Abstract
Background and Purpose:
Pityriasis versicolor (PV) is a common fungal skin infection caused by Malassezia species. Previous studies have shown that the prevalence of PV is influenced by geographic factors. The aim of the current study was to find the epidemiological characteristics of PV and distribution of Malassezia species in the secondary school students living in Hai Phong city, Vietnam.
Materials and Methods:
This study was conducted on 1357 students within the age range of 10 - 16 years selected from four secondary schools in Hai Phong city. The students were screened for PV skin lesions from August 2016 to December 2017. The isolates of Malassezia from PV patients were analyzed by performing direct microscopy and culturing on modified Dixon agar plates, containing gentamicin, at 32oC for 7 days. In the next stage, the fungal strains obtained from patients with positive fungal cultures were identified using the CHROMagarTM Malassezia medium, polymerase chain reaction-restriction fragment length polymorphism techniques, and D1/D2 rDNA genome sequencing.
Results:
Pityriasis versicolor was diagnosed in 305 (22.48%) students and confirmed by clinical appearance and direct examination. A total of 293 (96.07%) samples grew on modified Dixon agar. With regard to demographic characteristics, 50.49% of the PV cases were female, and 57.38% of cases resided in urban areas. Furthermore, 88.52% of the subjects had the illness duration of more than 6 months. Hypopigmented and erythematous skin lesions were also observed in the research participants, with hypopigmentation being the most frequent condition (97.05%). Most of the Malassezia fungal strains were isolated from the back (39.56%), face (23.99%), and chest (16.51%). Malassezia furfur and M. japonica accounted for PV in 96.25% and 3.75% of the cases, respectively. Furthermore, Malassezia furfur was distributed in both rural and urban areas, while M. japonica was found only in the urban areas.
Conclusion:
The findings of the present study were indicative of the high prevalence of Malassezia yeasts, mostly M. furfur, among the students in Hai Phong city, Vietnam
Keywords: Hai Phong city, Malassezia, Pityriasis versicolor, Students, Vietnam
Introduction
The lipophilic yeasts of genus Malassezia are the members of the resident skin microflora in humans and other warm-blooded animals [ 1 , 2 ]. These yeasts are associated with some skin diseases, such as pityriasis versicolor (PV), seborrheic dermatitis, scalp dandruff, atopic dermatitis, and folliculitis [ 3 , 4 ]. To date, 14 species have been identified in Malassezia genus [ 5 , 7 ], 10 cases of which have been isolated from humans. These species include M. dermatitis, M. furfur, M. globosa, M. japonica, M. obtusa, M. pachydermatis, M. restricta, M. slooffiae, M. sympodialis and M. yamatoensis [ 6 , 8 ].
Malassezia species are a normal part of human commensal skin flora; however, they could be responsible for cutaneous diseases, mainly PV [ 6 ]. The risk factors for PV are high temperature, high relative humidity, fatty skin, corticosteroid treatment, immunodeficiency diseases, and overcrowded households [ 9 ],[ 10 ]. In addition, some studies have shown that Malassezia may be associated with pachydermatis fungemia, cephalic pustulosis and fungal bloodstream infections, especially in neonates [ 6 , 11 , 12 ].
Malassezia species can be identified by phenotypic, biochemical, and physiological techniques [ 7 ]. However, the differentiation of these species, based on these methods is difficult because some species have very similar characteristics, especially for the newly identified species [ 6 , 7 ]. Because of the limitations of morphological and biochemical methods, several molecular approaches, using different targets, have been successfully used for the species identification of Malassezia. Some of these approaches include polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) based on ITS-rDNA regions and 26S genes [ 6 , 9 , 13 ], nested PCR [ 2 ], multiplex PCR [ 14 ], real-time PCR [ 7 ], and sequence analysis [ 15 ].
