CNSL feeding reduced enteric CH4 emission from Vietnamese cattle by 20%. CNSL feeding reduced the relative abundance of the major methanogens in the rumen. CNSL feeding also changed the whole rumen microbiome and its function, and the methanogen switched its bacterial partners by CNSL feeding.

Summary
The effects of cashew nut shell liquid (CNSL) feeding on the methane (CH4) emission and the ruminal microbiome of Lai Sind beef cattle were investigated. Changes in the methane production and rumen microbiome by CNSL feeding were monitored by a respiration chamber and 16S rRNA gene amplicon sequencing respectively. The results demonstrated that CNSL feeding mitigated 20.2%–23.4% of the CH4 emission in vivo without apparent adverse effects on feed intake and feed digestibility. The rumen fluid analysis revealed a significant increase in the proportion of propionate in the total short‐chain fatty acids. The relative abundance of methanogen (order Methanobacteriales) decreased significantly, indicating the direct inhibitory effect of CNSL on methanogens. The predicted function of the rumen microbiome indicated that carbohydrate and lipid metabolisms including propionate production were upregulated by CNSL feeding, whereas CH4 metabolism was downregulated. A network analysis revealed that methanogen changed its partner bacteria after CNSL feeding. The δ13C of CH4 ranged from −74.2‰ to −66.6‰ with significant fluctuation by CNSL feeding, in agreement with the shift of the rumen microbiome. Our findings demonstrate that CNSL feeding can mitigate the CH4 emission from local cattle production systems in South‐East Asia by modifying the rumen microbiome and its function.
Introduction
The livestock sector is a significant source of greenhouse gases (GHG), as it produces 5.6–7.5 GtCO2e year−1, which accounts for approx. 15% of the total anthropogenic GHG emission; the largest source of emission is enteric methane (CH4) (1.6–2.7 GtCO2e year−1), followed by nitrous oxide (N2O) emission associated with feed production (1.3–2.0 GtCO2e year−1) (Herrero et al., 2016). To achieve the goals of the Paris Agreement and to tackle global warming, the enteric CH4 emission from cattle is one of the key targets for GHG emission mitigation. The demand for meat is currently increasing worldwide. In developing countries that have recently experienced economic growth and changes in consumer preferences, the demand for meat is predicted to double by 2050 (Smith et al., 2018). Therefore, to minimize the increase of CH4 emissions by the larger cattle population, options for significantly mitigating enteric CH4 emissions are urgently needed.
One of the approaches for mitigating enteric CH4 emissions is to manipulate livestock diets with various additives. Extensive efforts have already been made in this direction, including studies using chemical compounds that specifically inhibit methanogens (Tomkins et al., 2009; Hristov et al., 2015), antibiotics (Grainger et al., 2008) or electron acceptors (Leng, 2008). Alternatives are natural compounds or agricultural by‐products, which are preferable from the viewpoint of environmental impact and circular agricultural production systems.
Several research groups have reported that the use of cashew nut shell liquid (CNSL) as a feed additive can mitigate enteric CH4 emissions by 18%–73% in vitro (Watanabe et al., 2010; Watanabe et al., 2010; Danielsson et al., 2014) and by 19.3%–38.3% in vivo (Shinkai et al., 2012; Konda et al., 2019). CNSL is a by‐product of cashew processing that consists mainly of anacardic acid, cardol and some other phenols; it is usually used for brake linings or surface coatings, or more recently for green diesel production (Lomonaco et al., 2013; Scaldaferri and Pasa, 2019), and it has an antimicrobial activity for several microbial species (Muroi and Kubo, 1993; Muroi and Kubo, 1996; Green et al., 2007; Rivero‐Cruz et al., 2011; da Silva et al., 2016).
The global cashew nut production in 2017 was 3.97 million t, with 50.6% of the total being produced in Asia, where Vietnam (21.7%) and India (18.8%) being the two largest producers on the Asian continent (FAOSTAT). The effective use of locally produced CNSL within local livestock farming is highly preferable from the point of view of sustainable food production.
Here, we evaluated the effects of CNSL on the CH4 emission by Lai Sind, a widely used breed of Vietnamese beef cattle, by a head‐cage respiration chamber technique. We also assessed feed digestibility to understand the effects of CNSL feeding on animal productivity. To determine the effects of CNSL feeding on the organic matter degradation in the rumen and the underlying mechanism, we measured the δ13C value of the produced CH4 and investigated the rumen microbiome by performing amplicon sequencing of the 16S rRNA gene.
Results
Methane emission, ruminal pH, SCFA concentrations and feed digestibility
The average CH4 emissions determined by the head‐cage chamber technique decreased significantly (P < 0.05) from 37.2 ± 4.4 l kg−1 dry matter intake (DMI) (in period 1 without CNSL feeding) to 28.1 ± 2.3 l kg−1 DMI in period 3 (24.5% reduction) and 28.3 ± 3.3 l kg−1 DMI in period 4 (23.9% reduction) with CNSL feeding (4 g/100 kg BW) in Run 1. This trend continued in Run 2 with the higher CNSL dose at 6 g/100 kg BW, with average CH4 emissions decreasing significantly (P < 0.05) from 35.6 ± 2.6 l kg−1 DMI (in period 1 without CNSL feeding) to 29.9 ± 3.7 l kg−1 DMI in period 3 (16.0% reduction) and 28.2 ± 2.5 l kg−1 DMI in period 4 (20.8% reduction; Fig. 1).
Fig. 1.
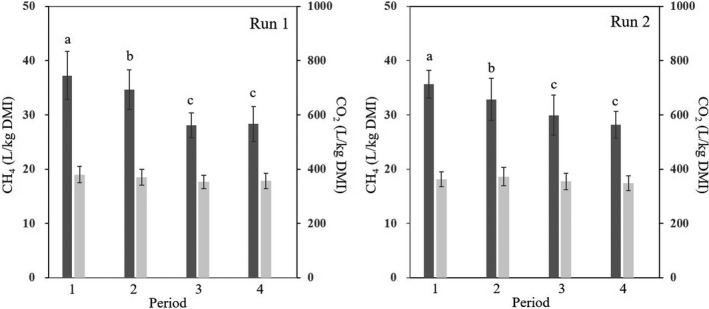
Enteric CH4 (dark grey) and CO2 (light grey) emissions per kg dry matter intake (DMI) from Lai Sind cattle with (periods 2–4) and without (period 1) CNSL feeding (n = 4). Error bars: standard deviation (SD). Different letters indicate significant differences (P < 0.05). Different doses of CNSL were set in Run 1 (4 g/100 kg BW) and Run 2 (6 g/100 kg BW).
The average CO2 emission was consistent in both experiments: it was not significantly affected by CNSL feeding. It ranged from 352.7 ± 25.1 l kg−1 DMI (period 3) to 379.9 ± 29.5 l kg−1 DMI (period 1) in Run 1 and from 347.9 ± 27.2 l kg−1 DMI (period 4) to 372.7 ± 34.2 l kg−1 DMI (period 2) in Run 2. The DMI was not significantly affected by CNSL feeding; it ranged from 4.06 ± 0.37 (period 1) to 4.25 ± 0.44 kg day−1 (period 4) in Run 1 and from 5.61 ± 0.58 (period 2) to 5.90 ± 0.55 kg day−1 (period 1) in Run 2.
The δ13C values of CH4 ranged from −74.2‰ to −66.6‰. The average δ13C values of CH4 in periods 1, 2, 3 and 4 were −71.1 ± 1.5‰ (n = 4, 1σ), −70.4 ± 1.8‰ (n = 4, 1σ), −69.7 ± 2.5‰ (n = 4, 1σ) and −72.0 ± 1.3‰ (n = 4, 1σ) respectively (Fig. 2). The δ13C value increased from period 1 to period 3 and decreased from period 3 to period 4. In contrast, the CH4 emission decreased from period 1 to period 3 and was almost unchanged from period 3 to period 4 (Fig. 1, Table 1). The variations of the δ13C value within individual cattle were higher in periods 2 and 3 compared with periods 1 and 4.
Fig. 2.
