Abstract
The safety of marketed drugs is an ongoing concern, with some of the more frequently prescribed medicines resulting in serious or life-threatening adverse effects in some patients. Safety-related information for approved drugs has been curated to include the assignment of toxicity class(es) based on their withdrawn status and/or black box warning information described on medicinal product labels. The ChEMBL resource contains a wide range of bioactivity data types, from early “Discovery” stage preclinical data for individual compounds through to postclinical data on marketed drugs; the inclusion of the curated drug safety data set within this framework can support a wide range of safety-related drug discovery questions. The curated drug safety data set will be made freely available through ChEMBL and updated in future database releases.
Introduction
ChEMBL (https://www.ebi.ac.uk/chembl) is a large-scale, open-access drug discovery resource containing information about bioactive molecules, their interaction with targets (e.g., molecular, cell- or tissue-based) and their biological effects.1,2 It broadly conforms to the FAIR data management principles (Findable, Accessible, Interoperable, and Reusable).3 ChEMBL (release 27) contains ∼13 000 approved drugs and drug candidates progressing through clinical trials, including manually curated information on many of their therapeutic targets and disease indications.2 It includes ∼1.9 million compounds with bioactivity data measured across a wide range of bioassays from individual protein interactions, through cell-, tissue-, or organ-based systems to whole animal models, as well as bioactivity data from large-scale toxicity data sets such as TG-GATES and DrugMatrix and other toxicity assays. As a result, ChEMBL provides a rich, high-quality resource for addressing a wide range of drug discovery-related questions.
The safety of marketed drugs to treat human disease is an ever-present concern, with some of our more frequently prescribed drugs resulting in serious or life-threatening adverse effects in a small number of cases. For example, anthracycline breast cancer treatments like doxorubicin may cause cardiotoxicity in up to 5% of patients,4,5 or the bipolar and epilepsy treatment valproic acid carries a dose-dependent risk of idiosyncratic hepatotoxicity.6 In some cases the risk is considered to outweigh the benefit so significantly that the drug has been withdrawn from the market.7
Medicinal product labels for approved drugs contain a rich amount of information that typically describes their efficacy, disease indications, target populations, drug–drug interactions, as well as adverse effects. However, the format of the available safety information differs between individual regulatory bodies. For example, safety information for United States Food and Drug Administration (FDA) drug approvals is contained within the Structural Product Labeling (SPL) standardized format.8 European Medicines Agency (EMA) regulated medicinal products contain adverse effect information in their ‘Summary of Product Characteristics’,9 while the Japanese Pharmaceuticals and Medical Devices Agency (PMDA) describe severe adverse events within the pink text description in the “Warnings” section on medicinal product labels (e.g., ref (10)).
Our work focused on the FDA medicinal product labels in the first instance because of the accessibility of the medicinal product labels described within their structured database. As background, the FDA has required submissions of medicinal product labels in an electronic form with standardized SPL data structures since 2005.8 More recently, the OpenFDA initiative has facilitated direct programmatic access to several public data sets, including the drug product label database.11 The database contains structured sections for each medicinal product label and is updated weekly. In addition, special database fields are annotated that assist in searching across standard terms like generic drug names or active ingredient(s). Any adverse event described on a medicinal product label is written in free text, although key phrases often have similar terms to that in medical vocabularies such as the MedDRA12 standardized medical terminology.8
FDA medicinal product labels can carry a boxed warning (also known as a black box warning) if the drug may cause a serious or life-threatening condition.13 A boxed warning is the most serious of the three adverse drug reaction sections that are described on FDA medicinal product labels (Boxed warning, Warnings and Precautions, and Adverse Reactions, in decreasing order of severity). An investigation of the three adverse drug reaction sections in FDA labels was performed by Wu et al.,14 who used data mining in combination with MedDRA to analyze their frequency, severity, and patterns. A data set of medicinal product labels for 200 FDA-approved drugs was used to develop text mining tools to annotate adverse events with MedDRA terms,15 and it was applied in deep learning architecture models.16 Trends in boxed warnings in medicinal product labels have been evaluated over time to see whether safety concerns could be predicted (e.g., refs (17 and 18)). However, the authors of this paper are not aware of any freely available resource that attempts to classify, at scale, the type of adverse effect described in boxed warnings on a per drug basis, as a means to facilitate an investigation of safety-related drug discovery questions.
A significant task has been undertaken to annotate serious or life-threatening safety-related information for approved drugs in ChEMBL. Toxicity class(es) have been assigned to approved drugs with boxed warning information described on medicinal product labels and to “withdrawn” drugs that have been approved but subsequently withdrawn from one, or more, markets in the world. Such curated toxicity information allows drugs that cause similarly reported toxicities to be easily grouped, analyzed, and visualized.
The scope of application for the annotated drug safety data set is broad and could be used to answer a wide range of safety-related drug discovery questions, especially given the unique capability of ChEMBL, which includes bioactivity data from early stage, preclinical dose−response data for individual compounds through to safety annotation of postclinical marketed drugs. For example, the curated drug safety data set could be used to predict potential toxicities for small molecules using Quantitative Structure–Activity Relationship (QSAR) or other machine-learning approaches. QSAR approaches have been widely used to predict numerous compound properties based on descriptors derived from the chemical structure and require a training data set of known outcomes (e.g., refs (19 and 20)). Overviews of relevant machine learning approaches to predict drug toxicity are available at, for example, refs (21 and 22) and can include adverse effect models for specific disease areas such as a drug-induced liver injury (e.g., ref (23)), together with broader predictions of adverse drug reactions (e.g., ref (24)). There is much value for data users to be able to access well-organized, clearly annotated information. To encourage usage the data set has initially been made available as flat files for download and will be included in the next release of ChEMBL.
There are three main parts to the paper:
First, the automated method to extract boxed warning descriptions for medicinal products that contain drugs that are described in ChEMBL is presented. The use of a script to perform this process allows the boxed warnings to be updated for future releases of ChEMBL in a straightforward manner. For example, the safety information will need periodic updating as new medicinal products are marketed, or if additional safety concerns are provided in boxed warning descriptions.
Second, a text classifier tool has been applied to assign toxicity class(es) to each boxed warning description. This required manual annotation of a representative subset of boxed warning descriptions with one (or more) classes of toxicity. The manually annotated boxed warning descriptions were used as the input data set to train a text classifier model across 17 toxicity classes. The trained classification model has been applied as part of the automated script; it annotates toxicity classes for the complete set of boxed warning descriptions. A separate task to manually annotate toxicity classes for drugs that have been withdrawn from the market had previously been completed.2 The overall curated drug safety data set comprises toxicity classes for drugs with boxed warnings, along with those for withdrawn drugs. To encourage use, the curated data set and its toxicity classification has been made freely available via ChEMBL. Example boxed warning descriptions have been retained to allow database users to drill down through the information “audit trail” to examine the source information.
Third, as a means to explore the curated drug safety data set, toxicity classes for drugs were compared with their quasi-equivalent therapeutic indications, and some illustrative drugs and their toxicity classification(s) are discussed.
Materials and Methods
Extraction of Boxed Warning Descriptions for U.S. Drugs
Boxed warning information is described within a black rectangle on a medicinal package insert, and the label descriptions are stored in the FDA’s Structured Product Label database. An example boxed warning for a medicinal product containing the active ingredient Oxaprozin that causes serious cardiovascular and gastrointestinal events can be viewed in refs (25 and 26). A medicinal product described in the SPL database may contain one or more active ingredient(s) that are often approved drug(s). One active ingredient can be present in multiple medicinal products due to the differences in regulatory applications, dosage forms, routes of administration, manufacturers, etc. The label for each medicinal product is assigned a unique identifier by the FDA (set id) that is stable across all versions or revisions. There are typically up to tens to hundreds of current medicinal products that contain a given active ingredient of interest, so to annotate the toxicity class(es) for an individual drug required examination of each medicinal product label and extraction and annotation of any boxed warning description. Although often fairly similar in wording, the boxed warning descriptions are not identical across different medicinal product labels, and therefore it is not possible to simply remove duplicate boxed warning descriptions to simplify the task. There are ∼8000 single ingredient and combination medicinal product labels for approved drugs in ChEMBL with boxed warnings described in the SPL database and an “effective date” prior to December 2019. It is noted that the SPL database is regularly updated as new medicinal products are approved, existing products are revised, and other medicinal products are discontinued (for a variety of different reasons that include manufacturing or other concerns such as loss of quality as well as safety or efficacy concerns).
The workflow to extract a boxed warning described on a medicinal product label for each approved drug in ChEMBL and assign its toxicity class is presented below (and in Figure 1).
