ABSTRACT
The well-studied members of the Trio family of proteins are Trio and kalirin in vertebrates, UNC-73 in Caenorhabditis elegans and Trio in Drosophila. Trio proteins are key regulators of cell morphogenesis and migration, tissue organization, and secretion and protein trafficking in many biological contexts. Recent discoveries have linked Trio and kalirin to human disease, including neurological disorders and cancer. The genes for Trio family proteins encode a series of large multidomain proteins with up to three catalytic activities and multiple scaffolding and protein–protein interaction domains. As such, Trio family proteins engage a wide array of cell surface receptors, substrates and interaction partners to coordinate changes in cytoskeletal regulatory and protein trafficking pathways. We provide a comprehensive review of the specific mechanisms by which Trio family proteins carry out their functions in cells, highlight the biological and cellular contexts in which they occur, and relate how alterations in these functions contribute to human disease.
KEY WORDS: Rho GTPase, Signal transduction, Trio family, Cytoskeleton, Neurodevelopmental disorders, Cell morphogenesis
Summary: A review of the widespread cellular roles of Trio family proteins in migration and morphogenesis whose alterations contribute to human disease.
Introduction
Trio family proteins are key regulators of cell motility and morphogenesis, tissue development, and protein trafficking and secretion in numerous biological contexts, including their prominent roles in developing nervous systems. These diverse roles are achieved through Trio protein interactions with membrane receptors, cytoskeleton-interacting proteins, lipids, endocytic machinery, kinases and Rho family GTPases in the cell (Fig. 1). Recent studies have linked mutations in the human genes TRIO and kalirin (KALRN) to neurological diseases (Paskus et al., 2020) and cancers (Schmidt and Debant, 2014), highlighting the need to understand the primary functions of Trio family proteins and underscoring the outstanding questions in the field. How are the different catalytic activities balanced within Trio proteins? How do the accessory domains in Trio proteins contribute to Trio function, and how does the primary function of Trio proteins differ based on its interactions with cellular binding partners?
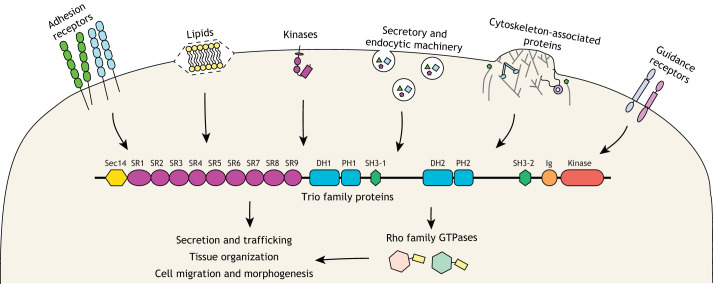
Fig. 1.
Trio family proteins integrate signaling from a wide array of interaction partners to impact cell behaviors. Trio family proteins relay signaling with adhesion receptors, lipids, kinases, secretory and endocytic machinery, cytoskeleton-associated proteins, and guidance receptors to regulate secretion and trafficking, tissue organization, and cell migration and morphogenesis. Much, but not all, of this signaling is achieved through Trio catalytic activities on Rho family GTPases. Domains of the Trio proteins: SR, spectrin repeats; DH, Dbl homology; PH, pleckstrin homology; GEF, guanine nucleotide exchange factor; SH3, Src homology 3; Ig, immunoglobulin-like.
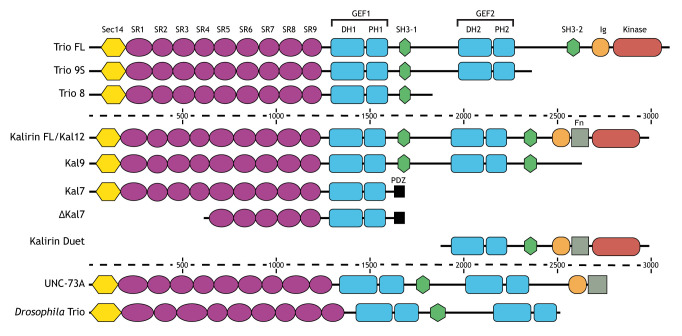
The Trio protein family has four well studied members – two vertebrate paralogs (Trio and kalirin) and two invertebrate orthologs (UNC-73 in Caenorhabditis elegans and Trio in Drosophila) (Rabiner et al., 2005). Trio family proteins are large proteins (up to 350 kDa), containing up to three catalytic domains, for which they are named; full-length (FL) isoforms of Trio family proteins contain two guanine nucleotide exchange factor (GEF) domains (GEF1 and GEF2), and the vertebrate paralogs contain an additional putative serine/threonine kinase domain (Rabiner et al., 2005). Trio family proteins also contain numerous accessory domains, which differ slightly across species, and whose functions are poorly understood. Furthermore, alternative splicing produces many different isoforms whose expression profiles vary across tissue and developmental stage (Miller et al., 2013) (Box 1).
Box 1. Isoforms of the Trio family proteins.
Trio family proteins are large multi-domain proteins that contain up to three catalytic domains and multiple accessory domains. All full-length (FL) Trio family proteins (Trio FL, kalirin FL/Kal12, UNC-73A, and Drosophila Trio) contain two catalytic GEF units (shown in blue in the figure) composed of tandem Dbl homology (DH) and pleckstrin homology (PH) domains. The vertebrate members, Trio and kalirin, also contain an additional putative kinase domain. The additional accessory domains vary slightly between species, but include: a Sec14 domain, nine spectrin repeats (SR1–SR9), one or two Src homology 3 (SH3) domains, and zero or one immunoglobulin-like (Ig) and fibronectin-like (Fn) domains (Debant et al., 1996; Alam et al., 1997; Steven et al., 1998; Awasaki et al., 2000). Trio, kalirin and UNC-73 are alternatively spliced to generate multiple isoforms. Only the isoforms mentioned in this review are shown in the figure; for a more comprehensive list, see Steven et al., (1998), McPherson et al. (2005), McPherson et al. (2002) and Steven et al. (1998). Amino acid numbers are marked on dotted line for scale.
While Trio and kalirin have nearly identical domain structure, they have different tissue-specific and temporal expression profiles (Miller et al., 2013). Trio is ubiquitously expressed, whereas kalirin is most highly expressed in the nervous system (Miller et al., 2013; Debant et al., 1996; Alam et al., 1997; Wu et al., 2013). Additionally, Trio is abundant in the developing brain (Ma et al., 2005), while kalirin predominates in the brain from postnatal development through adulthood (Ma et al., 2001). Finally, different isoforms of each vary in abundance in different contexts (McPherson et al., 2005, McPherson et al., 2002). An outstanding question in the field is how small differences between Trio and kalirin, or the usage of different isoforms, drives Trio family proteins to perform their vast set of distinct tasks within a cell.
Here, we review Trio family protein functions from a biochemical level to roles in neurodevelopment and disease. We first discuss the specific catalytic activities of Trio family proteins and how the different domains in Trio family proteins and interactions with signaling partners contribute to Trio family protein catalytic activity and function. We then discuss the specific cellular processes that are driven by these Trio activities. Finally, we address recent studies linking vertebrate Trio to human disease, and how specific disease-associated mutations and rare variants impact Trio function.
Trio family protein catalytic activities and membrane interactions
Vertebrate Trio family proteins contain two GEF domains and a kinase domain, each of which can activate discrete signaling outputs. Several domains within Trio also regulate Trio GEF activities and mediate membrane binding, which brings Trio in proximity to its membrane-bound Rho family GTPase substrates.
Trio family protein GEF domains
The Trio family protein GEF domains catalyze exchange of GDP for GTP on specific Rho family GTPases, master regulators of the cytoskeleton (Jaffe and Hall, 2005). They comprise a tandem catalytic Dbl-homology (DH) domain and a regulatory pleckstrin homology (PH) domain, together forming the functional GEF unit (Fig. 1). The Trio family GEF1 domain catalyzes GTP exchange on both Rac1 and RhoG in vitro (Penzes et al., 2000; Steven et al., 1998; Debant et al., 1996; Wu et al., 2002; Kubiseski et al., 2003), although vertebrate Trio GEF1 catalyzes faster exchange on RhoG compared to Rac1 (Blangy et al., 2000; Skowronek et al., 2004; Chhatriwala et al., 2007). By contrast, the Trio family GEF2 domain catalyzes GTP exchange on RhoA in vitro (Debant et al., 1996; Steven et al., 1998; Penzes et al., 2001), although vertebrate Trio GEF2 does so at a much slower rate than that its GEF1 exchanges GTP on RhoG (Bellanger et al., 2003). Vertebrate Trio GEF1 can also catalyze GTP exchange on membrane-anchored Cdc42 (Peurois et al., 2017), suggesting that the GEF1 and GEF2 domains may have additional Rho GTPase targets beyond just Rac1, RhoG and RhoA.
Loss of endogenous kalirin or Trio reduces activation of their GTPase targets in cells and tissues, demonstrating that they are major cellular activators of these GTPases (Yan et al., 2016; Peng et al., 2010; Backer et al., 2018; Valdivia et al., 2017). It is unknown how the Trio GEF1 and GEF2 activities are balanced in the cell, especially since Rac1 and RhoG typically have opposing signaling pathways and outputs in cells compared to RhoA (van Leeuwen et al., 1997; Sander et al., 1999). It is also unclear whether the GEF1 domain preferentially targets RhoG over Rac1 in cells; an assessment complicated by the fact that RhoG can activate Rac1 via the dedicator of cytokinesis protein 1 (Dock180; also known as DOCK1)–engulfment and cell motility protein 1 (Elmo1) complex during integrin-mediated cell spreading and other processes (Gauthier-Rouviere et al., 1998; Katoh and Negishi, 2003). Therefore, whether Trio GEF1 activates Rac1 directly or preferentially through activation of RhoG signaling through Dock180–Elmo1 is unknown.
GEF activity is modulated by accessory domains and phosphorylation
Accessory domains also modulate the Trio and kalirin GEF activities. For instance, the PH domain located within each GEF domain impacts catalysis by the adjacent DH domain. The DH1 and PH1 domains of Trio GEF1 coordinately engage Rac1 during GTP exchange, and these direct PH1–Rac1 interactions are critical for efficient exchange (Chhatriwala et al., 2007). The Trio and kalirin GEF1 domains share 88% sequence identity, so it is likely that the kalirin GEF1 also uses this regulation mechanism. Indeed, removal of the PH1 domain significantly impairs catalytic activity of the purified Trio or UNC-73 GEF1 domains, suggesting this mode of regulation is conserved (Liu et al., 1999; Bellanger et al., 2003; Kubiseski et al., 2003). In notable contrast, the Trio PH2 makes intramolecular inhibitory contacts with DH2 to block RhoA binding, explaining why loss of Trio PH2 enhances DH2 activity (Bellanger et al., 2003; Bandekar et al., 2019). Trio and kalirin only share 63% identity between their GEF2 domains, so it is less clear whether the kalirin GEF2 domain shares this mode of regulation. The Trio and kalirin spectrin repeats (SRs) also impact GEF1 interactions with Rac1 and catalytic activity. For example, Trio SRs 1 to 5 bind the GEF1 domain directly and inhibit the ability of Trio GEF1 to bind Rac1 in vitro (Chen et al., 2011). Similarly, kalirin fragments that contain portions of the SRs plus GEF1 exhibit reduced Rac1 exchange activity relative to that of the purified GEF1 domain alone (Schiller et al., 2006; Schiller et al., 2008). These findings indicate that the PH domains and SRs regulate Trio and kalirin GEF activities, but they raise questions of how these regulatory interactions are controlled to fine tune these activities in cells.