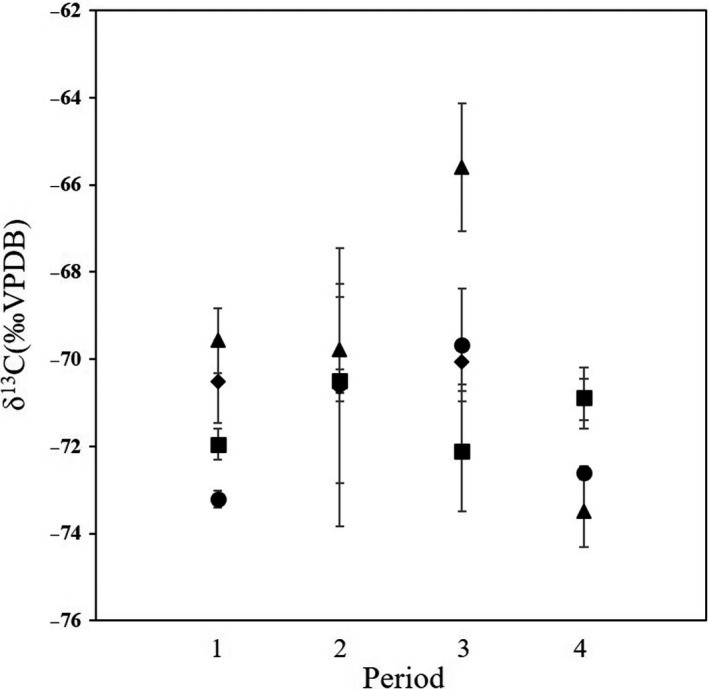
Changes in the δ13C values of enteric CH4 (Run 1). The δ13C values are expressed as relative to the VPDB (Vienna Pee Dee Belemnite). Each symbol (circle, triangle, square and diamond) indicates individual cattle. Error bar: SD (n = 3).
Table 1.
Nutrient digestibility of four Lai Sind cattle with and without cashew nut shell liquid (CNSL) feeding.
| Run | 1 | P | 2 | P | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Period | 1 | SD | 3 | SD | 4 | SD | 1 | SD | 3 | SD | 4 | SD | ||
| DM | 69.9 | 4.9 | 70.6 | 1.8 | 71.8 | 3.6 | 0.69 | 64.5 | 0.6 | 62.3 | 2.7 | 63.4 | 3.4 | 0.40 |
| OM | 72.9 | 4.1 | 73.5 | 1.6 | 74.4 | 3.6 | 0.73 | 67.2 | 0.3 | 65.8 | 1.2 | 67.2 | 3.0 | 0.39 |
| CP | 78.8 | 2.3 | 78.9 | 3.8 | 78.4 | 2.8 | 0.83 | 67.9a | 0.9 | 66.7ab | 1.6 | 64.3b | 3.2 | 0.02 |
| EE | 80.4 | 5.3 | 82.3 | 2.3 | 80.5 | 2.2 | 0.47 | 73.4 | 18.1 | 55.8 | 10.8 | 57.2 | 11.8 | 0.16 |
| NDF | 70.6 | 5.4 | 72.3 | 1.4 | 72.5 | 2.6 | 0.77 | 67.1 | 1.1 | 65.4 | 2.7 | 66.1 | 2.9 | 0.48 |
| GE | 69.9 | 4.3 | 71.4 | 2.0 | 71.8 | 3.3 | 0.76 | 64.1 | 0.5 | 63.8 | 1.7 | 62.6 | 2.8 | 0.47 |
The data are percentages. Different amounts of CNSL were fed in Run 1 (4 g/100 kg BW) and Run 2 (6 g/100 kg BW). Feed without CNSL was fed to cattle as control period 1, and CNSL was fed to the cattle for periods 3 and 4. The detailed experimental schedule can be found in Figure S1. Different superscript letters indicate that the means are significantly different between the periods (P < 0.05). CP, crude protein; DM, dry matter; EE, ether extract; GE, gross energy; NDF, neutral detergent fibre; OM, organic matter.
The changes in the rumen parameters (pH, total short‐chain fatty acids [SCFA] and ammonia‐N concentration) are illustrated in Figure S2. The rumen pH remained at a consistent level (6.9 ± 0.1) throughout Run 1, but it increased from 6.9 ± 0.1 to 7.3 ± 0.1 in Run 2. The total SCFA concentration remained at a mostly consistent level that ranged from 52.3 ± 0.5 mM to 68.6 ± 6.4 mM in Run 1, and it also remained at a consistent level in Run 2, but with the much higher variation of 40.5 ± 11.4 mM to 55.9 ± 23.8 mM among the four cattle in both runs; despite the mostly consistent levels of SCFA, small peaks were detected after CNSL feeding (Fig. S3).
We also determined the digestibility of the feeds, and we did not observe any significant effect of CNSL feeding on the dry matter (DM), organic matter (OM), crude protein (CP), ether extract (EE), neutral detergent fibre (NDF) and gross energy (GE) digestibility in Run 1, whereas a significant reduction in CP digestibility was observed in Run 2 (Table 1).
Diversity and composition of the rumen microbiome
After the quality filtering by a standard DADA2 procedure, 118 783 ± 25 839 reads were obtained for each sample. We evaluated the effects of the CNSL feeding on the rumen microbiome diversity by using six diversity indexes (Brillouin, Chao1, Evenness, Faith's PD, Shannon and Simpson) (Table 2). All six indexes decreased after CNSL feeding. Two indexes showed a significant decrease (P < 0.05) in Run 1 with the lower dose (Chao1: from 800.6 to 588.0; Faith's PD: from 53.03 to 45.51), and five indexes showed a significant decrease (P < 0.05) in Run 2 with the higher dose (Brillouin: from 5.22 to 4.72; Chao1: from 778.2 to 612.3; Evenness: from 0.787 to 0.738; Shannon: from 7.56 to 6.83; and Simpson: from 0.984 to 0.963), indicating that the CNSL feeding significantly reduced the rumen microbiome diversity.
Table 2.
Effect of CNSL feeding on the rumen microbiome diversity.
| Run 1 | Run 2 | |||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 1 | 2 | 3 | 4 | |
| Brillouin | 4.67 | 4.65 | 4.57 | 4.20 | 5.22ab | 5.35a | 5.04b | 4.72c |
| Chao1 | 800.6a | 741.3ab | 663.0ab | 588.0b | 778.2a | 760.1ab | 635.8ab | 612.3b |
| Evenness | 0.700 | 0.705 | 0.704 | 0.661 | 0.787a | 0.810a | 0.785a | 0.738b |
| Faith's PD | 53.03a | 51.3ab | 48.27ab | 45.51b | 46.34 | 45.18 | 42.79 | 41.88 |
| Shannon | 6.75 | 6.72 | 6.61 | 6.08 | 7.56ab | 7.74a | 7.30b | 6.83c |
| Simpson | 0.952 | 0.961 | 0.941 | 0.912 | 0.984a | 0.989a | 0.984a | 0.963b |
Different amounts of CNSL were fed in Run 1 (4 g/100 kg BW) and Run 2 (6 g/100 kg BW). Feed without CNSL was fed to cattle as the control at period 1, and CNSL was fed to the cattle for periods 2 through 4. The detailed experimental schedule is presented in Figure S1. Differing superscript letters indicate that the means are significantly different between the periods (P < 0.05).
The compositions of the rumen microbiome in the two runs are illustrated in Figure 3. The bar plots with order levels show that the CNSL feeding had a significant impact on the rumen microbiome (Fig. 3A). In the comparison between periods 1 and 4, the relative abundance of the following orders decreased in both runs: Erysipelotrichales (3.5 ± 2.1% to 0.6 ± 0.4% in Run 1; 6.2 ± 3.9% to 0.6 ± 0.3% in Run 2), Bacteroidales (58.4 ± 5.2% to 51.5 ± 9.5% in Run 1; 47.2 ± 5.8% to 37.5 ± 5.7% in Run 2) and Methanobacteriales (3.1 ± 0.9% to 1.4 ± 0.6 in Run 1; 4.2 ± 0.8% to 2.3 ± 0.6% in Run 2). The relative abundance of Clostridiales increased (26.2 ± 4.9% to 41.9 ± 11.4% in Run 1; 33.6 ± 2.3% to 46.4 ± 4.1% in Run 2).
Fig. 3.
