ABSTRACT
In our article published in Plant Physiology, we had reported tarani (tni) mutant in Arabidopsis, in which poly-ubiquitin hydrolysis is adversely affected, shows pleiotropic phenotypic defects including fewer lateral roots due to the stabilization of several AUX/IAAs and reduced auxin response. TNI encodes UBIQUITIN-SPECIFIC PROTEASE14 that maintains normal auxin response through ubiquitin recycling. Fewer lateral roots observed in tni could be due to defects in their primordia initiation or subsequent elongation post-initiation. Here we have tested this by marking the lateral root primordia with pCycB1;1::CycB1;1(DB):GUS reporter and counting the number of lateral root at various stages development of as a marker of lateral root primordium. The results suggest that TNI/UBP14 is required for LRP development, and a reduction in TNI activity causes a delay in LRP initiation and consequently shorter lateral roots in the tni seedlings.
ABBREVIATIONS: LRP, lateral root primordium; XPP, xylem pole pericycle; LRFC, lateral root founder cells
KEYWORDS: Lateral root, ubiquitin protease, auxin response, TARANI
Lateral root organogenesis is a post-embryonic developmental event that commences with the de-differentiation of a pair of xylem pole pericycle (XPP) cells in the basal meristem of the primary root.1,2 Following the commitment to de-differentiation, the XPP cells re-enter into cell cycle to specify lateral root founder cells (LRFCs). Thus, reactivation of cell cycle is the hallmark for lateral root initiation. An auxin maxima and robust auxin signaling in the LRFC are pre-requisites for re-initiating division of these cells.3 The activated LRFC eventually undergoes several rounds of anticlinal and periclinal divisions and form the lateral root primordium (LRP).1,4 LRPs, which are confined within the epidermal layer of the primary root, form the emerged lateral roots when they come out of the epidermis by disrupting it.4 The G2→M transition marker CycB1;1, a B-type cyclin, is strongly expressed in all the stages of lateral root development including the very first division of the LRFC and is widely used as a marker for lateral root initiation.5,6 Besides, several other cell-cycle regulators, including the D-type cyclins CYCD2;1, CYCD3;1 and CYCD4;1, are required for cell division in the LRPs.7 ICK2/KRP2 [INTERACTOR OF CYCLIN DEPENDANT KINASE 2/KINASE INHIBITORY PROTEIN (KIP) RELATED PROTEIN], a negative regulator of the G1→S phase transition, destabilizes CYCD2;1 and inhibits the formative division in LRP.8
Cell cycle check points are tightly regulated by auxin and perturbation in auxin biosynthesis, transport or signaling affects lateral root initiation.6,9,10 Auxin activity in LRPs reduces ICK2/KRP level to promote cell division in a CYCD2;1-dependant manner.8 Gain-of-function mutations that stabilize AUX/IAAs, including those in iaa3/shy2, iaa28, iaa18, iaa14/slr dominant mutants, exhibit defects in lateral root initiation, emergence, or elongation.11–14 This suggests that ubiquitin-mediated turn-over of AUX/IAA repressors by the 26S proteasome is crucial for auxin-dependent lateral root development.1,3,10
We previously reported that the tarani (tni) mutant in Arabidopsis, in which poly-ubiquitin hydrolysis is adversely affected, shows pleiotropic phenotypic defects including fewer lateral roots due to the stabilization of several AUX/IAAs and reduced auxin response.15,16 TNI encodes UBIQUITIN-SPECIFIC PROTEASE14 that maintains normal auxin response through ubiquitin re-cycling [16, 17, 18 and 19]. Fewer lateral roots observed in tni could be due to defects in their initiation or growth post-initiation. We have tested this by using the pCycB1;1::CycB1;1(DB):GUS reporter as a marker of lateral root primordium. The pCycB1;1::CycB1;1(DB):GUS line was crossed with tni and a double homozygous line of pCycB1;1::CycB1;1(DB):GUS tni genotype was established. The pCycB1;1::CycB1;1(DB):GUS signal was detected at various stages of the actively dividing LRPs in both Col-0 and tni seedlings (Figure 1z). The GUS-stained LRPs were counted in these seedlings at 5, 9 and 11 days after germination (DAG) and their average numbers were compared. The results show that, though fewer LRPs were observed in tni seedlings at 5 DAG compared to Col-0, this difference was not sustained at later stages of development (Figure 1b). This suggests a delay in LRP initiation in tni, perhaps accounting for a reduction in the emerged lateral root number reported earlier.16 A delay in LRP initiation is expected to result in shorter lateral roots especially at the early stages of seedling growth. We tested this by measuring the cumulative length of all lateral roots in a seedling and comparing the values between Col-0 and tni. At 9 DAG, the length of lateral roots was nearly 2.5-fold shorter in tni than in Col-0 (Figure 2a and 2b). As auxin is required for lateral root elongation, 3 impaired lateral root growth in tni could be due to reduced auxin signaling imposed by the stabilization of the AUX/IAA repressor proteins.10,16 We tested this by comparing the signal strength of DII:VENUS (DII denotes domain II of AUX/IAAs), a widely used inverse reporter of auxin activity, 18 between Col-0 and tni. Distinctly increased fluorescence signal was detected in the tni lateral roots compared to Col-0 (Figure 2c), suggesting reduced auxin activity in the mutant LRPs. Stabilization of multiple AUX/IAA proteins may have resulted in reduced auxin response in tni lateral roots.16 Taken together, these results suggest that TNI/UBP14 is required for LRP development, and a reduction in TNI activity causes a delay in LRP initiation and consequently shorter lateral roots in the tni seedlings. Further work is required to identify the specific targets of TNI that mediate this process.
