Abstract

Tuberculosis remains a leading cause of death from a single bacterial infection worldwide. Efforts to develop new treatment options call for expansion into an unexplored target space to expand the drug pipeline and bypass resistance to current antibiotics. Lipoamide dehydrogenase is a metabolic and antioxidant enzyme critical for mycobacterial growth and survival in mice. Sulfonamide analogs were previously identified as potent and selective inhibitors of mycobacterial lipoamide dehydrogenase in vitro but lacked activity against whole mycobacteria. Here we present the development of analogs with improved permeability, potency, and selectivity, which inhibit the growth of Mycobacterium tuberculosis in axenic culture on carbohydrates and within mouse primary macrophages. They increase intrabacterial pyruvate levels, supporting their on-target activity within mycobacteria. Distinct modalities of binding between the mycobacterial and human enzymes contribute to improved potency and hence selectivity through induced-fit tight binding interactions within the mycobacterial but not human enzyme, as indicated by kinetic analysis and crystallography.
Keywords: tuberculosis, mycobacteria, lipoamide dehydrogenase, residence time, slow binding, tight binding, inhibitor
Tuberculosis (TB) infected 10 million and killed 1.5 million people in 2018.1 It remains a worldwide health crisis due to rising drug resistance and emerging risk factors, such as diabetes. The resistance of Mycobacterium tuberculosis (Mtb) to all first-line anti-TB drugs is prevalent worldwide and calls for new strategies to develop effective therapeutics. The BPaL regimen recently approved by the FDA for the treatment of extensively drug-resistant (XDR) and nonresponsive multi-drug-resistant (MDR) TB features, for the first time in many decades, drugs against new Mtb targets and with novel modes of action. Nevertheless, more inhibitors against previously unexplored targets are urgently needed to sustain the TB drug pipeline and to shorten and diversify drug regimens, as resistance is already detected to components of BPaL.
Lipoamide dehydrogenase (Lpd) fulfills multiple metabolic functions and a unique antioxidant function in Mtb. It is a component of the pyruvate, α-ketoglutarate, and branched chain ketoacid dehydrogenase complexes2,3 involved in the breakdown of ketoacids coupled with the production of energy-rich acyl-CoA intermediates and NADH. In the reverse direction, Lpd supplies reducing equivalents from NADH through the lipoylated E2 cores of metabolic complexes to the thioredoxin-like adaptor AhpD and the peroxiredoxin AhpC to detoxify reactive nitrogen and oxygen intermediates.4,5 Mtb lacking Lpd fails to grow on carbohydrates as a sole carbon source in vitro, cannot establish TB infection in mice, is highly susceptible to reactive nitrogen intermediates, and accumulates an ∼100-fold excess of intracellular pyruvate, alanine, valine, leucine, isoleucine, and their corresponding ketoacids.3 The dependence of Mtb on Lpd for virulence and persistence provides genetic validation of Lpd as a target, but chemical validation remains to be achieved.
Bacterial enzymes with human homologues are often viewed as unattractive targets due to possible host toxicity. Although the three-dimensional structures of the mycobacterial and human enzymes align closely,6 mycobacterial Lpd is only 33% identical to the human homologue with pronounced differences in their substrate binding sites. This has allowed us to discover Lpd inhibitors with high species selectivity.7,8 These inhibitors were potent against the isolated enzyme but inactive against whole Mtb due to poor intrabacterial accumulation.
Here we report the development of improved sulfonamide-based Lpd inhibitors that exhibit acceptable Mtb permeability and phenocopy lpd genetic deletion in vitro. Our lead analogs are bacteriostatic to wild-type (WT) Mtb in a growing culture and bactericidal to WT Mtb under nitrosative stress in a carbohydrate-based medium. They elevate intracellular levels of pyruvate in Mtb, supporting their on-target activity against Lpd in the pyruvate dehydrogenase complex. They inhibit the growth of Mtb within primary mouse bone marrow macrophages, confirming their species selectivity and nontoxicity to these mammalian host cells. Their high efficacy, despite relatively low accumulation inside Mtb, is attributed to the tight binding interactions within the Mtb Lpd active site with a reduced dissociation rate constant and increased residence time on the Mtb enzyme. By contrast, mammalian Lpd is much less potently inhibited and exhibits rapid dissociation of the enzyme–inhibitor complex.
Results and Discussion
We previously reported the identification and initial SAR of sulfonamide-based Lpd inhibitors competitive with the lipoamide substrate at the Lpd’s active site.8 Those first-generation sulfonamide Lpd inhibitors, as exemplified by compounds 1a and 1b (Table 1), demonstrated low solubility and high metabolic instability when incubated with human liver microsomes. Poor physical properties and a propensity for metabolic clearance could have contributed to their low accumulation inside Mtb. Replacement of the pyridyl bromide by a methyl group (2, Table 1) improved aqueous solubility and allowed for measurable intrabacterial accumulation inside Mtb as determined by LC–MS analysis of Mtb extracts prepared from cultures exposed for 24 h to the 10 μM compound (Table 1). However, compound 2 did not demonstrate an inhibition of bacterial growth up to the maximum concentration tested, 100 μM (Table 2).
Table 1. Early Aminopyridine Analogsa.
ND, not detected. Intra-Mtb accumulation is reported as the percentage relative to the total parent compound in media during exposure.
Table 2. SAR Analysis, Antibacterial Activity, and ADME Profiling of Sulfonamide Analogsa.

| no. | R | Lpd IC50 (μM) | Mtb MIC90 (μM) | Sol (μg/mL, pH 6.8) | hLM (μL/min/mg) | log D |
|---|---|---|---|---|---|---|
| 2 | a | 0.104 | >100 | 4.6 | 115 | 1.85 |
| 3 | b | 1.70 | >100 | 0.15 | 113 | 1.94 |
| 4 | c | 0.11 | >100 | >110 | 2 | 1.69 |
| 5 | d | 0.128 | >100 | >120 | 15 | 2.12 |
| 6 | e | 0.056 | >100 | 20 | 18 | 2.12 |
| 7 | f | 0.196 | >100 | >100 | –14 | 1.37 |
| 8 | g | 0.331 | >100 | >97 | 6 | 1.32 |
| 9 | h | 2.75 | >100 | >95 | –18 | 0.4 |
| 10 | i | 0.165 | >100 | >97 | 13 | 0.45 |
| 11 | a | 0.035 | 25 | 18 | >768 | 2.26 |
| 12 | e | 0.029 | 12 | 23 | 195 | 2.55 |
| 13 | i | 0.045 | 3.1 | >110 | –11 | 0.83 |
IC50 values were determined by the DTNB assay with 50 nM Lpd.
