Abstract
Public databases provide a wealth of freely available information about chemicals, genes, proteins, biological networks, phenotypes, diseases, and exposure science that can be integrated to construct pathways for systems toxicology applications. Relating this disparate information from public repositories, however, can be challenging since databases use a variety of ways to represent, describe, and make available their content. The use of standard vocabularies to annotate key data concepts, however, allows the information to be more easily exchanged and combined for discovery of new findings. We explore some of the many public data sources currently available to support systems toxicology, and demonstrate the value of standardizing data to help construct chemical-induced outcome pathways.
Keywords: Public database, Systems toxicology, Data standard, Chemical, Gene, Disease
1. Introduction
Systems toxicology assembles information into pathways to describe a xenobiotic toxicant interacting with a mediator in a living system to set into motion a series of biological events that ultimately result in an outcome [1]. The toxicant is typically a chemical; the biological mediator can be any type of macromolecule, but is often studied as a gene-encoded product (e.g., receptor, transporter, transcription factor, enzyme); the series of biological events can be wide-ranging and include alterations in transcripts, protein expression patterns, metabolic pathways, interaction networks, and/or biological processes; and the outcome can be the perturbed biological system trying to return back to a normal state (resilience) or a new, and potentially, adverse state due to toxicity [2]. The toxicant may trigger multiple different pathways simultaneously by interacting with different biological mediators, complicating understanding about the specific connectivity between individual events, mediators, and outcomes. As well, in the real world, humans are typically exposed simultaneously to mixtures of chemicals and drugs, further complicating understanding of toxic mechanisms of actions [3,4].
The goal of systems toxicology is to discern and organize exposure and toxicogenomic events to help formalize an understanding for drug therapy and risk assessment [5–9]. Creating, testing, and validating systems toxicology pathways de novo can be an onerous task since they require information from diverse disciplines and data types, including environmental science, exposure science, chemistry, structural biology, pharmacokinetics, genomic influences, evolutionary biology, genetic and protein networks, high-throughput datasets (transcriptomics, proteomics, metabolomics, etc.), cellular and tissue physiology, phenotypes, diseases, medicine, epidemiology, global health, statistics, and computational modeling, among others. The intricacies of such events make it difficult for any one laboratory on its own to investigate and resolve a complete pathway without leveraging external data resources.
Conveniently, a surfeit of public databases applicable to systems toxicology readily provides information about many of these key interactions and pathway steps. These public data, made freely available and unrestricted to all people in all places at any time, can be leveraged to help generate information frameworks. Combining diverse information from a variety of resources is made easier when public databases use standardized terms, stable accession identifiers (IDs), and cross-reference IDs. Here, we review and explore a variety of public repositories that can be utilized and applied in the construction of systems toxicology pathways, with comparisons to how this is done at Comparative Toxicogenomics Database (CTD; http://ctdbase.org/), a public toxicology resource that advances understanding about environmental chemical exposures and their effects on human heath [10,11].
2. Finding public databases for systems toxicology
The primary literature can be perused to discover the most appropriate public databases for systems toxicology applications. Database: The Journal of Biological Databases and Curation (https://academic.oup.com/database) and the annual ‘Database issue’ of Nucleic Acids Research (https://academic.oup.com/nar) are popular journals where articles report new or updates to existing public databases and describe their resource, data standards, content, and features. As well, toxicology review articles are good sources for learning about public databases for systems toxicology applications [2,8,12–18]. In addition to print, researchers can use online catalogs to search for databases relevant to systems toxicology. The NAR Molecular Biology Database Collection (http://www.oxfordjournals.org/nar/database/a/), for example, features a categorized list of all the database resources featured in the Nucleic Acids Research annual reviews [19], and a search with ‘chemical’ retrieves over 65 available public repositories. One of the most extensive and comprehensive online catalogs is FAIRsharing (https://fairsharing.org/databases/), a manually curated resource of data standards and databases that implement the BioDBcore guidelines for core database descriptors [20,21]. Currently, over 1,200 repositories are listed at FAIRsharing, and each entry is annotated with subject and knowledge domain tags, allowing users to filter their searches; for example, a basic query with ‘chemical’ retrieves over 430 public resources, which can be further refined to include domains such as ‘protein interaction’, ‘phenotype’, and ‘disease’ to filter for resources geared towards systems toxicology. Searches for taxonomy, related databases, and data standards discover similarly themed and constructed public repositories. The accuracy, reliability, and effectiveness of this community-driven catalog, however, are dependent upon database creators reviewing and updating their entry page at the portal.
We surveyed over a hundred databases from selected articles and websites to identify those that are relevant to systems toxicology. To focus on current and truly public databases, we eliminated repositories that had since been decommissioned or now required a paid subscription, creation of a login account, licensing agreement, or software download and installation. In Supplemental Table 1, we provide a detailed matrix of these public databases with a short description, an updated hyperlink, a citation article, and nine sortable data fields applicable to systems toxicology: exposure information, chemical data (including drugs), gene data (including proteins, mRNA, etc.), chemical-gene interactions, protein-protein interactions, pathway/network data, phenotypes/diseases, anatomy, and human population-level information.
3. Leveraging public data
3.1. Data standards, terms, and accession identifiers
An important quality of any good public databases is the practice of defining the data object with a primary term and a stable accession ID, and then supplementing the primary term with a list of synonyms, abbreviations, and cross-reference accession IDs for the same data object in other public databases [22]. Use of controlled vocabularies and ontologies to represent and annotate data is critical, and many vocabularies exist for a wide variety of biological topics and are freely available for use at the Open Biological and Biomedical (OBO) Foundry, an open resource of vocabularies that adhere to core principles for ontology development, ensuring the resources are scientifically accurate, interoperable, and logical [23]. Choosing the most appropriate vocabulary for a biocuration initiative can be challenging; considerations include whether a particular vocabulary or ontology adequately address the specific content with respect to both breadth and depth for the resource’s goal. The stable accession ID allows information to be easily stored, identified, and exchanged with other databases. Database cross-reference links collate the same terms from various controlled vocabularies and act as a Rosetta stone to translate and unambiguously resolve terms and accession IDs from different vocabularies to the same biological concept. Using shared data standards, terms, and accession IDs help disparate databases speak the same language and enables their content to adhere to the FAIR principle, allowing information to be Findable, Accessible, Interoperable and Reusable [24], which in turn increases the value of the data by making it a more readily shareable asset [25].