Figure 1.
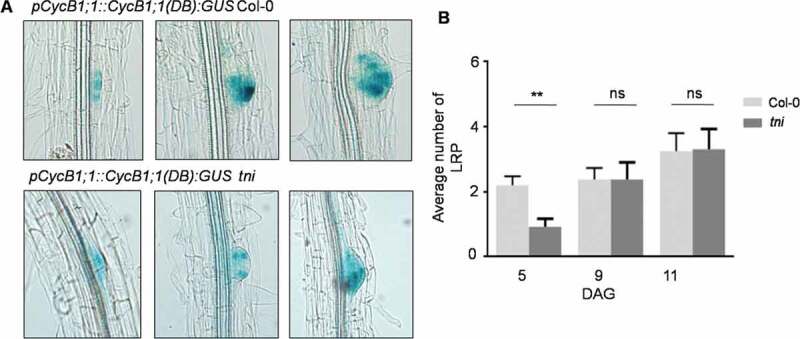
Comparison of the lateral root initiation in Col-0 and tni. (a) Light microscopic images of Col-0 (upper panel) and tni (lower panel) lateral root primordia (LRP) at various stages of initiation (from left to right) expressing the CYCB1;1:GUS signal (blue) in actively dividing cells at 5–9 days after germination (DAG). (b) The average number (N = 8–15) of LRPs per seedling at the indicated DAG. The pCycB1;1::CycB1;1(DB):GUS reporter was used as a marker to visualize LRPs. The error bars represent standard error of mean. Unpaired Student’s t-test was performed for significance analysis. ** denotes p = .0021; ns, not significant (p = .9876 and 0.9458 on 9 and 11 DAG, respectively)
Figure 2.
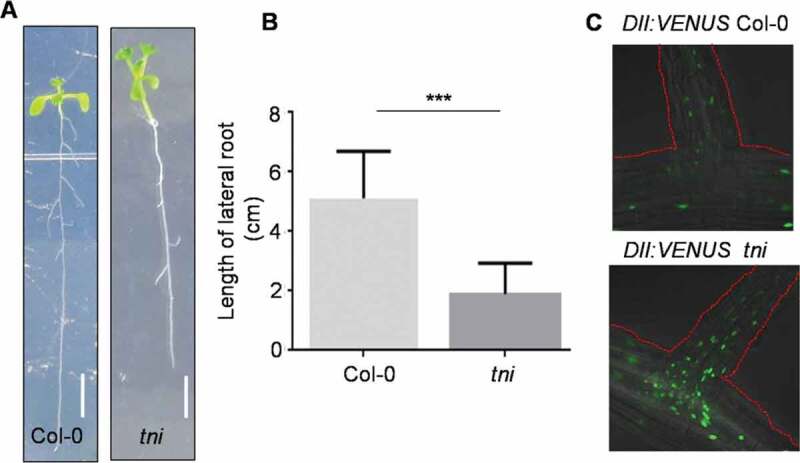
Reduced length of tni lateral roots. (a, b) 9 day-old seedlings showing the lateral roots (a) and their cumulative lengths per seedling (b). Scale bars, 5 mm. N = 33 (Col-0) and 34 (tni). Error bars represent standard deviation. Unpaired Student’s t-test was performed for significance test. *** denotes p < .0001. (c) DII:VENUS signal in the lateral roots of 9 day-old DII:VENUS Col-0 and DII:VENUS tni seedlings. The red dotted lines highlight the outline of lateral roots
Acknowledgments
We acknowledge MCB confocal microscope facility at the Indian Institute of Science, Bangalore for GFP experiments. We thank Ministry of Human Resource Development, Government of India for providing fellowship to PM and PK. UN is thankful to DST-FIST, UGC Centre for Advanced Study and DBT-IISc Partnership Program Phase-II at IISc (sanction No. BT/PR27952/INF/22/212/2018) for the funding and infrastructure support.