We reasoned that further modification of the physical properties may improve compound accumulation, resulting in an inhibition of bacterial growth. Compound 2 was docked into the Mtb Lpd lipoamide binding site (PDB 4M52) using the Schrodinger GLIDE program.10 The docking pose of 2 adopted the same configuration as co-crystallized compound 1a (Figure 1A). The extended aniline ring was oriented toward the solvent front providing a region that could be modified to adjust physical properties without negatively impacting binding. With this in mind we initiated a SAR campaign where compounds were profiled for enzymatic potency as well as aqueous solubility and stability against liver microsomes as a general measure of a compound’s metabolic stability. Mtb’s waxy cell wall is notorious for its poor permeability to small molecules and poses a major challenge for antibiotic development.11,12 Accordingly, we also profiled all compounds for the inhibition of bacterial growth in order to identify correlations with structure or physical properties that could be used for iterative compound design efforts. Replacement of the methoxy substituent with a cyano group in 3 resulted in a significant loss in potency and reduced aqueous solubility. Extended polar substitution, as exemplified by compounds 4–6, was well tolerated and provided improvements in both aqueous solubility and human liver microsomal stability. However, none of these compounds demonstrated the inhibition of bacterial growth. Similarly, replacement of the phenyl ring with heteroaromatic groups (7–10) provided compounds with desirable solubility and microsomal stability but with no inhibition of bacterial growth (Table 2). These results led us to believe that more extensive modifications would be required. As shown in Figure 1A, the sulfonamide sarcosine linker is involved in several critical hydrogen-bonding interactions, including that with the Arg93 present in Mtb but not in the human isoform. Therefore, we chose to leave that region untouched and to focus on the conserved aminopyridine core. To gain insight into the binding pocket, we characterized the binding site features using the Schrodinger SiteMap Program.9 The model revealed a hydrophobic area near the amino-pyridine ring suggesting that the binding pocket could be better filled (Figure 1B). In addition, only one of the amino-pyridine NH groups appeared to be important for hydrogen bonding with the enzyme (residue Ala381 in Figure 1) as well as forming an internal hydrogen bond with the sulfonamide oxygen. To this end, indazole 11 was designed by the cyclization of the amino group back onto the aromatic ring while retaining the key NH hydrogen bond donor. The profiling of 11 demonstrated an improvement in enzyme inhibition as well as whole cell activity with an MIC90 of 25 μM when tested against WT Mtb grown in a carbohydrate-based medium. In vitro ADME-tox profiling of 11 showed that while the aqueous solubility was maintained, the compound suffered from rapid microsomal clearance. Following the SAR generated for the aminopyridines, compounds 12 and 13 were synthesized. Compound 12 demonstrated an improvement in MIC compared to that of 11 but did not maintain the improved microsomal stability associated with the morpholine substitution (compound 6). However, N-methyl pyridone-substituted 13 maintained the improved properties and further increased the MIC potency to 3.1 μM in a carbohydrate-based medium.
Figure 1.

(A) Docking pose of compound 2 (pink) superimposed with Br analog 1a (green) in the Mtb Lpd co-crystal (PDB 4M52). The gray surface indicates the protein surface. (B) Overlaid docking pose of 13 (magenta) and crystal structure of 1a (green) with SiteMap9 predictions in the Lpd lipoamide site. Yellow indicates the hydrophobic region. Purple and red indicate hydrogen bond donor and acceptor regions, respectively.
The improved antibacterial activity of 11–13 prompted us to investigate the intrabacterial accumulation of these compounds and to measure the elevation in intracellular pyruvate. The latter serves as a biomarker of intrabacterial Lpd inhibition in accordance with the phenotype of lpd deletion in Mtb3 and Lpd’s function in Mtb’s pyruvate dehydrogenase.2Table 3 shows the fold increase in intrabacterial pyruvate levels observed after 24 h of exposure of Mtb to the 10 μM compound as well as the percentage of the parent compound associated with Mtb and remaining in the medium. The relative increase in intracellular pyruvate levels for 11–13 is correlated with increased efficacy for the inhibition of bacterial growth. Compound 12, although accumulated better in Mtb, was less efficacious than 13 in terms of intrabacterial target engagement, suggesting that other factors, such as association within the cell wall/membrane compartment or its lower metabolic stability may have further affected its intra-Mtb bioavailability. Compound 13 emerged as the most promising lead with the lowest MIC and highest pyruvate fold increase, suggesting significant gains in target engagement even at relatively low intra-Mtb accumulation levels. This prompted us to further profile the inhibition of Lpd by compound 13.
Table 3. Intrabacterial Pyruvate Levels and Compound Accumulation in Mtba.
| compound | 11 | 12 | 13 |
|---|---|---|---|
| pyruvate increase (fold) | 10.6 ± 0.16 | 25.6 ± 0.03 | 37.3 ± 0.05 |
| intra-Mtb accumulation (%) | 21 | 35 | 6 |
| compound remaining (%) | 41 | 39 | 85 |
Fold pyruvate is relative to levels observed in untreated Mtb. Intra-Mtb accumulation and the compound remaining is the percentage relative to the total parent compound in media at the initiation of the experiment.
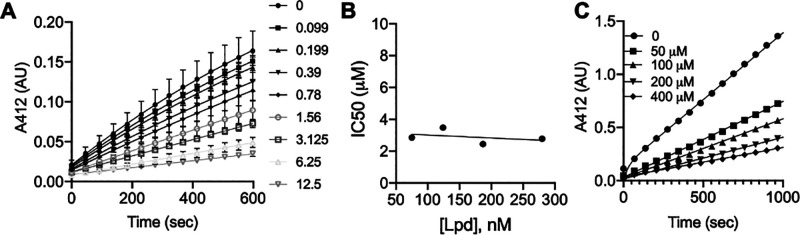
Compound 13 exhibited the time-dependent inhibition of purified recombinant Mtb Lpd following reaction initiation by the addition of enzyme (Figure 2A). Fitting the progress curve data to the first-order rate kinetics produced values of kobs that increased with an increasing molar excess of inhibitor to Lpd (Figure 2B). The relationship was not strictly linear, suggesting an induced fit model for 13 association with Lpd. Upon 30 min of preincubation, IC50 values tracked linearly with variable enzyme concentration, nearing the values for the concentrations of Lpd and suggesting a slow-onset, tight binding interaction (Figure 2C). Close tracking of IC50 values with the Lpd concentration for compound 13 suggested that the determination of true Ki values through the traditional Michaelis–Menten kinetics was not possible as the tight association of the inhibitor in the Lpd–inhibitor complex limited the free concentration of compound 13 in the assay. Thus, we proceeded to calculate Kiapp using the steady-state rate at fixed concentrations of Lpd and the lipoamide substrate (75 μM). This inhibition data, expressed as the fractional velocity at a given inhibitor concentration, was fitted to the Morrison equation for tight binding inhibitors to calculate Kiapp = 9 ± 6.9 nM. To determine the dissociation rate of compound 13 from the Lpd–13 complex, we preincubated Lpd with 13 and subsequently diluted the complex 500-fold into the assay mixture to follow the recovery of Lpd activity. We observed the rapid recovery of activity at subequimolar concentrations of inhibitor during preincubation and fractional recovery with a long lag period at a molar excess of 13 (Figure 2D). Fitting the recovery experimental data for compound 13 to the integrated rate equation to determine the dissociation rate constant13 produced a koff value of 0.084 min–1, which corresponded to a 12 min half-life (t1/2) for the Lpd–13 complex (Figure S1). At equimolar and higher concentrations of compound 13, the recovery slowed, with a >10-fold reduction in koff values corresponding to hours-long t1/2 (Figure S1). This sustained inhibition at higher concentrations likely reflected the continuous rebinding of the dissociated (and/or excess) inhibitor. Nevertheless, enzymatic data suggested an extended residence time of 13 on Mtb’s Lpd under conditions at which 13 is present in molar excess with respect to Lpd. Thus, achieving intra-Mtb levels of 13 in excess of Lpd intracellular concentration should result in sustained target engagement and drive the inhibition of Lpd activity in vivo.
