3D microscopy and deep learning classify adipose tissue immune microenvironments associated with obesity comorbidities.
Abstract
Crown-like structures (CLSs) are adipose microenvironments of macrophages engulfing adipocytes. Their histological density in visceral adipose tissue (VAT) predicts metabolic disorder progression in obesity and is believed to initiate obesity comorbidities. Here, we use three-dimensional (3D) light sheet microscopy and deep learning to quantify 3D features of VAT CLSs in lean and obese states. Obese CLS densities are significantly higher, composing 3.9% of tissue volume compared with 0.46% in lean tissue. Across the states, individual CLS structural characteristics span similar ranges; however, subpopulations are distinguishable. Obese VAT contains large CLSs absent from lean tissues, located near the tissue center, while lean CLSs have higher volumetric cell densities and prolate shapes. These features are consistent with inefficient adipocyte elimination in obesity that contributes to chronic inflammation, representing histological biomarkers to assess adipose pathogenesis. This tissue processing, imaging, and analysis pipeline can be applied to quantitatively classify 3D microenvironments across diverse tissues.
INTRODUCTION
Microenvironments are microscopic tissue domains composed of diverse cell types, extracellular matrix, and extracellular biochemicals that together are responsible for the emergent physiological functions of tissues (1, 2). Concerted efforts are being devoted to the classification and analysis of microenvironments of macrophages, cells that derive from myeloid lineage white blood cells and exhibit extreme levels of phenotypic and functional heterogeneity (3, 4). Macrophages play important roles in host defense by eliminating dead cells, debris, and pathogens by phagocytosis; modulating inflammatory responses by secreting cytokines, enzymes, and other factors; and triggering adaptive immunity by presenting antigens to lymphocytes (5). In some tissues, macrophages make up a major fraction of total cells and play a central role in development, homeostasis, wound repair, and disease progression (6, 7). Macrophage dysfunction is now recognized to elicit a wide range of pathologies, including osteoarthritis, chronic inflammatory bowel disorders, and obesity-driven metabolic disorder (8).
A macrophage microenvironment of high clinical and biological importance is the crown-like structure (CLS) in adipose tissue, in which macrophages encircle dead or dying adipocytes (9). It is believed that CLS microenvironments are necessary for the extreme capacity of adipose tissue for growth and pruning, an unusual feature for adult tissues that allows dynamic adaptation to changes in host energy consumption and dense energy storage (10). A high number of CLSs measured by two-dimensional (2D) imaging of tissue biopsies is associated with worse outcomes of obesity and related metabolic disorders for CLSs in visceral adipose tissue (VAT) (9, 11). In several studies, CLS presence in human adipose tissue has been found to correlate with hyperinsulinemia, insulin resistance, and vascular dysfunction, sometimes with stronger correlation than clinical obesity parameters such as body mass index or waist circumference (12, 13). Similarly, high CLS numbers in adipose tissues of the breast and prostate are associated with worse clinical outcomes of breast and prostate cancers, respectively (14). Because macrophage biology in VAT is typically evaluated by analytical methods that disrupt tissue (e.g., flow cytometry and single-cell sequencing), there is a knowledge gap regarding how CLS structures are organized and function in their native tissue locations, features that may further contribute to histological scoring for prediction of metabolic or other physiological conditions.
Recently, clearing techniques have been developed that render tissue optically transparent by eliminating refractive lipids and replacing aqueous solvents with refractive index–matched solutions (15). Current methods are classified on the basis of their use of organic solvents (16, 17), aqueous solvents (18), or hydrogel embedment (19). The concurrent advancement of light sheet microscopy, in which thick tissues can be imaged intact at or near the optical diffraction limit, has led to a new tool set for comprehensive imaging of intact tissues at subcellular resolution, which is currently revolutionizing our understanding of tissue microenvironments (20). The resulting 3D images comprise large datasets, which has prompted demand for advanced data analysis pipelines (21–23). Deep learning approaches using convolutional neural networks (CNNs) have been particularly successful in computer vision applications such as object detection and image segmentation because of the ability to learn spatial hierarchies of features automatically through backpropagation (24, 25). Most strategies focus on 2D images such as chromophore-labeled histopathological images (26) or objects with homogeneous labels (e.g., pulmonary nodules in computed tomography) (27). CLS structures are challenging objects to recognize within a 3D fluorescence image, comprising a low-contrast spot (an adipocyte) surrounded by a high-contrast concentric shell (labeled macrophages) (28), which may be discontinuous and interconnected across multiple CLSs, with diverse sizes and shapes. Moreover, adipose tissue is constantly reorganizing on the basis of the metabolic demands and energy availability of the host, with few anatomical markers of orientation and with high diversity across individuals, even in healthy adults. Therefore, a robust, automated pipeline could benefit CLS analysis and help contextualize the role of CLSs in adipose tissue structure and function.
Here, we developed a pipeline for 3D identification and comprehensive analysis of macrophage microenvironments in CLSs in intact adipose tissue through a combination of optical tissue clearing, 3D light sheet microscopy, deep learning–based segmentation, and multivariate analysis. We used C57BL/6 mice fed either a high-fat diet or a low-fat diet and comprehensively imaged and evaluated perirenal VAT depots. Tissues were fluorescently labeled and cleared using the immunolabeling-enhanced 3D imaging of solvent-cleared organs (iDISCO) method (29), and light sheet microscopy was used to collect 3D datasets. Adapted Mask R-CNNs (30) were used to automate image segmentation and data analysis. Multivariate evaluation of the structures and sizes revealed a new class of CLSs in obese tissue that are unusually large, and comparison by cell numbers and volume reveals that obese CLSs are underpopulated with cells. Lean and obese CLSs with equal volume are also globally distinct, with lean CLSs having higher density of cells and higher sphericity in shape. These features are consistent with less efficient elimination of dying adipocytes in the obese state, which can contribute to low-grade and chronic inflammation in adipose tissue that is associated with the numerous comorbidities of obesity.
RESULTS
Obese adipose tissue clearing and labeling
C57BL/6 mice (25 weeks old) fed a high-fat diet (60% kcal from fat) or low-fat diet (10% kcal from fat) were physiologically obese or lean, respectively. The composition of the chow in the lean group was typical of that used commonly in experimental mouse models. Mean animal weights were 45.9 and 33.1 g with respective total VAT weights of 2.56 and 0.67 g after sacrifice in the obese and lean groups, respectively. Perirenal VATs in the retroperitoneal space surrounding the kidneys with weights near 0.44 g in the obese state and 0.08 g in the lean state were collected for clearing. Perirenal fat was chosen because it is active in metabolism and adipokine secretion and is present in both mouse models and humans (31). We evaluated four clearing methods: Clear Lipid-exchanged Acrylamide-hybridized Rigid Imaging-compatible Tissue hYdrogel (CLARITY) (19), ClearT (32), Benzyl Alcohol-Benzyl Benzoate (BABB) (33), and iDISCO (16). Figure 1A shows punched adipose tissues with diameter of 5 mm and thickness of 2 to 3 mm used to evaluate the capacity of each technique to render VAT transparent. The organic solvent–based approaches BABB and iDISCO yielded transparent tissues unlike the hydrogel-based CLARITY method and aqueous ClearT method. The very high content of hydrophobic triglycerides in adipose tissue (34) could only be fully dissolved and eliminated with organic solvents such as dichloromethane and tetrahydrofuran. Because of the compatibility of iDISCO with immunolabeling, we used iDISCO to clear intact perirenal adipose tissue. Figure 1B shows a representative tissue from an obese mouse with a size near 2.7 cm by 2.1 cm by 0.6 cm, yielding a highly transparent material suitable for 3D light sheet microscopy.
Fig. 1. Adipose tissue clearing, labeling, and fluorescence microscopy for visualization of macrophage and CLS features.