3.2. The scientific literature
One of the most resourceful and ubiquitous data standards in public databases is the PubMed identifier (PMID), a short numerical accession ID to represent a published scientific document. PubMed (https://www.ncbi.nlm.nih.gov/pubmed/) is a user-friendly portal developed by the National Center for Biotechnology Information (NCBI) that assigns a unique PMID to over 28 million citations in life sciences, medicine, molecular biology, behavioral sciences, and chemistry [26]. The PMID simplifies the process of describing and communicating complex article citations and allows other public databases to associate their curated content to original source articles, providing both traceability and interoperability for bioinformatics analysis. Most scientific abstracts at PubMed are additionally indexed using Medical Subject Headings (MESH), an exhaustive, wide-ranging thesaurus of controlled terms that tag scientific documents with descriptors summarizing the principle concepts addressed in research paper [27]. MESH terms cover a vast knowledge landscape with over 275,000 terms arranged in hierarchical tree branches (https://meshb.nlm.nih.gov/treeView), several of which are highly relevant to systems toxicology, including ‘Chemicals and Drugs’, ‘Phenomena and Processes’, ‘Anatomy’, and ‘Diseases’. Importantly, MESH terms also have unique accession IDs (MESH:ID). Thus, a corpus of scientific articles can be represented as a series of PMIDs, with each PMID associated with a set of MESH:IDs indexing the content of each article. Compiling PMIDs that maximize the number of shared MESH:IDs efficiently organizes papers related to each other, allowing the literature to be triaged and meta-analyzed; for example, in a subset of documents, if one chemical MESH term consistently co-occurs with another chemical MESH term, then there is a potential relationship between the two toxicants [28].
3.3. Comparing and integrating data types relevant to systems toxicology
Other types of data, beyond article information, are represented in repositories integrating various types of vocabularies, ontologies, terms, and accession IDs. As evaluated in Supplemental Table 1, four of the key concepts in systems toxicology include chemicals (toxicant), gene-encoded products (typically, biological mediators that interact with the toxicant), phenotypes (i.e., induced events before the clinical manifestation of a disease), and diseases (toxic outcomes of a systems toxicology pathway).
We compared the data standards (primary terms, accession IDs, and cross-referencing IDs) used to annotate and represent these four concepts from six databases with content highly relevant to systems toxicology: CTD [10]; PharmGKB, a pharmacogenomics knowledgebase of genetic variants and drug responsiveness [29]; DrugBank, a drug-therapeutic target resource [30]; the human metabolome database HMDB [31]; Reactome, a pathways database [32]; and the Monarch Initiative, a multi-species, semantically integrated genotype-phenotype platform [33]. The data standards used to curate and report information for these four components vary greatly from database to database (Figure 1). Many resources create their own database-specific accession ID, but also associate their content with cross-referencing accession IDs. Cross-referencing accession IDs facilitate data harmonization and interoperability. For example, at HMDB the chemical arsenic trioxide has a unique accession ID (HMDB:0015300), but is also annotated with a Chemical Abstracts Service Registry Number (CASRN:1327-53-3) that in turn can be used to find arsenic trioxide at CTD, which is also represented by a unique MESH accession ID (MESH:D000077237).
Figure 1.
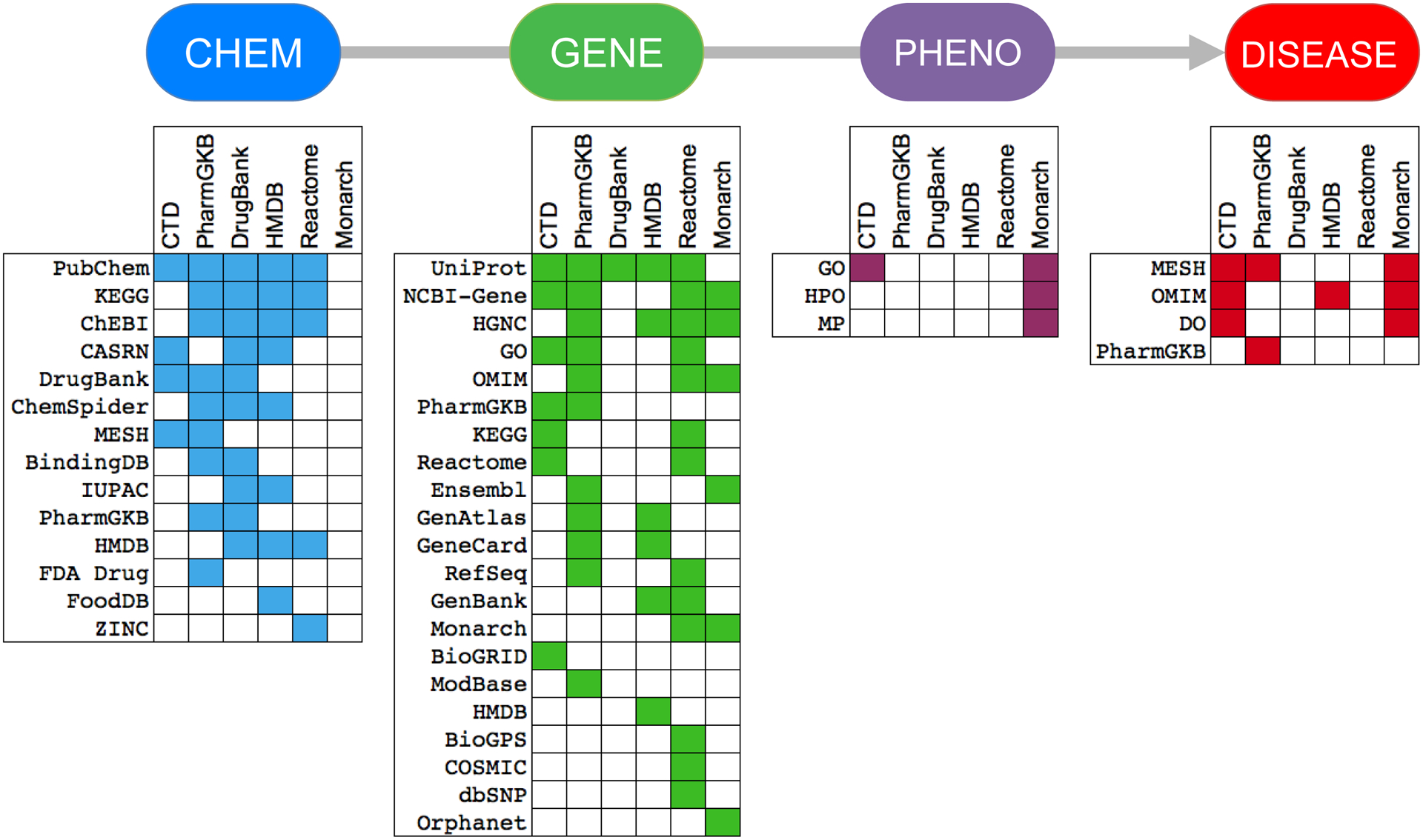
Four key components of a systems toxicology pathway include the toxicant chemical (CHEM), the interacting biological mediator (GENE), a series of pre-disease events (PHENO), and an adverse outcome (DISEASE). The data standards (identifying and/or cross-referencing terms and accession IDs) used by six top public resources for each key component are listed, and shared data types between the resources are highlighted by color in each grid. For example, PubChem, CASRN, and ChEBI terms/IDs are frequently used to depict chemical data at multiple public resources (blue blocks), while UniProt and NCBI Gene terms/IDs are used for genes (green blocks), etc. This shared use of common data standards allows seemingly disparate databases to combine information. For simplicity, not all of the data standards from all six resources are shown. Data standards listed in the grids are described in Supplemental Table 1.