Figure 2.
Compound 13 is a potent, time-dependent, slowly dissociating inhibitor of Mtb Lpd. (A) Compound 13 produces a time-dependent inhibition of Lpd activity. Pure recombinant Mtb Lpd (66 nM) was tested against variable concentrations of compound 13, and TNB product formation was followed over time at 412 nm. Each symbol corresponds to an indicated compound 13 concentration in nM. Symbols represent experimental data recorded every 30 s, and solid lines are best fits to the first-order association function. (B) First-order rate constant of inactivation (kobs) increases with the compound 13 concentration. Values of kobs were determined from the first-order association fit represented by a solid line in A. (C) IC50 values for compound 13 inhibition of Mtb Lpd depend on Lpd concentration. Lpd was tested at the indicated concentrations represented by different symbols (numbers for each symbol correspond to [Lpd] in nM) for inhibition by increasing concentrations of compound 13; the inset shows the plot of [Lpd] vs calculated IC50. (D) Recovery of Lpd activity upon dilution of the Lpd-13 complex. Lpd (10 μM) was preincubated with the indicated μM concentrations of 13 for 30 min at RT, diluted 500-fold into the reaction mixture with 75 μM lipoamide and monitored for TNB formation over time.
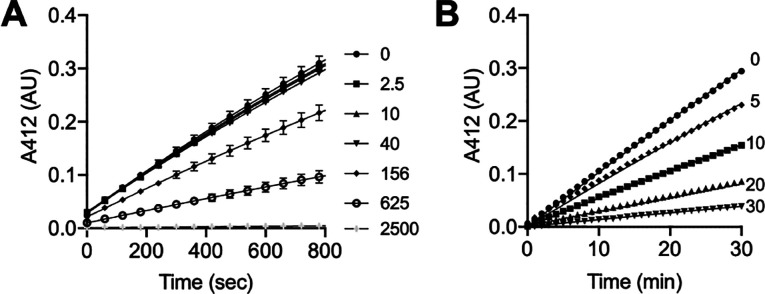
In contrast, when compound 13 was tested against the recombinant human Lpd in a similar manner, no time-dependent inhibition was observed (Figure 3A), the IC50 values did not change significantly with variable enzyme concentration (Figure 3B), and inhibition was rapidly reversible upon dilution, even at a high molar excess of inhibitor (Figure 3C). Taken together, these results demonstrate that 13 is a significantly less potent (Kiapp = 1.13 ± 0.18 μM) rapid equilibrium inhibitor of human Lpd.
Figure 3.
Compound 13 is a readily reversible inhibitor of human Lpd. (A) The activity of human Lpd (280 nM) was monitored over time in the presence of the indicated micromolar concentrations of 13. (B) IC50 values were calculated at variable Lpd (75, 124, 187, and 280 nM) and plotted against [Lpd]. (C) Human Lpd (10 μM) was preincubated with the indicated micromolar concentrations of 13 for 30 min at RT, diluted 125-fold into the reaction mixture with 75 μM lipoamide, and monitored for TNB formation over time.
Increased inhibition rates with excess 13 suggested the possibility of an induced fit model that may have contributed to the high affinity and slow dissociation of 13 when all active sites are occupied by an inhibitor. We examined the crystal structures of Mtb Lpd with and without bound 13 for conformational changes within the lipoamide binding pocket or in its vicinity upon the binding of 13 and identified the only significant structural change in the Arg93 conformation. The original crystal structure of Mtb Lpd6 identified Arg93 as being able to adopt two different conformations within the lipoamide channel by positioning its side chain to stack above His386′ for open access to the channel (open conformation) or to protrude into the channel and block access (closed conformation). The Lpd co-crystal with 13 adopted the open conformation6 to accommodate bound 13 (Figure 4). Compound 13 occupies each of the two lipoamide binding channels, which are formed between the protomers. The indazole ring sits deep within each channel and is in close proximity to the disulfide-based redox center adjacent to His443′ and Glu448′. A direct hydrogen bond is observed between the indazole NH and the Ala381′ backbone carbonyl. In addition, a water-mediated hydrogen bond forms between the indazole ring and the side-chain amide of Asn382′. The 5-position methyl group of 13 sits in a hydrophobic pocket pointing toward Pro13. Similar to the previous aminopyridine analog 1a,8 the Arg93 side chain swings out to adopt an open conformation (Figure S2). The amide group of 13 interacts with both the Arg93 guanidinium and the backbone carbonyl oxygen of Asn463′, which are believed to be important for imparting both potency and selectivity to human Lpd. At the solvent-exposed end of the channel, the pyridone carbonyl of 13 forms a water-mediated hydrogen bond with the Arg93 side chain, which orients the pyridone ring for π–π stacking with Phe99.
Figure 4.

Co-crystal structure of Lpd (green) with 13 (magenta) superimposed with the Lpd apo structure6 (PDB 2A8X, gray). Dotted lines indicate the distance in angstroms.
The crystal structure of Mtb Lpd suggested a critical role for Arg93 in maintaining access to the lipoamide binding site as it was able to adopt two different conformations.6 The co-crystal of Lpd with 13 identified Arg93 among contact residues and demonstrated that Arg93 adopts an open conformation upon 13 binding. Human Lpd has a Leu residue at that position, and the lack of coordination through Arg93 may have contributed to the lower affinity of 13 for the human homologue. We tested an Mtb Lpd mutant enzyme with single amino acid substitution R93A to determine if Arg93 contact contributes to the binding affinity and affects the dissociation rate of 13. No time-dependent inhibition of Mtb Lpd R93A activity in the presence of 13 was observed, progress curves with increasing 13 concentrations were linear from the onset (Figure 5A), and Kiapp was calculated to be 218 nM. Lpd R93A preincubation with 13 and dilution into the assay mixture to recover the activity resulted in linear progress curves without any lag, suggesting the rapid dissociation of 13 from Mtb Lpd R93A (Figure 5B).
Figure 5.
Compound 13 is a rapid equilibrium inhibitor of the Mtb Lpd R93A mutant. (A) Progress curves for Lpd R93A (200 nM) at the indicated nanomolar concentrations of 13. Reaction was initiated by enzyme addition. (B) Lpd R93A (10 μM) was preincubated with the indicated concentrations of 13 (in μM), diluted 125-fold into the reaction mixture with 75 μM lipoamide, and monitored for TNB formation over time.