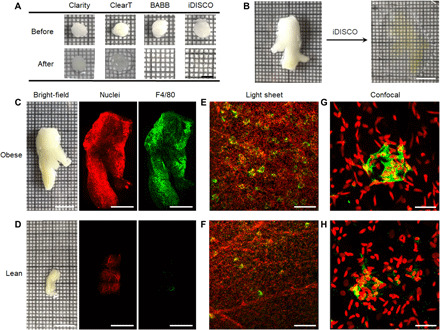
(A) Bright-field images of adipose tissue punches before and after treatment with four clearing techniques. All images share the same scale bar of 5 mm. (B) Bright-field images of intact perirenal adipose tissue before and after iDISCO clearing. Both images share the same scale bar of 1 cm. (C and D) Bright-field images of freshly dissected perirenal adipose tissue before clearing from both obese and lean mice and corresponding low-magnification light sheet microscopy images after iDISCO clearing. Tissues are stained with YO-PRO-1 nuclear dye (red) and a fluorescently labeled antibody against the macrophage F4/80 antigen (green). Scale bars, 10 mm for bright-field images and 5 mm for fluorescence images. (E and F) Zoomed light sheet microscopy images and (G and H) confocal fluorescence images for obese and lean tissues. Scale bars, 200 μm (E and F) and 20 μm (G and H). All fluorescence images are 3D projection images.
We immunolabeled macrophages using an antibody against the pan-macrophage membrane marker F4/80. In addition, we labeled cell nuclei with YO-PRO-1 to identify tissue contours and cell nucleus locations. The cleared and labeled adipose tissues were imaged with both light sheet fluorescence microscopy and confocal fluorescence microscopy. Cell nuclei [red in Fig. 1 (C to H)] and macrophage labels [green in Fig. 1 (C to H)] could be readily detected above background autofluorescence using both microscopy modalities. With F4/80 staining, CLSs are visibly recognizable in 3D as shell structures (Fig. 1, E and F, and movies S1 and S2) and in 2D slices as ring structures (see below). In addition, the complex and nonspherical macrophage orientation and organization within CLSs are apparent with zoomed light sheet microscopy images (Fig. 1, E and F) and confocal microscopy images (Fig. 1, G and H). It is apparent from visual inspection of equal tissue volumes that adipose tissues have more CLSs in obese mice (movie S1) compared with lean mice (movie S2), which is consistent with known correlations in 2D slices (35), and that many CLSs in adipose tissues from obese mice are interconnected (movie S3). Single-cell resolution could be easily achieved with light sheet microscopy at 2.5× magnification (see Materials and Methods), providing 3D images of macrophages within CLSs and macrophages excluded from CLSs.
Automated CLS detection through deep learning
A typical 2.5× perirenal adipose tissue generates a dataset with size near 70 gigabytes for obese mice and 30 gigabytes for lean mice. To identify CLS microenvironments in 3D, we developed an approach based on Mask R-CNN, a deep learning network for simultaneous object detection and instance segmentation (30). Mask R-CNN follows a two-stage recognition pipeline (Fig. 2A). In the first stage, image features are extracted through the convolutional backbone ResNet-101, and the Region Proposal Network (RPN) proposes candidate object bounding boxes. In the second stage, these proposals are classified as to whether they contain a CLS. Mask R-CNN outputs a binary mask for each region of interest in parallel to predicting the class and box offset.
Fig. 2. CLS segmentation with Mask R-CNN algorithm.

(A) Workflow of Mask R-CNN for CLS detection and segmentation. ROI, region of interest. (B) Free-response receiver operating characteristic (FROC) curve for testing results with Mask R-CNN with an average number of 100 ground truth CLSs. (C and D) Typical 3D macrophage-labeled light sheet images and respective segmentation images for adipose tissue from (C) obese and (D) lean mice. All images share the same scale bar of 200 μm. (E and F) Distributions and four-component Gaussian fits for CLSs in perirenal adipose tissues in (E) obese and (F) lean mice. Components are given in the legends together with fractional contribution to the total counts in parentheses. (G) CLS and (H) cell number densities across adipose tissue in both obese and lean mice (n = 4). Asterisks indicate the following: *P = 0.0143 and **P = 0.0034, Student’s t test.
Light sheet microscopy images were first manually annotated for CLSs by human experts using Microscopy Image Browser based on 2D images (Materials and Methods and fig. S1A). Because CLSs are defined by dead or dying adipocyte cells encircled by macrophages, the intensity distribution of a typical CLS is a dark hole because the adipocyte cell, composed primarily of a fat globule, is not labeled, whereas the shell of encircling macrophages is labeled by the F4/80 immunostain (Fig. 1, G and H). To allow Mask R-CNN to learn the comprehensive range of diverse CLS structures, we generated training sets that included instances spanning the broad space of both adipocyte cells and macrophage cells with CLSs that exhibit diverse shapes, sizes, and locations (fig. S1A). Through this process, we observed that multiple CLSs in adipose tissue from obese mice were frequently connected to one another. To train Mask R-CNN to learn this scenario, we manually separated the connected CLSs either from 2D or 3D images (fig. S1B). A total of 2792 raw images from different volumetric scans were used for training, containing 108,821 annotated CLSs. Note that Mask R-CNN proposes a bounding box for CLSs with a probability estimation so that we could tweak hyperparameters to optimize CLS identification (fig. S2A). To ensure that 2D segmentation masks generated by Mask R-CNN were also consistent in the third dimension, we performed a postprocessing CLS segmentation comparison to evaluate adjacent x-y planes to improve the performance of the automatic CLS detection and segmentation pipeline (see Materials and Methods).
The performance of the model for CLS detection and segmentation was first evaluated in 100 obese tissue images using a free-response receiver operating characteristic (FROC) curve (Fig. 2B) (36). A maximum sensitivity of 0.92 was obtained with 22 false positives per image at a detection threshold of 0.5, the lowest threshold tested for binary classification. We adopted a detection threshold of 0.85 to identify CLS structures across adipose tissue in both obese and lean tissues. Note that the algorithm identified CLS structures even in regions of low fluorescence intensity, which derives from the imperfect homogeneity of excitation using light sheet microscopy and inhomogeneities in staining. CLSs that were missed by human annotation were found to contribute substantially to false positives (fig. S2B). Mask R-CNN achieved an F1 score of 81%, which is much better than that obtained with traditional segmentation approaches such as the Surface function of Imaris software. Imaris had a very low precision for identifying CLS structures even with extensive efforts to manually optimize parameters to maximize performance (fig. S3). In addition, we compared the performance of our algorithm for CLS detection and segmentation across 220 obese and 220 lean tissues. In obese tissues, the precision was 0.74 and recall was 0.80 (F1 score, 0.76), and in lean tissues, values were similar, with 0.81 precision and 0.77 recall (F1 score, 0.78). We note that in these evaluations, CLS presence and obesity status were blinded during the image evaluation steps, but obesity status was not blinded during the tissue processing or imaging steps. Nevertheless, all tissues were processed using the same protocols.
While training sets using manually disconnected CLSs helped generate isolated segmentations of CLSs that were interconnected through Mask R-CNN, a small amount of overlap between the segments of neighboring CLSs, even in one individual image plane, resulted in connectivity in 3D segmented images. So that our characterizations of CLS number were accurate, morphological erosion of the masks was used to minimize overlap between CLSs in all 2D planes (fig. S2C). Using the CLS number differences before and after mask erosion, we calculated that approximately 4.6% of CLSs were dimers in adipose tissues in obese mice; the number of CLS dimers in lean mice was near zero. Because morphological erosion led to a reduction in size of all CLS masks, we used nonreduced CLS masks for analysis of cell identities with CLSs described below but used eroded masks for CLS counts.
