Abstract
Focal adhesions (FAs) are integrin‐containing protein complexes regulated by a network of hundreds of protein–protein interactions. They are formed in a spatiotemporal manner upon the activation of integrin transmembrane receptors, which is crucial to trigger cell adhesion and many other cellular processes including cell migration, spreading and proliferation. Despite decades of studies, a detailed molecular level understanding on how FAs are organized and function is lacking due to their highly complex and dynamic nature. However, advances have been made on studying key integrin activators, talin and kindlin, and their associated proteins, which are major components of nascent FAs critical for initiating the assembly of mature FAs. This review will discuss the structural and functional findings of talin and kindlin and their immediate interaction network, which will shed light upon the architecture of nascent FAs and how they act as seeds for FA assembly to dynamically regulate diverse adhesion‐dependent physiological and pathological responses.
Keywords: focal adhesions, integrin, kindlin, talin
1. INTRODUCTION
Cell‐extracellular matrix (ECM) and cell–cell adhesion are critically regulated by integrins, a class of cell surface receptors. Integrins are heterodimeric and consist of an α and a β subunit with each containing a large extracellular domain, a single‐helical transmembrane segment and typically a short cytoplasmic tail. There are 24 different integrins assembled from 18 α and 8 β subunits in vertebrates to control diverse physiological and pathological processes, such as blood clotting, immune response, wound‐healing and cancer metastasis. 1 , 2 Integrin is known to function via unique bi‐directional signaling, which has been studied extensively over the past three decades: (i) “Inside‐out” signaling where intracellular proteins, such as talin and kindlin, bind to the cytoplasmic face of an inactive integrin to trigger a conformational change of its extracellular domain, switching it to a high‐affinity ligand binding state (a process also known as “integrin activation”), and (ii) “Outside‐in” signaling where activated (ligand‐bound) integrin triggers assembly of large protein complexes called focal adhesions (FAs) that link integrin to the actin cytoskeleton, so as to promote firm cell adhesion and dynamic cell adhesive processes (e.g., cell spreading, migration and proliferation). 1 , 3 , 4 , 5 FAs have also been defined by different terms such as “integrin adhesome” or “integrin adhesion complexes (IACs)” to reflect the dynamic and complex nature of the protein network. 5 , 6 , 7 More than 150 proteins have been identified in IACs, which are connected by estimated 690 protein–protein interactions (PPIs) 6 . However, a detailed picture of how these proteins engage with one another spatiotemporally to form such complex networks remains obscure.
Talin and kindlin are the key activators of integrins, which also serve as seeds to recruit proteins to initiate FA assembly following their binding to and activation of integrin via “inside‐out” signaling. Indeed, talin and kindlin and their associated proteins such as vinculin, paxillin, focal adhesion kinase (FAK), and integrin‐linked kinase (ILK) are found in nascent FAs. 7 , 8 Extensive biophysical studies including structural characterization (via NMR spectroscopy, X‐ray crystallography and Cryo‐EM), combined with functional analyses (e.g., integrin activation assays, cell adhesion or spreading assays, and mice knock‐out and knock‐in studies) have been conducted to understand the regulation of these nascent FA proteins. They share some common features: (a) essential for embryonic development and integrin‐mediated FA assembly, cell spreading and migration (Table 1, Figure 1); and (b) all contain multiple domains or motifs allowing them to bind multiple proteins to trigger the growth of FAs. In this review, we attempt to summarize the role of talin and kindlin in regulating integrin activation, and important protein–protein interactions (PPIs) and binding interfaces that are mediated by each domain or motif of talin and kindlin. We propose that these interactions are critically involved in the initial network and the core of nascent FAs (Figure 1), which then expand into larger and mature FA complexes via cascades of PPIs in a spatiotemporal manner to promote dynamic cell adhesion processes.
TABLE 1.
Phenotypes caused by deletion of talin and kindlin and critically associated proteins
| FA assembly b | Cell function b | |||||
|---|---|---|---|---|---|---|
| KO mice (day) a | Number | Size | Spreading area | Migration | Ref. | |
| Talin‐1 (2,541 aa) | E8.5–9.5 | Decrease | Decrease | Decrease | Decrease | 8 , 9 , 10 |
| Kindlin‐2 (680 aa) | E7.5 | Decrease | Decrease | Decrease | Decrease | 8 , 11 , 12 |
| Paxillin (591 aa) | E9.5 | Increase | Decrease | Decrease | Decrease | 12 , 13 , 14 |
| Vinculin (1,134 aa) | E10.5 | Decrease | Decrease | Decrease | Increase | |
| ILK (452 aa) | E5.5–6.5 | Decrease c | N/A d | Decrease c | Decrease | 17 , 18 , 19 |
| FAK (1,052 aa) | E8.5 | Increase | N/A e | Decrease | Decrease | 20 , 21 , 22 |
FIGURE 1.
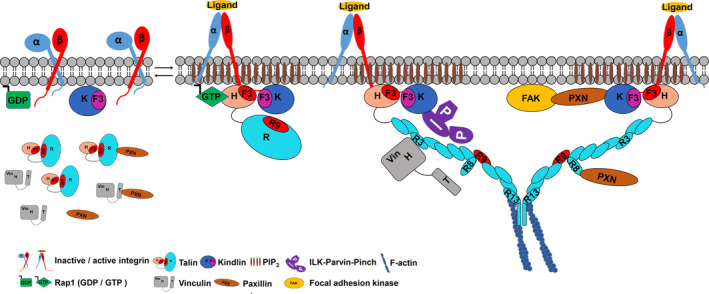
Initiation of nascent FAs upon the integrin activation by talin and kindlin via “inside‐out” signaling. Activated GTPase Rap1 recruits talin to the membrane site. Talin becomes activated by membrane PIP2 and exposes talin‐F3 domain to bind integrin. Kindlin also binds integrin via its F3 domain and cooperates with talin to fully activate integrin. Talin stretches in the presence of mechanical force and together with kindlin triggers multiple axes including “talin‐vinculin”, “talin‐paxillin” “kindlin‐paxillin‐FAK” and “kindlin‐ILK‐parvin‐pinch”, to initiate FA assembly, and regulate actin cytoskeleton remodeling and dynamic cell adhesion
2. TALIN ACTION AND ITS INTERACTION NETWORK
Talin was discovered in early 1980s 23 as a large cytoskeletal protein (MW: ~ 270 kDa) involved in cell adhesion. However, it only started to draw major attention two decades later when researchers realized that it can bind and activate integrin to trigger cell adhesion. 1 , 3 Since FAs are formed following integrin activation, talin and its network provide an excellent window for us to understand the initiation of FA assembly. Vertebrates encode two talin isoforms, talin‐1 and talin‐2, which share ~76% sequence identity. 24 Talin‐1 is ubiquitously expressed, whereas talin‐2 is tissue‐specific and most abundant in heart, brain and skeletal muscle. 25 Here we focus on discussing talin‐1 (hereafter talin refers to talin‐1), which has been extensively characterized and shown to be vital for life since its ablation in mice causes embryonic lethality (Table 1) and its tissue‐specific depletions cause severe phenotypes, such as impaired platelet aggregation and leukocyte trafficking, due to defective integrin activation. 26
Talin consists of an N‐terminal FERM (4.1/ezrin/radixin/moesin)‐like head domain (talin‐H), which is further divided into four subdomains, F0, F1, F2 and F3, and a C‐terminal rod domain (talin‐R), which is made up of 13 consecutive helical bundles (R1‐13), followed by a single‐helical dimerization domain (talin‐DD) (Figure 2(a)). Structural studies have shown that talin uses its F3 domain, a PTB (phosphotyrosine‐binding domain)‐like domain, to directly bind to integrin β cytoplasmic tail utilizing the membrane proximal helix and “NPxY” motif (Figure 2(b)). 27 , 28 , 29 It is well understood that such binding results in the separation of integrin α and β subunit at the cytoplasmic face, which further leads to the separation of the transmembrane α and β segments and the “bent‐to‐extension” conformational change of the extracellular domain leading to a “high‐affinity” ligand binding state (Figure 1). 29 , 30 Notably, the talin‐integrin interaction is tightly regulated. In resting‐state cells, intact talin is randomly distributed in the cytosol 31 exhibiting an auto‐inhibitory conformation and incapable of binding to integrin. 32 , 33 Membrane‐PIP2 has long been recognized as talin activator that binds and induces conformational change of talin, exposing its F3 domain to bind and activate integrin (Figure 1). 32 , 33 , 34 Talin exists in a monomer/dimer equilibrium in solution and the monomeric form of the inactive talin has been resolved recently by cryo‐EM (Figure 2(b)). 33 Consistent with previous domain‐based structural studies by NMR 35 and X‐ray crystallography, 34 the cryo‐EM structure of the intact talin revealed that the integrin binding site of talin‐F3 is masked by talin‐R9 (Figures 2(b) and S1B). The structure also revealed that the PIP2 binding site on talin‐F2F3, critical for the activation of talin, is occupied by talin‐R12 (Figure 2(b) and S1B), 33 thereby providing a basis for understanding how PIP2 sterically occludes talin‐R12 and conformationally activates talin. Talin autoinhibition‐activation is delicately balanced in cells. Mutation disrupting the auto‐inhibitory interface between talin‐F3 and talin‐R9 leads to integrin activation, resulting impaired adhesion dynamics and delayed wound healing in vivo. 36
FIGURE 2.