Enzyme kinetics suggested that compound 13 is a potent inhibitor of Mtb Lpd and that Arg93 is important for this binding. To learn more about what was driving this tight association, we performed ITC and SPR profiling for both the WT and the R93A Mtb Lpd as well as the human Lpd interaction with compound 13 (Figures S3–S6). We observed that the affinity of compound 13 for WT Mtb Lpd depended on the presence of NADH and was in good agreement with the Kiapp values calculated through enzyme inhibition assays. Thus, the Kd for the WT Lpd decreased more than 2-fold in the presence of NADH, suggesting a possible conformational adjustment within the lipoamide site upon the binding of NADH that resulted in a tighter association with compound 13 (Table 4). The NADH-dependent change in the shape of the lipoamide channel upon its binding may explain why we were unable to identify any additional differences except for the Arg93 conformation in the crystal structures upon 13 binding, as both structures were crystallized in the presence of NADH. The R93A mutant bound compound 13 in an NADH-independent manner with Kd = 227 ± 6.6 nM, which was in agreement with the Kiapp calculated from enzyme inhibition data. Thus, it appeared that R93 contact contributes to the binding affinity and may adjust its conformation upon 13 and/or NADH binding, which is consistent with its conformational flexibility in the Mtb Lpd crystal structure.6 The human Lpd bound compound 13 with a much lower affinity, and in some repeats we were not able to fit the data properly. The binding of compound 13 to the WT Mtb Lpd was mostly enthalphy-driven, suggesting hydrogen-bonding involvement (Figure S5).
Table 4. Characterization of 13 Binding to Lpd by ITC and SPRa.
| ITC | SPR |
||||
|---|---|---|---|---|---|
| Kd, nM | Kd, nM | kon, M–1 s–1 | koff, s–1 | t1/2, min | |
| Mtb WT | 63.9 ± 2.75 | 52.3 | 35 500 (1) | 0.0315 (1) | 0.5 (1) |
| 0.0177 (2) | 0.0011 (2) | 15.15 (2) | |||
| Mtb WT/NADH | 28.3 ± 1.74 | 44.5 | 18 500 (1) | 0.00083 (1) | 20.15 (1) |
| 0.00000222 (2) | 0.00158 (2) | 10.5 (2) | |||
| Mtb R93A | 231.4 ± 8.81 | ND | ND | ND | ND |
| Mtb R93A/NADH | 228.6 ± 2.44 | ND | ND | ND | ND |
| human WT | 592 | 7600 | 1170 | 0.00887 | 1.9 |
| human WT/NADH | ND | ND | 110 | 0.00628 | 2.7 |
ND, not determined. Kd values were calculated by using the stoichiometric equilibria model assuming A + B ↔ AB(ITC) and the two-state model assuming complex AB undergoes a conformational change: A + B ↔ AB ↔ AB* (SPR; kon(1) is the association rate constant for the formation of AB; koff(1) is the dissociation rate constant for complex AB; kon(2) is the association rate constant for the conversion of AB to AB*; and koff(2) is the dissociation rate constant for the conversion of AB to AB*). Results are representative of two independent repeats.
Similarly, the kinetic analysis of compound 13 binding to WT Mtb Lpd by SPR produced Kd values close to the ones observed by ITC, but we did not observe a significant dependence of the binding affinity on the NADH presence. This could reflect a difference in the Lpd native conformation as a dimer, which was preserved in ITC but could have been disrupted in the SPR by the covalent immobilization of Lpd. Best fitting of the kinetic binding data was achieved with the two-state model, which had fast on/off and slow on/off components in the absence of NADH. In the presence of NADH, the fast on/off component converted to the fast on/slow off site with slow dissociation corresponding to a 40-fold increase in the half-life of the Lpd–13 complex (Table 4, Figure S6). The SPR values for koff agreed well with the numbers calculated from rapid dilution experiments. Human Lpd protein produced Kd in the micromolar range and faster dissociation rates by SPR with a half-life of 2 min, confirming its lower affinity and 10-fold higher dissociation rates for compound 13. These results are consistent with our enzymatic data analysis and support the interpretation that Mtb’s Lpd binds 13 tightly through an induced fit model in the presence of NADH. This scenario will likely play out inside Mtb in vivo as the mycobacterium maintains high nanomolar to low micromolar NADH levels throughout different stages of its growth cycle.14−16 Human Lpd will be spared from inhibition due to its lower affinity for 13, aided by the rapid dissociation of the human Lpd–13 complex.
We speculated that the tight binding and relatively slow dissociation of 13 from Mtb Lpd drove the whole cell activity of 13 against Mtb even at relatively low intrabacterial accumulation levels. Accordingly, we decided to examine whether treatment with 13 could reproduce any of the other phenotypes associated with the lpd genetic deletion in Mtb. In accordance with the high susceptibility of Δlpd Mtb to nitrosative stress,3 where 99% of Δlpd Mtb is killed upon exposure to 3 mM NaNO2 at pH 5.5 (NaNO2 generates a flux of NO at acidic pH), 13 proved to be bactericidal in a concentration-dependent manner to WT Mtb at pH 5.5 in the presence of 3 mM NaNO2 but had no effect at the same concentrations when Mtb was exposed to pH 5.5 alone (Figure 6A). Mtb Δlpd does not replicate in naïve mouse bone marrow-derived macrophages (BMDM) and dies by 1 log10 in BMDM activated by interferon-γ (IFNγ).3 Compound 13 inhibited the growth of WT Mtb in naïve BMDM in a concentration-dependent manner, recapitulating the Mtb Δlpd phenotype. By day 6 after infection, treatment with 13 inhibited the growth of Mtb by 1 log10 in naïve BMDM and by 0.5 log10 in IFNγ-activated BMDM at the highest concentration tested, 30 μM.
Figure 6.
Compound 13 selectively kills Mtb under nitrosative stress and inhibits the growth of Mtb inside mouse BMDM. (A) WT Mtb was exposed to pH 5.5 or pH 5.5 plus 3 mM NaNO2 in the presence of 13 for 4 days and plated on agar to enumerate colony-forming units (CFU) of surviving bacteria. (B) Mouse BMDMs were infected with WT Mtb (multiplicity of infection (MOI) = 0.1) and exposed to the indicated μM concentrations of 13. At the indicated time points, BMDM were lysed and CFU were determined. (C) The same as in B, but BMDM were activated with 10 ng/mL IFNγ for 24 h prior to infection. Results are the means ± SD of triplicate wells in a single experiment representative of two independent experiments. P values were calculated with an unpaired t test.
No toxicity to mouse BMDM was observed at any concentration of 13 tested as assessed by the MTS assay, suggesting that the prolonged residence time of 13 on Mtb’s Lpd selectively contributed to increased vulnerability of Mtb to Lpd inhibition inside mammalian host cells. Similarly, the profiling of 13 in HEPG2 cells showed no signs of cytotoxicity up to 100 μM, consistent with the efficacy in the BMDM experiment being driven through on-target activity in Mtb.
