ABSTRACT
Air pollution is associated with early declines in lung function and increased levels of asthma-related cysteinyl leukotrienes (CysLT) but a biological pathway linking this rapid response has not been delineated. In this randomized controlled diesel exhaust (DE) challenge study of 16 adult asthmatics, increased exposure-attributable urinary leukotriene E4 (uLTE4, a biomarker of cysteinyl leukotriene production) was correlated (p = 0.04) with declines in forced expiratory volume in 1-second (FEV1) within 6 hours of exposure. Exposure-attributable uLTE4 increases were correlated (p = 0.02) with increased CysLT receptor 1 (CysLTR1) methylation in peripheral blood mononuclear cells which, in turn, was marginally correlated (p = 0.06) with decreased CysLTR1 expression. Decreased CysLTR1 expression was, in turn, correlated (p = 0.0007) with FEV1 declines. During the same time period, increased methylation of GPR17 (a negative regulator of CysLTR1) was observed after DE exposure (p = 0.02); this methylation increase was correlated (p = 0.001) with decreased CysLTR1 methylation which, in turn, was marginally correlated (p = 0.06) with increased CysLTR1 expression; increased CysLTR1 expression was correlated (p = 0.0007) with FEV1 increases. Collectively, these data delineate a potential mechanistic pathway linking increased DE exposure-attributable CysLT levels to lung function declines through changes in CysLTR1-related methylation and gene expression.
KEYWORDS: Asthma, cysteinyl leukotriene, diesel exhaust, epigenetics, GPR17, methylation
Background
Air pollution is associated with acute effects on asthma occurring within hours of exposure [1,2]. Previously we had reported on acute health effect responses to air pollution exposure in school children with asthma [1,3]. That multi-year repeated-measures study demonstrated that increased morning maximum ambient PM2.5 concentrations were consistently associated with increased unscheduled albuterol use and urinary leukotriene E4 levels beginning within hours of exposure [4]. The cysteinyl leukotriene C4 (LTC4) is released into the lungs within minutes of exposure to allergen, exercise, tobacco smoke and air pollution and binds to the cysteinyl leukotriene receptor 1 (CysLTR1) [5]; LTC4 can be measured in urine in terms of its stable end-product LTE4. In earlier studies, we reported that urinary LTE4 (uLTE4) levels were associated with lung function declines in these children with asthma [6]. These changes in uLTE4 and FEV1 occurred rapidly within hours of exposure, suggesting that rapid changes in gene expression of cysteinyl leukotriene (CysLT)-related genes are occurring.
One potential mechanism for such a rapid response to exposure is through deoxyribonucleic acid (DNA) methylation, as epigenetic changes can occur quickly and have been shown to be very sensitive to environmental exposures [7,8]. summarizes genes involved in the cysteinyl leukotriene (CysLT) pathway which were assessed in our study. The first leukotriene to be synthesized, leukotriene A4 (LTA4) is formed with the conversion of arachidonic acid located in membrane phospholipids to LTA4 by membrane-bound 5-lipoxygenase (ALOX5). LTC4 is subsequently converted by LTC4 Synthase (LTC4S) to LTD4 and then to the stable end product LTE4 that can be measured in urine (uLTE4). LTC4, LTD4 and LTE4 bind to CysLTR1, which is found in airway smooth muscle and mediates bronchoconstriction. CysLTR1 cell surface expression is reduced in the presence of increased CysLTs or G-Protein Coupled Receptor (GPR17) expression. LTE4 has been reported to bind to GPR99 leading to increased airway inflammation.
The aim of the present study was to extend our earlier epidemiological findings linking air pollution exposure to increases in CysLTs and acute asthma severity and determine if rapid changes in CysLT-related gene methylation and expression provide a mechanism explaining this association. Accordingly, we demonstrate the relationship between air pollution exposure, changes in methylation and expression of CysLTR1 and changes in FEV1 using an experimental diesel exhaust (DE) challenge study in 16 adults with asthma.
Results
Figures 1 and 2, along with Table 1, describe both significant and marginally significant correlations. Table 1 also shows how correction for cell type in our study was critical in determining true DNA methylation patterns. This is important because methylation changes in uncorrected data likely reflect, at least partly, changes in cell proportions after exposure (rather than changes in DNA methylation within a given cell type alone) [9]. Figure 3 illustrates the biological pathways suggested by the correlations. All correlations described below refer to exposure-attributable changes (in other words, changes from baseline measured 6 hours after exposure to DE minus changes over same time period after exposure to filtered air (FA).
Figure 1.