Due to their different scientific objectives, these public databases do not necessarily have information pertaining to all four key components; some (e.g., Monarch) do not contain chemical data, while others (e.g., DrugBank, HMDB, Reactome) invoke phenotype and disease information but only in a free-text format which limits computational possibilities, as a user must now first manually map each free-text description individually to a controlled term or accession ID in their phenotype/disease vocabulary of choice before performing any systematic integration. However, by combining and integrating data from disparate public resource, these public database groups have the opportunity to glean new information and fill in ‘knowledge gaps’. For instance, while the Monarch database does not contain direct chemical information, it does share data standards for gene, phenotype, and disease information with CTD. Leveraging those shared connections allows CTD chemical information to be brought into the Monarch framework (Figure 2). Thus, in Monarch the human gene HMOX1 (NCBIGene:3162) is associated with five phenotypes and 26 diseases, while in CTD human HMOX1 (NCBIGene:3162) is associated with 13 phenotypes, 63 diseases, and directly interacts with 628 chemicals that affect the expression, activity, localization, mRNA stability, ubiquitination, and DNA methylation of HMXO1 products. By integrating the content from both databases, potentially new combinations of chemical-phenotype and chemical-disease connections are discovered at both resources. Similarly, CTD phenotype and disease data can be brought into DrugBank by shared gene data standards, and new chemical-gene interactions can be expanded at both repositories (Figure 2). Integrating and harmonizing data across different platforms facilitates discovery of new connections at each individual resource and accommodates the rapid construction and expansion of potential systems toxicology pathways.
Figure 2.
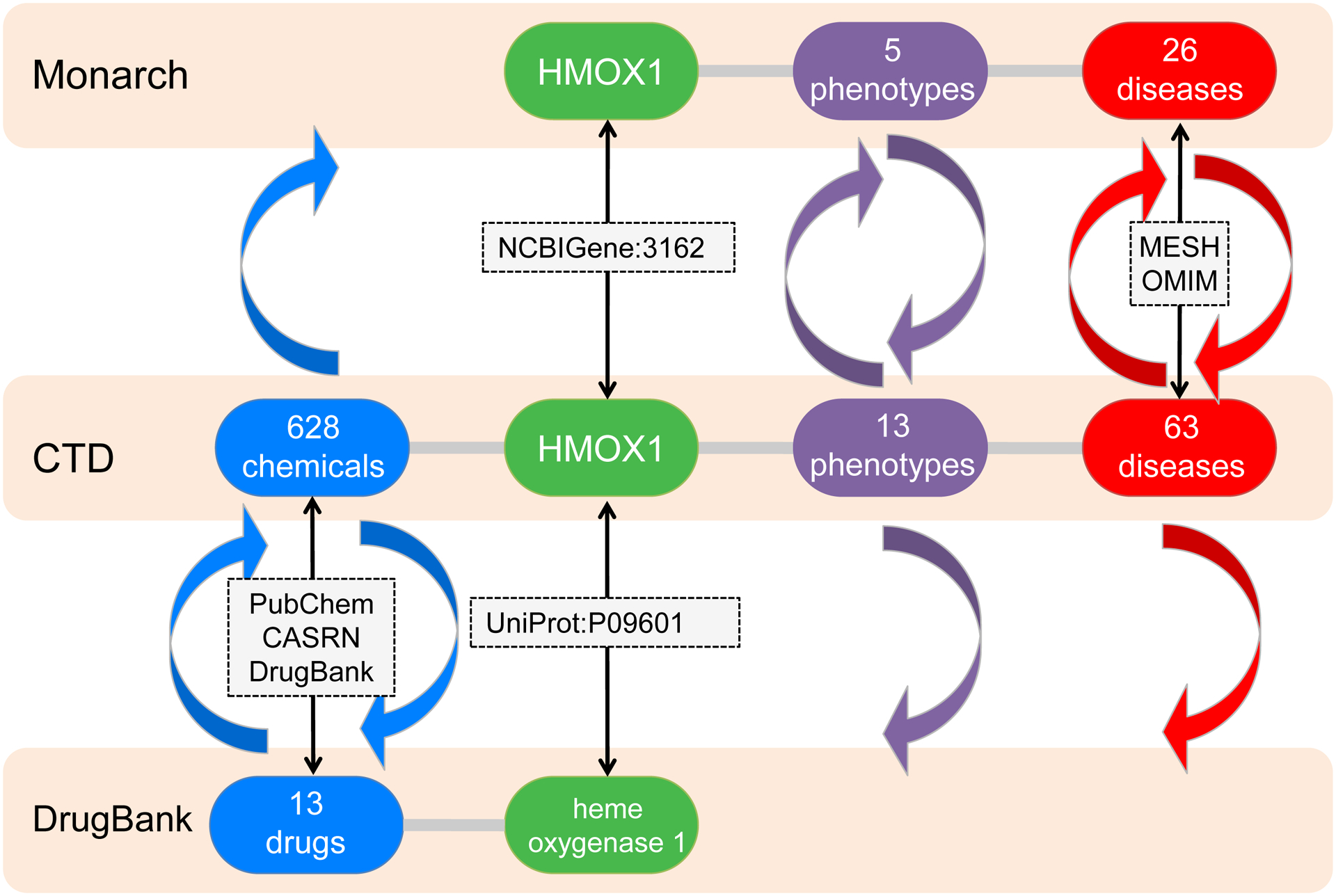
Shared data standards for the human gene HMOX1 (NCBIGene:3162 and UniProt:P09601) between three different public resources (Monarch, CTD, and DrugBank) allows novel chemical (blue arrows), phenotype (purple arrows), and disease (red arrows) information to be integrated and brought into each repository, expanding knowledge at each individual resource, as well as expanding and improving systems toxicology pathways.