Trio and kalirin are phosphorylated by a diverse set of kinases (Kawai et al., 1999; Kiraly et al., 2011; Sonoshita et al., 2015; Ma et al., 2014; Forsthoefel et al., 2005; Miller et al., 2017; Xin et al., 2008; DeGeer et al., 2013; Xie et al., 2007). Phosphorylation events within the GEF domains directly impact GEF activity in vitro, indicating phosphorylation as a key mode of regulation (Xin et al., 2008; Sonoshita et al., 2015). Phosphorylation of Trio and kalirin on sites outside the GEF domains also impact active Rac1 levels in cells (Xin et al., 2004; Xie et al., 2007; DeGeer et al., 2013), although how these phosphorylation events impact GEF1 activity is less clear. The context and outcome of these events are further discussed below.
Putative kinase domain
Several vertebrate Trio and kalirin splice isoforms contain a putative serine/threonine kinase domain that may have kinase activity (see Box 1). In support of this, the kalirin Duet isoform is phosphorylated in 3T3 cells, and mutating a predicted key catalytic residue (K2713A) disrupts this event, suggesting autophosphorylation (Kawai et al., 1999). Furthermore, expression of the kalirin kinase domain in cultured rat hippocampal neurons enhances neurite outgrowth, while a predicted catalytically inactive kalirin kinase mutant blocks neurite extension (Yan et al., 2014). These findings strongly suggest that Trio protein kinase activity has physiological roles, but the fundamental questions of how kinase activity is regulated and what substrates are targeted remain to be answered.
Membrane localization and regulation of Trio family proteins
Trio proteins contain a lipid-binding N-terminal Sec14 domain and two PH domains, each with the potential to bind phospholipids. Not surprisingly, Trio family proteins localize to diverse membrane regions, including membrane ruffles, cell–cell junctions and the trans-Golgi network (Bellanger et al., 2000; Seipel et al., 2000; Koo et al., 2007; Tao et al., 2019; Kroon et al., 2017; Medley et al., 2003; Sun et al., 2006). Interactions with lipids likely enable Trio proteins to interact with their Rho family GTPase targets, which are themselves targeted to the membrane by covalently attached isoprenoid moieties (Chenette and Der, 2011).
The kalirin Sec14 domain binds to phosphatidylinositols (PIs), including phosphatidylinositol (3,4)-bisphosphate [PI(3,4)P2], PI3P and PI4P (Schiller et al., 2008; Ma et al., 2014; Miller et al., 2015), which are found in the plasma membrane, endosomes, secretory granules and the trans-Golgi network. Loss of the Sec14 domain in Kal7 impairs its ability to promote changes in cell shape and dendritic spine length, indicating the importance of lipid interactions for Kal7 function (Schiller et al., 2008; Ma et al., 2014). In addition, the Trio GEF1 domain binds PIs in the presence of free RhoG, but not Rac1 (Skowronek et al., 2004), suggesting that membrane interactions may be impacted in conjunction with substrate recognition by Trio GEF1. Whether Trio or kalirin PH2 domains also bind to phospholipids has not been directly tested. Nevertheless, these observations indicate that lipids are critical regulators of Trio and kalirin function and may even regulate their catalytic activities.
Trio family proteins in cell migration
Considering their central roles in regulating Rho family GTPases, it is not surprising that Trio family proteins regulate cell migration. Knockdown of Trio in HeLa cells disrupts spreading on fibronectin and impairs chemotaxis towards serum (van Rijssel et al., 2012a). These defects are restored by expression of the Trio GEF1 domain, but not the GEF2 domain, indicating that Rac1 and/or RhoG signaling via Trio GEF1 may be the primary driver of this output (van Rijssel et al., 2012a). Similarly, loss of kalirin (KALRN) or chemical inhibition of kalirin GEF1 in smooth muscle cells significantly reduces serum-evoked cell migration, implicating kalirin GEF1 activity in this process (Wu et al., 2013). Since the process of cell migration involves both the extension of the leading edge of a cell (powered by Rac1), and retraction of the trailing edge (powered by RhoA) (Ridley, 2001), it is interesting that Trio-driven migration is mainly powered by Trio GEF1 activity. Whether this is due to different intrinsic activities of the Trio family GEF domains, differences in protein localization or protein–protein interactions is unknown.
Trio family proteins employ both GEF activities to regulate neuronal cell migration. UNC-73 coordinates the migration of neuronal precursor cells during C. elegans development. (Forrester and Garriga, 1997; Lundquist et al., 2001; Spencer et al., 2001; Wu et al., 2002; Vanderzalm et al., 2009; Zipkin et al., 1997). P cell neuronal precursors do not migrate normally to the ventral midline in unc-73 mutants, and optimal migration requires both the GEF1 and GEF2 activities of UNC-73 (Spencer et al., 2001; Lundquist et al., 2001; Wu et al., 2002; Zipkin et al., 1997). In addition, brain-specific ablation of Trio in mice with a nestin-Cre transgene (nestin-Trio−/− mice) also disrupts neuronal migration in the cerebellum (Peng et al., 2010). The activities of RhoA, Rac1 and Cdc42, key coordinators of cell migration, are all reduced in nestin-Trio−/− mice (Peng et al., 2010), providing in vivo evidence that Trio serves as a signal integrator to those three GTPases in the developing brain.
Less is known regarding cell surface receptors or intercellular signaling partners that may regulate Trio GEF activities during cell migration. One interesting candidate is Supervillin (SVIL) isoform 4 (Supervillin4), which links the actin cytoskeleton to the plasma membrane. Trio employs its SR6 and SR7 to bind Supervillin4 directly (Son et al., 2015). Supervillin4 expression in HeLa cells induces Rac1 activation, and depletion of Trio prevents this, suggesting Supervillin4 signals through Trio to activate Rac1. Furthermore, loss of Trio, Supervillin4, or expression of dominant-negative Trio that interacts with Supervillin4 but cannot activate Rac1, inhibits initial cell spreading, implicating the Supervillin4-Trio-Rac1 signaling axis in cell spreading (Son et al., 2015).
Trio family proteins in cell morphogenesis
Trio family proteins employ their GEF activities downstream of cell surface receptors to regulate cell morphogenesis. The ability of Trio proteins to regulate these processes also depends on a collection of interaction partners and substrates in different cell types. Here, we discuss the interaction partners and mechanisms by which Trio regulates changes in cell shape in different cell types.
Cytoskeletal reorganization and cell-edge protrusions
Trio family proteins employ their noncatalytic domains to engage actin- and microtubule (MT)-binding proteins, including Supervillin4, CARMIL (CRML-1 in C. elegans), Tara (also known as TRIOBP), filamin proteins, EB1 (also known as MAPRE1) and neuron navigator 1 (Nav1); this impacts their regulation of their target GTPases, thereby modulating cell morphology and behavior (van Rijssel et al., 2012a; Son et al., 2015; Chen et al., 2011; van Haren et al., 2014; Yano et al., 2011; Vanderzalm et al., 2009). It remains unknown whether Trio family proteins bind actin or MTs directly.
Overexpression of full-length Trio (Trio-FL) or the GEF1 domain alone decreases stress fiber formation in fibroblast cells and increases cortical actin filament numbers in HeLa cells (Seipel et al., 1999; van Rijssel et al., 2012a). These phenotypes match those obtained by expression of constitutively active Rac1 and RhoG, consistent with a role for the GEF1 activity in driving these changes (Blangy et al., 2006; Ferraro et al., 2007; Bouquier et al., 2009). In contrast, Trio GEF2 domain expression increases stress fiber abundance in cells, which phenocopies the constitutive RhoA activation (Bellanger et al., 1998; Seipel et al., 1999; van Rijssel et al., 2012a). It is unclear how the opposing outputs of the Trio GEF1 and GEF2 domains are balanced in the context of Trio-FL. However, GEF1 activity appears to dominate, since the phenotypes resulting from Trio-FL expression most closely those following expression of the GEF1 domain alone.
Trio family proteins also promote cell edge protrusions in various cell types. The Trio and kalirin GEF1 domains induce cell edge ruffling, protrusions, and/or lamellipodia formation, which are mediated by Rac1 or RhoG, depending on cell type and context (Bellanger et al., 1998; Seipel et al., 1999; Bellanger et al., 2000; Blangy et al., 2000; van Rijssel et al., 2012a; Steven et al., 1998; Schiller et al., 2005). For instance, Trio or kalirin GEF1 expression induces AtT20 cells, which are normally spindle-shaped, to adopt a flattened, round morphology, with uniform radial lamellipodia (Ferraro et al., 2007), characteristics also observed in AtT20 cells with constitutively active RhoG or Rac1 (Ferraro et al., 2007). In addition, UNC-73 GEF1 activity, which acts on CED-10 (Rac1) or MIG-2 (RhoG), promotes epithelial cell edge protrusions during intercalation of epidermal cells (Vanderzalm et al., 2009; Liang et al., 2009; Walck-Shannon et al., 2015). Finally, co-expression of a dominant-negative RhoG (F37A) with Trio GEF1 in fibroblasts eliminates Trio-induced lamellipodia (Blangy et al., 2000), whereas Rac1 knockdown reduces the ability of Trio GEF1 to induce membrane ruffles in HeLa cells (van Rijssel et al., 2012a). Hence, while Trio family protein GEF1 domains clearly promote cell edge protrusions, whether this occurs via distinct Rac1 or RhoG signaling, or by integrating activation of both, is unclear. Roles for the full-length Trio family proteins or the GEF2 domains in cell edge protrusion have not been extensively characterized.
Axon pathfinding
Significant changes in cell morphology occur in neurons as they elaborate axonal and dendritic processes to form connections with other neurons. Trio family proteins have widespread roles in regulating axon pathfinding. In the fly central nervous system, loss of Trio function results in mistargeting of individual axons both in central and peripheral neurons (Awasaki et al., 2000; Bateman et al., 2000; Newsome et al., 2000; Forsthoefel et al., 2005). Similarly, thalamocortical axons (TCAs) in Trio−/− mice stall and misroute in the ventral telencephalon and, ultimately, do not reach their cortical targets (Backer et al., 2018).