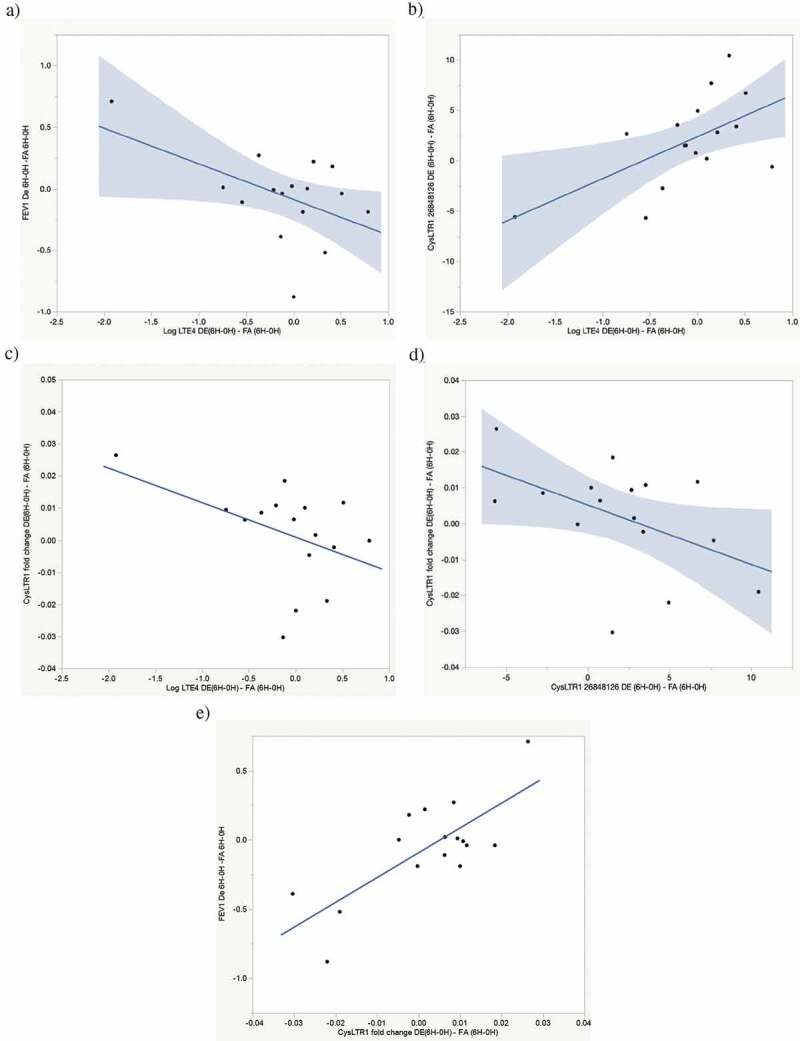
Exposure-attributable FEV1 decline pathway correlations. (a) Correlation between exposure-attributable changes (6H–0H) in FEV1 and log uLTE4 levels. r = −0.51, p = 0.04. (b) Correlation between exposure-attributable changes (6H–0H) in CysLTR1 cg26848126 and log uLTE4 levels. r = 0.59, p = 0.02. (c) Correlation between exposure-attributable changes (6H–0H) in CysLTR1 expression and log uLTE4 levels. r = −0.45, p = 0.08. (d) Correlation between exposure-attributable changes (6H–0H) in CysLTR1 expression and CysLTR1 cg26848126 methylation. r = −0.49, p = 0.06. (e) Correlation between exposure-attributable changes (6H-0H) in FEV1 and CysLTR1 expression r = 0.76, p = 0.0007
Figure 2.
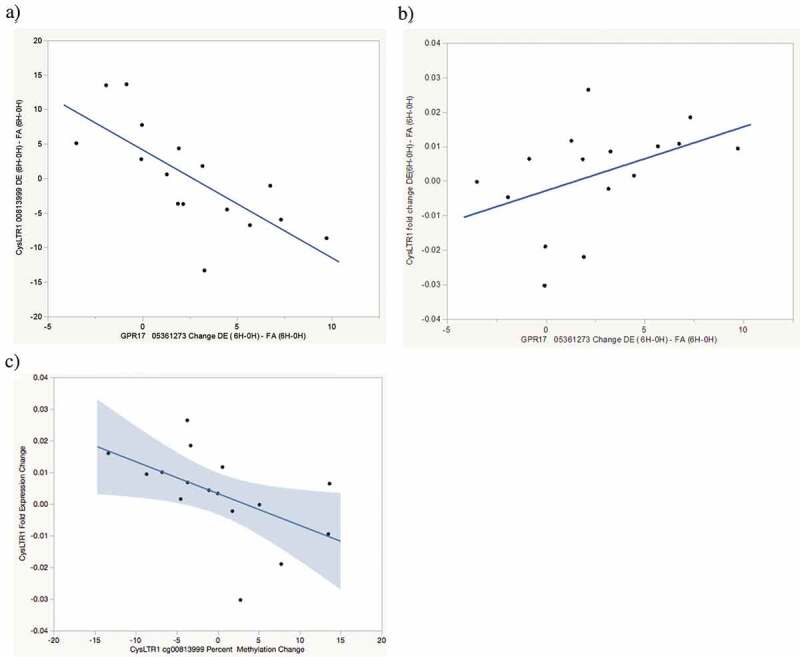
Exposure-attributable FEV1 increase pathway correlations. (a) Correlation between exposure-attributable changes (6H-0H) in CysLTR1 cg0081399 methylation and GPR17 cg05361273 methylation r = −0.73, p = 0.001. (b) Correlation between exposure-attributable changes (6H-0H) in CysLTR1 expression and GPR17 cg05361273 methylation r = 0.47, p = 0.07. 9 c) Correlation between exposure-attributable changes (6H-0H) in CysLTR1 expression and CysLTR1 cg0081399 methylation r = −0.49, p = 0.06
Table 1.
