Abstract
Canonical Wnt signaling is critical for melanocyte lineage commitment and melanoma development. RAD6B, a ubiquitin-conjugating enzyme critical for translesion DNA synthesis, potentiates β-catenin stability/activity by inducing proteasome-insensitive polyubiquitination. RAD6B expression is induced by β-catenin, triggering a positive feedback loop between the two proteins. RAD6B function in melanoma development/progression was investigated by targeting RAD6B using CrispR/Cas9 or an RAD6-selective small-molecule inhibitor #9 (SMI#9). SMI#9 treatment inhibited melanoma cell proliferation but not normal melanocytes. RAD6B knockout or inhibition in metastatic melanoma cells downregulated β-catenin, β-catenin–regulated microphthalmia-associated transcription factor (MITF), sex-determining region Y-box 10, vimentin proteins, and MITF-regulated melan A. RAD6B knockout or inhibition decreased migration/invasion, tumor growth, and lung metastasis. RNA-sequencing and stem cell pathway real-time RT-PCR analysis revealed profound reductions in WNT1 expressions in RAD6B knockout M14 cells compared with control. Expression levels of β-catenin–regulated genes VIM, MITF-M, melan A, and TYRP1 (a tyrosinase family member critical for melanin biosynthesis) were reduced in RAD6B knockout cells. Pathway analysis identified gene networks regulating stem cell pluripotency, Wnt signaling, melanocyte development, pigmentation signaling, and protein ubiquitination, besides DNA damage response signaling, as being impacted by RAD6B gene disruption. These data reveal an important and early role for RAD6B in melanoma development besides its bonafide translesion DNA synthesis function, and suggest that targeting RAD6B may provide a novel strategy to treat melanomas with dysregulated canonical Wnt signaling.
Melanomas develop from malignant transformation of pigment producing melanocytes predominantly found in skin, and less frequently in other tissues.1 Commitment of neural crest cells to the melanocyte lineage requires sex-determining region Y-box 10 (SOX10) and WNT/β-catenin–regulated microphthalmia-associated transcription factor (MITF).2,3 Unlike several epithelial cancers, β-catenin–activating mutations are uncommon in melanoma; rather, β-catenin is stabilized via nonmutational events.4
RAD6B, a ubiquitin-conjugating enzyme and a principal component of the translesion DNA synthesis pathway,5 stabilizes β-catenin via K63-linked β-catenin polyubiquitination.6,7 RAD6B depletion or inhibition decreases β-catenin levels, decreases transcriptional activity, and reverses epithelial-mesenchymal transition in breast cancer cells.6,8 Humans have two RAD6 paralogs, RAD6A (UBE2A) and RAD6B (UBE2B), that are situated on chromosomes Xq24-q25 and 5q23-q34, respectively, and encode proteins with 95% amino acid sequence identity and similar ubiquitinating activities.9, 10, 11 RAD6B is responsive to β-catenin–mediated transcription, generating a positive feedback loop between RAD6B and β-catenin.12 Although RAD6A and RAD6B are weakly expressed in normal melanocytes and normal skin, RAD6B has a more robust expression in melanomas13 and is coexpressed with β-catenin in primary and metastatic clinical melanomas.13,14 Although the role of canonical Wnt signaling in melanoma development is clear, its role in melanoma metastasis is contradictory. Some studies have shown that loss of β-catenin predicts for poor survival,15, 16, 17, 18 whereas others have shown a prometastatic role for β-catenin.19, 20, 21
To investigate RAD6B relevance in melanoma and its functional link with WNT/β-catenin signaling in melanoma development/progression, RAD6B function was disrupted by CrispR/Cas9 or by inhibition with an RAD6-selective small-molecule inhibitor 9 (SMI#9).22 RAD6B inhibition or knockout in metastatic melanoma cells inhibited proliferation, migratory/invasion potentials, tumor development, and lung metastasis. RAD6B knockout or inhibition in metastatic M14 melanoma cells decreased steady-state protein levels of β-catenin and β-catenin–regulated transcriptional targets MITF, SOX10, vimentin, and MITF-regulated melan A. RNA-sequencing and stem cell pathway real-time RT-PCR analysis revealed profound suppression of canonical WNT1, and up-regulation of stem cell pluripotency factors SOX2 and KLF17 in RAD6B knockout melanoma cells. RNA-sequencing and real-time RT-PCR analysis showed decreases in β-catenin–regulated VIM, tyrosinase-related protein 1 (TYRP1), MITF-M, and melan A expression levels without change in β-catenin expression in RAD6B knockout cells. Pathway analysis of RNA-sequencing transcriptome data identified gene networks regulating stem cell pluripotency, WNT signaling, melanocyte development/pigmentation, cell metabolism, and protein ubiquitination, besides DNA damage response signaling, as being significantly impacted by RAD6B loss. These results show a critical functional link between RAD6B and WNT/β-catenin signaling in melanoma development that could be exploited to inhibit canonical WNT signaling for melanoma therapy.
Materials and Methods
Cell Culture
Normal human epidermal melanocytes HEMa-LP and metastatic human melanoma A2058 cells were purchased from ATCC (Manassas, VA). Human melanoma M14 cells were obtained from the National Cancer Institute (Frederick, MD). Melanoma lines were maintained in Dulbecco’s modified Eagle’s medium/F12 medium supplemented with 5% fetal bovine serum. HEMa-LP cells were maintained in dermal cell basal medium supplemented with melanocyte growth supplements insulin (5 μg/mL), ascorbic acid (50 μg/mL), l-glutamine (6 mmol/L), epinephrine (1.0 μmol/L), calcium chloride (0.2 mmol/L), and M8 supplement (ATCC).14 The authenticated cell lines were used within 5 to 10 passages.
RAD6B CrispR/Cas9 Gene Editing
Plasmids encoding guide RNAs, Cas9 and green fluorescent protein (GFP) (sc-404361), and homology-directed DNA repair (HDR) templates (sc-404361-HDR) for targeting RAD6B were purchased from Santa Cruz Biotechnology (Dallas, TX). Guide RNAs (gRNAs) 1 and 2 target exon 2, and gRNA 3 targets exon 4 of human RAD6B (https://www.ncbi.nlm.nih.gov/nucleotide; accession number NM_003337). The HDR vectors contained the 5′ and 3′ homology donor arms (https://www.ncbi.nlm.nih.gov/nucleotide; accession number DQ090910) flanking a puromycin cassette and red fluorescent protein (RFP) gene. M14 cells were cotransfected with RAD6B CrispR/Cas9 and HDR plasmids (1:2 ratio) or HDR plasmids alone using Metafectine SI+ (Biontex Laboratories GmbH, München, Germany), and stable clones were selected with puromycin. Further selection was accomplished by fluorescence-activated cell sorting of GFP/RFP–positive cells. RAD6B knockout was verified by immunoblotting, RT-PCR, genomic DNA PCR, and sequence analysis. The gRNA sequences and primers for characterization of CrispR-edited cells are as follows: gRNA 1 (5′-GTTACAAGAGGACCCACCTG-3′) and gRNA 2 (5′-GCGCCACTGACACCCACAGG-3′) target exon 2, and gRNA 3 (5′-CAAATAAACCACCAACTGTT-3′) targets the exon 4 of RAD6B gene (accession number NM_003337). The primers used for determining RAD6B gene (accession number DQ090910) disruption by puromycin cassette integration were as follows: RAD6B HDR1, 5′-GAAAGAGGGATGATTCTCAAGC-3′ (forward) and 5′-CGTTGAGCTACTACTGTGCTG-3′ (reverse); RAD6B HDR3, 5′-CTTGCCAATGCTGCGTGTTAC-3′ (forward) and 5′-CATGGTGAAACCCCGTCTCTAC-3′ (reverse); and puromycin cassette, 5′-GAGTACCTGCAGGATCTGATC-3′ (reverse). Because gRNAs 1 and 2 both target exon 2, and the 5′ donor arms of HDR1 and HDR2 vectors are identical, PCR analysis was performed only for HDR1 and HDR3 homology directed repair.
Cell Survival Assays
Metastatic melanoma cells (M14 or A2058) or normal human melanocytes HEMa-LP (5 × 103) seeded in 96-well plates were treated with vehicle, 0 to 10 μmol/L SMI#9,23 SMI#9–gold nanoparticle (GNP; size, 32 to 40 nm), or blank-GNP. SMI#9-GNP was used in conjugation with GNP enhances intracellular SMI#9 solubility and stability, rendering it suitable for systemic in vivo application.23 Details of SMI#9-GNP synthesis, characterization, and functionality are previously described.24 Cell viability was measured at 72 to 96 hours by MTT assay, and results were expressed relative to vehicle or blank GNP-treated controls.
Western Blot Analysis
Whole cell lysates were prepared from M14 cells treated with 1 μmol/L SMI#9, SMI#9-GNP or vehicle, and RAD6B knockout or control M14 cells. Nuclear and cytoplasmic subfractions were prepared using a kit (MBL International, Woburn, MA). Samples were subjected to gradient SDS-PAGE and immunoblotted with antibodies to RAD6,25 β-catenin (Santa Cruz Biotechnology), melan A (Dako Corp., Santa Clara, CA), vimentin (Dako Corp.), MITF (Abcam, Cambridge, MA), SOX10 (Santa Cruz Biotechnology), Snail-1 (Abcam), β-actin (Sigma-Aldrich, St. Louis, MO), glyceraldehyde-3-phosphate dehydrogenase (GAPDH; Santa Cruz Biotechnology), or lamin A/C (Cell Signaling, Danvers, MA). Because RAD6A and RAD6B share approximately 95% amino acid sequence identity, our RAD6 antibody does not distinguish the two paralogs.25 Thus, the endogenous protein detected by our antibody is labeled as RAD6 rather than RAD6A or RAD6B. Protein levels relative to the loading control β-actin were quantified by ImageJ version 1.46 (NIH, Bethesda, MD; http://imagej.nih.gov/ij, last accessed July 20, 2020).
Immunofluorescence Analysis
M14 cells treated overnight with 1 μmol/L SMI#9, SMI#9-GNP, or vehicle, and RAD6B knockout cells were stained with antibodies to RAD6, β-catenin, vimentin, proliferating cell nuclear antigen (PCNA; Dako), or Snail-1, and developed with relevant fluorescein isothiocyanate– or Texas Red–conjugated secondary antibodies (Molecular Probes, Eugene, OR). F-actin was detected with Alexa Fluor 488 phalloidin (Molecular Probes). Nuclei were counterstained with DAPI. Slides were stained with isotype-matched nonimmune IgG or without primary antibody to assess specificity. Images were processed using CellSens version 2.1 software (Olympus, Tokyo, Japan).
Real-Time Quantitative PCR Array and RNA-Sequencing Analysis
Real-time RT-PCR analysis of RAD6A, RAD6B, β-catenin, MITF, MITF-M, melan A, TYRP1, VIM, and GAPDH reference control in control and RAD6B knockout M14 cells was performed using DNase I–treated total RNAs and Maxima SYBR Green/ROX qPCR mix (ThermoFisher, Waltham, MA). Primers for real-time RT-PCR analysis were as follows: RAD6A (https://www.ncbi.nlm.nih.gov/nucleotide; accession number NM_003336), 5′-GGATGGAACATTTAAACTTAC-3′ (forward) and 5′-TGCTGGACTATTGGGATTG-3′ (reverse); RAD6B (https://www.ncbi.nlm.nih.gov/nucleotide; accession number NM_003337), 5′-AGACTGACCGCGGGGCA-3′ (forward) and 5′-CAAAAGGTGTCCCTTCTGGTCC-3′ (reverse); GAPDH (https://www.ncbi.nlm.nih.gov/nucleotide; accession number NM_002046), 5′-AAATATGATGACACCAAGAAGG-3′ (forward) and 5′-TGAAGTCGGAGGAGACCAC-3′ (reverse); β-catenin (https://www.ncbi.nlm.nih.gov/nucleotide; accession number NM_001098209), 5′-ATACCACCCACTTGGCAGAC-3′ (forward) and 5′-GGAAGGTCTCCTTGGGACTC-3′ (reverse); TYRP1 (https://www.ncbi.nlm.nih.gov/nucleotide; accession number NM_000550), 5′-ATGGCAACACGCCACAATTTGAG-3′ (forward) and 5′-CCCGTTGCAAAATTCCAGTAAG-3′ (reverse); melan A (https://www.ncbi.nlm.nih.gov/nucleotide; accession number NM_005511), 5′-GATGCCAAGAGAAGATGCTCAC-3′ (forward) and 5′-GTCTCGCTGGCTCTTAAGGTGAA-3′ (reverse); and VIM (https://www.ncbi.nlm.nih.gov/nucleotide; accession number NM_003380.5), 5′-AGATGGCCCTTGACATTGAG-3′ (forward) and 5′-TGGAAGAGGCAGAGAAATCC-3′ (reverse). The forward primer sequences for MITF (5′-TCTACCGTCTCTCACTGGATTGGTGCC-3′) and MITF-M (5′-ATGCTGGAAATGCTAGAATATAATCACTATCAG-3′) were based on human MITF mRNA (accession number Z29678) (https://www.ncbi.nlm.nih.gov/nucleotide) and MITF transcript isoform 4 (accession number NM_000248) (https://www.ncbi.nlm.nih.gov/nucleotide), respectively. The reverse primer was 5′-AGCCATGGGGCTGTTGGGTGC-3′. Amplification was performed in StepOne Plus Real-Time PCR system (Applied Biosystems), and the cycling conditions were 10 minutes at 95°C, and 35 cycles of 15 seconds at 95°C, 1 minute at 57°C, and 1 minute at 68°C. Amplification of the relevant amplicon was confirmed by agarose gel electrophoresis.