CLS sizes and counts
Using the optimized Mask R-CNN identification and segmentation algorithm, we counted CLSs in 3D images using either Microscopy Image Browser (37) or Imaris. Typical light sheet fluorescence microscopy images of macrophages and CLS mask images are shown in Fig. 2 (C and D). We evaluated CLS volumes in VAT from four obese mice and four lean mice using Imaris and calculated the equivalent spherical diameter, with diameter distributions shown in Fig. 2 (E and F). On the basis of the centroids of the distributions alone, the mean CLS diameter in obese mice (74 ± 20 μm) (Fig. 2E) is only slightly larger than that of lean mice (65 ± 16 μm) (Fig. 2F). Akaike information criteria (AIC) showed that the obese CLS size distribution fit to four underlying components (fig. S4). The largest-size component of the distribution contributed a much larger proportion of the total population in the obese state (22%) compared to the lean state (2%). We observed that most of the larger CLSs were located in the center of the tissue, whereas smaller CLSs were located more toward the tissue periphery (movies S4 and S5 and fig. S5, A and B). In addition, the CLSs in the center were more spherical in shape as compared with those at the edge of tissue (fig. S5, C and D). CLS counts in obese mice were, on average, 5.2 times higher in obese mice than in lean mice (Fig. 2G), whereas the overall cell density was higher in lean tissue (Fig. 2H), which is consistent with studies in 2D slices (35). The density of CLSs was also linearly proportional to body weight across all animals (R2 = 0.84, P = 0.0012; fig. S6A). Considering both their volumetric density and size, obese VAT was composed of 3.9% CLS domains by volume, compared with 0.46% in lean tissues, differing by a factor of 8.7-fold.
Identification of macrophages
After we identified CLSs as densely stained F4/80-positive microenvironments that typically contain tens of cells, we next aimed to identify all macrophages in intact VAT. To do so, we used nuclear stain images to identify all cells and filtered the cells on the basis of F4/80 staining intensity as a pan-macrophage classifier. Cell nucleus centroids were easily identified from 3D nuclear images using the spot detection function in Imaris software (see Materials and Methods) with an estimated accuracy of more than 90% based on visual inspection, resulting in an ellipsoidal mask for each nucleus with a mean radius near 5 μm and long axis near 15 μm (Fig. 3A and movie S6). The F4/80 signal intensity registered with nuclear masks across more than 10 million cells yielded a consistent histogram distribution with discrete spikes of intensities, with a distinct difference in distribution components between obese and lean tissues (Fig. 3, B and C). Lean tissues exhibited two peaks with intensity lower than 100 U, whereas obese tissues exhibited an additional peak with intensity near 140 U and a broad high-intensity tail. We manually evaluated the distribution of cells categorized by intensity in representative tissue regions, revealing that low-intensity cells (<120 units; fig. S7) corresponded to many structures with linear features, consistent with vessels, which are unlikely to be macrophages, whereas the vast majority of cells within CLS structures were identified as being above the 120–intensity unit threshold. We fit the curves to six Gaussian components based on the AIC minima, with three components of intensity units greater than 120 classified as macrophages, yielding a classifier with 0.88 area under the curve of the ROC (fig. S8A). This metric showed that the macrophage cell population was, on average, 43% of total cells in obese VAT and 8% of cells in lean VAT (fig. S8B), which is consistent with flow cytometry results from lean and obese animals (fig. S8C), as well as previous reports using similar diets (38). Also consistent with previous findings, flow cytometry identified approximately 75% of obese adipose tissue macrophages as inflammatory (M1 polarization) on the basis of the CD11c marker, compared with 39% of macrophages in lean adipose tissue (fig. S8D). CLS macrophages stained positive for CD11c, but some non-CLS macrophages did as well (fig. S8E). In the future, we will optimize costains in intact 3D tissues to comprehensively enumerate and classify the microenvironments of different CLS subtypes.
Fig. 3. Macrophage identification in intact adipose tissues and in CLSs.
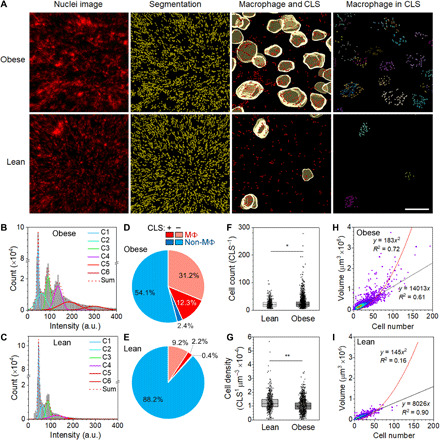
(A) Representative images of obese and lean adipose tissues showing raw nuclear image, nuclear segmentation, nuclei of cells classified as macrophages overlaid on CLS segments, and macrophages within CLSs. (B and C) Histograms show F4/80 intensity distribution across cells in obese and lean adipose tissues, in addition to six component Gaussian fittings. Plots show 10 million cells in obese adipose tissue and 5 million cells in lean adipose tissue. a.u., arbitrary units. (D and E) Cell populations in obese and lean adipose tissues, including macrophages (MΦ) and other cells (non-MΦ) within or not within CLSs. (F) Cell counts within CLSs from adipose tissues of lean and obese mice (*Mann-Whitney U < 0.0001). (G) Volumetric density of cells within each CLS calculated by dividing cell number by CLS volume (**Mann-Whitney U = 0.0038). In (F) and (G), boxes indicate 25th/75th percentile, black lines are mean values, whiskers are SDs. (H and I) Density plots of CLS volume versus cell number in adipose tissue of obese and lean mice. Linear and quadratic fits are shown with indicated coefficients of determination (R2). Linear fits only include data points up to 70 cells.
CLS classification by macrophage number
Using these methods to identify CLSs and macrophages within adipose tissues, we next identified macrophage populations inside and outside of CLSs. On the basis of the imposed F4/80 intensity threshold, we estimated that 83.7% of cells within obese VAT CLSs were macrophages and that 84.6% of cells in lean VAT CLSs were macrophages (Fig. 3, D and E), a similar fractional cell distribution. The average number of cells per obese CLS was 26 with a range between 6 and 278 compared with an average of 22 with a range between 9 and 107 for lean CLSs (Fig. 3F). The average volumetric cell density for CLSs was 1.3-fold higher in lean tissue compared with obese tissue (Fig. 3G) and the mean CLS cell density decreased linearly with increasing mouse body weight (R2 = 0.68, P = 0.011; fig. S6B). Although global cell counts and cell density differences between lean and obese CLSs were statistically significant, the mean differences were small (Fig. 3, F and G)) and not visually obvious in tissue or CLS images. Differences were more apparent when the two parameters were cross-correlated (Fig. 3, H and I). For a fixed cell number within a CLS, the CLS volume was 1.75 times larger on average for obese CLSs compared with lean VAT CLSs based on linear regression of the correlated parameters. However, the linear fit was much better in the case of the lean tissue (R2 = 0.90) compared with obese tissue (R2 = 0.61). For obese tissues, data for cell numbers higher than 70 matched better to a quadratic fit. These distinguishable fitting regimes are important, as the CLS volume would be expected to linearly depend on cell number if the size of the adipocyte globule only contributes a small fraction of the CLS volume, a regime that is expected to yield efficient adipocyte clearance. In contrast, large CLSs from obese VAT have a large volume fraction contributed by the adipocyte, which was apparent from light sheet images (Fig. 4, A and B). In this case, macrophages and other associated CLS cells only sparsely encircle an adipocyte and contribute a small monolayer shell volume that quadratically correlates with total CLS volume. On the basis of the current understanding that dead adipocyte cells are digested and eliminated by macrophage cells, a low CLS volume contributed by macrophages would be expected to be insufficient to efficiently eliminate these cells. This regime contrasts strongly with images of the smallest CLSs in lean tissue (Fig. 4, C to F), which contained large numbers of macrophages that are present in multiple layers (fig. S9, A and B) and contribute a large total volume of the CLS.
Fig. 4. Structural heterogeneity of CLSs.