Significant correlations with data corrected for cell-type (versus uncorrected data)
| Exposure attributable gene correlation | Uncorrected correlation and p-value | Correlation and p-value after correction for cell-type |
|---|---|---|
| Log uLTE4 and CysLTR1 cg26848126 methylation | r = 0.54, p = 0.03 | r = 0.59, p = 0.02 |
| CysLTR1 cg26848126 methylation and CysLTR1 expression | r = −0.63, p = 0.009 | r = −0.49, p = 0.06 |
| Log uLTE4 and CysLTR1 expression | r = −0.30, p = 0.26 | r = −0.45, p = 0.08 |
| CysLTR1 cg0081399 methylation and GPR17 cg05361273 methylation | r = −0.36, p = 0.20 | r = −0.73, p = 0.001 |
| CysLTR1 expression and GPR17 cg05361273 methylation | r = −0.22, p = 0.42 | r = 0.47, p = 0.07 |
| CysLTR1 expression and CysLTR1 cg0081399 methylation | r = 0.10, p = 0.72 | r = −0.49, p = 0.06 |
| FEV1 and CysLTR1 expression | r = 0.70, p = 0.003 | r = 0.76, p = 0.0007 |
Figure 3.
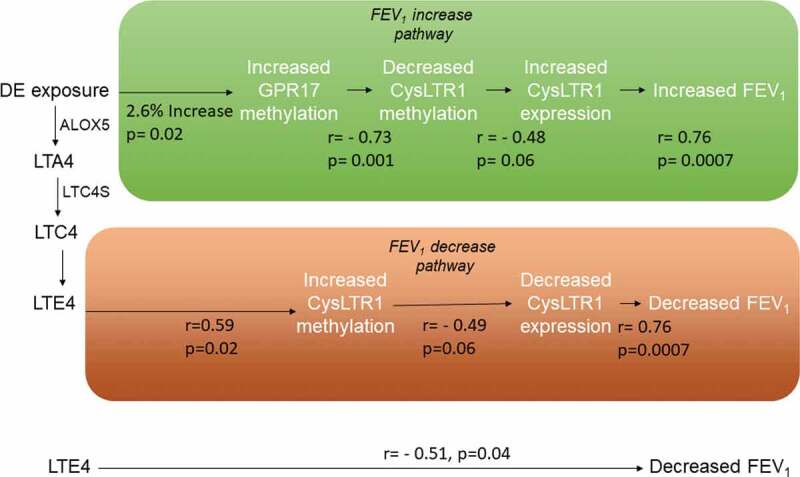
Biological pathways linking DE exposure to acute FEV1 changes through methylation and expression changes on CysLTR1-related genes
In the lung function decrease pathway, DE exposure-attributable cysteinyl leukotrienes bind to the CysLTR1 receptor resulting in increased CysLTR1 cg26848126 methylation and decreased CysLTR1 expression. Decreased CysLTR1 expression results in an attenuated antioxidant response and is correlated with FEV1 decline. In the lung function increase pathway, exposure attributable increases in GPR17 gene methylation are correlated with decreased CysLTR1 cg0081399 methylation which, in turn is correlated with increased CysLTR1 expression. Increased CysLTR1 expression leads to a greater antioxidant response thereby increasing FEV1.
Urinary LTE4 was correlated with FEV1 declines (r = −0.51, p = 0.04, Figure 1(a)). Exposure-attributable uLTE4 levels were also correlated (r = 0.59, p = 0.02) with increased CysLTR1 cg26848126 methylation (Figure 1(b)), and marginally correlated with decreased CysLTR1 expression (r = −0.49, p = 0.06, Figure 1(c)). This marginal significance becomes non-significant when the most outlying point is eliminated but, in that instance, the relationship between LTE4 and CysLTR1 methylation remains intact (p = 0.06). Acute increases in exposure attributable CysLTR1 cg26848126 methylation was marginally correlated (r = 0.49, p = 0.06) with decreased CysLTR1 expression (Figure 1(d)). Decreased CysLTR1 expression was correlated (r = 0.76, p = 0.0007) with FEV1 declines (Figure 1(e)). These findings suggest a pathway by which increased exposure attributable CysLTs as measured by uLTE4 bind to CysLTR1 mediating increased CysLTR1 cg26683198 methylation, decreased CysLTR1 expression and acute FEV1 decline (See Figure 3, lung function decrease pathway).