Both Trio family GEF1 and GEF2 activities mediate axon pathfinding processes that are often opposing, suggesting that specific Trio activities can be utilized depending on the needs of the cell. Trio acts downstream of the deleted in colorectal cancer (DCC) family of guidance receptors to mediate attractive growth of axons toward sources of the secreted guidance cue netrin (Fig. 2) (Bateman et al., 2000; Forsthoefel et al., 2005; Briançon-Marjollet et al., 2008; DeGeer et al., 2013; DeGeer et al., 2015). Deletion of Trio eliminates Rac1 activation by netrin in mouse cortical explants and blocks netrin-induced axon outgrowth in cortical, spinal cord and cerebellar explants (Briançon-Marjollet et al., 2008; Peng et al., 2010). Likewise, expression of DCC in neuroblastoma cells induces neurite outgrowth, but this is blocked by co-expression of a Trio9 (see Box 1) mutant lacking GEF1 activity (Briançon-Marjollet et al., 2008). Treatment of rat cortical explants with netrin also induces Trio phosphorylation at Y2622 by Fyn (DeGeer et al., 2013). Accordingly, loss of Trio in rat cortical axons causes axon outgrowth defects that cannot be rescued with a nonphosphorylatable Y2622F Trio mutant, suggesting that Trio phosphorylation is important for netrin signaling (DeGeer et al., 2013). Additionally, loss of Trio reduces surface levels of DCC, indicating interactions with Trio may be critical for proper receptor localization (DeGeer et al., 2013). It is not understood whether or how Trio binds DCC directly.
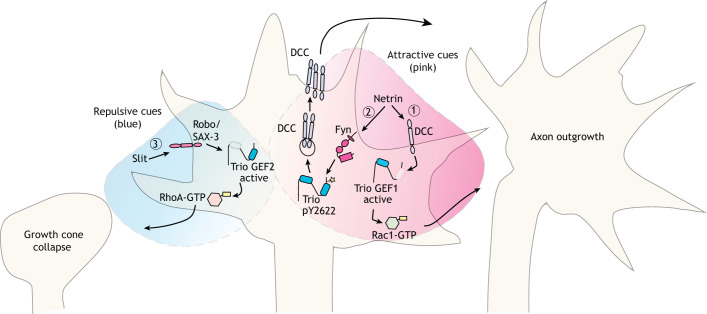
Fig. 2.
Trio family proteins respond to axon guidance cues through guidance receptors. Trio family proteins respond to both attractive and repulsive guidance cues, which induces either axon outgrowth or growth cone collapse. (1) In the presence of netrin, the netrin receptor DCC interacts with Trio – it is unknown if this interaction is direct. This interaction results in a Trio GEF1-mediated activation of Rac1, which is necessary for axon outgrowth. (2) Netrin stimulation also induces phosphorylation of Trio at Y2622 by the kinase Fyn (yellow star), which leads to increased surface levels of DCC. While the roles of Trio in pathways (1) and (2) have not been explicitly connected, Trio-dependent stimulation of DCC at the surface (2) likely constitutes a positive feedback loop to amplify more netrin-DCC signaling in (1). (3) In contrast, binding of Slit to the Robo/SAX-3 receptors results in Trio GEF2-mediated RhoA activation, which causes growth cone collapse.
Trio family proteins also mediate signaling by the Robo/SAX-3 family of receptors for repellent Slit ligands in C. elegans and vertebrates (Fig. 2) (Watari-Goshima et al., 2007; Li et al., 2013; Backer et al., 2018). Slit2 induces growth cone collapse in cultured neurons and this response is not observed in Trio−/− axons (Backer et al., 2018). Application of recombinant Slit2 to mouse fibroblasts activates RhoA, but this response is absent in Trio−/− mouse embryo fibroblasts, confirming a requirement for Trio GEF2 in mediating Slit2 signaling (Backer et al., 2018). A major unresolved question is how the netrin/DCC and Robo receptors engage Trio differently to elicit GEF1-mediated attractant responses or GEF2-mediated repellent responses, respectively.
Dendritic arbor formation and spine structure
Developing neurons form elaborate branched dendritic arbors studded with small protrusions called dendritic spines that serve as the receptive antennae for synaptic input. Disruption of Kalrn or Trio leads to significant reduction of dendritic arbor development in mouse cortical layer 5 pyramidal neurons (Yan et al., 2014; Katrancha et al., 2019). Reduced dendritic arbors are also observed in cultured neurons following knockdown or knockout of kalirin or Trio (May et al., 2002; Yan et al., 2014; Katrancha et al., 2019), demonstrating that both proteins act cell autonomously to control dendritic arbor development. Chemical inhibitors of Trio GEF1 activity reduce dendritic arbor development, suggesting a downstream requirement for activation of Rac1 or RhoG (Yan et al., 2014). Trio also regulates neurite morphology, using its SRs to impact Golgi-derived vesicle trafficking, discussed further below (Tao et al., 2019). Thus, Trio and kalirin act via multiple catalytic and scaffolding roles to regulate dendritic arbor structure.
While ablation of Trio from cortical excitatory neurons yields smaller dendritic arbors, the remaining dendrites have higher densities of dendritic spines. However, dendritic spines on Trio−/− cortical neurons in vivo are also smaller and thinner, having a more immature appearance (Katrancha et al., 2019). Interestingly, overexpression of Kal7 (see Box 1), the predominant kalirin isoform in the postnatal brain, is sufficient to drive dendritic spine formation in a manner that depends on its GEF1 activity (Ma et al., 2008). Kal7 is even capable of inducing dendritic- spine-like protrusions in inhibitory neurons, which normally lack them (Ma et al., 2008).
Adhesion at the synapse
The postsynaptic proteins EphB2 tyrosine kinase and neuroligin-1 engage their presynaptic partners ephrin B and neurexin, respectively, to mediate synapse formation in a manner that also depends on Trio family proteins. First, EphB2 phosphorylates the Kal7 kalirin isoform and recruits it to synaptic clusters in dendritic spines, and ephrin signaling through Rac1 is dependent on kalirin GEF1 activity in primary cultured hippocampal neurons (Penzes et al., 2001, 2003). Secondly, overexpression of neuroligin-1 in hippocampal organotypic slices increases dendritic spine density and functional synapses, which also requires kalirin (Paskus et al., 2019). Finally, UNC-73 regulates extension of muscle arms (Alexander et al., 2010), the postsynaptic contact at the C. elegans neuromuscular junction. Collectively, these findings demonstrate that Trio family proteins play evolutionarily conserved functions in regulating post-synaptic development.
Vertebrate Trio in tissue organization
Vertebrate Trio regulates tissue organization by mediating signaling from transmembrane adhesion receptors including cadherins and Notch1 (Fig. 3) (Timmerman et al., 2015; Polacheck et al., 2017; Kruse et al., 2018). Cadherins mediate cell–cell adhesions that are crucial for maintaining blood vessel wall integrity (Vestweber, 2008). Vascular endothelial (VE)-cadherins mediate homotypic interactions, or adherens junctions (AJs), between endothelial cells that comprise the vessel wall and provide a barrier to permeability (Vestweber, 2008). Two important processes regulate barrier permeability – (1) laminar flow and (2) heterotypic interactions with other non-endothelial cells (Vestweber, 2008; Henrique and Schweisguth, 2019; Kruse et al., 2018). Through various signaling mechanisms, these distinct inputs reinforce existing adhesion sites and induce Rac1 activation to recruit VE-cadherins to nascent adhesion sites (Timmerman et al., 2015). In the absence of flow or heterotypic cell interactions, endothelial cells lacking Trio make unstable and irregular AJs (Timmerman et al., 2015). Recovery of these deficits requires Trio GEF1 catalytic activity and Rac1 activity, suggesting that Trio directly activates Rac1 in regulating VE-cadherin based AJs (Timmerman et al., 2015). Importantly, Trio utilizes SR5 and SR6 to bind the VE-cadherin intracellular tail and colocalizes at AJs with VE-cadherin and sites of focally increased Rac1 activity (Timmerman et al., 2015). While these data suggest that VE-cadherin recruits Trio to AJs to induce local activation of Rac1 and reinforce AJs at cell–cell junctions with no stimulus, Trio also plays a role in strengthening AJs in response to laminar flow and heterotypic interactions.
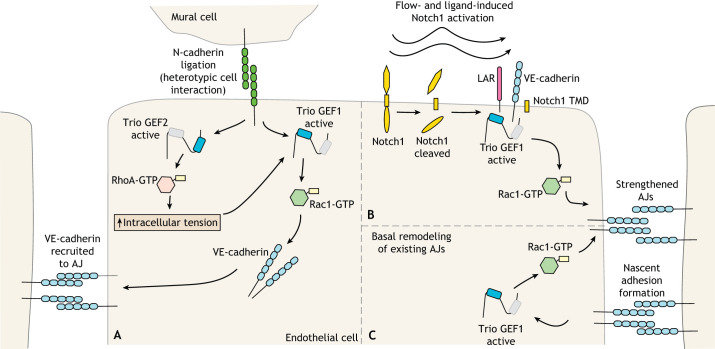
Fig. 3.
Trio family proteins utilize both GEF domains to promote endothelial adherens junctions. Trio family proteins are important for maintaining adherens junction (AJ) integrity between endothelial cells. AJ formation and integrity are impacted by interactions with other types of cells (mural cells), laminar flow, and the basal remodeling of AJs. (A) Neural (N)-cadherin ligation between a mural cell and an epithelial cell induces Trio GEF1 to activate Rac1, increasing vascular endothelial (VE)-cadherin recruitment to AJs between neighboring endothelial cells. How N-cadherin signals to Trio, and how Rac1 activation increases VE-cadherin recruitment to AJs in this context, are unknown. N-cadherin ligation also increases Trio GEF2-mediated RhoA activation, increasing intracellular tension, and serving as a positive-feedback loop to reinforce the activity of Trio GEF1. It is unclear how N-cadherin signals to Trio GEF2 and how intracellular tension influences Trio GEF1 activity. (B) Notch1 is cleaved upon interaction with ligand DLL4 under conditions of laminar flow, which allows the Notch1 transmembrane domain (TMD) to colocalize with protein-tyrosine phosphatase (LAR), VE-cadherin and Trio. By an unknown mechanism, assembly of this Notch1–LAR–VE-cadherin–Trio complex induces Trio GEF1-mediated activation of Rac1, which strengthens AJs. (C) In basal conditions, AJs are constantly remodeled. Ligation of VE-cadherins in the formation of new AJs recruits Trio GEF1 to nascent AJs and subsequent local Trio GEF1-mediated Rac1 activation to strengthen nascent adhesions. Trio binds directly to VE-cadherin so this interaction likely drives Trio recruitment to AJs.
Notch1 is a single-pass transmembrane protein essential for maintaining AJ integrity. In response to laminar flow, Notch1 is activated in a process that also depends on its extracellular interaction with ligand delta-like 4 (DLL4) (Polacheck et al., 2017; Henrique and Schweisguth, 2019). This activation is followed by the initial cleavage of the extracellular domain by metalloproteases and subsequent cleavage of the intracellular domain by the γ-secretase complex (Henrique and Schweisguth, 2019; Polacheck et al., 2017). The remaining transmembrane domain can then form a complex with protein-tyrosine phosphatase (LAR; also known as PTPRF in vertebrates), VE-cadherin and Trio (Polacheck et al., 2017) (Fig. 3). Formation of this complex is associated with increased Rac1 activity and strengthened VE-cadherin based AJs (Polacheck et al., 2017). While Trio, LAR and Notch1 are required for this process, it is unclear whether Trio GEF1 activity is directly responsible for the Rac1 activation that increases barrier strength in this context.