The secondary school students in Vietnam (10-16 years old) begin to go through puberty; therefore, they have highly active sebaceous glands. These students are at a playful age with excessive sweating. Based on the literature, Malassezia exists in different countries, including Vietnam [ 1 , 16 , 17 ]. Nevertheless, there are limited data on Malassezia infections in the secondary students in Hai Phong city, Vietnam. Hai Phong is located in the north of Vietnam with the all-year averaging temperature of 23-26oC and humidity of 80-85%. With this background in mind, the present study was conducted to determine the epidemiological characteristics of PV and distribution of Malassezia species in the secondary school students living in Hai Phong city, Vietnam.
Materials and Methods
Study population
A total of 1,357 students within the age range of 10-16 years were selected from four secondary schools, namely Vinh Niem (Le Chan district), Lac Vien (Ngo Quyen district), Quang Hung (An Lao district), and Doan Xa (Kien Thuy district), in Hai Phong city, Vietnam, from August 2016 to December 2017. All these students were screened for PV. A questionnaire was used to record the informative data about the epidemiology of each person.
Sampling
The specimens were taken by scraping the lesions with a sterile blade. In case of the presence of normal subjects or insufficient scales, the samples were taken by means of sellotape. In patients with PV having more than two lesion sites, all lesions were sampled, and a record was made regarding the affected body site.
Direct microscopy and culture
The diagnosis of PV was based on clinical appearance and direct microscopic examination in 20% potassium hydroxide (KOH) and staining with methylene blue. To this end, the skin specimens obtained from patients with clinically suspected PV were used for direct microscopic examination with 20% KOH (RedStar Co. Ltd, Vietnam) and staining with methylene (Merk, Germany). The PV was established based on the characteristic clusters of spores with short hyphae. Skin scale sampling was continued in the patients who had positive direct microscopy, and the samples were immediately cultured on modified Dixon agar plates, containing gentamicin (at a final concentration of 25 μg/ml).
The inoculated plates were incubated at 32°C and observed every day for a maximum of 7 days before negative results were noted. The colonies were sub-cultured on CHROMagarTM Malassezia medium (CHROMagar, France) to determine the co-infection rate of Malassezia species and distinguish between M. furfur and other Malassezia species according to the manufacturer’s instructions. If Malassezia species were the same in different body sites, it was considered to be a single isolate.
Molecular identification of the isolated species of Malassezia
DNA isolation
The pure cultures of all Malassezia isolates were homogenized in 100 μl of sterile water (Corning, USA) and incubated with sorbitol buffer (1M sorbitol; 100 mM sodium EDTA; 14 mM β-mercaptoethanol) and 200 Lyticase enzyme units (L2524, Sigma-Aldrich Co. Ltd., Poole, UK) at 30°C for 60 min to destroy fungal cell membranes. In the next stage, the DNA of each individual isolate was extracted using the biologic QIAamp DNA Mini Kit (No. 51304, Qiagen, Hilden, Germany), according to the manufacturer's guideline. The purified DNA was preserved with distilled water at -20°C until the implementation of the PCR reaction.
Polymerase chain reaction amplification
The PCR products (400-550 bp) of Malassezia, containing ITS2 rDNA, were amplified by ITS3 (5'-GCA TCG ATG AAG AAC GCA GC-3) and ITS4 (5'- TCC TCC GCT TAT TGA TAT GC-3') primers (Integrated DNA Technologies, USA) as described previously [ 9 ]. The total PCR reaction volume of 50 μl consisted of 5 µl of the DNA solution of Malassezia, 25 µl Master Mix 2X (Thermo Fisher Scientific, USA), 1 μl of each primer (0.2 μM), and 18.0 μl of deionized water. The PCR reaction was performed on the Thermo Mastercycler gradient cycler (Thermo Fisher Scientific, USA) with a thermal cycle at 94oC for 5 min. This was followed by 35 cycles of 94oC for 30 sec, 53oC for 30 sec, and 72oC for 45 sec and then one cycle of 72oC for 15 min and 1 cycle of 25oC for 10 min. After amplification, the products were stored at 4oC until being used.