The ‘substance_name’ field of medicinal product labels in the OpenFDA’s SPL database11 was searched using an exact match to the preferred name, or synonym, of each drug described in ChEMBL. This was performed for each drug form within a drug family. Note that ChEMBL uses a hierarchy of compounds whereby any specific drug form belongs to a family of compound structures that contains one parent (salt-stripped compound) and one or more salts.27 The ‘substance_name’ field was annotated by OpenFDA in the SPL database and is defined as “the list of active ingredients of a drug product”.
Assuming that one or more medicinal products containing a specified drug form could be identified within the SPL database, a further query checked whether a boxed warning field exists for each medicinal product, and if present, then the textual description of the boxed warning was extracted along with associated information for the FDA application number, FDA set id, FDA annotated substance name(s), and the date stamp of the medicinal product label (“effective time”). The presence of a single active ingredient within a medicinal product allows the direct assignment of a boxed warning description to an individual drug form. By contrast, the presence of two or more active ingredients within a medicinal product (a combination medicinal product) and a boxed warning description means that a boxed warning cannot be directly assigned to an individual drug form, but it may be possible subsequently to deconvolute the boxed warning descriptive signal if additional information is available from other medicinal product labels with different combinations of active ingredients. Typically, the boxed warning description for a combination medicinal product contains a portion of the boxed warning text for each active ingredient. For example, a combination medicinal product with a boxed warning described as “WARNING: HYPERSENSITIVITY REACTIONS AND EXACERBATIONS OF HEPATITIS B” relates to a warning of hypersensitivity reactions due to lamivudine and hepatitis B exacerbations due to abacavir sulfate, and it has been annotated with immune system toxicity and hepatotoxicity.28 As a result, the information for combination medicinal products has also been extracted and stored by the workflow.
Figure 1.
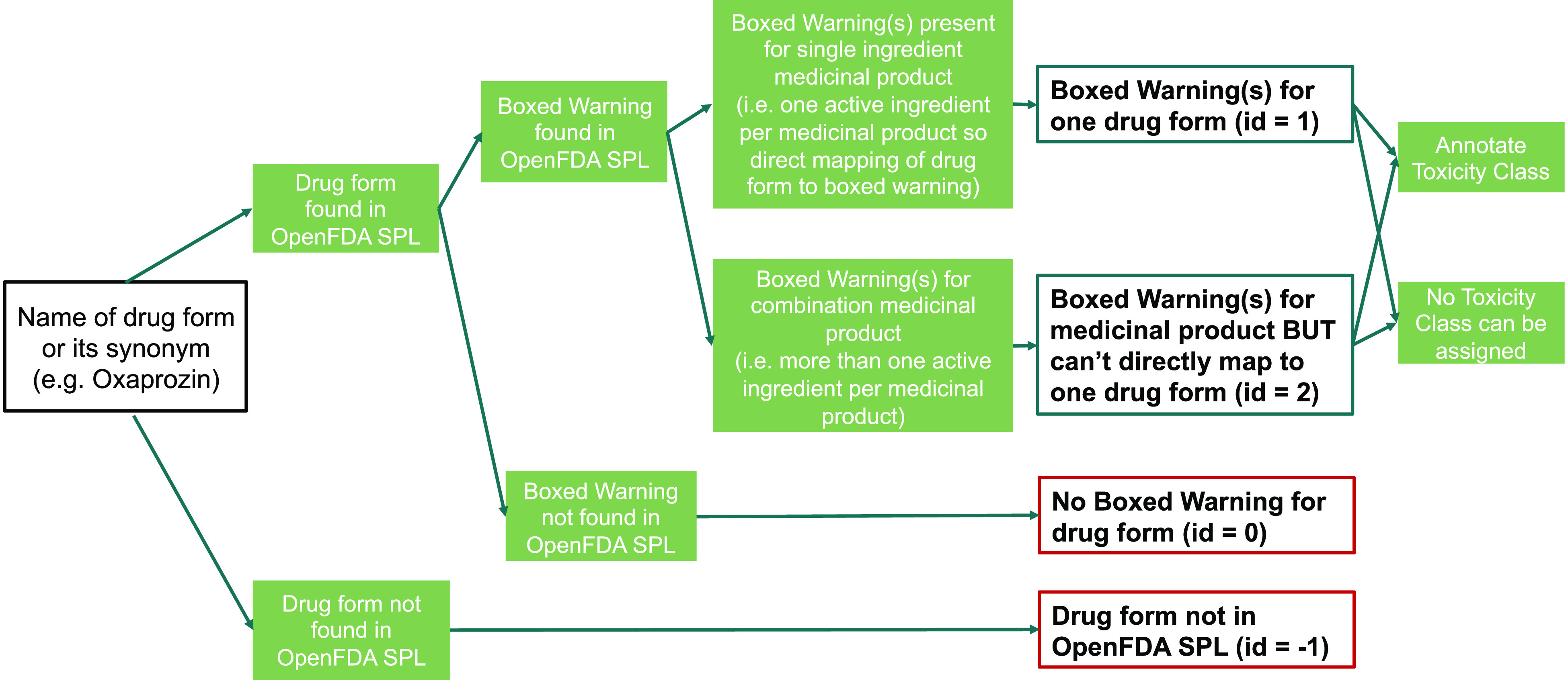
Workflow to extract boxed warning descriptions from medicinal product label(s) for approved drugs and assign one or more toxicity class(es). Different drug forms within one compound family would have their boxed warnings independently extracted using an exact name (or synonym) match (e.g., the parent drug pentazocine and its salt pentazocine hydrochloride are active ingredients in different combination medicinal products even if they were subsequently annotated with identical toxicity classes (for respiratory toxicity and misuse29,30). Note that a third drug is also matched as an active ingredient within each of these medicinal products (naloxone hydrochloride). However, none of the three drugs have been identified within single-ingredient medicinal products, so a potential deconvolution of the boxed warning information assigned to any one drug is not possible without additional safety information. A boxed warning description in the SPL database is matched to the prodrug capecitabin, although no boxed warning is extracted for its biologically active drug form fluorouracil. However, the capecitabine boxed warning describes a drug–drug interaction with warfarin and as a result is not assigned a toxicity class31 (see the next section for more detail).
The automated script extracts medicinal product labels with boxed warning information from the SPL database within a specified date range so that any new information can be periodically updated as part of each ChEMBL release cycle. Each boxed warning description is annotated with one or more toxicity class(es) (see sections below), and the script takes into account temporal changes to the boxed warning information. For example, if a drug form with medicinal product labels in the SPL database previously did not have any boxed warning (‘note_id’ = 0) but a new single ingredient medicinal product label includes a boxed warning description, then the script captures the new boxed warning information and its toxicity class(es) and amends the ‘note_id’ for the drug form to be equal to 1. Similarly, if a drug form has no medicinal product labels in the SPL database within a date range (note_id = −1) but a subsequent search with a later data range shows the presence of a medicinal product label, then the script updates the information (and sets ‘note_id’ to be equal to 0). Equally, if more recent single-ingredient or combination medicinal products are available for a drug form, then the boxed warning descriptions and their toxicity class(es) are captured and appended to the existing information.
Building the Manually Annotated Input Data Set Required for the Text Classifier Model
A representative subset of 3021 boxed warning descriptions was chosen by selecting one label per drug form per publication year for single-ingredient labels and for combination labels, with selected additional labels that represent boxed warnings that could not be assigned a toxicity class (see more detail below). The toxicity annotation of these labels was created by reading the boxed warning description, manually mapping key phrases that describe toxicity caused by the drug to terms in the MedDRA standardized medical terminology,12 and assigning a toxicity class. The manually annotated labels with their associated toxicity class(es) are provided in the Supporting Information. Seventeen toxicity classes were assigned, with class names that are based on the primary MedDRA System Organ Class (SOC). Note that MedDRA allocates only one primary SOC to each specific Lowest Level Term (LLT) even if the term is mapped to multiple SOC terms, and as a result, a key phrase described in boxed warning text can only be assigned to one toxicity class. For example, the key phrase “Cardiopulmonary arrest” described within a boxed warning description has been mapped to the primary MedDRA SOC term “Cardiac Disorders” (10007541, and not to the secondary MedDRA SOC term ‘Respiratory, Thoracic, and Mediastinal Disorders’), and therefore the boxed warning label can be annotated as “cardiotoxicity”. Similarly, the phrase ‘nephrogenic systemic fibrosis’ has been mapped to the primary MedDRA SOC term ‘Skin and Subcutaneous Tissue Disorders’, and the boxed warning has been annotated with a toxicity class of “dermatological toxicity”, even though secondary MedDRA SOC terms are available for ‘Immune System Disorders’, ‘Musculoskeletal and Connective Tissue Disorders’ and ‘Renal and Urinary Disorders’. The list of annotated toxicity classes is presented in Table 1, with some key phrase examples.
