Abstract
Background
The activation of tumor-associated macrophages (TAMs) facilitates the progression of gastric cancer (GC). Cell metabolism reprogramming has been shown to play a vital role in the polarization of TAMs. However, the role of methionine metabolism in function of TAMs remains to be explored.
Methods
Monocytes/macrophages were isolated from peripheral blood, tumor tissues or normal tissues from healthy donors or patients with GC. The role of methionine metabolism in the activation of TAMs was evaluated with both in vivo analyses and in vitro experiments. Pharmacological inhibition of the methionine cycle and modulation of key metabolic genes was employed, where molecular and biological analyses were performed.
Results
TAMs have increased methionine cycle activity that are mainly attributed to elevated methionine adenosyltransferase II alpha (MAT2A) levels. MAT2A modulates the activation and maintenance of the phenotype of TAMs and mediates the upregulation of RIP1 by increasing the histone H3K4 methylation (H3K4me3) at its promoter regions.
Conclusions
Our data cast light on a novel mechanism by which methionine metabolism regulates the anti-inflammatory functions of monocytes in GC. MAT2A might be a potential therapeutic target for cancer cells as well as TAMs in GC.
Keywords: tumor microenvironment, tumor escape, macrophages, immunomodulation, immune evation
Introduction
Gastric cancer (GC) is the fourth most frequent malignancy and the third leading cause of cancer-related mortality around the world.1 2 Despite the great advances in GC screening and therapy, the prognosis of patients with GC at advanced stages remains to be dismal.3 The advent of immunotherapy has greatly changed the landscape of cancer therapy.4 Cancer immunotherapies, including immune checkpoint inhibitors and adoptive cell transfer, have demonstrated therapeutic benefits in a variety of tumors.5 However, the proportion of patients who respond to immunotherapy remains modest (about 15%) because of resistance and tumor immune evasion.6 An ideal immunotherapy for GC would be relieving immunosuppression through remodeling the tumor microenvironment (TME).5 Thus, further research on the molecular mechanisms underlying tumor immune evasion is required.
Monocytes/macrophages constitute the major component of tumor-infiltrating immune cell population.7 Monocytes/macrophage are commonly educated by the TME and promote tumor progression via diversified mechanisms, including immune evasion.7–9 Previous studies have demonstrated that GC is highly infiltrated with monocytes with activated phenotype (mainly M2 subtype).10–12 The functional phenotype of monocytes/macrophage can be influenced by surrounding milieu.7–9 However, the underlying molecular mechanisms are largely unclear.
A growing volume of literature have demonstrated the important role of cell metabolism in the activity and function of immune cells, including macrophage,13–15 T cells16 17 and dendritic cells.18 19 Two main subtypes of macrophages have been defined with distinct metabolic profiles.20 The anti-inflammatory M2 macrophage is characterized by increased oxidative phosphorylation and fatty acid oxidation, while the classical proinflammatory M1 macrophage is characterized by enhanced glycolysis.21 These metabolic characteristics have been shown to be vital in sustaining the functional phenotypes of macrophages; however, the precise molecular mechanism remains to be explored.21
Cell metabolism provides energy and material basis for cellular signaling transduction.13 Methionine, an essential amino acid, has been shown to play a vital role in the maintenance of stemness of embryonic22 and pluripotent stem cells23 as well as carcinogenesis.24 S-adenosylmethionine (SAM) is generated from methionine by a family of conserved methionine adenosyltransferase (MAT) enzymes. The catalytic subunits of MAT enzymes are derived from two genes, the ubiquitously expressed methionine adenosyltransferase II alpha (MAT2A) genes and the liver-specific MAT1A.25 There has been an established link between methionine adenosyltransferases and the progression of cancer, including GC.26 27 However, whether MAT enzymes are also involved in tumor-associated macrophage (TAM) polarization remains unknown. Interestingly, Covarrubias et al28 performed unbiased metabolic profiling of M2 macrophages and found that methionine metabolism was among the top enriched pathways. This bioinformatics analysis result makes us consider the effects of methionine metabolism on TAM. SAM is a major biological methyl donor, which is required for methylation.24 Recent studies have shown that epigenetic modifications, including acetylation15 22 29 and methylation30–32 modulate the polarization and activation of macrophages. While much attention has been paid to the effect on glycolysis and lipid metabolism in the macrophage, little is known about the role of its methionine metabolism.
In this study, we have found that the upregulated MAT enzyme MAT2A regulates the cellular SAM levels and histone methylation pattern in monocytes/macrophages. MAT2A epigenetically induces RIP1 expression and a protumor phenotype in macrophages.
Materials and methods
Patients and clinical samples
The study was censored by the Clinical Research Committee of Yijishan Hospital, Wenzhou People’s Hospital and the Fudan University Shanghai Cancer Center. The GC tissues were confirmed pathologically. A cohort of patients involving 42 paired blood samples, GC tissues and normal tissues were obtained from Yijishan Hospital who underwent surgery in Yijishan Hospital between June 2019 and October 2019 (n=32) and October 3, 2020 and October 17, 2020 (n=4 from Yijishan Hospital, n=6 from Wenzhou People’s Hospital). These samples were used for the purification of peripheral blood-infiltrating and tumor-infiltrating leukocytes. Written consent was obtained from all patients. All of these patients were preoperative chemotherapy or radiotherapy naïve. No data that could identify patients were provided. Blood samples were collected from 31 healthy donors who worked at Yijishan Hospital. Heparin tubes (BD Biosciences, San Jose, California, USA) were used for the collection of blood samples, and all blood-related experiments were carried out with fresh cells. The clinicopathological characteristics of the patients with GC in this study are provided in online supplemental table S1.
jitc-2020-001364supp001.pdf (1.1MB, pdf)
Isolation of monocytes from peripheral blood and tissue samples
Leukocytes were isolated from the peripheral blood using Ficoll density gradient centrifugation assay (AS1114547; Axis-Shield) as previously described.13 33 GC tissues or paired normal tissues were cut into small pieces and digested in RPMI 1640 (HyClone, Invitrogen, Camarillo, California, USA) supplemented with 20% fetal bovine serum (FBS, HyClone, Invitrogen, Camarillo, California, USA), 0.05% collagenase IV (Sigma, St. Louis, Missouri, USA) and 0.005% DNase I (Roche Diagnostics, Indianapolis, Indiana, USA) for 1 hour at 37°C. The dispersed monocytes were filtered through anylon mesh of 150 mm pore size and separated by Ficoll centrifugation. The obtained monocytes were washed with phosphate buffered solution (PBS) three times and resuspended in RPMI 1640 supplemented with 10% FBS. CD14+ monocytes and autologous T lymphocytes were isolated using magnetic beads (130-050-201/130-095-130, Miltenyi Biotec, Bergisch Gladbach, Germany) following the manufacturer’s instructions and further verified for CD14 expression using flow cytometry analysis with a purity of >88% CD14+.
Preparation of methionine starvation medium
Methionine starvation media were generated from RPMI 1640 powder (US Biomedical) lacking methionine as described previously.24
Preparation of tumor culture supernatants (TSNs) from gastric cancer cells and tumor-conditioned monocytes
The origin and characterization (including DNA fingerprinting, cell vitality and mycoplasma detection) of human gastric cell lines (BGC823 and MGC803) used in this study were performed as described previously.34 The latest cell characterization was performed in September 2019. TSNs were prepared as described previously.13 5×106 GC cells in 10 mL Dulbecco’s modified Eagle’s medium (DMEM, Gibco BRL, Grand Island, New York, USA), were seeded into 100 mm tissue culture dishes for 24 hours thereafter changing the medium to complete medium supplemented with 10% FBS (HyClone, Invitrogen, Camarillo, CA, USA).
The supernatant was then collected, centrifuged (10,000 g, 15 min) and stored in aliquots at −80°C.
In vitro culture of monocytes
Purified CD14+ monocytes were cultured in DMEM supplemented with 10% FBS in the presence or absence of 30% TSN. In some cases, purified CD14+ monocytes were treated with medium or TSN in the presence or absence of FIDAS-5 (10 µM, Merck Millipore, USA), RIP1 inhibitor (GSK547, 50 nM, GlaxoSmithKline, Brentford, UK).