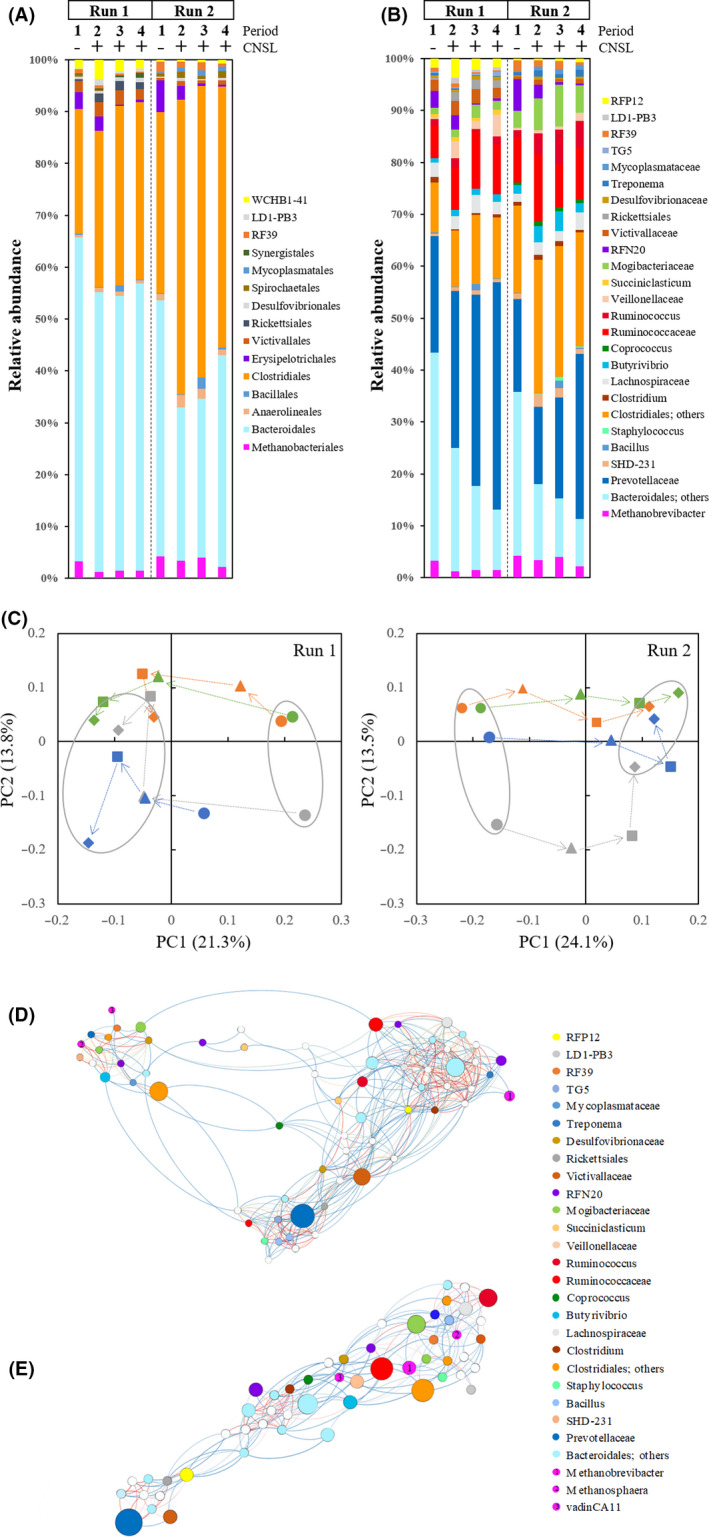
Changes in the ruminal microbiome over time. The ruminal total bacterial/archaeal composition is shown with the order level (A) and family/genus level (B) by the amplicon sequencing of partial 16S rRNA gene. ‘Period’ and ‘CNSL’ indicate the sampling period (after CNSL feeding; period 1 indicates just before CNSL feeding) of ruminal fluid with (+) or without (−) CNSL feeding. (C) Principal component analysis of the ruminal microbiome over time. Each colour indicates a different individual. Circle: period 1, triangle: period 2, square: period 3, diamond: period 4. (D, E) Network analysis of the rumen microbiome in periods 1 (D) and 4 (E). Data from both runs were analysed together. The relative abundance of each group is reflected in the size of the circles. Colours of the lines between each circle indicate the correlation coefficient, from low (red) to high (blue) correlation. Only significant correlations (r > 0.5) are shown.
Since some major orders such as Clostridiales and Bacteroidales consist of many important families with different functions, we broke these orders down to the family or genus level and generated another bar plot to observe the effects of the CNSL feeding in more detail (Fig. 3B). The CNSL feeding exerted a consistent effect on the rumen microbiome, although the variation among the individual cattle was high, with CNSL feeding increasing the abundance of the family Prevotellaceae (20.8 ± 19.7% to 40.3 ± 13.1% in Run 1; 17.1 ± 3.0% to 29.1 ± 8.0% in Run 2) and decreasing the abundance of other species belonging to Bacteroidales. Among the species in the order Clostridiales, the relative abundance of the following three families was significantly increased by CNSL feeding: those belonging to Ruminococcaceae (7.3 ± 2.0% to 10.4 ± 4.2% in Run 1; 9.6 ± 0.3% to 13.8 ± 1.5% in Run 2), Veillonellaceae (0.9 ± 0.3% to 5.0 ± 1.4% in Run 1; 0.4 ± 0.1% to 1.4 ± 0.3% in Run 2) and Mogibacteriaceae (1.1 ± 0.3% to 1.5 ± 0.7% in Run 1; 3.0 ± 0.5% to 4.9 ± 1.2% in Run 2). The relative abundance of the following was significantly decreased: the genus Methanobrevibacter (3.1 ± 0.9% to 1.4 ± 0.5% in Run 1; 4.0 ± 0.7% to 2.0 ± 0.5% in Run 2), which is the most abundant methanogen, and RFN20 (3.1 ± 1.9% to 0.4 ± 0.3% in Run 1; 5.9 ± 4.0% to 0.3 ± 0.4% in Run 2), which belongs to the order Erysipelotrichales.
The results of the principal component analysis confirmed these clear effects of CNSL feeding (Fig. 3C). The initial rumen microbiomes of the four cattle were located in the same area, and they all fell into another independent area in period 4 samples, which was consistent in both runs. We also conducted a network analysis based on the correlation coefficient between each species for periods 1 and 4 to compare the initial and final rumen microbiome (Fig. 3D). The loss of microbial diversity due to the CNSL feeding significantly changed the shape of the rumen microbiome and made it simpler in period 4. The network analysis data are illustrated based on their correlation and thus the connected species with lines indicating the functional partners that share or hand over the substrates and are functionally related in the micro‐ecosystems.
In period 1, the three detected methanogens fell into two clusters that had high correlations with Anaeroplasmatales and Acholeplasmatales, whereas in period 4, the methanogens all had high correlations with different species such as Mogibacteriaceae, Ruminococcaceae and Christensenellaceae as their potential functional partners. These results indicate that the CNSL feeding significantly affected the rumen microbiome ecosystem, including the activity of methanogens and their partners. Prevotellaceae belonged to different clusters in the two periods, indicating that the induction of propionate production and methanogenic activity are independent of each other.
Predicted functions and metabolic pathways of the rumen microbiome
To compare the functional potential of the rumen microbiome with and without CNSL feeding, we used PICRUSt to predict the function of the rumen microbiome (Fig. 4). The CNSL feeding significantly (P < 0.05) affected 34 features in Run 1 and 92 of the 328 features in total. The two runs showed similar trends, with CNSL feeding increasing the lipid, carbohydrate and vitamin metabolisms. However, these differences became significant (P < 0.01) only in Run 2 at the higher CNSL dose (Fig. 4A). In addition, the samples with and without CNSL feeding were well separated in the dendrogram (Fig. 4B), indicating that the predicted functions in the rumen microbiome with and without CNSL feeding were significantly different. The active features affected by the CNSL feeding included the CH4 metabolism (Run 1, P = 0.045; Run 2, P = 0.014). Thus, the CNSL feeding clearly affected the CH4 production and related metabolism by the rumen microbiome, which agrees well with the reduced CH4 production after CNSL feeding (Fig. 1).
Fig. 4.