Neural (N)-cadherins mediate heterotypic interactions between endothelial cells and vascular smooth muscle cells or pericytes, collectively called mural cells. These interactions ultimately increase the formation of AJs between endothelial cells (Kruse et al., 2018). When a mural cell adheres to an endothelial cell via N-cadherin, Trio GEF1 becomes activated as measured by its increased binding to nucleotide-free Rac1 and RhoA (Kruse et al., 2018). This increase in activity promotes VE-cadherin recruitment to AJs, thereby strengthening barrier function (Kruse et al., 2018) (Fig. 3).
Trio is also implicated in the process of transendothelial migration (van Rijssel et al., 2012b; Van Rijssel et al., 2013), where bloodstream leukocytes migrate between endothelial cells to enter tissues, and in the formation of muscular tissue (Charrasse et al., 2007). Overall, Trio has a hand in multiple adhesion pathways and plays a clear role in regulating tissue formation, often utilizing its GEF1 activity to do so. The impact of other Trio protein family members on tissue organization is less well understood.
Trio family proteins in secretion and intracellular trafficking
Kalirin was first identified through its association with the neuropeptide processing enzyme peptidylglycine α-amidating monooxygenase (PAM), which is secreted along with neuropeptides in dense core vesicles (DCVs) (Alam et al., 1997). Subsequent work has shown that Trio family proteins play widespread roles in regulating the secretion and trafficking of membrane-bound vesicles, which we review here.
Secretion and endocytosis
Trio and kalirin control both secretion and endocytosis in cells. Trio and kalirin both interact with PAM through their SRs, and co-expression of PAM with kalirin in AtT20 cells, adrenocorticotropic hormone (ACTH)-secreting pituitary tumor cells, increases ACTH secretion (Mains et al., 1999). Overexpression of the Trio or kalirin GEF1 domain alone in AtT20 cells stimulates secretion, and this requires GEF1 catalytic activity (Ferraro et al., 2007), although exactly how GEF1 activity promotes secretion is unknown. Cyclin-dependent kinase 5 (Cdk5) appears to be a key regulator of Trio in controlling secretion. The Cdk5 inhibitor roscovitine reduces active Rac1 levels in HEK293 cells that express both Trio-FL and PAM, and significantly reduces stimulated secretion of ACTH, prolactin, and growth hormone from cultured rat anterior pituitary cells (Xin et al., 2004). Kalirin and Trio are both phosphorylated by Cdk5 in vitro (Xin et al., 2004; Xin et al., 2008), suggesting that Cdk5 phosphorylates Trio to increase Rac1 activation and regulate secretion, although the key regulatory sites and mechanism of this regulation are not known.
Trio family proteins also regulate synaptic vesicle release in neurons. Glutamate release at excitatory synapses is deficient in Trio−/− neurons in vivo (Katrancha et al., 2019). In C. elegans, loss of unc-73 reduces the release of peptide neurotransmitters via DCVs (Hu et al., 2011), and genetic manipulation and rescue experiments indicate that signaling from UNC-73 GEF2 is necessary and sufficient to mediate DCV release and support normal locomotion in this context (Hu et al., 2011). It is not clear whether the vertebrate Trio family members regulate DCV release via a similar GEF2-dependent mechanism. These data suggest that Trio family proteins may act through multiple outputs to coordinate vesicle release. Interestingly, Trio and kalirin also regulate postsynaptic responses (Katrancha et al., 2019; Xie et al., 2007; Herring and Nicoll, 2016), and several studies have implicated Trio and kalirin in endocytosis (Xin et al., 2009; Schiller et al., 2008), indicating possible key functions for Trio family proteins on both sides of the synapse.
Intracellular vesicle trafficking
Trio regulates intracellular vesicle trafficking in cerebellar granule neurons (CGNs). Trio colocalizes with Golgi markers and uses its SRs to interact with Rabin8 (also known as RAB3IP), an activator of the Rab8 and Rab10 GTPases, which are key regulators of Golgi-derived vesicle trafficking (Tao et al., 2019). Loss of Trio in CGNs significantly impairs Rabin8 activity and Golgi-derived vesicle trafficking. Since trafficking of membrane-embedded cargo from Golgi outposts is essential for dendritic arbor development and maintenance (Lin and Koleske, 2010), it is no surprise that this impaired vesicle trafficking also correlates with deficits in neurite extension and reduced neurite length in Trio−/− CGNs (Tao et al., 2019). Indeed, a constitutively active Rab8 mutant rescues these neurite extension defects in Trio−/− CGNs (Tao et al., 2019). This provides an interesting connection between the role of Trio in vesicle trafficking and regulating cell morphology. Overall, it is still unclear whether the interactions between the Trio SRs and Rabin8 are sufficient for regulating trafficking, or if other Trio catalytic activities are required as well.
Disease-associated mutations and rare Trio variants
Disrupted Trio and kalirin function have been connected to human disease, including neurological disorders, cancer and vascular disease. While Trio and kalirin display high sequence similarity, their disease associations differ. Trio has been widely studied for its disease relevance in cancer (Schmidt and Debant, 2014) and neurodevelopmental disorders (NDDs) (Paskus et al., 2020). In contrast, kalirin has been implicated in a few instances of neurodevelopmental disorders (Hill et al., 2006; Kushima et al., 2010), but also in neurodegenerative disorders (Youn et al., 2007) and vascular disease (Wang et al., 2007; Krug et al., 2010; Rudock et al., 2011; Ikram et al., 2009). Interestingly, recent whole-exome sequencing studies have identified Trio, but not kalirin, as having mutations associated with autism spectrum disorder (ASD) (Sadybekov et al., 2017) and schizophrenia (SCZ) (Singh et al., 2020 preprint) (Table S1). Since Trio is more highly expressed during development than kalirin (McPherson et al., 2002; Ma et al., 2005), it is unsurprising that disruptions to Trio, but not kalirin, would be associated with neurodevelopmental disorders. However, the molecular mechanisms driving these changes remain poorly understood.
Changes in expression level or genetic variants in kalirin have been associated with SCZ, Alzheimer's disease (AD) and vascular disease. For instance, mRNA levels of KALRN are significantly decreased in the dorsolateral prefrontal cortex of SCZ patients (Hill et al., 2006). Re-sequencing analyses also revealed a significant association between a kalirin P2255T mutation and SCZ, although the specific effect of this mutation is unknown (Kushima et al., 2010). Kalirin has also been connected to AD pathology, as there are decreased levels of its mRNA in the hippocampus of patients with AD (Youn et al., 2007). Finally, single-nucleotide polymorphisms in kalirin are associated with risk of cardiovascular disease (Rudock et al., 2011), ischemic stroke (Krug et al., 2010; Ikram et al., 2009) and early-onset coronary artery disease (Wang et al., 2007), but the specific effects of the polymorphisms are unknown.
Increases in Trio gene expression and protein levels occur in numerous cancers. The Trio gene resides in a chromosomal region that is commonly amplified in cancer, increasing its gene copy number. Trio mRNA levels are increased in carcinomas of the bladder (Zheng et al., 2004), breast (Lane et al., 2008), liver (Wang et al., 2015), oral cavity (Baldwin et al., 2005; Chattopadhyay et al., 2010), lungs (Coe et al., 2005) and cervix (Hou et al., 2018), in soft tissue sarcomas (Adamowicz et al., 2006) and in glioblastoma (Salhia et al., 2008). Higher Trio expression correlates with poor prognosis in individuals with glioblastoma (Salhia et al., 2008), breast cancer (Lane et al., 2008) and hepatocellular carcinomas (Wang et al., 2015).
Elevation of Trio levels or signaling is associated with cancer progression. High Trio pY2681 levels correlate with poor prognosis in patients with colorectal cancer after surgery, supporting the idea that increased Trio-mediated signaling promotes cancer progression (Sonoshita et al., 2015). Trio promotes cell proliferation by integrating signals from Gαq to the transcription factor activator protein (AP)-1 and promoting DNA synthesis (Vaqué et al., 2013). Indeed, loss of Trio impairs the ability of HeLa cell tumors to grow in vivo (Vaqué et al., 2013). Finally, depletion of Trio in multiple cancer cell lines also reduces invasive cell migration (Salhia et al., 2008; Hou et al., 2018). Thus, Trio likely plays a role in several aspects of cancer progression, from invasive cell migration to cell proliferation.
De novo mutations and ultra-rare damaging variants in Trio are also associated with NDDs (Katrancha et al., 2017; Sadybekov et al., 2017; Barbosa et al., 2020; Singh et al., 2020 preprint; Pengelly et al., 2016). Many of these Trio variants are heterozygous nonsense mutations that reduce Trio protein levels or missense mutations that disrupt Trio function (Katrancha et al., 2017; Sadybekov et al., 2017; Barbosa et al., 2020; Singh et al., 2020 preprint). Behavioral and anatomical phenotypes related to loss of Trio protein, which mimics the nonsense mutations, have been thoroughly described previously (Katrancha et al., 2019). Interestingly, many of the Trio missense mutations associated with these disorders cluster in specific regions of the gene, and many directly impact Trio catalytic function (Fig. 4; Table S1).
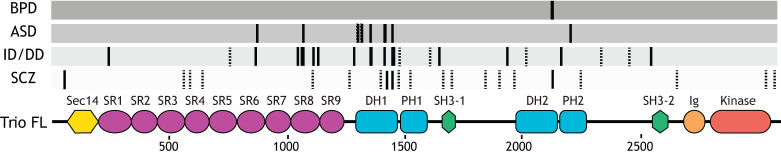
Fig. 4.
Trio family protein mutations and phosphorylation sites. Specific locations of Trio mutations associated with various neurodevelopmental disorders. Interestingly, mutations to Trio cluster in distinct regions of the gene, many clustering around the area encoding the first GEF domain. Solid lines indicate missense mutations; dotted lines indicate nonsense mutations. For a full list of known mutations, see Table S1. BPD, bipolar disorder; ASD, autism spectrum disorder; ID, intellectual disability; DD, developmental delay; SCZ, schizophrenia. Domains of Trio are as in Fig. 1.
Mutations in the Trio GEF1 or adjacent regions are associated with intellectual disability (ID), autism spectrum disorder (ASD), developmental delay (DD), SCZ and microcephaly (Katrancha et al., 2017; Barbosa et al., 2020; Pengelly et al., 2016; Sadybekov et al., 2017) (Fig. 4; Table S1). Many of these mutations disrupt highly conserved residues at the Rac1–Trio DH1 binding interface that impact GEF1 exchange activity (Katrancha et al., 2017; Sadybekov et al., 2017; Pengelly et al., 2016; Barbosa et al., 2020). Interestingly, some mutations increase GEF1 activity, while others impair it (Katrancha et al., 2017; Barbosa et al., 2020; Sadybekov et al., 2017). For instance, the Trio K1431M mutation found in ASD increases GEF1 activity by eight-fold (Katrancha et al., 2017; Sadybekov et al., 2017) and disrupts the ability of Trio to support normal synapse development (Sadybekov et al., 2017). Other variants in the GEF1 domain, like the mild ID- and microcephaly-associated E1299K, R1428Q and H1469K, compromise GEF1 activity (Barbosa et al., 2020). One variant associated with ID, D1368V, which lies in the DH1 domain but outside the Rac1–DH1 interface, instead hyperactivates Rac1 in HEK293 cells (Sadybekov et al., 2017). Thus, functional studies of these mutations suggest that both increased and decreased Rho GTPase signaling mediated by Trio contributes to the pathology of NDDs.