3.4. Integrating CTD public data to make systems toxicology pathways
Since 2004, CTD has leveraged the use of in-house curated content and public data integration to make numerous types of novel, putative discoveries [34]. CTD uses professional biocurators [35] to read the toxicology literature and manually curate chemical-gene-phenotype-disease-exposure interactions in a structured format using standardized, controlled vocabularies, ontologies, and accession IDs (e.g., Figure 1) adapted from other public databases [36]. This curation process has resolved and harmonized a tremendous amount of diverse, disparate data from more than 130,000 scientific articles published in over 4,850 different journals, going as far back as 1946, and making all of the information traceable, interoperable, and cohesive for developing testable hypotheses [37]. In total, CTD biocurators have used common data standards to annotate contextualized toxicological relationships for 1.8 million chemical-gene, 182,000 chemical-phenotype, 213,000 chemical-disease, and 38,100 gene-disease interactions for over 15,800 chemicals, 47,100 genes, 4,600 phenotypes, 7,100 diseases, 590 different species, and 810 anatomical sites (http://ctdbase.org/about/dataStatus.go), all publicly available to any user.
CTD direct interactions, in turn, are integrated to generate inferred relationships supportive of systems toxicology: if chemical C1 interacts with gene G1 (C1-G1), and independently gene G1 is associated with disease D1 (G1-D1), then chemical C1 can be said to have an inferred relationship to disease D1 via gene G1 (C1-G1-D1) [38]. Likewise, phenotype P1 can be inferred to disease D1 (P1-C1-D1) if chemical C1 has a directly curated relationship with each [39]. Common data nodes allow binary relationships to be linked together, and simple linear pathways emerge: C1-G1-P1-D1. When additional curated relationships are added for chemical 1 (C1) with other genes (G2, G3) and phenotypes (P2, P3), more complex and interconnecting systems toxicology pathways can be constructed (Figure 3). These computationally generated predictive adverse outcome pathways assemble and organize intricate information for biologically plausible pathways [40]. Towards that end, CTD content has been integrated in several recent independent bioinformatics studies to generate predictive systems toxicology pathways for pesticide-associated reproductive disorders [15], cadmium-induced neurological phenotypes related to Alzheimer disease [39], arsenic-induced alterations in glucose phenotypes preceding diabetes [39], AHR gene networks connected to glaucoma [41], piperonyl butoxide-induced pathway modulations associated with liver cirrhosis [42], chemical-induced androgen receptor activation leading to infertility [43], and lead exposure disrupting cortical plasticity in neurodevelopment [44], amongst others.
Figure 3.
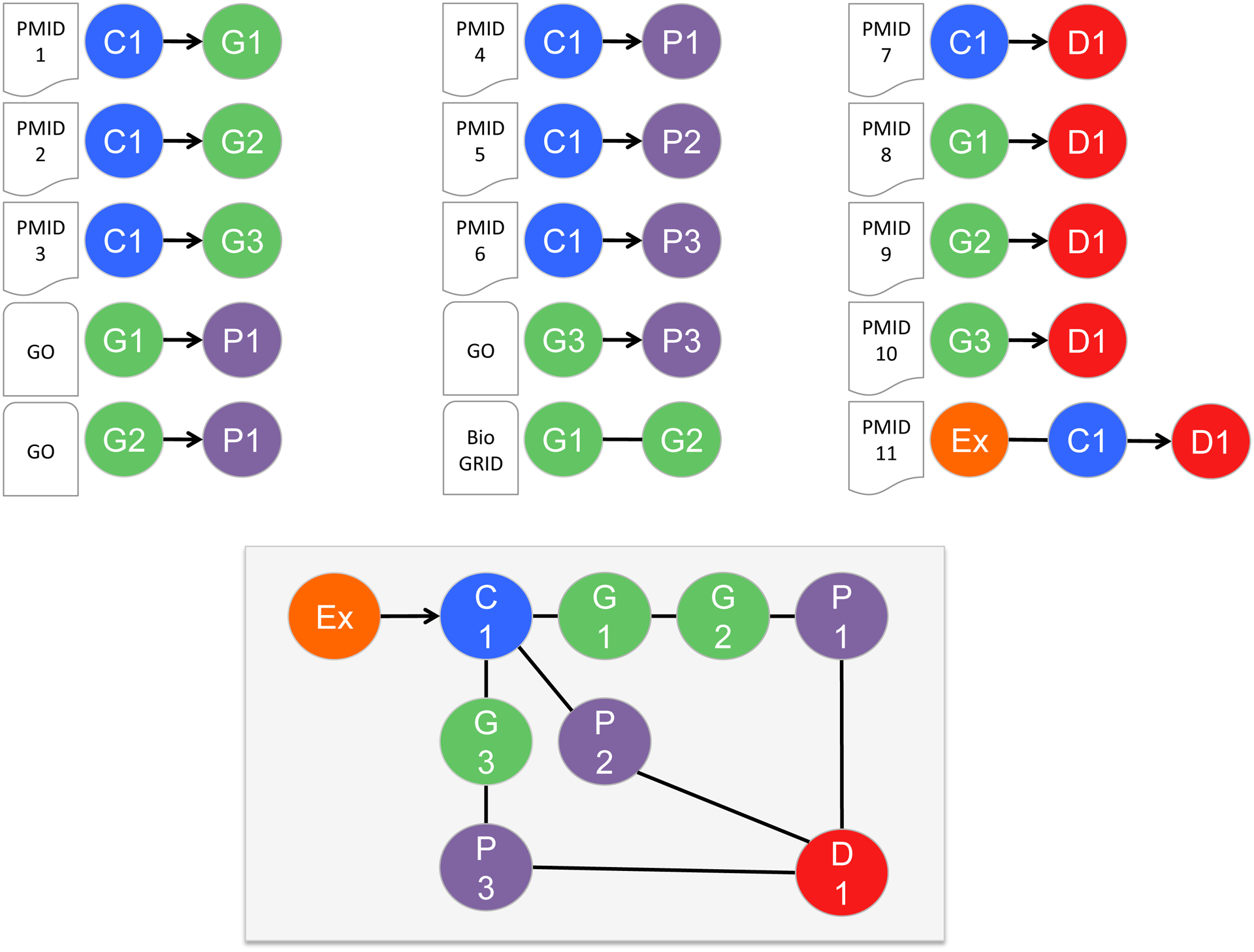
Integrating data from various public resources helps construct information pathways for systems toxicology. CTD manually curates the toxicology literature (PMID1–11) to capture contextualized relationships between chemicals (C1, blue), genes (G1–3, green), biological process phenotypes (P1–3, purple), diseases (D1, red), and exposure events (Ex, orange). As well, CTD imports Gene Ontology (GO) data (connecting genes to biological process phenotypes) and BioGRID data (for gene-gene interactions) from other public databases (Supplemental Table 1). Integrating these independent data modules generate a potential systems toxicology pathway (bottom box), linking an initial exposure event to a toxicant-mediator interaction, connecting genes and phenotypes, and resulting in a disease. Common accession terms and shared data standards allow systems toxicology pathways to be generated programmatically.
Additionally at CTD, C1-G1-P1-D1 data are integrated with real-world environmental exposure stressor and human biomarker measurements manually curated from the exposure science literature [11]. This content includes information for stressor sources, chemical stressor levels measured in the environment, human receptor descriptions (sex, race, smoking status), exposure media, biomarker measurements, assessed time period, geographical details, and associated outcomes [45]. This CTD data can be used to build aggregate exposure pathways [46], which, in turn, can be integrated with adverse outcome pathways to better inform risk assessment [47].