In other experiments, monocytes were pretreated with DMSO, 2-aminobicyclo-(2,2,1)-heptane-2-carboxylic acid (BCH) (5 mM, Sigma, St. Louis, Missouri, USA), Trichostatin A (TSA) (200 ng/mL, Sigma, St. Louis, Missouri, USA), chaetocin (0.5 µM, Sigma, St. Louis, Missouri, USA), OICR-9429 (5 µM, Sigma, St. Louis, Missouri, USA), BIX-01294 (5 µM, Sigma, St. Louis, Missouri, USA), OG-L002 (50 µM, Sigma, St. Louis, Missouri, USA) and DZNep (1 µM, Sigma, St. Louis, Missouri, USA) for 1 hour before their exposure to medium or TSN for indicated times. In some cases, monocytes were transfected with desired siRNA before exposure to medium or TSN.
ELISA
The concentration of the cytokines in the culture supernatants of monocytes was determined with the ELISA assay following the manufacturers’ instructions (eBioscience, Santiago, California, USA).
Coculture of monocytes and T cells
The coculture of monocytes and T cells was performed as described previously.13 33 The CD14+ cells purified from peripheral blood of healthy donors were treated with medium or TSN for 24 hours. Then, the CD14+ cells were washed with PBS three times and cocultured with autologous T cells at a 1:2 ratio for 24 hours. They were cultured in DMEM supplemented with anti-CD3 (2 µg/mL) and anti-CD28 antibodies (2 µg/mL).
RNA and protein extraction, real-time qPCR analysis, western blot analysis and chromatin immunoprecipitation (ChIP)
RNA and protein extraction, real-time qPCR analysis, western blot and ChIP analysis were performed as described previously.35 Primers used in this study are provided in online supplemental table S3. Information about antibodies is provided in online supplemental table S4.
UPLC-MS analysis
Untargeted liquid chromatograph-mass spectrometer (LC-MS) analysis of polar metabolites was performed by using an ultra performance liquid (UPLC) system (ACQUITY, Waters) interfaced with a mass spectrometer (LTQ-Orbitrap, Thermo Scientific) as described previously.24 Elution conditions for LC/MS analysis are provided in online supplemental table S5. Optimized compound-dependent MS parameters are provided in online supplemental table S6.
Tumor cell apoptosis assay
CD14+ cells purified from the peripheral blood were treated with medium (MO) or TSN (TMO) for 24 hours in the presence or absence of MAT2A inhibitor FIDAS-5 (10 µM, Merck Millipore, USA). The monocytes were then washed with cold PBS three times and exposed to autologous CD8+ T cells (with a 1:2 ratio) for 24 hours. These cells were collected and cocultured with Carboxyfluorescein diacetate, succinimidyl ester (CSFE)-labeled MGC803 cells (with a 10:1 ratio) for 12 hours. Flow cytometry assay was used to determine the apoptosis of tumor cell.
Quantitative histone methylation proteomics
Quantitative histone methylation proteomics were performed as described previously.36
Plasmid construction and cell transfection
Plasmid construction and cell transfections were performed as described previously.35
Cell viability assay
Cell viability was determined with the Cell Counting Kit 8 (CCK-8, Donjindo).
In vivo experiments
Nude mice (age 4–5 weeks) were purchased from the Shanghai Experimental Animal Center of the Chinese Academy of Sciences, Shanghai, and housed in a pathogen-free facility in the Experimental Animal Centre of Yijishan Hospital. All animal experiments of laboratory animals were performed by the Guide for the Care and Use published by the US NIH (NIH publication number 85–23, revised 1996). 1×106 MGC803 cells in 0.1 mL PBS were injected subcutaneously into unilateral flank areas of nude mice. 5×106 purified autologous T cells conditioned with monocytes or TSN treated monocytes in the presence or absence of MAT2A inhibitor were subsequently injected into the peritoneum in 0.1 mL PBS every 5 days 1 day after inoculation. Tumor growth was determined every 5 days for 30 days using calipers fitted with a vernier scale. Then, tumor-bearing mice were sacrificed. The tumors formed were surgically removed and weighed. The length (L) and width (W) were measured with calipers, and tumor volumes were calculated using the following formula: (L*W2)/2.
Statistical analysis
All statistical analyses were performed using SPSS software (V.17.0, Chicago, Illinois, USA). All data are presented as the mean±SD from three independent repeats. The correlation between MAT2A and RIP1 expression levels was evaluated with Spearman rank correlation test. The differences between two groups were analyzed using the Student’s t test. P values were two-sided, and a value of <0.05 was considered to be statistically significant. Detailed descriptions of Materials and methods can be found in the online supplemental file 2.
jitc-2020-001364supp002.pdf (153.9KB, pdf)
Results
Tumor-infiltrating monocytes/macrophages in GC had enhanced level of methionine cycle activity
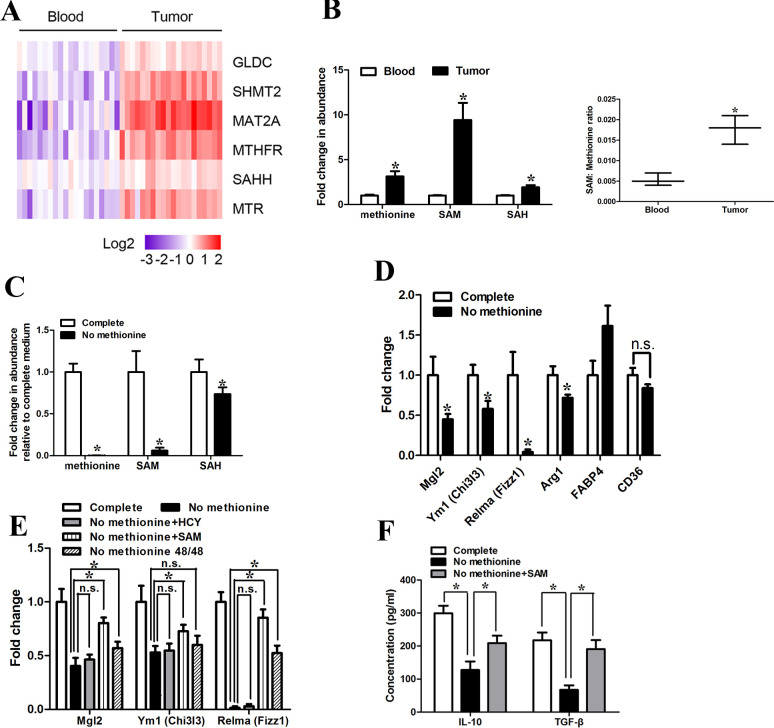
To explore the role of methionine metabolism in GC, we determined the expression of a panel of genes involved in the methionine metabolism (online supplemental figure S1A) with qRT-PCR in CD14+ cells purified from tumor tissues and peripheral blood of 15 patients with GC (online supplemental table S2). We found that the key methionine metabolism-related genes (SHMT2, MAT2A, MTHFR, MTR) were significantly dysregulated in CD14+ cells purified from GC tissues compared with that from paired peripheral blood in 15 patients with GC (figure 1A). Among the key methionine metabolism-related genes, MAT2A was consistently upregulated and demonstrated the highest fold-change. A significant correlation between MAT2A expression in CD14+ cells purified from GC tissues and from paired peripheral blood was observed (r2=0.592, p<0.001, online supplemental figure S1B). Moreover, CD14+ cells isolated from tumor tissues had elevated levels of methionine, S-adenosyl methionine (SAM) and S-adenosyl homocysteine (SAH) (figure 1B), and the ratio of SAM to methionine (the indicator of the capacity for methylation reactions) was increased in CD14+ cells derived from tumor tissues (figure 1B). To further characterize the phenotype of monocytes purified from GC tissues, we used CD86 (M1 marker) and CD163 (M2 marker).10–12 We found that the costimulatory molecule CD86 was almost absent, while strong appearance of CD163 was observed in monocytes isolated from GC tissues, indicating an overwhelming majority of the M2 phenotype. However, the expression of CD163 was significantly downregulated in CD14+ cells purified from peripheral blood of healthy donors (online supplemental figure S1C). To further investigate the role of methionine cycle metabolites in the function of the tumor-infiltrating monocytes/macrophages, we exposed the CD14+ cells purified from GC tissues to transient 48 hours methionine starvation. The methionine starvation for 48 hours reduced methionine cycle activity, as exemplified by a drastic decrease in SAM levels and a slight decrease in SAH levels (figure 1C). Forty-eight hours methionine starvation led to a significant decrease in the expression of M2 genes (Mgl2, Ym1 (Chi3I3), Relma (Fizz1), Arg1(marginally significant, p=0.045)), while CD36 and FABP4 were not affected or even inducible (figure 1D). It is worth noting that shorter-term starvation (24 hours) was sufficient to induce significant change in the expression of M2 genes (Mgl2, Ym1 (Chi3I3), Relma (Fizz1)) (online supplemental figure S1D), which highlighted the dependence of TAM on methionine. Furthermore, there was no significant difference in cell viability after 48 hours methionine starvation (online supplemental figure S1E).
