1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection has led to the outbreak of COVID-19 syndrome since December 2019, becoming prevalent worldwide afterward [1,2]. Unlike severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome (MERS), which cause severe symptoms and high mortality [1], the morbidity from COVID-19 is relatively low, especially in young people [3]. In China, the mortality rate was 1023 of 44,672 confirmed cases by the end of February11, 2020 [4]. However, there were no cases of death among 416 patients aged 0–9 years, and only 1 in 549 patients aged 10-19 years [4]. The exact reasons for this clinical phenomenon remain unclear.
It is widely accepted that viral infection triggers innate and adaptive immunity sequentially [5]. While innate immune systems such as monocytes, dendritic cells, and NK cells constitute the first line of the protection against the invading virus, viral-specific antibody and T cell responses are more efficient in eliminating virus and virus-infected host cells, combined with the induction of memory responses for long-term protection. It is well documented that antibodies with neutralizing activity targeting SARS-CoV-2 spike (S) protein exist in convalescent COVID-19 patients [6], [7], [8], which could be helpful for the resistance to viral re-infection in the long run. These antibodies can last for at least 6 months in the human body [9]. Cellular immune responses are also reported in recovered COVID-19 patients targeting viral S, nucleocapsid (N), and membrane (M) proteins [10]. Herein, we report a 15-year-old COVID-19 patient with mild symptoms and a deficiency in anti-viral antibody responses at the rehabilitation stage. We defined immunological profiles of peripheral blood mononuclear cells using single-cell RNA sequencing for a better understanding of potential protective mechanisms against SARS-CoV-2 infection.
2. Materials and methods
2.1. Patient and clinical manifestations
The COVID-19 adolescent was an inpatient at the Shanghai Public Health Clinical Center (Shanghai, China). Clinical manifestations were recorded at several time points. There included body temperature, treatments, total white blood cell count, neutrophil count, lymphocyte count, monocyte count, platelet count, and C-reactive protein (CRP) level. Computed tomography (CT) images were taken at different time points as needed. Reverse transcription polymerase chain reaction (RT-PCR) results for viral RNA from oropharyngeal swabs, feces, and urine were collected retrospectively. This study was approved by the Shanghai Ethics Committee for Clinical Research (SECCR/2020-04-01). Written informed consent was obtained from the parents of the included child.
2.2. Detection of SARS-CoV-2 RNA
A magnetic bead-based nucleic acid extraction kit was used in a fully automated nucleic acid extraction instrument (Master Biotechnology, China). Total RNA was extracted from 200 μL samples, and dual fluorescence PCR was performed according to the manufacturer's instructions (Zhijiang Co., Shanghai, China) using Applied Biosystems 7500 Real-Time PCR System (Foster City, CA, USA). A Ct value < 37 was defined as a positive result, and Ct > 40 was defined as a negative result.
2.3. Fluorescence ImmunoAssay (FIA) for detection of anti-spike and anti-nucleocapsid protein IgG and IgM
The FIA assay was performed using detection cards coated with fluorescence-labeled S and N protein (Sino Biological, Beijing, China) for IgG and IgM detection according to the manufacturer's instructions (Dialab ZJG Biotech Co, Suzhou, China) [11]. Briefly, 10 μL plasma was mixed with 990 μL dilution buffer. 80 μL diluted plasma was added to the sampling well of the detection cards. The fluorescence signal was captured by a DL300 Quantitative Immunofluorescent Analyzer within 15 min. Mouse anti-human IgG and mouse anti-human IgM antibodies for capturing fluorescent antigen-serum IgG/IgM complexes were purchased from Sigma-Aldrich (St. Louis, MO, USA). Anti-S and anti-N IgG and IgM levels were represented by fluorescence signal values. The cutoff value for IgG positivity was 15 RU/mL, while 3.4 RU/mL was the cutoff value for IgM positivity of anti-S and anti-N antibodies.
2.4. Isolation of peripheral blood mononuclear cells (PBMCs)
Whole blood was collected in a tube containing ethylene diamine tetraacetic acid (EDTA). PBMCs were isolated by Ficoll-Hypaque density gradient centrifugation with LymphoprepTM solution (AXIS-SHIELD Poc AS, Oslo, Norway) according to the manufacturer's recommendations. The mononuclear cell layer was carefully transferred to a new 15 mL conical tube and washed twice with RPMI 1640 medium (Hyclone, Logan, UT, USA) by centrifuging at 486 × g for 10 min at room temperature. PBMCs were resuspended in RPMI 1640 culture medium containing 10% fetal bovine serum (FBS) (Millipore, Danvers, MA, USA), 100 units/mL penicillin, and 100 µg/mL streptomycin (GIBCO, Grand Island, NY, USA).