Table 1. Annotated Toxicity Classes with Disease Examples and Equivalent MedDRA SOC Categories.
| toxicity class | examples of key phrases mapping boxed warning text to MedDRA SOCa | MedDRA SOC code | MedDRA SOC name |
|---|---|---|---|
| Carcinogenicity | Neoplasm, Lymphoma, Leukemia | 10029104 | Neoplasms benign, malignant and unspecified (incl cysts and polyps) |
| Cardiotoxicity | Cardiac arrhythmia, Heart failure, myocardial disorders | 10007541 | Cardiac disorders |
| Dermatological toxicity | Dermatitis, Stevens-Johnson syndrome, Urticaria | 10040785 | Skin and subcutaneous tissue disorders |
| Gastrotoxicity | Gastric perforation, Colitis, Inflammatory Bowel Disease | 10017947 | Gastrointestinal disorders |
| Hematological toxicity | Thrombocytopenia, Hemorrhagic disorder, Neutropenia | 10005329 | Blood and lymphatic system disorders |
| Hepatotoxicity | Cholestasis, Liver injury, Hepatitis, Jaundice | 10019805 | Hepatobiliary disorders |
| Immune system toxicity | Allergy, Anaphylactic shock, Graft versus host disease | 10021428 | Immune system disorders |
| Infections | Clostridium difficile associated diarrhea, Pseudomembranous colitis, (Serious) infection, Gangrene, Pneumonia Increased susceptibility to infection | 10021881 | Infections and infestations |
| (Energy) Metabolism toxicity | Lactic acidosis, Obesity, Hypochloremia, Polydipsia, Hyperkalemia, Diabetes mellitus (Type I and II), Hyperglycemia, Hypoglycemia, Hyperlipidaemia, Gout, Vitamin C deficiency | 10027433 | Metabolism and nutrition disorders |
| Misuse | Accidental Poisoning, Drug misuse, Overdose | 10022117 | Injury, poisoning and procedural complications |
| Musculoskeletal toxicity | Scleroderma, Reynold’s syndrome, Lupus erythematosus, Allergic arthritis, Osteoarthritis, Rheumatoid arthritis, Ankylosing spondylitis, Rhabdomyolysis | 10028395 | Musculoskeletal and connective tissue disorders |
| Nephrotoxicity | Urogenital disorder, Nephritis, Renal failure, Injury to Kidney, Nephrotoxicity | 10038359 | Renal and urinary disorders |
| Neurotoxicity | Stroke, Multiple sclerosis, Encephalopathy, Dementia, Alzheimer’s disease, Amnesia, Dyskinesia, Parkinson’s disease, Tremor, Convulsions, Guillain Barre syndrome | 10029205 | Nervous system disorders |
| Psychiatric toxicity | Anxiety, Depression, Schizophrenia, Sleep disorder, Suicide Ideation, Suicide attempt, Drug Dependence | 10037175 | Psychiatric disorders |
| Respiratory toxicity | Bronchospasm, Pulmonary toxicity, Pulmonary Hypertension, Pulmonary embolism, Asthma | 10038738 | Respiratory, thoracic and mediastinal disorders |
| Teratogenicity | Birth defects, Teratogenicity, Fetal toxicity | 10010331 | Congenital, familial and genetic disorders |
| Vascular toxicity | Hypertension, Hypotension, Thrombosis, Thromboembolism, Bleeding Risk | 10047065 | Vascular disorders |
Key phrases described in boxed warning text are checked against the primary MedDRA SOC and therefore can only be mapped to one toxicity class.
In some cases, a toxicity class was not manually assigned because the boxed warning description does not demonstrate that there is a direct link between the drug form and the adverse effect in all cases. For example, drug–drug interactions, or adverse effects that only apply to a subpopulation of patients, were not assigned a toxicity class. For example, boxed warnings for ritonavir (e.g., ref (32)) or ergotamine tartrate (e.g., ref (33)) that describe serious or life-threatening drug–drug interactions have not been assigned a toxicity class because the boxed warning cannot be directly ascribed to an individual drug. Equally if the adverse effect that is described in the boxed warning is only observed in a small subpopulation of patients then these have not been assigned a toxicity class. For example, a boxed warning for the schizophrenia drug, paliperidone, states “WARNING: INCREASED MORTALITY IN ELDERLY PATIENTS WITH DEMENTIA-RELATED PSYCHOSIS” and has not been assigned a toxicity class.34
In addition, boxed warning text that does not directly relate to a severe or life-threatening adverse reaction were labeled as such. In some cases, descriptive text delineated with a black box on the medicinal product label has been included as a boxed warning on a medicinal product label. For example, literal boxed warning descriptions such as ‘NOT FOR INTRATHECAL USE’,35 ‘BOXED WARNING:’,36 ‘WARNING PHYSICIANS SHOULD COMPLETELY FAMILIARIZE THEMSELVES WITH THE COMPLETE CONTENTS OF THIS LEAFLET BEFORE PRESCRIBING PRIMAQUINE PHOSPHATE’,37 or ‘WARNING BREVITAL should be used only in hospital or ambulatory care settings that provide for continuous monitoring···’ have not been assigned a toxicity classification.38
Binary Text Classification Models for Toxicity Annotation
For each medicinal product with a boxed warning, the textual description was extracted and annotated with a toxicity class. This was performed using a Natural Language Processing (NLP) binary toxicity classification approach, applying the SpaCy39 tool with the input data set of 3021 medicinal product labels containing a boxed warning description and one (or more) manually annotated toxicity classes (see previous section). An NLP text classification approach was chosen because simpler approaches such as regular expression text pattern matches were found to perform insufficiently well; ∼10% of the boxed warnings were assigned incorrect annotations (e.g., text describing patients with a liver transplant were incorrectly annotated with hepatotoxicity40). The boxed warning descriptions are free text, and although some are relatively short (mean length of 2556 characters for the extracted descriptions, e.g., ref (41)), other descriptions have substantial length and complexity (up to ∼17 000 characters) and can include concatenated descriptions for each active ingredient within a combination medicinal product (e.g., ref (42)). We found that the complexity and length of the boxed warning descriptions meant that an approach based on matching to regular expression text patterns did not deliver a curated data set with annotated toxicity classes of sufficient accuracy. As a result, the NLP text classification approach was explored and found to give improved performance (see Results and Discussion).
The manually annotated set of boxed warning descriptions was used as input to construct the binary toxicity classification models (Figure 2). For each toxicity class, the boxed warning descriptions were divided into a set of labels for model training and testing (∼66% of the positively annotated boxed warning descriptions per toxicity class, Table 2) and a validation set of labels. It was noted that the boxed warning descriptive text is relatively similar for medicinal products that contain the same active ingredient(s), and some active ingredients are described on many medicinal products labels over many years, resulting in unequal numbers of annotated boxed warning descriptions per drug form within the manually annotated labels. As a result, at least one boxed warning per drug form was randomly chosen from the single-ingredient medicinal product labels (or the combination product labels) for model training and testing. This approach gave better model performance than a purely random approach, probably because of the more comprehensive representation of the variety of boxed warning descriptions across the boxed warning space.
Figure 2.