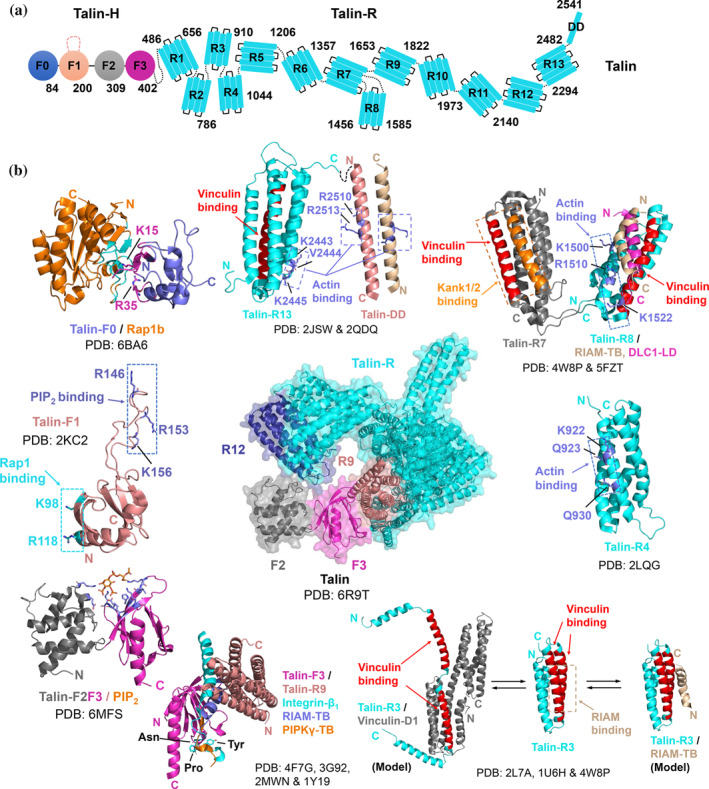
The binding interfaces of talin‐mediated interactions in integrin and FA signaling. (a) Domain organization of talin. (b) Reported structures of individual domains of talin in apo or complex form. The binding interfaces/sites for target proteins are indicated in the figure. Important residues involved in the binding interfaces or targeted in mutagenesis studies are shown in stick representation. The interface residues of talin‐F0 and Rap1b (within a cut‐off of 5 Å) are colored in magenta and cyan, respectively. The interface residues of talin‐F2F3 (within a cut‐off of 5 Å) that interact with PIP2 are colored in blue and shown in stick representation. N and C indicate the N‐terminus and C‐terminus of the protein molecule, respectively. PDB codes used to generate the figures are indicated
To activate integrin, talin needs to be recruited to the plasma membrane, a process that was not clearly understood for many years. One previously proposed pathway was that talin is recruited to membrane by a membrane‐anchored GTPase called Rap1 via its effector RIAM (referred to the “Rap1‐RIAM‐talin” pathway). 37 The RIAM binding to talin‐F3 (Figure 2(b)) was also shown to contribute to the activation of talin. 38 However, the function of the Rap1‐RIAM‐talin pathway appears to be leukocyte‐specific. 39 , 40 , 41 Recent breakthrough studies revealed that ubiquitin‐like talin F0 and F1 domains control the membrane recruitment of talin via direct binding to Rap1 (Figures 1 and 2(b)). 42 , 43 , 44 , 45 , 46 Mutations that abolished either Rap1/talin‐F0 or Rap1/talin‐F1 interaction resulted in reduced integrin activation in CHO cells, impaired FA assembly, cell spreading and adhesion in talin‐null fibroblast‐based assays, as well as defective platelet and leukocyte function in mouse models. 42 , 43 , 44 , 45 , 46 Disruption of both interactions led to more severe defects in FA assembly, cell spreading and adhesion, 45 and more intriguingly, embryonic lethality in mice and a platelet phenotype resembling that of talin or Rap1 knockout. 46 It is of particular interest to note that talin F0 and F1 both bind to Rap1 very weakly in solution (Table S1), which was initially overlooked. However, in a cellular context, talin‐F0 and F1 may bind simultaneously to two membrane‐anchored Rap1 molecules, resulting in high affinity binding and effective talin recruitment to membrane. 42 , 43 , 44 , 45 , 46 Talin‐F1 also contains a 30‐residue flexible loop, which was suggested to interact with membrane‐lipid mainly via R146, R153 and K156 (Figure 2(b)) to assist recruitment of talin to the membrane. 47 It was further suggested recently that the F1 loop may interfere with a salt‐bridge formed between the transmembrane domains of integrin α and β subunits to promote integrin clustering and activation. 48 As mentioned above, talin‐F2F3 domain bears a PIP2 binding pocket formed by K272 of F2, K316, K324 and K343 of F3 (Figure 2(b)). 49 The PIP2 binding may not only activate talin 32 , 33 , 34 but also cooperate with Rap1 to enhance the association of talin‐H with the cell membrane. 42 , 45 Interestingly, talin‐F3 also binds to the C‐terminal segment “WVYSPLHY” of PIPKIγ (Type I phosphatidylinositol phosphate kinase gamma) in a similar mode as it binds integrin‐β “NPxY” motif (Figure 2(b)). 50 , 51 The interaction allows PIPKIγ to be co‐localized with talin to promote local production of PIP2 at the membrane site that binds and activates talin. 52 , 53 It is important to clarify here conceptually that although PIPKIγ seems to block integrin from binding talin‐F3 (Figure 2(b)), only a very small fraction of talin, which is extremely abundant in cells, is needed to recruit PIPKIγ that is typically at very low concentration as a kinase but can enzymatically produce a large amount of PIP2 to bind and activate the large portion of PIPKIγ‐free talin.