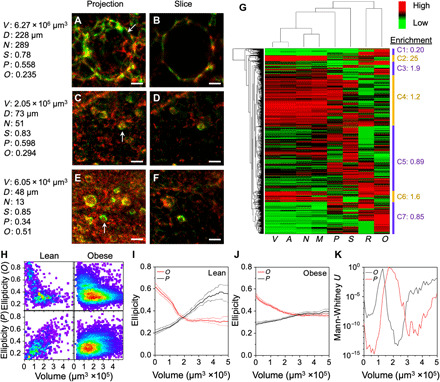
(A to F) Representative 3D light sheet microscopy images of CLSs in adipose tissue showing both 3D projections and slices. Scale bars, 50 μm. The CLS volume (V), diameter (D), cell number (N), sphericity (S), oblate ellipticity (O), and prolate ellipticity (P) are listed. (G) Unsupervised hierarchical clustering of the CLS parameters in adipose tissue of obese and lean mice. The corresponding cluster of the representative CLSs in (A to F) is (A and B), C4; (C and D), C5; and (E and F), C7. (H) Density plots of CLS ellipticity for oblate (O) and prolate (P) shape classification versus volume in adipose tissue of obese and lean mice. (I and J) Mean oblate and prolate ellipticity is shown with 95% confidence intervals (shaded lines) for indicated volumes of lean CLSs (I) and obese CLSs (J) with rolling average of 1.5 × 105 μm3. (K) Mann-Whitney U test for comparison of oblate and prolate ellipticity between obese and lean CLSs with rolling 1.5 × 105 μm3 increment data binning across the indicated volumes.
CLS classification by shape
On the basis of these findings of distinct populations of lean and obese CLSs extracted from cell parameters, we next evaluated structural parameters. The representative CLS structures shown in Fig. 4 (A to F) include extracted volume (V), sphericity (S), oblate ellipticity (O), and prolate ellipticity (P). These parameters, together with surface area (A), total cell number (N), macrophage number (M), and ratio of macrophage to total CLS cell number (R), were used to sort nearly 4000 CLSs from both lean and obese tissues by unsupervised hierarchical clustering. As shown in Fig. 4G, seven high level categories were identified. Category 1 CLSs were small with oblate shapes and were enriched fivefold in lean tissues. Category 2 CLSs were large and more spherical, enriched 25-fold in obese tissues, and located deep within the tissue center (fig. S5A). The global shift of CLS structures from the lean to obese state by principal component analysis (fig. S10, A to C) also showed these enriched features, with small, oblate CLSs in the lean state shifting to large CLSs in the obese state. Both of these bioinformatics pipelines support that these two CLS populations distinguished these two tissue states.
The other five CLS categories were more similarly represented across both lean and obese tissues, but with a higher degree of heterogeneity within each class. One clear trend that was evident across both lean and obese tissues was that in the progression from CLSs that were small (category 7) to medium (category 5) to large (category 4), the shape transitioned from more oblate (flattened) to spherical to prolate (elongated). Heatmaps in Fig. 4H show that with increasing volume, the prolate character in the lean VAT CLSs increases, while the smallest CLSs are clearly more oblate. These trends are reflected in moving average plots, showing a more distinguishable trend in lean tissues (Fig. 4, I and J). The slope of linear regression for prolate ellipticity with volume was positive in the lean tissues (P < 10−66) but insignificant for obese tissue CLSs (P > 0.35). Shape differences between the obese and lean tissues were significant across all volumes less than 5.2 × 105 μm3 for either oblate or prolate character (P < 0.001; Fig. 4K) and also reflected in the global sphericity parameter S (fig. S10, D to G).
These differences in shape are important for physiological processes in tissues, as prolate ellipsoids have overall higher curvature than equivalent volume ellipsoids that are spherical or oblate (39). These higher curvatures are expected to allow more rapid transport across a prolate surface (39). On the basis of coculture studies, macrophages digest dead adipose cells in CLSs through exocytosis of lysosomes at points of intercellular contact using local actin polymerization (28), further engulfing and processing the adipocyte contents and ultimately becoming foam cells. These processes would allow more efficient CLS clearance when transport is more rapid. Greater sphericity for the larger obese CLSs therefore implies less effective digestion of adipose cells in obese tissues due to reduced interfacial transport of macrophage lysosomes to adipocytes and adipocyte content to macrophages. Because of the inflammatory phenotype of CLS macrophages, slower clearance of unusually large CLSs may contribute to elevated tissue inflammation. The larger and more spherical size toward the center of adipose tissue may imply that this region could experience a slower rate of adipocyte efferocytosis, although it is not clear what the biological consequence of this effect might be for the tissue as a whole. For smaller CLSs, for which large numbers of macrophages and high cell number–to–volume ratios may already make transport efficient, it may be beneficial to instead adopt an oblate morphology to match the geometrical shapes of spatial voids that emerge within 3D tissues with hexagonal cell packing as the dying adipocyte shrinks below the mean size of its neighboring adipocytes (39). We further note that overlapping CLS segments may increase the CLS size parameter to some degree, particularly for adipose tissues of obese mice that have a higher CLS density. For the data analyzed here for Figs. 3 and 4, we manually separated interconnected CLSs to ensure that they would not affect CLS classifications. New object detection and segmentation algorithms that function for polydisperse and densely packed 3D objects [i.e., StarDist (40)] could soon provide a fully automated segmentation pipeline for interconnected objects.
DISCUSSION
Adipose tissue is perhaps the most structurally dynamic tissue in the adult human body. Its capacity to grow and shrink in size by large magnitudes is fundamental to human metabolism, health, fitness, and adaptation. Unraveling the mechanisms of this process has become increasingly important because of the growing obesity epidemic, afflicting more than 40% of the adult U.S. population and an increasing fraction of the worldwide population, including countries at all levels of development (41). Tissue microenvironments and macrophage cells are believed to be essential to adipose dynamics, with CLSs believed to be the histopathological inflammatory connection to obesity comorbidities, including metabolic disorders, glucose intolerance, and cardiovascular events (9). The counterintuitive necessity to pare and kill off cells to grow tissue in the obese state is not yet fully understood, partly because of our incomplete ability to correlate microscopic tissue structures with global tissue metabolism. Our current understanding has been limited to analyses from 2D adipose tissue sections with limited 3D structural information, and nonlinear changes have been observed in the progressive development of obesity (42). 3D analysis is needed to accurately assess CLS features such as cell composition, size, and shape, while rare events such as large CLS structures may not be possible to identify in 2D datasets. Whether these additional features will further improve correlation with clinical conditions or predict progression to comorbidities will be the focus of further studies. Time course microscopy in living tissues has revealed important dynamic processes in adipose tissues (43); however, there are considerable limitations to these techniques, as the spatial depth of confocal imaging only allows observations at the tissue periphery and with low throughput. Tissue clearing and deep learning–based image processing instead provide the capacity to comprehensively map all CLS structures and to further evaluate cell-cell interactions with much richer detail, albeit without the capacity for longitudinal analysis of individual structures (15, 44). Further studies will be needed to analyze cell subclassifications involved, as single-cell sequencing in lean and obese adipose tissues revealed seven distinct subclasses of macrophages (45). Adipocytes, while not directly probed in this work, can also be evaluated using related image analysis workflows, which may yield further insights into factors modulating organism-level metabolism, such as adipokines (e.g., adiponectin and leptin) or free fatty acids. Advanced forms of multiplexed immunolabeling, potentially applying technologies like quantum dots, will be necessary to identify numerous additional cell types and phenotype markers in heterogeneous tissue microenvironments (46) to help understand their spatial relationships, contributions to pathologies, and response to interventions. These workflows may similarly contribute to the development of novel bioengineering technologies in adipose tissue, which is a common source of stem cells, a target for novel biopharmaceuticals, and a potential depot for drug delivery (47, 48). While applications in the sciences are clear, there are still challenges that prevent the use of 3D imaging techniques in the clinic for histopathological analyses, such as long immunolabeling times and low-throughput analysis methods for terabyte-sized data (44). Deep learning in the most recent 10 years has led to streamlined procedures for image processing and analysis, with particularly impactful contributions for object recognition and localization, processes that have traditionally been manual bottlenecks in image analysis. Nevertheless, the coordinated development of tissue clearing and labeling approaches, microscopy modalities, and advanced deep learning algorithms can together drive solutions to the challenges in 3D structural biology of tissue.