Slowly reversible, tight binding enzyme inhibitors offer the benefits of both enhanced potency and prolonged on-target residence time, extending their pharmacologic effect in vivo.17 Our work reports on a first in class, slowly dissociating, high-affinity inhibitor of Mtb Lpd, which is whole cell active, recapitulates phenotypes of lpd genetic deletion in Mtb, and affords high selectivity over the human homologue due to its tight binding induced-fit interaction with Arg93 within the Mtb lipoamide substrate binding site but not the human enzyme. Tight binding of compound 13 to Mtb’s Lpd may be aided by NADH binding to Lpd and/or may reflect conformational adjustments within the lipoamide channel or its desolvation upon 13 binding to achieve tighter contacts.
In vitro, compound 13 demonstrated comparable potency against Lpd in singularity or as a component of a larger PDH complex (in the presence of recombinant E2 = DlaT and E1 = AceE proteins), suggesting that the Lpd conformation, or at least the architecture of the lipoamide channel, is similar under both conditions. However, in vitro data cannot fully predict how efficacious such an inhibitor will be when it encounters a target in its natural milieu, where physiological protein expression, turnover, and posttranslational modification levels (E2 cores of PDH and BCKDH present covalently attached lipoamide to Lpd) as well as other substrate levels (e.g., NAD+/NADH) may affect the affinity and on-target residence time to define the in vivo target vulnerability. Our studies on whole cell Mtb in axenic culture, under nitrosative stress or inside mouse BMDM, demonstrate that, despite its low accumulation inside Mtb, compound 13 effectively targets intracellular Lpd functions in PDH and BCKDH. This is demonstrated by the accumulation of pyruvate and branched-chain amino acids (isoleucine, leucine, and valine) inside Mtb exposed to compound 13, the selective killing of Mtb under nitrosative stress, and the inhibition of Mtb growth inside mouse BMDM. High levels of pyruvate accumulation inside Mtb despite limited 13 accumulation suggest that tight binding and slow dissociation features of 13 are likely to manifest under physiological intracellular levels of NAD+/NADH inside Mtb and that intracellular or local concentrations of compound 13 likely reach levels equimolar with or higher than intracellular Lpd levels to sustain on-target activity. Alternatively, Lpd assembly into the large intracellular complexes may aid the maintenance of highly concentrated local pools of dissociated inhibitor in close proximity to other Lpd active sites, promoting rebinding17 and sustained on-target residence.
One can argue that the sustained inhibition of enzymatic activity may lead to the generation of resistance mechanisms to overcome the inhibition through mutations within the Lpd protein or upregulation of bypass metabolic routes. We are in the process of determining the frequency of resistance for the indazole analogs and plan to systematically identify resistant mutants if such arise upon treatment of Mtb in culture or in mice with compound 13 or its improved analogs.
Our future analogs will further explore the binding site to identify water molecules which could be displaced to facilitate higher-potency interactions and improve on-target residence time. Our data demonstrates that Arg93 is critical to maintaining tight interactions within the binding site; therefore, we will preserve those contacts but explore the distal pocket to pick up new contacts. We will also target improved in vivo efficacy by identifying compounds that accumulate better inside Mtb to maintain full target occupancy as well as having improved PK. Testing such an inhibitor in an animal model of TB may offer the best way to confirm the vulnerability of Mtb to Lpd chemical inhibition in a mammalian host. Compound 13 was not tested for efficacy in the mouse model of TB because it is highly susceptible to mouse microsomal metabolism and most likely would not be able to achieve suitable target coverage over the course of the experiment. However, our present results support the rationale of efforts to improve bacterial accumulation and metabolic stability while preserving tight binding for Mtb Lpd to develop analogs suitable for testing in vivo.
Tight binding interactions have been explored in the development of antibiotics against other bacterial targets, such as dihydrofolate reductase,18 zinc-dependent deacetylase of lipid A biosynthesis,19 bacterial and insect trehalases,20 and proteases. The residence time of macrolide antibiotics on ribosomes was reported to define their antibacterial whole cell activity, with slowly dissociating macrolides being cidal and rapidly dissociating macrolides resulting in bacteriostasis.21 The residence time of macrolide antibiotics on the bacterial ribosome22 was recently correlated with their post-antibiotic effect to build a model that demonstrates the dependence of the post-antibiotic effect on the target residence time. A similar approach was undertaken to predict the in vivo efficacy of UDP-3-O-acyl-N-acetylglucosamine deacetylase inhibitors.23 These examples underscore the utility of tight binding and slowly dissociating inhibitors in antibiotic development for target vulnerability validation and the prediction of in vivo efficacy.
In summary, our results demonstrate that Mtb’s Lpd is vulnerable to chemical inhibition in axenic Mtb cultures and inside Mtb-infected mouse primary macrophages under the conditions where intrabacterial levels of reactants and protein turnover are maintained. Improved analogs with longer residence times could reveal further gains in in vivo efficacy, leading to studies on Mtb’s Lpd vulnerability to small-molecule inhibition in acute and chronic models of TB infection.
Methods
General
All chemicals were from Sigma. Recombinant protein production and purification (Mtb Lpd WT and Mtb Lpd R93A) was performed as reported previously.8 Purified recombinant human Lpd was a generous gift from Prof. M. Patel, University at Buffalo, SUNY. Lpd activity was measured by the DTNB assay according to previously published procedures.8 For the preincubation of Lpd with the inhibitor, Lpd was dispensed into the wells, and compounds were added at specified concentrations, mixed, and preincubated at RT for 30 min without any other components. Reaction was started by adding the assay mixture containing the substrates (lipoamide, NADH) and DTNB, and TNB production was recorded over time at RT in the SpectraMax plate reader at 412 nm. For time-dependent measurements, compounds were added to wells containing the assay mixture, the reaction was started by the addition of Lpd protein, and TNB production was followed over time in the SpectraMax plate reader at 412 nm. Final concentrations of components in the reaction mixture were as follows: 150 μM NADH, 150 μM DTNB, 40 μM NAD+, 75 μM lipoamide in 25 mM potassium phosphate, and 1 mM EDTA pH 7.0. The Lpd protein was tested at variable concentrations specified in the graphs. The protein concentration was determined by the Bradford assay kit (Sigma), the Lpd concentration is provided for the monomeric enzyme; the active species is a dimer and binds 2 molecules of compound per dimer. Progress curves were fitted to first-order association in GraphPad Prism to calculate kobs. Kiapp for tight binding inhibitors were calculated in GraphPad Prism by fitting the fractional velocity data to the Morrison Kiapp equation Y = Vo(1 – ((((Et + X + (Ki(1 + (S/Km)))) – (((Et + X + (Ki(1 + (S/Km))))2) – 4EtX)0.5))/(2Et))) where Et = 66 nM, S = 75 μM, and Km = 50 μM. The Kiapp values for R93A and human Lpd proteins were calculated by using the equation Ki = IC50/(S/Km + 1) for a competitive inhibitor.