The Human Stem Cell RT2 Profiler PCR array (PAHS-176Z; Qiagen, Germantown, MD) was used to quantitate expression of 84 genes linked to stem cell renewal and pluripotency in control and 2mRAD6Bb knockout M14 cells. cDNA synthesis and real-time RT-PCR analysis were conducted, according to the manufacturer's instructions. Data were normalized using reference genes Actb, B2m, GAPDH, Hprt1, and Rplp0. Relative changes in gene expression levels (fold change) were quantitated by the 2−ΔΔCt method, and P- values were calculated using two-tailed t-test.
RNA-sequencing libraries were prepared from DNase 1–treated total RNA (250 ng) using Lexogen's QuantSeq 3′mRNA-seq Library Prep Kit FWD for Illumina (San Diego, CA) with three independent replicates. Aliquots of the RNA were assessed by microfluidics using the ScreenTape for the Agilent 2200 TapeStation (Agilent Technologies, Santa Clara, California). The electrophoretogram, RNA integrity number, and the ratio of the 28S:18S RNA bands were collectively examined to determine overall quality of the RNA. Libraries were quantified on the Qubit and Agilent 2200 TapeStation using the DNA High Sensitivity Screen tape, and the size distribution was determined on a fragment analyzer. The barcoded libraries were multiplexed (in batches of 27) at equimolar concentrations and sequenced with 50-bp reads in rapid mode on Illumina HiSeq 2500. Data were demultiplexed using Illumina's CASAVA 1.8.2, and reads were mapped to the human reference genome (hg38).26 RNA-sequencing data were analyzed using the R/Bioconductor package edgeR version 3.28.0,27 and false discovery rate was computed using Benjamini-Hochberg method.28 Genes were retained with at least one count per million for the analysis, followed by normalization of read counts both within and between samples using the trimmed mean of M-values method. Differentially expressed genes (DEGs) in M14 RAD6B knockout versus control M14, M14 versus HEMa-LP, and M14 RAD6B knockout versus HEMa-LP were detected using 1% false discovery rate and fold change ≥2. Heat map and hierarchical clustering were performed using the R function heatmap.2 from gplots version 3.0.1.1. Canonical pathways impacted by RAD6B loss were identified by Ingenuity Pathway Analysis (Ingenuity Systems, Redwood City, CA). RNA-sequencing analysis was performed at the Wayne State University Applied Genomics Technology Center.
Dual-Luciferase Reporter Assay
Parental and RAD6B knockout M14 cells were cotransfected with 1 μg pTOP/FLASH or pFOP/FLASH (Upstate Biotech, Lake Placid, NY) and 100 ng of Renilla luciferase pRLTK (Promega, Madison, WI) vectors using metafectine (Biontex Laboratories GmbH). Luciferase activities were measured as described previously.6
Chemotaxis, Chemoinvasion, and Wound-Healing Analysis
M14 cells were treated with 0.5 to 2.5 μmol/L SMI#9 or SMI#9-GNP or the corresponding vehicles for 1 hour, and subjected to Boyden chamber chemotaxis assays, as previously described.22 For chemoinvasion assays, M14 RAD6B knockout or control cells (50 × 103 cells) were loaded onto Matrigel-coated chambers (8 μm; BD Biosciences, San Jose, California) in triplicate, and the invaded cells were fixed and quantitated as previously described.22 For wound-healing assays, M14 cells were seeded in wells containing inert silicone inserts to generate defined gaps for wound healing assessment. Cells were treated overnight with vehicle, 1 μmol/L SMI#9 or SMI#9-GNP, inserts were removed, and cells were imaged immediately at set positions. Cells were allowed to migrate and were re-imaged at the preset positions at various times, and processed using SlideBook 6 software (Intelligent Innovations Inc., Denver, Colorado).
Colony Formation Assay
RAD6B knockout or parental control M14 cells were seeded in quadruplicate at 100 cells, and 2 to 3 weeks later, colonies were stained with crystal violet and quantified using GelCount Oxford Optronix and CHARM algorithm, as previously described.22 Colonies with 100 μm set as the minimum diameter threshold were scored. Colony-forming efficiency was expressed relative to control cells.
Tumor Xenograft Modeling
RAD6B knockout or control M14 (1 × 106) cells suspended in Matrigel were bilaterally implanted subcutaneously into the flanks of 6-week–old female nude mice (n = 5). For evaluation of RAD6 inhibitor effects, mice bearing control M14 tumors were treated with SMI#9 or SMI#9-GNP when the tumors reached approximately 150 mm3. Mice were randomly assigned to vehicle, SMI#9, SMI#9-GNP, or blank-GNP groups (n = 5 mice per group) and treated twice weekly with intratumoral injections of SMI#9 (1.5 mg/kg body weight in phosphate-buffered saline/0.5% dimethyl sulfoxide) or vehicle, or i.p. injections of SMI#9-GNP (SMI#9 equivalent dose, 0.85 mg/kg body weight in phosphate-buffered saline) or the equivalent amount of blank-GNP.23 Bidimensional tumor size and body weights were measured twice weekly. Mice were sacrificed at 37 to 44 days after implantation, and the mass of harvested tumors was determined. Lungs were removed and evaluated for metastasis by gross analysis, hematoxylin and eosin staining, or culture of mechanically disrupted lung digests. The excised tumors and lungs were fixed in buffered formalin and paraffin embedded for histologic analysis. The in vivo studies were conducted in accordance with the Institutional Animal Care and Use Committee guidelines of Wayne State University.
Statistical Analysis
Group comparisons were performed using two-tailed independent Student's t-test or Mann-Whitney U-test. Correlations between RNA-sequencing and RT2 Profiler PCR data were analyzed using linear regression models with log-transformed data. Results are presented as means ± SD or SEM. Statistical significance was accepted at P < 0.05.
Data Availability
Data supporting the findings from this article have been deposited as GSE149390 with the National Center for Biotechnology Information Gene Expression Omnibus (http://ncbi.nlm.nih.gov/geo) database.
Results
RAD6B Is Essential for β-Catenin Activation and Melan A Expression
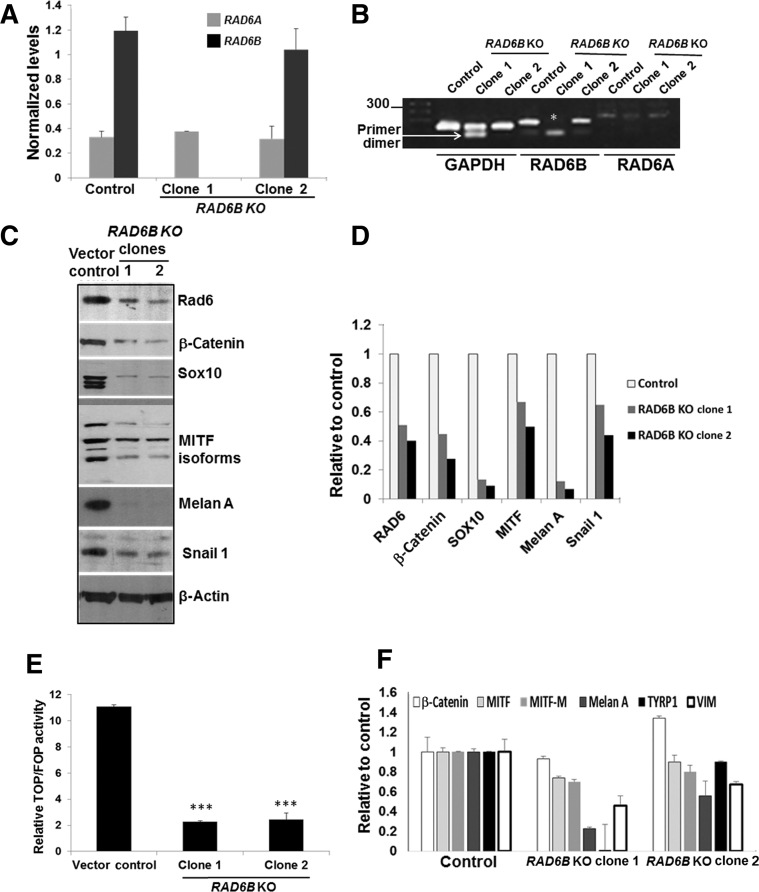
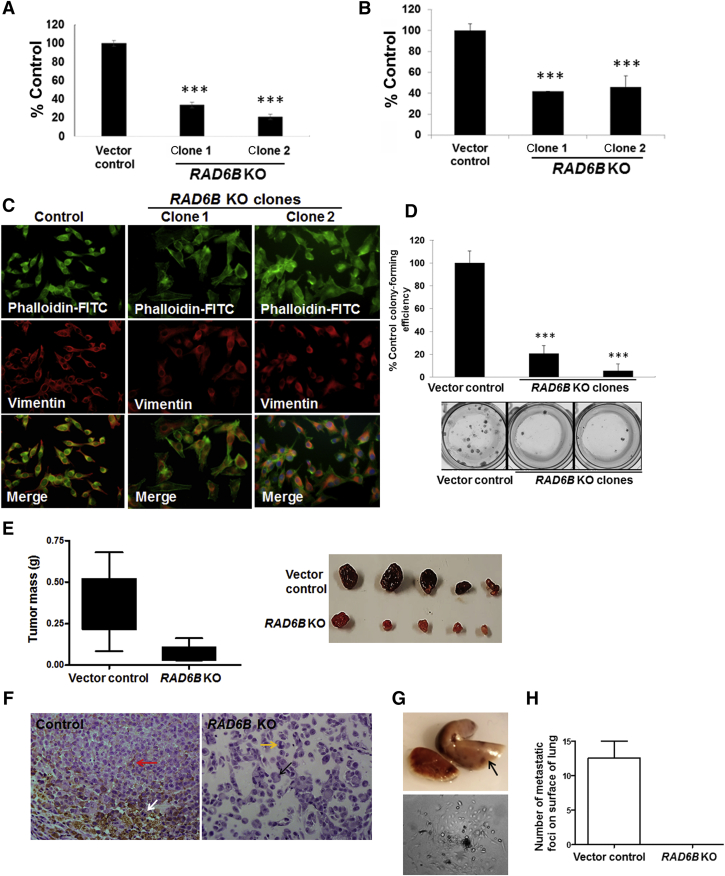
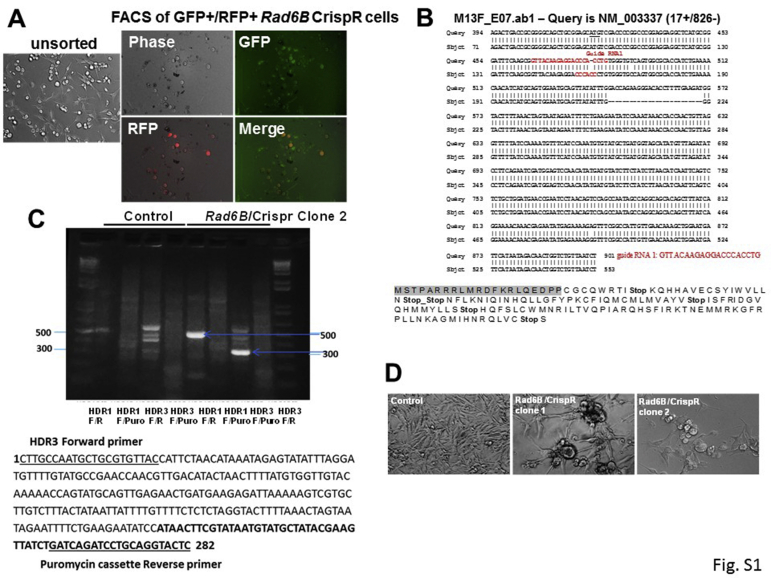
To determine the importance of RAD6B paralog in melanoma cell survival, the RAD6B gene was disrupted using CrispR/Cas9 strategy. Puromycin-resistant stable clones were screened for RAD6 protein levels. Because the antibody used does not distinguish RAD6A and RAD6B proteins, the immunoreactive RAD6 protein was labeled as RAD6 (Materials and Methods). Clones showing loss/decrease in RAD6 levels were purified by fluorescence-activated cell sorting (Supplemental Figure S1A), and RAD6A and RAD6B mRNA, and RAD6 protein expression in the clones were characterized by real-time RT-PCR, sequencing, and Western blot analysis. Real-time RT-PCR analysis of RAD6B using primers involving exons 1 and 2 showed absence of RAD6B amplification in clone 1 but not clone 2 (Figure 1, A and B). Sequence analysis revealed that RAD6B disruption in clone 1 resulted from gRNA 1–mediated Cas9 cleavage (Supplemental Figure S1B). Sequence analysis of clone 2 revealed that RAD6B gene editing resulted from gRNA 3–mediated Cas9 cleavage at exon 4 and puromycin cassette integration from the HDR3 donor vector (Supplemental Figure S1C). Real-time RT-PCR analysis showed that RAD6B expression levels were over fourfold higher than RAD6A in control cells, and that RAD6A expression levels were similar in control and RAD6B-edited clones (Figure 1, A and B). Western blot analysis revealed that RAD6 protein levels were decreased approximately twofold in the RAD6B knockout clones compared with control (Figure 1, C and D). These data suggest that the RAD6 protein band detected in the RAD6B knockout clones represents RAD6A-derived product (Figure 1, C and D). In both RAD6B knockout clones, the steady-state levels of β-catenin and β-catenin–regulated MITF, SOX10, Snail-1, and vimentin proteins were downregulated (Figure 1, C and D), and MITF-regulated melan A protein expression was lost (Figure 1, C and D). Unlike the control cells that displayed the characteristic mesenchymal phenotype, both the RAD6B knockout clones produced multicellular spheroids even under regular culture conditions (Supplemental Figure S1D). Multicellular spheroid formation depends on homotypic cell adhesions, which is consistent with downregulation of epithelial-mesenchymal transition activator β-catenin in RAD6B knockout cells. TOP/Flash reporter assays showed approximately fivefold lower levels (P < 0.001) of β-catenin–dependent transcriptional activity in both RAD6B knockout clones compared with control (Figure 1E). Consistent with reductions in β-catenin–mediated transcriptional activity, real-time RT-PCR analysis showed significant decreases in expressions of MITF-M, melan A, TYRP1, and VIM in both RAD6B knockout clones compared with control (P < 0.05) (Figure 1F). β-Catenin gene expression was not impacted by RAD6B loss, and MITF expression was significantly reduced (P = 0.0131) only in RAD6B knockout clone 1 (Figure 1F). These data suggest RAD6B selectivity in β-catenin–mediated transcription regulation of MITF and melan A, and melanoma phenotype.