MATERIALS AND METHODS
Materials
Unless specified, all organic solvents and chemicals were purchased from Sigma-Aldrich and used as received. Paraformaldehyde (PFA; 15710-S) was purchased from Electron Microscopy Science. Methanol (A412SK-4), YO-PRO-1 (catalog no. Y3603), F4/80 rat anti-mouse immunoglobulin G (IgG; catalog nos. 50-112-9624 for immunostaining and 50-167-59 for flow cytometry), Dulbecco’s modified Eagle’s medium (DMEM), penicillin-streptomycin stock solutions (P/S), fetal bovine serum (FBS), phosphate-buffered saline (PBS), Triton X-100, trypsin EDTA, l-glutamine, sodium pyruvate, formamide, glycine, dimethyl sulfoxide (DMSO), and methanol were acquired from Fisher Scientific. Normal donkey serum and Alexa Fluor 647–labeled donkey anti-rat IgG were purchased from Jackson ImmunoResearch.
Animals and diets
Mice were purchased as lean or obese from the Jackson laboratory at 23 weeks of age, having been fed either a high-fat diet or a low-fat diet since an age of 6 weeks. Upon arrival, mice were fed a high-fat diet (60% kcal fat; D12492, Research Diet Inc.) or a low-fat diet (10% kcal fat; D12450J, Research Diet Inc.), ad libitum, with adequate and unrestricted supply of water for approximately 2 weeks before experiments. All animals were housed in groups of four in standard shoebox cages in a temperature- and humidity-controlled environment, with a 12-hour light/12-hour dark cycle. The Institutional Animal Care and Use Committee of the University of Illinois at Urbana-Champaign approved all of the animal experiment procedures in this study before initiation.
Tissue collection
Mice were anesthetized by intraperitoneal injection of a ketamine and xylazine mixture according to their body weight. An incision through the skin with surgical scissors along the thoracic midline from just beneath the xiphoid process to the clavicle was made, and the diaphragm from the chest wall on both sides was separated. The heart was secured with forceps, and an incision of 1 to 2 mm was made on the left ventricle followed by insertion of a 24-gauge, 25.4-mm needle. The right atrium was incised for the sign of blood flow and infused with 10 ml of heparinized PBS, followed by 20 ml of 4% PFA in PBS. This was followed by removal of skin and dissection of fat depots. The collected fat pads were further fixed in 4% PFA for 24 hours at 4°C and rinsed with PBS three times for tissue clearing and immunostaining. For flow cytometry analysis, fat pads were collected from mice that were not perfused with PFA.
Flow cytometry
The collected adipose tissues were washed with cold PBS and cut into tiny pieces with volumes near 5 to 10 mm3 and immersed in collagenase buffer (1 mg/ml in DMEM without phenol red and serum, supplemented with P/S). After incubation at 37°C for 1 hour with agitation, collected cells were passed through a 70-μm filter. The collected cells were washed in fluorescence-activated cell sorter buffer (PBS supplemented with 2% FBS and 1% P/S). Staining of F4/80 was carried out in fluorescence-activated cell sorter buffer on ice for 30 min, then washed, and fixed for flow cytometry. Data were acquired on a LSRFortessa (Becton Dickinson Biosciences) and analyzed with FCS Express software (De Novo Software).
Tissue clearing
Four approaches including CLARITY, ClearT, 3DISCO, and iDISCO were tested for obese adipose tissue clearing. Similar tissue samples were obtained with a tissue punch (Miltex Biopsy Punch, Ted Pella Inc.) and were used for the different protocols.
CLARITY was performed following protocols described in previous reports (19). The punched tissue samples were incubated with hydrogel monomer solution containing 4% (w/v) acrylamide, 0.05% (w/v) bisacrylamide, 4% (w/v) PFA, and 0.25% (w/v) VA-004 initiator in PBS in a 2-ml Eppendorf tube at 4°C for 12 hours. Samples were degassed in a desiccator and purged with nitrogen. The samples were further embedded at 37°C in a water bath for 4 hours until the hydrogel was solid. The embedded sample was cleared through passive CLARITY clearing with the clearing solution containing 4% sodium dodecyl sulfate in 0.2 M boric acid buffer at pH 8.5 and 7°C. The solution was exchanged with freshly prepared clearing solution every 24 hours for 1 week. The adipose tissue remained opaque after 1 week of treatment.
ClearT was conducted according to protocols in previous publications (32). Formamide solutions in PBS were prepared with compositions of 20, 40, 80, and 95% (v/v). Each punched adipose tissue sample was sequentially incubated in solutions of increasing formamide fraction for 30 min, followed by a second 95% formamide solution for 1 hour. The transparency of the tissue was improved as compared with tissues treated with CLARITY.
BABB was performed by following protocols in previous publications (33). Punched adipose tissue was treated sequentially with aqueous solutions of ethanol with compositions of 50, 80, 96, 100, and 100% (v/v), each for 1 hour at room temperature. The tissues were further immersed in neat hexane for 1 hour and then transferred to a clearing solution composed of a 1:2 (v/v) mixture of benzyl alcohol and benzyl benzoate.
iDISCO was conducted following protocols in previous publications (16). The tissues were treated sequentially with aqueous solutions of methanol with compositions of 20, 40, 60, 80, 100, and 100% (v/v), each for 1 hour. Then, each sample was treated with a bleaching solution containing 2 ml of 30% hydrogen peroxide, 2 ml of DMSO, and 8 ml of methanol at 4°C overnight. The sample was then washed sequentially with methanol solutions with compositions of 100, 100, 80, 60, 40, and 20%, each for 1 hour, followed by two treatments with PBS for 1 hour. Next, the samples were washed twice with 0.2% Triton X-100 in PBS and permeabilized with a PBS solution of 0.2% Triton X-100, 20% DMSO, and 0.3 M glycine at 37°C for 1 day. Tissues were then blocked for 1 day with a PBS solution of 0.2% Triton X-100, 10% DMSO, and 6% donkey serum at 37°C. The samples were then labeled with a primary antibody against mouse F4/80 with 1:200 dilution in PTwH (PBS solution of 0.2% Tween 20 and 0.01% mg heparin) with 5% DMSO and 3% donkey serum for 2 days at 37°C. The samples were washed four times with PTwH for 6 hours each and incubated in secondary antibody with 1:100 dilution in PTwH with 3% donkey serum for 2 days at 37°C. YO-PRO-1 (1:500) was added 12 hours before completion of secondary antibody labeling to label nuclei. Samples were lastly washed four times in PTwH for 6 hours each. The labeled samples were then dehydrated sequentially with methanol/water solutions with compositions of 20, 40, 60, 80, 100, and 100%, each for 1 hour at room temperature. Tissues were lastly incubated in a solution of 66% dichloromethane and 33% methanol for 30 min at room temperature, followed by two incubations in 100% dichloromethane for 30 min each. The samples were transparent at this stage and were further incubated in dibenzyl ether for storage and imaging.
Confocal fluorescence microscopy
Confocal images were acquired using a Zeiss LSM 880 laser scanning microscope with Airyscan Superresolution. Excitation and emission wavelengths included the following: 488-nm excitation with 500- to 550-nm emission (YO-PRO-1) and 633-nm excitation with 650- to 750-nm emission (Alexa Fluor 647). A photomultiplier tube detector was used to scan volumetric thin sections using a 40× Plan Apochromat [1.4 numerical aperture (NA)] oil immersion objective in confocal mode with the Z-scan function with Zeiss software.
