ABSTRACT
Autophagy, a bulk degradation system conserved among most eukaryotes, is also involved in responses to viral infection in plant. In our previous study, a new host factor P3IP was identified to interact with RSV (rice stripe virus) p3 and mediate its autophagic degradation to limit the viral infection. Here, we further discovered that P3IP of Nicotiana benthamiana (NbP3IP) participated in regulation of autophagy. Overexpression of NbP3IP induced autophagy and down-regulation of NbP3IP reduced autophagy. Combined the functions of autophagy-mediated plant defense against plant virus and regulation autophagy, we indicate that P3IP participates in the regulation of autophagy.
KEYWORDS: NbP3IP, autophagy, ATG8, rice stripe virus
Autophagy is a highly conserved eukaryotic mechanism that leads to the degradation and recycling of cytoplasmic components and damaged organelles through lysosomal pathways. Macroautophagy (hereafter referred to as autophagy) is mediated by autophagosome, a de novo-formed double-membrane vesicles. This process involves multiple autophagy-related (ATG) proteins. Autophagy can be induced in various stress conditions including starvation, oxidative stress, drought, salt and pathogen invasion in plants.1–5 The recruitment of autophagy targets is mediated by cargo adaptor proteins like p62/SQSTM1 and NBR1 that interact with membrane-associated ATG8/LC3 through a conserved motif termed LC3-interacting region (LIR).6–8
In our previous paper, an uncharacterized plant protein P3IP was shown to interact with p3 and mediate its autophagic degradation to limit RSV infection. P3IP also interacted with ATG8f, indicating a potential selective autophagosomal cargo receptor role for P3IP.9 It is known that ATG8 family proteins execute important functions during autophagy in various species.10–13 To determine whether P3IP itself also possesses the ability to induce autophagy in N. benthamiana, we transiently co-expressed NbP3IP with CFP-NbATG8f to visualize autophagic structures and to monitor induced autophagic activity in N. benthamiana using a previously validated approach.14 The term relative autophagic activity is widely used in plant autophagy research. It means autophagy activity of samples relative to that of control. When we co-expressed CFP-NbATG8f with NbP3IP-Myc, we found an increase in the number of CFP-labeled autophagic structures compared to cells in which CFP-NbATG8f was co-expressed with a control protein GUSp-Myc (Figure 1a and b). Moreover, treating the cells with the lysosomal proteinase inhibitor E-64d, which blocks the vacuolar degradation of proteins, increased even more the number of CFP-ATG8f-labeled structures in both NbP3IP-Myc and GUSp-Myc co-expressed cells (Figure 1a and b), which is evidence that autophagic flux was normal in all the tested cells. In the presence of E-64d, both of autophagic bodies and autolysosomes could be observed. In addition, a GFP-ATG8 (green fluorescent protein fused to ATG8) processing assay was also performed to monitor autophagy, as indicated by the appearance of free GFP generated within the vacuole/lysosome.15–17 In the absence of E-64d, the ratio of GFP:GFP-ATG8 in NbP3IP-Myc containing leaves was increased compared to that in GUSp-Myc containing leaves (Figure 1c). Furthermore, the ratio of GFP:GFP-ATG8 in NbP3IP-Myc samples treated with E-64d was also higher than that in NbP3IP-Myc samples without E-64d treatment, indicating that the vacuolar processing of GFP-ATG8 could be blocked by the protease inhibitor (Figure 1c). These results reveal that overexpression of NbP3IP-Myc induces autophagy without affecting the autophagic flux. Transmission electron microscopy was also used to verify the autophagy activation. Compared to the control plants, we could clearly observed increased numbers of autophagic structures in leaves with transient expression of NbP3IP (Figure 1d). There was about a twofold increase in the number of visible structures typical of autophagosomes in the cytoplasm (Figure 1e).
Figure 1.