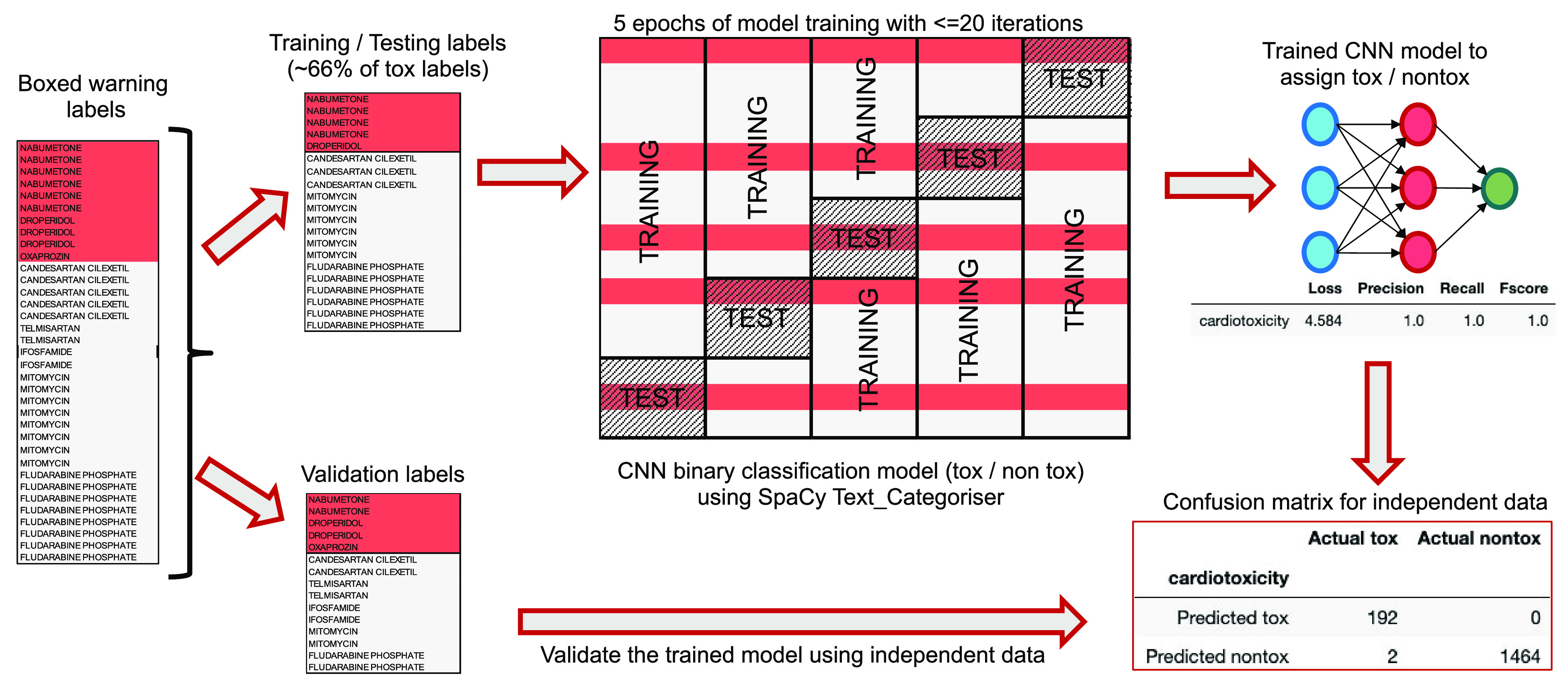
Training the NLP toxicity classification model: an example for cardiotoxicity using a training/testing set of manually annotated medicinal product labels with boxed warning descriptions. Note that the NLP model was performed for each toxicity class described in Table 1.
Table 2. Toxicity Annotation Model Statisticsa.
| Toxicity class | Total number of positively annotated tox labelsb | Number of tox labels in train/test modelc | Number of drug forms in train/test modeld | Loss | Precision | Sensitivity | F1-Score |
|---|---|---|---|---|---|---|---|
| Carcinogenicity | 287 | 154 | 75 | 0.576 | 1 | 1 | 1 |
| Cardiotoxicity | 482 | 316 | 119 | 4.584 | 1 | 1 | 1 |
| Dermatological toxicity | 77 | 46 | 27 | 0.011 | 1 | 1 | 1 |
| Gastrotoxicity | 317 | 208 | 65 | 0.881 | 0.99 | 0.99 | 0.99 |
| Hematological toxicity | 195 | 136 | 61 | 0.954 | 1 | 1 | 1 |
| Hepatotoxicity | 301 | 209 | 78 | 0.977 | 0.99 | 0.99 | 0.99 |
| Immune system toxicity | 136 | 98 | 53 | 7.137 | 0.98 | 0.98 | 0.98 |
| Infections | 97 | 71 | 39 | 0.973 | 0.99 | 0.99 | 0.99 |
| (Energy) Metabolism toxicitye | 130 | 88 | 33 | 0.016 | 1 | 1 | 1 |
| Misusef | 383 | 256 | 132 | 3.599 | 0.99 | 0.99 | 0.99 |
| Musculoskeletal toxicity | 85 | 60 | 14 | 0.038 | 1 | 1 | 1 |
| Nephrotoxicity | 79 | 58 | 24 | 0.874 | 0.99 | 0.99 | 0.99 |
| Neurotoxicity | 394 | 252 | 94 | 0.888 | 0.99 | 0.99 | 0.99 |
| Psychiatric toxicityf | 274 | 157 | 62 | 0.8 | 1 | 1 | 1 |
| Respiratory toxicity | 291 | 192 | 95 | 2.799 | 0.98 | 0.98 | 0.98 |
| Teratogenicity | 357 | 246 | 78 | 0.898 | 1 | 1 | 1 |
| Vascular toxicity | 236 | 151 | 86 | 0.701 | 0.98 | 0.98 | 0.98 |
The performance statistics are given for the trained model; see definitions in Abbreviations.
The total number of positively annotated toxicity labels for each toxicity class. These labels were then divided into a set of training and testing labels and a validation set of labels.
The number of positively annotated toxicity labels used in the model training and testing. At least one positively annotated toxicity label per active ingredient was selected for the model training and testing, since this represents the broadest diversity of boxed warning text descriptions.
The number of drug forms (i.e., active ingredients) for positively annotated toxicity labels used in the model training and testing. Examples of diseases for each toxicity class are given in Table 1.
The class of “metabolism toxicity” is toxicity due to energy metabolism processes.
Convolutional neural network (CNN) model training using the TextCategorizer function of SpaCy39 (version 2) was performed on the training/testing labels over five epochs where the whole training/testing data set was seen by the CNN model. The TextCategorizer function assigns one label to each “document” (in this case a description of a boxed warning) with the simple CNN model where token vectors are mean pooled and used as features in a feed-forward network.39 As a result, the importance of specific words or phrases within the boxed warning description cannot be individually deconvoluted from the overall document. Other SpaCy parameters were set to their default values, which gave the desired level of model performance, so further examination of a range of model parameters was not performed. Within each epoch, the network weights were optimized iteratively using default parameters. The batch size was initially set to 1 and increased to a maximum size of 64 using the optimization function recommended in the documentation.43 To prevent overtraining, the training iteration was stopped when the F1-score is constant over the three previous iterations (F1-score = 2*TP/(2*TP + FP + FN)) (where TP indicates true positive counts, FP indicates false positive counts, and FN indicates false negative counts) and the loss is less than 1, or 20 iterations of the model training had been performed. The loss applied in the SpaCy TextCategorizer function uses multilabel log loss where the logistic function is applied to each neuron in the output layer independently.44 The small BioMedical SciSpaCy NLP model45 was used initially (version 0.2.3), and the optimized CNN model was saved at the end of each epoch, before being read in for the start of the next epoch. The model performance statistics are presented in Table 2.
The trained binary toxicity classification models were applied to annotate toxicity class(es) for the complete set of boxed warning descriptions, and the annotated data set has been made available via the ChEMBL resource (see section on ‘Access to the curated data set’).
The toxicity annotation was not performed for medicinal product labels that suggest endocrine toxicity, ophthalmic toxicity, ototoxicity, and reproductive system toxicity because the manually annotated input data set of boxed warning labels had sparse positive annotation of toxicity (with less than 50 toxicity labels per toxicity class out of the manually annotated boxed warning label input data set). These toxicity classes may be included in the future if sufficient manually annotated boxed warning labels are available.
To assist users in cases of uncertainty or a potential misclassified toxicity class, full descriptions of selected boxed warnings were flagged and exposed in the curated data set in order to maintain the information “audit trail”. Therefore, one exemplar boxed warning description has been randomly flagged per drug form per toxicity class per year.
Toxicity Classification for Withdrawn Drugs
Withdrawn drugs were manually assigned a toxicity class using the same toxicity classification that has been applied to drugs with a boxed warning (Figure 3). A withdrawn drug is an approved drug contained in a medicinal product that subsequently had been removed from the market. The reasons for withdrawal may include toxicity, lack of efficacy, or other reasons such as an unfavorable risk-to-benefit ratio following approval and marketing of the drug. ChEMBL considers an approved drug to be withdrawn only if all medicinal products that contain the drug as an active ingredient have been withdrawn from one (or more) regions of the world. Note that all medicinal products for a drug can be withdrawn in one region of the world while still being marketed in other jurisdictions. The manually assigned toxicity class was based on the reason(s) for withdrawal that had previously been manually curated in ChEMBL,2 typically citing information described in refs (46−48).
Figure 3.