Figure 1.
Tumor-infiltrating macrophages of GC had enhanced levels of methionine metabolism activity. (A) CD14+ cells were purified from the tumor tissues and peripheral blood of five patients with GC. The expression levels of methionine metabolism-related gene were quantified by qRT-PCR. (B) (Left) LC-MS was used to determine abundance of intracellular primary methionine cycle metabolites. Values were normalized to that in CD14+ cells purified from peripheral blood. (Right) Ratio of SAM to methionine levels in CD14+ cells purified from peripheral blood and tumor tissues. (C) LC-MS was used to determine abundance of methionine cycle metabolites in CD14+ cells purified from tumor tissues 48 hours after methionine starvation. Values were normalized to that in the complete condition. (D) qPCR analysis of the M2-associated genes Relma, Mgl2, Ym1, Fabp4, Arg1 and CD36 48 hours after methionine starvation. (E) CD14+ cells purified from tumor tissues were starved for 48 hours for methionine but supplemented with homocysteine (HCY; 250 µM), SAM (500 µM) or replated into complete medium for the next 48 hours (48/48). qPCR analysis of the M2-associated genes Relma, Mgl2, Ym1. (F) CD14+ cells purified from tumor tissues were starved for 48 hours for methionine but supplemented with SAM (500 µM). The levels of IL-10 and TGF-β production were determined by ELISA. Data are presented as the mean±SD; *p<0.05. N=3 biologically independent experiments. GC, gastric cancer; SAM, S-adenosylmethionine.
To further clarify the role of methionine cycle activity in the maintenance of the phenotype of tumor-infiltrating macrophages, we attempted to rescue the methionine-starved cells with homocysteine (HCY, 250 µM), SAM (500 µM) or non-starvation reversal. As would have been anticipated, homocysteine supplementation (250 µM) was unable to attenuate the effects of methionine starvation, which suggested that tumor-infiltrating monocytes/macrophages required exogenous methionine for phenotype maintenance (figure 1E). SAM supplementation strategy was used to bypass the requirement of methionine for tumor-infiltrating macrophages. The downregulation of M2 genes induced by methionine starvation was greatly ameliorated with SAM supplementation (500 µM) (figure 1E). However, the attenuation level of M2 gene expression when macrophages were recovered in complete medium was not as dramatic as that with SAM supplementation (figure 1E). Surprisingly, we found that while methionine starvation markedly decreased the production of IL-10 and TGF-β, SAM supplementation significantly rescued the production of IL-10 and TGF-β in tumor-infiltrating monocytes/macrophages (figure 1F).
MAT2A modulates methionine metabolism-induced RIP1 expression on monocytes
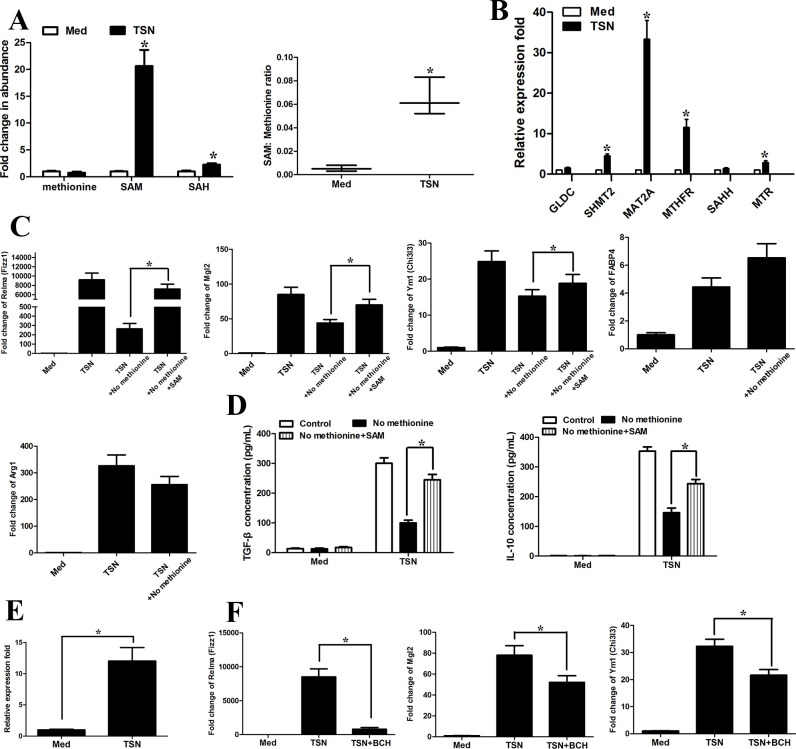
To further confirm the relationship between increased methionine cycle activity and cancer microenvironment, we treated with monocytes purified from healthy peripheral blood with GC cell line MGC803 derived tumor cell supernatants (TSN). TSN treatment resulted in significantly enhanced levels of SAM, SAH and SAM:methionine ratio compared with those in the control medium-treated monocytes (figure 2A). TSN-exposed monocytes also demonstrated higher transcripts levels of key methionine metabolism-related genes (figure 2B). As anticipated, TSNs derived from BGC823 cells markedly increased the intracellular levels of SAM, SAH and SAM:methionine ratio (online supplemental figure 2A).
Figure 2.
Methionine mediates tumor-induced M2 phenotype of monocytes. CD14+ cells were isolated from peripheral blood of healthy donors. CD14+ cells were left untreated or treated with TSN from MGC803 cells for 20 hours. (A) (Left) LC-MS was used to determine the abundance of methionine cycle metabolites in CD14+ cells purified from tumor tissues 48 hours after methionine starvation. Values were normalized to that of CD14+ cells in the absence of TSN. (Right) Ratio of SAM to methionine levels in CD14+ cells. (B) The expression levels of methionine metabolism-related gene with qRT-PCR. (C, D) Levels of the M2-associated genes Relma, Mgl2, Ym1, Fabp4, Arg1 by qPCR (C) and IL-10 and TGF-β production (D) by ELISA. (E) qRT-PCR analysis of the expression of amino acid transporter SLC7A5 on TSN treatment. (F) CD14+ cells were pretreated with DMSO or system L transporters (BCH, 5 mM) for 1 hour and then were exposed to medium (Med) or TSN for 20 hours. qRT-PCR analysis of the M2-associated genes Mgl2, Ym1 (Chi3I3) and Relma (Fizz1). Data are presented as the mean±SD; *p<0.05. N=3 biologically independent experiments. SAM, S-adenosylmethionine; TSN, tumor culture supernatant.
As TAMs have been reported to promote immune escape within the cancer microenvironment via diverse mechanisms,21 29 we would like to explore the role of methionine metabolism in TAM-associated antitumor immunity. TSN induced marked upregulation of Mgl2, Ym1 (Chi3I3) and Relma (Fizz1) (figure 2C) as well as IL-10 and TGF-β production (figure 2D), while transient methionine 24 hours starvation before TSN treatment significantly blunted the effects of TSN (figure 2C, D). As expected, SAM supplementation attenuated the suppressive effect of methionine starvation (figure 2C, D). Furthermore, the expression of amino acid transporter SLC7A537 was increased on TSN treatment (figure 2E). The competitive blocker for system L transporters (BCH, 5 mM) significantly blunted the promoting effects of TSN on Mgl2, Ym1 (Chi3I3) and Relma (Fizz1) (figure 2F).