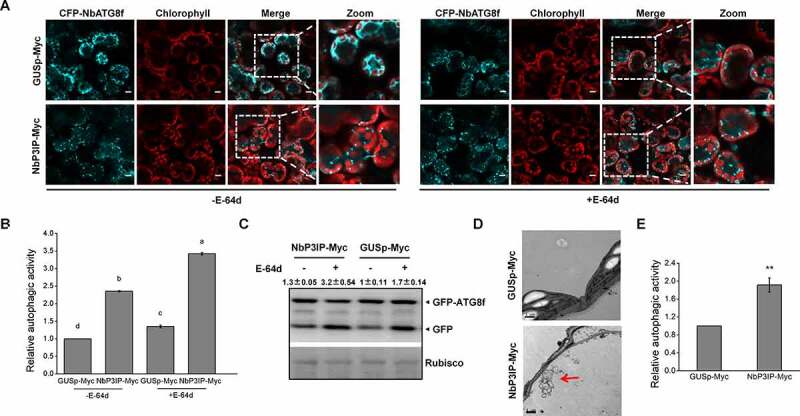
NbP3IP overexpression activates autophagy. (a) Co-expression of NbP3IP-Myc with CFP-NbATG8f increases the appearance of autophagosomes and autophagic vesicles compared to expression of CFP-NbATG8f with a control protein (GUSp-Myc) with or without (left) E-64d. Images were collected at 48 hpi. 50 μM E-64d was pre-infiltrated at 10 h ahead of imaging. Bars, 10 μm. The picture of chlorophyll is used to show the shape of leave cells. (b) Quantification of increase of autophagic activity in cells imaged in panel A. The autophagic activity was calculated in relation to GUSp-Myc with or without E-64d treated plants. Autophagic bodies were counted from approximately 150 cells for each treatment in three independent experiments. Values represent the mean ± SD. Different letters indicate significant difference (ANOVA, P < 0.05). (c) GFP-ATG8 processing assay in control and NbP3IP-Myc-expressing leaves with or without E-64d treatment. Arrowheads indicate the free GFP band and GFP-ATG8f band respectively. The value represents ratio of GFP/GFP-ATG8f relative to rubisco, and the relative levels were calculated in relation to GUSp-Myc without E-64d treated. GFP-ATG8f 40 kD, GFP 27 kD. (d) Examination of autophagic vesicle production by TEM of leaf cells from plants infiltrated with GUSp-Myc or NbP3IP-Myc. Samples collected for processing at 60 hpi. Typical autophagic structures are indicated with red arrows. Bars, 1 μm. (e) Quantification of autophagic vesicles from approximately 20 cells present in TEM images. The autophagic activity is calculated relative to GUSp-Myc-treated plants. The value represents the mean ± SD from three independent experiments. Double asterisks indicate P < 0.01 of significant difference between GUSp-Myc and NbP3IP-Myc treatments (Student’s t-test, two-sided)
In addition, we employed expression of CFP-NbATG8f to monitor autophagy in NbP3IP-silenced plants (TRV:NbP3IP). Confocal microscopy showed that no matter treated with E-64d or not, there were fewer autophagosomes as represented by CFP-NbATG8f puncta in TRV:NbP3IP plants compared with control plants, respectively (Figure 2a and b). Compared to TRV:00 control plants, the mRNA levels of ATGs were down-regulated in NbP3IP-silenced plants (Figure 2c). The accumulation of Joka2/NBR1, which has been demonstrated to be a selective autophagy substrate and also a suitable autophagy marker for autophagic flux analysis in plants,18,19 was examined in NbP3IP-silenced plants. The protein level of Joka2 increased in NbP3IP-silenced leaves compared to non-silenced (TRV:00) leaves, similar to what was observed in ATG5 and ATG7-silenced leaves (Figure 2d), suggesting that autophagy is suppressed in NbP3IP-silenced plants. Taken together, these data suggest that NbP3IP is a new player in the regulation of autophagy in plants.
Figure 2.
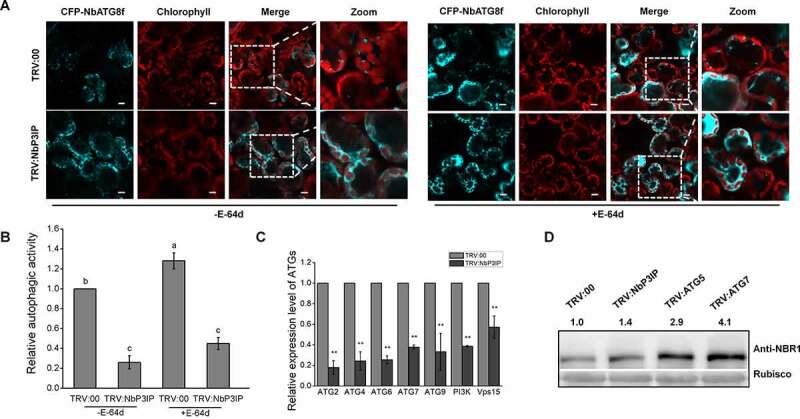
Silencing of NbP3IP inhibits autophagy. (a) Silencing of the endogenous NbP3IP gene reduces the number of autophagosomes with or without (upper) E-64d. Confocal images showing NbP3IP-silenced (TRV:NbP3IP) or control (TRV:00) plants transiently expressing CFP-NbATG8f at 48 hpi. 50 μM E-64d was pre-infiltrated at 10 h ahead of imaging. Bars, 10 μm. (b) Relative autophagic activity in NbP3IP-silenced plants. The autophagic activity was calculated in comparison to TRV:00 plants with or without E-64d. Autophagic bodies were counted from approximately 150 cells for each treatment in three independent experiments. Values represent the mean ± SD. Different letters indicate significant difference (ANOVA, P < 0.05). (c) Real-time RT-PCR analysis of the relative expression of ATGs in TRV:NbP3IP plants compared to TRV:00 plants. Values represent means ± SD from three independent experiments. Double asterisks indicate P < 0.01 of significant difference between TRV:00 control plants and TRV:NbP3IP silenced plants. (Student’s t-test, two-sided). (d) Joka2 was accumulated in NbP3IP-silenced plants. Joka2 was detected using Anti-NBR1 antibody. The value represents protein accumulation relative to rubisco, and the relative levels were calculated in relation to the TRV:00 treatment. Tests were repeated three independent times. Representative result was displayed. Anti-NBR1 120 kD
Moreover, OsP3IP, an NbP3IP homolog in rice, which was reported to interact with p3, OsATG8b and mediate the degradation of p3 protein, could also induce autophagy in our previous study. All these data suggest that P3IP has function as regulator in autophagy pathway.