Figure 1.
CrispR/Cas9 knockout of RAD6B downregulates β-catenin and β-catenin–regulated melanoma genes. A: Levels of RAD6A and RAD6B gene expression. B: Visualization of the products amplified by real-time RT-PCR by agarose gel electrophoresis. Note the absence of RAD6B amplification in clone 1, indicated by asterisk (details provided in Supplemental Figure S1B). B:RAD6B is amplified in clone 2 using primers spanning exons 1 and 2, as its disruption occurs downstream (details in Supplemental Figure S1C). C: Western blot analysis of the indicated proteins in RAD6B knockout clones of M14 cells. D: Quantitation. E: β-Catenin–regulated transcriptional activity, measured by TOP/Flash (wild-type TCF) and FOP/Flash (mutant TCF) reporter assays. Relative TOP/FOP activities were normalized against Renilla luciferase, and results are expressed from three independent measurements. F: Real-time RT-PCR analysis of β-catenin–regulated target expression levels. mRNA expression levels were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Results represent data from two independent experiments performed in duplicate and expressed relative to control. RAD6B knockout (KO) clone 1 shows significant decreases in expression levels of microphthalmia-associated transcription factor (MITF; P = 0.0131), MITF-M (P = 0.0025), melan A (P = 0.0002), tyrosinase-related protein 1 (TYRP1; P = 0.0016), and vimentin (VIM; P = 0.0204), and RAD6B KO clone 2 shows significant decreases in MITF-M (P = 0.027), melan A (P = 0.0210), TYRP1 (P = 0.0174), and VIM (P = 0.0474) compared with control. Results are expressed means ± SEM (E and F). ∗∗∗P < 0.001.
RAD6 Inhibition Sensitizes Melanoma Cells
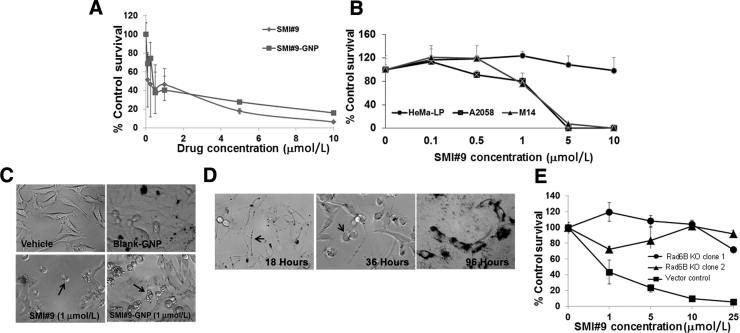
RAD6B stabilizes and activates β-catenin by inducing proteasome-insensitive K63-linked ubiquitin modifications.6,7 To verify the importance of RAD6B/β-catenin link in melanoma survival, the effects of SMI#9 (a small-molecule inhibitor that inhibits the catalytic activities of both RAD6A and RAD6B) on normal human HEMa-LP melanocytes, metastatic A2058, or M14 human melanoma cells, or RAD6B knockout on M14 cell survival were analyzed by MTT assays. Cells were treated with SMI#9 or with SMI#9-GNP as GNP conjugation enhances SMI#9 solubility and stability.23 Previous studies have shown that both SMI#9 and SMI#9-GNP similarly inhibit proliferation and clonogenic potentials, and induce apoptosis of triple-negative breast cancer cells.23 SMI#9 or SMI#9-GNP treatments inhibited M14 and A2058 (IC50, 0.5 to 1.0 μmol/L) (Figure 2, A and B) cell proliferations but not HEMa-LP cells (Figure 2B). The apparent retention of 20% cells in the SMI#9-GNP treated group (Figure 2A) reflects residual absorbance from GNP aggregates rather than surviving cells, as determined by phase-contrast microscopy (Figure 2C) and trypan blue staining. SMI#9-GNP uptake allowed visualization of time-dependent morphologic alterations from the spindle shape at 18 hours (Figure 2D) to a rounded phenotype (Figure 2D) by 36 hours, and loss of cellular integrity and viability by 96 hours (Figure 2D) compared with blank-GNP treated cells that were unaffected (Figure 2C). These data show SMI#9 and SMI#9-GNP similarly inhibit melanoma cell proliferation. Proliferation of RAD6B knockout M14 clones was not affected by SMI#9 compared with control (Figure 2E), suggesting that RAD6B, rather than RAD6A, is the major player in melanoma cell survival and is consistent with the low levels of RAD6A expression (Figure 1A).
Figure 2.
Treatment with RAD6 small-molecule inhibitor 9 (SMI#9) decreases metastatic melanoma cell survival. A: Gold nanoparticle (GNP) conjugated or unconjugated SMI#9 effects on metastatic M14 melanoma cells by MTT assay. Results are expressed relative to cells treated with vehicle or 10 μmol/L blank-GNP. B: SMI#9 sensitivities of HEMa-LP normal melanocytes and metastatic melanoma cells. C: Phase-contrast micrographs of M14 cells treated with vehicle, blank-GNP, SMI#9, or SMI#9-GNP. Arrows indicate sensitized cells. D: Time course monitoring of SMI#9-GNP uptake. Left panel: Arrow indicates SMI#9-GNP uptake at 18 hours. Middle panel: Arrow indicates loss of epithelial-mesenchymal transition visualized by cell rounding at 36 hours. Right panel: Loss of cell integrity by 96 hours. E: M14 RAD6B knockout (KO) clones exhibit decreased sensitivity to SMI#9 compared with control. The results represent data from at least three independent experiments performed in triplicate and are expressed relative to control. Results are expressed as means ± SD (E). Original magnification, ×20 (D).
RAD6 Inhibitor Induces β-Catenin Nuclear Exclusion and Downregulation of β-Catenin Transcriptional Targets
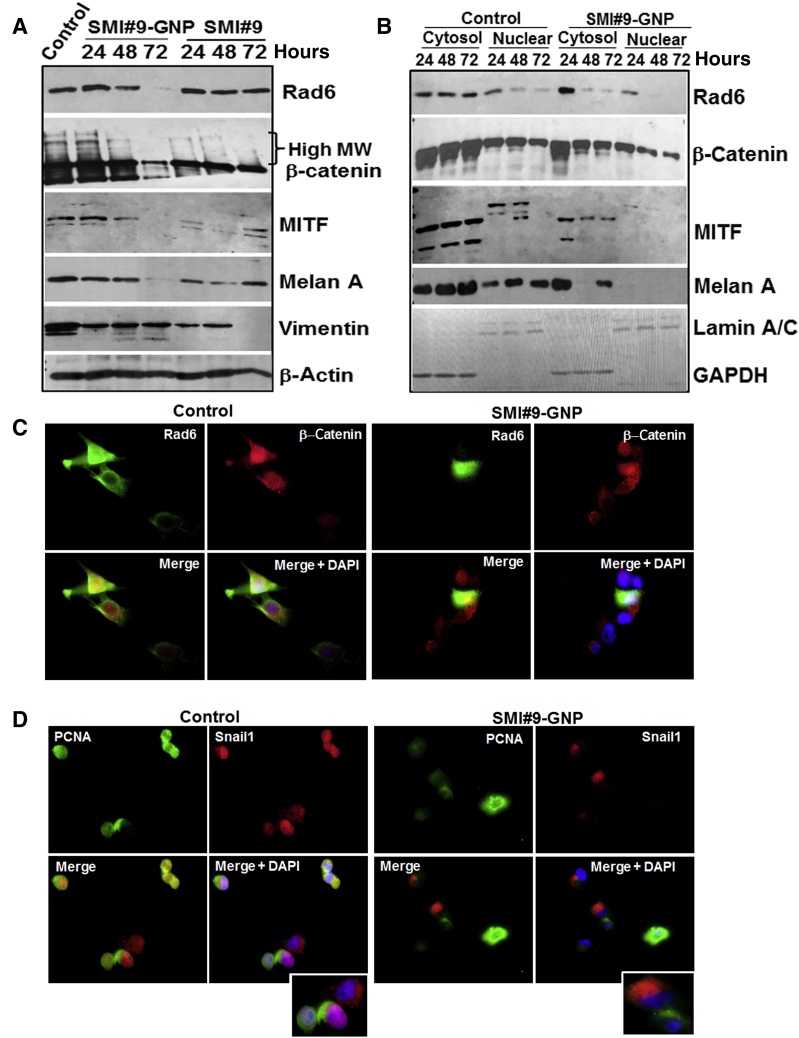
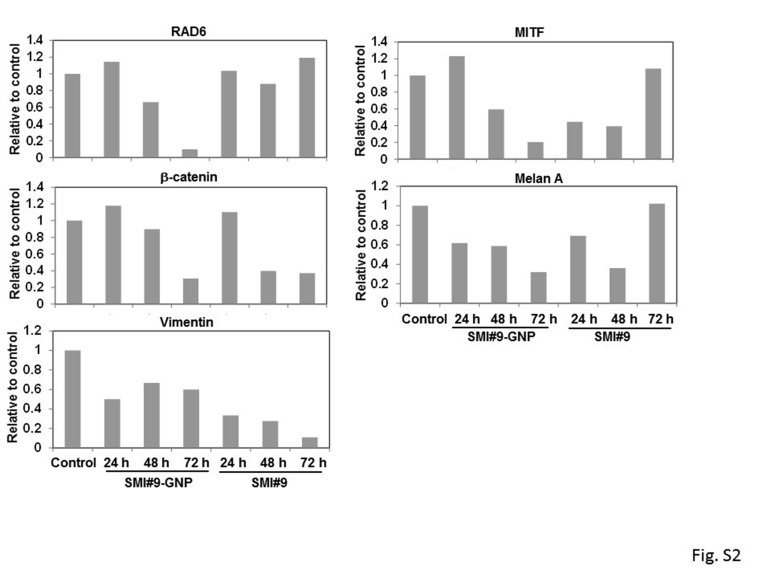
To further verify RAD6B/β-catenin link in melanoma cell survival, the molecular effects of SMI#9 or SMI#9-GNP on the steady-state levels of RAD6, β-catenin, MITF, melan A, and vimentin proteins were analyzed in M14 cells. Since the RAD6 antibody does not distinguish RAD6A and RAD6B proteins, the immunoreactive protein is indicated as RAD6. Compared with control, nascent and high-molecular-weight β-catenin forms, reflective of β-catenin polyubiquitination, were decreased in both SMI#9- and SMI#9-GNP–treated cells, albeit more strongly in the latter (Figure 3A and Supplemental Figure S2). Consistent with potent β-catenin lowering effects, RAD6 levels were noticeably decreased by 48 hours in SMI#9-GNP–treated cells but not in SMI#9- treated cells. MITF levels were reduced, although more durably in SMI#9-GNP–treated cells (Figure 3A), and decreases in melan A, a key marker of melanocyte differentiation,29 mirrored MITF regulation (Figure 3A and Supplemental Figure S2). Compared with control cells that expressed several anti-vimentin immunoreactive proteins, SMI#9- and SMI#9-GNP–treated cells expressed mainly the higher molecular weight, approximately 50-kDa form, albeit at approximately 50% reduced levels (Figure 3A and Supplemental Figure S2). To confirm these results, the impact of RAD6 inhibition on the expression levels of these proteins in the cytoplasmic and nuclear subfractions was analyzed. Cells were treated with SMI#9-GNP as it produced more durable effects compared with SMI#9. SMI#9-GNP treatment decreased cytoplasmic and nuclear β-catenin and RAD6 levels compared with controls (Figure 3B). Distinct MITF isoforms were detected in the cytoplasm and nucleus (Figure 3B), and commensurate with β-catenin levels, MITF was reduced in both compartments of SMI#9-GNP–treated cells compared with controls. Consistent with MITF’s role in melan A regulation, melan A levels were downregulated by SMI#9-GNP (Figure 3B). These data further support a critical role for RAD6B in regulation of β-catenin transcriptional activity and melanoma phenotype.