Restriction fragment length polymorphism analysis
The RFLP was performed according to the method described by Rudramurthy et al. [ 9 ] to distinguish Malassezia species. Digestion was performed in a final reaction volume of 16 μl, consisting of 5 μl of PCR product, 1 µl (10U) of each restriction enzyme (i.e., AluI, BanI, and MspA1I) (Thermo Fisher Scientific, USA), 1 µl of 10X Tango buffer solution, and 9 μl of deionized water (Thermo Fisher Scientific, USA). After 3 h of incubation at 37oC, the enzyme was inactivated at 65oC for 15 min. For analyzing the digestion products, 6 μl of each product in addition to 1 μl of loading dye buffer was separated by 2% agarose gel in 1X TBE buffer for about 1.5 h at 90 V. The ethidium bromide staining was the visualized by ultraviolet illumination (UVP, Canada). The size of each band was determined by a 100-bp Plus Ladder molecular weight marker (Thermo Fisher Scientific, USA).
D1/D2 26S rDNA gene sequencing
The D1/D2 domain region of 26S rRNA genes was amplified with the primers NL-1 (5’-GCA TAT CAA TAA GCG GAG GAA AAG-3’) and NL-4 (5’-GGT CCG TGT TTC AAG ACG G-3’) (Integrated DNA Technologies, USA). The PCR products of seven strains of Malassezia were sent to the Apical Scientific Sdn Bhd (Seri Kembangan 43300, Selangor, Malaysia) for purification and automatic sequencing with the same primers being used for PCR. The sequences were read on the ABI 3130 Genetic Analyzer software (SeqScape Software, version 2.1).
Data and sequence analysis
The SPSS statistics software (Chicago, IL, USA; version 20.0) was used for processing the data in our study. A P-value less than 0.05 was considered statistically significant. The obtained sequences were then compared to the available data in the NCBI database, using the BLAST guidelines (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Ethical considerations
The purpose and benefits of the study were explained to the students, as well as their parents/guardians and head teachers. The inclusion criteria were: 1) willingness to participate in the study, 2) a written informed consent, and 3) informed consent of the parents/guardians. The study protocol was approved by the Scientific and Ethical Committee of the National Institute of Malariology, Parasitology, and Entomology (Hanoi, Vietnam) in November 2015 (ethics code: 1212/QĐ-VSR). Furthermore, the study was conducted in accordance with the Declaration of Helsinki Principles.
Results
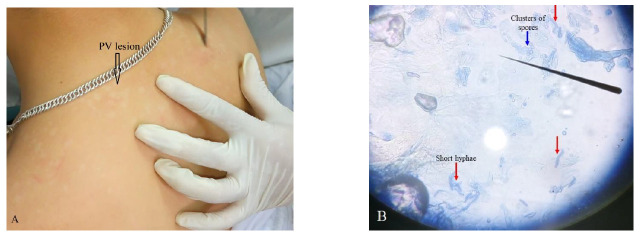
A total of 1,357 students aged 10-16 years were chosen to be screened for PV skin lesions (Figure 2 A). The mean age at the onset of PV was 13.5 years. The research population consisted of 690 males (age range: 10-16 years) and 667 females (age range: 11-16 years). The demographic characteristics of the subjects are shown in Table 1. Based on the clinical appearance and direct microscopic examination in 20% KOH and staining with methylene (Figure 2B), a total of 305 (22.48%) patients were diagnosed with PV. There were no statistically significant differences between the two genders or between the urban and rural residents in terms of the prevalence of PV (P=0.59 and P=0.99, respectively).
Figure 2.
Pityriasis versicolor lesion (A) and clusters of spores with short hyphae (B) of Malassezia stained with methylene blue (40X)
Table 1.
Demographic characteristics of the research population
| Demographic characteristics | Total (n = 1,357) |
|---|---|
| n (%) | |
| Gender | |
| Male | 690 (50.85) |
| Female | 667 (49.15) |
| Place of residence | |
| Rural | 578 (42.59) |
| Urban | 779 (57.41) |
Figure 1.