We note that although previous crystallographic studies of talin‐H revealed a linear conformation different from cloverleaf‐like folding of a canonical FERM domain (Figure S1A), 49 , 54 SAXS data suggested that talin‐H in solution is not as linear as in the crystal structure. 48 , 54 In line with the latter, Rangarajan et al. recently solved the crystal structure of intact talin2‐H in which the F0‐F1‐F2 domain adopted a cloverleaf‐like fold (Figure S1A). 55 However, both of these conformations contrast with the recent cryo‐EM structure of full‐length talin wherein the densities of F0 and F1 domains were not observed (Figure 2(b)). 33 These findings suggest that talin‐H, which is critical for binding and activating integrin, is highly dynamic in nature likely due to the existence of a long linker between F1 and F2 subdomain (Figure S1A). It remains to be investigated if the presence of membrane‐anchored Rap1 and membrane‐PIP2 would stabilize talin‐H to a rigid conformation that favors integrin binding.
While talin‐H is directly involved in talin membrane recruitment and integrin activation, talin‐R is thought to be indispensable for transmitting mechanical force between integrin and actin cytoskeleton to support cell adhesion and spreading. Expression of talin‐H in talin‐null fibroblasts was sufficient to support integrin activation but not cell spreading and migration. 10 , 56 All 13 talin‐R subdomains are an assembly of 4–5 alpha‐helices (Figure 2(a)) and bear binding sites for a variety of adhesion proteins, such as vinculin, paxillin, RIAM, DLC‐1 (deleted in liver cancer 1) and Kank‐1/2, as well as two major actin binding sites, ABS2 and ABS3 (Note that talin‐H also contains one actin binding site ABS1 in talin‐F2F3, see ref. 57 , 58 ). ABS2 and ABS3 locate at R4‐R8 (R4 and R8 are the major binding sites) and R13‐DD respectively (Figure 2(b)). 59 , 60 The binding of ABS3 to actin is crucial for initial force transduction, which promotes the talin binding to vinculin and initiates FA assembly. Deletion or mutations of ABS3 to block actin binding resulted in loss of FA formation, and defective cell spreading, polarization and migration. 10 , 59 In contrast, mutations of ABS2 led to smaller size of FAs and reduced number of actin stress fibers, suggesting its role in traction force generation for FA maturation and stabilization. 59
At least 9 talin‐R subdomains (R1, R2, R3, R6, R7, R8, R10, R11 and R13) interact with vinculin (~116 kDa), which contains a head domain (vinculin‐H) composed of D1‐D4 subdomains, followed by a flexible proline‐rich neck region and a tail D5 domain (vinculin‐T). 61 The binding of vinculin to talin is essential for force transmission and mechanosensing, which are important for the growth and maturation of FAs. 59 , 62 , 63 As many as 11 potential single‐helical vinculin binding sites (VBS) were identified in talin‐R (Figures 2(b) and S1B, Table S1), and they are all capable of binding to the hydrophobic pocket of the D1 domain (Vd1) of vinculin‐H (Figure 2(b)), 64 which is otherwise masked by vinculin‐T via an auto‐inhibitory mechanism (Figure 1). 65 However, these binding sites are not accessible in a folded rod subdomain (Figures 2(b) and S1B). Studies showed that mechanical force can unfold talin‐R subdomains into a linear conformation to allow vinculin binding (Figure 2(b)). 66 , 67 , 68 Among the talin‐R subdomains, talin‐R3 is the first domain to unfold to bind vinculin due to its having the highest susceptibility to mechanical force. 67 , 69 This interpretation was also supported by mutagenesis studies which showed that the initial talin/vinculin engagement is highly dependent on the stability of talin‐R3. 62 , 70 The binding of vinculin to talin‐R2R3 triggered a conformational change that regulates actin binding to ABS2 to promote maturation of FAs linked to actin filaments. 59 The FA localization of vinculin is highly dependent on talin‐R subdomains, as the intensity of FA‐localized vinculin correlates with the number of VBS in talin constructs expressed in talin‐null fibroblasts. 10 , 59 Lack of VBS in talin severely impairs cell spreading, adhesion and migration. 10 , 71 Likewise, a vinculin mutant (A50I) that does not bind talin failed to promote the growth of FAs. 62 , 63 The unfolding of talin‐R to bind vinculin may be assisted by mechanical force but initial engagement of talin and vinculin is likely to be independent of mechanical force as supported by several lines of evidence: (a) Atherton et al. showed that talin and vinculin are able to co‐localize in force‐free mitochondria, 62 (b) Kelley et al. applied an in‐vitro synthetic membrane system to show that membrane PIP2 is sufficient to trigger the activation of talin and its engagement with vinculin to induce actin bundling in the absence of force, 72 (c) active vinculin or vinculin‐H can form complex with open‐conformation of talin or talin‐R fragments in solution, 33 , 71 and (d) vinculin‐H directly causes the unfolding of single talin‐R subdomains in NMR studies. 73 On the other hand, the stoichiometry of active vinculin binding to open‐conformation talin in solution is 1:1, 33 suggesting that the engagement of all 11 VBS of talin‐R subdomains with vinculin may be assisted by mechanical force. Overall, both force‐independent and dependent processes are involved in “talin‐vinculin” engagement.
In addition to vinculin binding, talin was also shown to interact with other target proteins (e.g., RIAM, DLC‐1, Kank‐1/2, and paxillin). Unlike vinculin, these proteins bind to folded talin‐R subdomains via a similar interacting mode. The groove formed by two helices of a talin‐R subdomain provides a potential binding site for a helical LD or LD‐like motif of the target proteins (Figures 2(b) and S1B). Talin‐R2, R3, R8, and R11 (R3 and R8 are most potent) are all capable of binding to the N‐terminal talin binding (TB) region of RIAM (Figures 2(b) and S1B), which assists talin membrane recruitment for integrin activation 37 , 71 , 74 and forms “sticky fingers” with talin to regulate leukocyte migration. 75 Talin‐R8 also interacts with paxillin and DLC‐1 via the LD1 and LD‐like motif respectively in a similar fashion but with different binding affinities (Figure 2(b), Table S1). 76 Talin‐R8/DLC‐1 interaction is required for the FA targeting and tumor suppressor activity of DLC‐1, 77 and mechanistically, acts like a force‐controlled molecular switch to regulate the mechanotransduction of talin so as to regulate FA assembly and cell migration. 76 , 78 Paxillin binding to talin‐R8 is not well characterized at structural level but may act similarly to DLC‐1. 76 , 78 This interaction may account for the association of talin/paxillin to form initial adhesion complex, as suggested recently. 62 In addition, paxillin could also engage vinculin in the absence of talin to form initial adhesion complex, 62 suggesting an interesting interplay among these three proteins that remains to be further explored. Talin‐R7 specifically interacts with the KN domain of Kank‐1/2 which also contains a LD‐like motif (Figure 2(b)). The interaction was suggested to regulate talin and integrin activation, the force transduction between integrin‐talin and actomyosin, as well as the recruitment of cortical microtubule stabilizing complexes to FAs. 79 , 80 Interestingly, these binding events (vinculin included) mediated by talin‐R subdomains are mutually exclusive and likely regulated spatiotemporally or in a cell‐type specific manner.
The cryo‐EM structure of monomeric inactive talin reveals that talin‐R subdomains and talin‐H fold into a compact conformation. 33 It is conceivable that upon activation, talin would engage with the above‐mentioned target proteins in a spatiotemporal manner to trigger the formation of nascent FAs. It is important to note that fully active talin likely exists as a dimer at physiological condition, 33 so one can imagine that dimeric talin may provide a broad platform for initiating the networking of nascent FAs (Figure 1). Future structural and functional studies may provide more detailed insights into the operation of such a talin‐based platform.