4. Evaluating other public data standards
4.1. CTD’s approach
Ultimately, data exchange between different resources for systems toxicology applications could be even more seamless if all public databases used the same standards and vocabularies. To keep abreast of data standards, CTD frequently reviews available ontologies and controlled vocabularies used by other public resources, determining their suitability for migration from our existing curation. For example, CTD’s initial phenotype-related curation was completed as part of a CTD/Pfizer collaboration [48], and employed terms from the ‘Phenomena and Process’ branch of MESH; new and existing phenotype curation was later migrated to terms in the Gene Ontology (GO) [49] to enable biocurators to more accurately represent the outcomes reported in the toxicology literature and improve interoperability with other external data-sets [39]. Other vocabularies that CTD is very interested in monitoring and potentially migrating to include the Disease Ontology (DO) [50] and Monarch Disease Ontology (MONDO) [33] for disease content and Chemical Entities of Biological Interest (ChEBI) [51] for chemical terms.
To aid in this analysis, CTD developed an in-house tool called the Computational Vocabulary Evaluation Tool (CVET) to computationally evaluate proposed vocabularies. When contemplating converting to a new vocabulary, the most important question is: how would it affect existing CTD curation? Consequently, rather than evaluate complete vocabularies against one another, the tool focuses on existing curation coverage, i.e., what percentage of CTD existing curation would be lost due to conversion? CVET uses an algorithm to evaluate each curated term. First, does the proposed vocabulary provide a direct cross-reference accession ID to the curated term? This is considered by CTD as the most effective means of resolving a term, because it is essentially the vocabulary provider unambiguously mapping their term to another vocabulary. Next, CVET runs through a series of comparisons looking to see if there is a term in the proposed vocabulary that directly matches the CTD curated term (term-term), a synonym in the proposed vocabulary that directly matches the CTD curated term (synonym-term), a term in the proposed vocabulary that directly matches a synonym for the CTD curated term (term-synonym), or a synonym in the proposed vocabulary that directly matches a synonym for the CTD curated term (synonym-synonym). As imagined, attempting to match terms across controlled vocabularies can be complicated. Even using clear, unambiguous cross-references computationally can yield results that must be manually reconciled. Consequently, CTD biocurators manually determine the appropriate mapping, in some cases referencing the source material to identify the term that most closely reflects the authors’ intent based on the context in which it was used. Using CVET, CTD currently is able to achieve a 53% curated chemical term match to ChEBI, a 62% curated disease term match to DO, and an 88% curated disease term match rate to MONDO. CTD will continue to periodically monitor and evaluate these and other promising vocabularies to improve our content for systems toxicology applications.
Supplementary Material
Supplemental Table 1. A sample of over 100 public repositories with data content that support systems toxicology applications.
Acknowledgements
This work was supported by the National Institute of Environmental Health Sciences [R01 ES014065, R01 ES023788, P30 ES025128]. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Conflicts of Interest
The authors declare no conflict of interest.
References
Papers of particular interest, published within the period of review have been highlighted as:
* of special interest
- 1.Plant NJ: An introduction to systems toxicology. Toxicol Res 2014, 4:9–22. 10.1039/C4TX00058G [DOI] [Google Scholar]
- 2.Hartung T, FitzGerald RE, Jennings P, Mirams GR, Peitsch MC, Rostami-Hodjegan A, Shah I, Wilks MF, Sturla SJ: Systems toxicology: real world applications and opportunities. Chem Res Toxicol 2017, 30:870–882. 10.1021/acs.chemrestox.7b00003 [DOI] [PMC free article] [PubMed] [Google Scholar]; * The authors provide a good introduction to systems toxicology, with discussion on emerging technologies and methods, and the importance of linking in vitro network data to toxic phenotypes.