GPR17 has been reported to be a negative regulator of CysLTR1 activation [10]. We therefore examined correlations between exposure-attributable changes in GPR17 cg05361273 and CysLTR1 DNA methylation and expression. Statistically significant DE exposure-attributable increases in GPR17 cg05361273 methylation were observed both with methylation measured by pyrosequencing (2.6% increase, p = 0.02 Table 2. Increased DE exposure- attributable GPR17 cg05361273 methylation was correlated (r = −0.73, p = 0.001) with decreased CysLTR1 cg0081399 (Figure 2(a)) methylation and marginally correlated with decreased CysLTR1 expression (r = 0.47, p = 0.07, Figure 2(b)). Decreased cg0081399 methylation was, in turn, marginally correlated (r = −0.49, p = 0.06) with decreased CysLTR1 expression (Figure 2(c)). As noted above, increased CysLTR1 expression is correlated with increased FEV1 (Figure 1(e)). These findings suggest a pathway by which acute air pollution exposure results in increased GPR17 methylation, decreased CysLTR1 methylation, increased CysLTR1 expression and FEV1 increase (See Figure 3, lung function increase pathway). There were no significant correlations observed between any gene methylation changes and ALOX5 or LTA4 expression nor were changes in methylation of any genes other than GPR17 and CysLTR1 correlated with changes in FEV1. Although all methylation changes are summarized in Table 2, lack of correlation suggests that methylation changes on genes other than CysLTR1 and GPR17 are likely not clinically relevant to acute changes in lung function.
Table 2.
CpG percent methylation by pyrosequencing at baseline and changes after DE, FA
| Gene CpG | Percent methylation difference (DE-FA) at 0H, 6H and 6H-0H |
Percent methylation at 0H, 6H and 6H-0H after DE exposure |
Percent methylation at 0H, 6H and 6H-0H after FA exposure |
|---|---|---|---|
| ALOX5 cg24302529 | 0H: −1.06 6H: 0.28% 6H-0H: 1.3% (p = 0.13) |
0H: 73.9% 6H: 75.8% 6H-0H: 1.9% (p = 0.02) |
0H: 75.0% 6H: 75.6% 6H-0H: 0.6% (p = 0.28) |
| GPR99 cg24663970 | 0H: −0.56% 6H: −0.23% 6H-0H: 0.32% (p = 0.66) |
0H: 73.9% 6H: 75.8% 6H-0H: 1.9% (p = 0.02) |
0H: 75.0% 6H: 75.6% 6H-0H: 0.6% (p = 0.28) |
| CysLTR1 cg26848126 | 0H: −0.77% 6H: 1.2% 6H-0H: 1.8% (p = 0.17) |
0H: 35.2% 6H: 39.6% 6H-0H:4.4% (p = 0.0007) |
0H: 35.7% 6H: 37.8% 6H-0H: 2.1% (p = 0.04) |
| CysLTR1 cg0081399 | 0H: −0.06% 6H: 0.40% 6H-0H: 0.2% (p = 0.91) |
0H: 29.1% 6H: 31.4% 6H-0H: 2.3% (p = 0.13) |
0H: 28.6% 6H: 30.2% 6H-0H: 1.6% (p = 0.30) |
| LTC4 S cg14674859 | 0H: 0.02% 6H: −0.84% 6H-0H: −0.9% (p = 0.80) |
0H: 70.9% 6H: 65.7% 6H-0H: 5.2% (p = 0.006) |
0H: 71.7% 6H: 65.7% 6H-0H: 6.1% (p = 0.07) |
| LTC4 S cg26683398 | 0H: −2.43% 6H: 1.6% 6H-0H: 4.1% (p = 0.01) |
0H: 73.3% 6H: 77.4% 6H-0H: 4.1% (p = 0.0007) |
0H: 75.8% 6H: 75.8% 6H-0H:0.0003% (p = 0.99) |
| LTC4 S cg16361890 | 0H: 8.2% 6H: −1.6% 6H-0H: −9.8% (p < 0.001) |
0H: 65.7% 6H: 58.8% 6H-0H: −6.9% (p < 0.0001) |
0H: 57.5% 6H: 60.5% 6H-0H: 3.0% (p = 0.002) |
| GPR17 cg05361273 | 0H: −2.0% 6H: 0.62 6H-0H: 2.6% (p = 0.02) * |
0H: 52.6% 6H: 57.5% 6H-0H: 4.9% (p < 0.0001) |
0H: 54.6% 6H: 56.9% 6H-0H: 2.3% (p = 0.001) |
Bold denotes statistically significant ‘exposure-attributable’ changes by pyrosequencing. *Denotes statistically significant ‘exposure-attributable’ changes by Illumina array.