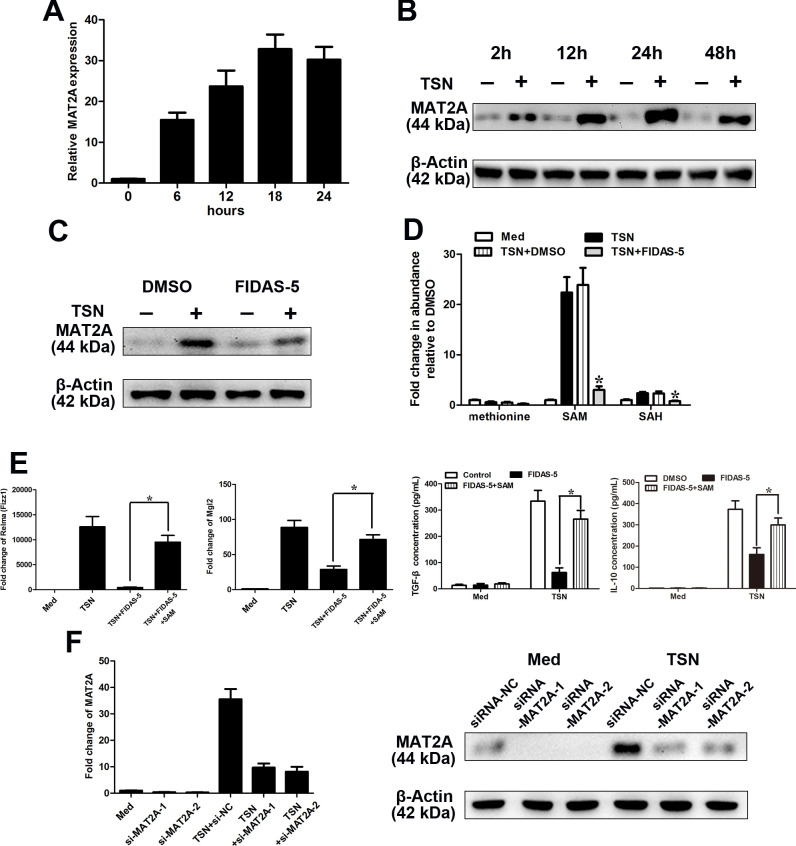
Among the key methionine metabolism-related genes dysregulated in tumor-infiltrating or TSN-exposed monocytes, MAT2A demonstrated the highest fold-change (figure 2B). TSN increased the expression of MAT2A in a time-dependent manner, which peaked at 18 hours (figure 3A). Furthermore, TSN also induced a significant increase in the protein level of MAT2A (figure 3B). To investigate whether MAT2A played a role in the establishment of the phenotype of TAM, we treated CD14+ cells derived from normal blood with TSN in the presence or absence of MAT2A inhibitor FIDAS-5.38 FIDAS-5 treatment (10 µM) significantly attenuated the increase in the MAT2A protein level in TSN-exposed monocytes (figure 3C). Furthermore, we observed that the MAT2A inhibitor FIDAS-5 (10 µM) obviously downregulated the intracellular levels of SAM and SAH (figure 3D). Transient exposure to FIDAS-5 did not result in significant differences in cell viability in TSN-treated CD14+ cells (online supplemental figure 2B). As anticipated, FIDAS-5 inhibitor significantly attenuated the IL-10, TGF-β, Mgl2 and Relma (Fizz1) production/expression in/on TSN-exposed monocytes (figure 3E), while supplementation with SAM (500 µM) in the context of FIDAS-5 treatment markedly rescued the IL-10, TGF-β, Mgl2 and Relma (Fizz1) production/expression (figure 3E). To further confirm the role of MAT2A in the TAM phenotype, we employed the RNA interference strategy to modulate the expression level of MAT2A. As illustrated in figure 3F, si-MAT2A obviously abrogated the upregulation of MAT2A in TSN-exposed monocytes. MAT2A silencing led to suppression on M2 markers, which could be phenocopied by SAM supplementation (online supplemental figure 3).
Figure 3.
MAT2A modulates the M2-phenotype of monocytes. (A–F) CD14+ cells were isolated from peripheral blood of healthy donors. (A, B) CD14+ cells were treated with MGC803 TSN for the indicated times, and the levels of MAT2A expression were determined by qRT-PCR (A) and western blotting (B) analysis. (C, D) CD14+ cells were left treated or untreated with MGC803 TSN for 20 hours in the presence or absence of FIDAS-5 treatment (10 µM). (C) The expression level of MAT2A was determined by western blotting. (D) LC-MS was used to determine the abundance of methionine cycle metabolites in CD14+ cells. Values were normalized to that in the absence of TSN. (E) CD14+ cells were left untreated or treated with MGC803 TSN for 20 hours in the presence or absence of FIDAS-5 treatment (10 µM) or SAM supplementation (500 µM). Levels of the M2-associated genes Relma, Mgl2 by qPCR and IL-10 and TGF-β production by ELISA. (F) CD14+ cells were transfected with siRNA-NC or siRNA-MAT2A and then untreated or treated with TSN for 28 hours. The expression levels of MAT2A were determined by qRT-PCR (left panel) and western blotting (right panel). Data are presented as the mean±SD; *p<0.05. N=3 biologically independent experiments. MAT2A, methionine adenosyltransferase II alpha; SAM, S-adenosylmethionine; TSN, tumor culture supernatant.
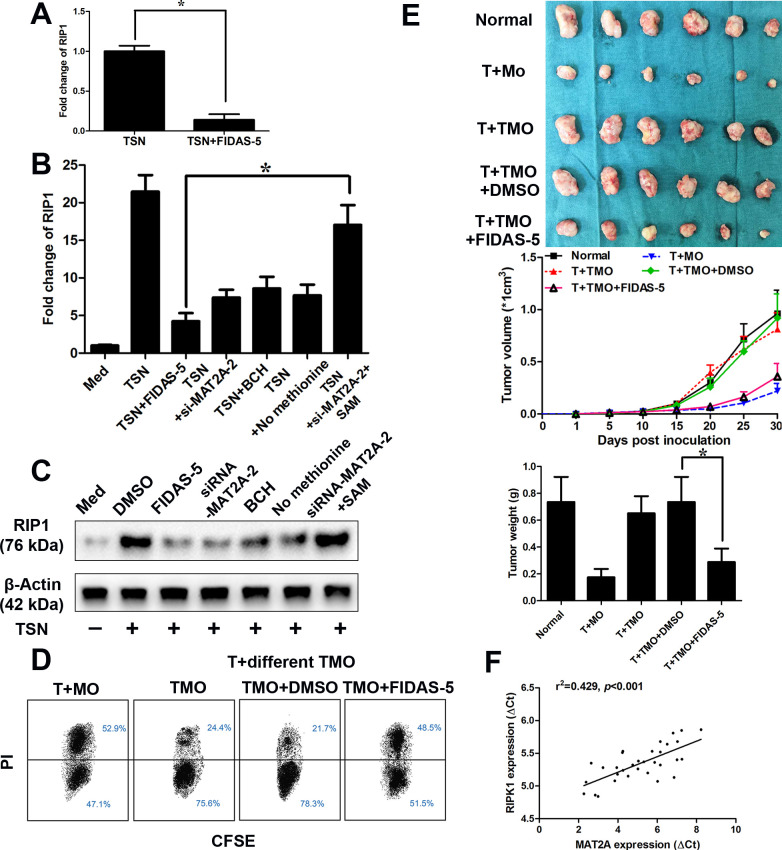
As TSN treatment induced the antitumor immunity in monocytes, we then screened the reported the regulators of the immunosuppressive effects of TAM (online supplemental table S7). Among the dysregulated regulators of the immunosuppressive effects of TAM, RIP1 exhibited the most significant fold-change in TSN-exposed monocytes pretreated with MAT2A inhibitor (figure 4A). The increase in the mRNA level of RIP1 in response to TSN could be significantly attenuated by MAT2A inhibitor (FIDAS-5, 10 µM), siRNA mediated-MAT2A silencing, the competitive blocker for system L transporters (BCH, 5 mM) and transient methionine starvation. However, SAM supplementation could rescue the decrease induced by MAT2A inhibition (figure 4B, C). MAT2A suppression also abrogated the RIP1 expression on monocytes exposed to TSN from BGC823 cells (online supplemental figure 4A). Furthermore, RIP1 inhibitor39 (GSK547, 50 nM) obviously suppressed the upregulation/production of M2 gene in response to TSN (online supplemental figure 4B, C). Functionally, MAT2A inhibitor antagonized the suppressive effect on the antitumor activity of autologous CD8+ T lymphocytes exerted by TSN (figure 4D, E).