Light sheet microscopy
Light sheet image stacks were acquired using an UltraMicroscope II (LaVision BioTec) equipped with a 2× MVPLAPO objective (0.5 NA) coupled to an Olympus MVX10 zoom body, which provides a zoom range from 0.63× to 6.3×. Lower-magnification images (0.63×) were acquired with a light sheet width set to 100% with 0.038 NA, exposure time of 100 ms, and laser power of 2% for 488 nm (YO-PRO-1) with 500- to 550-nm emission and 15% for 633 nm with 650- to 750-nm emission (Alexa Fluor 647). Tile scans with 20% overlap were acquired with 5-μm z step. Higher-magnification images (2.5×) were acquired with light sheet width set to 50% with 0.101 NA, exposure time of 100 ms, and laser power of 2% for 488-nm laser and 20% for 633 nm. Tile scans with 20% overlap were acquired using a 3-μm z step. Each image size was 2560 pixels in height and 2160 pixels in width.
Image processing
The Alexa Fluor 647 channel from light sheet 3D volumetric images including four obese adipose tissues and four lean adipose tissues was deconvolved using AutoQuant X3 (Media Cybernetics), using a minimal voxel size of 500 μm by 500 μm by 500 μm that was cropped from the center of the original images to estimate the point spread function. The deconvolved images were annotated using Microscopy Image Browser (version 2.51) (37), combining both intensity and size thresholds to isolate CLSs. The CLS signal was first identified on the basis of an intensity threshold to generate a rough mask of each CLS. Then, an area size threshold was applied to eliminate objects with areas smaller than 100 μm2. CLS contours were filled to generate quasi-spherical masks as training samples (fig. S1A). Interconnected CLSs were manually disconnected to provide a more accurate training data for CLS identification (fig. S1B). To stitch together the title scan images to reconstruct the entire dataset, TeraStitcher was used with default settings (49).
CLS identification with mask R-CNN
Machine learning–based CLS identification was based on an implementation by Matterport Inc. released under a Massachusetts Institute of Technology license based on the open source libraries Keras and TensorFlow (50). Annotated images were used for Mask R-CNN training. A total of 2792 light sheet microscopy images from one obese tissue were used for training and 100 were held out for testing. Obese tissue rather than lean tissue was used for training because of its higher density of CLSs and high spatial heterogeneity, characteristics important for efficient and robust training. The models were initialized with the ResNet-101 backbone, pretrained on the Microsoft Common Objects in COntext (MS COCO) dataset. The models were trained using stochastic gradient descent with momentum of 0.9. The network training was arranged in three stages. First, network heads were trained for 25 epochs with a learning rate of 0.001. Then, the ResNet stage 4 and upper layers were trained with the same learning rate for 25 epochs. Last, the whole network was trained end to end with a learning rate of 0.0001 for 15 epochs. In view of the large size of images, we used a batch size of 1 and trained the network on a single NVIDIA Tesla V100 graphics processing unit. Gradient was clipped to 5.0, and weight decay factor was set to 0.0001. The anchors of the RPN span five scales of [16, 32, 64, 128, 256] targeting the size variations of CLSs in this task, which showed the best performance in our dataset.
To ensure that 2D segmentation masks generated by Mask R-CNN were also consistent in the third dimension, a postprocessing strategy was designed to improve the performance in our automatic CLS detection and segmentation pipeline. A “sliding window” of a width of six continuous image slices was defined. To refine segmentation results, each image slice was compared with one preceding slice and one of four succeeding slices in order. If a missing mask for CLS was detected according to the adjacent slices within the sliding window, then a mask was added back to the current segmentation map and the detection results were updated for postprocessing of the next image slice. The relevant code of CLS identification pipeline with Mask R-CNN is publicly available on https://github.com/CaelynCheung1996/CLS-Mask_R-CNN.
The generated masks were further analyzed with Microscopy Image Browser (37) or Imaris to measure their volume, sphericity, and other information. When the images were analyzed manually, we observed that a small fraction of CLSs were interconnected as single CLSs because of a small portion of mask overlap (fig. S2C). To obtain an accurate analysis of the number of CLSs in intact tissues, morphological erosion was used to shrink overlapping masks from Mask R-CNN to eliminate CLS overlap in three dimensions.
Performance evaluation
The performance of the model was evaluated using FROC analysis (36). The FROC curve is defined as a plot of true positive fraction versus mean number of false positives per image. A detection and segmentation result is considered as correct when intersection over union (IoU) between the predicted instance mask and ground truth mask is larger than 0.5. The IoU is defined as
To compute the FROC curve, we swept over the detection confidence thresholds and plotted the average true positive (TP) sensitivity per image versus the average number of false positives (FP) per image, in which the precision, recall, and F1 score are defined as
To compare the applicability and sensitivity of Mask R-CNN for CLS detection and segmentation in adipose tissues from both lean and obese mice, the detection threshold was fixed to 0.90 and an IoU threshold of 0.5 was used to identify correct predictions in the testing stage. In addition, 220 images of obese adipose tissue and 220 images of lean adipose tissue were further used to evaluate the detection and segmentation performance of the algorithm.
Nucleus segmentation and macrophage identification
Because fluorescence intensity is heterogeneous across light sheet images, image regions were cropped from the original light sheet images with sizes of 1000 μm by 1000 μm by 1000 μm to generate 36 voxels for adipose tissue from obese mice and 24 voxels for adipose tissue from lean mice. The cell nuclei were then identified by 3D spot detection using Imaris software. The summed intensity from the F4/80 channel was calculated within each nuclear mask. The intensity histogram was then fit to a Gaussian mixture model using the least squares approximation, and AIC was calculated according the following equation
where nbin is the number of bins used to construct the intensity histogram, RSS is the residual sum of square, and nGauss is the number of Gaussians used to fit the intensity histogram. Because six Gaussians yielded the lowest AIC value, we fit each histogram to six functions, of which three were set below the 120 intensity threshold corresponding to non–macrophage cell types and three were set above the threshold for macrophage cell types. ROC curves were generated by assuming the three non-macrophage Gaussian curves and the three macrophage Gaussian curves as ground truth. The intensity histograms from lean mice were also fit with six Gaussians with the same peak center and widths. In these datasets, we also manually separated interconnected CLS in the cropped tissue sections. The fraction of macrophage cells in CLSs was calculated using Imaris XTension of spots split into surface objects. CLSs were identified with the Surface function, and the nuclei were identified with the Spots function.
Geometric parameter extraction
CLS geometric parameters including volume, sphericity, oblate ellipticity, and prolate ellipticity were determined on the basis of surface contours using the Surface function of Imaris. Sphericity (S) is the ratio of the surface area of a sphere with equivalent volume of the object (V) to the surface area of the object (A)
which spans values from 1 for a perfect sphere to 0 for an object with infinite surface area.
CLS contours were also fit to ellipsoids or axially deformed spheres with axis lengths of a, b, and c, where a ≤ b ≤ c. Prolate ellipticity describes the degree to which a single sphere axis is elongated and is defined as
which spans values from 1 for a spheroid with infinite length relative to its width (e.g., a = b and c = ∞) to 0 for a perfect sphere or oblate ellipsoid with infinite width relative to its length (e.g., b = c = ∞).
Oblate ellipticity describes the degree to which a single sphere axis is flattened and is defined as
which spans values from 1 for a spheroid with infinite width relative to its length (e.g., b = c = ∞) to 0 for a perfect sphere or prolate ellipsoid with infinite length relative to its width (e.g., a = b and c = ∞).
These metrics were correlated with CLS cell numbers, macrophage numbers, and macrophage per total cell ratio in MATLAB (MathWorks). Significance analysis of various CLS parameters between obese and lean mice was performed with the Mann-Whitney U test.
Cluster analysis
Principal component analysis of CLS parameters was performed with results displayed as biplots using MATLAB (MathWorks). Unsupervised hierarchical clustering analysis of CLS parameters was performed using the clustergram function of MATLAB (MathWorks). The resulting dendrograms and heatmaps were manually evaluated to interpret relatedness between parameters and CLSs.