Figure 3.
RAD6 inhibition downregulates β-catenin and β-catenin–regulated melanoma genes. A: Western blot analysis of the indicated proteins in M14 cells following treatment with SMI#9 or SMI#9–gold nanoparticle (GNP). Quantitation results are shown in Supplemental Figure S2. B: Western blot analysis of the indicated proteins in cytoplasmic and nuclear subfractions of control and SMI#9-GNP–treated M14 cells. C and D: Dual-immunofluorescence analysis of RAD6 and β-catenin (C) and Snail-1 and proliferating cell nuclear antigen (PCNA; D) in control and SMI#9-GNP–treated M14 cells. Insets: Enlarged images of cells in D. Note the loss of nuclear β-catenin, PCNA, and Snail-1 in SMI#9-GNP–treated cells. Original magnification, ×100 (C and D). GAPDH, glyceraldehyde-3-phosphate dehydrogenase; MITF, microphthalmia-associated transcription factor; MW, molecular weight.
Immunofluorescence analysis was performed to determine the impact of SMI#9-GNP treatment on expression/localization of RAD6, β-catenin, Snail-1, and PCNA. Compared with control cells that show abundant nuclear β-catenin (reflective of its transcriptional activity), SMI#9-GNP treatment caused exclusion of β-catenin from the nucleus (Figure 3C). Robust RAD6 staining in the cytoplasm, nucleus, and lamellipodia of control cells was diminished by SMI#9-GNP (Figure 3C). Similarly, strong nuclear PCNA and Snail-1 staining, observed in control cells, was decreased by SMI#9-GNP, which was accompanied by cytoplasmic Snail-1 localization, a characteristic of canonical Wnt signaling inhibition30 (Figure 3D). These data support RAD6B’s role in melanoma cell survival via activation of canonical Wnt signaling and the potential benefit of targeting RAD6B catalytic activity.
RAD6 Inhibitor Decreases Melanoma Cell Migration
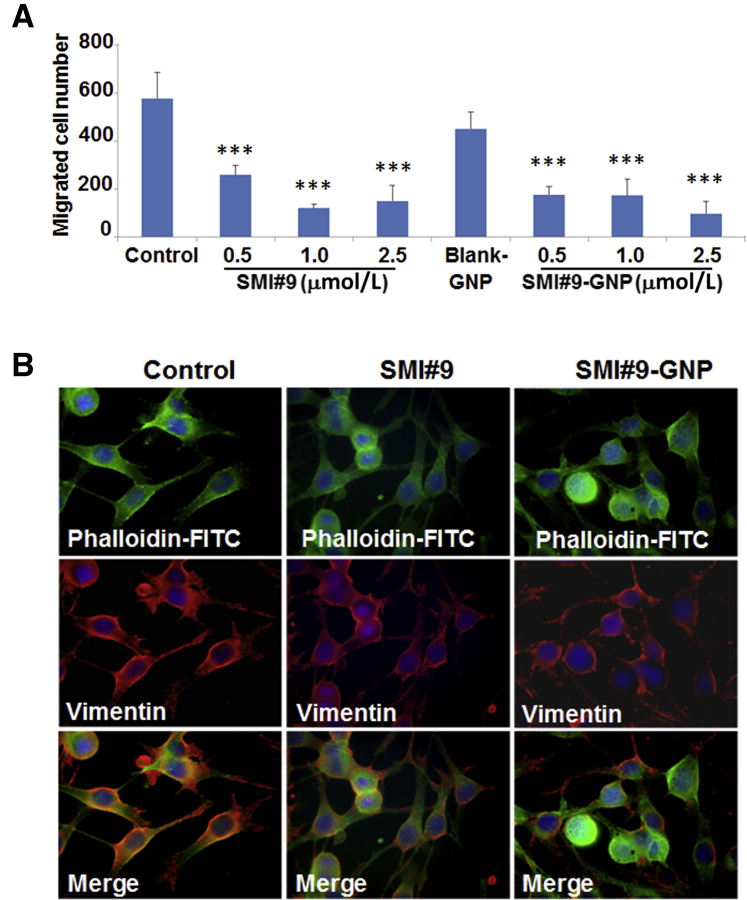
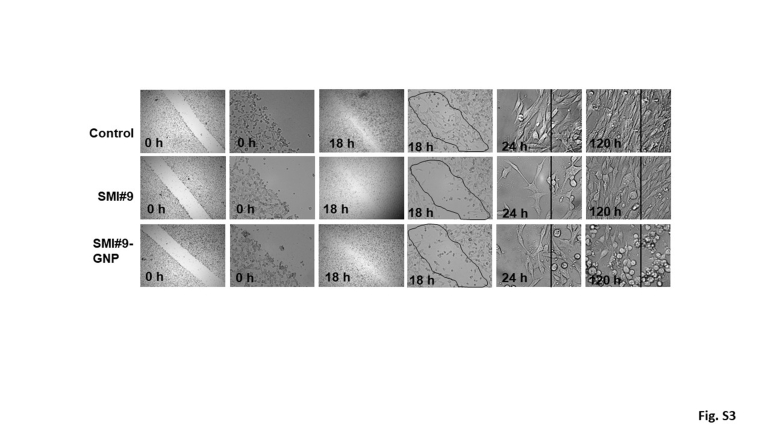
Next, the effects of SMI#9 and SMI#9-GNP on melanoma cell migration were investigated. Compared with the cells treated with vehicle or blank-GNP, both SMI#9 and SMI#9-GNP potently inhibited M14 cell migration in Boyden chamber assays (P < 0.001) (Figure 4A). SMI#9 or SMI#9-GNP effects on population migration were determined by wound-healing assays. Compared with control cells that migrated efficiently, the wound site was less populated in SMI#9-GNP– or SMI#9-treated cells at 18 hours (Supplemental Figure S3). Because of the prolonged stability of GNP-conjugated SMI#9, these cells retained their rounded and nonmotile phenotype even at 120 hours compared with SMI#9-treated cells that showed notable recovery of their migratory potential (Supplemental Figure S3). Cancer cells utilize lamellipodia to invade, which are dependent on actin/vimentin filament networks.31,32 To determine if RAD6 inhibitor-induced antimigratory effects involve alterations in actin/vimentin networks, dual-fluorescence imaging of vimentin and F-actin networks was performed using anti-vimentin antibody and fluorescein isothiocyanate–labeled phalloidin, respectively. Although controls showed prominent vimentin/actin stained lamellipodia, SMI#9 and SMI#9-GNP treatment induced loss of vimentin at the leading edges and lamellipodial contraction (Figure 4B). The decrease in migratory potentials is consistent with the decreased levels of vimentin expressed in RAD6-inhibited cells (Figure 3A).
Figure 4.
RAD6 inhibition reduces melanoma cell migration. A: Small-molecule inhibitor 9 (SMI#9) or SMI9–gold nanoparticle (GNP) effects on M14 cell migration by Boyden chamber assays. B: Immunofluorescence analysis of vimentin (Texas Red) and F-actin [fluorescein isothiocyanate (FITC)–phalloidin] filaments in control, SMI#9, and SMI#9-GNP–treated M14 cells. Note the loss or decrease in vimentin from lamellipodia and rounding of cells in SMI#9-GNP–treated cells. ∗∗∗P < 0.001.
RAD6B Is Necessary for Melanoma Cell Migration/Invasion and Tumor Development
RAD6 inhibitor decreased M14 cell migration and vimentin/actin filament formation (Figure 4, A and B). To confirm if RAD6B plays an important role in melanoma development and tumor progression, the migratory/invasion and tumorigenic potentials of RAD6B knockout M14 clones was analyzed. Compared with control, both migration and invasion potentials were impaired >60% in RAD6B knockout clones (P < 0.001) (Figure 5, A and B), and RAD6B knockout clones showed similar loss of vimentin in lamellipodia (Figure 5C) as SMI#9- or SMI#9-GNP–treated cells (Figure 4B). RAD6B loss also reduced M14 colony-forming efficiency by >80% (P < 0.001) (Figure 5D).
Figure 5.
RAD6B knockout decreases migration, invasion, and tumorigenic potentials. A and B: Migratory (A) and invasive (B) potentials. Assays were performed in triplicate, and results are expressed relative to control from two independent experiments. C: Immunofluorescence analysis of vimentin (Texas Red) and F-actin [fluorescein isothiocyanate (FITC)–phalloidin] filaments. Note the selective loss of vimentin from the lamellipodia of RAD6B knockout (KO) clones compared with controls. D: Colony formation. Colony-forming efficiency was expressed relative to control cells. Assays were performed in triplicate, and results are expressed from two independent experiments. E: Vertical scatter plots of excised tumor mass at time of sacrifice. Data analyzed by two-tailed t test (P = 0.0096). Note the characteristic melanoma pigmentation in control xenografts and its loss in RAD6B knockout xenografts. F: Hematoxylin and eosin analysis. Giant multinucleated cells characteristic of mitotic catastrophe (black arrow) and cells with aberrant mitotic shape (yellow arrow) in RAD6B KO clone are indicated. Mitotic (white arrow) and pigment-producing (red arrow) cells in control tumors are indicated. G: Pigmented metastatic foci (arrow) in lungs (top panel) and tumor cells derived from lung digests (bottom panel) of control M14 tumor-bearing mice. H: Quantification of metastatic foci on the surfaces of lungs. Results are expressed as means ± SD (A, B, D, and H). N = 3 (H). ∗∗∗P < 0.001. Original magnification, ×40 (F).