3. KINDLINS AND KINDLIN NETWORK
Kindlins have also attracted extensive attention since a decade ago due to their essential ability to enhance talin‐mediated integrin activation. 4 , 26 , 81 There is growing evidence that kindlins may be bound to integrin prior to integrin activation, 8 , 82 but how the integrin‐bound kindlins cooperate with talin to activate integrin is still not well understood. Kindlin family contains three members, namely kindlin‐1, kindlin‐2, and kindlin‐3, which are encoded by separate genes. Kindlin‐2 is ubiquitously expressed, while kindlin‐1 and 3 are primarily expressed in epithelial and hematopoietic cells, respectively, 81 , 83 , 84 although there are exceptions to this as kindlin‐3 is found in endothelial cells 85 and some tumor cells. 86 All kindlins are physiologically important. While kindlin‐2 is essential for embryonic development, loss of kindlin‐1 in humans and in mice causes skin fragility and blistering (known as “Kindler Syndrome”), and kindlin‐3 deficiency can lead to bleeding disorder and immune defects (a disease known as “leukocyte adhesion deficiency type III, LAD‐III”). 81 , 83 , 84 Kindlins (MW: ~ 77 kDa) adopt a FERM (4.1/ezrin/radixin/moesin)‐like domain which consists of F1, F2, and F3 subdomains with a pleckstrin homology (PH) domain inserted into F2, and a preceding ubiquitin‐like F0 domain (Figure 3(a)). Similar to talin‐H, kindlins also use its F3 domain to bind integrin β cytoplasmic tail but at a distinct site known as distal “NxxY” or “NPxY/F" motif (Figure 3(a)). Despite only the involvement of the F3 domain in direct binding to integrin, the integrity of kindlin is required for its integrin activation function. 87 All the other domains of kindlin have been shown to mediate different binding events to regulate FA assembly.
FIGURE 3.
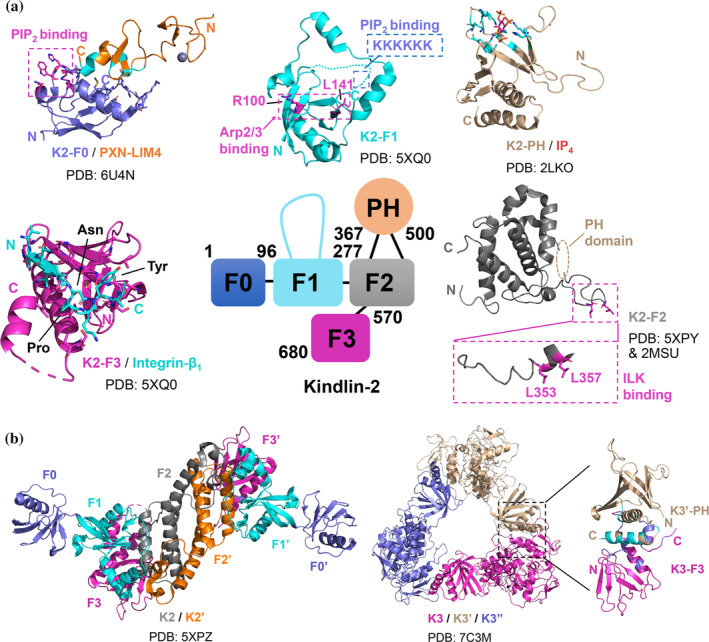
The binding interfaces of kindlin‐mediated interactions in integrin and FA signaling. (a) Domain organization of kindlin‐2 and reported structures of individual domains of kindlin‐2 in apo or complex form. The binding interfaces/sites for target proteins are indicated in the figure. Important residues involved in the binding interfaces or targeted in mutagenesis studies are shown in stick representation. The interface residues of paxillin LIM4 (within a cut‐off of 5 Å) that interact with kindlin‐2 F0 are colored in cyan. The interface residues of kindlin‐2 PH (within a cut‐off of 5 Å) that interact with IP4 are colored in cyan. (b) Reported crystal structures of truncated kindlin‐2 and full‐length kindlin‐3 in respective dimeric and trimeric conformation. The interface residues of kindlin‐3 F3 and kindlin‐3’ PH (within a cut‐off of 5 Å) are colored in blue and cyan, respectively. PXN, paxillin. K2, kindlin‐2. K3, kindlin‐3. N and C indicate the N‐terminus and C‐terminus of the protein molecule, respectively. PDB codes used to generate the figures are indicated
The F0 domain of kindlin was initially suggested to contribute to the membrane association of kindlin via a cluster of positive charged residues binding to negatively charged membrane lipid PIP2 (Figure 3(a)), so as to regulate integrin activation. 88 Recently, kindlins have drawn a lot of attentions due to their newly identified ability to bind paxillin. Studies by different groups gave rise to an interesting “kindlin‐paxillin‐FAK” pathway where kindlin recruits paxillin to regulate FAK activation, FA assembly and dynamic cell adhesion. 8 , 12 , 89 NMR studies revealed a modest affinity (~200 μM) of kindlin‐2/paxillin interaction mediated by a unique interface between kindlin‐2 F0 and paxillin LIM4 (Figure 3(a)). 12 The interaction is highly conserved between kindlin family and paxillin superfamily, 12 , 90 but the functional importance could be cell‐type specific. Disruption of kindlin‐3 binding to paxillin and leupaxin in macrophages resulted in enhanced cell spreading and migration, 90 in sharp contrast to the effects of kindlin‐2/paxillin interaction in fibroblasts. 12 , 89 Moreover, Klapproth et al. showed that the recruitment of leupaxin but not paxillin to podosomes (another type of adhesive structure) in hematopoietic cells is dependent on kindlin‐3, 91 suggesting the binding of kindlin‐3 to paxillin or leupaxin is spatiotemporally regulated. It is via this interaction that led Gao et al to suggest that paxillin may bridge kindlin and talin (paxillin also binds talin‐H shown in the study) to promote integrin activation. 92 Since paxillin was found in nascent FAs, these studies highlight the physiological importance of the F0 domain of kindlins binding to paxillin superfamily in regulating nascent FA assembly in different types of cells upon integrin activation by talin and kindlins. To further understand the potential phenotype caused by the deficiency of the interactions between the two protein families, mutagenesis‐based knock‐in mice could be generated for detailed investigation in the future.
F1 domain of kindlin adopts similar folding to F0 but contains a surprisingly long unstructured loop, 108 residues for kindlin‐1/2 and 83 residues for kindlin‐3. 93 , 94 A conserved stretch of six positive charged lysines (polylysine motif) locates at the beginning of the loop and was shown to be capable of associating with membrane lipids (Figure 3(a)). 93 Deletion or mutation of this polylysine motif diminished the FA targeting of kindlin as well as the integrin activation in CHO cells. 93 Interestingly, the replacement of the kindlin F1 loop by talin F1 loop which is also known to bind membrane lipids resulted in the same defects, 93 suggesting that the intact F1 loop is required for kindlin’s action and some other unknown function of F1 loop may also be important. For example, the tyrosine 193 of F1 loop could be phosphorylated by tyrosine kinase Src, and the phosphorylation was shown to be important for the FA localization of kindlin and migfilin as well as cell spreading. 95 Note that this residue is not conserved in kindlin‐3, indicating the function of F1 loop may vary among kindlins. Other than the loop region, the binding partners of F1 domain are less identified, except a recent study suggesting that kindlin F1 can complex with Arp2/3 (Figure 3(a)) to regulate cell spreading. 89
F2 domain of kindlin associates tightly (~110 nM) with another essential adhesion protein, integrin‐linked kinase (ILK), which forms complex with pinch and parvin, namely “ILK‐pinch‐parvin (IPP)” complex, to regulate actin cytoskeleton remodeling and dynamic cell adhesion. 96 , 97 Kindlin F2 binds to the kinase‐like or pseudokinase domain (KLD) of ILK. Although the exact nature of the binding interface is not yet defined, the key residues of both proteins involved in the binding have been identified via mutagenesis (Figure 3(a)). 96 , 98 Mutations that disrupt the interaction resulted in significant defects in the FA localization of kindlin and cell spreading, but interestingly had little effect on integrin activation in CHO cells, 96 suggesting the interaction is likely involved in early integrin “outside‐in” signaling. Like canonical PH domains, kindlin PH domain harbors a binding site for membrane‐lipids including PIP2 and PIP3 (Figure 3(a)) to assure the membrane localization of kindlin for integrin activation and cell adhesion. 99 , 100 The cooperative membrane binding of kindlin and talin likely promotes a stable active conformation of integrin allowing potent ligand binding and clustering (Figure 1).