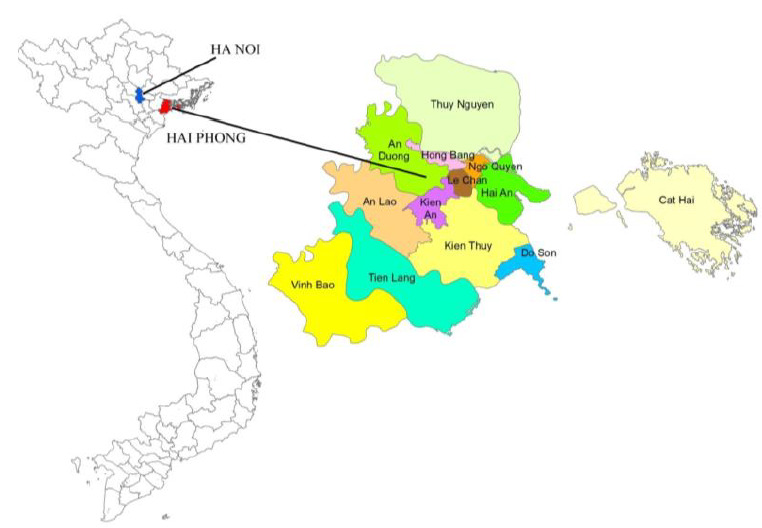
Map of Vietnam (a) Hai Phong city, north of Vietnam (b)
Based on the results, 5.25% of subjects who had PV lesion in two locations. Furthermore, out of three types of pigmentations, two types were seen in the study participants, with hypopigmentation being the most frequent one. The clinical characteristics of the subjects are presented in Table 2. Most of the specimens were collected from the lesions on the back (39.56%), face (23.99%), and chest (16.51%). Out of the 305 samples collected from PV lesions, 293 cases grew on modified Dixon agar (96.07% growth rate). Based on the results of CHROMagarTM Malassezia medium, PCR-RFLP, and gene sequencing methods, out of 293 Malassezia isolates, 282 (96.25%) and 11 (3.75%) cases were identified as M. furfur and M. japonica, respectively; however, other Malassezia species were not detected in the specimens (Figure 3).
Table 2.
Distribution of clinical characteristics of the research population
| Clinical characteristics | Total [n (%)] |
|---|---|
| Gender (n = 305) | |
| Male | 151 (49.51) |
| Female | 154 (50.49) |
| Residence (n = 305) | |
| Rural | 130 (42.62) |
| Urban | 175 (57.38) |
| Duration of illness (months) (n = 305) | |
| < 3 | 28 (9.18) |
| 3 - 6 | 7 (2.30) |
| > 6 | 270 (88.52) |
| Pruritus (n = 305) | |
| Yes | 77 (25.25) |
| No | 228 (74.75) |
| Number of affected sites (n = 305) | |
| 1 | 289 (94.75) |
| ≥ 2 | 16 (5.25) |
| Pigmentation (n = 305) | |
| Hypo-pigmentation | 296 (97.05) |
| Hyper-pigmentation | 0 (0) |
| Erythema | 9 (2.95) |
| Combination | 0 (0) |
| Lesion location (n = 321) | |
| Face | 77 (23.99) |
| Neck | 44 (13.71) |
| Chest | 53 (16.51) |
| Back | 127 (39.56) |
| Arm | 7 (2.18) |
| Stomach | 13 (4.05) |
Figure 3.
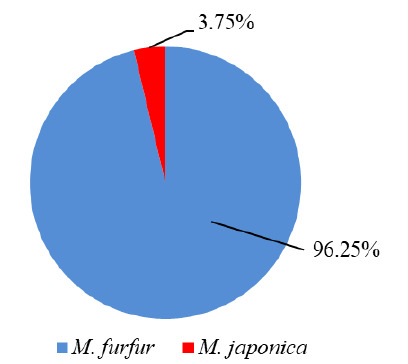
Distribution of Malassezia species
The prevalence of the co-colonization of the two identified Malassezia species was 0.34% (Table 3). These two species were discovered in both males and females. In addition, Malassezia furfur was found in both rural and urban areas, while M. japonica was only found in urban areas (data not shown). Figure 4 depicts the agarose gel electrophoresis of PCR products of the two isolates after digestion with the AluI, BanI, and MspA1I restriction enzymes.