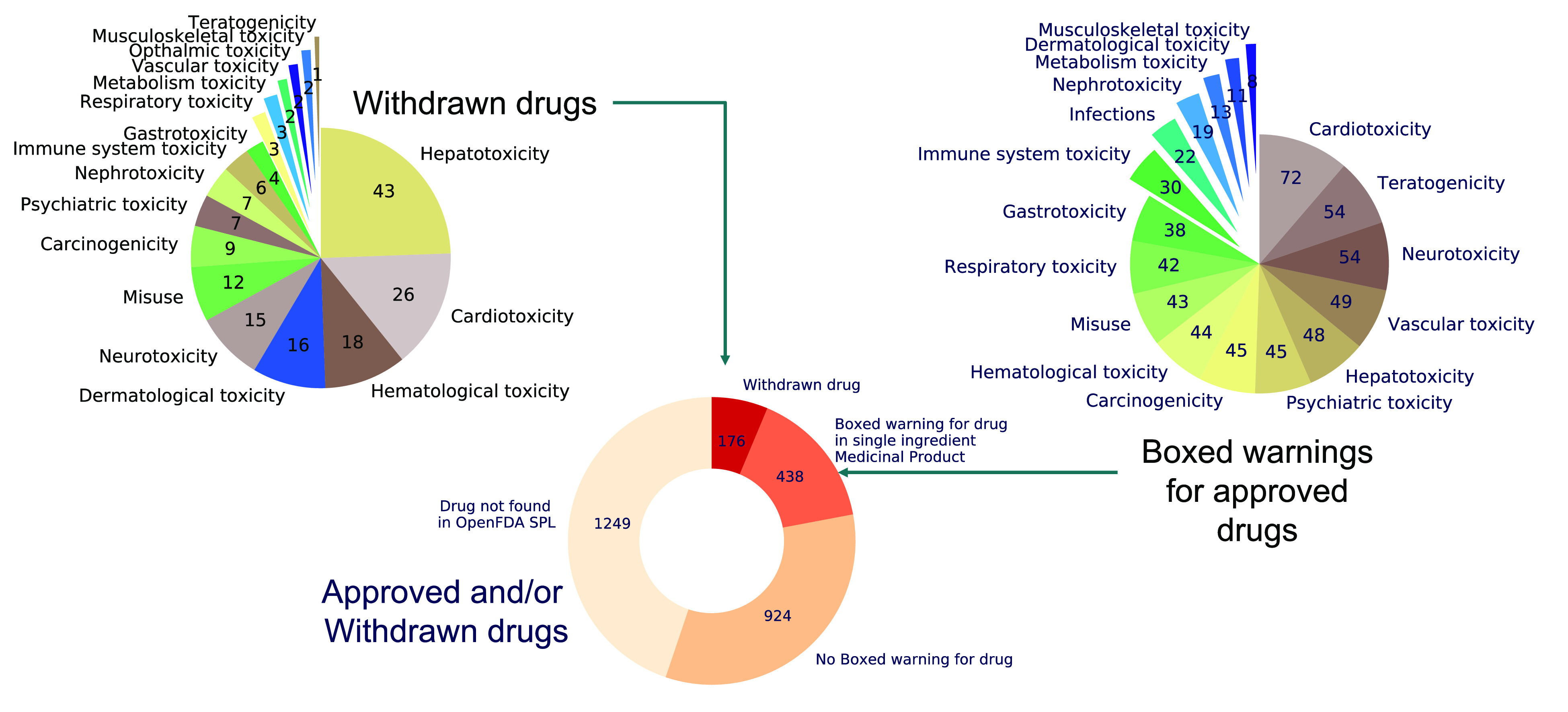
Summary of the curated toxicity data set for approved parent drugs in ChEMBL.
Comparison of Assigned Toxicity Classes and Therapeutic Indications for Drugs
We were interested to explore to what extent the adverse effect of an individual drug would be in the same or a different class to its quasi-equivalent therapeutic indication. Therefore, parent drugs in single-ingredient medicinal products for each toxicity class in the curated drug safety data set were compared against the list of approved drugs with the quasi-equivalent disease indications from the ChEMBL database (Figure 4). For example, parent drugs with boxed warnings assigned as hepatotoxic were compared against drugs with therapeutic indications for Liver Diseases (Medical Subject Heading thesaurus, MeSH49 tree number: C06.552 as described in ChEMBL), or the parent drugs with boxed warnings assigned as neurotoxic were compared to drugs with therapeutic indications for Nervous System Diseases (MeSH tree number: C10 as described in ChEMBL). The mapping table between the quasi-equivalent toxicity class and therapeutic indication is provided in the Supporting Information, along with a list of approved drugs in ChEMBL that have a therapeutic indication and/or toxicity class.
Figure 4.
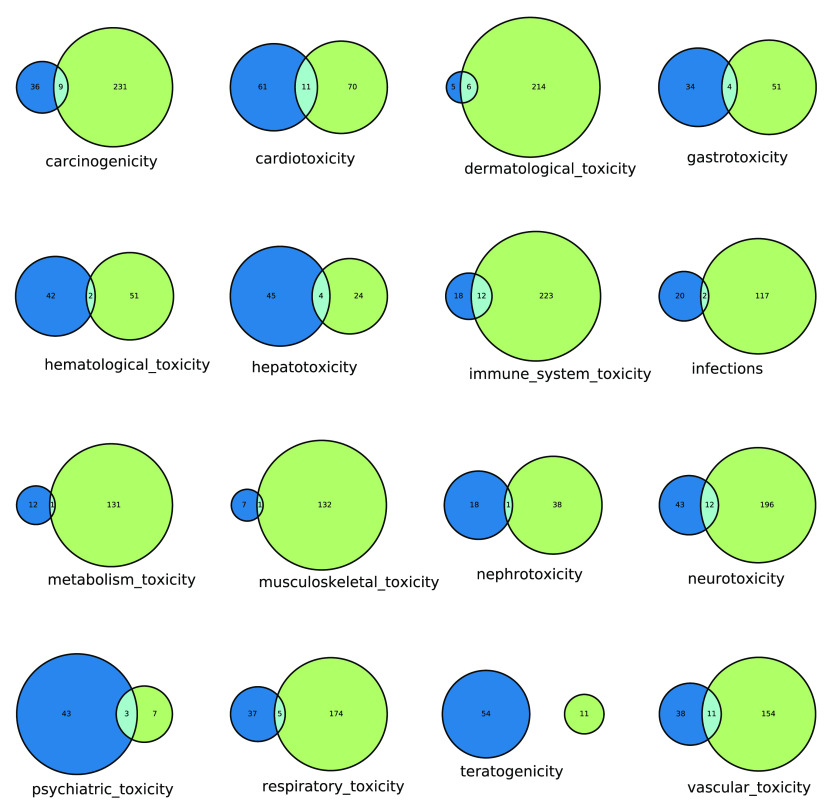
Comparison of the number of drugs that have been assigned a similar therapeutic disease indication and/or toxicity classification. The blue (left-hand circles) indicate the number of drugs in single-ingredient medicinal products that carry a boxed warning with an assigned toxicity class, while the green (right-hand circles) indicate the number of approved drugs in ChEMBL with therapeutic disease indications in quasi-equivalent classes.
In addition to the direct manual inspection of the curated drug safety data set (see Results and Discussion), this comparison of toxicity classes and therapeutic indications also provides a useful way to assess drugs within the curated drug safety data set where both the toxicity and therapeutic effect are aligned. Therefore, for each toxicity class, the boxed warning descriptions for all drugs in the intersection of the Venn diagram (Figure 4) were examined in detail to check that the assigned toxicity classification was consistent with a quasi-equivalent therapeutic class.
Results and Discussion
First, the assignment of toxicity class(es) to each boxed warning description using the NLP text classification models is discussed, followed by a summary of the overall curated drug safety data set, which comprises toxicity classes for drugs with boxed warnings, along with those for withdrawn drugs. Second, the toxicity classes are explored by comparison with their quasi-equivalent therapeutic indications.
Binary Text Classification Model Performance
Each boxed warning description was assigned one (or more) toxicity class(es) using the NLP text classification models. The performance of the trained model is summarized in Table 2. Each trained model was also validated against manually labeled data that had not been used in the model training and testing. The resulting confusion matrices typically showed low numbers of false positive and false negative results; an example for cardiotoxicity is given in Figure 2, with the validation performance statistics in the Supporting Information. Very good model performance was observed across all toxicity classes and was considered to be a result of:
the significant manual effort to annotate 3021 medicinal product labels with a boxed warning across all 17 toxicity classes, which represents ∼38% of the total medicinal product labels extracted. The manually annotated labels were applied in the model training and testing. During the course of the work, significant care was taken to identify boxed warning descriptions with incorrectly predicted toxicity classes and to re-examine and correct manually annotated training/testing labels as required, before rerunning the updated NLP text classification model for the toxicity class under consideration.
a relatively high similarity of boxed warning text for individual active ingredients, which facilitates good NLP model performance, although the complexity of the boxed warning description means that the NLP model approaches significantly outperform simple regular expression text pattern matching. Typically, the boxed warning text for a single-ingredient medicinal product with a later date is very similar to an earlier single-ingredient medicinal product containing the same drug, with a slight rewording of individual sentences, or differences in spaces, commas, or other punctuation, which suggests that the authors often reuse existing text, writing an updated description based on existing knowledge. In addition, combination medicinal product labels often use a concatenation of descriptive phrases for the boxed warning that are very similar to relevant single-ingredient medicinal product labels. The similarity of text descriptions from different boxed warning labels that describe the same drug form lends itself to the NLP text classification model approach to result in the correct assignment of a specified toxicity class(es) in most cases.