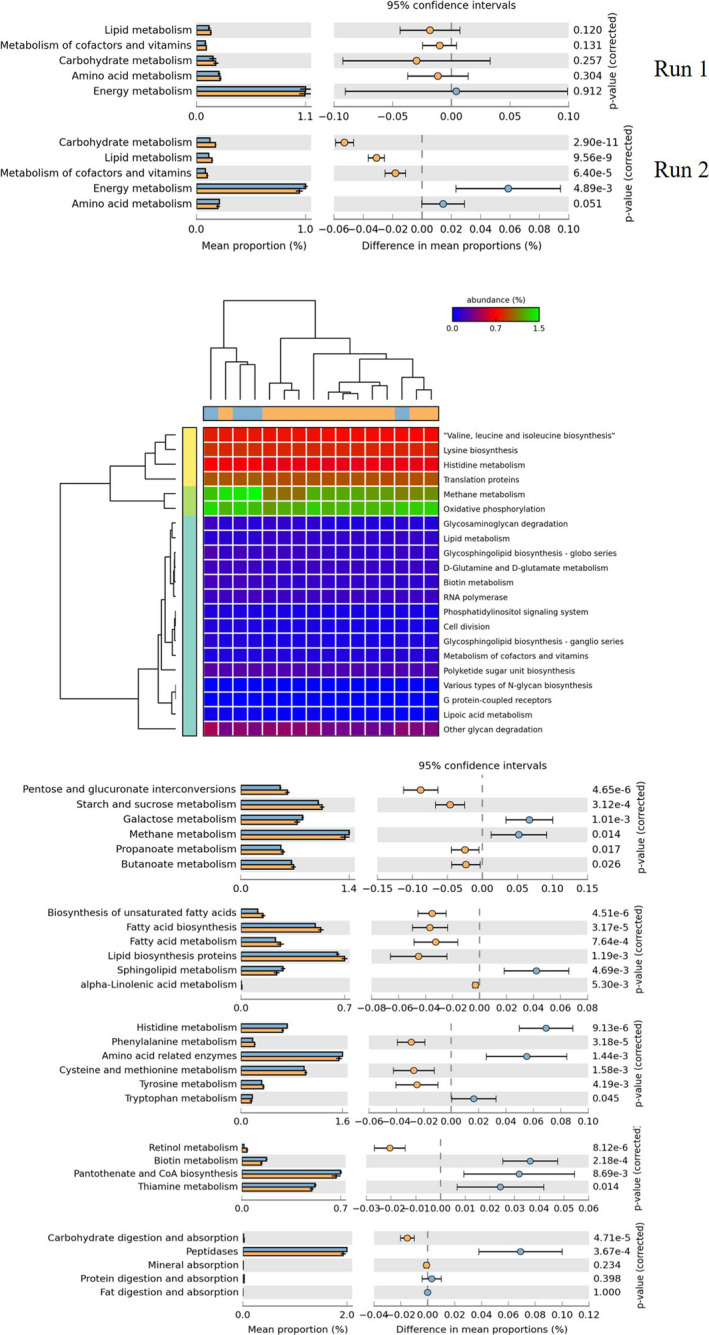
Predicted functions of the ruminal microbiome by PICRUSt.
A. The effect of CNSL feeding on ruminal functions related to feed digestion. Only higher hierarchies in KEGG pathways were selected.
B. Significantly changed features by CNSL feeding in Run 1. Orange and light blue at the bottom of the dendrogram indicate the results with (periods 2–4) and without (period 1) CNSL feeding.
C. Significantly changed features by CNSL feeding in Run 2. The lower hierarchy in KEGG pathways that was related to feed digestion and nutrient intake was selected.
Other metabolisms related to feed degradation and important metabolisms for cattle nutrients are summarized in Figure 4C. This prediction indicates that CNSL feeding might activate the metabolism of starches (P < 0.01), including cellulose, and the metabolism and biosynthesis of fatty acids, including propionate or butyrate (P < 0.05). In regard to the metabolism of amino acids, the effect of the CNSL feeding differed among the amino acid species. Histidine (P < 0.01) and tryptophan (P < 0.05) metabolism appeared to be suppressed by the CNSL feeding, whereas the phenylalanine, cysteine, methionine and tyrosine metabolisms were activated (P < 0.01).
Discussion
The world cattle population increased by 13.4% over the last 20 years, from 1.32 billion in 1998 to 1.49 billion in 2018 (FAOSTAT). In South‐East Asia, the rate of increase within the same time frame was much higher at 37.5%. Greater numbers of cattle can lead to greater environmental pollution, including GHG emissions, and thus, there is an urgent need for a feasible GHG mitigation strategy for local production systems. We focused herein on the GHG mitigation potential of the locally available feed resource CNSL, and our results demonstrated that the feeding of CNSL to Lai Sind, a breed of Vietnamese local cattle, successfully reduced the CH4 emission per kg DMI by 20.8%–23.9% (Fig. 1).
We fed the cattle CNSL at two different doses, but the reduction rate was higher at the lower dose, indicating that this level of ~ 20% is the maximum mitigation potential. Since we had a sufficiently long buffer time with 505 days between runs, we suspect that there was a minimum carryover effect for Run 2. This reduction rate is comparable to the rate of 19.3% determined by respiration chamber measurements in an in vivo study of Holstein dairy cattle (Shinkai et al., 2012) with the same CNSL dose level. More recently, a similar in vitro experiment using incubation of rumen fluid from Thai native cattle and swamp buffalo revealed a CH4 mitigation potential of 53%–73% (Konda et al., 2019). Although it is in vitro study that did not use a respiration chamber and therefore a direct comparison with our study is not possible, these previous results and our present findings indicate that CNSL can be effective regardless of the genetic background of the cattle.
The reduction rate in the present study was lower than that in studies using the chemical inhibitor 3NOP (25%–32%; Hristov et al., 2015) or nitrate (11.8%–29.4%; Newbold et al., 2014) but significantly higher than that in a study using monensin (7–9%; Odongo et al., 2007), which is widely used in the beef production system in the United States, and also significantly higher than those in studies using other natural resources such as tannin (0%–11%; Beauchemin et al., 2007a; Bhatta et al., 2013) or lipid sources (17%; Beauchemin et al., 2007b). More recently, some seaweed species were shown to be effective to mitigate ruminal CH4 emission (26.4%–67.1%), but the feed intake was also significantly reduced (10.8%–38.0%), which may reduce the animals' productivity (Maia et al., 2016; Kinley et al., 2016; Roque et al., 2019). Vietnam is the world's largest producer of cashew, and the production of this crop has increased in recent years, with a cultivation area of 283 thousand ha and production at 2.6 million t in 2018 (FAOSTAT). The cashew shell as a by‐product is abundantly available with low cost for transportation, which is promising for its use as a feed additive in the 5.8 million cattle in Vietnam.
The results of our CH4 isotopic analysis indicated that the CNSL feeding also affected the δ13C values of the emitted CH4 (Fig. 2). The range of the δ13C values (−74.2‰ to −66.6‰) fell within the range of earlier investigations (Rust, 1981; Levin et al., 1993; Bilek et al., 2001; Klevenhusen et al., 2009, 2011), indicating that the main methanogenic pathway is CO2 reduction (Whiticar, 1999). This agrees well with the results of our microbiome analysis, which demonstrated that the major part of the ruminal methanogens was Methanobrevibacter (Fig. 3), since it is known as a hydrogenotrophic methanogen (Leahy et al., 2010). Temporary high variations and enriched δ13C values were observed after the CNSL feeding (periods 2 and 3), which was the opposite of the result in a study of methanogenesis inhibition by 3‐NOP (Lopes et al., 2016). Although we did not measure the δ13C of precursor CO2, these results indicate that the CNSL feeding affected the feed digestion and its isotope fractionation from feed to CO2 and further to CH4 in some individual cattle. This might have been due to the significant disturbance of the rumen microbiome (Fig. 3) by the CNSL feeding, but this is only circumstantial evidence, and much more detailed measurements (including those of precursors) are required in future studies.
We observed that the effects of the CNSL feeding on the ruminal total SCFA, ammonia concentration and pH were small. The pH tended to increase slightly only in Run 2, whereas the total SCFA concentration tended to decrease in both runs with very high variation among the individual cattle (Fig. S2). Moreover, the CNSL feeding affected the proportion of SCFA, and it significantly increased the proportion of propionate (Fig. S3), which agrees with other reports of CNSL feeding experiments (Shinkai et al., 2012; Konda et al., 2019). The effect of the CNSL feeding on feed digestibility was also small. We measured the DM, OM, CP, EE, NDF and GE and observed no significant effect on any of them in Run 1 with the lower dose, indicating that CNSL feeding does not have a negative effect on feed digestion in the rumen. Since this experiment was performed only for short periods (i.e. 48 days for each run), another experiment with longer periods is needed to determine whether this effect can be sustained.
In Run 2 with the 50% higher dose, the digestibility of CP was significantly reduced (Table 1). Moreover, although the difference was not significant, the digestibility of EE in the CNSL‐fed period (periods 3 and 4) also tended to be lower than that of the CNSL‐free period 1. Since the CH4 mitigation potential was in the same range, the lower dose in Run 1 is preferable for practical use. These results indicate that CNSL feeding can modify the rumen fermentation characteristics and change the SCFA profiles without adversely affecting the feed digestion efficiency under the optimum dose, which is very important from the point of view of animal production.