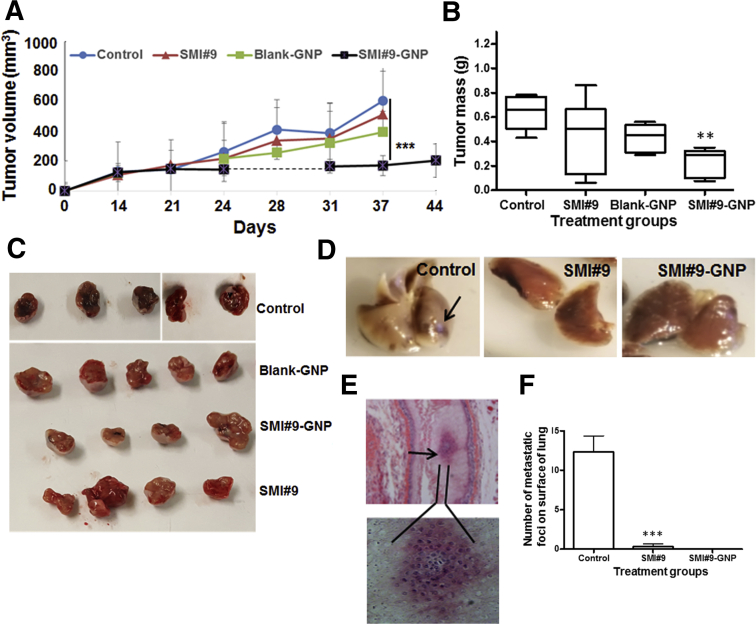
To validate their in vivo relevance, the impact of RAD6B loss on tumor development was analyzed. Compared with controls that produced robustly pigmented tumors (mean mass, 0.3572 ± 0.0711 g), RAD6B knockout significantly inhibited tumor development (mean mass, 0.06484 ± 0.024 g; P = 0.0096) and pigmentation (Figure 5E). Hematoxylin and eosin analysis revealed giant multinucleated cells characteristic of mitotic catastrophe (Figure 5F) and cells with aberrant mitosis (Figure 5F) in the RAD6B knockout group compared with mitotically active (Figure 5F) and pigment-producing (Figure 5F) cells in control tumors. Control tumor-bearing mice also demonstrated overt lung metastasis, marked by pigmented spots (Figure 5, G and H) and confirmed by tumor cell recovery from lung digests (Figure 5G). To assess the in vivo therapeutic effects of SMI#9 and SMI#9-GNP, mice bearing control tumors were randomly assigned for twice weekly treatments with vehicle, SMI#9, SMI#9-GNP, or blank-GNP. SMI#9 (or vehicle) and SMI#9-GNP (or blank-GNP) were administered intratumorally or intraperitoneally, respectively. All groups initially tracked similar growth; however, continued treatment with SMI#9-GNP significantly decreased tumor growth (P < 0.001 and P = 0.0196) (Figure 6, A–C). Although SMI#9 treatment had minimal effect on primary tumor growth, lung metastasis was inhibited in both SMI#9- and SMI#9-GNP–treated mice. Metastatic lesions were detected in the lungs of control mice by gross tissue (Figure 6, D and F) and histologic (Figure 6E) analysis; however, lung metastasis was rare or not detectable in SMI#9- and SMI#9-GNP–treated mice (Figure 6F). The body weights were unaffected by treatment, suggesting absence of overt toxicity (Supplemental Figure S4). Taken together, these data show that knockout or inhibition of RAD6B in M14 suppresses migration/invasion, melanoma development, and lung metastasis.
Figure 6.
Small-molecule inhibitor 9 (SMI#9)–gold nanoparticle (GNP) treatment inhibits melanoma growth and lung metastasis. M14 cells were bilaterally implanted subcutaneously in the flanks, and when the tumors reached approximately 150 mm3, mice were randomly assigned to vehicle control, SMI#9, SMI#9-GNP, or blank-GNP group. Mice were treated twice weekly with SMI#9 (or vehicle) administered intratumorally or SMI#9-GNP (or blank-GNP) administered intraperitoneally. A: Tumor volumes. B: Vertical scatter plots of mass of excised tumors at the time of sacrifice at 37 to 44 days. Data were analyzed by one-way analysis of variance and two-tailed t test. C: Representative excised tumors. D: Lungs from control, SMI#9, and SMI#9-GNP–treated mice. D: Arrow shows a pigmented metastatic lesion on the surface of control lungs. E: Top panel: Metastatic melanoma in control tumor-bearing lung tissue revealed by hematoxylin and eosin staining (arrow). Bottom panel: The enlarged image of the metastatic lesion. F: Quantification of metastatic foci detected on surface of lungs in control and treated mice. Results are expressed as means ± SD (A and F). N = 3 (F). ∗∗P < 0.01, ∗∗∗P < 0.001. Original magnification, ×10 (E, top panel).
RAD6B Gene Disruption Downregulates WNT/β-Catenin–Regulated Melanoma Genes
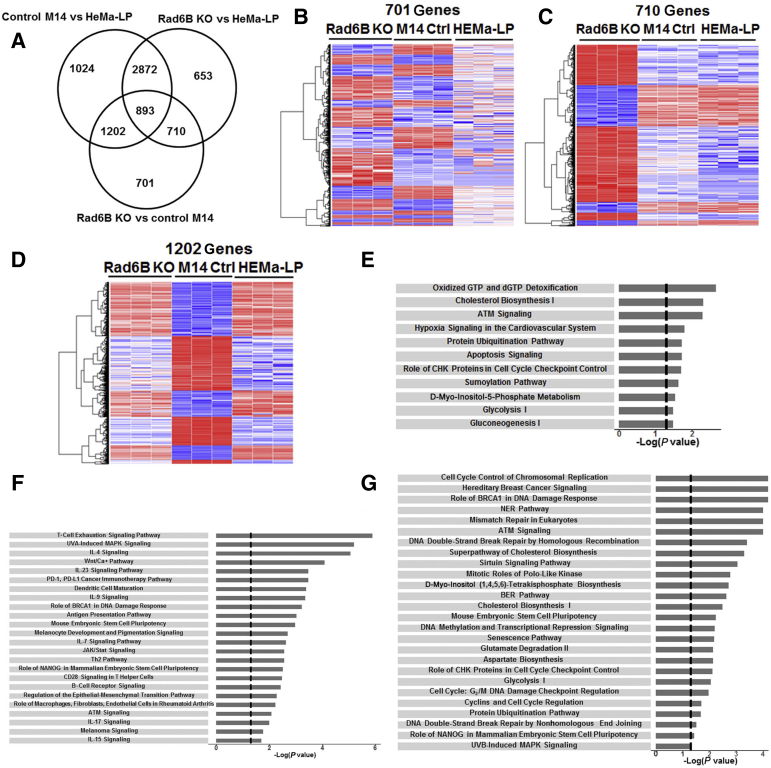
To delineate the genes impacted by RAD6B loss, RNA-sequencing analysis of control and RAD6B knockout M14 models and normal HEMa-LP melanocytes was performed. Using STAR software (https://github.com/alexdobin/STAR), all samples showed >80% alignment to hg38.26 On the basis of a fold change ≥2 and 1% false discovery rate, 3506 DEGs were identified between RAD6B knockout and parental control M14 cells, 5128 DEGs were identified between RAD6B knockout M14 and HEMa-LP cells, and 5991 DEGs were identified between control M14 and HEMa-LP cells (Supplemental Figure S5). Meta-analysis and hierarchical cluster analysis were performed to assess the differences between RAD6B knockout, parental control M14, and HEMa-LP models (Figure 7, A–D), and clustering of these samples validated the reproducibility of the biological replicates and the robustness of the data (Figure 7, B–D). Venn diagram (Figure 7A) and hierarchical cluster analysis showed that among the 3506 DEGs in RAD6B knockout cells, 710 transcripts are shared by M14 and HEMa-LP models, 1202 are shared by HEMa-LP and RAD6B knockout M14, and 701 are differentially expressed in RAD6B knockout cells compared with parental control M14 and HEMa-LP cells (Figure 7, A–D). Ingenuity Pathway Analysis of 701, 710, and 1202 DEGs revealed three pathway networks (Figure 7, E–G) that were impacted by RAD6B disruption. The network related to 701 DEGs is concerned with cholesterol metabolism, glycolysis, protein ubiquitination, and apoptosis (Figure 7E), and includes 48 focus molecules (Supplemental Table S1). Genes included in this network are NUDT1, SQLE, MSMO1, ATR, CHEK1, SMC1A, SMC3, PTEN, UBE2H, UBE2W, BCL2L10, PARP1, ALDOA, ENO4, and GAPDH. The network related to 710 DEGs is enriched for pathways concerned with immune cell signaling, stem cell pluripotency, WNT signaling, melanoma signaling, and melanocyte development and pigmentation (Figure 7F), and includes 58 focus molecules (Supplemental Table S2). Genes included in this network are FZD3, FZD6, FZD8, ATF2, ATM, IL1B, B2M, HLA-B, HLA-DOA, HLA-DOB, JAK3, STAT1, RASD1, AKT3, CD19, NFAT5, and PIK3CD. The network connected with 1202 DEGs is related to cell cycle control, DNA replication, DNA repair, and damage response and senescence (Figure 7G), and includes 163 focus molecules or genes (Supplemental Table S3). Genes included in this network are BLM, BRCA2, FANCA, FANCC, FANCD2, FANCM, H2AX, RAD50, RAD51, TUBG1, CCNB1, PCNA, POLD1, POLE, POLK, ATRX, AURKA, JAK2, and BIRC5. These data reveal distinct roles for RAD6B in melanoma development besides its bonafide translesion DNA synthesis function.33
Figure 7.
RNA-sequencing analysis of control and RAD6B knockout M14 melanoma cells and normal HEMa-LP melanocytes. A: Venn diagram of differentially expressed genes (DEGs; 1% false discovery rate and fold change ≥2) and overlap in RAD6B knockout (KO) compared with control (Ctrl) M14 and HEMa-LP cells. B–D: Hierarchical cluster maps of 701, 710, and 1202 genes, respectively, differentially expressed in RAD6B KO cells compared with control M14 and HEMa-LP cells. E–G: Canonical pathways identified by Ingenuity Pathway Analysis of 701, 710, and 1202 DEGs, respectively. BER, base excision repair; NER, nucleotide excision repair.
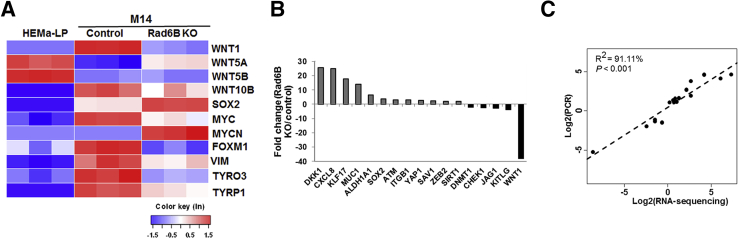
Heat map analysis of the transcriptome data for WNT/β-catenin–regulated genes implicated in melanoma (Figure 8A) identified, among the canonical WNT family members, WNT1 (>15-fold decrease; P = 9.36 × 10−188) and WNT10B (approximately twofold decrease; P = 2.55 × 10−5) expression levels to be significantly downregulated by RAD6B knockout. Concordantly, expression of β-catenin–regulated melanoma genes TYRP1 (5.6-fold; P = 2.6 × 10−48), TYRO3 (3.8-fold; P = 1.72 × 10−38), FOXM1 (5.8-fold; P = 3.54 × 10−120), and c-MYC (approximately twofold; P = 1.29 × 10−14) were all decreased in RAD6B knockout cells relative to parental control (Figure 8A). SOX2 and KLF1734 (transcription factors important for stem cell pluripotency) were increased approximately 2.6-fold (P = 2.2 × 10−38) and 6.0-fold (P = 1.9 × 10−5), respectively, and MYCN (a marker of neural stem cell differentiation)35 was increased approximately 15.6-fold (P = 8.92 × 10−20) in RAD6B knockout cells compared with control cells (Figure 8A). These data suggest potential acquisition of stem cell property in RAD6B-disrupted cells. The noncanonical WNT5A and WNT5B ligands were strongly expressed in HEMa-LP cells, and WNT5A was increased approximately fivefold (P = 2.88 × 10−84) in the RAD6B knockout cells relative to parental control (Figure 8A). These data suggest that noncanonical WNT5A and WNT5B ligands may be important in normal melanocytes, and that RAD6B-mediated regulation of β-catenin activity in melanoma utilizes canonical WNT1.