2.5. Interferon gamma (IFN-γ)-releasing assay (IGRA)
Antigen-specific IFN-γ release was detected using an enzyme-linked immunospot (ELISpot) assay according to the manufacturer's instructions (U-CyTech, Utrecht, Netherlands). Briefly, 96-well PVDF plates (Millipore) were coated with an anti-human IFN-γ coating antibody overnight at 4°C. PBMCs (0.25 × 106) were added to each well and stimulated with the recombinant receptor-binding domain of S protein (S-RBD), N (nucleocapsid protein), envelope protein (E) (Novoprotein, Shanghai, China) (both at 20 μg/mL), or tuberculin purified protein derivative (PPD) (20 μg/mL) (Statens Serum Institut, SSI, Copenhagen, Denmark) for 20 h at 37°C. RPMI 1640 culture medium served as a negative control, and treatment with 2.5 μg/mL phytohemagglutinin (PHA) (Sigma-Aldrich) was used as a positive control. After incubation for 20 h at 37°C, the plates were incubated with a biotin-labeled detection antibody at 37°C for 1 h, and subsequently HRP-conjugated streptavidin working solution for an additional 1 h. The AEC substrate solution was added to each well for 30 min in the dark at room temperature (RT). Color development was stopped by thoroughly rinsing both sides of the PVDF membrane with demineralized water. Plates were dried in the dark at RT. The spots were counted using a C.T.L. ImmunoSpotⓇ S6 Ultra Analyzer (Cellular Technology Limited, OH, USA). The number of antigen-specific IFN-γ-producing cells was calculated based on the number of spot-forming units (SFUs) per 2.5 × 105 PBMCs after deducting the background SFUs of the paired negative control wells.
2.6. Single-cell RNA library preparation and sequencing
Isolated PBMCs were diluted as a single-cell suspension with cell viability exceeding 90%. Sequencing libraries of PBMCs were prepared following the Drop-seq methodology. Single-cell RNA (scRNA) libraries were prepared according to the guidelines of the Chromium SingleCell 5′v3 chemistry (10x Genomics, Cat No. 120237). Libraries were sequenced using an Illumina NovaSeqS6000 device (Genergy Biotechnology, Shanghai, China).
2.7. Processing FASTQ reads of single-cell transcriptomic data into gene expression matrices
scRNA sequencing reads were subjected to quality control based on FastQC software v0.11.9 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Cell Ranger software (version 3.1.0) was used to process, align, and summarize sequencing data. De-multiplexed FASTQ reads were aligned to the human reference genome (GRCh38–3.0.0). Reads with low base-calling quality scores and assigned cell barcodes were filtered out. Unique molecular identifier (UMI) counts of each transcript were quantified. The UMI matrix was converted to Seurat objects using the R package Seurat v3. Further analysis was performed using the R package. Raw UMI count matrices were filtered to remove genes expressed in fewer than three cells, cells with either fewer than 200 or more than 6000 genes, or with high percentages of mitochondrial genes (more than 15%). A total of 9340 cells were used for further analysis. To account for the differences in sequencing depth across cells, UMI count was normalized by a global-scaling method, converted with a scaling factor (10,000 by default), and log-transformed with the LogNormalize function in Seurat for downstream analysis.
2.8. Dimension reduction and clustering analysis
FindVariableFeaturesfunction in R package Seurat v3 was used to scale the data with the top 2000 most variable genes. Principal component analysis (PCA) was subsequently performed based on these variable genes. FindNeighbors in Seurat was used to obtain nearest neighbors for graph clustering based on PCs, and FindCluster in Seurat to divide cell subsets. Cells were divided into myeloid, NK/T, and B cells based on their type-specific gene signatures, then visualized with the uniform manifold approximation and projection (UMAP) algorithm. Signature scores were calculated as the mean log2(LogNormalizedUMI+1) across the related gene signatures. Each cluster was assigned to the compartment of its maximal score, and all cluster assignments were manually checked to ensure the accurate partition of the cells.
2.9. Identification of differentially expressed genes and gene annotation
To identify differentially expressed genes (DEGs) across different immune cell types, we used the FindAllMarkers function in Seurat based on normalized data. Non-parametric Wilcoxon rank-sum tests were used to obtain p-values for comparisons. p-value adjustment was performed using Bonferroni correction based on the total number of genes in the dataset. DEGs with adjusted p-values < 0.05 and an average expression log2-fold-change > 0.25 were kept for further analysis.