MIC Assay
MIC values were determined on the WT Mycobacterium tuberculosis H37Rv strain in 96-well plates in 200 μL of Middlebrook 7H9 medium at pH 6.6 with 0.2% glycerol, 0.02% tyloxapol, and 10% ADN (5% fatty acid-free BSA (Roche), 2% dextrose, and 0.85% NaCl). The starting bacterial inoculum was 0.01 (OD580). Inhibitors were tested at 2-fold serial dilutions from 100 to 0.1 μM. Growth was evaluated by the change in OD580 after 10 days of incubation at 37 °C in 5% CO2, 95% humidified air. Blank wells (0% growth) contained 200 μL of medium; control wells without inhibitor were defined as having 100% growth. Growth in experimental wells with inhibitors was calculated as the percentage relative to no inhibitor controls. MICs were defined as compound concentrations that inhibited bacterial growth of >90%.
Determination of Inhibitor Mtb Intracellular Accumulation by Metabolic Profiling
WT H37Rv Mtb was grown in 7H9 Middlebrook medium supplemented with 0.2% glycerol, 0.2% dextrose, 0.5% BSA, 0.085% NaCl, and 0.02% tyloxapol to an OD580 of 1.0. One milliliter of culture at OD580 = 1.0 was filtered through a nitrocellulose membrane (0.22 μM; Millipore GSWP 02500). Mtb-laden membranes were placed atop “swimming pools” constructed from inverted 15 mL falcon tube caps filled with media. Samples were incubated at 37 °C in 5% CO2 for 7 days to accumulate biomass and checked daily to ensure that membranes were in constant contact with the growth medium. Membranes were then transferred to fresh media-based pools with or without 10 μM compound for 24 h. Filters were flipped into a 1 mL ice-cold 40:40:20 solution of methanol/acetonitrile/water. Mtb was scraped off the filters and lysed via bead beating three times. Small-molecule extracts were mixed 1:1 with an acetonitrile/0.2% formic acid solvent for MS analysis. To determine compound intracellular accumulation, a standard curve was generated by spiking a mycobacterial small-molecule extract with known concentrations of compound to correct for the ion suppressive effects of lysate. To assess the metabolite abundance, protein was measured using a Pierce BCA kit, and ion counts of specified metabolites were normalized to protein concentration.
BMDM
BMDM were differentiated as reported24 for 6 to 7 days in complete DMEM (4.5 g/L glucose, 10% FBS, 1% HEPES, 1% sodium pyruvate, and 1% l-glutamine) supplemented with 20% L929 cell-conditioned medium (LCM). Cells were collected in 0.5 mM EDTA in PBS, washed with 10% LCM in complete DMEM, counted with a hemocytometer, and plated in 48-well plates [(2.0–2.5) × 105 cells/well] in 0.5 mL 10% LCM in complete DMEM. No antibiotics were used at any stage in any experiments. All cultures were incubated at 37 °C in a mixture of 5% CO2 and 95% humidified air. Isolation of bone marrow macrophages from mice was performed in accordance with NIH guidelines for the housing and care of laboratory animals and WCM institutional regulations after protocol review and approval by the Institutional Animal Care and Use Committee (IACUC protocol 2013-0001).
Infection of BMDM with Mtb
BMDM were plated in 48-well plates [(2.0–2.5) × 105 cells/well] in 0.5 mL of 10% LCM in complete DMEM. Where indicated, mouse IFNγ (final 10 ng/mL, Roche) was added and cells were left to adhere overnight. A single-cell suspension of Mtb H37Rv in PBS with 0.02% tyloxapol was added in 50 μL to achieve an MOI of 0.1. Four hours later, the medium was removed, the monolayers were washed twice with warm PBS, and 0.5 mL of fresh 10% LCM in complete DMEM was replaced. BMDM were observed daily, and photomicrographs were recorded. Only wells with intact BMDM monolayers were used for the determination of CFU. Test agents were added 24 h after Mtb infection by replacing the medium in wells with BMDM with 10% LCM in complete DMEM containing the indicated final concentrations of inhibitors. Data are presented according to the time after treatment as opposed to the time after infection. At each time point, the medium was removed, and monolayers washed with warm PBS and BMDM were lysed with 100 μL of 0.5% Triton X100 in PBS. PBS (400 μL) was immediately added to wells to dilute the Triton X100 to 0.1% to minimize any impact on Mtb viability. Samples were serially diluted, and 10 μL was plated on 7H11 agar plates incubated at 37 °C for CFU enumeration 3 weeks later.
Crystallization and Structure Determination
Mtb Lpd expression and purification for crystallographic analysis were performed as reported.6,8 Lpd (200 μM), inhibitor 13 (300 μM), and 600 μM NADH were mixed and incubated on ice for 10 min prior to crystallization at 18 °C by hanging drop vapor diffusion against a well solution containing 100 mM Tris at pH 8.5, 50 mM NaCl, 15% PEG 10000 (w/v), and 14% glycerol. Crystals were cryo-protected by the addition of 25% glycerol and flash cooled in liquid nitrogen. A single crystal was diffracted and data were collected at NE-CAT beamline 24-IDC at 0.9791 Å to obtain a data set at 2.2 Å resolution. The structure of Lpd bound to 13 was determined by molecular replacement using programs contained within CCP4 and our previously determined structure of Lpd.6 The crystal lattice included four dimers in the asymmetric unit and a model comprising Lpd amino acids 1–464 and an additional N-terminal Ser residue that remained from the cleavage tag (Gly-Ser). This model was refined using Phenix25 after manual rebuilding in Coot.26 Water molecules (1560) and 13 were added to the model and refined to an R/Rfree of 0.189/0.221 at 2.2 Å. Densities within the NADH binding site could not be modeled reliably with NADH, so densities were fit with solvent atoms. The model has excellent geometry, with 96.5 and 3.5% of residues in favored and allowed regions of Ramachandran space with MolProbity scores for all atom contacts and protein geometry in the 100th percentile for both regions.27 Atomic coordinates and structure factors have been deposited in the Protein Data Bank with accession code 7KMY.
ITC Measurements
All measurements were performed on a MicroCal Auto-ITC200 instrument. Lpd was used at 10 or 20 μM, and compound 13, at 250 μM. Measurements were performed in a mixture of 25 mM potassium phosphate buffer at pH 7.0 and 2.5% DMSO ∓ 100 μM NADH or in a mixture of 20 mM triethanolamine buffer at pH 7.8 and 2.5% DMSO ∓ 100 μM NADH. Data analysis was performed with Affinimeter software using the stoichiometric equilibria model assuming free species ↔ A1B1. Kd was calculated as a reciprocal of Ka. The free energy was calculated with ΔG = RT ln Kd, where R = 1.9858775 cal M–1 K–1 and T = 298.15 K. TΔS was calculated with
SPR
All measurements were performed on the eight-needle high-sensitivity SPR Biacore 8K system (Cytiva). A CM5 sensor chip was used for all experiments. Mtb and human WT proteins were immobilized on the matrix via amine coupling at pH 5.0 to achieve comparable RUs (11 342 ± 58.96 for Mtb and 12 735 ± 125.49 for human Lpd). Parallel kinetics was run in a mixture of 25 mM potassium phosphate at pH 7.0, 0.05% surfactant P20, and 1% DMSO ∓ 100 μM NADH with four variable concentrations of compound 13. The contact time was 120 s, the dissociation time was 300 s, and the flow rate was 50 μL/min. Data evaluation and fitting were performed with Biacore Insight Evaluation Software.