References
- 1.De Smet I. Lateral root initiation: one step at a time. New Phytol. 2012;193:1–3. doi: 10.1111/j.1469-8137.2011.03996.x. [DOI] [PubMed] [Google Scholar]
- 2.Laskowski MJ, Williams ME, Nusbaum HC, Sussex IM.. Formation of lateral root meristems is a two-stage process. Development. 1995;121:3303–3310. [DOI] [PubMed] [Google Scholar]
- 3.Du Y, Scheres B. Lateral root formation and the multiple roles of auxin. J Exp Bot. 2018;69:155–167. doi: 10.1093/jxb/erx223. [DOI] [PubMed] [Google Scholar]
- 4.Malamy JE, Benfey PN. Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development. 1997;124:33–44. [DOI] [PubMed] [Google Scholar]
- 5.Beeckman T, Burssens S, Inze D. The peri-cell-cycle in Arabidopsis. J Exp Bot. 2001;52:403–411. [DOI] [PubMed] [Google Scholar]
- 6.Himanen K, Boucheron E, Vanneste S, de Almeida Engler J, Inzé D, Beeckman T. Auxin-mediated cell cycle activation during early lateral root initiation. Plant Cell. 2002;14:2339–2351. doi: 10.1105/tpc.004960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nieuwland J, Maughan S, Dewitte W, Scofield S, Sanz L, Murray JA. The D-type cyclin CYCD4;1 modulates lateral root density in Arabidopsis by affecting the basal meristem region. Proc Natl Acad Sci U S A. 2009;106:22528–22533. doi: 10.1073/pnas.0906354106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanz L, Dewitte W, Forzani C, Patell F, Nieuwland J, Wen B, Quelhas P, De Jager S, Titmus C, Campilho A, et al. The Arabidopsis D-type cyclin CYCD2;1 and the inhibitor ICK2/KRP2 modulate auxin-induced lateral root formation. Plant Cell. 2011;23:641–660. doi: 10.1105/tpc.110.080002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casimiro I, Marchant A, Bhalerao RP, Beeckman T, Dhooge S, Swarup R, Graham N, Inze D, Sandberg G, Casero PJ, et al. Auxin transport promotes Arabidopsis lateral root initiation. Plant Cell. 2001;13:843–852. doi: 10.1105/tpc.13.4.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leyser O. Auxin Signaling. Plant Physiol. 2018;176:465–479. doi: 10.1104/pp.17.00765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fukaki H, Tameda S, Masuda H, Tasaka M. Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant J. 2002;29:153–168. [DOI] [PubMed] [Google Scholar]
- 12.Rogg LE, Lasswell J, Bartel B. A gain-of-function mutation in IAA28 suppresses lateral root development. Plant Cell. 2001;13:465–480. doi: 10.1105/2020.13.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tian Q, Reed JW. Control of auxin-regulated root development by the Arabidopsis thaliana SHY2/IAA3 gene. Development. 1999;126:711–721. [DOI] [PubMed] [Google Scholar]
- 14.Uehara T, Okushima Y, Mimura T, Tasaka M, Fukaki H. Domain II mutations in CRANE/IAA18 suppress lateral root formation and affect shoot development in Arabidopsis thaliana. Plant Cell Physiol. 2008;49:1025–1038. doi: 10.1093/pcp/pcn079. [DOI] [PubMed] [Google Scholar]
- 15.Karidas P, Challa KR, Nath U. The tarani mutation alters surface curvature in Arabidopsis leaves by perturbing the patterns of surface expansion and cell division. J Exp Bot. 2015;66:2107–2122. doi: 10.1093/jxb/erv015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Majumdar P, Karidas P, Siddiqi I, Nath U. The ubiquitin-specific protease TNI/UBP14 functions in ubiquitin recycling and affects auxin response. Plant Physiol. 2020a;184:1–15. doi: 10.1104/pp.20.00689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Majumdar P, Nath U. De-ubiquitinases on the move: an emerging field in plant biology. Plant Biol. 2020b;22:563–572. doi: 10.1111/plb.13118. [DOI] [PubMed] [Google Scholar]
- 18.Xu Y, Jin W, li N, Zhang W, Liu C, Li C, Li Y. UBIQUITIN-SPECIFIC PROTEASE 14 interacts with ULTRAVIOLET-B INSENSITIVE 4to regulate endoreduplication and cell and organ growth in Arabidopsis. Plant Cell 2016; 28:1200-1214. doi: 10.1080/15592324.2020.1860386 [DOI] [PMC free article] [PubMed]
- 19.Brunoud G, Wells DM, Oliva M, Larrieu A, Mirabet V, Burrow AH, Beeckman T, Kepinski S, Traas J, Bennett MJ, et al. A novel sensor to map auxin response and distribution at high spatio-temporal resolution. Nature. 2012;482:103–106. doi: 10.1038/nature10791. [DOI] [PubMed] [Google Scholar]