Figure 4.
MAT2A modulates the RIP1 expression on monocytes. (A–E) CD14+ cells were isolated from peripheral blood of healthy donors. (A) CD14+ cells were treated with MGC803 TSN for 20 hours in the presence or absence of FIDAS-5 treatment (10 µM). qRT-PCR analysis of RIP1. (B, C) CD14+ cells transfected with siRNA-NC or siRNA-MAT2A were left untreated or treated with MGC803 TSN for 20 hours in the presence or absence of FIDAS-5 treatment (10 µM), system L transporters (BCH, 5 mM), starved for 48 hours for methionine or SAM supplementation (500 µM). qRT-PCR analysis (B) and western blotting of RIP1 (C). (D, E) CD14+ cells were isolated from peripheral blood of healthy donors and left untreated (MO) or treated with MGC803 TSN (TMO) for 20 hours in the presence or absence of FIDAS-5 treatment (10 µM). These cells were then washed with PBS and exposed to autologous CD8+ T cells for 20 hours (with a 1:2 ratio), (D) before the latter were harvested and cocultured with CSFE-labeled MGC803 cells for another 12 hours (with a 10:1 ratio). The death of tumor cells was analyzed through flow cytometry. T+MO: Tumor cells treated with MO-primed T cells; T+different TMO: tumor cells treated with different TMO-primed T cells. (E) These cells (treated as described in D) were then washed and injected into the established NOD/SCID mice bearing MGC803-derived gastric cancer with autologous CD8+ T cells. Top left, representative images of tumors formed (n=6 per group). Top middle, tumor growth curves. Top right, tumor weights. (F) The correlation between MAT2A and RIP1 mRNA levels was measured in CD14+ cells isolated from tumor tissues of 32 patients with GC. The ΔCt values (normalized to β-actin) were subjected to Spearman rank-correlation analysis. Data are presented as the mean±SD; *p<0.05. N=3 biologically independent experiments. GC, gastric cancer; MAT2A, methionine adenosyltransferase II alpha; NOD/SCID, non-obese diabetic/severe combined immunodeficiency; SAM, S-adenosylmethionine; TSN, tumor culture supernatant.
Higher expression levels of MAT2A/RIP1 are correlated with a significantly poorer overall survival and progression-free survival in GC according to data from the KMPlot database (online supplemental figure 5A, B). According to data from TIMER (http://cistrome.org/TIMER/), a significant correlation between MAT2A and RIP1 expression was observed in GC (online supplemental figure 5C). Correlation results between MAT2A and RIP1 in GEPIA (http://gepia.cancer-pku.cn/) were similar to that in TIMER (online supplemental figure 5D). Such a correlation could also be found in other cancers (online supplemental figure 6A, B). A positive correlation between the transcript levels of MAT2A and RIP1 was observed in monocytes purified from GC tissues (n=32, R2=0.501, p<0.001, figure 4F).
MAT2A leads to epigenetic activation of RIP1 expression on monocytes
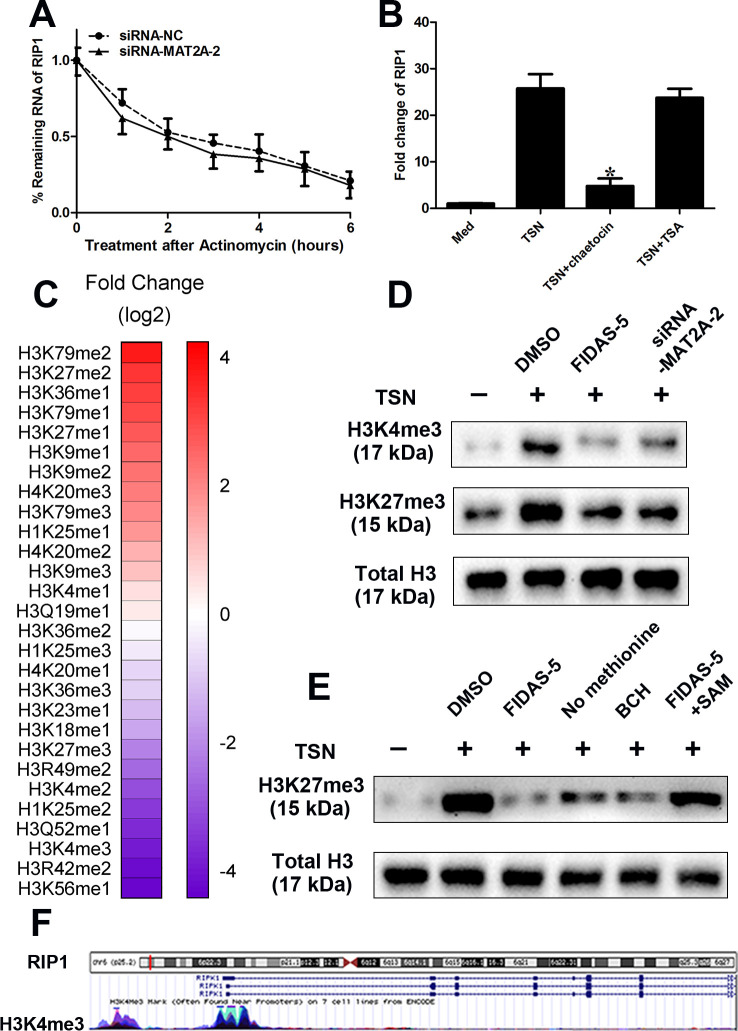
It was well known that cellular SAM levels could affect the histone methylation. Thus, we hypothesized that induction of MAT2A drives gene expression changes and acquisition of M2 phenotype through epigenetic activation/repression of vital regulators. To further clarify the contribution of epigenetic mechanisms, we transfected CD14+ monocytes purified from healthy peripheral blood with siRNA-MAT2A or siRNA-NC, then exposed to TSN for 14 hours, a time course for RNA stability was started after treatment with actinomycin D (0.5 mg/mL). Our data showed that the degradation rate of RIP1 appeared to be similar to MAT2A silencing (figure 5A). The CD14+ monocytes purified from healthy peripheral blood were pretreated with histone methyltransferase (HMT) inhibitor (chaetocin, 0.5 µM) or histone deacetylases inhibitor (TSA, 200 ng/mL) for 1 hour before exposure to TSN. We found that HMT inhibitor significantly abolished the induction of RIP1 by TSN, while TSA had no such effect (figure 5B). Furthermore, chaetocin treatment greatly suppressed the induction of M2 genes induced by TSN, which directly supported the role of histone methylation in the regulation of TAM (online supplemental figure 7). Altogether, these data suggested that histone methylation might play a role in the modulation of RIP1, which was consistent with the previous studies that methionine metabolism could influence histone methylation of the epigenome.40
Figure 5.