Mutations in other Trio domains, including the SRs and GEF2 domains, are also associated with NDDs (Fig. 4; Table S1). A cluster of mutations in SR8, including T1075I and R1078W/G/Q, are associated with distinct phenotypes in individuals with DD and macrocephaly (Barbosa et al., 2020). These mutations increase Trio GEF1-dependent activation of Rac1 (Barbosa et al., 2020), but it is not known how this gain-of-function allele contributes to macrocephaly. Finally, a single de novo mutation in the GEF2 domain, associated with bipolar disorder (M2145T), increases GEF2 exchange activity four-fold, highlighting the importance of both GEF activities of the Trio protein (Katrancha et al., 2017). Taken together, these findings are clear examples that alterations in Trio catalytic activities lead to distinct NDD phenotypes.
Remaining questions and future challenges
Genetic, biochemical and cell-based studies of Trio family proteins have revealed many important functions for these proteins, cellular contexts in which they act, and their key roles in human disease. However, many questions remain. Some of the biggest unresolved questions center on the catalytic functions of Trio. Why do many Trio isoforms contain two distinct GEF domains, especially when they act on distinct substrates that often have opposing cellular roles? Are the distinct GEF activities coordinated in cells and, if so, how? How do the accessory domains in Trio alter GEF activity, through phosphorylation, interactions with cellular binding partners or autoregulation? Finally, does the Trio kinase domain have substrates and significant signaling outputs in cells? While some of these questions are addressable with current biochemical approaches, some will require advances in single-molecule enzyme assays and/or single molecule live-cell imaging. Addressing these questions will reveal how distinct cell receptors and intracellular binding partners differentially impact Trio catalytic activities and unveil the possible distinct functions of the various isoforms of each gene. With the exception of individual domains, the structures of the entire Trio proteins are largely unknown, in particular with regard to how their domains are arranged in three dimensions, the extent to which domain–domain interactions are regulated and how they are impacted by interactions with other cellular partners. Advances in electron cryo-microscopy and electron cryo-tomography should facilitate the three-dimensional reconstructions of specific Trio proteins and enable the field to study their structure and interactions in their native cellular context.
Overall, Trio family proteins play incredibly diverse roles in cells, and their disruption is associated with cancer progression and neurodevelopmental disorders. The ubiquitous roles of these proteins in biological systems highlights the need to fully understand their exact function, and why disruption of their functions impacts development and yields disease.
Supplementary Material
Acknowledgements
We thank Ellen Corcoran and Amanda Jeng for critical input on the manuscript.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
Work in our laboratory is supported by National Institutes of Health (NIH) grants NS105640, NS11212, MH115939, pilot grants from the Simons Foundation and the Kavli Institute for Neuroscience at Yale, and by a Target Practice Grant from the Stanley Center for Psychiatric Research. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at https://jcs.biologists.org/lookup/doi/10.1242/jcs.248393.supplemental
References
- Adamowicz, M., Radlwimmer, B., Rieker, R. J., Mertens, D., Schwarzbach, M., Schraml, P., Benner, A., Lichter, P., Mechtersheimer, G. and Joos, S. (2006). Frequent amplifications and abundant expression of Trio, NKD2, and IRX2 in soft tissue sarcomas. Genes Chromosomes Cancer 45, 829-838. 10.1002/gcc.20343 [DOI] [PubMed] [Google Scholar]
- Alam, M. R., Johnson, R. C., Darlington, D. N., Hand, T. A., Mains, R. E. and Eipper, B. A. (1997). Kalirin, a cytosolic protein with spectrin-like and GDP/GTP exchange factor-like domains that interacts with peptidylglycine α-amidating monooxygenase, an integral membrane peptide-processing enzyme. J. Biol. Chem. 272, 12667-12675. 10.1074/jbc.272.19.12667 [DOI] [PubMed] [Google Scholar]
- Alexander, M., Selman, G., Seetharaman, A., Chan, K. K. M., D'Souza, S. A., Byrne, A. B. and Roy, P. J. (2010). MADD-2, a homolog of the Opitz syndrome protein MID1, regulates guidance to the midline through UNC-40 in Caenorhabditis elegans. Dev. Cell 18, 961-972. 10.1016/j.devcel.2010.05.016 [DOI] [PubMed] [Google Scholar]
- Awasaki, T., Saito, M., Sone, M., Suzuki, E., Sakai, R., Ito, K. and Hama, C. (2000). The Drosophila trio plays an essential role in patterning of axons by regulating their directional extension. Neuron 26, 119-131. 10.1016/S0896-6273(00)81143-5 [DOI] [PubMed] [Google Scholar]
- Backer, S., Lokmane, L., Landragin, C., Deck, M., Garel, S. and Bloch-Gallego, E. (2018). Trio GEF mediates RhoA activation downstream of Slit2 and coordinates telencephalic wiring. Development 145, dev153692 10.1242/dev.153692 [DOI] [PubMed] [Google Scholar]
- Baldwin, C., Garnis, C., Zhang, L., Rosin, M. P. and Lam, W. L. (2005). Multiple microalterations detected at high frequency in oral cancer. Cancer Res. 65, 7561-7567. 10.1158/0008-5472.CAN-05-1513 [DOI] [PubMed] [Google Scholar]
- Bandekar, S. J., Arang, N., Tully, E. S., Tang, B. A., Barton, B. L., Li, S., Gutkind, J. S. and Tesmer, J. J. G. (2019). Structure of the C-terminal guanine nucleotide exchange factor module of Trio in an autoinhibited conformation reveals its oncogenic potential. Sci. Signal. 12, eaav2449 10.1126/scisignal.aav2449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa, S., Greville-Heygate, S., Bonnet, M., Godwin, A., Fagotto-Kaufmann, C., Kajava, A. V., Laouteouet, D., Mawby, R., Wai, H. A., Dingemans, A. J. M.et al. (2020). Opposite modulation of RAC1 by mutations in TRIO is associated with distinct, domain-specific neurodevelopmental disorders. Am. J. Hum. Genet 106, 338-355. 10.1016/j.ajhg.2020.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman, J., Shu, H. and van Vactor, D. (2000). The guanine nucleotide exchange factor trio mediates axonal development in the Drosophila embryo. Neuron 26, 93-106. 10.1016/S0896-6273(00)81141-1 [DOI] [PubMed] [Google Scholar]
- Bellanger, J.-M., Lazaro, J.-B., Diriong, S., Fernandez, A., Lamb, N. and Debant, A. (1998). The two guanine nucleotide exchange factor domains of Trio link the Rac1 and the RhoA pathways in vivo. Oncogene 16, 147-152. 10.1038/sj.onc.1201532 [DOI] [PubMed] [Google Scholar]
- Bellanger, J.-M., Astier, C., Sardet, C., Ohta, Y., Stossel, T. P. and Debant, A. (2000). The Rac1- and RhoG-specific GEF domain of Trio targets filamin to remodel cytoskeletal actin. Nat. Cell. Biol. 2, 888-892. 10.1038/35046533 [DOI] [PubMed] [Google Scholar]
- Bellanger, J.-M., Estrach, S., Schmidt, S., Briançon-Marjollet, A., Zugasti, O., Fromont, S. and Debant, A. (2003). Different regulation of the Trio Dbl-Homology domains by their associated PH domains. Biol. Cell 95, 625-634. 10.1016/j.biolcel.2003.10.002 [DOI] [PubMed] [Google Scholar]
- Blangy, A., Vignal, E., Schmidt, S., Debant, A., Gauthier-Rouviere, C. and Fort, P. (2000). TrioGEF1 controls Rac- and Cdc42-dependent cell structures through the direct activation of rhoG. J. Cell Sci. 113, 729-739. [DOI] [PubMed] [Google Scholar]
- Blangy, A., Bouquier, N., Gauthier-Rouvière, C., Schmidt, S., Debant, A., Leonetti, J.-P. and Fort, P. (2006). Identification of TRIO-GEFD1 chemical inhibitors using the yeast exchange assay. Biol. Cell 98, 511-522. 10.1042/BC20060023 [DOI] [PubMed] [Google Scholar]
- Bouquier, N., Vignal, E., Charrasse, S., Weill, M., Schmidt, S., Léonetti, J.-P., Blangy, A. and Fort, P. (2009). A cell active chemical GEF inhibitor selectively targets the Trio/RhoG/Rac1 signaling pathway. Chem. Biol. 16, 657-666. 10.1016/j.chembiol.2009.04.012 [DOI] [PubMed] [Google Scholar]
- Briançon-Marjollet, A., Ghogha, A., Nawabi, H., Triki, I., Auziol, C., Fromont, S., Piché, C., Enslen, H., Chebli, K., Cloutier, J.-F.et al. (2008). Trio mediates netrin-1-induced Rac1 activation in axon outgrowth and guidance. Mol. Cell. Biol. 28, 2314-2323. 10.1128/MCB.00998-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrasse, S., Comunale, F., Fortier, M., Portales-Casamar, E., Debant, A. and Gauthier-Rouvière, C. (2007). M-Cadherin Activates Rac1 GTPase through the Rho-GEF Trio during Myoblast Fusion. Mol. Biol. Cell 18, 1734-1743. 10.1091/mbc.e06-08-0766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay, I., Singh, A., Phukan, R., Purkayastha, J., Kataki, A., Mahanta, J., Saxena, S. and Kapur, S. (2010). Genome-wide analysis of chromosomal alterations in patients with esophageal squamous cell carcinoma exposed to tobacco and betel quid from high-risk area in India. Mutat. Res. 696, 130-138. 10.1016/j.mrgentox.2010.01.001 [DOI] [PubMed] [Google Scholar]
- Chen, S.-Y., Huang, P.-H. and Cheng, H.-J. (2011). Disrupted-in-Schizophrenia 1-mediated axon guidance involves TRIO-RAC-PAK small GTPase pathway signaling. Proc. Natl. Acad. Sci. USA 108, 5861-5866. 10.1073/pnas.1018128108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenette, E. J. and Der, C. J. (2011). 5-lipid modification of Ras superfamily GTPases: not just membrane glue. In The Enzymes (ed. Tamanoi F., Hrycyna C. A. and Bergo M. O.), pp. 59-95, Academic Press. [Google Scholar]