Recent structural studies have suggested that kindlins may undergo oligomerization to regulate the integrin function, but the precise mechanism remains controversial. Li et al. in 2017 first reported crystal structures of truncated kindlin‐2 with deletion of PH domain and F1 loop, which revealed a monomeric conformation as well as a domain‐swapped dimer conformation (Figure 3(b)). The dimer interface is mediated by F2 domain (Figure 3(b)) and was shown to contribute to integrin activation in CHO cells (20%–30% reduction by mutations that disrupt dimer formation). 101 However, in sharp contrast, the crystal structure of full‐length kindlin‐3 solved by Bu et al. revealed an auto‐inhibitory homotrimer where the integrin binding site on F3 domain of one molecule is masked by the PH domain of another (Figure 3(b)). 102 The authors also provided evidence to show that disruption of the trimer interface promotes integrin‐mediated cell adhesion and spreading. 102 Moreover, Kadry et al. in a recent study suggested that both kindlin‐2 and kindlin‐3 can self‐associate into 2–4 molecule oligomers via their F2PH domains but with an interface different from that in the truncated kindlin‐2 dimer. 103 The F3 domain is clearly not involved since deletion of F3 domain increased the self‐association of kindlin molecules. 103 Given that the majority of either isolated kindlin‐2 or kindlin‐3 is monomeric as shown in all these studies, careful investigation is needed to examine if or when kindlin oligomerization occurs in physiological context.
While all kindlins share conserved domain architecture, they are not fully compensatory to one another. Kindlin‐1 and 2 are more alike by sharing 64% identity and 79% similarity in primary sequence, while kindlin‐3 and 2 share 52% identity and 72% similarity. In adherent cells, overexpressed kindlin‐1 or 2 but not kindlin‐3 targets to FAs and also kindlin‐3 is not able to substitute for kindlin‐2 in supporting the activation of β1 integrin. 104 One explanation for these differences may arise from the observation that kindlin‐3 binds much less potently to ILK than kindlin‐2. However, such explanation cannot explain why kindlin‐1 or 2 but not kindlin‐3 robustly enhances integrin activation when co‐expressed with talin‐H in CHO cells. 96 , 105 Interestingly, kindlin‐3 fused to a GST tag, which forms an artificial dimer, is able to enhance talin‐H mediated integrin activation, 105 suggesting that some unknown factor(s) may play a role in the oligomerization of kindlin1/2 which is not operative for kindlin‐3 in non‐hematopoietic cells. This hypothesis remains to be tested in the future. In addition, some phosphorylation sites are not fully conserved among kindlins. As mentioned earlier, the Y193 of kindlin‐2 F1 loop is conserved in kindlin‐1 but not kindlin‐3. Conversely, kindlin‐3 possesses a unique phosphorylation site (T482 or S484) that is not conserved in kindlin‐1 and 2. The phosphorylation is crucial for regulating integrin activation and cell spreading in hematopoietic cells. 106
4. CONCLUSION
As summarized in this review, talin and kindlin are centrally involved in the regulation of integrin activation and early FA assembly via mediating dozens of binding events. The affinities of the interactions range from nM to mM (Tables S1 and S2), indicating the dynamic nature of interactions during cell adhesion. Since one single domain or motif of the proteins is often capable of mediating diverse binding events, we envision that these interactions occur in a spatiotemporal or tissue‐specific manner. Our analysis also suggests that domain‐deletion based functional assays are not sufficient as an exclusive approach to pinpoint the function of a specific interaction as deletion of a domain may disrupt many interactions and potentially the overall folding of these proteins. Continuous efforts by employing combination of structural tools (Cryo‐EM, X‐ray crystallography and NMR spectroscopy) and interface‐based mutagenesis studies will allow us to gain more precise insight into the regulation of such complex and dynamic network. Currently, many drugs including monoclonal antibodies, peptide‐mimetics and small compounds have been developed to target the extracellular domains of integrin receptors to treat related human diseases. 107 , 108 Intracellular interactions summarized here also play pivotal roles in the regulation of integrin signaling. Therefore, membrane permeable therapeutics could be developed to target these intracellular interfaces to intervene integrin‐related human diseases.
AUTHOR CONTRIBUTIONS
Liang Zhu: conceptualization; writing‐review and editing. Edward F. Plow and Jun Qin: funding acquisition; supervision; writing‐review and editing.
Supporting information
Figure S1 (A) Structural comparison of the head domains of talin‐1 and talin‐2. Talin head domain could be highly dynamic and adopt different conformations due to the presence of a long linker between talin‐F1 and F2 subdomain. (B) Reported structures of talin‐R subdomains in apo or complex form. The binding interfaces/sites for target proteins are indicated in the figure. The interface residues of talin‐R9 (within a cut‐off of 5 Å) that interact with talin‐F3 are colored in cyan. The interface residues of talin‐R12 and talin‐F2F3 (within a cut‐off of 5 Å) are colored in orange and blue respectively, and shown in stick representation. Also note that talin‐R5 and R6 are part of ABS2 and contribute to actin binding. Talin‐R11 was shown to bind RIAM as well but the exact interface is not known. N and C indicate the N‐terminus and C‐terminus of the protein molecule respectively. PDB codes used to generate the figures are indicated.
Table S1 Talin‐mediated interactions in FAs.
Table S2 Kindlins‐mediated interactions in FAs a.
ACKNOWLEDGMENTS
This work was supported by NIH grants to J.Q. (R01HL058758 and R01GM062823) and E.F.P. (P01HL073311 and R01HL096062). We acknowledge all the individuals in the field who have contributed to the understanding of molecular mechanisms of integrin signaling and FA assembly.