- 3.Boberg J, Dybdahl M, Petersen A, Hass U, Svingen T, Vinggaard AM: A pragmatic approach for human risk assessment of chemical mixtures. Curr Opin Toxicol 2019, 15:1–7. 10.1016/j.cotox.2018.11.004 [DOI] [Google Scholar]
- 4.Zheng Y, Peng H, Zhang X, Zhao Z, Yin J, Li J: Predicting adverse drug reactions of combined medication from heterogeneous pharmacologic databases. BMC Bioinformatics 2018, 19:517 10.1186/s12859-018-2520-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krewski D, Acosta D, Andersen M, Bailar JC, Boekelheide K, Brent R, Chamley G, Cheung VG, Green S, Kelsey KT, Kerkvliet NI, Li AA, McCray L, Meyer O, Patterson RD, Pennie W, Scala RA, Solomon GM, Stephens M, Yager J, Zeise L: Toxicity testing in the 21st century: a vision and a strategy. J Toxicol Environ Health B Crit Rev 2010, 13:51–138. 10.1080/10937404.2010.483176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kiyosawa N, Manabe S: Data-intensive drug development in the information age: applications of systems biology/pharmcaology/toxicology. J Toxicol Sci 2016, 41:SP15–SP25. 10.2131/jts.41.SP15 [DOI] [PubMed] [Google Scholar]
- 7.Mortensen HM, Chamberlin J, Joubert B, Angrish M, Sipes N, Lee JS, Euling SY: Leveraging human genetic and adverse outcome pathway (AOP) data to inform susceptibility in human health risk assessment. Mamm Genome 2018, 29:190–204. 10.1007/s00335-018-9738-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alexander-Dann B, Pruteanu LL, Oerton E, Sharma N, Berindan-Neagoe I, Modos D, Bender A: Developments in toxicogenomics: understanding and predicting compound-induced toxicity from gene expression data. Mol Omics 2018, 14:218–236. 10.1039/c8mo00042e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Z, Huang R, Roberts R, Tong W: Toxicogenomics: a 2020 vision. Trends Pharmacol Sci 2018, pii: S0165-6147(18)30226-8 10.1016/j.tips.2018.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]; * The authors describe the significance of in vitro toxicogenomics, and how the use of this technology can be leveraged for analyzing and assessing human risk, as well as combined with other methods (i.e., machine learning) to generate predictive models and adverse outcome pathways for systems toxicology.
- 10.Davis AP, Grondin CJ, Johnson RJ, Sciaky D, McMorran R, Wiegers J, Wiegers TC, Mattingly CJ: The Comparative Toxicogenomics Database: update 2019. Nucleic Acids Res 2018, 47:D948–D954. 10.1093/nar/gky868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grondin CJ, Davis AP, Wiegers TC, Bing BL, Wiegers JA, Reif DM, Hoppin JA, Mattingly CJ: Advancing exposure science through chemical data curation and integration in the Comparative Toxicogenomics Database. Environ Health Perspect 2016, 124:1592–1599. 10.1289/EHP174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mattingly CJ: Chemical databases for environmental health and clinical research. Toxicol Lett 2009, 186:62–65. 10.1016/j.toxlet.2008.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Judson R: Public databases supporting computational toxicology. J Toxicol Environ Health B Crit Rev 2010, 13:218–231. 10.1080/10937404.2010.483937 [DOI] [PubMed] [Google Scholar]
- 14.Sturla SJ, Boobis AR, FitzGerald RE, Hoeng J, Kavlock RJ, Schirmer K, Whelan M, Wilks MF, Peitsch MC: Systems toxicology: from basic research to risk assessment. Chem Res Toxicol 2014, 27:314–329. 10.1021/tx400410s [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kongsbak K, Hadrup N, Audouze K, Vinggaard AM: Applicability of computational systems biology in toxicology. Basic Clin Pharmacol Toxicol 2014, 115:45–46. 10.1111/bcpt.12216 [DOI] [PubMed] [Google Scholar]
- 16.Oki NO, Nelms MD, Bell SM, Mortensen HM, Edwards SW: Accelerating adverse outcome pathway development using publicly available data sources. Curr Environ Health Rep 2016, 3:53–63. 10.1007/s40572-016-0079-y [DOI] [PubMed] [Google Scholar]
- 17.Zhang W, Zhang H, Yang H, Li M, Xie Z, Li W: Computational resources associating diseases with genotypes, phenotypes and exposures. Brief Bioinform 2018. 10.1093/bib/bby071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu Z, Li W, Liu G, Tang Y: Network-based methods for prediction of drug-target interactions. Front Pharmacol 2018, 9:1134 10.3389/fphar.2018.01134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Galperin MY, Rigden DJ, Fernandez-Suarez XM: The 2015 Nucleic Acids Research database issue and molecular biology database collection. Nucleic Acids Res 2015, 43:D1–D5. 10.1093/nar/gku1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McQuilton P, Gonzalez-Beitran A, Rocca-Serra P, Thurston M, Lister A, Maguire E, Sansone SA: BioSharing: curated and crowd-sourced metadata standards, databases and data policies in the life sciences. Database 2016, 2016:baw075 10.1093/database/baw075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaudet P, Bairoch A, Field D, Sansone SA, Taylor C, Attwood TK, Bateman A, Blake JA, Bult CJ, Cherry JM, Chisholm RL, Cochrane G, Cook CE, Eppig JT, Galperin MY, Gentleman R, Goble CA, Gojobori T, Hancock JM, Howe DG, Imanishi T, Kelso J, Landsman D, Lewis SE, Mizrachi IK, Orchard S, Ouellette BF, Ranganathan S, Richardson L, Rocca-Serra P, Schofield PN, Smedley D, Southan C, Tan TW, Tatusova T, Whetzel PL, White O, Yamasaki C: Towards BioDBcore: a community-defined information specification for biological databases. Nucleic Acids Res 2011, 39:D7–D10. 10.1093/nar/gkq1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McMurry JA, Juty N, Blomberg N, Burdett T, Conlin T, Conte N, Courtot M, Deck J, Dumontier M, Fellows DK, Gonzalez-Beltran A, Gormanns P, Grethe J, Hastings J, Heriche JK, Hermjakob H, Ison JC, Jimenez RC, Jupp S, Kunze J, Laibe C, Le Novere N, Malone J, Martin MJ, McEntyre JR, Morris C, Muilu J, Muller W, Rocca-Serra P, Sansone SA, Sariyar M1, Snoep JL, Soiland-Reyes S, Stanford NJ, Swainston N, Washington N, Williams AR, Wimalaratne SM, Winfree LM, Wolstencroft K, Goble C, Mungall CJ, Haendel MA, Parkinson H: Identifiers for the 21st century: how to design, provision, and reuse persistent identifiers to maximize utility and impact of life science data. PLoS Biol Res 2017, 15:e2001414 10.1371/journal.pbio.2001414 [DOI] [PMC free article] [PubMed] [Google Scholar]; * An excellent discussion on the necessity of correctly designing stable accession identifiers for biological data; while this may seem a mundane topic, it is paramount for streamlining database interoperability and ensuring data is findable, accessible, and reusable.