- Chhatriwala, M. K., Betts, L., Worthylake, D. K. and Sondek, J. (2007). The DH and PH domains of Trio coordinately engage Rho GTPases for their efficient activation. J. Mol. Biol. 368, 1307-1320. 10.1016/j.jmb.2007.02.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coe, B. P., Henderson, L.-J., Garnis, C., Tsao, M.-S., Gazdar, A. F., Minna, J., Lam, S., Macaulay, C. and Lam, W. L. (2005). High-resolution chromosome arm 5p array CGH analysis of small cell lung carcinoma cell lines. Genes Chromosomes Cancer 42, 308-313. 10.1002/gcc.20137 [DOI] [PubMed] [Google Scholar]
- Debant, A., Serra-Pages, C., Seipel, K., O'brien, S., Tang, M., Park, S. H. and Streuli, M. (1996). The multidomain protein Trio binds the LAR transmembrane tyrosine phosphatase, contains a protein kinase domain, and has separate rac-specific and rho-specific guanine nucleotide exchange factor domains. Proc. Natl. Acad. Sci. USA 93, 5466-5471. 10.1073/pnas.93.11.5466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGeer, J., Boudeau, J., Schmidt, S., Bedford, F., Lamarche-Vane, N. and Debant, A. (2013). Tyrosine phosphorylation of the Rho guanine nucleotide exchange factor Trio regulates netrin-1/DCC-mediated cortical axon outgrowth. Mol. Cell. Biol. 33, 739-751. 10.1128/MCB.01264-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGeer, J., Kaplan, A., Mattar, P., Morabito, M., Stochaj, U., Kennedy, T. E., Debant, A., Cayouette, M., Fournier, A. E. and Lamarche-Vane, N. (2015). Hsc70 chaperone activity underlies Trio GEF function in axon growth and guidance induced by netrin-1. J. Cell Biol. 210, 817-832. 10.1083/jcb.201505084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraro, F., Ma, X.-M., Sobota, J. A., Eipper, B. A. and Mains, R. E. (2007). Kalirin/Trio Rho guanine nucleotide exchange factors regulate a novel step in secretory granule maturation. Mol. Biol. Cell 18, 4813-4825. 10.1091/mbc.e07-05-0503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrester, W. C. and Garriga, G. (1997). Genes necessary for C. elegans cell and growth cone migrations. Development 124, 1831-1843. [DOI] [PubMed] [Google Scholar]
- Forsthoefel, D. J., Liebl, E. C., Kolodziej, P. A. and Seeger, M. A. (2005). The Abelson tyrosine kinase, the Trio GEF and Enabled interact with the Netrin receptor Frazzled in Drosophila. Development 132, 1983-1994. 10.1242/dev.01736 [DOI] [PubMed] [Google Scholar]
- Gauthier-Rouviere, C., Vignal, E., Mériane, M., Roux, P., Montcourier, P. and Fort, P. (1998). RhoG GTPase controls a pathway that independently activates Rac1 and Cdc42Hs. Mol. Biol. Cell 9, 1379-1394. 10.1091/mbc.9.6.1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrique, D. and Schweisguth, F. (2019). Mechanisms of Notch signaling: a simple logic deployed in time and space. Development 146, dev172148 10.1242/dev.172148 [DOI] [PubMed] [Google Scholar]
- Herring, B. E. and Nicoll, R. A. (2016). Kalirin and Trio proteins serve critical roles in excitatory synaptic transmission and LTP. Proc. Natl. Acad. Sci. USA 113, 2264-2269. 10.1073/pnas.1600179113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, J. J., Hashimoto, T. and Lewis, D. A. (2006). Molecular mechanisms contributing to dendritic spine alterations in the prefrontal cortex of subjects with schizophrenia. Mol. Psychiatry 11, 557-566. 10.1038/sj.mp.4001792 [DOI] [PubMed] [Google Scholar]
- Hou, C., Zhuang, Z., Deng, X., Xu, Y., Zhang, P. and Zhu, L. (2018). Knockdown of Trio by CRISPR/Cas9 suppresses migration and invasion of cervical cancer cells. Oncol. Rep. 39, 795-801. 10.3892/or.2017.6117 [DOI] [PubMed] [Google Scholar]
- Hu, S., Pawson, T. and Steven, R. M. (2011). UNC-73/Trio RhoGEF-2 activity modulates Caenorhabditis elegans motility through changes in neurotransmitter signaling upstream of the GSA-1/GαS pathway. Genetics 189, 137-151. 10.1534/genetics.111.131227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikram, M. A., Seshadri, S., Bis, J. C., Fornage, M., Destefano, A. L., Aulchenko, Y. S., Debette, S., Lumley, T., Folsom, A. R., van den Herik, E. G.et al. (2009). Genomewide association studies of stroke. N. Engl. J. Med. 360, 1718-1728. 10.1056/NEJMoa0900094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe, A. B. and Hall, A. (2005). Rho GTPases: biochemistry and biology. Annu. Rev. Cell Dev. Biol. 21, 247-269. 10.1146/annurev.cellbio.21.020604.150721 [DOI] [PubMed] [Google Scholar]
- Katoh, H. and Negishi, M. (2003). RhoG activates Rac1 by direct interaction with the Dock180-binding protein Elmo. Nature 424, 461-464. 10.1038/nature01817 [DOI] [PubMed] [Google Scholar]
- Katrancha, S. M., Wu, Y., Zhu, M., Eipper, B. A., Koleske, A. J. and Mains, R. E. (2017). Neurodevelopmental disease-associated de novo mutations and rare sequence variants affect TRIO GDP/GTP exchange factor activity. Hum. Mol. Genet. 26, 4728-4740. 10.1093/hmg/ddx355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katrancha, S. M., Shaw, J. E., Zhao, A. Y., Myers, S. A., Cocco, A. R., Jeng, A. T., Zhu, M., Pittenger, C., Greer, C. A., Carr, S. A.et al. (2019). Trio haploinsufficiency causes neurodevelopmental disease-associated deficits. Cell Rep. 26, 2805-2817.e9. 10.1016/j.celrep.2019.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai, T., Sanjo, H. and Akiran, S. (1999). Duet is a novel serine/threonine kinase with Dbl-Homology (DH) and Pleckstrin-Homology (PH) domains. Gene 227, 249-255. 10.1016/S0378-1119(98)00605-2 [DOI] [PubMed] [Google Scholar]
- Kiraly, D. D., Stone, K. L., Colangelo, C. M., Abbott, T., Wang, Y., Mains, R. E. and Eipper, B. A. (2011). Identification of kalirin-7 as a potential post-synaptic density signaling hub. J. Proteome Res. 10, 2828-2841. 10.1021/pr200088w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo, T. H., Eipper, B. A. and Donaldson, J. G. (2007). Arf6 recruits the Rac GEF Kalirin to the plasma membrane facilitating Rac activation. BMC Cell Biol. 8, 29 10.1186/1471-2121-8-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroon, J., Heemskerk, N., Kalsbeek, M. J. T., de Waard, V., van Rijssel, J. and van Buul, J. D. (2017). Flow-induced endothelial cell alignment requires the RhoGEF Trio as a scaffold protein to polarize active Rac1 distribution. Mol. Biol. Cell 28, 1745-1753. 10.1091/mbc.e16-06-0389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krug, T., Manso, H., Gouveia, L., Sobral, J., Xavier, J. M., Albergaria, I., Gaspar, G., Correia, M., Viana-Baptista, M., Simões, R. M.et al. (2010). Kalirin: a novel genetic risk factor for ischemic stroke. Hum. Genet. 127, 513-523. 10.1007/s00439-010-0790-y [DOI] [PubMed] [Google Scholar]
- Kruse, K., Lee, Q. S., Sun, Y. J., Klomp, J., Yang, X., Huang, F., Sun, M. Y., Zhao, S., Hong, Z., Vogel, S.et al. (2018). N-cadherin signaling via Trio assembles adherens junctions to restrict endothelial permeability. J. Cell Biol. 218, 299-316. 10.1083/jcb.201802076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubiseski, T. J., Culotti, J. and Pawson, T. (2003). Functional analysis of the Caenorhabditis elegans UNC-73B PH domain demonstrates a role in activation of the Rac GTPase in vitro and axon guidance in vivo. Mol. Cell. Biol. 23, 6823-6835. 10.1128/MCB.23.19.6823-6835.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushima, I., Nakamura, Y., Aleksic, B., Ikeda, M., Ito, Y., Shiino, T., Okochi, T., Fukuo, Y., Ujike, H., Suzuki, M.et al. (2010). Resequencing and association analysis of the KALRN and EPHB1 genes and their contribution to Schizophrenia suscepitibility. Schizophr. Bull. 38, 552-560. 10.1093/schbul/sbq118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane, J., Martin, T. A., Mansel, R. E. and Jiang, W. G. (2008). The expression and prognostic value of the guanine nucleotide exchange factors (GEFs) Trio, Vav1 and TIAM-1 in human breast cancer. Int. Semin. Surg. Oncol. 5, 23 10.1186/1477-7800-5-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., Pu, P. and Le, W. (2013). The SAX-3 receptor stimulates axon outgrowth and the signal sequence and transmembrane domain are critical for SAX-3 membrane localization in the PDE neuron of C. elegans. PLoS ONE 8, e65658 10.1371/journal.pone.0065658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, Y., Niederstrasser, H., Edwards, M., Jackson, C. E. and Cooper, J. A. (2009). Distinct roles for CARMIL isoforms in cell migration. Mol. Biol. Cell 20, 5037-5313. 10.1091/mbc.e08-10-1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, Y.-C. and Koleske, A. J. (2010). Mechanisms of synapse and dendrite maintenance and their disruption in psychiatric and neurodegenerative disorders. Annu. Rev. Neurosci. 33, 349-378. 10.1146/annurev-neuro-060909-153204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X., Wang, H., Eberstadt, M., Schnuchel, A., Olejniczak, E., Meadows, R. P., Schkeryantz, J. M., Janowick, D. A., Harlan, J. E., Harris, E. A. S.et al. (1999). NMR structure and mutagenesis of the N-terminal Dbl homology domain of the nucleotide exchange factor trio. Cell 95, 269-277. 10.1016/S0092-8674(00)81757-2 [DOI] [PubMed] [Google Scholar]
- Lundquist, E. A., Reddien, P. W., Hartwieg, E., Horvitz, H. R. and Bargmann, C. I. (2001). Three C. elegans Rac proteins and several alternative Rac regulators control axon guidance, cell migration, and apoptotic cell phagocytosis. Development 128, 4475-4488. [DOI] [PubMed] [Google Scholar]
- Ma, X.-M., Huang, J.-P., Eipper, B. A. and Mains, R. E. (2005). Expression of Trio, a member of the Dbl family of Rho GEFs in the developing rat brain. J. Comp. Neurol. 482, 333-348. 10.1002/cne.20404 [DOI] [PubMed] [Google Scholar]
- Ma, X.-M., Miller, M. B., Vishwanatha, K. S., Gross, M. J., Wang, Y., Abbott, T., Lam, T. K. T., Mains, R. E. and Eipper, B. A. (2014). Nonenzymatic domains of Kalirin7 contribute to spine morphogenesis through interactions with phosphoinositides and Abl. Mol. Biol. Cell 25, 1458-1471. 10.1091/mbc.e13-04-0215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, X. M., Johnson, R. C., Mains, R. E. and Eipper, B. A. (2001). Expression of kalirin, a neuronal GDP/GTP exchange factor of the trio family, in the central nervous system of the adult rat. J. Comp. Neurol. 429, 388-402. [DOI] [PubMed] [Google Scholar]