The whole‐genome sequence information of ruminal bacteria has been substantially improved, enabling functional estimations by using the next‐generation sequencing data of marker genes (Langille et al., 2013; Seshadri et al., 2018). High‐throughput sequencing of partial bacterial/archaeal 16S rRNA genes reveals the effects of CNSL feeding on the composition of the rumen microbiome and the metabolic functions in detail. Here, the CNSL feeding clearly reduced the relative abundance of the dominant methanogen Methanobrevibacter, which directly explains the 20% reduction in CH4 emission. The bacterial community in the Lai Sind cattle rumen is dominated by two orders, Bacteroidales and Clostridiales, which together account for > 80% of the whole sequences. CNSL feeding affected the balance of two dominant orders, increasing the Clostridiales abundance and decreasing the Bacteroidales order (Fig. 3A).
More specifically, while the abundance of the order Bacteroidales was decreased by CNSL feeding, the relative abundance of Prevotellaceae, the major group within Bacteroidales, was significantly increased (Fig. 3B). This group is known to be able to produce propionate through fumarate reduction and succinate production pathway (Louis and Flint, 2017), which is a pathway that requires vitamin B12 and consumes hydrogen (Strobel, 1992; Ungerfeld, 2013). Our present analyses revealed that the CNSL feeding also significantly increased the abundance of Ruminococcaceae; some species belonging to this family produce succinate as their end product (Macfarlane and Gibson, 1997). The abundance of another minor group, Veillonellaceae, which is also known to produce propionate through the succinate pathway (Reichardt et al., 2014), was also significantly increased after the CNSL feeding.
The predicted functions of the rumen microbiome also suggest that the CH4 metabolism was significantly downregulated and the propionate or butyrate metabolism was upregulated by the CNSL feeding (Fig. 4B and C). Although the proportion of butyrate in the total SCFA was not changed in our measurements (Fig. S2), all of these results well underpin the concept that with the feeding of CNSL, the cattle's CH4 emission was reduced, while the propionate proportion in total SCFA was increased. We propose the following mechanism to explain these results.
First, the CNSL feeding directly inhibited the methanogenesis by the major methanogen Methanobrevibacter and reduced the relative abundance and activity of this species. Second, the inhibition of methanogenesis led to a greater excess of hydrogen in the rumen (not measured in this study), since methanogenesis is a hydrogen‐consuming process that uses CO2 as an electron acceptor (Lyu et al., 2018). Third, some of the excess hydrogen that was not consumed by methanogenesis was consumed by an alternative hydrogen sink – namely the production of propionate through the succinate pathway (or the biohydrogenation of unsaturated fatty acids) – and the bacterial species that have enzymes that are required for these reactions proliferated in the rumen ecosystem.
The CNSL feeding also affected other bacterial species, increasing the relative abundance of Butyrivibrio. One of the functions of Butyrivibrio in the rumen is the biohydrogenation of unsaturated fatty acids (Maia et al., 2010; Dewanckele et al., 2018). Although it is estimated to be a minor pathway, biohydrogenation can be an alternative hydrogen sink, which might be activated in the methanogenesis‐inhibited rumen ecosystem by CNSL feeding.
The results of our comparison of the network analysis outputs between periods 1 (without CNSL feeding) and 4 (Fig. 3D) indicate that the CNSL feeding significantly changed the structure of the rumen microbiome, making it simpler, which agrees with the results of the diversity index (Table 2). In both periods, Prevotella, which was estimated to be the main propionate producer whose abundance was increased after CNSL feeding, was completely apart from all three methanogens. Although this result was not surprising, this strongly suggests that the methanogenesis and the propionate production by Prevotella are functionally independent. Prevotella tended to have a close relationship with Victivallaceae, Rickettsiales, Acinetobacter and TG5 (Synergistales) in both periods 1 and 4. Previously, Pybus and Onderdonk (1998) showed that Prevotella bivia can provide amino acids to Peptostreptococcus species, which cannot grow in pure culture. Based on this, it may be possible that Prevotella species have this kind of symbiotic relationship with these groups stated above in the complex rumen ecosystem. These results suggest that the function and partners of Prevotella are stable irrespective of CNSL feeding.
In contrast, the most abundant methanogen, Methanobrevibacter, changed its partners after the CNSL feeding. In period 1, two of the three bacterial species that had a significant relationship with Methanobrevibacter belonged to the orders Anaeroplasmatales and Acholeplasmatales, both of which belong to the class Mollicutes. Although their functions are not well understood, these species can be free‐living and have some potential to be parasites of ruminal fungi or protozoa (Joblin and Naylor, 2002). The reconstructed genome of their close relatives revealed that these two species are fermenters and that they utilize simple sugars and possess hydrogenase (Skennerton et al., 2016), indicating that they have some potential to supply hydrogen to the methanogens. The relative abundance of these species was very low (0.1%–0.4%), and their abundance dropped significantly (< 0.1%) after CNSL feeding.
In period 4, species with a significant correlation with Methanobrevibacter changed to other groups such as Mogibacteriaceae, Ruminococcaceae and Christensenellaceae (Fig. 3D). They also had a close relationship with the minor methanogens Methanosphaera and Methanomassillicoccaceae in period 1, indicating that the CNSL feeding forced Methanobrevibacter to switch its partner. These results also suggest that the hydrogen supply from Mollicutes species to Methanobrevibacter was inhibited and that Methanobrevibacter changed and shared its metabolic partners with other methanogens in the CNSL‐fed rumen. This change agrees well with the reduced CH4 emission and Methanobrevibacter population in the CNSL‐fed rumen with lower diversity.
The prediction of the ruminal microbiome function by PICRUSt showed that CNSL feeding can affect not only important features of feed digestion (such as carbohydrate or lipid metabolisms) but also many fundamental processes of the microbes (such as cell division, oxidative phosphorylation and translation proteins) (Fig. 4B). Among them, it is noteworthy that metabolism of carbohydrate, lipid, cofactors and vitamins tended to be upregulated, especially in Run 2 with the higher CNSL dose (Fig. 4A). In the detailed KEGG pathways, the metabolisms of starch and sucrose (including cellulose degradation) were significantly upregulated by the CNSL feeding, whereas the protein digestion and peptidase tended to be higher in the ruminal microbiome without CNSL (Fig. 4C).
These predictions partly agree with the digestibility of the feed; the CP digestibility in Run 2 was significantly affected by CNSL feeding (Table 1), and these predictions indicate that CNSL feeding has some potential for better feed digestion or utilization by the significantly changed rumen microbiome. In addition, the prediction results regarding the ruminal microbiome function biosynthesis of fatty acids (including unsaturated fatty acids) and their metabolism were also significantly upregulated (Fig. 4C), in contrast to the trend for total SCFA (Fig. S2), which had high variation but tended to be decreased by CNSL feeding. Another study also showed that the total SCFA was sometimes significantly lower than in the control (Shinkai et al., 2012). Since the SCFA produced in the rumen can be absorbed from the rumen, these opposite results should be further examined with future experiments.
In conclusion, the feeding of CNSL successfully reduced the enteric CH4 emission from the Vietnamese cattle breed Lai Sind with high reproducibility. The hydrogen consumption in the rumen was switched from methanogenesis to other pathways such as the propionate production pathway or other minor hydrogen sinks. The feed digestion efficiency was not affected by the CNSL feeding, but the prediction of ruminal microbiome function indicated that some microbial pathways for fibre or lipid metabolisms might be upregulated. Since CNSL is abundant in the local agricultural production system in Vietnam, these results pave the way for the efficient utilization of cashew nutshells as a by‐product of the world's primary cashew industry. This approach can contribute to the mitigation of the enteric CH4 emissions in South‐East Asian countries.
Experimental procedures
Animals, diet composition, CNSL feeding and CH4 emission measurement
A CNSL feeding experiment was performed twice on the experimental farm at Can Tho University (CTU). Four male Lai Sind cattle with the mean body weights of 246.1 ± 22.6 kg (Run 1, low dose) and 375 ± 36 kg (Run 2, high dose) were used for the CNSL feeding experiments with 48 days (14 days for adaptation, 7 days of feeding without CNSL and 27 days of feeding with CNSL) for each run (Fig. S1). There was a buffer time (505 days) between the runs to minimize the carryover effect of CNSL feeding. During the experimental period, the cattle were kept individually in their stalls and fed a typical local feed based on rice straw and concentrate (60:40). The cattle were fed 1.7% of dry matter (DM) per kg body weight per day. Details of the feeding composition are summarized in Table S1.