MAT2A leads to epigenetic activation of RIP1 on monocytes. (A–F) CD14+ cells were isolated from peripheral blood of healthy donors. (A) CD14+ cells were transfected with siRNA-NC or siRNA-MAT2A and then treated with TSN for 14 hours. Then, a time course for RNA stability was started by adding actinomycin D (0.5 mg/mL). Cells were harvested at the indicated time points. RIP1 mRNA levels determined by qPCR were normalized to “0 hour.” (B) The monocytes were pretreated with histone methyltransferase inhibitor (chaetocin, 0.5 µM) or histone deacetylases inhibitor (TSA, 200 ng/mL) for 1 hour before exposure to TSN for 20 hours. The expression levels of MAT2A were determined by western blotting. (C) CD14+ cells were treated with or without FIDAS-5 treatment (10 µM) and then treated with TSN for 20 hours. Quantitative histone methylation proteomics following chromatin extraction. (D) Cells were treated with MGC803 TSN for 20 hours in the presence or absence of FIDAS-5 treatment (10 µM) or siRNA-MAT2A. Western blot analyses of H3K4me3. Total histone H3 was used as a loading control. (E) UCSC Genome Bioinformatics Site (http://genome.ucsc.edu/) showed high enrichment of H3K4me3 at the promoter of RIP1. (F) Cells transfected with desired vectors were treated with MGC803 TSN for 20 hours in the presence or absence of FIDAS-5 treatment (10 µM), methionine starvation and system L transporters (5 mM) or SAM supplementation (500 µM). Western blot analyses of H3K4me3. Total histone H3 was used as a loading control. Data are presented as the mean±SD; *p<0.05. N=3 biologically independent experiments. MAT2A, methionine adenosyltransferase II alpha; SAM, S-adenosylmethionine; TSN, tumor culture supernatant.
To explore how methionine metabolism affected histone methylation in TAM, we used targeted histone proteomic analyses using a multireaction monitoring approach to quantify relative levels of histone lysine and arginine methylation.34 MAT2A inhibitor decreased the histone methylation at residues associated with transcriptional modulation, including H3K4 and H3K27 trimethylation (me3) (figure 5C). Our data were consistent with the previous studies,22 41 42 which indicated that methionine metabolism, especially the concentration of SAM, regulated histone methylation. It mainly had effect on trimethylation at lysine-4 on histone H3 (H3K4me3) and at lysine-27 on histone H3 (H3K27me3). Immunoblot analysis confirmed that H3K4me3 and H3K27me3 were decreased with MAT2A suppression (figure 5D). Moreover, we found high enrichment of H3K4me3 at the promoter region of RIP1 according to University of California (UCSC, Santa Cruz) Genome Bioinformatics Site (http://genome.ucsc.edu/) (figure 5E). It prompted us to determine whether it was H3K4me3 that was responsible for the transactivation of RIP1. As predicted, TSN exposure greatly increased global H3K4me3 levels in CD14+ monocytes derived from healthy peripheral blood, while MAT2A suppression, methionine starvation and system L transporters markedly decreased the H3K4me3 levels (figure 5F). SAM supplementation rescued the effects of MAT2A suppression on histone methylation (figure 5F).
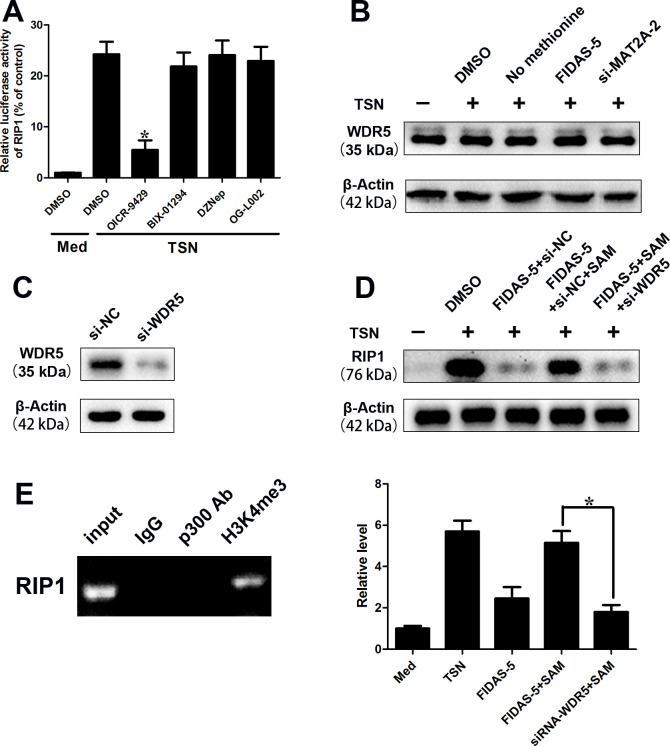
WDR5, a vital component of SET/MLL (SET-domain/mixed-lineage leukemia) histone-methyltransferase complexes, played a key role in H3K4me3 and subsequent transactivation of target genes.32 We constructed the luciferase reporter vector containing the 5’ flanking DNA fragment (~3 kbp) of RIP1 promoter region. We found that TSN treatment enhanced the luciferase activity of pGL3 enhancer plasmid of RIP1 (figure 6A). To determine the specific type of HMT that took part in the transcriptional regulation of RIP1, we pretreated monocyte cells with three pharmacological HMT/demethylases inhibitors: OICR-9429 (an inhibitor of WDR5, 5 µM), BIX-01294 (an inhibitor of G9a HMT, 5 µM), DZNep (an inhibitor of EZH2,1 μM) and OG-L002 (an inhibitor of LSD1, 50 µM). We observed that OICR-9429 treatment ameliorated the elevated luciferase activity of pGL3 plasmid of RIP1 induced by TSN exposure in monocytes (figure 6A). It indicated that WDR5 might take part in the regulation of RIP1. Furthermore, none of the TSN exposure, methionine starvation, MAT2A suppression had any significant effect on the protein level of WDR5 (figure 6B). We found that WDR5 silencing (figure 6C) abolished the effect of SAM supplementation on and RIP1 levels (figure 6D). The chromatin IP (ChIP) assay illustrated that obvious enrichment of H3K4me3 was found at the promoter region of RIP1 (figure 6E). TSN treatment greatly increased the H3K4me3 levels at the promoter region, while MAT2A suppression abolished these effects (figure 6E). SAM attenuated the effects of MAT2A suppression; however, such effects disappeared with WDR5 silencing (figure 6E). These data suggest that MAT2A leads to epigenetic activation of RIP1 expression on monocytes.
Figure 6.
MAT2A-mediated H3K4me3 modification regulates RIP1 expression. (A–E) CD14+ cells were isolated from peripheral blood of healthy donors. (A) Luciferase reporter vector was constructed by inserting the promoter region (−2500 to 500 bp) of the RIP1 gene into pGL3 enhancer plasmid. The reporter vectors were then cotransfected into CD14+ cells. Cells were pretreated with DMSO or OICR-9429 (5 µM) or BIX-01294 (5 µM) or DZNep (1 µM) or OG-L002 (50 µM) for 1 hour before exposure to TSN for 20 hours. Cells were harvested and subjected to luciferase activity assay. (B) Western blotting analysis of WDR5 in CD14+ cells after TSN exposure, methionine starvation, MAT2A suppression. (C) Western blot was performed to detect WDR5 expression after transfection of WDR5 siRNA in CD14+ cells. (D) Western blotting analysis of RIP1 in CD14+ cells treated with TSN in presence or absence of MAT2A inhibitor or SAM supplementation or WDR5 silencing. (E) (Left panel) The RIP1 DNA was detected in the chromatin sample immunoprecipitated from CD14+ cells using an antibody against H3K4me3. (Right panel) Real-time PCR of the ChIP samples shows the binding efficacy of H3K4me3 to the RIP1 gene promoter treated with TSN in presence or absence of MAT2A inhibitor or SAM supplementation or WDR5 silencing. Data are presented as the mean±SD; *p<0.05. N=3 biologically independent experiments. ChIP, chromatin immunoprecipitation; GC, gastric cancer; MAT2A, methionine adenosyltransferase II alpha; SAM, S-adenosylmethionine; TSN, tumor culture supernatant.
Discussion
The polarization of monocytes/macrophages requires distinct transcriptional profiles, in which epigenetic regulation plays an important role. It is well known that cell metabolism is vital for the polarization of macrophages. Metabolic reprogramming may support macrophage polarization by increasing uptake of certain nutrients (glutamine, glucose and lipid) and reprogramming of cellular biosynthetic and biogenetic pathways.20 21 In this study, we established a link between methionine metabolism and epigenetic reprogramming to support M2 polarization. We found that depletion of exogeneous methionine or inhibition of methionine adenosyltransferase enzyme (MAT2A) impaired the polarization of macrophages. Methylation patterns demonstrated great dependence on SAM content, with genes involved in M2 polarization showing decreased levels of H3K4me3 on the promoter region on SAM starvation. The effect of methionine metabolism on immune surveillance was confirmed by both in vitro and ex vivo findings.