- Ma, X. M., Kiraly, D. D., Gaier, E. D., Wang, Y., Kim, E. J., Levine, E. S., Eipper, B. A. and Mains, R. E. (2008). Kalirin-7 is required for synaptic structure and function. J. Neurosci. 28, 12368-12382. 10.1523/JNEUROSCI.4269-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mains, R. E., Alam, M. R., Johnson, R. C., Darlington, D. N., Bäck, N., Hand, T. A. and Eipper, B. A. (1999). Kalirin, a multifunctional PAM COOH-terminal domain interactor protein, affects cytoskeletal organization and ACTH secretion from AtT-20 cells. J. Biol. Chem. 274, 2929-2937. 10.1074/jbc.274.5.2929 [DOI] [PubMed] [Google Scholar]
- May, V., Schiller, M. R., Eipper, B. A. and Mains, R. E. (2002). Kalirin Dbl-homology guanine nucleotide exchange factor 1 domain initiates new axon outgrowths via RhoG-mediated mechanisms. J. Neurosci. 22, 6980-6990. 10.1523/JNEUROSCI.22-16-06980.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson, C. E., Eipper, B. A. and Mains, R. E. (2002). Genomic organization and differential expression of Kalirin isoforms. Gene 284, 41-51. 10.1016/S0378-1119(02)00386-4 [DOI] [PubMed] [Google Scholar]
- McPherson, C. E., Eipper, B. A. and Mains, R. E. (2005). Multiple novel isoforms of Trio are expressed in the developing rat brain. Gene 347, 125-135. 10.1016/j.gene.2004.12.028 [DOI] [PubMed] [Google Scholar]
- Medley, Q. G., Buchbinder, E. G., Tachibana, K., Ngo, H., Serra-Pagès, C. and Streuli, M. (2003). Signaling between focal adhesion kinase and trio. J. Biol. Chem. 278, 13265-13270. 10.1074/jbc.M300277200 [DOI] [PubMed] [Google Scholar]
- Miller, M. B., Yan, Y., Eipper, B. A. and Mains, R. E. (2013). Neuronal Rho GEFs in synaptic physiology and behavior. Neuroscientist 19, 255-273. 10.1177/1073858413475486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, M. B., Vishwanatha, K. S., Mains, R. E. and Eipper, B. A. (2015). An N-terminal amphipathic helix binds phosphoinositides and enhances Kalirin Sec14 domain-mediated membrane interactions. J. Biol. Chem. 290, 13541-13555. 10.1074/jbc.M115.636746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, M. B., Yan, Y., Machida, K., Kiraly, D. D., Levy, A. D., Wu, Y. I., Lam, T. K. T., Abbott, T., Koleske, A. J., Eipper, B. A.et al. (2017). Brain region and isoform-specific phosphorylation alters Kalirin SH2 domain interaction sites and calpain sensitivity. ACS Chem. Neurosci. 8, 1554-1569. 10.1021/acschemneuro.7b00076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newsome, T. P., Schmidt, S., Dietzl, G., Keleman, K., Åsling, B., Debant, A. and Dickson, B. J. (2000). Trio combines with dock to regulate Pak activity during photoreceptor axon pathfinding in Drosophila. Cell 101, 283-294. 10.1016/S0092-8674(00)80838-7 [DOI] [PubMed] [Google Scholar]
- Paskus, J. D., Tian, C., Fingleton, E., Shen, C., Chen, X., Li, Y., Myers, S. A., Badger, J. D., Bemben, M. A., Herring, B. E.et al. (2019). Synaptic Kalirin-7 and Trio interactomes reveal a GEF protein-dependent neuroligin-1 mechanism of action. Cell Rep. 29, 2944-2952.e5. 10.1016/j.celrep.2019.10.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paskus, J. D., Herring, B. E. and Roche, K. W. (2020). Kalirin and Trio: RhoGEFs in synaptic transmission, plasticity, and complex brain disorders. Trends Neurosci. 43, 505-518. 10.1016/j.tins.2020.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, Y.-J., He, W.-Q., Tang, J., Tao, T., Chen, C., Gao, Y.-Q., Zhang, W.-C., He, X.-Y., Dai, Y.-Y., Zhu, N.-C.et al. (2010). Trio is a key guanine nucleotide exchange factor coordinating regulation of the migration and morphogenesis of granule cells in the developing cerebellum. J. Biol. Chem. 285, 24834-24844. 10.1074/jbc.M109.096537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pengelly, R. J., Greville-Heygate, S., Schmidt, S., Seaby, E. G., Jabalameli, M. R., Mehta, S. G., Parker, M. J., Goudie, D., Fagotto-Kaufmann, C., Mercer, C.et al. (2016). Mutations specific to the Rac-GEF domain of TRIO cause intellectual disability and microcephaly. J. Med. Genet. 53, 735-742. 10.1136/jmedgenet-2016-103942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penzes, P., Johnson, R. C., Alam, M. R., Kambampati, V., Mains, R. E. and Eipper, B. A. (2000). An isoform of kalirin, a brain-specific GDP/GTP exchange factor, is enriched in the postsynaptic density fraction. J. Biol. Chem. 275, 6395-6403. 10.1074/jbc.275.9.6395 [DOI] [PubMed] [Google Scholar]
- Penzes, P., Johnson, R. C., Kambampati, V., Mains, R. E. and Eipper, B. A. (2001). Distinct roles for the two Rho GDP/GTP exchange factor domains of kalirin in regulation of neurite growth and neuronal morphology. J. Neurosci. 21, 8426-8434. 10.1523/JNEUROSCI.21-21-08426.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penzes, P., Beeser, A., Chernoff, J., Schiller, M. R., Eipper, B. A., Mains, R. E. and Huganir, R. L. (2003). Rapid induction of dendritic spine morphogenesis by trans-synaptic ephrinB-EphB receptor activation of the Rho-GEF kalirin. Neuron 37, 263-274. 10.1016/S0896-6273(02)01168-6 [DOI] [PubMed] [Google Scholar]
- Peurois, F., Veyron, S., Ferrandez, Y., Ladid, I., Benabdi, S., Zeghouf, M., Peyroche, G. and Cherfils, J. (2017). Characterization of the activation of small GTPases by their GEFs on membranes using artificial membrane tethering. Biochem. J. 474, 1259-1272. 10.1042/BCJ20170015 [DOI] [PubMed] [Google Scholar]
- Polacheck, W. J., Kutys, M. L., Yang, J., Eyckmans, J., Wu, Y., Vasavada, H., Hirschi, K. K. and Chen, C. S. (2017). A non-canonical Notch complex regulates adherens junctions and vascular barrier function. Nature 552, 258-262. 10.1038/nature24998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabiner, C. A., Mains, R. E. and Eipper, B. A. (2005). Kalirin: a dual Rho guanine nucleotide exchange factor that is so much more than the sum of its many parts. Neuroscientist 11, 148-160. 10.1177/1073858404271250 [DOI] [PubMed] [Google Scholar]
- Ridley, A. (2001). Rho GTPases and cell migration. J. Cell Sci. 114, 2713-2722. [DOI] [PubMed] [Google Scholar]
- Rudock, M. E., Cox, A. J., Ziegler, J. T., Lehtinen, A. B., Connelly, J. J., Freedman, B. I., Carr, J. J., Lengefeld, C. D., Hauser, E. R., Horne, B. D.et al. (2011). Cigarette smoking status has a modifying effect on the association between polymorphisms in KALRN and measures of cardiovascular risk in the diabetes heart study. Genes Genomics 33, 483 10.1007/s13258-011-0069-2 [DOI] [Google Scholar]
- Sadybekov, A., Tian, C., Arnesano, C., Katritch, V. and Herring, B. E. (2017). An autism spectrum disorder-related de novo mutation hotspot discovered in the GEF1 domain of Trio. Nat. Commun. 8, 601 10.1038/s41467-017-00472-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salhia, B., Tran, N. L., Chan, A., Wolf, A., Nakada, M., Rutka, F., Ennis, M., McDonough, W. S., Berens, M. E., Symons, M.et al. (2008). The guanine nucleotide exchange factors Trio, Ect2, and Vav3 mediate the invasive behavior of glioblastoma. Am. J. Pathol. 173, 1828-1838. 10.2353/ajpath.2008.080043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander, E. E., ten Klooster, J. P., van Delft, S., van der Kammen, R. A. and Collard, J. G. (1999). Rac downregulates Rho activity: reciprocal balance between both GTPases determines cellular morphology and migratory behavior. J. Cell Biol. 147, 1009-1022. 10.1083/jcb.147.5.1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller, M. R., Blangy, A., Huang, J., Mains, R. E. and Eipper, B. A. (2005). Induction of lamellipodia by Kalirin does not require its guanine nucleotide exchange factor activity. Exp. Cell Res. 307, 402-417. 10.1016/j.yexcr.2005.03.024 [DOI] [PubMed] [Google Scholar]
- Schiller, M. R., Chakrabarti, K., King, G. F., Schiller, N. I., Eipper, B. A. and Maciejewski, M. W. (2006). Regulation of RhoGEF activity by intramolecular and intermolecular SH3 domain interactions. J. Biol. Chem. 281, 18774-18786. 10.1074/jbc.M512482200 [DOI] [PubMed] [Google Scholar]
- Schiller, M. R., Ferraro, F., Wang, Y., Ma, X.-M., Mcpherson, C. E., Sobota, J. A., Schiller, N. I., Mains, R. E. and Eipper, B. A. (2008). Autonomous functions for the Sec14p/spectrin-repeat region of Kalirin. Exp. Cell Res. 314, 2674-2691. 10.1016/j.yexcr.2008.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, S. and Debant, A. (2014). Function and regulation of the Rho guanine nucleotide exchange factor Trio. Small GTPases 5, e29769 10.4161/sgtp.29769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seipel, K., Medley, Q. G., Kedersha, N. L., Zhang, X. A., O'brien, S. P., Serra-Pages, C., Hemler, M. E. and Streuli, M. (1999). Trio amino-terminal guanine nucleotide exchange factor domain expression promotes actin cytoskeleton reorganization, cell migration, and anchorage-independent cell growth. J. Cell Sci. 112, 1825-1834. [DOI] [PubMed] [Google Scholar]
- Seipel, K., O'brien, S. P., Iannotti, E., Medley, Q. G. and Streuli, M. (2000). Tara, a novel F-actin binding protein, associates with the Trio guanine nucleotide exchange factor and regulates actin cytoskeletal organization. J. Cell Sci. 114, 389-399. [DOI] [PubMed] [Google Scholar]
- Singh, T., Poterba, T., Curtis, D., Akil, H., AL Eissa, M., Barchas, J. D., Bass, N., Bigdeli, T. B., Breen, G., Daly, M. J. and et al. (2020). Exome sequencing identifies rare coding variants in 10 genes which confer substantial risk for schizophrenia. medRxiv, 2020.09.18.20192815 10.1101/2020.09.18.20192815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowronek, K. R., Guo, F., Zheng, Y. and Nassar, N. (2004). The C-terminal basic tail of RhoG assists the guanine nucleotide exchange factor trio in binding to phospholipids. J. Biol. Chem. 279, 37895-37907. 10.1074/jbc.M312677200 [DOI] [PubMed] [Google Scholar]