The cattle were fed at 09:00 and 17:00 each day and provided with clean drinking water. Cashew nut shell liquid (CNSL) at the low dose of 4 g/100 kg body weight (BW) for Run 1 and at the high dose of 6 g/100 kg BW for Run 2 was provided by Idemitsu Kosan (Japan) as a mixture with silica powder and was mixed with molasses and concentrate before feeding. Only silica powder was fed during the control period. The cattle were cared for in accordance with institutional guidelines and the Guidelines for Proper Conduct of Animal Experiments (Science Council of Japan, June 1, 2006).
The emission of CH4 was measured by the ventilated hood system installed at the CTU experimental farm (Sakai et al., 2016). Briefly, blowers delivered a constant airflow from the hood into the measurement system, and the concentrations of CO2 and CH4 were measured continuously by a non‐dispersive infrared sensor (IR‐200; Yokogawa Electric, Tokyo, Japan). The detailed schedule of the experiment is presented in Figure S1.
Briefly, experimental feed was fed to all four cattle for 3 weeks for adaptation. Four CH4 measurement periods consisting of 5 days each were used: periods 1 to 4. The cattle were always placed in the chamber system for the entire days during each period. The cattle were fed the control feed (without CNSL) only during Period 1, and throughout periods 2 to 4, they were fed CNSL‐containing feed. During the experimental period, feed intake was recorded based on the given amount of feed after correction of leftovers, and rumen fluid (collected after the morning feeding, around 10 am), total faeces and urine were collected, weighed and stored in a freezer (−20°C) for further analysis.
Stable isotopic analysis of enteric CH4
Gas samples for isotopic signature measurement were taken 2 h after the morning feeding. Each period had three sampling times (Fig. S1). The stable carbon isotope ratio of CH4 was measured by gas chromatography combustion isotope ratio mass spectrometry (GC‐C‐IRMS). Methane in the samples was separated from interfering components (CO) using a column packed with a 5A molecular sieve (10 mm ID × 500 mm length, 30/60 mesh; GL Sciences, Tokyo) before it was concentrated in a cryofocusing trap. The δ13C value was calculated as shown below:
| (1) |
Here, VPDB stands for Vienna Pee Dee Belemnite, the international standard for the 13C/12C ratio. We used 1000 ppm CH4 in He (Taiyo Nissan, Japan) with δ13C = −39.56% as the working standard (Yamada et al., 2005). The precision of the δ13C value of CH4 was estimated as < 0.4% (n = 10, 1σ) based on repeated analyses of the standards. The differences between the measured concentrations and the δ13C of duplicate air samples were 0.1%–0.4%.
Chemical analyses of the feed, faeces, urine and rumen fluid samples
For the analyses of the feed, faeces and urine, the dry matter was measured after drying the samples at 105°C for 24 h. The levels of crude protein (CP), ether extract (EE) and crude ash were determined according to the standard method (Chemists and Horwitz, 1990). The gross energy was determined by an automatic adiabatic bomb calorimeter (C6000; IKA‐Werke, Staufen, Germany). The neutral detergent fibre exclusive of residual ash without inclusion of sodium sulfite (NDF) was determined according to the method of (Van Soest et al., 1991). The apparent digestibilities for nutrients were calculated according to the formula: (nutrient consumed − nutrient in faeces)/nutrient consumed.
Rumen fluid samples were taken orally by using the stomach tube. Ruminal pH was measured with a calibrated electrode (Horiba, Fukuoka, Japan). The ammonium concentration of the rumen fluid was determined by a standard colorimetric method using a spectrophotometer (SmartSpec Plus; Bio‐Rad, Hercules, CA) (O'dell, 1993). The short‐chain fatty acids (SCFAs) of the rumen fluid were measured by high‐performance liquid chromatography (Ultimate 3000; Thermo Fisher Scientific, Waltham, MA) (Guerrant et al., 1982).
DNA extraction and 16S rRNA gene amplicon sequencing
DNA was extracted from 2 ml of rumen fluid samples using Isofecal for Beads Beating (Nippon Gene, Tokyo), quantified by NanoDrop Lite (Thermo Fisher) and stored at −20°C until further analysis.
Partial fragments of the 16S rRNA gene (the V4 hypervariable region) were amplified by two‐step polymerase chain reaction (PCR). Primers 515F and 806R (Caporaso et al., 2011) with Illumina adapter overhang sequences were used for the first‐round PCR with 20 cycles, and indexes were attached to the amplicon with eight additional cycles. Each 20‐µl PCR mixture contained 0.2 µl TaKaRa Ex Taq HS DNA polymerase (TaKaRa Bio, Shiga, Japan) with 2 µl of buffer (10 × buffer), 1.6 µl of 2.5 mM dNTP mix, 1 µl of each forward and reverse primer (10 mM), and 1 µl of template DNA.
The first‐round PCR conditions were as follows: 94°C for 2 min; 20 cycles of 94°C for 30 s, 50°C for 30 s and 72°C for 30 s; and a final round of 72°C for 5 min. The PCR products were purified using an Agencourt AMPure XP purification system (Beckman Coulter, Indianapolis, IN) and used for the second‐round PCR with the following conditions: 94°C for 2 min; 8 cycles of 94°C for 30 s, 60°C for 30 s and 72°C for 30 s; and a final round of 72°C for 5 min. Tag‐indexed PCR products were purified again, and their quality and quantity were checked by an Agilent 2100 Bioanalyzer (Agilent, Santa Clara, CA) and a Qubit 2.0 Fluorometer and dsDNA HS Assay Kit (Life Technologies, Carlsbad, CA) respectively. Qualified amplicons were pooled in equal amounts and sequenced with a 250‐bp paired‐end sequencing protocol (Illumina, San Diego, CA).
Raw sequence reads were processed by qiime 2‐2019.7 (Bolyen et al., 2019). Paired‐end sequences were merged and quality‐filtered by dada2 (Callahan et al., 2016), and the denoised feature table and amplicon sequence variants (ASVs) were used for the taxonomic diversity analysis. Taxonomic classifications were assigned using a naïve Bayes classifier trained on the Greengenes 13_8_99% database, and mitochondria or chloroplast sequences were removed (Bokulich et al., 2018). The statistical analyses for the diversity metrics and the principal component analysis were performed through QIIME 2 (diversity ‘core‐metrics‐phylogenetic’). We used ‘Sparse Cooccurrence Network Investigation for Compositional data’ (SCNIC) in qiime 2 (q2‐SCNIC) to perform the network analysis. The correlation network was built using SparCC; the network was built using edges with a correlation coefficient of at least 0.5, and network was visualized by Gephi.
PICRUSt was used for predicting the function of the rumen microbiome (Langille et al., 2013). The closed‐reference OTUs were normalized by copy number, and a new matrix of predicted functional categories was created with the KEGG database. We used STAMP to analyse the PICRUSt output file (Parks et al., 2014). The DNA sequences from this study were deposited in the DDBJ Sequence Read Archive (accession numbers: DRR225289 to DRR225230).
Statistical analyses
All data were analysed by a one‐way analysis of variance (ANOVA) using the general linear model procedure described by SAS (SAS Institute, 2001nstitute, 2001). The statistical model included treatment, cattle and period. Tukey's multiple range comparison test was used to separate the means. A P‐value < 0.05 was considered significant.
Conflicts of interest
None declared.
Supporting information
Fig. S1. Detailed schedule of the feeding experiment. Check marks indicate that sampling or measurements were done. Grey columns indicate the five‐day metabolic tests with CH4 emission measurements. Body weights were measured at the beginning and the end of each metabolic test. Methane emission was measured continuously with head cage chambers. Feces and urine samples were analyzed for the digestibility measurements. Rumen fluid samples were used for pH, NH4 +‐N, and SCFA concentration measurements.
Fig. S2. Change of the rumen parameters (pH, SCFA and NH4 +‐N concentrations) during the experimental period. Circles indicate total SCFA concentrations, triangles indicate the NH4 +‐N concentrations, and squares indicate the pH. Error bars indicate the standard deviations (n = 4). Arrows indicate the starting time point of CNSL feeding.
Fig. S3. Change of the composition of total SCFA (in % of total SCFA concentration). Circles indicate the acetate, squares indicate the propionate, and triangles indicate the butyrate. Error bars indicate the standard deviation (n = 4). Arrows indicate the starting time point of CNSL feeding.
Table S1. Feed composition (Run1).
Table S2. Feed composition (Run2).