2.10. Flow cytometry
Freshly isolated PBMCs were washed with phosphate-buffered saline (PBS) containing 2% FBS. Cells were incubated for 30 min at 4°C with the Live/Dead Fixable Viability Stain 575V (ThermoFisher Scientific, USA) and fluorescent conjugated antibodies to surface markers, including anti-CD66b (clone G10F5), anti-CD56 (clone NCAM16.2), anti-CD3 (clone UCHT1), anti-CD16 (clone 3G8), anti-CD8 (clone SK1), anti-CD4 (clone SK3), anti-CD14 (clone MφP9), anti-HLA-DR (clone G46-6), anti-CD19 (clone SJ25C1), anti-CD11c (clone B-ly6), anti-CD123 (clone 7G3) (all from BD Bioscience, USA). To determine cytokine production, PBMCs were stimulated with Lipopolysaccharides (LPS) or Phorbol-12-myristate-13-acetate(PMA)/Ionomycin (both from BD Bioscience) for 4 h in the presence of GolgiStop (BD Bioscience). The following antibodies, including anti-IFN-γ (clone B27), anti-TNF-α (clone MAb11), anti-GM-CSF (clone BVD2-21C11), anti-IL-2 (clone MQ1-17H12), anti-IL-17A (clone N49-653) (all from BD Bioscience), anti-Granzyme B (GzmB) (clone GB12), anti-IL-1β (clone CRM56), anti-IL-6 (clone MQ2-13A5) (all from Invitrogen, USA) were used in intracellular cytokine staining with the Cytofix/Cytoperm Fixation /Permeabilization kit (BD Biosciences) according to the manufacturer's instructions. Cells were acquired using a BD FACSymphony sorter (BD Biosciences), and data were analyzed using FlowJo software (Version 10.0.8, Tree Star Inc., Ashland, OR, USA).
3. Results
3.1. Clinical manifestation and treatments
A 15-year-old male adolescent developed symptoms on February 4, 2020, including fever with a body temperature of 37.7°C the next day, accompanied by nasal congestion, runny nose, cough, and white sputum. However, no other typical COVID-19 symptoms (headache, dizziness, chest tightness, chest pain, hemoptysis, nausea, vomiting, abdominal pain, diarrhea, or bloody stool) were observed. The patient was diagnosed at Huashan Hospital (Shanghai, China). In terms of epidemiological history, the patient denied the travel to Hubei province (including Wuhan), a history of contact with the Huanan seafood market, and wild animals. However, he had contacted a confirmed COVID-19 patient displaying fever, cough, and other symptoms. Nucleic acid detection of SARS-CoV-2 using oropharyngeal swabs was negative on February 5 and positive after the second RT-PCR detection on February 6 (Fig. 1 A). The patient was then admitted to the Shanghai Public Health Clinical Center (SPHCC) for further treatment. A typical pneumonia lesion was observed in the dorsal segment of the right lower lobe and the outer basal segment of the right lung on the same day based on chest CT scans (Fig. 1B). Blood C-reaction protein (CRP) was 11.4 mg/L (normal range: 0–5 mg/L). The adolescent was diagnosed as COVID-19. He had received recombinant human Interferon-α2b spray at admission and LianhuaQingwen granules on February 10. Vitamin C tablets were administered daily. No antibiotics, steroids, or antiviral agents were administered during hospitalization. SARS-CoV-2 detection was performed using oropharyngeal swabbing, as well as stool and urine samples after admission. However, all detection results were negative (Fig. 1A). On February 10, the inflammation lesion in the lung began to be absorbed. With negligible lesion in the right lower lobe (Fig. 1B), normal CRP value (Fig. 1C), and two successive negative results for SARS-CoV-2 virus (with a 24 h sampling interval), he was discharged on February 15. During hospitalization, the counts of peripheral white blood cells (5.24–6.63 × 109cells/L (range: 3.5–9.5 × 109/L)), neutrophils (2.49–3.59 × 109/L (range: 1.8–6.3 × 109/L)), lymphocytes (2.29–3.17 × 109/L (range: 1.10–3.20 × 109 /L)), monocytes (0.35–0.57 × 109 cells/L (range: 0.10–0.6 × 109/L)), and platelets (230–286 × 109 /L (range: 125–350 × 109/L)) were within normal ranges (Extended Data Fig. 1).
Fig. 1.