- Son, K., Smith, T. C. and Luna, E. J. (2015). Supervillin binds the Rac/Rho-GEF Trio and increases Trio-mediated Rac1 activation. Cytoskeleton 72, 47-64. 10.1002/cm.21210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoshita, M., Itatani, Y., Kakizaki, F., Sakimura, K., Terashima, T., Katsuyama, Y., Sakai, Y. and Taketo, M. M. (2015). Promotion of colorectal cancer invasion and metastasis through activation of NOTCH-DAB1-ABL-RHOGEF protein TRIO. Cancer Discov. 5, 198-211. 10.1158/2159-8290.CD-14-0595 [DOI] [PubMed] [Google Scholar]
- Spencer, A. G., Orita, S., Malone, C. J. and Han, M. (2001). A RHO GTPase-mediated pathway is required during P cell migration in Caenorhabditis elegans. Pnas 98, 13132-13137. 10.1073/pnas.241504098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steven, R., Kubiseski, T. J., Zheng, H., Kulkarni, S., Mancillas, J., Morales, A. R., Hogue, C. W. V., Pawson, T. and Culotti, J. (1998). UNC-73 activates the Rac GTPase and is required for cell and growth cone migrations in C. elegans. Cell 92, 785-795. 10.1016/S0092-8674(00)81406-3 [DOI] [PubMed] [Google Scholar]
- Sun, Y.-J., Nishikawa, K., Yuda, H., Wang, Y.-L., Osaka, H., Fukazawa, N., Naito, A., Kudo, Y., Wada, K. and Aoki, S. (2006). Solo/Trio8, a membrane-associated short isoform of Trio, modulates endosome dynamics and neurite elongation. Mol. Cell. Biol. 26, 6923-6935. 10.1128/MCB.02474-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao, T., Sun, J., Peng, Y., Li, Y., Wang, P., Chen, X., Zhao, W., Zheng, Y.-Y., Wei, L., Wang, W.et al. (2019). Golgi-resident TRIO regulates membrane trafficking during neurite outgrowth. J. Biol. Chem. 294, 10954-10968. 10.1074/jbc.RA118.007318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmerman, I., Heemskerk, N., Kroon, J., Schaefer, A., van Rijssel, J., Hoogenboezem, M., van Unen, J., Goedhart, J., Gadella, T. W. J., Yin, T.et al. (2015). A local VE-cadherin and Trio-based signaling complex stabilizes endothelial junctions through Rac1. J. Cell Sci. 128, 3041-3054. 10.1242/jcs.168674 [DOI] [PubMed] [Google Scholar]
- Valdivia, A., Goicoechea, S. M., Awadia, S., Zinn, A. and Garcia-Mata, R. (2017). Regulation of circular dorsal ruffles, macropinocytosis, and cell migration by RhoG and its exchange factor, Trio. Mol. Biol. Cell 28, 1768-1781. 10.1091/mbc.e16-06-0412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Haren, J., Boudeau, J., Schmidt, S., Basu, S., Liu, Z., Lammers, D., Demmers, J., Benhari, J., Grosveld, F., Debant, A.et al. (2014). Dynamic microtubules catalyze formation of navigator-TRIO complexes to regulate neurite extension. Curr. Biol. 24, 1778-1785. 10.1016/j.cub.2014.06.037 [DOI] [PubMed] [Google Scholar]
- van Leeuwen, F. N., Kain, H. E. T., van der Kammen, R. A., Michiels, F., Kranenburg, O. W. and Collard, J. G. (1997). The guanine nucleotide exchange factor Tiam1 affects neuronal morphology; opposing roles for the small GTPases Rac and Rho. J. Cell Biol. 139, 797-807. 10.1083/jcb.139.3.797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rijssel, J. and van Buul, J. D. (2012). The many faces of the guanine-nucleotide exchange factor trio. Cell Adh. Migr. 6, 482-487. 10.4161/cam.21418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rijssel, J., Hoogenboezem, M., Wester, L., Hordijk, P. L. and van Buul, J. D. (2012a). The N-terminal DH-PH domain of Trio induces cell spreading and migration by regulating lamellipodia dynamics in a Rac1-dependent fashion. PLoS ONE 7, e29912 10.1371/journal.pone.0029912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rijssel, J., Kroon, J., Hoogenboezem, M., van Alphen, F. P. J., de Jong, R. J., Kostadinova, E., Geerts, D., Hordijk, P. L. and van Buul, J. D. (2012b). The Rho-guanine nucleotide exchange factor Trio controls leukocyte transendothelial migration by promoting docking structure formation. Mol. Biol. Cell 23, 2831-2844. 10.1091/mbc.e11-11-0907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rijssel, J., Timmerman, I., van Alphen, F. P., Hoogenboezem, M., Korchynskyi, O., Geerts, D., Geissler, J., Reedquist, K. A., Niessen, H. W. and van Buul, J. D. (2013). The Rho-GEF Trio regulates a novel pro-inflammatory pathway through the transcription factor Ets2. Biol. Open 2, 569-579. 10.1242/bio.20134382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderzalm, P. J., Pandey, A., Hurwitz, M. E., Bloom, L., Horvitz, H. R. and Garriga, G. (2009). C. elegans CARMIL negatively regulates UNC-73/Trio function during neuronal development. Development 136, 1201-1210. 10.1242/dev.026666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaqué, J. P., Dorsam, R. T., Feng, X., Iglesias-Bartolome, R., Forsthoefel, D. J., Chen, Q., Debant, A., Seeger, M. A., Ksander, B. R., Teramoto, H.et al. (2013). A Genome-wide RNAi screen reveals a Trio-regulated Rho GTPase circuitry transducing mitogenic signals initiated by G protein-coupled receptors. Mol. Cell 49, 94-108. 10.1016/j.molcel.2012.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vestweber, D. (2008). VE-cadherin the major endothelial adhesion molecule controlling cellular junctions and blood vessel formation. Arterioscler. Thromb. Vasc. Biol. 28, 223-232. 10.1161/ATVBAHA.107.158014 [DOI] [PubMed] [Google Scholar]
- Walck-Shannon, E., Reiner, D. and Hardin, J. (2015). Polarized Rac-dependent protrusions drive epithelial intercalation in the embryonic epidermis of C. elegans. Development 142, 3549-3560. 10.1242/dev.127597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L., Hauser, E. R., Shah, S. H., Pericak-Vance, M. A., Haynes, C., Crosslin, D., Harris, M., Nelson, S., Hale, A. B., Granger, C. B.et al. (2007). Peakwide mapping on chromosome 3q13 identifies kalirin gene as a novel candidate gene for coronary artery disease. Am. J. Hum. Genet. 80, 650-663. 10.1086/512981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, B., Fang, J. Q., Qu, L., Cao, Z., Zhou, J. G. and Deng, B. (2015). Upregulated TRIO expression correlates with a malignant phenotype in human hepatocellular carcinoma. Tumour Biol. 36, 6901-6908. 10.1007/s13277-015-3377-3 [DOI] [PubMed] [Google Scholar]
- Watari-Goshima, N., Ogura, K.-I., Wolf, F. W., Goshima, Y. and Garriga, G. (2007). C. elegans VAB-8 and UNC-73 regulate the SAX-3 receptor to direct cell and growth-cone migrations. Nat. Neuro 10, 169-176. 10.1038/nn1834 [DOI] [PubMed] [Google Scholar]
- Wu, Y.-C., Cheng, T.-W., Lee, M.-C. and Weng, N.-Y. (2002). Distinct Rac activation pathways control Caenorhabditis elegans cell migration and axon outgrowth. Dev. Biol 250, 145-155. 10.1006/dbio.2002.0785 [DOI] [PubMed] [Google Scholar]
- Wu, J.-H., Fanaroff, A. C., Sharma, K. C., Smith, L. S., Brian, L., Eipper, B. A., Mains, R. E., Freedman, N. J. and Zhang, L. (2013). Kalirin promotes neointimal hyperplasia by activating Rac in smooth muscle cells. Arterioscler. Thromb. Vasc. Biol. 33, 702-708. 10.1161/ATVBAHA.112.300234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Z., Srivastava, D. P., Photowala, H., Kai, L., Cahill, M. E., Woolfrey, K. M., Shum, C. Y., Surmeier, D. J. and Penzes, P. (2007). Kalirin-7 controls activity-dependent structural and functional plasticity of dendritic spines. Neuron 56, 640-656. 10.1016/j.neuron.2007.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin, X., Ferraro, F., Back, N., Eipper, B. A. and Mains, R. E. (2004). Cdk5 and Trio modulate endocrine cell exocytosis. J. Cell Sci. 117, 4739-4748. 10.1242/jcs.01333 [DOI] [PubMed] [Google Scholar]
- Xin, X., Wang, Y., Ma, X.-M., Rompolas, P., Keutmann, H. T., Mains, R. E. and Eipper, B. A. (2008). Regulation of Kalirin by Cdk5. J. Cell Sci. 121, 2601-2611. 10.1242/jcs.016089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin, X., Rabiner, C. A., Mains, R. E. and Eipper, B. A. (2009). Kalirin12 interacts with dynamin. BMC Neurosci. 10, 61 10.1186/1471-2202-10-61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, Y., Eipper, B. A. and Mains, R. E. (2014). Kalirin-9 and Kalirin-12 play essential roles in dendritic outgrowth and branching. Cereb. Cortex 25, 3487-3501. 10.1093/cercor/bhu182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, Y., Eipper, B. A. and Mains, R. E. (2016). Kalirin is required for BDNF-TrkB stimulated neurite outgrowth and branching. Neuropharmacology 107, 227-238. 10.1016/j.neuropharm.2016.03.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano, T., Yamazaki, Y., Adachi, M., Okawa, K., Fort, P., Uji, M., Tsukita, S. and Tsukita, S. (2011). Tara up-regulates E-cadherin transcription by binding to the Trio RhoGEF and inhibiting Rac signaling. J. Cell Biol. 193, 319-332. 10.1083/jcb.201009100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn, H., Jeoung, M., Koo, Y., Ji, H., Markesbery, W. R., Ji, I. and Ji, T. H. (2007). Kalirin is Under-Expressed in Alzheimer's Disease Hippocampus. J. Alzheimers Dis. 11, 385-397. 10.3233/JAD-2007-11314 [DOI] [PubMed] [Google Scholar]
- Zheng, M., Simon, R., Mirlacher, M., Maurer, R., Gasser, T., Forster, T., Diener, P. A., Mihatsch, M. J., Sauter, G. and Schraml, P. (2004). TRIO amplification and abundant mRNA expression is associated with invasive tumor growth and rapid tumor cell proliferation in urinary bladder cancer. Am. J. Pahol. 165, 63-69. 10.1016/S0002-9440(10)63275-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipkin, I. D., Kindt, R. M. and Kenyon, C. J. (1997). Role of a new rho family member in cell migration and axon guidance in C. elegans. Cell 90, 883-894. 10.1016/S0092-8674(00)80353-0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.