Acknowledgements
This work was implemented under the Japan International Research Center for Agricultural Sciences project 'Climate Change Measures in Agricultural Systems' and supported by a grant‐in‐aid from MEXT, Japan (17H06105), to N.Y.
Microbial Biotechnology (2021) 14(1), 277–290
Funding information This work was implemented under the Japan International Research Center for Agricultural Sciences project ‘Climate Change Measures in Agricultural Systems’ and supported by a grant‐in‐aid from MEXT, Japan (17H06105), to N.Y.
References
- Beauchemin, K. , McGinn, S. , Martinez, T. , and McAllister, T. (2007a) Use of condensed tannin extract from quebracho trees to reduce methane emissions from cattle. J Anim Sci 85: 1990–1996. [DOI] [PubMed] [Google Scholar]
- Beauchemin, K.A. , McGinn, S.M. , and Petit, H.V. (2007b) Methane abatement strategies for cattle: Lipid supplementation of diets. Can J Anim Sci 87: 431–440. [Google Scholar]
- Bhatta, R. , Enishi, O. , Yabumoto, Y. , Nonaka, I. , Takusari, N. , Higuchi, K. , et al (2013) Methane reduction and energy partitioning in goats fed two concentrations of tannin from Mimosa spp. J Agr Sci 151: 119–128. [Google Scholar]
- Bilek, R. , Tyler, S. , Kurihara, M. , and Yagi, K. (2001) Investigation of cattle methane production and emission over a 24‐hour period using measurements of δ13C and δD of emitted CH4 and rumen water. J Geophys Res Atmos 106: 15405–15413. [Google Scholar]
- Bokulich, N.A. , Kaehler, B.D. , Rideout, J.R. , Dillon, M. , Bolyen, E. , Knight, R. , et al (2018) Optimizing taxonomic classification of marker‐gene amplicon sequences with QIIME 2's q2‐feature‐classifier plugin. Microbiome 6: 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolyen, E. , Rideout, J.R. , Dillon, M.R. , Bokulich, N.A. , Abnet, C.C. , Al‐Ghalith, G.A. , et al (2019) Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37: 852–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan, B.J. , McMurdie, P.J. , Rosen, M.J. , Han, A.W. , Johnson, A.J.A. , and Holmes, S.P. (2016) DADA2: High‐resolution sample inference from Illumina amplicon data. Nat Methods 13: 581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso, J.G. , Lauber, C.L. , Walters, W.A. , Berg‐Lyons, D. , Lozupone, C.A. , Turnbaugh, P.J. , et al (2011) Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA 108: 4516–4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chemists, A.A. , and Horwitz, W. (1990) Official methods of analysis. Vol. I, 15th edn Arlington, VA: AOAC, p. 489. [Google Scholar]
- Danielsson, R. , Werner‐Omazic, A. , Ramin, M. , Schnürer, A. , Griinari, M. , Dicksved, J. , and Bertilsson, J. (2014) Effects on enteric methane production and bacterial and archaeal communities by the addition of cashew nut shell extract or glycerol—An in vitro evaluation. J Dairy Sci 97: 5729–5741. [DOI] [PubMed] [Google Scholar]
- Dewanckele, L. , Vlaeminck, B. , Hernandez‐Sanabria, E. , Ruiz‐González, A. , Debruyne, S. , Jeyanathan, J. , and Fievez, V. (2018) Rumen biohydrogenation and microbial community changes upon early life supplementation of 22: 6n–3 enriched microalgae to goats. Front Microbiol 9: 573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Food and Agriculture Organization Corporate Statistical Database (FAOSTAT) . http://www.fao.org/faostat/en/#home
- Grainger, C. , Auldist, M. , Clarke, T. , Beauchemin, K. , McGinn, S. , Hannah, M. , et al (2008) Use of monensin controlled‐release capsules to reduce methane emissions and improve milk production of dairy cows offered pasture supplemented with grain. J Dairy Sci 91: 1159–1165. [DOI] [PubMed] [Google Scholar]
- Green, I.R. , Tocoli, F.E. , Lee, S.H. , Nihei, K.‐I. , and Kubo, I. (2007) Molecular design of anti‐MRSA agents based on the anacardic acid scaffold. Bioorg Med Chem 15: 6236–6241. [DOI] [PubMed] [Google Scholar]
- Guerrant, G. , Lambert, M. , and Moss, C.W. (1982) Analysis of short‐chain acids from anaerobic bacteria by high‐performance liquid chromatography. J Clin Microbiol 16: 355–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero, M. , Henderson, B. , Havlík, P. , Thornton, P.K. , Conant, R.T. , Smith, P. , et al (2016) Greenhouse gas mitigation potentials in the livestock sector. Nat Clim Change 6: 452–461. [Google Scholar]
- Hristov, A.N. , Oh, J. , Giallongo, F. , Frederick, T.W. , Harper, M.T. , Weeks, H.L. , et al (2015) An inhibitor persistently decreased enteric methane emission from dairy cows with no negative effect on milk production. Proc Natl Acad Sci USA 112: 10663–10668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joblin, K. , and Naylor, G. (2002) The ruminal mycoplasmas: a review. J Appl Anim Res 21: 161–179. [Google Scholar]
- Kinley, R.D. , de Nys, R. , Vucko, M.J. , Machado, L. , and Tomkins, N.W. (2016) The red macroalgae Asparagopsis taxiformis is a potent natural antimethanogenic that reduces methane production during in vitro fermentation with rumen fluid. Anim Prod Sci 56: 282–289. [Google Scholar]
- Klevenhusen, F. , Bernasconi, S.M. , Hofstetter, T.B. , Bolotin, J. , Kunz, C. , and Soliva, C.R. (2009) Efficiency of monolaurin in mitigating ruminal methanogenesis and modifying C‐isotope fractionation when incubating diets composed of either C3 or C4 plants in a rumen simulation technique (Rusitec) system. Br J Nutr. 102: 1308–1317. [DOI] [PubMed] [Google Scholar]
- Klevenhusen, F. , Bernasconi, S.M. , Kreuzer, M. , and Soliva, C.R. (2011) Corrigendum to: Experimental validation of the Intergovernmental Panel on Climate Change default values for ruminant‐derived methane and its carbon‐isotope signature. Anim Prod Sci 51: 974. [Google Scholar]
- Konda, S. , Onodera, R. , Kanchanasatit, E. , Boonsaen, P. , Sawanon, S. , Nagashima, K. , et al (2019) Effect of cashew nut shell liquid feeding on fermentation and microbiota in the rumen of Thai native cattle and swamp buffaloes. Livest Sci 226: 99–106. [Google Scholar]
- Langille, M.G. , Zaneveld, J. , Caporaso, J.G. , McDonald, D. , Knights, D. , Reyes, J.A. , et al (2013) Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31: 814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leahy, S.C. , Kelly, W.J. , Altermann, E. , Ronimus, R.S. , Yeoman, C.J. , Pacheco, D.M. , et al (2010) The genome sequence of the rumen methanogen Methanobrevibacter ruminantium reveals new possibilities for controlling ruminant methane emissions. PLoS One 5: e8926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng, R.A. (2008) The potential of feeding nitrate to reduce enteric methane production in ruminants. A report to the department of climate change. Commonwealth Government of Australia, Canberra. Available from http://www.penambulbooks.com
- Levin, I. , Bergamaschi, P. , Dörr, H. , and Trapp, D. (1993) Stable isotopic signature of methane from major sources in Germany. Chemosphere 26: 161–177. [Google Scholar]
- Lomonaco, D. , Maia, F.J.N. , and Mazzetto, S.E. (2013) Thermal evaluation of cashew nutshell liquid as new bioadditives for poly (methyl methacrylate). J Therm Anal Calor 111: 619–626. [Google Scholar]
- Lopes, J. , de Matos, L. , Harper, M. , Giallongo, F. , Oh, J. , Gruen, D. , et al (2016) Effect of 3‐nitrooxypropanol on methane and hydrogen emissions, methane isotopic signature, and ruminal fermentation in dairy cows. J Dairy Sci 99: 5335–5344. [DOI] [PubMed] [Google Scholar]
- Louis, P. , and Flint, H.J. (2017) Formation of propionate and butyrate by the human colonic microbiota. Environ Microbiol 19: 29–41. [DOI] [PubMed] [Google Scholar]
- Lyu, Z. , Shao, N. , Akinyemi, T. , and Whitman, W.B. (2018) Methanogenesis. Curr Biol 28: R727–R732. [DOI] [PubMed] [Google Scholar]
- Macfarlane, G.T. , and Gibson, G.R. (1997) Carbohydrate fermentation, energy transduction and gas metabolism in the human large intestine In Gastrointestinal Microbiology. Chapman & Hall Microbiology Series. Mackie, R.I. , White, B.A. , (eds). Boston, MA, USA: Springer; 10.1007/978-1-4615-4111-0_9 [DOI] [Google Scholar]
- Maia, M.R. , Chaudhary, L.C. , Bestwick, C.S. , Richardson, A.J. , McKain, N. , Larson, T.R. , et al (2010) Toxicity of unsaturated fatty acids to the biohydrogenating ruminal bacterium, Butyrivibrio fibrisolvens . BMC Microbiol 10: 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maia, M.R. , Fonseca, A.J. , Oliveira, H.M. , Mendonça, C. , and Cabrita, A.R. (2016) The potential role of seaweeds in the natural manipulation of rumen fermentation and methane production. Sci Rep 6: 32321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muroi, H. , and Kubo, I. (1993) Bactericidal activity of anacardic acids against Streptococcus mutans and their potentiation. J Agric Food Chem 41: 1780–1783. [Google Scholar]
- Muroi, H. , and Kubo, I. (1996) Antibacterial activity of anacardic acid and totarol, alone and in combination with methicillin, against methicillinresistant Staphylococcus aureus . J Appl Bacter 80: 387–394. [DOI] [PubMed] [Google Scholar]
- Newbold, J. , Van Zijderveld, S. , Hulshof, R. , Fokkink, W. , Leng, R. , Terencio, P. , et al (2014) The effect of incremental levels of dietary nitrate on methane emissions in Holstein steers and performance in Nelore bulls. J Anim Sci 92: 5032–5040. [DOI] [PubMed] [Google Scholar]
- O'Dell, J.W. (1993) Determination of ammonia nitrogen by semi‐automated colorimetry. Method 350.1. US EPA, Cincinnati, OH.