Zhu L, Plow EF, Qin J. Initiation of focal adhesion assembly by talin and kindlin: A dynamic view. Protein Science. 2021;30:531–542. 10.1002/pro.4014
Funding information National Institutes of Health, Grant/Award Numbers: P01HL073311, R01HL096062, R01GM062823, R01HL058758
REFERENCES
- 1. Hynes RO. Integrins: Bidirectional allosteric signaling machines. Cell. 2002;110:673–687. [DOI] [PubMed] [Google Scholar]
- 2. Bachmann M, Kukkurainen S, Hytonen VP, Wehrle‐Haller B. Cell adhesion by integrins. Physiol Rev. 2019;99:1655–1699. [DOI] [PubMed] [Google Scholar]
- 3. Qin J, Vinogradova O, Plow EF. Integrin bidirectional signaling: A molecular view. PLoS Biol. 2004;2:e169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Moser M, Legate KR, Zent R, Fassler R. The tail of integrins, Talin, and kindlins. Science. 2009;324:895–899. [DOI] [PubMed] [Google Scholar]
- 5. Humphries JD, Chastney MR, Askari JA, Humphries MJ. Signal transduction via integrin adhesion complexes. Curr Opin Cell Biol. 2019;56:14–21. [DOI] [PubMed] [Google Scholar]
- 6. Zaidel‐Bar R, Itzkovitz S, Ma’ayan A, Iyengar R, Geiger B. Functional atlas of the integrin adhesome. Nat Cell Biol. 2007;9:858–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Horton ER, Byron A, Askari JA, et al. Definition of a consensus integrin adhesome and its dynamics during adhesion complex assembly and disassembly. Nat Cell Biol. 2015;17:1577–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Theodosiou M, Widmaier M, Bottcher RT, et al. Kindlin‐2 cooperates with Talin to activate integrins and induces cell spreading by directly binding paxillin. Elife. 2016;5:e10130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Monkley SJ, Zhou XH, Kinston SJ, et al. Disruption of the Talin gene arrests mouse development at the gastrulation stage. Dev Dyn. 2000;219:560–574. [DOI] [PubMed] [Google Scholar]
- 10. Rahikainen R, Ohman T, Turkki P, Varjosalo M, Hytonen VP. Talin‐mediated force transmission and Talin rod domain unfolding independently regulate adhesion signaling. J Cell Sci. 2019;132:jcs226514. [DOI] [PubMed] [Google Scholar]
- 11. Montanez E, Ussar S, Schifferer M, et al. Kindlin‐2 controls bidirectional signaling of integrins. Genes Dev. 2008;22:1325–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhu L, Liu H, Lu F, Yang J, Byzova TV, Qin J. Structural basis of paxillin recruitment by kindlin‐2 in regulating cell adhesion. Structure. 2019;27:1686–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hagel M, George EL, Kim A, et al. The adaptor protein paxillin is essential for normal development in the mouse and is a critical transducer of fibronectin signaling. Mol Cell Biol. 2002;22:901–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wade R, Bohl J, Vande Pol S. Paxillin null embryonic stem cells are impaired in cell spreading and tyrosine phosphorylation of focal adhesion kinase. Oncogene. 2002;21:96–107. [DOI] [PubMed] [Google Scholar]
- 15. Xu W, Baribault H, Adamson ED. Vinculin knockout results in heart and brain defects during embryonic development. Development. 1998;125:327–337. [DOI] [PubMed] [Google Scholar]
- 16. Saunders RM, Holt MR, Jennings L, et al. Role of vinculin in regulating focal adhesion turnover. Eur J Cell Biol. 2006;85:487–500. [DOI] [PubMed] [Google Scholar]
- 17. Sakai T, Li S, Docheva D, et al. Integrin‐linked kinase (ilk) is required for polarizing the epiblast, cell adhesion, and controlling Actin accumulation. Genes Dev. 2003;17:926–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Elad N, Volberg T, Patla I, et al. The role of integrin‐linked kinase in the molecular architecture of focal adhesions. J Cell Sci. 2013;126:4099–4107. [DOI] [PubMed] [Google Scholar]
- 19. Kunschmann T, Puder S, Fischer T, Perez J, Wilharm N, Mierke CT. Integrin‐linked kinase regulates cellular mechanics facilitating the motility in 3d extracellular matrices. Biochim Biophys Acta Mol Cell Res. 1864;2017:580–593. [DOI] [PubMed] [Google Scholar]
- 20. Ilic D, Furuta Y, Kanazawa S, et al. Reduced cell motility and enhanced focal adhesion contact formation in cells from fak‐deficient mice. Nature. 1995;377:539–544. [DOI] [PubMed] [Google Scholar]
- 21. Sieg DJ, Hauck CR, Schlaepfer DD. Required role of focal adhesion kinase (fak) for integrin‐stimulated cell migration. J Cell Sci. 1999;112:2677–2691. [DOI] [PubMed] [Google Scholar]
- 22. Deramaudt TB, Dujardin D, Noulet F, et al. Altering fak‐paxillin interactions reduces adhesion. migration and invasion processes PLoS One. 2014;9:e92059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Burridge K, Connell L. Talin: A cytoskeletal component concentrated in adhesion plaques and other sites of Actin‐membrane interaction. Cell Motil. 1983;3:405–417. [DOI] [PubMed] [Google Scholar]
- 24. Gough RE, Goult BT. The tale of two talins ‐ two isoforms to fine‐tune integrin signalling. FEBS Lett. 2018;592:2108–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Debrand E, El Jai Y, Spence L, et al. Talin 2 is a large and complex gene encoding multiple transcripts and protein isoforms. FEBS J. 2009;276:1610–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Calderwood DA, Campbell ID, Critchley DR. Talins and kindlins: Partners in integrin‐mediated adhesion. Nat Rev Mol Cell Biol. 2013;14:503–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Anthis NJ, Wegener KL, Ye F, et al. The structure of an integrin/Talin complex reveals the basis of inside‐out signal transduction. EMBO J. 2009;28:3623–3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Garcia‐Alvarez B, de Pereda JM, Calderwood DA, et al. Structural determinants of integrin recognition by Talin. Mol Cell. 2003;11:49–58. [DOI] [PubMed] [Google Scholar]
- 29. Wegener KL, Partridge AW, Han J, et al. Structural basis of integrin activation by Talin. Cell. 2007;128:171–182. [DOI] [PubMed] [Google Scholar]
- 30. Vinogradova O, Velyvis A, Velyviene A, Hu B, Haas T, Plow E. Qin J. a structural mechanism of integrin alpha(iib)beta(3) "inside‐out" activation as regulated by its cytoplasmic face. Cell. 2002;110:587–597. [DOI] [PubMed] [Google Scholar]
- 31. Beckerle MC, Miller DE, Bertagnolli ME, Locke SJ. Activation‐dependent redistribution of the adhesion plaque protein, Talin. in intact human platelets J Cell Biol. 1989;109:3333–3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Goksoy E, Ma YQ, Wang X, et al. Structural basis for the autoinhibition of Talin in regulating integrin activation. Mol Cell. 2008;31:124–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dedden D, Schumacher S, Kelley CF, et al. The architecture of talin1 reveals an autoinhibition mechanism. Cell. 2019;179:120–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Song X, Yang J, Hirbawi J, et al. A novel membrane‐dependent on/off switch mechanism of Talin ferm domain at sites of cell adhesion. Cell Res. 2012;22:1533–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Goult BT, Bate N, Anthis NJ, et al. The structure of an interdomain complex that regulates Talin activity. J Biol Chem. 2009;284:15097–15106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Haage A, Goodwin K, Whitewood A, et al. Talin autoinhibition regulates cell‐ecm adhesion dynamics and wound healing in vivo. Cell Rep. 2018;25:2401–2416. [DOI] [PubMed] [Google Scholar]