Docking and Binding Site Characterization
All calculations were performed using the Schrödinger Suite molecular modeling package (release 2019-2, Maestro, Schrödinger, LLC, New York, NY). The crystal structure of Mtb Lpd in complex with 1a (PDB 4M52) was used for docking studies.8 The structure was prepared using the Protein Preparation Wizard with default settings. Waters with fewer than three hydrogen bonds to nonwaters were removed. A receptor grid centered at the centroid of co-crystallized compound 1a, which can accommodate ligands with lengths of up to 20 Å, was generated. New ligands were prepared using LigPrep and docked using Glide XP. The binding site region of 1a plus a 6 Å buffer area was evaluated by SiteMap.
Acknowledgments
We thank our colleagues Leigh Baxt, Stacia Kargman, Robert Myers, and Gang Lin for their critical comments and discussions. For the expert technical help with ITC and SPR, we thank Carolina Adura Alcaino, Lavoisier Ramos-Espiritu, Fraser Glickman, and the Rockefeller University HTSRC. For the SPR data analysis, we thank Michael Murphy (Cytiva). We thank Yan Ling for help with protein purification and Daniel Pfau for the isolation of mouse primary bone marrow macrophages.
Glossary
Abbreviations
- ADME
absorption, distribution, metabolism, and excretion
- ADN
albumin, dextrose, NaCl media supplement
- AhpC
alkyl hydroperoxide reductase subunit C
- AhpD
alkyl hydroperoxide reductase subunit D
- BMDM
bone-marrow-derived macrophage
- BPaL
bedaquiline, pretomanid, and linezolid regimen
- CFU
colony-forming units
- CoA
coenzyme A
- DTNB
5,5′-dithiobis-2-nitrobenzoic acid
- HEPG2
human hepatocyte cell line
- hLM
human liver microsomes
- IFNγ
interferon γ
- IC50
half-maximal inhibitory concentration
- ITC
isothermal titration calorimetry
- LC–MS
liquid chromatography–mass spectrometry
- Lpd
lipoamide dehydrogenase
- MIC
minimal inhibitory concentration
- MDR
multi-drug-resistant
- MOI
multiplicity of infection
- Mtb
Mycobacterium tuberculosis
- PDH
pyruvate dehydrogenase
- PK
pharmacokinetics
- RT
room temperature
- SAR
structure–activity relationship
- SPR
surface plasmon resonance
- MTS
tetrazolium-based cell toxicity assay
- TB
tuberculosis
- TNB
5-thio-2-nitrobenzoic acid
- WT
wild type
- XDR
extensively drug-resistant
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsinfecdis.0c00788.
Additional experimental data for Mtb Lpd recovery at different concentrations of 13; Lpd co-crystal structure with 13 superimposed on the co-crystal with 1a; ITC and SPR data; X-ray crystallography data collection and refinement statistics for the Mtb Lpd/compound 13 co-crystal; and details on chemical synthesis and compound purity (PDF)
Author Present Address
□ (R.J.) Department of Microbiology, Radboud University, Nijmegen 6525 AJ, The Netherlands. Email: robert.jansen@ru.nl.
This work was supported by NIH grant R01 AI064768 to CN and the Abby and Howard P. Milstein Program in Chemical Biology and Translational Medicine at WCM. This work was also supported in part by NIH grant R35 GM118080 (CDL) and NIH National Cancer Institute-Cancer Center Support Grant P30 CA008748. The work presented here was also based in part on research conducted at the NE-CAT beamlines (NIH NIGMS Grant P41 GM103403 and NIH Office of Research Infrastructure Programs High-End Instrumentation Grant S10 RR029205). Beamline research used resources of the APS, a US Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract DE-AC02–06CH11357. The content is solely the responsibility of the authors and does not represent the official views of the NIH. CDL. is an Investigator of the Howard Hughes Medical Institute. The Department of Microbiology & Immunology is supported by the William Randolph Hearst Trust. The authors gratefully acknowledge the generous support to this project (not to the non-TDI laboratories) provided by the Tri-Institutional Therapeutics Discovery Institute (TDI), a 501(c)(3) organization. TDI receives financial support from Takeda Pharmaceutical Company, TDI’s parent institutes (Memorial Sloan Kettering Cancer Center, The Rockefeller University and Weill Cornell Medicine) and from a generous contribution from Mr. Lewis Sanders and other philanthropic sources.
The authors declare no competing financial interest.
Supplementary Material
References
- WHO . Global Tuberculosis Reports; WHO: https://www.who.int/tb/publications/global_report/en/, 2019.