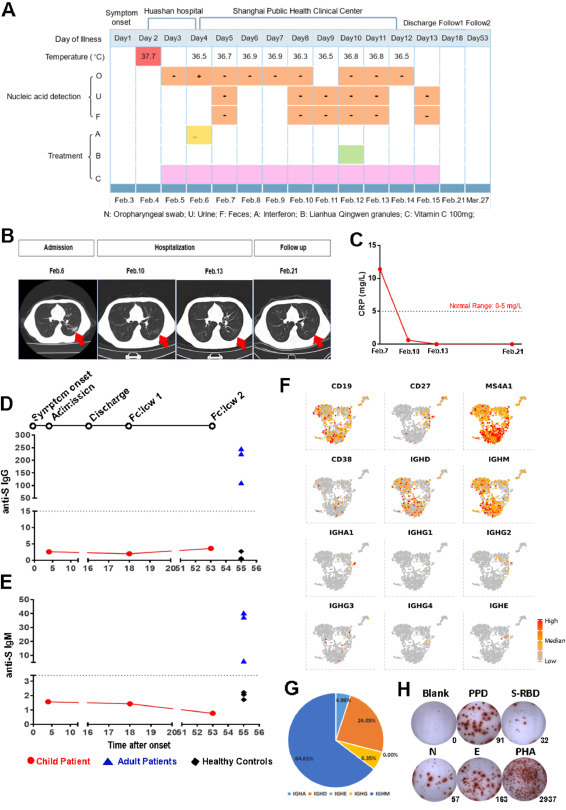
Clinical manifestations and humoral immune responses of the COVID-19 adolescent patient during the disease process. (A) Timeline of the COVID-19 adolescent patient during the disease process, including body temperature; RT-PCR results of SARS-CoV-2 virus from oropharyngeal swabbing, urine, and feces; and the treatments. (B) Computed tomography (CT) on admission day (Feb 6), day 8 and day 11 (hospitalization in the Shanghai Public Health Clinic Center), and day 21 (the first follow-up visit). Red arrows indicate the inflammation lesion in the lung. (C) Dynamics of blood C-reactive protein (CRP) levels from admission to the follow-up visit. (D-E) Dynamics of SARS-CoV-2 spike (S) protein-specific IgG (D) and IgM (E) in the plasma of the adolescent patient. Limit of detection (LOD): anti-S IgG15 RU/mL, anti-S IgM3.4 RU/mL. Three adult convalescent COVID-19 patients (blue triangles) and three healthy controls (black diamonds) were used as controls. (F) UAMP plots of B cell gene signatures, including CD19, CD27, MS4A1, CD38, IGHD, IGHM, IGHA1, IGHG1, IGHG2, IGHG3, IGHG4, and IGHE. (G) Distribution percentages of each isotype of immunoglobulin. (H) Antigen-specific IFN-γ releasing cells in PBMCs were determined by an ELISpot assay upon stimulation with tuberculin pure protein derivative (PPD), SARS-CoV-2 spike protein fragment (S-RBD), nucleocapsid protein (N), and envelope protein (E). PHA: positive control; culture medium (blank): negative control.
3.2. SARS-CoV-2-specific antibody and T cell responses in the recovered COVID-19 adolescent
Since anti-S antibody is most commonly detected after recovery and likely to play critical roles in neutralizing SARS-CoV-2 virus [6,7], we performed anti-S antibody detection in the serum using an FIA assay at the first follow-up visit. Surprisingly, both anti-S IgG and IgM were below the limit of detection (LOD) of the assay when compared to the samples from convalescent COVID-19 adults (Fig. 1D and1E). Anti-S IgG and IgM were also absent at the second follow-up visit (Day 53). We traced both back to the day of admission, and neither anti-S IgG and IgM were detected.
A scRNA-seq was further performed using freshly isolated PBMCs from the recovered COVID-19 adolescent to dissect the immune cell distribution with gene signatures. Three lineages, including myeloid, NK/T, and B cells, were clustered in 9340 PBMCs using t-distributed stochastic neighbor embedding (t-SNE) analysis (Extended Fig. 2 A and 2B) according to gene signatures (Extended Fig. 2C and 2D). B cells were defined by the signatures of CD19 and CD20 (MS4A1), and were 10.7% of the total (Fig. 1F). Based on immunoglobulin heavy chain (IGH) gene expression in B cells, it was found that the percentages of IGHD- and IGHM-positive B cells were the most frequent, whereas IGHA1-, IGHG1-, IGHG2-, IGHG3-, and IGHG4-expressing B cells were very few, with a proportion of 6.35% for IGHG (including IGHG1, IGHG2, IGHG3 and IGHG4), and 4.99% for IGHA expression in total B cells (Fig. 1G).
Fig. 2.