Figure 8.
RNA-sequencing analysis of Wnt and Wnt/β-catenin–regulated gene expression levels and stem cell PCR array analysis of control and RAD6B knockout (KO) M14 melanoma cells and normal HEMa-LP melanocytes. A: Heat map of WNT and β-catenin–regulated gene expression levels from RNA-sequencing analysis. B: RT2 Stem Cell real-time PCR analysis of control and RAD6B KO M14 cells. C: Pearson correlation analysis of genes identified by RT2 Profiler real-time PCR and RNA-sequencing analysis (R2 = 91.11%; P < 0.001). The dotted line indicates the fitted linear regression line.
The data in Figures 1, 3, and 8A show that RAD6B loss or inhibition downregulates canonical WNT signaling and melanoma-specific MITF, melan A, and TYRP1 expression levels. To determine whether RAD6B loss affects melanoma cell programming, RT2 Profiler stem cell real-time PCR arrays were used to quantitate expression of 84 genes linked to stem cell proliferation, renewal, and pluripotency. WNT1 and the WNT antagonist DKK1 were decreased >35-fold (P = 0.04) and increased 25-fold (P = 0.0015), respectively, in RAD6B knockout cells relative to parental control (Figure 8B and Supplemental Table S4), authenticating the RNA-sequencing data. Concordant with the RNA-sequencing results (Figure 8A), RT2 Profiler real-time PCR analysis showed 3.9- and 17-fold increases in SOX2 and KLF17 expression levels, respectively, in RAD6B knockout cells compared with control (Figure 8B and Supplemental Table S4). Pearson correlation analysis using linear regression established strong concordance between the RNA-sequencing and RT2 Profiler real-time PCR data (R2 = 91.11%; P < 0.001) (Figure 8C and Supplemental Table S4), strengthening the validity of transcriptome analysis and supporting a critical role for RAD6B in WNT/β-catenin activation and melanoma programming.
Discussion
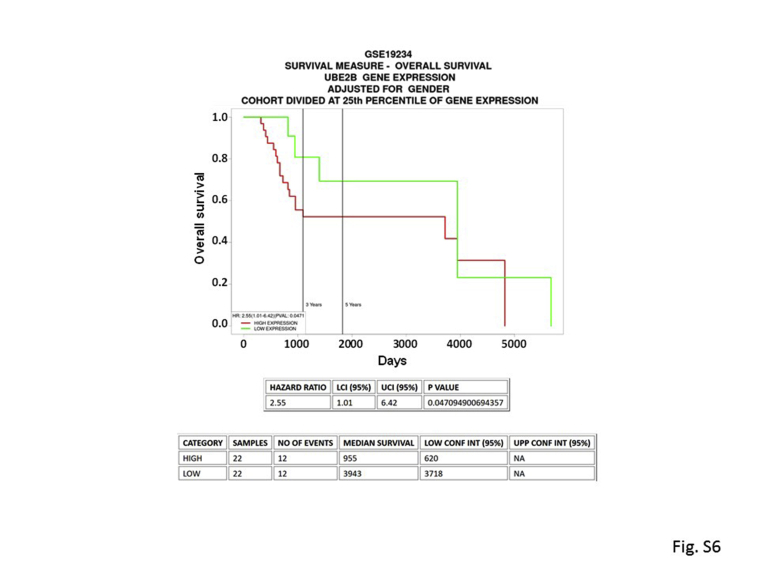
This study provides evidence for a principal role for RAD6B function in melanoma development/progression. Although RAD6A and RAD6B paralogs have similar ubiquitin conjugating activity–dependent functions in translesion DNA synthesis,10 the use of CrispR/Cas9 and SMI#9 inhibition of RAD6 activity23 showed that the RAD6B paralog is important in melanoma development. Despite maintenance of RAD6A expression, loss of RAD6B in metastatic melanoma line causes downregulation of β-catenin transcriptional activity and β-catenin–regulated MITF, SOX10, melan A, VIM, WNT1, TYRP1, and TYRO3 gene expression levels. Although β-catenin protein levels are decreased in RAD6B knockout cells, β-catenin gene expression is unaffected. These data are consistent with the role of RAD6B in stabilizing/activating β-catenin protein via K63-linked polyubiquitination.6 Concomitant down-regulation of SOX10, MITF, and melan A transcripts and proteins in RAD6B knockout cells is consistent with the direct links between SOX10 and WNT signaling in MITF transcription,36,37 and the importance of SOX10 and canonical WNT-1 and WNT-3A in expansion and early specification of neural crest stem cells to the melanocytic lineage.38,39 TYRO3, a TAM tyrosinase kinase receptor, up-regulates MITF expression by enabling SOX10 nuclear localization.40 MITF is important for melanocyte proliferation and differentiation.41, 42, 43 A MITFlow state has, however, been associated with drug resistance44, 45, 46 and tumor initiation capacity.47 This is not supported by our findings as both RAD6B knockout and RAD6-inhibited melanoma cells display MITF and melan A downregulation, and lack migratory/invasion potentials and metastatic abilities. Although both SMI#9 and SMI#9-GNP treatments inhibited lung metastasis, primary tumor growth was significantly inhibited by SMI#9-GNP but not SMI#9. It is possible that the therapeutic efficacy of the intratumorally delivered SMI#9 was reduced because of decreased distribution within the tumor mass. Alternatively, it is possible that the poor stability of SMI#9 allows for recovery of the sensitized tumor cells. The data from RAD6B knockout experiments establish an important role for RAD6B in melanoma development as these cells have substantially diminished tumor-forming potentials and lack the characteristic pigmentation of control melanomas. Loss of pigmentation is supported by downregulation of β-catenin–regulated TYRP1, a melanosomal enzyme that belongs to the tyrosinase family and plays an important role in the melanin biosynthetic pathway.48 These findings suggest an early requirement for RAD6B in melanoma development and a continued role for RAD6B in melanoma progression via WNT/β-catenin signaling. Using The Cancer Genome Atlas skin cutaneous melanoma database, no significant association was found between RAD6B and overall survival. However, when a similar analysis was performed using the GSE19234 data set (http://ncbi.nlm.nih.gov/geo) developed from metastatic cutaneous melanomas,49 a significant inverse association (P = 0.047; hazard ratio, 2.55) was found between high expression of RAD6B and overall survival (Supplemental Figure S6). This association was seen when cohorts were divided at 25th percentile, at median of RAD6B gene expression, and adjusted for sex. Because there was no consistency between multiple data sets of the same disease, this indicates that the effects observed are database-specific. This may be due to diverse collection of melanoma patients or differences in therapy or treatment protocols between the patient cohorts. The discrepancies in survival results between the data sets could also result from variability in gene expression measurements, lack of accuracy, and/or completeness of clinical information.
Ingenuity Pathway Analysis of DEGs from RNA-sequencing analysis revealed three networks that were impacted by RAD6B gene disruption: i) a 701 gene network involved in cholesterol metabolism, glycolysis, protein ubiquitination, and apoptosis; ii) a 710 gene network enriched for pathways involved in immune cell signaling, stem cell pluripotency, WNT signaling, melanoma signaling, and melanocyte development and pigmentation; and iii) a 1202 gene network involved in cell cycle control, DNA replication, DNA repair, and damage response. These data reveal novel RAD6B functions besides its recognized role in the translesion DNA synthesis pathway. Because β-catenin is frequently activated during melanomagenesis via nonmutational alterations,4 the reported data suggest the importance of RAD6B-mediated β-catenin activation in melanoma signaling. RAD6B disruption most profoundly affected WNT1 expression, suggesting that canonical WNT1 is the major β-catenin activator in M14 cells. Besides WNT1 downregulation, these data also showed increases in noncanonical WNT5A expression in RAD6B knockout cells, resembling normal melanocytes. WNT5A is implicated in melanoma invasion19,50,51; however, this is not supported by our data. Conflicting roles for canonical WNT signaling in the context of melanoma development and progression have also been reported, with some showing correlations between β-catenin loss and poor survival,15, 16, 17, 18 whereas others reporting a prometastatic role for β-catenin.19, 20, 21,52,53 Our findings are consistent with the latter role, demonstrating a direct role for RAD6B/WNT/β-catenin in melanoma development/progression. Using a variety of analysis methods, it was shown that RAD6B knockout or inhibition decreases β-catenin transcriptional activity and expression of β-catenin–regulated transcriptional targets. However, the direct impact of β-catenin targeting could not be assessed because transfection with β-catenin siRNA caused loss of melanoma cell viability. A major obstacle to achieving selective inhibition of β-catenin is the overlap of β-catenin binding sites for T-cell factor 4(TCF), adenomatous polyposis coli (APC), and E-cadherin.54 Although β-catenin interaction with E-cadherin is essential for cell adhesion and stem cell function, binding of β-catenin with APC is required for its degradation, and β-catenin binding with TCF is required for its transcriptional activity. Similarly, the β-catenin binding sites for coactivators CREB-binding protein (CBP) and p300 also overlap, also making it difficult to selectively target β-catenin.
Neural crest cells are multipotent stem cells that differentiate into neurons, glia, melanocytes, and cartilage.55 Melanocytic lineage specification and commitment are initiated by TCF/β-catenin induction of SOX10 and MITF.56 Because RAD6B knockout results in SOX10, MITF, and melan A protein down-regulation, we posit that under conditions of RAD6B loss, the melanoma cells undergo reprogramming or transdifferentiation to nontumorigenic states. Expression of SOX2, a transcription factor essential for self-renewal and maintenance of pluripotency,57 is increased in RAD6B knockout cells compared with parental control M14 and HEMa-LP cells. SOX2 is also required for directing differentiation to neural progenitors and maintenance of neural progenitor stem cells.58 In this context, although β-catenin–regulated c-MYC expression is reduced, MYCN expression is increased approximately 15-fold in the RAD6B knockout cells compared with parental M14 and HEMa-LP cells. MYCN expression is generally excluded in neural crest stem cells; however, its expression along with SOX2 biases the stem cells toward neural progenitor expansion and maintenance of neural fate.59, 60, 61 Consistent with these data, the proneural gene ASCL162,63 is overexpressed approximately 12-fold (P = 1.14 × 10−5) in RAD6B knockout cells compared with parental control and HEMa-LP cells, suggesting potential lineage alteration. RNA-sequencing analysis also showed significant decreases in SOX15 (approximately threefold; P = 0.0083) and SOX18 (approximately 10-fold; P = 3.21 × 10−23) expression levels in the RAD6B knockout cells compared with control. Although the functional relationships of these SOX family members in melanoma programming remain to be investigated, SOX family members can act as oncogenes, tumor suppressor genes, or both, depending on the cellular context.64 Rather than reprogramming to a pluripotent state, it is also possible that alterations caused by RAD6B disruption result from dedifferentiation or transdifferentiation of melanoma to alternative or intermediate nontumorigenic states. In this context, forced expression of reprogramming factors SOX2, OCT3/4, or KLF4 in metastatic A2058 melanoma cells resulted in mesenchymal-epithelial–like transdifferentiation and attenuation of malignancy in vitro and in vivo.65 This reprogramming-induced loss of malignancy was accompanied by reductions in β-catenin and epithelial-mesenchymal transition–related genes, and loss of MITF, SOX10, and TYR expression levels.65 Interestingly, because RAD6B gene disruption induces similar changes, we posit an important regulatory role for RAD6B in melanoma programming and tumor development.
Additional studies are needed to establish the trajectory of melanoma reprogramming in RAD6B-disrupted cells; nonetheless, our data reveal a novel and significant role for RAD6B via WNT/β-catenin signaling in melanoma development. Our data suggest that RAD6B targeting can provide a new approach to force aggressive melanoma cells to nontumorigenic nonmelanotic cell types.
Author Contributions
M.P.S. conceived and designed the study, conducted experiments, analyzed the results, and coordinated the project; M.P.S. wrote the article, with input from S.K. and A.G.; G.M. synthesized and characterized gold nanoparticle conjugated RAD6 inhibitor; A.S. and A.G. conducted the experiments and analyzed data; M.P.S. performed CrispR/Cas9 gene editing; P.N.M. conducted the migration assays and assisted M.P.S. with in vivo experiments and analysis; and S.K. and K.G. assisted with RNA-sequencing data analysis.
Acknowledgment
We thank Sarah Petit for technical assistance.