- 37. Lee HS, Lim CJ, Puzon‐McLaughlin W, Shattil SJ, Ginsberg MH. Riam activates integrins by linking Talin to ras gtpase membrane‐targeting sequences. J Biol Chem. 2009;284:5119–5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yang J, Zhu L, Zhang H, et al. Conformational activation of Talin by riam triggers integrin‐mediated cell adhesion. Nat Commun. 2014;5:5880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Stritt S, Wolf K, Lorenz V, et al. Rap1‐gtp‐interacting adaptor molecule (riam) is dispensable for platelet integrin activation and function in mice. Blood. 2015;125:219–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Klapproth S, Sperandio M, Pinheiro EM, et al. Loss of the rap1 effector riam results in leukocyte adhesion deficiency due to impaired beta2 integrin function in mice. Blood. 2015;126:2704–2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Su W, Wynne J, Pinheiro EM, et al. Rap1 and its effector riam are required for lymphocyte trafficking. Blood. 2015;126:2695–2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhu L, Yang J, Bromberger T, et al. Structure of rap1b bound to Talin reveals a pathway for triggering integrin activation. Nat Commun. 2017;8:1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bromberger T, Klapproth S, Rohwedder I, et al. Direct rap1/talin1 interaction regulates platelet and neutrophil integrin activity in mice. Blood. 2018;132:2754–2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gingras AR, Lagarrigue F, Cuevas MN, et al. Rap1 binding and a lipid‐dependent helix in Talin f1 domain promote integrin activation in tandem. J Cell Biol. 2019;218:1799–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bromberger T, Zhu L, Klapproth S, Qin J, Moser M. Rap1 and membrane lipids cooperatively recruit Talin to trigger integrin activation. J Cell Sci. 2019;132:jcs235531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lagarrigue F, Paul DS, Gingras AR, et al. Talin‐1 is the principal platelet rap1 effector of integrin activation. Blood. 2020;136:1180–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Goult BT, Bouaouina M, Elliott PR, et al. Structure of a double ubiquitin‐like domain in the Talin head: A role in integrin activation. EMBO J. 2010;29:1069–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kukkurainen S, Azizi L, Zhang P, et al. The f1 loop of the Talin head domain acts as a gatekeeper in integrin activation and clustering. J Cell Sci. 2020;133:jcs239202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chinthalapudi K, Rangarajan ES, Izard T. The interaction of Talin with the cell membrane is essential for integrin activation and focal adhesion formation. Proc Natl Acad Sci U S A. 2018;115:10339–10344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. de Pereda JM, Wegener KL, Santelli E, et al. Structural basis for phosphatidylinositol phosphate kinase type igamma binding to Talin at focal adhesions. J Biol Chem. 2005;280:8381–8386. [DOI] [PubMed] [Google Scholar]
- 51. Kong X, Wang X, Misra S, Qin J. Structural basis for the phosphorylation‐regulated focal adhesion targeting of type igamma phosphatidylinositol phosphate kinase (pipkigamma) by Talin. J Mol Biol. 2006;359:47–54. [DOI] [PubMed] [Google Scholar]
- 52. Ling K, Doughman RL, Firestone AJ, Bunce MW, Anderson RA. Type i gamma phosphatidylinositol phosphate kinase targets and regulates focal adhesions. Nature. 2002;420:89–93. [DOI] [PubMed] [Google Scholar]
- 53. Di Paolo G, Pellegrini L, Letinic K, et al. Recruitment and regulation of phosphatidylinositol phosphate kinase type 1 gamma by the ferm domain of Talin. Nature. 2002;420:85–89. [DOI] [PubMed] [Google Scholar]
- 54. Elliott PR, Goult BT, Kopp PM, et al. The structure of the Talin head reveals a novel extended conformation of the ferm domain. Structure. 2010;18:1289–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rangarajan ES, Primi MC, Colgan LA, Chinthalapudi K, Yasuda R, Izard T. A distinct talin2 structure directs isoform specificity in cell adhesion. J Biol Chem. 2020;295:12885–12899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhang X, Jiang G, Cai Y, Monkley SJ, Critchley DR, Sheetz MP. Talin depletion reveals independence of initial cell spreading from integrin activation and traction. Nat Cell Biol. 2008;10:1062–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lee HS, Bellin RM, Walker DL, et al. Characterization of an Actin‐binding site within the Talin ferm domain. J Mol Biol. 2004;343:771–784. [DOI] [PubMed] [Google Scholar]
- 58. Ciobanasu C, Wang H, Henriot V, et al. Integrin‐bound Talin head inhibits Actin filament barbed‐end elongation. J Biol Chem. 2018;293:2586–2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Atherton P, Stutchbury B, Wang DY, et al. Vinculin controls Talin engagement with the actomyosin machinery. Nat Commun. 2015;6:10038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Gingras AR, Bate N, Goult BT, et al. The structure of the c‐terminal Actin‐binding domain of Talin. EMBO J. 2008;27:458–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Carisey A, Ballestrem C. Vinculin, an adapter protein in control of cell adhesion signalling. Eur J Cell Biol. 2011;90:157–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Atherton P, Lausecker F, Carisey A, et al. Relief of Talin autoinhibition triggers a force‐independent association with vinculin. J Cell Biol. 2020;219:e201903134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Humphries JD, Wang P, Streuli C, Geiger B, Humphries MJ, Ballestrem C. Vinculin controls focal adhesion formation by direct interactions with Talin and Actin. J Cell Biol. 2007;179:1043–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Gingras AR, Ziegler WH, Frank R, et al. Mapping and consensus sequence identification for multiple vinculin binding sites within the Talin rod. J Biol Chem. 2005;280:37217–37224. [DOI] [PubMed] [Google Scholar]
- 65. Bakolitsa C, Cohen DM, Bankston LA, et al. Structural basis for vinculin activation at sites of cell adhesion. Nature. 2004;430:583–586. [DOI] [PubMed] [Google Scholar]
- 66. del Rio A, Perez‐Jimenez R, Liu R, Roca‐Cusachs P, Fernandez JM, Sheetz MP. Stretching single Talin rod molecules activates vinculin binding. Science. 2009;323:638–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Yao M, Goult BT, Klapholz B, et al. The mechanical response of Talin. Nat Commun. 2016;7:11966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Tapia‐Rojo R, Alonso‐Caballero A, Fernandez JM. Direct observation of a coil‐to‐helix contraction triggered by vinculin binding to Talin. Sci Adv. 2020;6:eaaz4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Yao M, Goult BT, Chen H, Cong P, Sheetz MP, Yan J. Mechanical activation of vinculin binding to Talin locks Talin in an unfolded conformation. Sci Rep. 2014;4:4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Rahikainen R, von Essen M, Schaefer M, et al. Mechanical stability of Talin rod controls cell migration and substrate sensing. Sci Rep. 2017;7:3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Goult BT, Zacharchenko T, Bate N, et al. Riam and vinculin binding to Talin are mutually exclusive and regulate adhesion assembly and turnover. J Biol Chem. 2013;288:8238–8249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kelley CF, Litschel T, Schumacher S, Dedden D, Schwille P, Mizuno N. Phosphoinositides regulate force‐independent interactions between Talin. vinculin, and actin Elife. 2020;9:e56110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Gingras AR, Vogel KP, Steinhoff HJ, et al. Structural and dynamic characterization of a vinculin binding site in the Talin rod. Biochemistry. 2006;45:1805–1817. [DOI] [PubMed] [Google Scholar]