Statistical analysis
Data are presented as means ± SD unless stated otherwise. Statistical significance was determined using either the Student’s t test or one-way analysis of variance (ANOVA) using Origin Pro 2019 software or the Mann-Whitney U test, as indicated in the text. Asterisks (*) indicate P or U values defined in figure captions.
Acknowledgments
Funding: This work was supported by a grant from the NIH (R01 DK112251) and funds from the Cancer Center at Illinois. J.G. was supported by the Beckman Institute for Advanced Science and Technology in the form of a postdoctoral fellowship. Author contributions: J.G. and A.M.S. designed the project and analyzed data, with the exception of machine learning–based analyses. J.G. performed all experiments, generated the training instances, and developed the data analysis methods. X.Z. and M.A.A. developed and ran the training and segmentation algorithms. E.R.N. and K.S.S. performed all animal protocols. S.P. and J.G. performed tissue dissections. S.H.S. performed flow cytometry. J.G. and A.M.S. wrote the manuscript, with input from all authors. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. All materials and reagents used to collect the data are available through commercial vendors. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/8/eabe2480/DC1
REFERENCES AND NOTES
- 1.Sun Y., You S., Tu H., Spillman D. R. Jr., Chaney E. J., Marjanovic M., Li J., Barkalifa R., Wang J., Higham A. M., Luckey N. N., Cradock K. A., Liu Z. G., Boppart S. A., Intraoperative visualization of the tumor microenvironment and quantification of extracellular vesicles by label-free nonlinear imaging. Sci. Adv. 4, eaau5603 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sadtler K., Estrellas K., Allen B. W., Wolf M. T., Fan H., Tam A. J., Patel C. H., Luber B. S., Wang H., Wagner K. R., Powell J. D., Housseau F., Pardoll D. M., Elisseeff J. H., Developing a pro-regenerative biomaterial scaffold microenvironment requires T helper 2 cells. Science 352, 366–370 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lavin Y., Winter D., Blecher-Gonen R., David E., Keren-Shaul H., Merad M., Jung S., Amit I., Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell 159, 1312–1326 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vitale I., Manic G., Coussens L. M., Kroemer G., Galluzzi L., Macrophages and metabolism in the tumor microenvironment. Cell Metab. 30, 36–50 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Mosser D. M., Edwards J. P., Exploring the full spectrum of macrophage activation. Nat. Rev. Immunol. 8, 958–969 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krzyszczyk P., Schloss R., Palmer A., Berthiaume F., The role of macrophages in acute and chronic wound healing and interventions to promote pro-wound healing phenotypes. Front. Physiol. 9, 419 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Labonte A. C., Tosello-Trampont A.-C., Hahn Y. S., The role of macrophage polarization in infectious and inflammatory diseases. Mol. Cells 37, 275–285 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murray P. J., Wynn T. A., Protective and pathogenic functions of macrophage subsets. Nat. Rev. Immunol. 11, 723–737 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quail D. F., Dannenberg A. J., The obese adipose tissue microenvironment in cancer development and progression. Nat. Rev. Endocrinol. 15, 139–154 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee Y.-H., Petkova A. P., Granneman J. G., Identification of an adipogenic niche for adipose tissue remodeling and restoration. Cell Metab. 18, 355–367 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haka A. S., Barbosa-Lorenzi V. C., Lee H. J., Falcone D. J., Hudis C. A., Dannenberg A. J., Maxfield F. R., Exocytosis of macrophage lysosomes leads to digestion of apoptotic adipocytes and foam cell formation. J. Lipid Res. 57, 980–992 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Apovian C. M., Bigornia S., Mott M., Meyers M. R., Ulloor J., Gagua M., McDonnell M., Hess D., Joseph L., Gokce N., Adipose macrophage infiltration is associated with insulin resistance and vascular endothelial dysfunction in obese subjects. Atertio. Thromb. Vasc. Biol. 28, 1654–1659 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farb M. G., Bigornia S., Mott M., Tanriverdi K., Morin K. M., Freedman J. E., Joseph L., Hess D. T., Apovian C. M., Vita J. A., Gokce N., Reduced adipose tissue inflammation represents an intermediate cardiometabolic phenotype in obesity. J. Am. Coll. Cardiol. 58, 232–237 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mullooly M., Yang H. P., Falk R. T., Nyante S. J., Cora R., Pfeiffer R. M., Radisky D. C., Visscher D. W., Hartmann L. C., Carter J. M., Degnim A. C., Stanczyk F. Z., Figueroa J. D., Garcia-Closas M., Lissowska J., Troester M. A., Hewitt S. M., Brinton L. A., Sherman M. E., Gierach G. L., Relationship between crown-like structures and sex-steroid hormones in breast adipose tissue and serum among postmenopausal breast cancer patients. Breast Cancer Res. 19, 8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Richardson D. S., Lichtman J. W., Clarifying tissue clearing. Cell 162, 246–257 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Renier N., Wu Z., Simon D. J., Yang J., Ariel P., Tessier-Lavigne M., iDISCO: A simple, rapid method to immunolabel large tissue samples for volume imaging. Cell 159, 896–910 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Ertürk A., Becker K., Jährling N., Mauch C. P., Hojer C. D., Egen J. G., Hellal F., Bradke F., Sheng M., Dodt H.-U., Three-dimensional imaging of solvent-cleared organs using 3DISCO. Nat. Protoc. 7, 1983–1995 (2012). [DOI] [PubMed] [Google Scholar]
- 18.Hama H., Hioki H., Namiki K., Hoshida T., Kurokawa H., Ishidate F., Kaneko T., Akagi T., Saito T., Saido T., Miyawaki A., ScaleS: An optical clearing palette for biological imaging. Nat. Neurosci. 18, 1518–1529 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Chung K., Wallace J., Kim S.-Y., Kalyanasundaram S., Andalman A. S., Davidson T. J., Mirzabekov J. J., Zalocusky K. A., Mattis J., Denisin A. K., Pak S., Bernstein H., Ramakrishnan C., Grosenick L., Gradinaru V., Deisseroth K., Structural and molecular interrogation of intact biological systems. Nature 497, 332–337 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Y., Shen Q., White S. L., Gokmen-Polar Y., Badve S., Goodman L. J., Three-dimensional imaging and quantitative analysis in CLARITY processed breast cancer tissues. Sci. Rep. 9, 5624 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Renier N., Adams E. L., Kirst C., Wu Z., Azevedo R., Kohl J., Autry A. E., Kadiri L., Venkataraju K. U., Zhou Y., Wang V. X., Tang C. Y., Olsen O., Dulac C., Osten P., Tessier-Lavigne M., Mapping of brain activity by automated volume analysis of immediate early genes. Cell 165, 1789–1802 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liebmann T., Renier N., Bettayeb K., Greengard P., Tessier-Lavigne M., Flajolet M., Three-dimensional study of Alzheimer’s disease hallmarks using the iDISCO clearing method. Cell Rep. 16, 1138–1152 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pan C., Schoppe O., Parra-Damas A., Cai R., Todorov M. I., Gondi G., von Neubeck B., Böğürcü-Seidel N., Seidel S., Sleiman K., Veltkamp C., Förstera B., Mai H., Rong Z., Trompak O., Ghasemigharagoz A., Reimer M. A., Cuesta A. M., Coronel J., Jeremias I., Saur D., Acker-Palmer A., Acker T., Garvalov B. K., Menze B., Zeidler R., Ertürk A., Deep