There has been an established link between methionine adenosyltransferases and the progression of cancer, including GC.26 27 However, the role of methionine metabolism in TAM remains unknown. Yu et al43 reported that SAM generation maintains a high SAM:methionine ratio to support histone H3K36me3 for macrophage IL-1b production. The discrepancy between our study and the study by Yu et al43 may be due to differences in stimulation conditions (ie, lipopolysaccharide (LPS) by Yu et al). The two studies may suggest that methionine metabolism is important to the M1 and M2 polarization of macrophages. Yu et al43 demonstrate that MAT2A inhibitor exerts the greatest change on H3K36me3 on LPS stimulation. It is known that HMTs have different affinities for SAM. For example, due to the lower Km of H3K36me3 methyltransferase SETD2 compared with H3K4me3 methyltransferase WDR5/MLL1,44 H3K4me3 abundance would be lower in the competition between these enzymes for limited SAM availability. It suggests that the impact of SAM bioavailability on methylation pattern depends both on stimulation conditions and Km of HMTs.
Although this study concentrates on the effect of methionine metabolism on histone methylation pattern, it is worth noting that SAM availability affects a plethora of cellular processes, including DNA/RNA methylation, which may contribute to the M2 polarization induced by MAT2A inhibition. It is reported that SAM depletion reduced the N6-methyladenosine (m6A) in the 3' Untranslated Regions (UTR) of MAT2A, leading to mRNA stabilization.45 46 To exclude the possibility that m6A might be involved in the regulation of RIP1, we tested the effect of MAT2A silencing on the stability of RIP1 mRNA. We found that MAT2A silencing had no effect on the stability of RIP1 mRNA, thus excluding the possibility that m6A is involved in the regulation of RIP1 transcript. Roy et al47 found that acute methionine starvation (6 hours) had limited impact on the level of lineage-specific transcription factors (eg, RORγt expression by Th17 cells). However, we found that methionine starvation (48 hours) suppressed the expression of M2 markers. It still requires investigation whether extended exposure (>6 hours) to methionine starvation of established Th17 cells would result in changes in methylation markers (ie, H3K4me3) that affect Th cell lineage stability.
Previous studies demonstrate that TAMs within the TME (5%–10% of the tumor stromal mass) prominently exhibit an M2-like phenotype and promote tumor growth and metastasis via different mechanisms, including tumor inmmune evasion.7–9 Thus, modulating macrophage function represents an attractive approach. Inhibitors targeting MAT2A would modulate the TAM and cancer cells with high methionine metabolism rate, which would provide more ideal TMEs for chemotherapies or immune-therapies.
In conclusion, our study showed the detailed mechanistic insight of MAT2A-WDR5-RIP1 axis in TAM. This study suggests a novel cancer immune modulatory strategy.
Footnotes
YZ, HY and JZ contributed equally.
Correction notice: This paper has been updated since first published to amend author name 'Kun Lv'. Additionally, the provenance and peer review statement has been added.
Contributors: MM, YZ, JZ, HY, YH and XY acquired data, drafted the manuscript, and critically revised the manuscript for important intellectual content. MM, YZ, HY, PW, YH and YRH performed in vitro and in vivo assays. MM, YH and XY analyzed and interpreted data and statistical analysis. YZ, YH and LK provided technical or material support. MM, XY, YH and LK approved the final version of the manuscript.
Funding: This work was supported by the National Nature Science Foundation of China (Grant Nos. 81972213 for MMZ, Nos. 81702388 for YRH, Nos.81802503 for HY, Nos. 81 772 180 for KL), the Key University Science Research Project of Anhui Province (KJ2020A0594 for YZ, 1808085MH271 for JZ), Shanghai Training and Support Program for Young Physician (for MMZ), Fudan University Shanghai Cancer Center for Outstanding Youth Scholars Foundation (YJYQ201803 for MMZ), Shanghai Sailing Program (18YF1412700 for PW), Peak Training Program for Scientific Research of Yijishan Hospital, Wannan Medical College (GF2019T01 for KL, GF2019G15 for HY).
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available in a public, open access repository. It is available.
Ethics statements
Patient consent for publication
Not required.
References
- 1.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394–424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115–32. 10.3322/caac.21338 [DOI] [PubMed] [Google Scholar]
- 3.Salati M, Orsi G, Smyth E, et al. Gastric cancer: translating novels concepts into clinical practice. Cancer Treat Rev 2019;79:101889. 10.1016/j.ctrv.2019.101889 [DOI] [PubMed] [Google Scholar]
- 4.Fritz JM, Lenardo MJ. Development of immune checkpoint therapy for cancer. J Exp Med 2019;216:1244–54. 10.1084/jem.20182395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saleh R, Elkord E. Acquired resistance to cancer immunotherapy: role of tumor-mediated immunosuppression. Semin Cancer Biol 2020;65:13–27. 10.1016/j.semcancer.2019.07.017 [DOI] [PubMed] [Google Scholar]
- 6.Goldberg MS. Improving cancer immunotherapy through nanotechnology. Nat Rev Cancer 2019;19:587–602. 10.1038/s41568-019-0186-9 [DOI] [PubMed] [Google Scholar]
- 7.Hinshaw DC, Shevde LA. The tumor microenvironment Innately modulates cancer progression. Cancer Res 2019;79:4557–66. 10.1158/0008-5472.CAN-18-3962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hill BS, Sarnella A, D'Avino G, et al. Recruitment of stromal cells into tumour microenvironment promote the metastatic spread of breast cancer. Semin Cancer Biol 2020;60:202–13. 10.1016/j.semcancer.2019.07.028 [DOI] [PubMed] [Google Scholar]
- 9.DeNardo DG, Ruffell B. Macrophages as regulators of tumour immunity and immunotherapy. Nat Rev Immunol 2019;19:369–82. 10.1038/s41577-019-0127-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang H-C, Chen C-W, Yang C-L, et al. Tumor-Associated macrophages promote epigenetic silencing of gelsolin through DNA methyltransferase 1 in gastric cancer cells. Cancer Immunol Res 2017;5:885–97. 10.1158/2326-6066.CIR-16-0295 [DOI] [PubMed] [Google Scholar]
- 11.Harada K, Dong X, Estrella JS, et al. Tumor-associated macrophage infiltration is highly associated with PD-L1 expression in gastric adenocarcinoma. Gastric Cancer 2018;21:31–40. 10.1007/s10120-017-0760-3 [DOI] [PubMed] [Google Scholar]
- 12.Yamaguchi T, Fushida S, Yamamoto Y, et al. Tumor-associated macrophages of the M2 phenotype contribute to progression in gastric cancer with peritoneal dissemination. Gastric Cancer 2016;19:1052–65. 10.1007/s10120-015-0579-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen D-P, Ning W-R, Jiang Z-Z, et al. Glycolytic activation of peritumoral monocytes fosters immune privilege via the PFKFB3-PD-L1 axis in human hepatocellular carcinoma. J Hepatol 2019;71:333–43. 10.1016/j.jhep.2019.04.007 [DOI] [PubMed] [Google Scholar]