The impact of similar boxed warning descriptions for one drug form in both the model training/test data and the validation data was examined by excluding all drug forms from the training/test data if they were present in validation data and rerunning the binary text classification model training (results shown in the Supporting Information). It was concluded that the assignment of a toxicity class performs reasonably well for unseen descriptions of boxed warnings, but there is a significant improvement to the correct assignment of toxicity/nontoxicity when the text classifier model has been trained on labels with very similar wording (e.g., sensitivity of 0.88 for fully independent validation labels for the hepatotoxicity text classification model vs 0.99 for validation labels that include a similar wording from other labels for the same drug seen by the trained model). By contrast, the text classification tool performs particularly well to distinguish text from a boxed warning that did not relate to the toxicity class under examination (e.g., specificity of 1 when assigning a nonhepatotoxic label for a binary hepatotoxic/nonhepatotoxic classification model for both independent and normal validation labels). In an ideal world, the final curated data set would have 100% accuracy of annotated toxicity classes, and therefore the approach to include validation labels that contain similar wording to those that the text classification model was trained on is considered to be appropriate because it results in higher accuracy.
Overall, the NLP text classification models provide a method to assign toxicity classes to the boxed warning text with good performance. The automated approach provides a high-quality annotation of a large number of boxed warnings descriptions (∼8000) that would not be viable to manually curate without significant effort on a regular basis. In addition, an automated process minimizes potential human errors such as those that can occur in text transcription. Looking forward, it is clear that, to maintain the good performance of the text classification models, the manually annotated input data set of labels will need to be updated as new medicinal product labels are produced. This will be particularly important for active ingredients that have not previously had a boxed warning, especially if they are not chemically similar to a drug with a current boxed warning, which may manifest in significantly different adverse effects to those described in existing boxed warnings.
The Curated Data Set of Toxicity Class(es) for Drugs with Boxed Warnings, along with Those for Withdrawn Drugs
A summary of the toxicity classes assigned to approved drugs with boxed warnings is presented in Figure 3, and the annotated data set has been made available via the ChEMBL resource (see section on ‘Access to the curated data set’). Of the 2715 approved parent drugs described in ChEMBL, there are 438 approved drugs with one or more boxed warnings for single-ingredient medicinal products, 102 drugs with one or more boxed warnings for combination medicinal products containing the active ingredient and other ingredients, that is, where the boxed warning cannot be unambiguously assigned to a specific drug, and 924 approved drugs with no boxed warning described in the FDA’s SPL database. Most of the 8053 extracted medicinal product labels that carry a boxed warning refer to single ingredients (7084 labels), and therefore the boxed warning can be directly assigned to an individual approved drug. The remaining 969 labels are for combination medicinal products that refer to one or more active ingredients.
There are 10 withdrawn drugs that also have a boxed warning for a single-ingredient medicinal product (bromfenac, celecoxib, gemtuzumab ozogamicin, methamphetamine, oxycodone, potassium chloride, rosiglitazone, thioridazine, tolcapone, and triazolam). For example, rosiglitazone (CHEMBL121) has been withdrawn from the European Union for cardiotoxicity, but single-ingredient medicinal products containing this drug continue to be marketed in other regions of the world and carry a boxed warning in the SPL database (with a cardiotoxicity annotation). 1249 approved drugs described in ChEMBL were not found in the SPL database, typically because they are not marketed in the United States. Most of the 438 marketed parent drugs with a boxed warning have one (or more) annotated therapeutic target(s) and indication(s) recorded in ChEMBL: 411 parent drugs have at least one annotated target, and 364 have at least one annotated indication, with 357 having both annotated target(s) and indications(s).
A medicinal product label with a boxed warning description may have one, or multiple, toxicity annotations, and a drug form may occur in many different medicinal products leading to the extraction of boxed warning descriptions in multiple medicinal products in some cases. Typically, if a boxed warning is present, there is a median of three single-ingredient product labels per drug form, although there may be up to several hundred labels for different single-ingredient product labels containing the same drug form. For example, lisinopril (a high blood pressure medication) is the active substance in 209 product labels with a typical boxed warning for fetal toxicity (148 single-ingredient medicinal products and 61 combination medicinal products), while bupropion hydrochloride (a smoking cessation aid) is the active ingredient in 190 single-ingredient medicinal product labels (and 4 combination medicinal product labels) that describe a typical boxed warning for suicidal thoughts and behaviors.
Any boxed warnings for prodrugs have been approached in a similar manner to other drug forms, that is, by an exact match of the name of the (pro) drug form, or its synonym, to the SPL database. However, Figure 3 does not aggregate different (pro) drug forms because ChEMBL does not currently consider the inactive prodrug form and its biologically active drug form within its hierarchy of compound families.
Comparison of Assigned Toxicity Classes and Therapeutic Indications for Drugs
The comparison of adverse effect class for each individual drug (using the toxicity class(es) assigned by our work) and their therapeutic indication (as described in ChEMBL) is presented in Figure 4. For each class in the toxicity classification, there is little overlap in the number of drugs that have a quasi-equivalent therapeutic indication and a boxed warning with an assigned toxicity class, as would be expected when any toxic side effects are distinct from those driving the therapeutic benefit. This suggests that the target(s) and biological mechanisms responsible for the toxicity are different than those driving the therapeutic benefit, which is a useful observation given that there is often little mechanistic evidence to explain off-target effects. However, there are some exceptions where both the toxicity and therapeutic effect are aligned, and for these cases, the boxed warnings were examined in detail. For example, it was observed that some drugs provide therapeutic benefit within a certain dose range but may cause adverse effects at higher doses due to exaggerated pharmacology at the therapeutic target:
antiarrhythmia drugs such as amiodarone and quinidine may exhibit paradoxical pro-arrhythmic effects at supra-therapeutic doses, for example, refs (50 and 51) and carry a boxed warning assigned as cardiotoxicity. Equally, the beta-blocker Metoprolol has a phase IV therapeutic indication for cardiovascular diseases, angina pectoris, myocardial infarction, hypertension, and heart failure but a cardiotoxicity warning for ischemic heart disease following abrupt cessation of the therapy.
anticoagulants like Warfarin carry a boxed warning assigned as vascular toxicity due to their potential risk of causing major or fatal bleeding.52
long-acting beta2 adrenergic agonists, such as Salmeterol xinafoate or Indacaterol maleate, typically have a therapeutic indication for obstructive lung diseases or chronic bronchitis but also carry a boxed warning for increased risk of asthma-related death and have been assigned a respiratory toxicity class.
Access to the Curated Data Set
ChEMBL provides a number of mechanisms to search and retrieve relevant information (https://www.ebi.ac.uk/chembl/). Withdrawn drugs and their toxicity classification are available via the compounds webpage (https://www.ebi.ac.uk/chembl/g/#browse/compounds) or drugs webpage (i.e., for parent drugs that have been assigned withdrawn information from their family of drug forms https://www.ebi.ac.uk/chembl/g/#browse/drugs). The boxed warning flags for drugs using the updated workflow described in this paper are currently available via ChEMBL, with their toxicity classification available for download (see Data Citation 1 in the Supporting Information). The toxicity classification for boxed warnings will be made available as part of a later release of ChEMBL, updated for subsequent releases, and will also be made accessible via the web interface or web services (https://www.ebi.ac.uk/chembl/ws).
Users should always be aware that, although our best effort has been made to accurately annotate safety information within ChEMBL, we cannot guarantee that there are no errors, and it is always prudent to consult the source medicinal product label to ascertain further details. To this end, example references of representative medicinal product labels have been retained as part of the curated data set for information audit purposes (see Materials and Methods).