- 74. Chang YC, Zhang H, Franco‐Barraza J, et al. Structural and mechanistic insights into the recruitment of Talin by riam in integrin signaling. Structure. 2014;22:1810–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Lagarrigue F, Vikas Anekal P, Lee HS, et al. A riam/lamellipodin‐Talin‐integrin complex forms the tip of sticky fingers that guide cell migration. Nat Commun. 2015;6:8492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Zacharchenko T, Qian X, Goult BT, et al. Ld motif recognition by Talin: Structure of the Talin‐dlc1 complex. Structure. 2016;24:1130–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Li G, Du X, Vass WC, Papageorge AG, Lowy DR, Qian X. Full activity of the deleted in liver cancer 1 (dlc1) tumor suppressor depends on an ld‐like motif that binds Talin and focal adhesion kinase (fak). Proc Natl Acad Sci U S A. 2011;108:17129–17134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Haining AWM, Rahikainen R, Cortes E, et al. Mechanotransduction in Talin through the interaction of the r8 domain with dlc1. PLoS Biol. 2018;16:e2005599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Bouchet BP, Gough RE, Ammon YC, et al. Talin‐kank1 interaction controls the recruitment of cortical microtubule stabilizing complexes to focal adhesions. Elife. 2016;5:e18124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Sun Z, Tseng HY, Tan S, et al. Kank2 activates Talin, reduces force transduction across integrins and induces central adhesion formation. Nat Cell Biol. 2016;18:941–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Plow EF, Qin J, Byzova T. Kindling the flame of integrin activation and function with kindlins. Curr Opin Hematol. 2009;16:323–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Bachir AI, Zareno J, Moissoglu K, Plow EF, Gratton E, Horwitz AR. Integrin‐associated complexes form hierarchically with variable stoichiometry in nascent adhesions. Curr Biol. 2014;24:1845–1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Meves A, Stremmel C, Gottschalk K, Fassler R. The kindlin protein family: New members to the club of focal adhesion proteins. Trends Cell Biol. 2009;19:504–513. [DOI] [PubMed] [Google Scholar]
- 84. Malinin NL, Plow EF, Byzova TV. Kindlins in ferm adhesion. Blood. 2010;115:4011–4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Bialkowska K, Ma YQ, Bledzka K, et al. The integrin co‐activator kindlin‐3 is expressed and functional in a non‐hematopoietic cell, the endothelial cell. J Biol Chem. 2010;285:18640–18649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Sossey‐Alaoui K, Pluskota E, Davuluri G, et al. Kindlin‐3 enhances breast cancer progression and metastasis by activating twist‐mediated angiogenesis. FASEB J. 2014;28:2260–2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Xu Z, Gao J, Hong J, Ma YQ. Integrity of kindlin‐2 ferm subdomains is required for supporting integrin activation. Biochem Biophys Res Commun. 2013;434:382–387. [DOI] [PubMed] [Google Scholar]
- 88. Perera HD, Ma YQ, Yang J, Hirbawi J, Plow EF, Qin J. Membrane binding of the n‐terminal ubiquitin‐like domain of kindlin‐2 is crucial for its regulation of integrin activation. Structure. 2011;19:1664–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Bottcher RT, Veelders M, Rombaut P, et al. Kindlin‐2 recruits paxillin and arp2/3 to promote membrane protrusions during initial cell spreading. J Cell Biol. 2017;216:3785–3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Liu H, Zhu L, Dudiki T, et al. Macrophage migration and phagocytosis are controlled by kindlin‐3's link to the cytoskeleton. J Immunol. 2020;204:1954–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Klapproth S, Bromberger T, Turk C, Kruger M, Moser M. A kindlin‐3‐leupaxin‐paxillin signaling pathway regulates podosome stability. J Cell Biol. 2019;218:3436–3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Gao J, Huang M, Lai J, et al. Kindlin supports platelet integrin alphaiibbeta3 activation by interacting with paxillin. J Cell Sci. 2017;130:3764–3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Bouaouina M, Goult BT, Huet‐Calderwood C, et al. Calderwood DA. A conserved lipid‐binding loop in the kindlin ferm f1 domain is required for kindlin‐mediated alphaiibbeta3 integrin coactivation. J Biol Chem. 2012;287:6979–6990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Chua GL, Tan SM, Bhattacharjya S. Nmr characterization and membrane interactions of the loop region of kindlin‐3 f1 subdomain. PLoS One. 2016;11:e0153501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Liu Z, Lu D, Wang X, Wan J, Liu C, Zhang H. Kindlin‐2 phosphorylation by src at y193 enhances src activity and is involved in migfilin recruitment to the focal adhesions. FEBS Lett. 2015;589:2001–2010. [DOI] [PubMed] [Google Scholar]
- 96. Fukuda K, Bledzka K, Yang J, Perera HD, Plow EF, Qin J. Molecular basis of kindlin‐2 binding to integrin‐linked kinase pseudokinase for regulating cell adhesion. J Biol Chem. 2014;289:28363–28375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Vaynberg J, Fukuda K, Lu F, et al. Non‐catalytic signaling by pseudokinase ilk for regulating cell adhesion. Nat Commun. 2018;9:4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Kadry YA, Huet‐Calderwood C, Simon B, Calderwood DA. Kindlin‐2 interacts with a highly conserved surface of ilk to regulate focal adhesion localization and cell spreading. J Cell Sci. 2018;131:jcs221184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Liu J, Fukuda K, Xu Z, et al. Structural basis of phosphoinositide binding to kindlin‐2 protein pleckstrin homology domain in regulating integrin activation. J Biol Chem. 2011;286:43334–43342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Hart R, Stanley P, Chakravarty P, Hogg N. The kindlin 3 pleckstrin homology domain has an essential role in lymphocyte function‐associated antigen 1 (lfa‐1) integrin‐mediated b cell adhesion and migration. J Biol Chem. 2013;288:14852–14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Li H, Deng Y, Sun K, et al. Structural basis of kindlin‐mediated integrin recognition and activation. Proc Natl Acad Sci U S A. 2017;114:9349–9354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Bu W, Levitskaya Z, Loh ZY, et al. Structural basis of human full‐length kindlin‐3 homotrimer in an auto‐inhibited state. PLoS Biol. 2020;18:e3000755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Kadry YA, Maisuria EM, Huet‐Calderwood C, Calderwood DA. Differences in self‐association between kindlin‐2 and kindlin‐3 are associated with differential integrin binding. J Biol Chem. 2020;295:11161–11173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Huet‐Calderwood C, Brahme NN, Kumar N, et al. Differences in binding to the ilk complex determines kindlin isoform adhesion localization and integrin activation. J Cell Sci. 2014;127:4308–4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Sun J, Xiao D, Ni Y, et al. Structure basis of the ferm domain of kindlin‐3 in supporting integrin alphaiibbeta3 activation in platelets. Blood Adv. 2020;4:3128–3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Bialkowska K, Byzova TV, Plow EF. Site‐specific phosphorylation of kindlin‐3 protein regulates its capacity to control cellular responses mediated by integrin alphaiibbeta3. J Biol Chem. 2015;290:6226–6242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Ley K, Rivera‐Nieves J, Sandborn WJ, Shattil S. Integrin‐based therapeutics: Biological basis, clinical use and new drugs. Nat Rev Drug Discov. 2016;15:173–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Zheng Y, Leftheris K. Insights into protein‐ligand interactions in integrin complexes: Advances in structure determinations. J Med Chem. 2020;63:5675–5696. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 (A) Structural comparison of the head domains of talin‐1 and talin‐2. Talin head domain could be highly dynamic and adopt different conformations due to the presence of a long linker between talin‐F1 and F2 subdomain. (B) Reported structures of talin‐R subdomains in apo or complex form. The binding interfaces/sites for target proteins are indicated in the figure. The interface residues of talin‐R9 (within a cut‐off of 5 Å) that interact with talin‐F3 are colored in cyan. The interface residues of talin‐R12 and talin‐F2F3 (within a cut‐off of 5 Å) are colored in orange and blue respectively, and shown in stick representation. Also note that talin‐R5 and R6 are part of ABS2 and contribute to actin binding. Talin‐R11 was shown to bind RIAM as well but the exact interface is not known. N and C indicate the N‐terminus and C‐terminus of the protein molecule respectively. PDB codes used to generate the figures are indicated.
Table S1 Talin‐mediated interactions in FAs.
Table S2 Kindlins‐mediated interactions in FAs a.