- Odongo, N. , Bagg, R. , Vessie, G. , Dick, P. , Or‐Rashid, M. , Hook, S. , et al (2007) Long‐term effects of feeding monensin on methane production in lactating dairy cows. J Dairy Sci 90: 1781–1788. [DOI] [PubMed] [Google Scholar]
- Parks, D.H. , Tyson, G.W. , Hugenholtz, P. , and Beiko, R.G. (2014) STAMP: Statistical analysis of taxonomic and functional profiles. Bioinformatics 30: 3123–3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pybus, V. , and Onderdonk, A.B. (1998) A commensal symbiosis between Prevotella bivia and Peptostreptococcus anaerobius involves amino acids: potential significance to the pathogenesis of bacterial vaginosis. FEMS Immunol Med Microbiol 22: 317–327. [DOI] [PubMed] [Google Scholar]
- Reichardt, N. , Duncan, S.H. , Young, P. , Belenguer, A. , Leitch, C.M. , Scott, K.P. , et al (2014) Phylogenetic distribution of three pathways for propionate production within the human gut microbiota. ISME J 8: 1323–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivero‐Cruz, B.E. , Esturau, N. , Sánchez‐Nieto, S. , Romero, I. , Castillo‐Juárez, I. , and Rivero‐Cruz, J.F. (2011) Isolation of the new anacardic acid 6‐[16' Z‐nonadecenyl]‐salicylic acid and evaluation of its antimicrobial activity against Streptococcus mutans and Porphyromonas gingivalis . Nat Prod Res 25: 1282–1287. [DOI] [PubMed] [Google Scholar]
- Roque, B.M. , Salwen, J.K. , Kinley, R. , and Kebreab, E. (2019) Inclusion of Asparagopsis armata in lactating dairy cows' diet reduces enteric methane emission by over 50 percent. J Clean Prod 234: 132–138. [Google Scholar]
- Rust, F. (1981) Ruminant methane delta (13C/12C) values: relation to atmospheric methane. Science 211: 1044–1046. [DOI] [PubMed] [Google Scholar]
- Sakai, T. , Kim, C. , Nguyen, V.T. , Suzuki, T. , Hayashi, K. , and Higuchi, K. (2016) Ventilated hood system for measurements of enteric methane emissions in Vietnam. JIRCAS Working Rep 84: 10–14. [Google Scholar]
- SAS Institute (2001) SAS/STAT User's Guide. Cary, NC: SAS Institute. [Google Scholar]
- Scaldaferri, C. , and Pasa, V. (2019) Green diesel production from upgrading of cashew nut shell liquid. Renew Sustain Energy Rev 111: 303–313. [Google Scholar]
- Seshadri, R. , Leahy, S.C. , Attwood, G.T. , Teh, K.H. , Lambie, S.C. , Cookson, A.L. , et al (2018) Cultivation and sequencing of rumen microbiome members from the Hungate1000 Collection. Nat Biotechnol 36: 359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinkai, T. , Enishi, O. , Mitsumori, M. , Higuchi, K. , Kobayashi, Y. , Takenaka, A. , et al (2012) Mitigation of methane production from cattle by feeding cashew nut shell liquid. J Dairy Sci 95: 5308–5316. [DOI] [PubMed] [Google Scholar]
- da Silva, R.A. , Liberio, S.A. , do Amaral, F.M. , do Nascimento, F.R.F. , Torres, L.M.B. , Neto, V.M. , and Guerra, R.N. (2016) Antimicrobial and antioxidant activity of Anacardium occidentale L. flowers in comparison to bark and leaves extracts. J Biosci Med 4: 87–99. [Google Scholar]
- Skennerton, C.T. , Haroon, M.F. , Briegel, A. , Shi, J. , Jensen, G.J. , Tyson, G.W. , and Orphan, V.J. (2016) Phylogenomic analysis of Candidatus 'Izimaplasma'species: Free‐living representatives from a Tenericutes clade found in methane seeps. ISME J 10: 2679–2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, S.B. , Gotoh, T. , and Greenwood, P.L. (2018) Current situation and future prospects for global beef production: overview of special issue. Asian‐Australas J Anim Sci 31: 927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobel, H.J. (1992) Vitamin B12‐dependent propionate production by the ruminal bacterium Prevotella ruminicola 23. Appl Environ Microbiol 58: 2331–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomkins, N. , Colegate, S. , and Hunter, R. (2009) A bromochloromethane formulation reduces enteric methanogenesis in cattle fed grain‐based diets. Anim Prod Sci 49: 1053–1058. [Google Scholar]
- Ungerfeld, E.M. (2013) A theoretical comparison between two ruminal electron sinks. Front Microbiol 4: 319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Soest, P.V. , Robertson, J. , and Lewis, B. (1991) Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. J Dairy Sci 74: 3583–3597. [DOI] [PubMed] [Google Scholar]
- Watanabe, Y. , Suzuki, R. , Koike, S. , Nagashima, K. , Mochizuki, M. , Forster, R. , and Kobayashi, Y. (2010) In vitro evaluation of cashew nut shell liquid as a methane‐inhibiting and propionate‐enhancing agent for ruminants. J Dairy Sci 93: 5258–5267. [DOI] [PubMed] [Google Scholar]
- Whiticar, M.J. (1999) Carbon and hydrogen isotope systematics of bacterial formation and oxidation of methane. Chem Geol 161: 291–314. [Google Scholar]
- Yamada, K. , Yoshida, N. , Nakagawa, F. , and Inoue, G. (2005) Source evaluation of atmospheric methane over western Siberia using double stable isotopic signatures. Org Geochem 36: 717–726. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Detailed schedule of the feeding experiment. Check marks indicate that sampling or measurements were done. Grey columns indicate the five‐day metabolic tests with CH4 emission measurements. Body weights were measured at the beginning and the end of each metabolic test. Methane emission was measured continuously with head cage chambers. Feces and urine samples were analyzed for the digestibility measurements. Rumen fluid samples were used for pH, NH4 +‐N, and SCFA concentration measurements.
Fig. S2. Change of the rumen parameters (pH, SCFA and NH4 +‐N concentrations) during the experimental period. Circles indicate total SCFA concentrations, triangles indicate the NH4 +‐N concentrations, and squares indicate the pH. Error bars indicate the standard deviations (n = 4). Arrows indicate the starting time point of CNSL feeding.
Fig. S3. Change of the composition of total SCFA (in % of total SCFA concentration). Circles indicate the acetate, squares indicate the propionate, and triangles indicate the butyrate. Error bars indicate the standard deviation (n = 4). Arrows indicate the starting time point of CNSL feeding.
Table S1. Feed composition (Run1).
Table S2. Feed composition (Run2).