Conclusion
A data set of safety information has been curated for drugs with boxed warnings and withdrawn drugs, including the annotation of toxicity classes described in boxed warning text for single-ingredient or combination medicinal products. The curated drug safety data set has the potential to progress our understanding of safety-related issues that arise as part of the drug discovery process. The availability of a consistent, formalized annotation of severe or life-threatening adverse events from boxed warning labels facilitates further analysis and modeling. The curated data set provides a structured means to access toxicity information on a per-drug basis and can be linked to other relevant bioactivity data in a straightforward manner within the broader framework of ChEMBL. Further work to extend the safety-related drug information and its curation and annotation is ongoing.
Acknowledgments
The helpful comments of R. Brennan are acknowledged.
Glossary
Abbreviations
- CNN
convolutional neural network
- FDA
U.S. Food and Drug Administration
- NLP
natural language processing
- SPL
structured product label
Model Performance Parameters
- Sensitivity
(TP/(TP+FN))
- Specificity
(TN/(TN+FN))
- Precision
(TP/(TP+FP))
- F1-score
(2*TP/(2*TP+FP+FN))
- Matthews correlation coefficient (MCC)
(TP*TN–FP*FN)/√(TP+FP)(TP+FN)(TN+FP)(TN+FN)
- and the correct classification rate (CCR: ((Sensitivity + Specificity)/2))
where TP are true positive counts, TN are true negative counts, FP are false positive counts, and FN are false negative counts.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.chemrestox.0c00296.
Manually annotated boxed warning labels with their associated toxicity class(es): Modelling_Analysis_SupportingInformation.xlsx, worksheet ’tox_label_training_subset’ (XLSX)
CNN text classifier model performance for validation labels: Modelling_Analysis_SupportingInformation.xlsx, worksheet: ‘nlp_indep_validation’ XLSX
CNN text classifier model performance for independent validation labels excluding parent drugs in the training/test set: Modelling_Analysis_SupportingInformation.xlsx, worksheet: ‘nlp_indep_validation _excl_Pare’ XLSX
Mapping between toxicity classification and quasi-equivalent therapeutic MeSH disease indications: Modelling_Analysis_SupportingInformation.xlsx, worksheet: ‘equivalent_classes’ XLSX
Toxicity classes and therapeutic MeSH disease indications for parent drug ChEMBL identifiers: Modelling_Analysis_SupportingInformation.xlsx, worksheet: ‘tox_class_therapeutic_indication’ (XLSX)
Full curated drug safety data set: Boxed_warnings_for_drugs_CompToxSI.csv.zip with accompanying README explanation of spreadsheet column headings (TXT, ZIP)
Data Citations: (1) Hunter, F. M. I., ChEMBL (2020) 10.6019/CHEMBL.boxedwarning.
Author Contributions
F.H. set up the method to extract and annotate boxed warnings with a toxicity classification, checked the toxicity classification results, compared therapeutic and toxicity classifications, and applied the data set to predict the toxicity class of novel compound. A.P.B. amended the ChEMBL release process to include the updated safety annotation data set. All authors contributed ideas and support during the work. All authors have given approval to the final version of the manuscript.
The research leading to these results has received funding from (i) TransQST: This work has received funding from the Innovative Medicines Initiative 2 Joint Undertaking under grant agreement No. 116030. This Joint Undertaking receives support from the European Union’s Horizon 2020 research and innovation programme and EFPIA, (ii) a Strategic Award from the Wellcome Trust [104104/A/14/Z], (iii) a Biomedical Resources Grant from the Wellcome Trust [218244/Z/19/Z], and (iv) Member States of the European Molecular Biology Laboratory.
The authors declare no competing financial interest.
Supplementary Material
References
- Papadatos G.; Gaulton A.; Hersey A.; Overington J. (2015) Activity, assay and target data curation and quality in the ChEMBL database. J. Comput.-Aided Mol. Des. 29, 885–896. 10.1007/s10822-015-9860-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez D.; Gaulton A.; Bento A.; Chambers J.; De Veij M.; Felix E.; Magarinos M.; Mosquera J.; Mutowo P.; Nowotka M.; Gordillo-Maranon M.; Hunter F.; Junco L.; Mugumbate G.; Rodriguez-Lopez M.; Atkinson F.; Bosc N.; Radoux C.; Segura-Cabrera A.; Hersey A.; Leach A. (2019) ChEMBL: towards direct deposition of bioassay data. Nucleic Acids Res. 47, D930–D940. 10.1093/nar/gky1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson M., Dumontier M., Aalbersberg I., Appleton G., Axton M., Baak A., Blomberg N., Boiten J., Santos L., Bourne P., Bouwman J., Brookes A., Clark T., Crosas M., Dillo I., Dumon O., Edmunds S., Evelo C., Finkers R., Gonzalez-Beltran A., Gray A., Groth P., Goble C., Grethe J., Heringa J., ’t Hoen P., Hooft R., Kuhn T., Kok R., Kok J., Lusher S., Martone M., Mons A., Packer A., Persson B., Rocca-Serra P., Roos M., van Schaik R., Sansone S., Schultes E., Sengstag T., Slater T., Strawn G., Swertz M., Thompson M., van der Lei J., van Mulligen E., Velterop J., Waagmeester A., Wittenburg P., Wolstencroft K., Zhao J., and Mons B. (2016) Comment: The FAIR Guiding Principles for scientific data management and stewardship. Scientific Data 3, www.go-fair.org/fair-principles/. [DOI] [PMC free article] [PubMed]
- Volkova M.; Russell R. (2012) Anthracycline Cardiotoxicity: Prevalence, Pathogenesis and Treatment. Curr. Cardiol. Rev. 7, 214–220. 10.2174/157340311799960645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriksen P. (2018) Anthracycline cardiotoxicity: an update on mechanisms, monitoring and prevention. Heart 104, 971–977. 10.1136/heartjnl-2017-312103. [DOI] [PubMed] [Google Scholar]
- Snodgrass W. R., and Hsu C. W. (2017) Valproic Acid. In Critical Care Toxicology: Diagnosis and Management of the Critically Poisoned Patient (Brent J., Burkhart K., Dargan P., Hatten B., Megarbane B., Palmer R., and White J., Eds.) pp 1083–1094, Springer International Publishing, Cham, Switzerland. [Google Scholar]
- McNaughton R., Huet G., and Shakir S. (2014) An investigation into drug products withdrawn from the EU market between 2002 and 2011 for safety reasons and the evidence used to support the decisionmaking. BMJ Open 4.e004221. 10.1136/bmjopen-2013-004221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang H.; Harris S.; Liu Z.; Zhou G.; Zhang G.; Xu J.; Rosario L.; Howard P.; Tong W. (2016) FDA drug labeling: rich resources to facilitate precision medicine, drug safety, and regulatory science. Drug Discovery Today 21, 1566–1570. 10.1016/j.drudis.2016.06.006. [DOI] [PubMed] [Google Scholar]
- EC . (2009) A Guideline on Summary of Product Characteristics (SmPC), European Commission, https://ec.europa.eu/health//sites/health/files/files/eudralex/vol-2/c/smpc_guideline_rev2_en.pdf.
- PMDA . (2020) Example medicinal product label for Amiodarone Hydrochloride; Pharmaceuticals and Medical Devices Agency, www.pmda.go.jp/PmdaSearch/iyakuDetail/ResultDataSetPDF/270428_2129010F1049_1_16.
- FDA . (2019) OpenFDA Structured Product Labeling, Food and Drug Administration, https://open.fda.gov/apis/drug/label/.
- ICH . (2019) MedDRA: the Medical Dictionary for Regulatory Activities terminology is the international medical terminology developed under the auspices of the International Council for Harmonisation of Technical Requirements for Pharmaceuticals for Human Use, www.meddra.org.
- FDA . (2011) Warnings and Precautions, Contraindications, and Boxed Warning Sections of Labeling for Human Prescription Drug and Biological Products — Content and Format, pp FDA-2011-D-0694, Center for Drug Evaluation and Research (CDER) Center for Biologics Evaluation and Research (CBER), https://www.fda.gov/media/71866/download.
