deamidation: a regulatory hourglass. Mech Ageing Dev 2004, 125 (4), 255–7. [DOI] [PubMed] [Google Scholar]
- 36.Yang H; Zubarev RA, Mass spectrometric analysis of asparagine deamidation and aspartate isomerization in polypeptides. Electrophoresis 2010, 31 (11), 1764–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu Z; Frutos S; Bick MJ; Vila-Perello M; Debelouchina GT; Darst SA; Muir TW, Structure of the branched intermediate in protein splicing. Proc Natl Acad Sci U S A 2014, 111 (23), 8422–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Regulski K; Courtin P; Kulakauskas S; Chapot-Chartier MP, A novel type of peptidoglycan-binding domain highly specific for amidated D-Asp cross-bridge, identified in Lactobacillus casei bacteriophage endolysins. J Biol Chem 2013, 288 (28), 20416–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bouhss A; Josseaume N; Severin A; Tabei K; Hugonnet JE; Shlaes D; Mengin-Lecreulx D; Van Heijenoort J; Arthur M, Synthesis of the L-alanyl-L-alanine cross-bridge of Enterococcus faecalis peptidoglycan. J Biol Chem 2002, 277 (48), 45935–41. [DOI] [PubMed] [Google Scholar]
- 40.Maidhof H; Reinicke B; Blumel P; Berger-Bachi B; Labischinski H, femA, which encodes a factor essential for expression of methicillin resistance, affects glycine content of peptidoglycan in methicillin-resistant and methicillin-susceptible Staphylococcus aureus strains. J Bacteriol 1991, 173 (11), 3507–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Henze U; Sidow T; Wecke J; Labischinski H; Berger-Bachi B, Influence of femB on methicillin resistance and peptidoglycan metabolism in Staphylococcus aureus. J Bacteriol 1993, 175 (6), 1612–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Monteiro JM; Covas G; Rausch D; Filipe SR; Schneider T; Sahl HG; Pinho MG, The pentaglycine bridges of Staphylococcus aureus peptidoglycan are essential for cell integrity. Sci Rep 2019, 9 (1), 5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stranden AM; Ehlert K; Labischinski H; Berger-Bachi B, Cell wall monoglycine cross-bridges and methicillin hypersusceptibility in a femAB null mutant of methicillin-resistant Staphylococcus aureus. J Bacteriol 1997, 179 (1), 9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tschierske M; Mori C; Rohrer S; Ehlert K; Shaw KJ; Berger-Bachi B, Identification of three additional femAB-like open reading frames in Staphylococcus aureus. FEMS Microbiol Lett 1999, 171 (2), 97–102. [DOI] [PubMed] [Google Scholar]
- 45.Waxman DJ; Strominger JL, Penicillin-binding proteins and the mechanism of action of beta-lactam antibiotics. Annu Rev Biochem 1983, 52, 825–69. [DOI] [PubMed] [Google Scholar]
- 46.Wu S; Piscitelli C; de Lencastre H; Tomasz A, Tracking the evolutionary origin of the methicillin resistance gene: cloning and sequencing of a homologue of mecA from a methicillin susceptible strain of Staphylococcus sciuri. Microb Drug Resist 1996, 2 (4), 435–41. [DOI] [PubMed] [Google Scholar]
- 47.Jensen A; Valdorsson O; Frimodt-Moller N; Hollingshead S; Kilian M, Commensal streptococci serve as a reservoir for beta-lactam resistance genes in Streptococcus pneumoniae. Antimicrob Agents Chemother 2015, 59 (6), 3529–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Magnet S; Arbeloa A; Mainardi JL; Hugonnet JE; Fourgeaud M; Dubost L; Marie A; Delfosse V; Mayer C; Rice LB; Arthur M, Specificity of L D -transpeptidases from gram-positive bacteria producing different peptidoglycan chemotypes. J Biol Chem 2007, 282 (18), 13151–9. [DOI] [PubMed] [Google Scholar]
- 49.Arbeloa A; Hugonnet JE; Sentilhes AC; Josseaume N; Dubost L; Monsempes C; Blanot D; Brouard JP; Arthur M, Synthesis of mosaic peptidoglycan cross-bridges by hybrid peptidoglycan assembly pathways in gram-positive bacteria. J Biol Chem 2004, 279 (40), 41546–56. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