Table 3.
Co-colonization of Malassezia species in pityriasis versicolor patients
| Malassezia species | Number (%) |
|---|---|
| M. furfur | 281 (96.23) |
| M. japonica | 10 (3.43) |
| M. furfur - M. japonica | 1 (0.34) |
| Total | 292 (100) |
Figure 4.
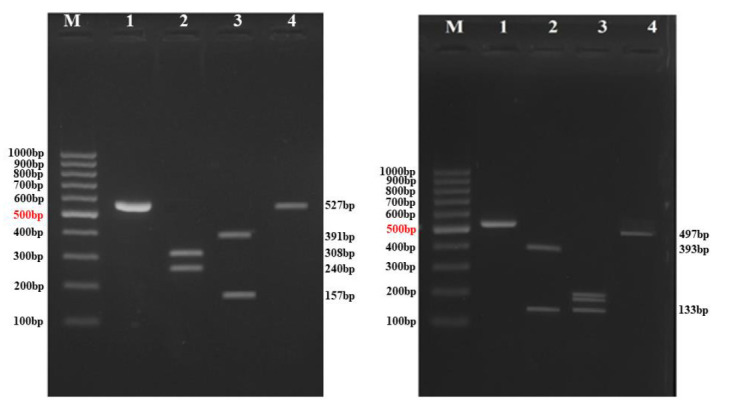
Polymerase chain reaction (PCR) product and restriction fragment length polymorphism pattern of PCR products of M. furfur (a) and M. japonica (b) after digestion with AluI, BanI, and MspA1I enzymes; lane M) 100-bp ladder molecular weight marker, land 1 [a, b]: PCR product of approximately 550bp, and lanes 2, 3 and 4 [a, b] digestion of this product with different restriction enzymes (i.e., AluI, BanI and MspA1I)
The representative sequences (D1/D2 rDNA regions) of seven isolates, including three M. furfur isolates and four M. japonica isolates, were deposited in the NCBI database (GenBank, USA) with the accession numbers of MF595845.1 to MF595847.1 and MG890324.1 to MG890327.1, respectively (Table 4).
Table 4.
Accession number of some strains subjected to D1/D2 region sequencing and GenBank
| Name of strain | Accession number | Name of species |
|---|---|---|
| VN-DX9C26 | MF595845 | Malassezia furfur |
| VN-DX7A35 | MF595846 | Malassezia furfur |
| VN-LV6A239 | MF595847 | Malassezia furfur |
| LV9D2-37-THAN | MG890324 | Malassezia japonica |
| LV8C8-12-MAT | MG890325 | Malassezia japonica |
| LV9D3-43-THAN | MG890326 | Malassezia japonica |
| VN8B3-15-THAN | MG890327 | Malassezia japonica |
Discussion
Pityriasis versicolor is observed more commonly among teenagers and young adults, especially in tropical and temperate regions [ 13 , 18 ]. The prevalence of PV is higher in tropical climates (nearly 30-40%), compared to that in temperate temperatures (1-4%) [ 16 ]. The relationship among disease, environment, and host factors has not been clearly described yet [ 1 ]. Moreover, there are no clear data on the pathogenesis of skin condition and the association of new Malassezia species with PV lesions [ 18 ]. With regard to the host, the prevalence of Malassezia infection depends on various factors, such as age, gender, body position, environmental, and endogenous factors [ 1 ]. Based on a body of evidence, the prevalence and distribution of Malassezia species depend on the identification techniques, location, and local micro-environmental variation [ 6 , 19 ].
In our study, 100% of the specimens were positive in the direct microscopic examination in 20% KOH and staining with methylene blue and showed clusters of yeast cells with short hyphae. The prevalence of PV was found to be higher in the secondary students (22.48%) investigated in this study, compared to the values reported for other countries, such as Iran [ 6 ] and Nigeria [ 20 ]. However, out of the 305 samples collected from PV lesions, a positive growth rate only accounted for 96.07% (n=293) of the cases. In the same vein, previous studies have shown that Malassezia growth rates are usually lower in the culture method than in the direct microscopic examination [ 1 , 21 ].