- 23.Smith B, Ashburner M, Rosse C, Bard J, Bug W, Ceusters W, Goldberg LJ, Eilbeck K, Ireland A, Mungall CJ, Leontis N, Rocca-Serra P, Ruttenberg A, Sansone SA, Scheuermann RH, Shah N, Whetzel PL, Lewis S: The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nat Biotechnol 2007, 25:1251–1255. 10.1038/nbt1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilkinson MD, Dumontier M, Aalbersberg IJ, Appleton G, Axton M, Baak A, Blomberg N, Boiten JW, da Silva Santos LB, Bourne PE, Bouwman J, Brookes AJ, Clark T, Crosas M, Dillo I, Dumon O, Edmunds S, Evelo CT, Finkers R, Gonzalez-Beltran A, Gray AJ, Groth P, Goble C, Grethe JS, Heringa J, ‘t Hoen PA, Hooft R, Kuhn T, Kok R, Kok J, Lusher SJ, Martone ME, Mons A, Packer AL, Persson B, Rocca-Serra P, Roos M, van Schaik R, Sansone SA, Schultes E, Sengstag T, Slater T, Strawn G, Swertz MA, Thompson M, van der Lei J, van Mulligen E, Velterop J, Waagmeester A, Wittenburg P, Wolstencroft K, Zhao J, Mons B: The FAIR Guiding Principles for scientific data management and stewardship. Sci Data 2016, 3:160018 10.1038/sdata.2016.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.International Society for Biocuration: Biocuration: distilling data into knowledge. PLoS Biol 2018, 16:e2002846 10.1371/journal.pbio.2002846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sayers EW, Agarwala R, Bolton EE, Brister JR, Canese K, Clark K, Connor R, Fiorini N, Funk K, Hefferon T, Holmes JB, Kim S, Kimchi A, Kitts PA, Lathrop S, Lu Z, Madden TL, Marchler-Bauer A, Phan L, Schneider VA, Schoch CL, Pruitt KD, Ostell J: Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 2018, 47:D23–D28. 10.1093/nar/gky1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baumann N: How to use the medical subject headings (MeSH). Int J Clin Pract 2016, 70:171–174. 10.1111/ijcp.12767 [DOI] [PubMed] [Google Scholar]
- 28.Lu Y, Figler B, Huang H, Tu YC, Wang J, Cheng F: Characterization of the mechanism of drug-drug interactions from PubMed using MeSH terms. PLoS One 2017, 12:e0173548 10.1371/journal.pone.0173548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whirl-Carrillo M, McDonagh EM, Hebert JM, Gong L, Sangkuhl K, Thorn CF, Altman RB, Klein TE: Pharmacogenomics knowledge for personalized medicine. Clin Pharmacol Ther 2012, 92:414–417. 10.1038/clpt.2012.96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wishart DS, Feunang YD, Guo AC, Lo EJ, Marcu A, Grant JR, Sajed T, Johnson D, Li C, Sayeeda Z, Assempour N, Iynkkaran I, Liu Y, Maciejewski A, Gale N, Wilson A, Chin L, Cummings R, Le D, Pon A, Knox C, Wilson M: DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res 2018, 46:D1074–D1082. 10.1093/nar/gkx1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wishart DS, Feunang YD, Marcu A, Guo AC, Liang K, Vazquez-Fresno R, Sajed T, Johnson D, Li C, Karu N, Sayeeda Z, Lo E, Assempour N, Berjanskii M, Singhal S, Arndt D, Liang Y, Badran H, Grant J, Serra-Cayuela A, Liu Y, Mandal R, Neveu V, Pon A, Knox C, Wilson M, Manach C, Scalbert A: HMDB 4.0: the human metabolome database for 2018. Nucleic Acids Res 2018, 46:D608–D617. 10.1093/nar/gkx1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fabregat A, Jupe S, Matthews L, Sidiropoulos K, Gillespie M, Garapati P, Haw R, Jassal B, Korninger F, May B, Milacic M, Roca CD, Rothfels K, Sevilla C, Shamovsky V, Shorser S, Varusai T, Viteri G, Weiser J, Wu G, Stein L, Hermjakob H, D’Eustachio P: The Reactome pathway knowledgebase. Nucleic Acids Res 2018, 46:D649–D655. 10.1093/nar/gkx1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mungall CJ, McMurry JA, Kohler S, Balhoff JP, Borromeo C, Brush M, Carbon S, Conlin T, Dunn N, Engelstad M, Foster E, Gourdine JP, Jacobsen JO, Keith D, Laraway B, Lewis SE, NguyenXuan J, Shefchek K, Vasilevsky N, Yuan Z, Washington N, Hochheiser H, Groza T, Smedley D, Robinson PN Haendel MA: The Monarch Initiative: an integrative data and analytic platform connecting phenotypes to genotypes across species. Nucleic Acids Res 2017, 45:D712–D722. 10.1093/nar/gkw1128 [DOI] [PMC free article] [PubMed] [Google Scholar]; *An excellent example of a public resource that semantically integrates data types, sources, and ontologies across public platforms to great effect, especially with respect to genes, phenotypes, and disease curated content.