- Wu L., Ingle T., Liu Z., Zhao-Wong A., Harris S., Thakkar S., Zhou G., Yang J., Xu J., Mehta D., Ge W., Tong W., and Fang H. (2019) Study of serious adverse drug reactions using FDA-approved drug labeling and MedDRA. BMC Bioinf. 20. 10.1186/s12859-019-2628-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demner-Fushman D., Shooshan S., Rodriguez L., Aronson A., Lang F., Rogers W., Roberts K., and Tonning J. (2018) A dataset of 200 structured product labels annotated for adverse drug reactions. Sci. Data 5. 10.1038/sdata.2018.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiftikci M., Ozgur A., He Y., and Hur J. (2019) Machine learning-based identification and rule-based normalization of adverse drug reactions in drug labels. BMC Bioinf. 20. 10.1186/s12859-019-3195-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solotke M.; Dhruva S.; Downing N.; Shah N.; Ross J. (2018) New and incremental FDA black box warnings from 2008 to 2015. Expert Opin. Drug Saf. 17, 117–123. 10.1080/14740338.2018.1415323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schick A.; Miller K.; Lanthier M.; Dal Pan G.; Nardinelli C. (2017) Evaluation of Pre-marketing Factors to Predict Post-marketing Boxed Warnings and Safety Withdrawals. Drug Saf. 40, 497–503. 10.1007/s40264-017-0526-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong H., Chen M., Ng H. W., and Tong W. (2016) QSAR Models at the US FDA/NCTR, in In Silico Methods for Predicting Drug Toxicity (Benfenati E., Ed.) pp 431–459, Springer New York, New York, NY. [Google Scholar]
- Chen M.; Hong H.; Fang H.; Kelly R.; Zhou G.; Borlak J.; Tong W. (2013) Quantitative Structure-Activity Relationship Models for Predicting Drug-Induced Liver Injury Based on FDA-Approved Drug Labeling Annotation and Using a Large Collection of Drugs. Toxicol. Sci. 136, 242–249. 10.1093/toxsci/kft189. [DOI] [PubMed] [Google Scholar]
- Yang H., Sun L., Li W., Liu G., and Tang Y. (2018) In Silico Prediction of Chemical Toxicity for Drug Design Using Machine Learning Methods and Structural Alerts. Front. Chem. 6. 10.3389/fchem.2018.00129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo A.; Van Vleet T.; Gupta R.; Liguori M.; Rao M. (2020) An Overview of Machine Learning and Big Data for Drug Toxicity Evaluation. Chem. Res. Toxicol. 33, 20–37. 10.1021/acs.chemrestox.9b00227. [DOI] [PubMed] [Google Scholar]
- Hammann F.; Schoning V.; Drewe J. (2019) Prediction of clinically relevant drug-induced liver injury from structure using machine learning. J. Appl. Toxicol. 39, 412–419. 10.1002/jat.3741. [DOI] [PubMed] [Google Scholar]
- Dey S., Luo H., Fokoue A., Hu J., and Zhang P. (2018) Predicting adverse drug reactions through interpretable deep learning framework. BMC Bioinf. 19. 10.1186/s12859-018-2544-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DailyMed . (2020) Drug label example for oxaproxin showing a boxed warning description, https://dailymed.nlm.nih.gov/dailymed/lookup.cfm?setid=ea1de47e-3101-4414-817c-0a098af8988c.
- FDA . (2020) OpenFDA SPL Drug label example for Oxaproxin showing a boxed warning description, https://api.fda.gov/drug/label.json?search=set_id:ea1de47e-3101-4414-817c-0a098af8988c.
- Gaulton A.; Bellis L.; Bento A.; Chambers J.; Davies M.; Hersey A.; Light Y.; McGlinchey S.; Michalovich D.; Al-Lazikani B.; Overington J. (2012) ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Res. 40, D1100–D1107. 10.1093/nar/gkr777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DailyMed . (2004) Drug label example for abacavir sulfate and lamivudine, https://dailymed.nlm.nih.gov/dailymed/lookup.cfm?setid=8c6b1125-0ed7-4cb6-b0ad-6ac63dbb4109.
- DailyMed . (2018) Drug label example for pentazocine and naloxone, https://dailymed.nlm.nih.gov/dailymed/lookup.cfm?setid=41ebdaaf-3bbc-419f-b996-0341efc14623.
- DailyMed . (2020) Drug label example for pentazocine hydrochloride and naloxone hydrochloride, https://dailymed.nlm.nih.gov/dailymed/lookup.cfm?setid=017ef042-40aa-44c9-baa8-037f06356845.
- DailyMed . (2019) Drug label example for capecitabine, https://dailymed.nlm.nih.gov/dailymed/lookup.cfm?setid=e3b1a2b5-f1ce-45ea-afd8-68aac53665fa.
- DailyMed . (2020) Drug label for ritonavir, https://dailymed.nlm.nih.gov/dailymed/lookup.cfm?setid=11f757f1-cf48-47a8-925e-918359cbd2d4.
- DailyMed . (2020) Drug label for ergotamine tartrate, https://dailymed.nlm.nih.gov/dailymed/lookup.cfm?setid=dac9637f-3326-4f25-b7b9-f9f54b738232.
- DailyMed . (2018) Drug label for paliperidone, https://dailymed.nlm.nih.gov/dailymed/lookup.cfm?setid=0dc3a9b2-2edf-4fa2-a949-459b4218e763.
- DailyMed . (2019) Drug label for iothalamate meglumine, https://dailymed.nlm.nih.gov/dailymed/lookup.cfm?setid=77dd7fb3-18c4-4acb-af07-f929dff03903.
- DailyMed . (2020) Drug label for tadalafil, https://dailymed.nlm.nih.gov/dailymed/lookup.cfm?setid=8a78579b-98a6-44de-ad4e-503ab47ab56b.
- DailyMed . (2017) Drug label for primaquine phosphate, https://dailymed.nlm.nih.gov/dailymed/lookup.cfm?setid=5c920cdf-5c1e-43f7-a275-c72040005715.
- DailyMed . (2020) Drug label for methohexital sodium, https://dailymed.nlm.nih.gov/dailymed/lookup.cfm?setid=eccd8340-ead3-4363-8902-0c19d33aa2ac.
- Honnibal M., and Montani I. (2017) SpaCy 2: Natural language understanding with Bloom embeddings, convolutional neural networks and incremental parsing, https://spacy.io/api/textcategorizer.
- DailyMed . (2018) Drug label for belatacept, https://dailymed.nlm.nih.gov/dailymed/lookup.cfm?setid=c16ac648-d5d2-9f7d-8637-e2328572754e.
- DailyMed . (2019) Drug label example for metformin hydrochloride, https://dailymed.nlm.nih.gov/dailymed/lookup.cfm?setid=1c101aba-83a6-494f-b976-3f1c8494680a.
- DailyMed . (2020) Drug label example for promethazine hydrochloride and codeine phosphate, https://dailymed.nlm.nih.gov/dailymed/drugInfo.cfm?setid=168951c0-773d-4f3d-9745-cc6de2063b61.
- Honnibal M., and Montani I.. SpaCy batch size optimization, https://spacy.io/usage/training#tips-batch-size.
- Honnibal M., and Montani I.. SpaCy multi-label log loss, https://github.com/explosion/spaCy/blob/master/spacy/_ml.py.
- Neumann M., King D., Beltagy I., and Ammar W. (2019) ScispaCy: Fast and Robust Models for Biomedical Natural Language Processing. Sigbiomed Workshop on Biomedical Natural Language Processing; (Bionlp 2019), 319–327https://allenai.github.io/scispacy/.
- (2016) Additions and Modifications to the List of Drug Products That Have Been Withdrawn or Removed From the Market for Reasons of Safety or Effectiveness. Final Rule. Federal Register 81, 69668–69677. [PubMed] [Google Scholar]
- Qureshi Z.; Seoane-Vazquez E.; Rodriguez-Monguio R.; Stevenson K.; Szeinbach S. (2011) Market withdrawal of new molecular entities approved in the United States from 1980 to 2009. Pharmacoepidemiol. Drug Saf. 20, 772–777. 10.1002/pds.2155. [DOI] [PubMed] [Google Scholar]
- Fung M.; et al. (2001) Evaluation of the characteristics of safety withdrawal of prescription drugs from worldwide pharmaceutical markets-1960 to 1999. Drug Inf. J. 35, 293–317. 10.1177/009286150103500134. [DOI] [Google Scholar]
- NLM . (2019) Medical Subject Headings (MeSH); National Library of Medicine, https://www.nlm.nih.gov/mesh.
- Coughtrie A.; Behr E.; Layton D.; Marshall V.; Camm A.; Shakir S. (2017) Drugs and life-threatening ventricular arrhythmia risk: results from the DARE study cohort. BMJ Open 7, e016627. 10.1136/bmjopen-2017-016627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echt D.; Ruskin J. (2020) Use of Flecainide for the Treatment of Atrial Fibrillation. Am. J. Cardiol. 125, 1123–1133. 10.1016/j.amjcard.2019.12.041. [DOI] [PubMed] [Google Scholar]
- Shoeb M.; Fang M. (2013) Assessing bleeding risk in patients taking anticoagulants. J. Thromb. Thrombolysis 35, 312–319. 10.1007/s11239-013-0899-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.