Malassezia fungi can be found in the wrinkled sites of the body. The existence of these fungi depends on some factors such as, humidity and the amount and composition of skin lipids [ 1 ]. The influence of gender on the propensity to developing PV is still unclear [ 6 ]. Our results regarding the influence of gender on PV are in line with those of the previous studies. However, other authors reported a higher incidence in women. The reasons for such differences in the rate of Malassezia between males and females may be attributable to the extra attention of women to their beauty and skin hygiene [ 22 , 23 ]. On the other hand, males are more involved in outdoor activities which place them at a high risk of exposure to some predisposing factors, such as high temperature and humidity [ 24 ].
In the currents study, the frequency of hypopigmentation lesions accounted for 97.05% of the PV lesions, which were more common than other lesions. In a study performed on 139 PV cases in Mumbai, India, Shah et al. [ 25 ] reported that 84.17%, 8.63%, and 7.19% of the PV lesions had hypopigmentation, hyperpigmentation, and both hyperpigmentation and hypopigmentation, respectively. Likewise, in a study conducted on 98 PV patients in Indonesia, as the neighboring country of Vietnam, hypopigmentation was found to be the most common lesion (64.3%) [ 16 ]. The predominance of M. furfur in tropical climates is probably explained by its pityriacitrin production (i.e., an ultraviolet-absorbing indole alkaloid from M. furfur) [ 13 , 16 ]. This agent has the ability to protect fungus against ultraviolet exposure and induce the apoptosis of human melanocytes [ 16 ] that may explain over 90% hypo-pigmentation observed in our study.
Conclusion
As the findings indicated, the prevalence of PV was higher in the students living in Hai Phong city than in those residing in other parts of the world. In the present study, M. furfur and M. japonica were identified as the etiological agents, with M. furfur being more common than M. japonica. However, no other Malassezia species was detected.
Acknowledgement
This study was collaboratively supported by the National Institute of Malariology, Parasitology, and Entomology (Ha Noi, Viet Nam), Military Medical University (Ha Noi, Viet Nam), and Hai Phong University of Medicine and Pharmacy (Hai Phong, Vietnam). We gratefully thank all the staff of the Department of Parasitology (Hai Phong University of Medicine and Pharmacy) who kindly contributed to patient handling and sample preparation. We are also indebted to the Department of Parasitology and Entomology (Vietnam Military Medical University) for providing the equipment used for the molecular analysis of the samples. The authors are grateful to Mr. Tran Anh Minh for revising the English text.
Author’s contribution
N. D. B., V. T. T. H., and D. N. A. designed the study. D. T. T. M, V. V. T., L. T.T.T., N. T. M., B. T. H. A., T. V. K., C. B. L., and H. C. S. collected the samples and clinical data. V. T. T. H., T. T. S., L. T. K. D, N. T. M., L. T. A., and N. K. L. identified the species of the responsible agents. N. D. B., V. T. T. H., and D. N. A. drafted the manuscript. All authors read and approved the final manuscript.
Conflict of Interest: The authors declare that they have no conflicts of interest.
Financial disclosure
The authors have no relevant financial interests in this manuscript.