Discussion
Our earlier time-series studies of children had shown that changes in PM2.5 ambient concentrations and personal exposures were associated with increased uLTE4, a marker of total cysteinyl leukotriene production and excretion and that uLTE4 levels were associated with FEV1 decline all within hours of exposure. In the present study, increased uLTE4 levels were correlated with decreased FEV1 as well, now using a completely different design (randomized controlled challenge study) and population (adults with mild asthma). Here we report that changes in CysLTR1 methylation and expression soon after DE exposure might explain the association between CysLT levels and acute asthma worsening within hours after exposure.
This study implicates CysLTR1 in the maintenance of lung function homoeostasis after DE exposure. A potential biological mechanism for CysLTR1–dependent responses to reactive oxygen species has been previously described in a model of irritant-induced asthma in mice [11]. In this study, mice exposed to chlorine for 5 minutes developed airway neutrophilia, increased CysLT production and airway hyperresponsiveness to methacholine. CysLTR1-deficient (CysLTR1(-/-)) mice demonstrated significantly greater airway hyperresponsiveness, airway neutrophilia and bronchial epithelial cell apoptosis than wild-type (WT) BALB/c mice. WT mice also exhibited increased antioxidant and nuclear factor (erythroid derived 2) like-2 (Nrf2) gene expression, Nrf2 nuclear translocation in bronchial epithelial cells, and increases in reduced glutathione/oxidized glutathione following chloride exposure whereas CysLTR1(-/-) mice did not. The authors concluded that loss of functional CysLTR1 resulted in aberrant antioxidant response and increased susceptibility to oxidative injury via CysLTR1-dependent impairment of Nrf2 function. Similarly, in the present study, decreased CysLTR1 expression after DE exposure was correlated with lung function decline. In this FEV1 decline pathway (Figure 3), DE exposure increases uLTE4 levels which was correlated with increased CysLTR1 methylation and decreased CysLTR1 expression. The correlation of increased uLTE4 with decreased CysLTR1 expression is consistent with a previous report demonstrating that high levels of circulating cysteinyl leukotrienes result in a reduction of cell surface CysLTR1 receptors [12]. We hypothesize that decreased uLTE4-related CysLTR1 expression after exposure is leading to an attenuated anti-oxidant response to DE exposure and a greater FEV1 decline (Figure 3).
This study is also consistent with earlier studies identifying GPR17 as a negative regulator of CysLTR1. GPR17 is activated by uracil nucleotides, which accumulate in microenvironments with high levels of oxidative stress [13]. Maekawa et al. [10]. and Qi et al. [14]. reported that co-expression of GPR17 with CysLTR1 inhibited leukotriene-induced calcium influx and extracellular signal–regulated kinase phosphorylation compared with responses in cells expressing the CysLTR1 receptor alone. A later study by the same group reported constitutively increased CysLTR1 expression in the absence of GPR17 [15]. Similarly, the in-vitro study by Qi et al. reported a 50% decrease in CysLTR1 expression with co-expression of GPR17. Although we were unable to measure GPR17 expression in our PBMC assays, the clinical significance of increased GPR17 methylation after DE exposure is supported by the literature reporting that GPR17 is a negative regulator of CysLTR1 and the significant correlations with decreased CysLTR1 methylation and increased CysLTR1 in our study. There are also a number of studies demonstrating that the leukotriene receptor 1(CysLTR1) antagonist Montelukast (commonly used in asthma) blocks GPR17 upregulation suggesting interaction [16]. As such, the literature supports GPR17 and CysLTR1 acting as complementary negative regulators.
We therefore speculate that as reactive oxygen products such as uracil nucleotides accumulate in the lung microenvironment after air pollution exposure, increased GPR17 methylation results in decreased GPR17 expression. This presumed decrease in GPR17 expression results in greater CysLTR1 expression and anti-oxidant activity thereby ‘protecting’ lung function from the negative effects of oxidative stress mediated through CysLTs (Figure 3, lung function increase pathway).