NK/T cell profiles in the periphery of the COVID-19 adolescent. (A) A UMAP plot showed three clusters, including NK cells, CD4+ and CD8+ T cells, according to the expression of marker genes. Cells were color-coded according to gene signatures. (B) The features of UMAP plots defined NK/T cell types with specific genes, including CD3E, CD4, CD8A, NCAM1, and FCGR3A. (C-D) UMAP plots (C) and violin plots (D) showed the expression of cytokines across NK/T cell subtypes, including IFNG, TNF, GZMB, CSF2, IL2, and IL17RA. (E-G) Cytokine expression upon 12-myristate 13-acetate (PMA) and ionomycin stimulation in CD3+CD8- cells (E) CD3+CD8+ T cells (F), and CD3-CD56+ NK cells (G), including IFN-γ, IL-2, TNF-α, GM-CSF, GzmB, and IL-17. (H) Feature plot of KLRC2 expression.
Nevertheless, virus-specific cellular responses were apparent at the first follow-up visit. IFN-γ release in PBMCs upon in vitro stimulation of SARS-CoV-2 S-RBD, N, and E proteins separately was obvious, together with the positivity of PPD-specific cellular response (Fig. 1H). These data indicate that anti-SARS-CoV-2 humoral responses are absent, whereas SARS-CoV-2-specific T cell responses are still detectable in this COVID-19 recovered adolescent.
3.3. Presence of cytotoxic NK cells and T cells in the recovered COVID-19 adolescent
Based on scRNA-seq data, NK/T cell lineages (75.7% in total PBMCs) were further sub-grouped into CD4+ T cells (34.1%, co-expressing CD3E and CD4 genes), CD8+ T cells (21.9%, co-expressing CD3E and CD8 genes), and NK cells (44.4%, co-expressing NCAM1 and FCGR3A genes) (Fig. 2A and 2B). The proportion of NK cells in the NK/T cell subsets was extremely high (Fig. 2A), which was verified by flow cytometry (Extended Fig. 3 ). The percentage of NK cells in total lymphocytes was 29.9%, which was higher than that of CD4+ T cells (20.9%) or CD8+ T cells (20.45%). UMAP and violin plot analyses indicated that IFNG, TNF, and GZMB were highly expressed in NK cells and CD8+ T cells (Fig. 2C and 2D). We also determined cytokine secretion in CD8+ T cells and NK cells by flow cytometry. Upon PMA/ionomycin stimulation in vitro, it was obvious that CD8+ T cells produced robust IFN-γ (23.1%), TNF-α (14.1%), and GzmB (38.7%). The percentages of IFN-γ-, TNF-α-, and GzmB-positive NK cells were 36.6%, 18.1%, and 79.1%, respectively, higher than corresponding values for CD8+ T cells (Fig. 2F and 2G). Furthermore, the UMAP plot showed that a portion of NK cells highly expressed KLRC2 (Fig. 2H), an important memory-like marker [12], suggesting the existence of memory-like NK cells in the periphery of the recovered patient. Our results thus indicate that there exist functional CD8+ T cells and NK cells in the periphery of the recovered adolescent, with greater expansion of NK cells.
Fig. 3.

Characterization of myeloid cell subsets in the periphery of the COVID-19 child (A) A UMAP plot showed five clusters among 1120 myeloid cells based on the expression of marker genes. Cells were color-coded according to defined myeloid subtypes. (B) The feature plots of UMAP showed subtype-specific gene expression for myeloid cells, including CD14, FCGR3A, HLA-DRA, ITGAX, CD1C, and IL3RA. (C-D) UMAP (C) and violin plots (D) showed cytokine expression in different myeloid subtypes, including IFNAR1, IFNAR2, IL1B, IFNG, TNF, CSF2, and IL6. (E-F) Cytokine production in monocyte subsets after LPS stimulation, including IL-1β, GM-CSF, IL-6, IFN-γ, and TNF-α.(G-H) Co-expression of three cytokines (IL-1β, IL-6, TNF-α) in CD14low (G) and CD14hi (H) monocytes.
3.4. Profiles of myeloid subsets in the recovered COVID-19 adolescent
Myeloid cells, including monocytes and dendritic cells, are crucial for virus clearance at the early stage and engaged in the antigen-presenting process to initiate adaptive immunity. According to scRNA-seq analysis, myeloid cells were divided into five populations, including classical CD14++CD16− monocytes (CD14-high and FCGR3A-negative), intermediate CD14++CD16+ monocytes (CD14-high and FCGR3A-positive), non-classical CD14+CD16++ monocytes (CD14- and FCGR3A-positive), CD1C+ myeloid dendritic cells (mDCs) (expressing CD1C), and CD123+ plasmacytoid dendritic cells (pDCs) (expressing IL3RA) (Fig. 3A and 3B). The UMAP plots ( Fig. 3C) showed that TNF was expressed in all cell subsets, while IL-1β was highly expressed in monocytes and mDCs. In addition, IFNG was mainly expressed in CD14++CD16+ monocytes. However, IL-6 and CSF2 were rarely expressed in all subsets. Type I interferon (IFNI) plays an important role in inhibiting viral replication in both infected and non-infected cells. Although IFNA expression was not detectable, some monocytes and DCs expressed IFNAR1 and IFNAR2, two genes encoding IFN-α receptors ( Fig. 3D). Upon LPS stimulation, CD14hi monocytes expressed high levels of IL-1β, TNF-α, and IL-6, whereas CD14medium monocytes produced fewer cytokines (Fig. 3E and 3F). In particular, CD14hi monocytes co-expressed IL-1β andIL-6 more apparently, while IL-1β and TNFα were co-expressed to a less extent (Fig. 3G and 3H). Our results thus illustrate that peripheral CD14++ monocytes secrete high levels of pro-inflammatory cytokines in the recovered COVID-19 adolescent and may function in a more extensive manner.