- 14.Divakaruni AS, Hsieh WY, Minarrieta L, et al. Etomoxir inhibits macrophage polarization by disrupting CoA homeostasis. Cell Metab 2018;28:490–503. 10.1016/j.cmet.2018.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Langston PK, Nambu A, Jung J, et al. Glycerol phosphate shuttle enzyme GPD2 regulates macrophage inflammatory responses. Nat Immunol 2019;20:1186–95. 10.1038/s41590-019-0453-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones N, Vincent EE, Cronin JG, et al. Akt and STAT5 mediate naïve human CD4+ T-cell early metabolic response to TCR stimulation. Nat Commun 2019;10:2042. 10.1038/s41467-019-10023-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pacella I, Procaccini C, Focaccetti C, et al. Fatty acid metabolism complements glycolysis in the selective regulatory T cell expansion during tumor growth. Proc Natl Acad Sci U S A 2018;115:E6546–55. 10.1073/pnas.1720113115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hansen IS, Krabbendam L, Bernink JH, et al. FcαRI co-stimulation converts human intestinal CD103+ dendritic cells into pro-inflammatory cells through glycolytic reprogramming. Nat Commun 2018;9:863. 10.1038/s41467-018-03318-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thwe PM, Pelgrom LR, Cooper R, et al. Cell-Intrinsic glycogen metabolism supports early glycolytic reprogramming required for dendritic cell immune responses. Cell Metab 2017;26:558–67. 10.1016/j.cmet.2017.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jha AK, Huang SC-C, Sergushichev A, et al. Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization. Immunity 2015;42:419–30. 10.1016/j.immuni.2015.02.005 [DOI] [PubMed] [Google Scholar]
- 21.Vitale I, Manic G, Coussens LM, et al. Macrophages and metabolism in the tumor microenvironment. Cell Metab 2019;30:36–50. 10.1016/j.cmet.2019.06.001 [DOI] [PubMed] [Google Scholar]
- 22.Shiraki N, Shiraki Y, Tsuyama T, et al. Methionine metabolism regulates maintenance and differentiation of human pluripotent stem cells. Cell Metab 2014;19:780–94. 10.1016/j.cmet.2014.03.017 [DOI] [PubMed] [Google Scholar]
- 23.Chandrasekaran S, Zhang J, Sun Z, et al. Comprehensive mapping of pluripotent stem cell metabolism using dynamic genome-scale network modeling. Cell Rep 2017;21:2965–77. 10.1016/j.celrep.2017.07.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Z, Yip LY, Lee JHJ, et al. Methionine is a metabolic dependency of tumor-initiating cells. Nat Med 2019;25:825–37. 10.1038/s41591-019-0423-5 [DOI] [PubMed] [Google Scholar]
- 25.Frau M, Feo F, Pascale RM. Pleiotropic effects of methionine adenosyltransferases deregulation as determinants of liver cancer progression and prognosis. J Hepatol 2013;59:830–41. 10.1016/j.jhep.2013.04.031 [DOI] [PubMed] [Google Scholar]
- 26.Maldonado LY, Arsene D, Mato JM, et al. Methionine adenosyltransferases in cancers: mechanisms of dysregulation and implications for therapy. Exp Biol Med 2018;243:107–17. 10.1177/1535370217740860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang T, Zheng Z, Liu Y, et al. Overexpression of methionine adenosyltransferase II alpha (MAT2A) in gastric cancer and induction of cell cycle arrest and apoptosis in SGC-7901 cells by shRNA-mediated silencing of MAT2A gene. Acta Histochem 2013;115:48–55. 10.1016/j.acthis.2012.03.006 [DOI] [PubMed] [Google Scholar]
- 28.Covarrubias AJ, Aksoylar HI, Yu J, et al. Akt-mTORC1 signaling regulates Acly to integrate metabolic input to control of macrophage activation. Elife 2016;5:e11612. 10.7554/eLife.11612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lauterbach MA, Hanke JE, Serefidou M, et al. Toll-like receptor signaling rewires macrophage metabolism and promotes histone acetylation via ATP-citrate lyase. Immunity 2019;51:997–1011. 10.1016/j.immuni.2019.11.009 [DOI] [PubMed] [Google Scholar]
- 30.Lecoeur H, Prina E, Rosazza T, et al. Targeting macrophage histone H3 modification as a Leishmania strategy to dampen the NF-κB/NLRP3-Mediated inflammatory response. Cell Rep 2020;30:1870–82. 10.1016/j.celrep.2020.01.030 [DOI] [PubMed] [Google Scholar]
- 31.Kimball AS, Davis FM, denDekker A, et al. The histone methyltransferase Setdb2 modulates macrophage phenotype and uric acid production in diabetic wound repair. Immunity 2019;51:258–71. 10.1016/j.immuni.2019.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li T, Garcia-Gomez A, Morante-Palacios O, et al. SIRT1/2 orchestrate acquisition of DNA methylation and loss of histone H3 activating marks to prevent premature activation of inflammatory genes in macrophages. Nucleic Acids Res 2020;48:665–81. 10.1093/nar/gkz1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuang D-M, Xiao X, Zhao Q, et al. B7-H1-expressing antigen-presenting cells mediate polarization of protumorigenic Th22 subsets. J Clin Invest 2014;124:4657–67. 10.1172/JCI74381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma M, Zhang Y, Weng M, et al. lncRNA GCAWKR promotes gastric cancer development by scaffolding the chromatin modification factors WDR5 and KAT2A. Mol Ther 2018;26:2658–68. 10.1016/j.ymthe.2018.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xiang H, Ramil CP, Hai J, et al. Cancer-associated fibroblasts promote immunosuppression by inducing ROS-generating monocytic MDSCs in lung squamous cell carcinoma. Cancer Immunol Res 2020;8:436–50. 10.1158/2326-6066.CIR-19-0507 [DOI] [PubMed] [Google Scholar]
- 36.Eckert MA, Coscia F, Chryplewicz A, et al. Proteomics reveals NNMT as a master metabolic regulator of cancer-associated fibroblasts. Nature 2019;569:723–8. 10.1038/s41586-019-1173-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sinclair LV, Howden AJ, Brenes A, et al. Antigen receptor control of methionine metabolism in T cells. Elife 2019;8:e44210. 10.7554/eLife.44210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang W, Sviripa V, Chen X, et al. Fluorinated N, N-dialkylaminostilbenes repress colon cancer by targeting methionine S-adenosyltransferase 2A. ACS Chem Biol 2013;8:796–803. 10.1021/cb3005353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang W, Marinis JM, Beal AM, et al. Rip1 kinase drives macrophage-mediated adaptive immune tolerance in pancreatic cancer. Cancer Cell 2018;34:757–74. 10.1016/j.ccell.2018.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sanderson SM, Gao X, Dai Z, et al. Methionine metabolism in health and cancer: a nexus of diet and precision medicine. Nat Rev Cancer 2019;19:625–37. 10.1038/s41568-019-0187-8 [DOI] [PubMed] [Google Scholar]
- 41.Mentch SJ, Mehrmohamadi M, Huang L, et al. Histone methylation dynamics and gene regulation occur through the sensing of one-carbon metabolism. Cell Metab 2015;22:861–73. 10.1016/j.cmet.2015.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bai J, Gao Y, Chen L, et al. Identification of a natural inhibitor of methionine adenosyltransferase 2A regulating one-carbon metabolism in keratinocytes. EBioMedicine 2019;39:575–90. 10.1016/j.ebiom.2018.12.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu W, Wang Z, Zhang K, et al. One-carbon metabolism supports S-adenosylmethionine and histone methylation to drive inflammatory macrophages. Mol Cell 2019;75:1147–60. 10.1016/j.molcel.2019.06.039 [DOI] [PubMed] [Google Scholar]
- 44.Mentch SJ, Locasale JW. One-carbon metabolism and epigenetics: understanding the specificity. Ann N Y Acad Sci 2016;1363:91–8. 10.1111/nyas.12956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shima H, Matsumoto M, Ishigami Y, et al. S-Adenosylmethionine Synthesis Is Regulated by Selective N6-Adenosine Methylation and mRNA Degradation Involving METTL16 and YTHDC1. Cell Rep 2017;21:3354–63. 10.1016/j.celrep.2017.11.092 [DOI] [PubMed] [Google Scholar]
- 46.Doxtader KA, Wang P, Scarborough AM, et al. Structural basis for regulation of METTL16, an S-adenosylmethionine homeostasis factor. Mol Cell 2018;71:1001–11. 10.1016/j.molcel.2018.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roy DG, Chen J, Mamane V, et al. Methionine metabolism shapes T helper cell responses through regulation of epigenetic reprogramming. Cell Metab 2020;31:250–66. 10.1016/j.cmet.2020.01.006 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2020-001364supp001.pdf (1.1MB, pdf)
jitc-2020-001364supp002.pdf (153.9KB, pdf)
Data Availability Statement
Data are available in a public, open access repository. It is available.