A recently published summary paper highlighted the fact that small but statistically significant changes in DNA methylation (typically below 10% between exposed and unexposed and often below 5%) are typically seen in environmental studies including those that assess acute DNA methylation changes of air pollution in asthmatics [17]. Importantly, these small changes in DNA methylation after environmental exposures have been associated with robust effects on transcriptional activity, suggesting that they may be biologically relevant. Although GPR17 and CysLTR1 gene methylation and CysLTR1 expression changes in this study were measured in peripheral blood cells, they were correlated with acute changes in lung function suggesting that gene changes in the lung are reflected in the periphery. Similarly, studies in asthmatics have demonstrated that systemic changes reflected by uLTE4 levels are correlated with changes in lung function [6].
Conclusions
This study identified a mechanistic pathway through which DE exposure may rapidly induce changes in lung function by initiating changes in CysLTR1 methylation and expression. Consistent with our previous reports linking CysLTs to air pollution exposure and asthma, in this randomized challenge study of adult asthmatics, we observed that increases in DE exposure-attributable uLTE4 were correlated with FEV1 declines. This uLTE4- related FEV1 decline pathway was related to increased CysLTR1 cg26848126 methylation and decreased CysLTR1 expression. During the same early time period, exposure attributable GPR17 gene methylation increased which correlated with decreased CysLTR1 cg0081399 methylation and increased CysLTR1 expression. As such, exposure attributable FEV1 changes related to CysLTR1 expression appear to reflect the balance of two opposing CysLTR1 methylation pathways triggered soon after air pollution exposure.
Methods
Sixteen adult (19-to-35 years) non-smokers with mild asthma, described in Table 3, participated in this study performed at the University of British Columbia. This study followed a double-blinded crossover design in which each subject was exposed to filtered air (FA) or diesel exhaust (DE, nominally, 300 μg/m3 of PM2.5) for 2 hours on two separate occasions, at least two weeks apart. The DE exposure is similar to that intermittently experienced by travellers to Southeast Asia [18]; most particles inhaled from this ultrafine-predominant aerosol are expected to be retained in the lungs. The sequence of FA or DE exposure was randomized and counterbalanced. Blood was collected immediately before and 6 hours after exposure and peripheral blood mononuclear cells were isolated within 4 hours of collection. Urine was collected at these same time points and uLTE4 was measured as previously described [19].
Table 3.
Participant characteristics
| Variable | Participants clinically reported to have asthma (n = 16) |
|---|---|
| Male gender, subjects with | 7 [44%] |
| Age (Years) | 28.7 ± 6.7 [range: 19.2–46.5] |
| Ethnicity | Asian 2 [13%] Caucasian 12 [75%] Middle Eastern 1 [6%] South Asian 1 [6%] |
| Height (cm) | 170.0 ± 12.2 [151.0–185.5] |
| Weight (kg) | 71.9 ± 15.5 [53.0–104.6] |
| BMI | 24.8 ± 3.9 [19.8–34.7] |
| FEV1% predicted | 90.3 ± 14.1 [66.0–120.0] |
| Atopy, subjects with | 9 [56%] |
To assess gene expression, ribonucleic acid (RNA) was reverse-transcribed with the QuantiTect Reverse Transcription Kit (Qiagen, Inc.) according to the manufacturer’s protocol. Quantitative polymerase change reaction (qPCR) was performed on a Rotor-Gene 6000 (Corbett Research) with PerfeCTa SYBR Green FastMix (Quanta Biosciences) using the primers detailed in Table 4. Using LinRegPCR software, cycle threshold values of triplicates were computed after normalizing to Beta-Actin [20]. Gene expression levels were determined for leukotriene A4 hydrolase (LTA4 H), 5-lipoxygenase (ALOX5) and CysLT receptor 1 (CysLTR1).
Table 4.
Primers used for gene expression analysis
| Primer | Sequence |
|---|---|
| CysLTR1 Forward | TTTGGTGACTTCTTGTGCCG |
| CysLTR1 Reverse | CCTACACACACAAACCTGGC |
| LTA4 H Forward | ACCCTGCTACCTGATTGCTT |
| LTA4 H Reverse | CCCATACATACGGTCCTCCC |
| ALOX5 Forward | ACTGGAAACACGGCAAAAAC |
| ALOX5 Reverse | TTTCTCAAAGTCGGCGAAGT |
| ACTB1 Forward | GCCCTGAGGCACTCTTCCA |
| ACTB1 Reverse | CCAGGGCAGTGATCTCCTT |
DNA methylation of candidate CysLT-related CPGs were first assessed using the Illumina 450 K methylation array as previously described [21] and selected for pyrosequencing including real-time qPCR (RT-qPCR) [8]. PyroMark Assay Design 2.0 (Qiagen, Inc.) software was used to design the bisulphite pyrosequencing primers. DNA was subjected to bisulphite conversion using the EZ DNA Methylation Kit (Zymo Research). HotstarTaq DNA polymerase kit (Qiagen, Inc.) was used to amplify the target regions using the biotinylated primer set in Table 5 with the following PCR conditions: 15 minutes at 95°C, 45 cycles of 95°C for 30 s, 58°C for 30 s, and 72°C for 30 s, and a 5 minute 72°C extension step. Streptavidin-coated sepharose beads were bound to the biotinylated-strand of the PCR product and then washed and denatured to yield single-stranded DNA. This study was approved by the University of British Columbia’s Institutional Review Board.