- 34.Davis AP, Grondin CJ, Lennon-Hopkins K, Saraceni-Richards C, Sciaky D, King BL, Wiegers TC, Mattingly CJ: The Comparative Toxicogenomics Database’s 10th year anniversary: update 2015. Nucleic Acids Res 2015, 43:D914–D920. 10.1093/nar/gku935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bourne PE, McEntyre J: Biocurators: contributors to the world of science. PLoS Comput Biol 2006, 2:e142 10.1371/journal.pcbi.0020142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davis AP, Wiegers TC, Rosenstein MC, Murphy CG, Mattingly CJ: The curation paradigm and application tool used for manual curation of the scientific literature at the Comparative Toxicogenomics Database. Database 2011, 2011:bar034 10.1093/database/bar034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davis AP, Murphy CG, Saraceni-Richards CA, Rosenstein MC, Wiegers TC, Mattingly CJ: Comparative Toxicogenomics Database: a knowledgebase and discovery tool for chemical-gene-disease networks. Nucleic Acids Res 2009, 37:D786–D792. 10.1093/nar/gkn580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.King BL, Davis AP, Rosenstein MC, Wiegers TC, Mattingly CJ: Ranking transitive chemical-disease inferences using local network topology in the Comparative Toxicogenomics Database. PLoS One 2012, 7:e46524 10.1371/journal.pone.0046524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davis AP, Wiegers TC, Wiegers J, Johnson RJ, Sciaky D, Grondin CJ, Mattingly CJ: Chemical-induced phenotypes at CTD help inform the predisease state and construct adverse outcome pathways. Toxicol Sci 2018, 65:145–156. 10.1093/toxsci/kfy131 [DOI] [PMC free article] [PubMed] [Google Scholar]; * The authors describe a novel curation paradigm that generates over 165,000 chemical-phenotype (non-disease) interactions; this unique, freely available public data-set can be integrated with other data types to quickly generate toxicant-mediated pathways for systems toxicology applications.
- 40.Ankley GT, Edwards SW: The adverse outcome pathway: a multifaceted framework supporting 21st century toxicology. Curr Opin Toxicol 2018, 9:1–7. 10.1016/j.cotox.2018.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]; * A concise introduction to and historical perspective of adverse outcome pathways and how they play an important role in systems toxicology, with detailed case examples.
- 41.Oki NO, Edwards SW: An integrative data mining approach to identifying adverse outcome pathway signatures. Toxicology 2016, 350–352:49–61. 10.1016/j.tox.2016.04.004 [DOI] [PubMed] [Google Scholar]
- 42.Oki NO, Farcal L, Abdelaziz A, Florean O, Doktorova TY, Exner T, Kohonen P, Grafström R, Hardy B: Integrated analysis of in vitro data and the adverse outcome pathway framework for prioritization and regulatory applications: an exploratory case study using publicly available data on piperonyl butoxide and liver models. Toxicol In Vitro 2019, 54:23–32. 10.1016/j.tiv.2018.09.002 [DOI] [PubMed] [Google Scholar]
- 43.Pittman ME, Edwards SW, Ives C, Mortensen HM: AOP-DB: a database resource for the exploration of adverse outcome pathways through integrated association networks. Toxicol Appl Pharmacol 2018, 343:71–83. 10.1016/j.taap.2018.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith MR, Yevoo P, Sadahiro M, Austin C, Amarasiriwardena C, Awawda M, Arora M, Dudley JT, Morishita H: Integrative bioinformatics identifies postnatal lead (Pb) exposure disrupts developmental cortical plasticity. Sci Rep 2018, 8:16388 10.1038/s41598-018-34592-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grondin CJ, Davis AP, Wiegers TC, Wiegers JA, Mattingly CJ: Accessing an expanded exposure science module at the Comparative Toxicogenomics Database. Environ Health Perspect 2018, 126:014501 10.1289/EHP2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Teeguarden JG, Tan YM, Edwards SW, Leonard JA, Anderson KA, Corley RA, Kile ML, Simonich SM, Stone D, Tanguay RL, Waters KM, Harper SL, Williams DE: Completing the link between exposure science and toxicology for improved environmental health decision making: the aggregate exposure pathway framework. Environ Sci Technol 2016, 50:4579–4586. 10.1021/acs.est.5b05311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tan YM, Leonard JA, Edwards S, Teeguarden J, Paini A, Egeghy P: Aggregate exposure pathways in support of risk assessment. Curr Opin Toxicol 2018, 9:8–13. 10.1016/j.cotox.2018.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]; * A nice introduction to the concept of aggregate exposure pathways and how they can be combined with adverse outcome pathways to build a more informative knowledge environment for chemical exposure and risk assessment.
- 48.Davis AP, Wiegers TC, Roberts PM, King BL, Lay JM, Lennon-Hopkins K, Sciaky D, Johnson R, Keating H, Greene N, Hernandez R, McConnell KJ, Enayetallah AE, Mattingly CJ: A CTD-Pfizer collaboration: manual curation of 88,000 scientific articles text mined for drug-disease and drug-phenotype interactions. Database 2013, 2013:bat080 10.1093/database/bat080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G: Gene ontology: tool for the unification of biology. Nat Genet 2000, 25:25–29. 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schriml LM, Mitraka E, Munro J, Tauber B, Schor M, Nickle L, Felix V, Jeng L, Bearer C, Lichenstein R, Bisordi K, Campion N, Hyman B, Kurland D, Oates CP, Kibbey S, Sreekumar P, Le C, Giglio M, Greene C: Human Disease Ontology 2018 update: classification, content and workflow expansion. Nucleic Acids Res 2019, 47:D955–D962. 10.1093/nar/gky1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hastings J, Owen G, Dekker A, Ennis M, Kale N, Muthukrishnan V, Turner S, Swainston N, Mendes P, Steinbeck C: ChEBI in 2016: Improved services and an expanding collection of metabolites. Nucleic Acids Res 2016, 44:D1214–D1219. 10.1093/nar/gkv1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1. A sample of over 100 public repositories with data content that support systems toxicology applications.