learning reveals cancer metastasis and therapeutic antibody targeting in the entire body. Cell 179, 1661–1676.e19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moen E., Bannon D., Kudo T., Graf W., Covert M., Van Valen D., Deep learning for cellular image analysis. Nat. Methods 16, 1233–1246 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kamnitsas K., Ledig C., Newcombe V. F. J., Simpson J. P., Kane A. D., Menon D. K., Rueckert D., Glocker B., Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med. Image Anal. 36, 61–78 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Mobadersany P., Yousefi S., Amgad M., Gutman D. A., Barnholtz-Sloan J. S., Vega J. E. V., Brat D. J., Cooper L. A. D., Predicting cancer outcomes from histology and genomics using convolutional networks. Proc. Natl. Acad. Sci. U.S.A. 115, E2970–E2979 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Causey J. L., Zhang J., Ma S., Jiang B., Qualls J. A., Politte D. G., Prior F., Zhang S., Huang X., Highly accurate model for prediction of lung nodule malignancy with CT scans. Sci. Rep. 8, 9286 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cinti S., Mitchell G., Barbatelli G., Murano I., Ceresi E., Faloia E., Wang S. P., Fortier M., Greenberg A. S., Obin M. S., Adipocyte death defines macrophage localization and function in adipose tissue of obese mice and humans. J. Lipid Res. 46, 2347–2355 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Chi J., Wu Z., Choi C. H. J., Nguyen L., Tegegne S., Ackerman S. E., Crane A., Marchildon F., Tessier-Lavigne M., Cohen P., Three-dimensional adipose tissue imaging reveals regional variation in beige fat biogenesis and PRDM16-dependent sympathetic neurite density. Cell Metab. 27, 226–236.3 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Esteva A., Kuprel B., Novoa R. A., Ko J., Swetter S. M., Blau H. M., Thrun S., Dermatologist-level classification of skin cancer with deep neural networks. Nature 542, 115–118 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu B.-X., Sun W., Kong X.-Q., Perirenal fat: A unique fat pad and potential target for cardiovascular disease. Angiology 70, 584–593 (2019). [DOI] [PubMed] [Google Scholar]
- 32.Kuwajima T., Sitko A. A., Bhansali P., Jurgens C., Guido W., Mason C., ClearT: A detergent- and solvent-free clearing method for neuronal and non-neuronal tissue. Development 140, 1364–1368 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dodt H.-U., Leischner U., Schierloh A., Jährling N., Mauch C. P., Deininger K., Deussing J. M., Eder M., Zieglgänsberger W., Becker K., Ultramicroscopy: Three-dimensional visualization of neuronal networks in the whole mouse brain. Nat. Methods 4, 331–336 (2007). [DOI] [PubMed] [Google Scholar]
- 34.Bartelt A., Bruns O. T., Reimer R., Hohenberg H., Ittrich H., Peldschus K., Kaul M. G., Tromsdorf U. I., Weller H., Waurisch C., Eychmüller A., Gordts P. L. S. M., Rinninger F., Bruegelmann K., Freund B., Nielsen P., Merkel M., Heeren J., Brown adipose tissue activity controls triglyceride clearance. Nat. Med. 17, 200–205 (2011). [DOI] [PubMed] [Google Scholar]
- 35.Lumeng C. N., DelProposto J. B., Westcott D. J., Saltiel A. R., Phenotypic switching of adipose tissue macrophages with obesity is generated by spatiotemporal differences in macrophage subtypes. Diabetes 57, 3239–3246 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bunch P. C., Hamilton J. F., Sanderson G. K., Simmons A. H., Free response approach to the measurement and characterization of radiographic-observer performance. J. Appl. Photogr. Eng. 4, 166–171 (1978). [Google Scholar]
- 37.Belevich I., Joensuu M., Kumar D., Vihinen H., Jokitalo E., Microscopy image browser: A platform for segmentation and analysis of multidimensional datasets. PLOS Biol. 14, e1002340 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weisberg S. P., McCann D., Desai M., Rosenbaum M., Leibel R. L., Ferrante A. W., Obesity is associated with macrophage accumulation in adipose tissue. J. Clin. Invest. 112, 1796–1808 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.L. Karp-Boss, E. Boss, The Elongated, the Squat and the Spherical: Selective pressures for phytoplankton shape, in Aquatic Microbial Ecology and Biogeochemistry: A Dual Perspective, P. M. Glibert, T. M. Kana, Eds. (Springer International Publishing, 2016), pp. 25–34. [Google Scholar]
- 40.M. Weigert, U. Schmidt, R. Haase, K. Sugawara, G. Myers, Star-convex Polyhedra for 3D object detection and segmentation in microscopy (2019); https://ui.adsabs.harvard.edu/abs/2019arXiv190803636W.
- 41.Bluher M., Obesity: Global epidemiology and pathogenesis. Nat. Rev. Endocrinol. 15, 288–298 (2019). [DOI] [PubMed] [Google Scholar]
- 42.Strissel K. J., Stancheva Z., Miyoshi H., Perfield J. W. II, De Furia J., Jick Z., Greenberg A. S., Obin M. S., Adipocyte death, adipose tissue remodeling, and obesity complications. Diabetes 56, 2910–2918 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Nishimura S., Manabe I., Nagasaki M., Seo K., Yamashita H., Hosoya Y., Ohsugi M., Tobe K., Kadowaki T., Nagai R., Sugiura S., In vivo imaging in mice reveals local cell dynamics and inflammation in obese adipose tissue. J. Clin. Invest. 118, 710–721 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ueda H. R., Ertürk A., Chung K., Gradinaru V., Chédotal A., Tomancak P., Keller P. J., Tissue clearing and its applications in neuroscience. Nat. Rev. Neurosci. 21, 61–79 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vijay J., Gauthier M.-F., Biswell R. L., Louiselle D. A., Johnston J. J., Cheung W. A., Belden B., Pramatarova A., Biertho L., Gibson M., Simon M.-M., Djambazian H., Staffa A., Bourque G., Laitinen A., Nystedt J., Vohl M.-C., Fraser J. D., Pastinen T., Tchernof A., Grundberg E., Single-cell analysis of human adipose tissue identifies depot- and disease-specific cell types. Nat. Metab. 2, 97-+ (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cleary A. S., Leonard T. L., Gestl S. A., Gunther E. J., Tumour cell heterogeneity maintained by cooperating subclones in Wnt-driven mammary cancers. Nature 508, 113–117 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wen D., Wang J., Van Den Driessche G., Chen Q., Zhang Y., Chen G., Li H., Soto J., Liu M., Ohashi M., Wang Z., Abdou P., Hu Q., Dotti G., Li S., Fourches D., Gu Z., Adipocytes as anticancer drug delivery depot. Matter 1, 1203–1214 (2019). [Google Scholar]
- 48.Ma L., Liu T.-W., Wallig M. A., Dobrucki I. T., Dobrucki L. W., Nelson E. R., Swanson K. S., Smith A. M., Efficient targeting of adipose tissue macrophages in obesity with polysaccharide nanocarriers. ACS Nano 10, 6952–6962 (2016). [DOI] [PubMed] [Google Scholar]
- 49.Bria A., Iannello G., TeraStitcher–A tool for fast automatic 3D-stitching of teravoxel-sized microscopy images. Bmc Bioinformatics 13, 316 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.M. Abadi, A. Agarwal, P. Barham, E. Brevdo, Z. Chen, C. Citro, G. S. Corrado, A. Davis, J. Dean, M. Devin, S. Ghemawat, I. Goodfellow, A. Harp, G. Irving, M. Isard, Y. Jia, R. Jozefowicz, L. Kaiser, M. Kudlur, J. Levenberg, D. Mane, R. Monga, S. Moore, D. Murray, C. Olah, M. Schuster, J. Shlens, B. Steiner, I. Sutskever, K. Talwar, P. Tucker, V. Vanhoucke, V. Vasudevan, F. Viegas, O. Vinyals, P. Warden, M. Wattenberg, M. Wicke, Y. Yu, X. Zheng, TensorFlow: Large-scale machine learning on heterogeneous distributed systems. arXiv e-prints (2016); https://ui.adsabs.harvard.edu/abs/2016arXiv160304467A.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/8/eabe2480/DC1