Table 5.
Primers used for methylation analysis
| Primer | Sequence |
|---|---|
| CysLTR1 Forward | GAAAGAGGAAGAAGAATTTGTATATTTG |
| CysLTR1 Reverse – biotin | AACTTATCCCCAAACTCCTAAAAATT |
| CysLTR1 cg0081399 Sequencing | TTGGAGTTTGTTTTTGAG |
| CysLTR1 cg26848126 Sequencing | ATAGGGTATTAAGATATTGTGTA |
| ALOX5 cg1559007 Forward | GTGGGTATTTTGGTGGGTAAG |
| ALOX5 cg1559007 Reverse – biotin | TCTACCTCTCCCTCCCTATAC |
| ALOX5 cg1559007 Sequencing | AAGGTTTTTTTTTTATTTTAAGATT |
| ALOX5 cg2430202529 Forward | GATGTTTAGAGTTTATTTTTGGTGTTATAT |
| ALOX5 cg2430202529 Reverse – biotin | CTCCTTCTAAAACCTTTTCTATTTCC |
| ALOX5 cg2430202529 Sequencing | AGGTAGGGTAGATATTGATTTT |
| GPR99 cg24663970 Forward – biotin | GGTTGAGGTTTAGAGAAGAGAA |
| GPR99 cg24663970 Reverse | CCAAAAATTTCACCACTCTCC |
| GPR99 cg24663970 Sequencing | ATATCCAATTACAACCCT |
| LTC4 S Forward | ATTGTTTAGATTATAGGGTTTGAGTG |
| LTC4 S Reverse – biotin | TCCCCATCCAAACTATTCCTAACA |
| LTC4 S cg14674859 Sequencing | AGTGTGGGGAGGGTA |
| LTC4 S cg1631890 Sequencing | TGTGGGGTTTAGTGT |
| LTC4 S cg26683398 Sequencing | GGTATTGGGAAGGGT |
| GPR17 cg0561273 Forward | GGTTTTTTTGGAGGAAGTGG |
| GPR17 cg0561273 Reverse – biotin | CAACCCCCAAAACTCTTTAACA |
| GPR17 cg0561273 Sequencing | TGGTTTGAGTTGAGTTT |
In both expression and DNA methylation data sets, cell type is an important contributing factor. Data from the Illumina 450 K array [20] was used to predict white blood cell type proportions in each PBMC specimen using established methods [21,22]. These cell type predictions were used to regress out effects of cell type on pyrosequenced methylation and gene expression data on the genes of interest [10]. All methylation values presented are measured by pyrosequencing and cell type corrected unless otherwise indicated.
The exposure-attributable methylation change was tested for statistical significance using a repeated-measures paired t-test analysis. Pairwise linear correlations were assessed between exposure attributable changes in FEV1, log uLTE4, gene expression and DNA methylation (measured by pyrosequencing) with imputation of missing values for each comparison. JMP (version 13; Chicago, IL) was used to fit models. P-values below p = 0.05 were considered significant and <0.1 marginally significant.
Acknowledgments
The authors would like to thank Dusty Christian and Agnes Yuen for their administrative help, including formatting the manuscript.
Funding Statement
This work was supported by Canadian Institutes of Health Research CIHR; AllerGen Network Centre of Excellence. Funding agencies had no participation in the design of the study, nor were they a part of the collection, analysis, interpretation of the data, or preparation of the manuscript.
Authors contributions
All authors generated data and gave constructive criticism of the study manuscript. CC and NR constructed the study design and interpreted the data. MJ, AF, MS, NG and AS performed the statistical analysis. JM, AM, DL, and PR implemented the genetic assays with oversight by MJ and MK. NR and CC wrote the manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Ethics approval and consent to participate
Written consent was obtained from all study participants prior to participation in the study. The study was approved by the University of British Columbia’s institutional review board.
Availability of data and materials
The data set analyzed during the current study are available from the corresponding authors on reasonable request.