References
- 1.Zeinali E, Sadeghi G, Yazdinia F, Shams-Ghahfarokhi M, Razzaghi-Abyaneh M. Clinical and epidemiological features of the genus Malassezia in Iran. Iran J microbiol. 2014; 6(5):354–60. [PMC free article] [PubMed] [Google Scholar]
- 2.Oh BH, Song YC, Lee YW, Choe YB, Ahn KJ. Comparison of Nested PCR and RFLP for Identification and Classification of Malassezia Yeasts from Healthy Human Skin. Ann Dermatol. 2009; 21(4):352–7. doi: 10.5021/ad.2009.21.4.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Affes M, Ben Salah S, Makni F, Sellami H, Ayadi A. Molecular identification of Malassezia species isolated from dermatitis affections. Mycoses. 2009; 52(3):251–6. doi: 10.1111/j.1439-0507.2008.01589.x. [DOI] [PubMed] [Google Scholar]
- 4.Honnavar P, Chakrabarti A, Dogra S, Handa S, Rudramurthy SM. Phenotypic and molecular characterization of Malassezia japonica isolated from psoriasis vulgaris patients. J Med Microbiol. 2015; 64(Pt 3):232–6. doi: 10.1099/jmm.0.000011. [DOI] [PubMed] [Google Scholar]
- 5.Gaitanis G, Magiatis P, Hantschke M, Bassukas ID, Velegraki A. The Malassezia genus in skin and systemic diseases. Clin Microbiol Rev. 2012; 25(1):106–41. doi: 10.1128/CMR.00021-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moallaei H, Namazi MJ, Bouchara JP, Pourhammed S. Malassezia species in students from universities of Sabzevar, Northeastern Iran. J Mycol Med. 2018; 28(1):70–5. doi: 10.1016/j.mycmed.2017.12.004. [DOI] [PubMed] [Google Scholar]
- 7.Ilahi A, Hadrich I, Neji S, Trabelsi H, Makni F, Ayadi A. Real-Time PCR Identification of Six Malassezia Species. Curr Microbiol. 2017; 74(6):671–7. doi: 10.1007/s00284-017-1237-7. [DOI] [PubMed] [Google Scholar]
- 8.Castella G, Coutinho SD, Cabanes FJ. Phylogenetic relationships of Malassezia species based on multilocus sequence analysis. Med Mycol. 2014; 52(1):99–105. doi: 10.3109/13693786.2013.815372. [DOI] [PubMed] [Google Scholar]
- 9.Rudramurthy SM, Honnavar P, Dogra S, Yegneswaran PP, Handa S, Chakrabarti A. Association of Malassezia species with dandruff. Indian J Med Res. 2014; 139(3):431–7. [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao Y, Li L, Wang JJ, Kang KF, Zhang QQ. Cutaneous malasseziasis: four case reports of atypical dermatitis and onychomycosis caused by Malassezia. Int J Dermatol. 2010; 49(2):141–5. doi: 10.1111/j.1365-4632.2009.04178.x. [DOI] [PubMed] [Google Scholar]
- 11.Iatta R, Battista M, Miragliotta G, Boekhout T, Otranto D, Cafarchia C. Blood culture procedures and diagnosis of Malassezia furfur bloodstream infections: Strength and weakness. Med Mycol. 2018; 56(7):828–33. doi: 10.1093/mmy/myx122. [DOI] [PubMed] [Google Scholar]
- 12.Benito-Leon J, Laurence M. Malassezia in the central nervous system and multiple sclerosis. Infection. 2019; 47(1):135–6. doi: 10.1007/s15010-018-1196-3. [DOI] [PubMed] [Google Scholar]
- 13.Ibekwe PU, Ogunbiyi AO, Besch R, Ruzicka T, Sardy M. The spectrum of Malassezia species isolated from students with pityriasis vesicolor in Nigeria. Mycoses. 2015; 58(4):203–8. doi: 10.1111/myc.12298. [DOI] [PubMed] [Google Scholar]
- 14.Vuran E, Karaarslan A, Karasartova D, Turegun B, Sahin F. Identification of Malassezia species from pityriasis versicolor lesions with a new multiplex PCR method. Mycopathologia. 2014; 177(1-2):41–9. doi: 10.1007/s11046-013-9704-6. [DOI] [PubMed] [Google Scholar]
- 15.Lian CH, Shen LL, Gao QY, Jiang M, Zhao ZJ, Zhao JJ. Identification of Malassezia species in the facial lesions of Chinese seborrhoeic dermatitis patients based on DNA sequencing. Mycoses. 2014; 57(12):759–64. doi: 10.1111/myc.12229. [DOI] [PubMed] [Google Scholar]