Footnotes
Supported by 3 Balls Racing (M.P.V.S.) and Bridge grant (M.P.V.S.) from the Office of the Vice President of Research at Wayne State University. The Genome Sciences Core and Biostatistics Core facilities are supported by NIH Center grant P30 CA022453 (Karmanos Cancer Institute at Wayne State University).
A.S. and A.G. contributed equally to this work.
Disclosures: None declared.
Current address of G.M., University of New South Wales, Sydney, New South Wales, Australia; of A.S, Penrose Therapeutx, Plymouth, MI.
Supplemental material for this article can be found at http://doi.org/10.1016/j.ajpath.2020.10.015.
Supplemental Data
Supplemental Figure S1.
Characterization of M14 RAD6B knockout clones produced by CrispR/Cas9 strategy. A: Green fluorescent protein (GFP)+ (expressing guide RNA/Cas9) and red fluorescent protein (RFP)+ (indicative of homology-directed vector integration) cells were isolated by fluorescence-activated cell sorting (FACS) analysis. B: RAD6B knockout clone 1, resulting from guide RNA 1, mediated Cas9 cleavage, and the guide RNA 1 sequence is indicated in red. Full-length RAD6B cDNA was amplified, and sequence was aligned with human RAD6B mRNA (https://www.ncbi.nlm.nih.gov/nucleotide; accession number NM_003337). The effect of Cas9 gene editing on the translated product is shown below. The gray shaded sequence represents the amino terminal end of the normal Rad6B amino acid sequence. C: RAD6B loss in clone 2 was screened by PCR analysis of genomic DNA using forward (F) and reverse (R) primers specific for homology-directed repair (HDR) vectors that are selective for guide RNA 1 (HDR1) or guide RNA 3 (HDR3), and a reverse primer within the puromycin integration cassette (Puro) inserted in the HDR vectors. Note amplification of 282-bp band with HDR3 forward and Puro reverse primers in RAD6B knockout clone 2, indicating RAD6B gene disruption by puromycin cassette integration. Below, sequence analysis of clone 2 for integration of RFP-puromycin cassette. The positions of HDR3 forward and Puro reverse primers used for PCR amplification are shown as underlined sequences. The bolded sequence indicates integrated puromycin cassette. D: Phase contrast microscopy of the RAD6B knockout clones. Note spheroid cluster formation in the knockout clones compared with control cells with mesenchymal phenotype.
Supplemental Figure S2.
Effects of small-molecule inhibitor 9 (SMI#9) or SMI#9–gold nanoparticle (GNP) on the steady-state levels of RAD6, β-catenin, and β-catenin–regulated target proteins. The steady-state levels of proteins normalized against β-actin are expressed relative to control. MITF, microphthalmia-associated transcription factor.
Supplemental Figure S3.
Treatment with RAD6 inhibitor small-molecule inhibitor 9 (SMI#9) or gold nanoparticle (GNP)–conjugated SMI#9 decreases migratory potential of M14 melanoma cells in wound-healing assays. Images were captured at the indicated time points. The wound area is delineated in the ×10 images captured at 18 hours, and the long-term effects of drug treatment compared with control from the migratory front (vertical line) are shown in ×40 images captured at 5 days after treatment. Note that the decreased capacities of SMI#9 and SMI#9-GNP treated cells correlate with their rounded morphology (more visible and persistent in SMI#9-GNP at 120 hours after treatment) compared with control with spindle or mesenchymal phenotype. Original magnification, ×4; ×10; ×40. 0 h (left) and 0 h (right) are at 4× and 10×, respectively; 18 h (left) and 18 h (right) are at 4× and 10×, respectively; and 24 h at 40×.
Supplemental Figure S4.
Body weight (wt) assessment in control [vehicle or blank–gold nanoparticle (GNP)] and treated [unconjugated and conjugated small-molecule inhibitor 9 (SMI#9)] mice. Average body weights are shown. Error bars not shown because they were small. n = 5 mice per group.
Supplemental Figure S5.
Heat map of differentially expressed transcripts in normal HEMa-LP melanocytes and control and RAD6B knockout (KO) M14 cells. Significant genes were detected using false discovery rate ≤1% and fold change ≥2. A: The number of significant genes is 5991. B: The number of significant genes is 5128. C: The number of significant genes is 3506.
Supplemental Figure S6.
High expression of RAD6B is associated with decreased overall survival of cutaneous melanoma patients. Kaplan-Meier survival curve of 44 metastatic melanoma samples using the GSE19234 data set (http://ncbi.nlm.nih.gov/geo) was generated using the PROGgeneV2 database (http://genomics.jefferson.edu/proggene) based on low (green) and high (red) RAD6B gene expression levels. CONF INT, confidence interval; HR, hazard ratio; LCI, lower CI; PVAL, P value; UCI, upper CI; UPP, upper.
References
- 1.Shain A.H., Bastien B.C. From melanocytes to melanomas. Nat Rev Cancer. 2016;16:345–358. doi: 10.1038/nrc.2016.37. [DOI] [PubMed] [Google Scholar]
- 2.Mull A.N., Zolekar A., Wang Y.-C. Understanding melanocyte stem cells for disease modeling and regenerative medicine applications. Int J Mol Sci. 2015;16:30458–30469. doi: 10.3390/ijms161226207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Agathocleous M., Iordanova I., Willardsen M.I., Xue X.Y., Vetter M.L., Harris W.A., Moore K.B. A directional Wnt/β-catenin-Sox2-proneural pathway regulates the transition from proliferation to differentiation in the Xenopus retina. Development. 2009;136:3289–3299. doi: 10.1242/dev.040451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sinnberg T., Menzel M., Kaesler S., Biedermann T., Sauer B., Nahnsen S., Schwarz M., Garbe C., Schittek B. Suppression of casein kinase 1α in melanoma cells induces a switch in β-catenin signaling to promote metastasis. Cancer Res. 2010;70:6999–7009. doi: 10.1158/0008-5472.CAN-10-0645. [DOI] [PubMed] [Google Scholar]
- 5.Prakash S., Sung P., Prakash L. DNA repair genes and proteins of Saccharomyces cerevisiae. Ann Rev Genet. 1993;27:33–70. doi: 10.1146/annurev.ge.27.120193.000341. [DOI] [PubMed] [Google Scholar]
- 6.Shekhar M.P., Gerard B., Pauley R.J., Williams B.O., Tait L. Rad6B is a positive regulator of beta-catenin stabilization. Cancer Res. 2008;68:1741–1750. doi: 10.1158/0008-5472.CAN-07-2111. [DOI] [PubMed] [Google Scholar]
- 7.Gerard B., Sanders M.A., Visscher D.W., Tait L., Shekhar M.P. Lysine 394 is a novel Rad6B-induced ubiquitination site on beta-catenin. Biochim Biophys Acta. 2012;1823:1686–1696. doi: 10.1016/j.bbamcr.2012.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerard B., Tait L., Nangia-Makker P., Shekhar M.P. Rad6B acts downstream of Wnt signaling to stabilize β-catenin: implications for a novel Wnt/β-catenin target. J Mol Signal. 2011;6:6. doi: 10.1186/1750-2187-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koken M.H., Reynolds P., Jaspers-Dekker I., Prakash L., Prakash S., Bootsma D., Hoeijmakers J.H. Structural and functional conservation of two human homologs of the yeast DNA repair gene RAD6. Proc Natl Acad Sci U S A. 1991;88:8865–8869. doi: 10.1073/pnas.88.20.8865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koken M.H., Smit E.M., Jaspers-Dekker I., Oostra B.A., Hagemeijer A., Bootsma D., Hoeijmakers J.H. Localization of two human homologs, HHR6A and HHR6B, of the yeast DNA repair gene RAD6 to chromosomes Xq24-q25 and 5q23-q31. Genomics. 1992;12:447–453. doi: 10.1016/0888-7543(92)90433-s. [DOI] [PubMed] [Google Scholar]
- 11.Jentsch S., McGrath J.P., Varshavsky A. The yeast DNA repair gene RAD6 encodes a ubiquitin conjugating enzyme. Nature. 1987;329:131–134. doi: 10.1038/329131a0. [DOI] [PubMed] [Google Scholar]
- 12.Shekhar M.P., Tait L., Gerard B. Essential role of T-cell factor/beta-catenin in regulation of Rad6B: a potential mechanism for Rad6B overexpression in breast cancer cells. Mol Cancer Res. 2006;4:729–745. doi: 10.1158/1541-7786.MCR-06-0136. [DOI] [PubMed] [Google Scholar]
- 13.Gajan A., Martin C., Kim S., Joshi M., Michelhaugh S., Sloma I., Mittal S., Firestine S., Shekhar M.P. Alternative splicing of Rad6B and not Rad6A is selectively increased in melanoma: identification and functional characterization. Cells. 2019;8:1375. doi: 10.3390/cells8111375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosner K., Adsule S., Haynes B., Kirou E., Kato I., Mehregan D.R., Shekhar M.P. Rad6 is a potential early marker of melanoma development. Transl Oncol. 2014;7:384–392. doi: 10.1016/j.tranon.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arozarena I., Bischof H., Gilby D., Belloni B., Dummer R., Wellbrock C. In melanoma, beta-catenin is a suppressor of invasion. Oncogene. 2011;30:4531–4543. doi: 10.1038/onc.2011.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kageshita T., Hamby C.V., Ishihara T., Matsumoto K., Saida T., Ono T. Loss of β-catenin expression associated with disease progression in malignant melanoma. Br J Dermatol. 2011;145:210–216. doi: 10.1046/j.1365-2133.2001.04336.x. [DOI] [PubMed] [Google Scholar]
- 17.Mælandsmo G.M., Holm R., Nesland J.M., Fodstad Ø., Flørenes V.A. Reduced β-catenin expression in the cytoplasm of advanced-stage superficial spreading malignant melanoma. Clin Cancer Res. 2003;9:3383–3388. [PubMed] [Google Scholar]
- 18.Chien A.J., Moore E.C., Lonsdorf A.S., Kulikauskas R.M., Rothberg B.G., Berger A.J., Major M.B., Hwang S.T., Rimm D.L., Moon R.T. Activated Wnt/beta-catenin signaling in melanoma is associated with decreased proliferation in patient tumors and a murine melanoma model. Proc Natl Acad Sci U S A. 2009;106:1193–1198. doi: 10.1073/pnas.0811902106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grossmann A.H., Yoo J.H., Clancy J., Sorensen L.K., Sedgwick A., Tong Z., Ostanin K., Rogers A., Grossmann K.F., Tripp S.R., Thomas K.R., D'Souza-Schorey C., Odelberg S.J., Li D.Y. The small GTPase ARF6 stimulates β-catenin transcriptional activity during WNT5A-mediated melanoma invasion and metastasis. Sci Signal. 2013;6:ra14. doi: 10.1126/scisignal.2003398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Damsky W.E., Curley D.P., Santhanakrishnan M., Rosenbaum L.E., Platt J.T., Gould Rothberg B.E., Taketo M.M., Dankort D., Rimm D.L., McMahon M., Bosenberg M. β-Catenin signaling controls metastasis in Braf-activated Pten-deficient melanomas. Cancer Cell. 2011;20:741–754. doi: 10.1016/j.ccr.2011.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zuidervaart W., Pavey S., van Nieuwpoort F.A., Packer L., Out C., Maat W., Jager M.J., Gruis N.A., Hayward N.K. Expression of Wnt5a and its downstream effector beta-catenin in uveal melanoma. Melanoma Res. 2007;17:380–386. doi: 10.1097/CMR.0b013e3282f1d302. [DOI] [PubMed] [Google Scholar]