- Tian J.; Bryk R.; Shi S.; Erdjument-Bromage H.; Tempst P.; Nathan C. (2005) Mycobacterium tuberculosis appears to lack alpha-ketoglutarate dehydrogenase and encodes pyruvate dehydrogenase in widely separated genes. Mol. Microbiol. 57 (3), 859–68. 10.1111/j.1365-2958.2005.04741.x. [DOI] [PubMed] [Google Scholar]
- Venugopal A.; Bryk R.; Shi S.; Rhee K.; Rath P.; Schnappinger D.; Ehrt S.; Nathan C. (2011) Virulence of Mycobacterium tuberculosis depends on lipoamide dehydrogenase, a member of three multienzyme complexes. Cell Host Microbe 9 (1), 21–31. 10.1016/j.chom.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryk R.; Griffin P.; Nathan C. (2000) Peroxynitrite reductase activity of bacterial peroxiredoxins. Nature 407 (6801), 211–5. 10.1038/35025109. [DOI] [PubMed] [Google Scholar]
- Bryk R.; Lima C. D.; Erdjument-Bromage H.; Tempst P.; Nathan C. (2002) Metabolic enzymes of mycobacteria linked to antioxidant defense by a thioredoxin-like protein. Science 295 (5557), 1073–7. 10.1126/science.1067798. [DOI] [PubMed] [Google Scholar]
- Rajashankar K. R.; Bryk R.; Kniewel R.; Buglino J. A.; Nathan C. F.; Lima C. D. (2005) Crystal structure and functional analysis of lipoamide dehydrogenase from Mycobacterium tuberculosis. J. Biol. Chem. 280 (40), 33977–83. 10.1074/jbc.M507466200. [DOI] [PubMed] [Google Scholar]
- Bryk R.; Arango N.; Venugopal A.; Warren J. D.; Park Y. H.; Patel M. S.; Lima C. D.; Nathan C. (2010) Triazaspirodimethoxybenzoyls as selective inhibitors of mycobacterial lipoamide dehydrogenase. Biochemistry 49 (8), 1616–27. 10.1021/bi9016186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryk R.; Arango N.; Maksymiuk C.; Balakrishnan A.; Wu Y. T.; Wong C. H.; Masquelin T.; Hipskind P.; Lima C. D.; Nathan C. (2013) Lipoamide channel-binding sulfonamides selectively inhibit mycobacterial lipoamide dehydrogenase. Biochemistry 52 (51), 9375–84. 10.1021/bi401077f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halgren T. (2007) New method for fast and accurate binding-site identification and analysis. Chem. Biol. Drug Des. 69 (2), 146–8. 10.1111/j.1747-0285.2007.00483.x. [DOI] [PubMed] [Google Scholar]
- Friesner R. A.; Banks J. L.; Murphy R. B.; Halgren T. A.; Klicic J. J.; Mainz D. T.; Repasky M. P.; Knoll E. H.; Shelley M.; Perry J. K.; Shaw D. E.; Francis P.; Shenkin P. S. (2004) Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 47 (7), 1739–49. 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- Tanner L.; Denti P.; Wiesner L.; Warner D. F. (2018) Drug permeation and metabolism in Mycobacterium tuberculosis: Prioritising local exposure as essential criterion in new TB drug development. IUBMB Life 70 (9), 926–937. 10.1002/iub.1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T. C.; Aldridge B. B. (2019) Targeting drugs for tuberculosis. Science 364 (6447), 1234–1235. 10.1126/science.aay0211. [DOI] [PubMed] [Google Scholar]
- Anderson K.; Lai Z.; McDonald O. B.; Stuart J. D.; Nartey E. N.; Hardwicke M. A.; Newlander K.; Dhanak D.; Adams J.; Patrick D.; Copeland R. A.; Tummino P. J.; Yang J. (2009) Biochemical characterization of GSK1070916, a potent and selective inhibitor of Aurora B and Aurora C kinases with an extremely long residence time1. Biochem. J. 420 (2), 259–65. 10.1042/BJ20090121. [DOI] [PubMed] [Google Scholar]
- Gopinathan K. P.; Sirsi M.; Ramakrishnan T. (1963) Nicotin-amide-adenine nucleotides of Mycobacterium tuberculosis H37Rv. Biochem. J. 87, 444–8. 10.1042/bj0870444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boshoff H. I.; Xu X.; Tahlan K.; Dowd C. S.; Pethe K.; Camacho L. R.; Park T. H.; Yun C. S.; Schnappinger D.; Ehrt S.; Williams K. J.; Barry C. E. (2008) Biosynthesis and recycling of nicotinamide cofactors in mycobacterium tuberculosis. An essential role for NAD in nonreplicating bacilli. J. Biol. Chem. 283 (28), 19329–41. 10.1074/jbc.M800694200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S. P.; Alonso S.; Rand L.; Dick T.; Pethe K. (2008) The protonmotive force is required for maintaining ATP homeostasis and viability of hypoxic, nonreplicating Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 105 (33), 11945–50. 10.1073/pnas.0711697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland R. A.; Pompliano D. L.; Meek T. D. (2006) Drug-target residence time and its implications for lead optimization. Nat. Rev. Drug Discovery 5 (9), 730–9. 10.1038/nrd2082. [DOI] [PubMed] [Google Scholar]
- Srinivasan B.; Skolnick J. (2015) Insights into the slow-onset tight-binding inhibition of Escherichia coli dihydrofolate reductase: detailed mechanistic characterization of pyrrolo [3,2-f] quinazoline-1,3-diamine and its derivatives as novel tight-binding inhibitors. FEBS J. 282 (10), 1922–38. 10.1111/febs.13244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClerren A. L.; Endsley S.; Bowman J. L.; Andersen N. H.; Guan Z.; Rudolph J.; Raetz C. R. (2005) A slow, tight-binding inhibitor of the zinc-dependent deacetylase LpxC of lipid A biosynthesis with antibiotic activity comparable to ciprofloxacin. Biochemistry 44 (50), 16574–83. 10.1021/bi0518186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardona F.; Goti A.; Parmeggiani C.; Parenti P.; Forcella M.; Fusi P.; Cipolla L.; Roberts S. M.; Davies G. J.; Gloster T. M. (2010) Casuarine-6-O-alpha-D-glucoside and its analogues are tight binding inhibitors of insect and bacterial trehalases. Chem. Commun. (Cambridge, U. K.) 46 (15), 2629–31. 10.1039/b926600c. [DOI] [PubMed] [Google Scholar]
- Svetlov M. S.; Vázquez-Laslop N.; Mankin A. S. (2017) Kinetics of drug-ribosome interactions defines the cidality of macrolide antibiotics. Proc. Natl. Acad. Sci. U. S. A. 114 (52), 13673–13678. 10.1073/pnas.1717168115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davoodi S.; Daryaee F.; Chang A.; Walker S. G.; Tonge P. J. (2020) Correlating Drug-Target Residence Time and Post-antibiotic Effect: Insight into Target Vulnerability. ACS Infect. Dis. 6 (4), 629–636. 10.1021/acsinfecdis.9b00484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walkup G. K.; You Z.; Ross P. L.; Allen E. K.; Daryaee F.; Hale M. R.; O’Donnell J.; Ehmann D. E.; Schuck V. J.; Buurman E. T.; Choy A. L.; Hajec L.; Murphy-Benenato K.; Marone V.; Patey S. A.; Grosser L. A.; Johnstone M.; Walker S. G.; Tonge P. J.; Fisher S. L. (2015) Translating slow-binding inhibition kinetics into cellular and in vivo effects. Nat. Chem. Biol. 11 (6), 416–23. 10.1038/nchembio.1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryk R.; Gold B.; Venugopal A.; Singh J.; Samy R.; Pupek K.; Cao H.; Popescu C.; Gurney M.; Hotha S.; Cherian J.; Rhee K.; Ly L.; Converse P. J.; Ehrt S.; Vandal O.; Jiang X.; Schneider J.; Lin G.; Nathan C. (2008) Selective killing of nonreplicating mycobacteria. Cell Host Microbe 3 (3), 137–45. 10.1016/j.chom.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams P. D.; Afonine P. V.; Bunkoczi G.; Chen V. B.; Davis I. W.; Echols N.; Headd J. J.; Hung L. W.; Kapral G. J.; Grosse-Kunstleve R. W.; McCoy A. J.; Moriarty N. W.; Oeffner R.; Read R. J.; Richardson D. C.; Richardson J. S.; Terwilliger T. C.; Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr., Sect. D: Biol. Crystallogr. 66 (Pt 2), 213–221. 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P.; Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr., Sect. D: Biol. Crystallogr. 60, 2126–2132. 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Chen V. B.; Arendall W. B. III; Headd J. J.; Keedy D. A.; Immormino R. M.; Kapral G. J.; Murray L. W.; Richardson J. S.; Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr., Sect. D: Biol. Crystallogr. 66 (Pt 1), 12–21. 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.