- 16.Krisanty RI, Bramono K, Made Wisnu I. Identification of Malassezia species from pityriasis versicolor in Indonesia and its relationship with clinical characteristics. Mycoses. 2009; 52(3):257–62. doi: 10.1111/j.1439-0507.2008.01593.x. [DOI] [PubMed] [Google Scholar]
- 17.Cam VT, Van TN, Hau KT, Huu DL, Minh PPT, Huu SN, et al. Distribution of Malassezia Species from Scales of Patient with Pityriasis Versicolor by Culture in Vietnam. Open access Maced J Med Sci. 2019; 7(2):184–6. doi: 10.3889/oamjms.2019.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giusiano G, Sosa Mde L, Rojas F, Vanacore ST, Mangiaterra M. Prevalence of Malassezia species in pityriasis versicolor lesions in northeast Argentina. Rev Iberoam Micol. 2010; 27(2):71–4. doi: 10.1016/j.riam.2009.12.005. [DOI] [PubMed] [Google Scholar]
- 19.Xie Z, Ran Y, Zhang H, Zhang M, Wan H, Li C. An analysis of the Malassezia species distribution in the skin of patients with pityriasis versicolor in Chengdu, China. Sci World J. 2014; 2014:182596. doi: 10.1155/2014/182596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oke OO, Onayemi O, Olasode OA, Omisore AG, Oninla OA. The Prevalence and Pattern of Superficial Fungal Infections among School Children in Ile-Ife, South-Western Nigeria. Dermatol Res Pract. 2014; 2014:842917. doi: 10.1155/2014/842917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chaudhary R, Singh S, Banerjee T, Tilak R. Prevalence of different Malassezia species in pityriasis versicolor in central India. Indian J Dermatol Venereol Leprol. 2010; 76(2):159–64. doi: 10.4103/0378-6323.60566. [DOI] [PubMed] [Google Scholar]
- 22.Rasi A, Naderi R, Behzadi AH, Falahati M, Farehyar S, Honarbakhsh Y, et al. Malassezia yeast species isolated from Iranian patients with pityriasis versicolor in a prospective study. Mycoses. 2010; 53(4):350–5. doi: 10.1111/j.1439-0507.2009.01727.x. [DOI] [PubMed] [Google Scholar]
- 23.Tarazooie B, Kordbacheh P, Zaini F, Zomorodian K, Saadat F, Zeraati H, et al. Study of the distribution of Malassezia species in patients with pityriasis versicolor and healthy individuals in Tehran, Iran. BMC Dermatol. 2004; 4:5. doi: 10.1186/1471-5945-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharma A, Rabha D, Choraria S, Hazarika D, Ahmed G, Hazarika NK. Clinicomycological profile of pityriasis versicolor in Assam. Indian J Pathol & Microbiol. 2016; 59(2):159–65. doi: 10.4103/0377-4929.182027. [DOI] [PubMed] [Google Scholar]
- 25.Shah A, Koticha A, Ubale M, Wanjare S, Mehta P, Khopkar U. Identification and speciation of Malassezia in patients clinically suspected of having pityriasis versicolor. Indian J Dermatol. 2013; 58(3):239. doi: 10.4103/0019-5154.110841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee YW, Byun HJ, Kim BJ, Kim DH, Lim YY, Lee JW et al. Distribution of Malassezia species on the scalp in Korean seborrheic dermatitis patients. Ann Dermatol. 2011; 23(2):156–61. doi: 10.5021/ad.2011.23.2.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Awad AK, Al-Ezzy AI, Jameel GH. Phenotypic Identification and Molecular Characterization of Malassezia spp. isolated from Pityriasis versicolor patients with special emphasis to risk factors in Diyala province, Iraq. Open access Maced J Med Sci. 2019; 7(5):707–14. doi: 10.3889/oamjms.2019.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saxena R, Mittal P, Clavaud C, Dhakan DB, Hegde P, Veeranagaiah MM, et al. Comparison of healthy and dandruff scalp microbiome reveals the role of commensals in Scalp Health. Front Cell Infect Microbiol. 2018; 8:346. doi: 10.3389/fcimb.2018.00346. [DOI] [PMC free article] [PubMed] [Google Scholar]