- 22.Sanders M.A., Brahemi G., Nangia-Makker P., Balan V., Morelli M., Kothayer H., Westwell A.D., Shekhar M.P.V. Novel inhibitors of Rad6 ubiquitin conjugating enzyme: design, synthesis, identification, and functional characterization. Mol Cancer Ther. 2013;12:373–383. doi: 10.1158/1535-7163.MCT-12-0793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saadat N., Liu F., Haynes B., Nangia-Makker P., Bao X., Li J., Polin L.A., Gupta S., Mao G., Shekhar M.P. Nan-delivery of RAD6/translesion synthesis inhibitor SMI#9 for triple-negative breast cancer therapy. Mol Cancer Ther. 2018;12:2586–2597. doi: 10.1158/1535-7163.MCT-18-0364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haynes B., Zhang Y., Liu F., Li J., Petit S., Kothayer H., Bao X., Westwell A.D., Mao G., Shekhar M.P.V. Gold nanoparticle conjugated Rad6 inhibitor induces cell death in triple negative breast cancer cells by inducing mitochondrial dysfunction and PARP-1 hyperactivation: synthesis and characterization. Nanomedicine. 2016;12:745–757. doi: 10.1016/j.nano.2015.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shekhar M.P., Lyakhovich A., Visscher D.W., Heng H., Kondrat N. Rad6 overexpression induces multinucleation, centrosome amplification, abnormal mitosis, aneuploidy, and transformation. Cancer Res. 2002;62:2115–2124. [PubMed] [Google Scholar]
- 26.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robinson M.D., McCarthy D.J., Smyth G. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- 29.Seberg H.E., Otterloo E.V., Cornell R.A. Beyond MITF: multiple transcription factors directly regulate the cellular phenotype in melanocytes and melanoma. Pigment Cell Melanoma Res. 2017;30:454–466. doi: 10.1111/pcmr.12611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim J.Y., Kim Y.M., Yang C.H., Cho S.K., Lee J.W., Cho M. Functional regulation of Slug/Snail2 is dependent on GSK-3β-mediated phosphorylation. FEBS J. 2012;279:2929–2939. doi: 10.1111/j.1742-4658.2012.08674.x. [DOI] [PubMed] [Google Scholar]
- 31.Chung B.-M., Rotty J.D., Coulombe P.A. Networking galore: intermediate filaments and cell migration. Curr Opin Cell Biol. 2013;25:600–612. doi: 10.1016/j.ceb.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pallari H.-M., Eriksson J.E. Intermediate filaments as signaling platforms. Sci STKE. 2006;19:pe53. doi: 10.1126/stke.3662006pe53. [DOI] [PubMed] [Google Scholar]
- 33.Sanders M.A., Haynes B., Nangia-Makker P., Polin L.A., Shekhar M.P. Pharmacological targeting of RAD6 enzyme-mediated translesion synthesis overcomes resistance to platinum-based drugs. J Biol Chem. 2017;292:10347–10363. doi: 10.1074/jbc.M117.792192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bialkowska A.B., Yang V.W., Mallipattu S.K. Krüppel-like factors in mammalian stem cells and development. Development. 2017;144:737–774. doi: 10.1242/dev.145441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang J.T., Weng Z.H., Tsang K.S., Tsang L.L., Chan H.C., Jiang X.H. MycN is critical for the maintenance of human embryonic stem cell-derived neural crest stem cells. PLoS One. 2016;11:e0148062. doi: 10.1371/journal.pone.0148062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aoki Y., Saint-Germain N., Gyda M., Magner-Fink E., Lee Y.H., Credidio C., Saint-Jeannet J.P. Sox10 regulates the development of neural crest-derived melanocytes in Xenopus. Dev Biol. 2003;259:19–33. doi: 10.1016/s0012-1606(03)00161-1. [DOI] [PubMed] [Google Scholar]
- 37.Shibahara S., Takeda K., Yasumoto K., Udono T., Watanabe K., Saito H., Takahashi K. Microphthalmia-associated transcription factor (MITF): multiplicity in structure, function, and regulation. J Invest Dermatol Symp Proc. 2001;6:99–104. doi: 10.1046/j.0022-202x.2001.00010.x. [DOI] [PubMed] [Google Scholar]
- 38.Honore S.M., Aybar M.J., Mayor R. Sox 10 is required for the early development of the prospective neural crest in Xenopus embryos. Dev Biol. 2003;260:79–96. doi: 10.1016/s0012-1606(03)00247-1. [DOI] [PubMed] [Google Scholar]
- 39.Ikeya M., Lee S.M., Johnson J.E., McMahon A.P., Takada S. Wnt signalling required for expansion of neural crest and CNS progenitors. Nature. 1997;389:966–970. doi: 10.1038/40146. [DOI] [PubMed] [Google Scholar]
- 40.Zhu S., Wurdak H., Wang Y., Galkin A., Tao H., Li J., Lyssiotis C.A., Yan F., Tu B.P., Miraglia L., Walker J., Sun F., Orth A., Schultz P.G., Wu X. A genomic screen identifies TYRO3 as a MITF regulator in melanoma. Proc Natl Acad Sci U S A. 2009;106:17025–17030. doi: 10.1073/pnas.0909292106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garraway L.A., Widlund H.R., Rubin M.A., Getz G., Berger A.J., Ramaswamy S., Beroukhim R., Milner D.A., Granter S.R., Du J., Lee C., Wagner S.N., Li C., Golub T.R., Rimm D.L., Meyerson M.L., Fisher D.E., Sellers W.R. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–122. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- 42.Carreira S., Goodall J., Denat L., Rodriguez M., Nuciforo P., Hoek K.S., Testori A., Larue L., Goding C.R. Mitf regulation of Dia1 controls melanoma proliferation and invasiveness. Genes Dev. 2006;20:3426–3439. doi: 10.1101/gad.406406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cheli Y., Ohanna M., Ballotti R., Bertolotto C. Fifteen-year quest for microphthalmia-associated transcription factor target genes. Pigment Cell Melanoma Res. 2010;23:27–40. doi: 10.1111/j.1755-148X.2009.00653.x. [DOI] [PubMed] [Google Scholar]
- 44.Dugo M., Nicolini G., Tragni G., Bersani I., Tomassetti A., Colonna V., Del Vecchio M., De Braud F., Canevari S., Anichini S., Sensi M. A melanoma subtype with intrinsic resistance to BRAF inhibition identified by receptor tyrosine kinases gene-driven classification. Oncotarget. 2015;6:5118–5133. doi: 10.18632/oncotarget.3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Konieczkowski D.J., Johannessen C.M., Abudayyeh O., Kim J.W., Cooper Z.A., Piris A., Frederick D.T., Barzily-Rokni M., Straussman R., Haq R., Fisher D.E., Mesirov J.P., Hahn W.C., Flaherty K.T., Wargo J.A., Tamayo P., Garraway L.A. A melanoma cell state distinction influences sensitivity to MAPK pathway inhibitors. Cancer Discov. 2014;4:816–827. doi: 10.1158/2159-8290.CD-13-0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rambow F., Marine J.-C., Goding C.R. Melanoma plasticity and phenotypic diversity: therapeutic barriers and opportunities. Gene Dev. 2019;33:1295–1318. doi: 10.1101/gad.329771.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cheli Y., Giuliano S., Fenouille N., Allegra M., V Hofman V., Hofman P., Bahadoran P., Lacou J-Pr, Tartare-Deckert S., Bertolotto C., Ballotti R. Hypoxia and MITF control metastatic behaviour in mouse and human melanoma cells. Oncogene. 2012;31:2461–2470. doi: 10.1038/onc.2011.425. [DOI] [PubMed] [Google Scholar]
- 48.Lapedriza A., Petratou K., Kelsh R.N. Neural crest cells and pigmentation. In: Trainer P.A., editor. Neural Crest Cells: Evolution, Development and Disease. Academic Press, Cambridge, Massachusetts; 2014. pp. 287–311. [Google Scholar]
- 49.Bogunovic D., O'Neill D.W., Belitskaya-Levy I., Vacic V., Yu Y.L., Adams S., Darvishian F., Berman R., Shapiro R., Pavlick A.C., Lonardi S., Zavadil J., Osman I., Bhardwaj N. Immune profile and mitotic index of metastatic melanoma lesions enhance clinical staging in predicting patient survival. Proc Natl Acad Sci U S A. 2009;106:20429–20434. doi: 10.1073/pnas.0905139106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weeraratna A.T., Jiang Y., Hostetter G., Rosenblatt K., Duray P., Bittner M., Trent J.M. Wnt5a signaling directly affects cell motility and invasion of metastatic melanoma. Cancer Cell. 2002;1:279–288. doi: 10.1016/s1535-6108(02)00045-4. [DOI] [PubMed] [Google Scholar]
- 51.Dissanayake S.K., Wade M., Johnson C.E., O'Connell M.P., Leotlela P.D., French A.D., Shah K.V., Hewitt K.J., Rosenthal D.T., Indig F.E., Jiang Y., Nickoloff B.J., Taub D.D., Trent J.M., Moon R.T., Bittner M., Weeraratna A.T. The Wnt5A/protein kinase C pathway mediates motility in melanoma cells via the inhibition of metastasis suppressors and initiation of an epithelial to mesenchymal transition. J Biol Chem. 2007;282:17259–17271. doi: 10.1074/jbc.M700075200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Murakami T., Toda S., Fujimoto M., Ohtsuki M., Byers H.R., Etoh T., Nakagawa H. Constitutive activation of Wnt/beta-catenin signaling pathway in migration-active melanoma cells: role of LEF-1 in melanoma with increased metastatic potential. Biochem Biophys Res Commun. 2001;288:8–15. doi: 10.1006/bbrc.2001.5719. [DOI] [PubMed] [Google Scholar]
- 53.Vaid M., Prasad R., Sun Q., Katiyar S.K. Silymarin targets b-catenin signaling in blocking migration/invasion of human melanoma cells. PLoS One. 2011;6:e23000. doi: 10.1371/journal.pone.0023000. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 54.Spink K.E., Fridman S.G., Weis W.I. Molecular mechanisms of β-catenin recognition by adenomatous polyposis coli revealed by the structure of an APC–β-catenin complex. EMBO J. 2001;20:6203–6212. doi: 10.1093/emboj/20.22.6203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kerosuo L., Bronner-Fraser M. What is bad in cancer is good in the embryo: importance of EMT in neural crest development. Semin Cell Dev Biol. 2012;23:320–332. doi: 10.1016/j.semcdb.2012.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tudrej K.B., Czepielewska E., Kozłowska-Wojciechowska M. Sox10-Mitf activity in melanoma cells. Arch Med Sci. 2017;13:1493–1503. doi: 10.5114/aoms.2016.60655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sarkar A., Hochedlinger K. The sox family of transcription factors: versatile regulators of stem and progenitor cell fate. Cell Stem Cell. 2013;12:15–30. doi: 10.1016/j.stem.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Graham V., Khudyakov J., Ellis P., Pevny L. SOX2 functions to maintain neural progenitor identity. Neuron. 2003;39:749–765. doi: 10.1016/s0896-6273(03)00497-5. [DOI] [PubMed] [Google Scholar]
- 59.Knoepfler P.S., Cheng P.F., Eisenman R.N. N-myc is essential during neurogenesis for the rapid expansion of progenitor cell populations and the inhibition of neuronal differentiation. Genes Dev. 2002;16:2699–2712. doi: 10.1101/gad.1021202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sawai S., Shimono A., Wakamatsu Y., Palmes C., Hanaoka K., Kondoh H. Defects of embryonic organogenesis resulting from targeted disruption of the N-myc gene in the mouse. Development. 1993;117:1445–1455. doi: 10.1242/dev.117.4.1445. [DOI] [PubMed] [Google Scholar]
- 61.Zinin N., Adameyko I., Wilhelm M., Fritz N., Uhlén P., Ernfors P., Henriksson M.A. MYC proteins promote neuronal differentiation by controlling the mode of progenitor cell division. EMBO Rep. 2014;15:383–391. doi: 10.1002/embr.201337424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bertrand N., Castro D.S., Guillemot F. Proneural genes and the specification of neural cell types. Nat Rev Neurosci. 2002;3:517–530. doi: 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- 63.Ross S.E., Greenberg M.E., Stiles C.D. Basic helix–loop–helix factors in cortical development. Neuron. 2002;39:13–25. doi: 10.1016/s0896-6273(03)00365-9. [DOI] [PubMed] [Google Scholar]
- 64.Grimm D., Bauer J., Wise W., Krüger M., Simonsen U., Wehland M., Infanger M., Corydon T.J. The role of SOX family members in solid tumours and metastasis. Sem Cancer Biol. 2020;67:122–153. doi: 10.1016/j.semcancer.2019.03.004. [DOI] [PubMed] [Google Scholar]
- 65.Takaishi M., Tarutani M., Takeda J., Sano S. Mesenchymal to epithelial transition induced by reprogramming factors attenuates the malignancy of cancer cells. PLoS One. 2016;11:e0156904. doi: 10.1371/journal.pone.0156904. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data supporting the findings from this article have been deposited as GSE149390 with the National Center for Biotechnology Information Gene Expression Omnibus (http://ncbi.nlm.nih.gov/geo) database.