4. Discussion
The immune system plays critical roles in defending against SARS-CoV-2 virus infection by inducing innate immunity, and antigen-specific T cell and antibody responses. Occasionally, a cytokine storm is induced in certain cases and causes acute respiratory distress syndrome (ARDS), leading to multiple organ failure and mortality [13]. However, the vast majority of infected children do not develop ARDS and do not require respiratory support or intensive care [3]. Overall, SARS-CoV, MERS-CoV, and SARS-CoV-2 infections have less fatal effects on children compared to adults with mild clinical symptoms, low mortality, and a good prognosis [14], [15], [16], [17].
In the present study, we have reported a COVID-19 adolescent with no humoral responses after the recovery. He typically had mild symptoms, such as a transient increase in body temperature and CRP, no respiratory failure or acute respiratory distress syndrome, and a short period of hospitalization. The unexpected observation is that anti-S protein as well as anti-N protein (Extended Figure 4) IgG and IgM were absent from admission to discharge, implying that the adolescent exhibited a deficiency in humoral immune response upon SARS-CoV-2 infection. Nevertheless, based on the ELISpot assay an antigen-specific cellular immune response is still detectable in the recovered COVID-19 child, implying that COVID-19 adolescents displays unique immune profiles different from thethe adults. Interestingly, results from the ELISpot assay targeting S, N, and E proteins indicated that the immunoreactivity to E protein was the strongest (Fig. 1H). Due to their location and biological function, S and N proteins have been extensively investigated for their humoral and cellular immunogenicity [18], [19], [20], as well as for their applications in the diagnosis and antibody development [21,22]. E protein is a structural protein that exists in both monomeric and homo-pentameric forms. It is reported to be involved in the onset of viral infection, replication, and dissemination within host cells [23,24]. Its immunogenic peptides have been computationally predicted [25,26], but not clinically validated. Our data thus provide direct evidence of its capacity to induce cellular immune responses. The higher cellular immunoreactivity to E protein we observed in our case also implies the possibility that viral proteins display distinct immunogenicity related to their protein properties. The humoral responses targeting S protein prevent interaction with its receptor (ACE2), blocking entry of the virus into host cells, while cellular immune responses targeting other viral proteins are more inclined to eliminate virus-infected cells. Our data present the possibility that E protein exerts long-term protection against SARS-CoV-2 re-infection.
In addition to apparent T cell responses, the most striking immunological properties in this case are the exaggerations of innate immunity even at the rehabilitation stage. Several studies on scRNA-seq analysis of adult COVID-19 patients have been reported [27], [28], [29]. Both at the acute and convalescent stages, innate lymphocyte signatures are well defined, including cytokine and chemokine enrichment. In our case, NK cells accounted for nearly 30% of total lymphocytes with CD56dimCD16bright NK cells as the main group with high cytolytic ability. In the myeloid proportion, 90% of monocytes are CD14++CD16− classical monocytes. The remaining monocytes include CD14++CD16+ intermediate monocytes and CD14+CD16++non-classical monocytes [30]. Intermediate monocytes play an important role in viral infections, such as dengue fever [30]. Interestingly, the proportion of intermediate monocytes and non-classical monocytes increased in the COVID-19 child. It was previously reported that the “loss” of intermediate monocytes and non-classical monocytes is the result of down-regulation after stimulation [31], which is consistent with our results. Moreover, peripheral monocytes are able to secrete inflammatory cytokines, including IL-1β, TNF-α, and IL-6, even after the recovery, among which CD14++ monocytes co-expressed multiple cytokines. Therefore, we deduce that in the process of SARS-CoV-2 infection, innate immune responses are likely to play important roles when humoral immune responses are attenuated.
In conclusion, the case we report here indicates that although antibody response is absent after viral infection, CD8+ T cells and innate immune cells, including NK cells and monocytes, exhibit functional properties with cytokine secretion. The induction of strong cellular immunity also implies the possibility of rapid virus clearance after infection, and likely contributes to the mild symptoms and a good prognosis of younger COVID-19 patients.