References
- [1].Rabinovitch N, Strand M, Gelfand EW.. Particulate levels are associated with early asthma worsening in children with persistent disease. Am J Respir Crit Care Med. 2006;173. DOI: 10.1164/rccm.200509-1393OC [DOI] [PubMed] [Google Scholar]
- [2].Zheng X, Ding H, Jiang L, et al. Association between air pollutants and asthma emergency room visits and hospital admissions in time series studies: a systematic review and meta-analysis. PLoS One. 2015;10:e0138146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Rabinovitch N, Silveira L, Gelfand EW, et al. The response of children with asthma to ambient particulate is modified by tobacco smoke exposure. Am J Respir Crit Care Med. 2011;184:1350–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Rabinovitch N, Adams CD, Strand M, et al. Within-microenvironment exposure to particulate matter and health effects in children with asthma: a pilot study utilizing real-time personal monitoring with GPS interface. Environ Heal. 2016;15:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Hoffman BC, Rabinovitch N.. Urinary leukotriene E4 as a biomarker of exposure, susceptibility, and risk in asthma: an update. Immunol Allergy Clin North Am. 2018;38:599–610. [DOI] [PubMed] [Google Scholar]
- [6].Rabinovitch N, Zhang L, Gelfand EW. Urine leukotriene E4 levels are associated with decreased pulmonary function in children with persistent airway obstruction. J Allergy Clin Immunol. 2006;118:635–640. [DOI] [PubMed] [Google Scholar]
- [7].Mostafavi N, Vermeulen R, Ghantous A, et al. Acute changes in DNA methylation in relation to 24 h personal air pollution exposure measurements: A panel study in four European countries. Environ Int. 2018;120:11–21. [DOI] [PubMed] [Google Scholar]
- [8].Li Z, Li N, Guo C, et al. The global DNA and RNA methylation and their reversal in lung under different concentration exposure of ambient air particulate matter in mice. Ecotoxicol Environ Saf. 2019;172:396–402. [DOI] [PubMed] [Google Scholar]
- [9].Jones MJ, Islam SA, Edgar RD, et al. Adjusting for cell type composition in DNA methylation data using a regression-based approach. Methods Mol Biol. 2017;1589:99–106. [DOI] [PubMed] [Google Scholar]
- [10].Maekawa A, Balestrieri B, Austen KF, et al. GPR17 is a negative regulator of the cysteinyl leukotriene 1 receptor response to leukotriene D4. Proc Natl Acad Sci U S A. 2009;106. DOI: 10.1073/pnas.0905364106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].McGovern T, Goldberger M, Chen M, et al. CysLT1 receptor is protective against oxidative stress in a model of irritant-induced asthma. J Immunol. 2016;197:266–277. [DOI] [PubMed] [Google Scholar]
- [12].Agier J, Różalska S, Wódz K, et al. Leukotriene receptor expression in mast cells is affected by their agonists. Cell Immunol. 2017;317:37–47. [DOI] [PubMed] [Google Scholar]
- [13].Fumagalli M, Lecca D, Coppolino GT, et al. Pharmacological properties and biological functions of the GPR17 receptor, a potential target for neuro-regenerative medicine. Adv Exp Med Biol. 2017;1051:169–192. [DOI] [PubMed] [Google Scholar]
- [14].Qi A-D, Harden TK, Nicholas RA. Is GPR17 a P2Y/leukotriene receptor? Examination of uracil nucleotides, nucleotide sugars, and cysteinyl leukotrienes as agonists of GPR17. J Pharmacol Exp Ther. 2013;347:38–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Maekawa A, Xing W, Austen KF, et al. GPR17 regulates immune pulmonary inflammation induced by house dust mites. J Immunol. 2010;185:1846–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Marschallinger J, Schäffner I, Klein B, et al. Structural and functional rejuvenation of the aged brain by an approved anti-asthmatic drug. Nat Commun. 2015;6:8466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Breton CV, Marsit CJ, Faustman E, et al. Small-magnitude effect sizes in epigenetic end points are important in children’s environmental health studies: the children’s environmental health and disease prevention research center’s epigenetics working group. Environ Health Perspect. 2017;125:511–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Vilcassim MJR, Thurston GD, Chen L-C, et al. Exposure to air pollution is associated with adverse cardiopulmonary health effects in international travellers. J Travel Med. 2019;26. DOI: 10.1093/jtm/taz032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Armstrong M, Liu AH, Harbeck R, et al. Leukotriene-E4 in human urine: comparison of on-line purification and liquid chromatography-tandem mass spectrometry to affinity purification followed by enzyme immunoassay. J Chromatogr B Analyt Technol Biomed Life Sci. 2009;877:3169–3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ramakers C, Ruijter JM, Deprez RHL, et al. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett. 2003;339:62–66. [DOI] [PubMed] [Google Scholar]
- [21].Jiang R, Jones MJ, Sava F, et al. Short-term diesel exhaust inhalation in a controlled human crossover study is associated with changes in DNA methylation of circulating mononuclear cells in asthmatics. Part Fibre Toxicol. 2014;11:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Houseman EA, Accomando WP, Koestler DC, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data set analyzed during the current study are available from the corresponding authors on reasonable request.