Acknowledgments
Supports for this work are from the National Natural Science Foundation of China (31772239), the National Key Research and Development Program of China (2017YFA0503401), the Transgenic Science and Technology Program of China (2016ZX08001-002).
Funding Statement
This work was supported by the National Natural Science Foundation of China [31772239]; Transgenic Science and Technology Program of China [2016ZX08001-002]; National Key Research and Development Program of China [2017YFA0503401].
References
- 1.Han S, Yu B, Wang Y, Liu Y.. Role of plant autophagy in stress response. Protein & Cell. 2011;2:1–3. doi: 10.1007/s13238-011-1104-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu Y, Xiong Y, Bassham DC.. Autophagy is required for tolerance of drought and salt stress in plants. Autophagy. 2009;5:954–963. doi: 10.4161/auto.5.7.9290. [DOI] [PubMed] [Google Scholar]
- 3.Bassham DC. Plant autophagy–more than a starvation response. Curr Opin Plant Biol. 2007;10:587–593. doi: 10.1016/j.pbi.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 4.Bozhkov PV. Plant autophagy: mechanisms and functions. J Exp Bot. 2018;69:1281–1285. doi: 10.1093/jxb/ery070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hayward AP, Dinesh-Kumar SP. What can plant autophagy do for an innate immune response? Annu Rev Phytopathol. 2011;49:557–576. doi: 10.1146/annurev-phyto-072910-095333. [DOI] [PubMed] [Google Scholar]
- 6.Zaffagnini G, Martens S. Mechanisms of Selective Autophagy. Journal of Molecular Biology. 2016;428:1714–1724. doi: 10.1016/j.jmb.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stolz A, Ernst A, Dikic I. Cargo recognition and trafficking in selective autophagy. Nat Cell Biol. 2014;16:495–501. doi: 10.1038/ncb2979. [DOI] [PubMed] [Google Scholar]
- 8.Svenning S, Johansen T. Selective autophagy. Essays Biochem. 2013;55:79–92. [DOI] [PubMed] [Google Scholar]
- 9.Jiang L, Lu Y, Zheng X, Yang X, Chen Y, Zhang T, Zhao X, Wang S, Zhao X, Song X, et al. The plant protein NbP3IP directs degradation of Rice stripe virus p3 silencing suppressor protein to limit virus infection through interaction with the autophagy-related protein NbATG8. New Phytol. 2020. doi: 10.1111/nph.16917. [DOI] [PubMed] [Google Scholar]
- 10.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. Embo J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoshimoto K, Hanaoka H, Sato S, Kato T, Tabata S, Noda T, Ohsumi Y. Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell. 2004;16:2967–2983. doi: 10.1105/tpc.104.025395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakatogawa H, Ichimura Y, Ohsumi Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell. 2007;130:165–178. doi: 10.1016/j.cell.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 13.Xie Z, Nair U, Klionsky DJ. Atg8 controls phagophore expansion during autophagosome formation. Mol Biol Cell. 2008;19:3290–3298. doi: 10.1091/mbc.e07-12-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haxim Y, Ismayil A, Jia Q, Wang Y, Zheng X, Chen T, Qian L, Liu N, Wang Y, Han S, et al. Autophagy functions as an antiviral mechanism against geminiviruses in plants. eLife. 2017;6. doi: 10.7554/eLife.23897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng X, Wu M, Li X, Cao J, Li J, Wang J, Huang S, Liu Y, Wang Y. Actin filaments are dispensable for bulk autophagy in plants. Autophagy. 2019;15(12):2126–2141. doi: 10.1080/15548627.2019.1596496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ni HM, Bockus A, Wozniak AL, Jones K, Weinman S, Yin XM, Ding WX. Dissecting the dynamic turnover of GFP-LC3 in the autolysosome. Autophagy. 2011;7:188–204. doi: 10.4161/auto.7.2.14181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van der Vaart A, Griffith J, Reggiori F. Exit from the Golgi is required for the expansion of the autophagosomal phagophore in yeast Saccharomyces cerevisiae. Mol Biol Cell. 2010;21:2270–2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu G, Wang S, Han S, Xie K, Wang Y, Li J, Liu Y. Plant Bax Inhibitor-1 interacts with ATG6 to regulate autophagy and programmed cell death. Autophagy. 2017;13(7):1161–1175. doi: 10.1080/15548627.2017.1320633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kirkin V, Lamark T, Sou YS, Bjorkoy G, Nunn JL, Bruun JA, Shvets E, McEwan DG, Clausen TH, Wild P, et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell. 2009;33:505–516. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]