Declaration of Competing Interest
The funders had no role in study design, data collection, analysis, decision to publish or preparation of the manuscript. All the authors declare no conflict of interests.
Acknowledgments
Acknowledgments
This work was supported by the Chinese National Mega Science and Technology Program on Infectious Diseases (2018ZX10731301-001-004), Science and Technology Commission of Shanghai Municipality (20JC1410200), Shanghai Academic Research Leader Project (2018XD1403300), Shanghai Municipal Commission of Economic and Informatization(GYQJ-2020-FY-06) and the Emergency Project of Shanghai Institute of Immunology.
Author contributions
YW and BS conceived the project and designed the experiments. YDL, YYC, and JPS performed the experiments. YL, JX and HZL collected the samples and processed clinical data. YW, LS, YQY, and BS analyzed the data. YDL, YYC, YW, and BS wrote the manuscript. HZL, YW, and BS are senior authors.
Biographies

Yang-dian Lai obtained her B.S. degree in biomedicine engineering from Fujian Medical University of China in 2019. She is now a Master of Science candidate at Shanghai Institute of Immunology in Shanghai Jiao Tong University. Her major research interest focuses on infection immunity against tuberculosis and emerging infectious disease.

Hong-zhou Lu is the Professor at Shanghai Clinical Public Health Center affiliated to Fudan University. His research focuses on diagnosis and treatment of infectious diseases and pathogenic mechanisms.

Bing Su is the Professor in Immunology at Shanghai Institute of Immunology of Shanghai Jiao Tong University. His research focuses on immune regulation on mucosal immunity by MAPK and mTOR signaling pathways.

Ying Wang is the Professor in Immunology at Shanghai Institute of Immunology of Shanghai Jiao Tong University. Her research focuses on immune regulatory mechanisms of infectious diseases.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.fmre.2021.02.004.
Appendix. Supplementary materials
References
- 1.Prompetchara E., Ketloy C., Palaga T. Immune responses in COVID-19 and potential vaccines: Lessons learned from SARS and MERS epidemic. Asian Pac J Allergy Immunol. 2020;38(1):1–9. doi: 10.12932/AP-200220-0772. [DOI] [PubMed] [Google Scholar]
- 2.Ren L.L., Wang Y.M., Wu Z.Q., et al. Identification of a novel coronavirus causing severe pneumonia in human: a descriptive study. Chin Med J (Engl) 2020 doi: 10.1097/CM9.0000000000000722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu Y., Li X., Zhu B., et al. Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nat Med. 2020;26(4):502–505. doi: 10.1038/s41591-020-0817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Control, C.C.f.D. and P.E.W.G.f.N.E.R.J.Z.l.x.b.x.z.z.Z.l. zazhi, The epidemiological characteristics of an outbreak of 2019 novel coronavirus diseases (COVID-19) in China.2020. 41(2): p. 145–151. [DOI] [PubMed]
- 5.Aoshi T., Koyama S., Kobiyama K., et al. Innate and adaptive immune responses to viral infection and vaccination. Curr Opin Virol. 2011;1(4):226–232. doi: 10.1016/j.coviro.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 6.Tian X., Li C., Huang A., et al. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg Microbes Infect. 2020;9(1):382–385. doi: 10.1080/22221751.2020.1729069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dhama K., Sharun K., Tiwari R., et al. COVID-19, an emerging coronavirus infection: advances and prospects in designing and developing vaccines, immunotherapeutics, and therapeutics. Hum Vaccin Immunother. 2020:1–7. doi: 10.1080/21645515.2020.1735227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tian Y., Lian C., Chen Y., et al. Sensitivity and specificity of SARS-CoV-2 S1 subunit in COVID-19 serology assays. Cell Discovery. 2020;6(1) doi: 10.1038/s41421-020-00224-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gudbjartsson D.F., Norddahl G.L., Melsted P., et al. Humoral Immune Response to SARS-CoV-2 in Iceland. N Engl J Med. 2020;383(18):1724–1734. doi: 10.1056/NEJMoa2026116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ni L., Ye F., Cheng M.L., et al. Detection of SARS-CoV-2-Specific Humoral and Cellular Immunity in COVID-19 Convalescent Individuals. Immunity. 2020;52(6):971–977. doi: 10.1016/j.immuni.2020.04.023. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bao Y., Ling Y., Chen Y.Y., et al. Dynamic anti-spike protein antibody profiles in COVID-19 patients. Int J Infect Dis. 2020;103:540–548. doi: 10.1016/j.ijid.2020.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee J., Zhang T., Hwang I., et al. Epigenetic Modification and Antibody-Dependent Expansion of Memory-like NK Cells in Human Cytomegalovirus-Infected Individuals. Immunity. 2015;42(3):431–442. doi: 10.1016/j.immuni.2015.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi Y., Wang Y., Shao C., et al. COVID-19 infection: the perspectives on immune responses. Cell Death Differ. 2020;27(5):1451–1454. doi: 10.1038/s41418-020-0530-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zimmermann P., Curtis N. Coronavirus Infections in Children Including COVID-19: An Overview of the Epidemiology, Clinical Features, Diagnosis, Treatment and Prevention Options in Children. Pediatr Infect Dis J. 2020;39(5):355–368. doi: 10.1097/INF.0000000000002660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hong H., Wang Y., Chung H.T., et al. Clinical characteristics of novel coronavirus disease 2019 (COVID-19) in newborns, infants and children. Pediatr Neonatol. 2020;61(2):131–132. doi: 10.1016/j.pedneo.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Memish Z.A., Al-Tawfiq J.A., Assiri A., et al. Middle East respiratory syndrome coronavirus disease in children. Pediatr Infect Dis J. 2014;33(9):904–906. doi: 10.1097/INF.0000000000000325. [DOI] [PubMed] [Google Scholar]
- 17.Jiang, y., 儿童新型冠状病毒感染诊断,治疗和预防专家共识(第二版).中华实用儿科临床杂志, 2020.
- 18.Chi X., Yan R., Zhang J., et al. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. 2020;369(6504):650–655. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grifoni A., Weiskopf D., Ramirez S.I., et al. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell. 2020;181(7):1489–1501. doi: 10.1016/j.cell.2020.05.015. e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Le Bert N., Tan A.T., Kunasegaran K., et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584(7821):457–462. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- 21.Ji T., Liu Z., Wang G., et al. Detection of COVID-19: a review of the current literature and future perspectives. Biosens. Bioelectron. 2020;166 doi: 10.1016/j.bios.2020.112455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yüce M., Filiztekin E., Özkaya K.G. COVID-19 diagnosis —a review of current methods. Biosens. Bioelectron. 2021;172 doi: 10.1016/j.bios.2020.112752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stodola J.K., Dubois G., Coupanec A.Le, et al. The OC43 human coronavirus envelope protein is critical for infectious virus production and propagation in neuronal cells and is a determinant of neurovirulence and CNS pathology. Virology. 2018;515:134–149. doi: 10.1016/j.virol.2017.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nieto-Torres J.L., Dediego M.L., Alvarez E., et al. Subcellular location and topology of severe acute respiratory syndrome coronavirus envelope protein. Virology. 2011;415(2):69–82. doi: 10.1016/j.virol.2011.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tilocca B., Britti D., Urbani A., et al. Computational immune proteomics approach to target COVID-19. J. Proteome. Res. 2020;19(11):4233–4241. doi: 10.1021/acs.jproteome.0c00553. [DOI] [PubMed] [Google Scholar]
- 26.Abdelmageed M.I., Abdelmoneim A.H., Mustafa M.I., et al. Design of a Multiepitope-Based Peptide Vaccine against the E Protein of Human COVID-19: an immunoinformatics approach. Biomed Res Int. 2020;2020 doi: 10.1155/2020/2683286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wen W., Su W., Tang H., et al. Immune cell profiling of COVID-19 patients in the recovery stage by single-cell sequencing. Cell Discov. 2020;6:31. doi: 10.1038/s41421-020-0168-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang J.Y., Wang X.M., Xing X., et al. Single-cell landscape of immunological responses in patients with COVID-19. Nat. Immunol. 2020;21(9):1107–1118. doi: 10.1038/s41590-020-0762-x. [DOI] [PubMed] [Google Scholar]
- 29.Guo C., Li B., Ma H., et al. Single-cell analysis of two severe COVID-19 patients reveals a monocyte-associated and tocilizumab-responding cytokine storm. Nat. Commun. 2020;11(1):3924. doi: 10.1038/s41467-020-17834-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ozanska A., Szymczak D., Rybka J. Pattern of human monocyte subpopulations in health and disease. Scand J Immunol. 2020:e12883. doi: 10.1111/sji.12883. [DOI] [PubMed] [Google Scholar]
- 31.Waller K., James C., de Jong A., et al. ADAM17-Mediated Reduction in CD14(++)CD16(+) Monocytes ex vivo and reduction in intermediate monocytes with immune paresis in acute pancreatitis and acute alcoholic hepatitis. Front Immunol. 2019;10:1902. doi: 10.3389/fimmu.2019.01902. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


